Open Access Article
Open Access ArticleDesign, green synthesis, and quorum sensing quenching potential of novel 2-oxo-pyridines containing a thiophene/furan scaffold and targeting a LasR gene on P. aeruginosa†
Yousry A. Ammar‡
a,
Ahmed Ragab‡ *a,
M. A. Migahedb,
S. Al-Sharbasyc,
Mohamed A. Salem*d,
Omnia Karem M. Riad*e,
Heba Mohammed Refat M. Selim*ef,
Gehad A. Abd-elmaksoudc and
Moustafa S. Abusaifa
*a,
M. A. Migahedb,
S. Al-Sharbasyc,
Mohamed A. Salem*d,
Omnia Karem M. Riad*e,
Heba Mohammed Refat M. Selim*ef,
Gehad A. Abd-elmaksoudc and
Moustafa S. Abusaifa
aDepartment of Chemistry, Faculty of Science (boys), Al-Azhar University, 11884 Nasr City, Cairo, Egypt. E-mail: ahmed_ragab@azhar.edu.eg; ahmed_ragab7@ymail.com
bEgyptian Petroleum Research Institute (EPRI), 11727 Nasr City, Cairo, Egypt
cDepartment of Chemistry, Faculty of Science (girls), Al-Azhar University, 11884 Nasr City, Cairo, Egypt
dDepartment of Chemistry, Faculty of Science and Arts, King Khalid University, Mohail, Assir, Saudi Arabia. E-mail: masalem@kku.edu.sa
eDepartment of Microbiology and Immunology, Faculty of Pharmacy (Girls), Al-Azhar University, Nasr City, Cairo, Egypt. E-mail: dr_omniakarem@azhar.edu.eg; hebarefat1983@gmail.com
fDepartment of Pharmaceutical Sciences, Faculty of Pharmacy, Al-Maarefa University, Diriyah, 13713, Riyadh, Saudi Arabia
First published on 13th September 2023
Abstract
The current trend in fighting bacteria is attacking the virulence and quorum-sensing (QS) signals that control bacterial communication and virulence factors, especially biofilm formation. This study reports new Schiff bases and tetracyclic rings based on a pyridine pharmacophore by two methods: a green approach using CAN and a conventional method. The structure of designed derivatives was confirmed using different spectroscopies (IR and 1H/13C NMR) and elemental analysis. The designed derivatives exhibited good to moderate inhibition zones against bacterial and fungal pathogens. In addition, six compounds 2a,b, 3a,b, and 6a,b displayed potency against tested pathogens with eligible MIC and MBC values compared to standard antimicrobial agents. Compound 2a displayed MIC values of 15.6 μg mL−1 compared to Gentamicin (MIC = 250 μg mL−1 against K. pneumoniae), while compound 6b exhibited super-potent activity against P. aeruginosa, and K. pneumoniae with MIC values of 62.5 and 125 μg mL−1, as well as MBC values of 31.25 and 15.6 μg mL−1 compared to Gentamicin (MIC = 250 and 125 μg mL−1 and MBC = 62.5 μg mL−1), respectively. Surprisingly, these six derivatives revealed bactericidal and fungicidal potency and remarkable anti-biofilm activity that could significantly reduce the biofilm formation against MRSA, E. coli, P. aeruginosa, and C. albicans. Furthermore, the most active derivatives reduced the LasR gene's production between 10–40% at 1/8 MICs compared with untreated P. aeruginosa. Besides, they demonstrated promising safety profile on Vero cells (normal cell lines) with IC50 values ranging between (175.17 ± 3.49 to 344.27 ± 3.81 μg mL−1). In addition, the in silico ADMET prediction was carried out and the results revealed that these compounds could be used with oral bioavailability with low toxicity prediction when administered as a candidate drug. Finally, the molecular docking simulation was performed inside LasR and predicted the key binding interactions responsible for the activity that corroborated the biological results.
1. Introduction
Bacterial infections impose a burden on the public health system, and this issue has been exacerbated by the excessive use of antibiotics, leading to the emergence of multidrug-resistant bacterial strains (MDR).1,2 Each year, over 700![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 people lose their lives as a result of drug-resistant strains, and if this current rate continues to increase, it might rise to be 10 million per year.3,4 Humans spend more time in contact with microorganisms in their daily lives, leading to life-threatening infectious diseases caused by multidrug-resistant bacterial and fungal pathogens.5,6
000 people lose their lives as a result of drug-resistant strains, and if this current rate continues to increase, it might rise to be 10 million per year.3,4 Humans spend more time in contact with microorganisms in their daily lives, leading to life-threatening infectious diseases caused by multidrug-resistant bacterial and fungal pathogens.5,6
In general, antibiotics and anti-infective medications are commonly used to treat microbial infections by different modes of action with the ability to inhibit or stop the growth of pathogenic bacteria through inhibiting many factors such as protein and nucleic acids synthesis, cell membrane synthesis, ATP synthase, metabolic pathway, and membrane function and so on.7–9 Evaluating the potential for biofilm formation and antibiotic resistance is also essential for understanding the epidemiology of the disease and, as a result, establishing appropriate preventative and management strategies for the local environment.10,11 In biofilms, bacterial cells adhere to biotic and abiotic surfaces and form an organic polymeric matrix, the exopolysaccharide, surrounding them.12,13 These structures provide bacteria defense against antibiotics, ultraviolet radiation, dehydration, oxidation, environmental stresses, and immune responses from the host.14,15 In vitro studies have shown that both pathogenic and saprophytic microorganisms e.g., leptospires are capable of forming biofilms.16 Moreover, several studies have observed bacterial aggregates in mammalian hosts' proximal renal tubules,17,18 and enhanced biofilm formation in the presence of catheters.19
The Pseudomonas aeruginosa is one of the serious pathogens that develops resistance to almost all known conventional antimicrobial drugs, the human population is now in extreme danger, especially immune-compromised patients.20,21 There is an increasing need to develop numerous antibiotics to overcome the worrisome rise in resistant bacterial infections.22 Targeting infectious agents' virulence is a modern strategy that is getting much attention for combating antibiotic resistance.23 Using anti-virulence tactics that interfere with bacterial quorum sensing (QS) processes is feasible. QS is the capacity to detect the density of nearby microorganisms (of the same or different species); hence it is responsible for bacterial communication and virulence, especially biofilm formation.24 System heterogeneities, which are variations in the physical or chemical environment, significantly impact the ability of QS systems to coordinate gene expression and behavior.24,25 QS and biofilm formation are highly affected by heterogeneous conditions, including nutrients and growth media, bacterial numbers, and even the constant flow that can wash autoinducers away, thus attenuating QS.26
As QS is mediated by autoinducers released from bacterial cells for communication, targeting signaling pathways, such as the transcriptional proteins; transcriptional regulators (LasR, RhlR), Pseudomonas quinolone signal (Pqs), and quorum-sensing-control repressor (QScR), would aid in inhibiting the QS circuit of P. aeruginosa, to diminish bacterial pathogenicity.27 LasR is an auto-inducer gene related to QS that can affect virulence in P. aeruginosa.28 LasR binds specifically to DNA sequences to activate genes involved in virulence and biofilm development.29 Inhibition of LasR activity can reduce the expression of virulence factors and other pathogenic traits in P. aeruginosa. Several antibiotics have been investigated for their potential to inhibit quorum sensing in P. aeruginosa by targeting LasR, such as Ciprofloxacin, halogenated furanone, and Azithromycin.30
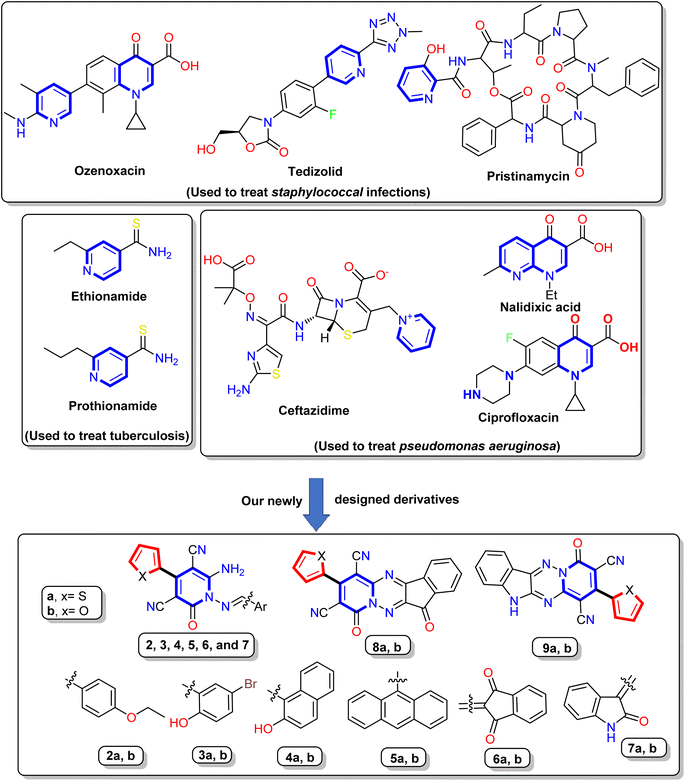
In recent years, nitrogen heterocycles have gained increasing attention because of their diverse pharmacological and biological properties.31 Among these nitrogenous compounds, pyridine is a widely distributed nucleus in the vitamin B6 coenzyme family, several alkaloids, and many drugs. The pyridine derivatives exhibited many activities, such as anti-oxidation, anti-inflammation, and anti-microbial defense.32 In addition, the pyridinone nucleus displayed in a numerous of drugs, such as Gimeracil (DPD inhibitor that used in treatment of cancer), Deferiprone (an iron chelator), Pirfenidone (antifibrotic agent used to treat idiopathic pulmonary fibrosis (IPF)), Doravirine (a drug used to treat HIV), and Ciclopirox (broad spectrum antifungal agent).33–36 Interest is increased toward antibiotics containing the pyridine nucleus, such as Pristinamycin, Ozenoxacin, and Tedizolid (used to treat Staphylococcal infections), Prothionamide and Ethionamide (used to treat tuberculosis (TB)), Nalidixic acid, Levofloxacin, Ciprofloxacin, and Ceftazidime (used to treat Pseudomonas aeruginosa infections)37–39 Fig. 1.
Furthermore, cyanopyridone is a fundamental structural component in many organic and synthetic compounds that exhibit potential as antibacterial,40 anti-inflammatory,41 and antioxidant properties.42 Subsequently, it has been shown that 3-cyano-2-oxo-pyridines can induce a variety of biological actions, such as antidepressant,43 antibacterial,44 and cardiotonic.45 Besides, the Milrinone and Olprinone are well-known medications available on the market and are utilized in treating congestive heart failure.46 Moreover, Schiff bases compounds exhibit various biological actions, including antibacterial, anticancer, antimalarial, anti-inflammatory, antioxidant, analgesic, antiviral, antifungal,47–49 and free radical scavenging capabilities.50
As described in the above findings, the authors in this work aimed to synthesize a new novel N-amino pyridinone Schiff's base and tetracyclic structures known as indeno[1,2-e]pyrido[1,2-b][1,2,4]triazine and pyrido[1′,2′:2,3][1,2,4]triazino[5,6-b]indole derivatives using ceric ammonium nitrate (CAN-catalyst) and conventional method. The synthesized compounds were screened for their antibacterial and antifungal activities against seven standard microbial strains, including two Gram-positive bacteria, four standard Gram-negative strains, and one fungal isolate (Candida albicans) by well diffusion, MIC, and MBC methods. To evaluate the anti-virulence and anti-QS activity of the most active compounds, the microtiter plate method was used to determine the biofilm inhibitory activity. Moreover, the real-time polymerase chain reaction (real-time PCR) was used to assess their ability to decrease the expression of LasR gene, the key regulatory gene for QS in P. aeruginosa. Besides, the cytotoxic activity against normal cell line (Vero cells) was assessed to ensure the safety of the synthesized compounds. The potential LasR inhibitory activity was examined using in silico molecular docking simulation. Besides, the in silico drug-likeness and toxicity prediction were predicted.
2. Results and discussion
2.1. Chemistry
The synthetic pathways for synthesis pyridine and 1,2,4-triazine derivatives were illustrated in Scheme 1 and 2 using 1,6-diamino-2-oxo-4-(thiophen-2-yl)-1,2-dihydropyridine-3,5-dicarbonitrile (1a) and 1,6-diamino-2-oxo-4-(furan-2-yl)-1,2-dihydropyridine-3,5-dicarbonitrile (1b) as starting materials. The 1,6-diamino-2-oxo-pyridine derivatives 1a and 1b were used as reactive intermediates due to their included two center of reaction, N-amino and C-amino centers, which may be able to react as mono or bi-nucleophile reagent to produce various heterocyclic molecules.51–53 Our work was designed to form condensation reaction by synthetized new Schiff bases through treatment of 1,6-diamino-2-oxo-pyridine derivatives 1a,b with different aldehydes through the N-amino center or condensation with di-ketones through N-amino and C-amino centers to afford polycyclic compounds containing pyridine in its core. Moreover, to keep pace with the development in the preparation of organic materials, the condensation reaction takes place with conventional method and green chemistry method using a clean and rapid ceric ammonium nitrate CAN-catalyst instead of hazardous catalysts, such as acetic acid. The advantage of the reaction in the case of using CAN as a catalyst is that the reaction has occurred by stirring at room temperature for a short time and with good yield compared to the other hazardous methods.54–56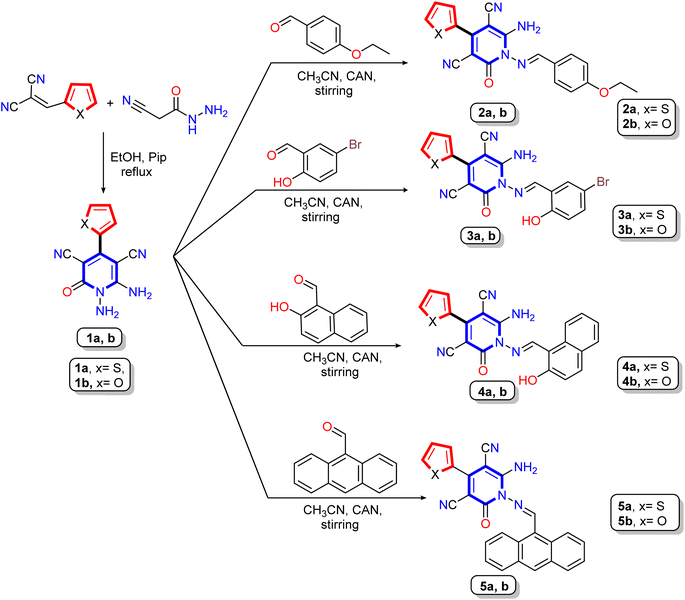 | ||
| Scheme 1 Synthetic pathway for reaction of the starting materials 1,6-diamino-2-oxo-pyridine derivatives 1a,b with different aldehydes. | ||
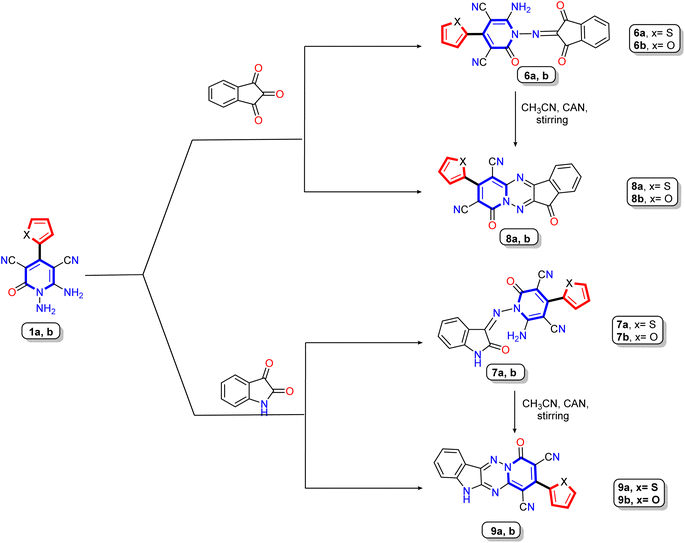 | ||
| Scheme 2 Synthesis of Schiff's base derivatives 6a,b and 8a,b and their cyclization to produce tetracyclic compounds 7a,b and 9a,b. | ||
Treatment of 1,6-diamino-2-oxo-pyridine derivatives 1a,b with different aldehydes such as 4-ethoxybenzaldehyde, 5-bromo-2-hydroxybenzaldehyde, 2-hydroxynaphthaldehyde, and 9-anthraldehyde in either refluxing solution in the presence of acid catalyst (acetic acid) or stirring the solution in the presence of cerium ammonium nitrate at room temperature to produce the corresponding Schiff base derivatives 2a,b, 3a,b, 4a,b, and 5a,b, respectively (Scheme 1). The structures of Schiff base derivatives were confirmed using elemental (CHN) and spectral analysis (IR, 1H NMR, and 13C NMR). Thus, the IR spectrum of compounds 2a,b, as an example, showed absorption bands between ν 3438 to 3300 cm−1 for amino groups and cyano group at ν 2214 and 2213 cm−1, as well as bands for carbonyl group between ν 1670–1668 cm−1, and Schiff base imine group (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) at ν 1628 and 1627 cm−1, respectively. Additionally, the 1H NMR spectra of compound 2a revealed two signals in the upfield region at δ 1.36 and 4.15 ppm as triplet and quartet referred to the ethoxy group, and the seven aromatic protons appeared as four doublets at δ 7.10, 7.45, 7.56, and 7.96 ppm and one triplet signals at δ 7.28 ppm. As well, a broad singlet signal appeared at δ 8.46 ppm assignable to two amino protons (NH2), besides the methine protons (–CH
N) at ν 1628 and 1627 cm−1, respectively. Additionally, the 1H NMR spectra of compound 2a revealed two signals in the upfield region at δ 1.36 and 4.15 ppm as triplet and quartet referred to the ethoxy group, and the seven aromatic protons appeared as four doublets at δ 7.10, 7.45, 7.56, and 7.96 ppm and one triplet signals at δ 7.28 ppm. As well, a broad singlet signal appeared at δ 8.46 ppm assignable to two amino protons (NH2), besides the methine protons (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–) at δ 8.79 ppm. The 13C NMR spectrum of compound 2b displayed specific signals at δ 14.99 and 64.11 ppm attributed to the ethoxy, signals at δ 84.04, 159.64, 163.06, 164.07, and 172.21 ppm related to carbon linked to the cyano group, methine carbon (–CH
N–) at δ 8.79 ppm. The 13C NMR spectrum of compound 2b displayed specific signals at δ 14.99 and 64.11 ppm attributed to the ethoxy, signals at δ 84.04, 159.64, 163.06, 164.07, and 172.21 ppm related to carbon linked to the cyano group, methine carbon (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–), carbon linked attached to the amino group and ethoxy group, and carbonyl group (C
N–), carbon linked attached to the amino group and ethoxy group, and carbonyl group (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), respectively.
O), respectively.
By the same manner, the IR spectrum of compound 4a,b revealed absorption bands between 3393 and 3315 cm−1 assigned to amino (–NH2) and hydroxyl groups (–OH), as well as the cyano group bands appear at ν 2213 and 2207 cm−1, besides two bands related to carbonyl group (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) and methine-CH (–CH
O) and methine-CH (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–) displayed in the range of ν 1655–1640 cm−1 and 1631–1599 cm−1, respectively. The 1H NMR spectra of compound 4a exhibited three specific singlet signals at δ = 8.47, 9.09 and 10.94 ppm assigned to amino protons (–NH2), methine-CH, and hydroxyl proton (–OH), respectively. In addition to nine aromatic protons between δ 7.29–8.25 ppm as five doublet and one triplet. Moreover, the 13C NMR spectrum of Schiff base 4a assigned three singlet signals at δ 155.07, 157.55, and 164.92 ppm due to carbon linked to amino, hydroxyl groups, and carbonyl carbons, respectively. In addition, four significant signals at δ 84.28, 116.34, 116.94, and 154.02 related to carbon attached to cyano group, two nitrile functions, and methine-carbon (C
N–) displayed in the range of ν 1655–1640 cm−1 and 1631–1599 cm−1, respectively. The 1H NMR spectra of compound 4a exhibited three specific singlet signals at δ = 8.47, 9.09 and 10.94 ppm assigned to amino protons (–NH2), methine-CH, and hydroxyl proton (–OH), respectively. In addition to nine aromatic protons between δ 7.29–8.25 ppm as five doublet and one triplet. Moreover, the 13C NMR spectrum of Schiff base 4a assigned three singlet signals at δ 155.07, 157.55, and 164.92 ppm due to carbon linked to amino, hydroxyl groups, and carbonyl carbons, respectively. In addition, four significant signals at δ 84.28, 116.34, 116.94, and 154.02 related to carbon attached to cyano group, two nitrile functions, and methine-carbon (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) respectively, as well as the aromatic carbons ranged from δ 113.50 to 146.84 ppm.
N) respectively, as well as the aromatic carbons ranged from δ 113.50 to 146.84 ppm.
Furthermore, our work extended to the synthesis of polycyclic compounds; the starting materials 1a,b was subjected to react with cyclic-ketone containing poly carbonyl groups as ninhydrin and or/isatin to produce the isolable intermediate Schiff base derivatives 6a,b and 8a,b, respectively (Scheme 2). The elemental and spectral data confirmed the structure of the prepared compounds. The 1H NMR of Schiff base derivative 8a showed two significant singlet signals at δ 8.4 and 11.02 ppm due to amino protons (NH2) and –NH proton of the isatin molecule, besides the seven aromatic protons ranging from δ 6.78 to 7.56 ppm, where the thiophene protons appear as one doublet at δ 7.33 ppm with coupling constant J = 3.6 Hz and two doublet signals at δ 6.78 and 6.89 ppm with coupling constant J = 4.9 and 3.2 Hz, respectively. The four aromatic protons related to the isatin nucleus displayed as two doublets at δ 7.06 and 7.48 ppm with coupling constant J = 7.2 Hz and one doublet signal with integration two at 7.56 ppm. The 13C NMR spectrum of the same compound revealed characteristic signals at δ 109.15, 114.10, 155.08, 166.09, and 172.12 ppm due to two cyano groups, imino carbon (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), and two carbonyl carbons, respectively.
N), and two carbonyl carbons, respectively.
In medicinal chemistry, polycyclic ring structures or fused rings serve as useful building blocks for developing multifunctional drugs and improving pharmacokinetics.57,58 The target tetracyclic compounds 7a,b and 9a,b were synthesized by the reaction of 1,6-diamino-pyridine derivatives 1a,b as bi-nucleophile with bi-electrophile molecules (ninhydrin and isatin) and therefore occurred condensation reaction. The structure of polycyclic compounds 7a,b and 9a,b was confirmed chemically by heating or stirring of Schiff base derivatives 6a,b and 8a,b in the presence of specific catalyst (acid or CAN) to produce the target polycyclic compounds 7a,b and 9a,b via cyclization reaction (Scheme 2). For more details, the target polycyclic compounds were prepared using four different methods (see Experimental section). Compared to the conventional methods, the best yield was obtained when using cerium ammonium nitrate as a green catalyst at room temperature. Elemental and spectral data confirmed the structure of compounds. The IR spectrum of 7a,b displayed stretching absorption bands due to two carbonyl and cyano groups at the exact frequencies. Also, 1H NMR of spectrum of compound 7a showed seven aromatic protons ranging between δ 7.42–8.36 ppm classified as three protons for thiophene nucleus appear as doublet signal at δ 7.82 ppm with coupling constant J = 2.7 Hz and integration two and one triplet signal at δ 7.42 ppm with coupling constant J = 4.3 Hz and integration one, while the others four protons related to phenyl ring of indeneone core and appear as one multiplet and two doublet signals at δ 8.12–8.15, 8.20, and 8.36 ppm. The 1H NMR of spectrum of compound 7b displayed only signals for aromatic protons ranging between δ 7.25 to 8.21 ppm with three signals at δ 7.25, 7.51, and 7.80 ppm with coupling constant J = 3.6, 3.2, and 4.0 Hz corresponding to furan bioactive core. Moreover, the 13C NMR of spectrum of compound 7a assigned significant signals at δ 91.50 and 99.17 ppm due to two carbons attached to nitrile groups, two singlet signals at δ 114.68 and 115.67 ppm for two cyano groups, besides signals for imino carbon (–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) and two carbonyl functions appeared at δ 156.20, 160.58, and 183.62 ppm, respectively. Similarly, the IR structure of compound 9a revealed significant absorption bands at ν 3173, 2213, and 1647 cm−1 attributed to NH, nitrile (CN), and carbonyl functions, respectively. Moreover, the 1H NMR spectra of the same compounds displayed one exchangeable singlet signal at δ 11.79 ppm for NH-isatin, besides seven aromatic protons ranging from δ 7.08 to 7.88 ppm. In addition, the 13C NMR spectrum of compound 9a revealed characteristic signals at δ 156.35, 163.69, and 168.83 ppm for two imino carbons (–C
N) and two carbonyl functions appeared at δ 156.20, 160.58, and 183.62 ppm, respectively. Similarly, the IR structure of compound 9a revealed significant absorption bands at ν 3173, 2213, and 1647 cm−1 attributed to NH, nitrile (CN), and carbonyl functions, respectively. Moreover, the 1H NMR spectra of the same compounds displayed one exchangeable singlet signal at δ 11.79 ppm for NH-isatin, besides seven aromatic protons ranging from δ 7.08 to 7.88 ppm. In addition, the 13C NMR spectrum of compound 9a revealed characteristic signals at δ 156.35, 163.69, and 168.83 ppm for two imino carbons (–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), and carbonyl group, respectively.
N), and carbonyl group, respectively.
2.2. Antimicrobial evaluation
| Cpd no. | Bacteria | Fungi | |||||
|---|---|---|---|---|---|---|---|
| Gram +ve | Gram −ve | ||||||
| Enterobacteriaceae | Non-Enterobacteriaceae | ||||||
| MSSA | MRSA | E. coli | K. pneumoniae | P. aeruginosa | A. baumannii | C. albicans | |
| a Well diameter = 8 mm, MSSA = methicillin sensitive S. aureus (ATCC 25923), MRSA = methicillin resistant S. aureus (ATCC 43300); Escherichia coli = E. coli (ATCC-25922) and K. pneumoniae = Klebsiella pneumoniae (ATCC-700603); P. aeruginosa = Pseudomonas aeruginosa (ATCC-2785), and A. baumannii = Acinetobacter baumannii (ATCC-19606); C. albicans = Candida albicans (ATCC-10231) Gen. = Gentamicin; FCZ = Fluconazole. | |||||||
| 2a | 18.3 ± 0.57 | 13.4 ± 0.57 | 13.3 ± 0.57 | 25.3 ± 0.57 | 18.3 ± 0.57 | 22.3 ± 0.57 | 16.3 ± 0.57 |
| 2b | 16.3 ± 0.57 | 13.3 ± 0.57 | 17.3 ± 0.57 | 21.3 ± 0.57 | 18.3 ± 0.57 | 22.6 ± 0.57 | 18.3 ± 0.57 |
| 3a | 20.3 ± 0.57 | 15.3 ± 0.57 | 15.3 ± 0.57 | 20.3 ± 0.57 | 20.3 ± 0.57 | 20.3 ± 0.57 | 16.3 ± 0.57 |
| 3b | 23.3 ± 0.57 | 20.3 ± 0.57 | 16.7 ± 0.57 | 20.3 ± 0.57 | 18.3 ± 0.57 | 21 ± 1.00 | 14.3 ± 0.57 |
| 4a | 15.3 ± 0.57 | 15.3 ± 0.57 | 17.3 ± 0.57 | 20.3 ± 0.57 | 18.3 ± 0.57 | 23.3 ± 0.57 | 16.6 ± 0.57 |
| 4b | 20.3 ± 0.57 | 16.3 ± 0.57 | 17.6 ± 0.57 | 20.3 ± 0.57 | 15.3 ± 0.57 | 20.3 ± 0.57 | 13 ± 1.00 |
| 5a | 18.3 ± 0.57 | 14.3 ± 0.57 | 17.3 ± 0.57 | 20.3 ± 0.57 | 15.3 ± 0.57 | 21 ± 1.00 | 15.3 ± 0.57 |
| 5b | 18.3 ± 0.57 | 16.3 ± 0.57 | 16.6 ± 0.57 | 22.3 ± 0.57 | 16.3 ± 0.57 | 21 ± 1.00 | 15.3 ± 0.57 |
| 6a | 18.3 ± 0.57 | 18.3 ± 0.57 | 17.3 ± 0.57 | 20.3 ± 0.57 | 17.3 ± 0.57 | 30.3 ± 0.57 | 15.3 ± 0.57 |
| 6b | 17.3 ± 0.57 | 16.3 ± 0.57 | 18.3 ± 0.57 | 20.3 ± 0.57 | 18.3 ± 0.57 | 16 ± 1.00 | 15.6 ± 0.57 |
| 7a | 20.3 ± 0.57 | 17.3 ± 0.57 | 14.6 ± 0.57 | 19 ± 1.00 | 12.3 ± 0.57 | 26.3 ± 0.57 | 15.3 ± 0.57 |
| 7b | 15.3 ± 0.57 | — | 15.3 ± 0.57 | 20.3 ± 0.57 | 16 ± 1.00 | 25.3 ± 0.57 | 13 ± 1.00 |
| 8a | 20.3 ± 0.57 | 15.3 ± 0.57 | 15.3 ± 0.57 | 18.3 ± 0.57 | 15.3 ± 0.57 | 23.3 ± 0.57 | 15.3 ± 0.57 |
| 8b | 13.3 ± 0.57 | 14.3 ± 0.57 | 16.6 ± 0.57 | 22 ± 1.00 | 16.3 ± 0.57 | 23.3 ± 0.57 | 14.3 ± 0.57 |
| 9a | 16 ± 1.00 | 15.3 ± 0.57 | 15.3 ± 0.57 | 19 ± 1.00 | 15 ± 1.00 | 23.3 ± 0.57 | 12.3 ± 0.57 |
| 9b | 17.3 ± 0.57 | 15.3 ± 0.57 | 14.6 ± 0.57 | 18.3 ± 0.57 | 20.3 ± 0.57 | 18.3 ± 0.57 | 15.3 ± 0.57 |
| Gen. | 19.6 ± 0.57 | 22.3 ± 0.57 | 18.3 ± 1.52 | 18.3 ± 0.57 | 17.3 ± 0.57 | 22 ± 1.00 | NA |
| FCZ | NA | NA | NA | NA | NA | NA | 16.3 ± 1.15 |
For methicillin-sensitive Staphylococcus aureus (MSSA) as a Gram-positive bacterial, the pyridine derivatives 2, 6, 7, and 8 that included thiophene moiety in position four displayed inhibition zones ranging from 18.3 ± 0.57 mm to 20.3 ± 0.57 mm higher than their analog furan-pyridine derivatives. In contrast, the combination of furan with pyridine showed higher activity in the case of compounds 3, 4, 5, and 9, with a zone of inhibition ranging from 17.3 ± 0.57 mm to 23.3 ± 0.57 mm compared with thiophene-pyridine derivatives and Gentamicin as a positive control (IZ = 19.6 ± 0.57 mm). The most active compounds against MSSA strain were revealed by furan–pyridine derivative 3b by IZ = 23.3 ± 0.57 mm. Additionally, for methicillin-resistant Staphylococcus aureus (MRSA), the synthesized pyridine derivatives 2–9 showed zone of inhibition ranging from 13.3 ± 0.57 mm to 20.3 ± 0.57 mm in comparison to Gentamicin (IZ = 22.3 ± 0.57 mm). The highest activity of 2-oxo-4-(thiophen-2-yl)-1,2-dihydropyridine derivative 3b against MSSA and MRSA may be attributed to the presence of the hydroxy group in position two and bromo atom in position five in the benzylidene ring at N1 of 1,6-diaminopyridine motif. Moreover, replacing the bromo atom with a fused phenyl ring to form N1-(2-hydroxynaphthalene)pyridine derivatives 4a,b don't enhance the activity indicating that the bromo atom has an important effect on antibacterial activity.
For Gram-negative strains from family of Enterobacteriaceae, all the designed pyridine derivatives 2, 3, 4, 5, 6, and 8 and tetracyclic derivatives 7 and 9 revealed higher activity against Klebsiella pneumoniae with zone of inhibition ranging from 18.3 ± 0.57 mm to 25.3 ± 0.57 mm compared to Gentamicin as a positive control (IZ = 18.3 ± 0.57 mm). Moreover, for Klebsiella pneumoniae the 1-((4-ethoxybenzylidene)amino)-2-oxo-4-(thiophen-2-yl)-pyridine derivative 2a showed the highest width for zone of inhibition (IZ = 25.3 ± 0.57 mm) and that may be due to the presence of ethoxy group attached to benzylidene at N1 that donating electron to N1 and therefore to pyridine nucleus. In addition, for Escherichia coli strain, the 6-amino-1-((1,3-dioxo-1,3-dihydro-2H-inden-2-ylidene)amino)-4-(furan-2-yl)-2-oxo-pyridine derivative 6b and 6-amino-4-(furan-2-yl)-1-(((2-hydroxynaphthalen-1-yl)methylene)amino)-2-oxo-pyridine derivative 4b displayed the most active members among these group with zone of inhibition (IZ = 18.3 ± 0.57 and 17.6 ± 0.57 mm), respectively, and compared with Gentamicin (IZ = 18.3 ± 1.52 mm). In contrast, the other derivatives exhibited very close zone of inhibition (13.3 ± 0.57 to 17.3 ± 0.57 mm).
For other Gram-negative strains outside family of Enterobacteriaceae (including, Pseudomonas aeruginosa and Acinetobacter baumannii) the tested pyridines and tetracyclic-containing pyridines appeared to effective with good diameter for zone of inhibitions indicating a broad-spectrum activity. Surprisingly, most of the pyridine derivatives exhibited super activity with a higher zone of inhibitions against non-Enterobacteriaceae strains compared with Gentamicin. The designed pyridine derivatives 2, 3, 4, 5, 6, and 8 exhibited zone of inhibitions (IZ) ranging between (15.3 ± 0.57 to 20.3 ± 0.57 mm) and (16 ± 1.00 to 30.3 ± 0.57 mm) compared with Gentamicin (IZ = 17.3 ± 0.57 and 22 ± 1.00 mm) against P. aeruginosa and A. baumannii strains, respectively. Additionally, the 9-(furan-2-yl)-7-oxo-[1,2,4]triazino[5,6-b]indole derivative 9b and 6-amino-1-((5-bromo-2-hydroxybenzylidene)-amino)-2-oxo-4-(thiophen-2-yl)-pyridine derivative 3a displayed equipotent activity with the highest zone of inhibition (IZ = 20.3 ± 0.57 mm) against to P. aeruginosa and higher than Gentamicin (IZ = 17.3 ± 0.57 mm), as well as higher than other tetracyclic derivatives 7a, 7b, and 9a that showed IZ = 12.3 ± 0.57, 16 ± 1.00, and 15 ± 1.00 mm, respectively. On the other hand, the 9-(furan-2-yl)-7-oxo-[1,2,4]triazino[5,6-b]indole derivative 9b revealed the lowest activity against A. baumannii with the zone of inhibitions (IZ = 18.3 ± 0.57 mm) compared with others tetracyclic derivatives 7a, 7b, and 9a that showed IZ = 26.3 ± 0.57, 25.3 ± 0.57, 23.3 ± 0.57 mm, respectively and compared with the Gentamicin (IZ = 23.3 ± 0.57 mm). For antifungal activity (Candida albicans), four derivatives 2a, 2b, 3a, and 4a among the sixteen derivatives that synthesized displayed zone of inhibition IZ = 16.3 ± 0.57, 18.3 ± 0.57, 16.3 ± 0.57 and 16.6 ± 0.57 mm higher than Fluconazole (IZ = 16.3 ± 1.15 mm). In addition, the rest of the other derivatives showed good antifungal activity with a zone of inhibitions ranging between 12.3 ± 0.57–15.6 ± 0.57 mm.
Finally, it can be concluded that the designed pyridine or tetracyclic containing pyridine derivatives with furan or thiophene moiety exhibited good to moderate activity against bacteria strains and yeast (Candida albicans). In addition, the conversion of the amino group at N1 to imino group (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) conjugated with benzylidene moiety or 1,3-dioxo-1,3-dihydro-2H-inden-2-ylidene derivatives showed broad spectrum among all the tested strains with a zone of inhibition in most cases higher than Gentamicin and Fluconazole as positive antibiotic controls compared with 2-hydroxynaphthalen-1-ylene, anthracen-9-ylmethylene, and tetracyclic derivatives. Moreover, the designed derivatives' activity increases by increasing the number of oxygen atoms attached to the arylidine-imino group attached to N1 of 1,6-diamino-2-oxo-pyridine derivatives.
N) conjugated with benzylidene moiety or 1,3-dioxo-1,3-dihydro-2H-inden-2-ylidene derivatives showed broad spectrum among all the tested strains with a zone of inhibition in most cases higher than Gentamicin and Fluconazole as positive antibiotic controls compared with 2-hydroxynaphthalen-1-ylene, anthracen-9-ylmethylene, and tetracyclic derivatives. Moreover, the designed derivatives' activity increases by increasing the number of oxygen atoms attached to the arylidine-imino group attached to N1 of 1,6-diamino-2-oxo-pyridine derivatives.
| Cpd no. | Bacteria | Fungi | ||||
|---|---|---|---|---|---|---|
| Gram +ve | Gram −ve | |||||
| Enterobacteriaceae | Non-Enterobacteriaceae | |||||
| MRSA | E. coli | K. pneumoniae | P. aeruginosa | A. baumannii | C. albicans | |
| 2a | 500 | 250 | 500 | 250 | 3.9 | 250 |
| 2b | 125 | 125 | 15.6 | 250 | 3.9 | 250 |
| 3a | 125 | 250 | 250 | 250 | 31.25 | 250 |
| 3b | 62.5 | 250 | 250 | 250 | 125 | 250 |
| 6a | 250 | 250 | 125 | 250 | 3.9 | 15.6 |
| 6b | 125 | 250 | 125 | 62.5 | 15.6 | 250 |
| Gen. | 125 | 250 | 250 | 250 | 1.95 | NA |
| FCZ | NA | NA | NA | NA | NA | 62.5 |
Furthermore, we further evaluated the MIC values for the most active derivatives against Enterobacteriaceae strains (E. coli and K. pneumoniae). The 1-((4-ethoxybenzylidene)amino)-2-oxo-4-(furan-2-yl)-pyridine derivative 2b represented MIC values 15.6 against Klebsiella pneumoniae with higher inhibitory activity with (MIC = 250–500 μg mL−1 and 16–32 folds) than the rest of pyridine derivatives and sixteen folds higher than Gentamicin (MIC = 250 μg mL−1). In addition, for activity against E. coli, the pyridine derivatives 2a, 3a, 3b, 6a, and 6b showed similar MIC 250 μg mL−1, while compound 2b displayed the lowest MIC 125 μg mL−1 and still demonstrated two-folds higher than Gentamicin (MIC = 250 μg mL−1).
Regarding Candida albicans, all compounds showed a similar MIC of 250 μg mL−1, except 2-oxo-pyridine derivative 6b, which displayed the lowest MIC of 15.6 μg mL−1 by sixteen folds than other derivatives and four folds than Fluconazole (MIC = 62.5 μg mL−1). Based on the above results, the 2-oxo-pyridine derivative 2b had the best antimicrobial activity against three of the tested microorganisms (A. baumannii, K. pneumoniae, and E. coli) with the lowest MIC values of 3.9, 15.6, and 125 μg mL−1 compared to Gentamicin (MIC = 1.95, 250 and 250 μg mL−1), respectively. At the same time, the pyridine derivative 6a that containing 1,3-dioxo-1,3-dihydro-2H-inden-2-ylidene and thiophene pharmacophore exhibited the best inhibitory activity against C. albicans with MIC value 15.6 μg mL−1 compared to other derivatives (MIC = 250 μg mL−1) and Fluconazole (MIC = 62.5 μg mL−1).
| Cpd no. | Bacteria | Fungi | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gram +ve | Gram −ve | |||||||||||
| Enterobacteriaceae | Non-Enterobacteriaceae | |||||||||||
| MRSA | E. coli | Klebsiella pneumoniae | P. aeruginosa | A. baumannii | Candida albicans | |||||||
| MBC | R | MBC | R | MBC | R | MBC | R | MBC | R | MFC | R | |
| a Gen. = Gentamicin; FCZ = Fluconazole. | ||||||||||||
| 2a | 125 | <4 | 250 | <4 | 125 | <4 | 125 | <4 | 1.95 | <4 | 62.5 | <4 |
| 2b | 62.5 | <4 | 125 | <4 | 1.95 | <4 | 125 | <4 | 3.9 | <4 | 62.5 | <4 |
| 3a | 62.5 | <4 | 125 | <4 | 250 | <4 | 125 | <4 | 7.8 | <4 | 125 | <4 |
| 3b | 15.6 | <4 | 125 | <4 | 31.25 | <4 | 250 | <4 | 125 | <4 | 62.5 | <4 |
| 6a | 250 | <4 | 62.5 | <4 | 125 | <4 | 125 | <4 | 1.95 | <4 | 15.6 | <4 |
| 6b | 15.6 | <4 | 125 | <4 | 15.6 | <4 | 31.25 | <4 | 15.6 | <4 | 125 | <4 |
| Gen. | 31.25 | <4 | 62.5 | <4 | 62.5 | <4 | 62.5 | <4 | 1.95 | <4 | NA | NA |
| FCZ | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 62.5 | 1 |
Moreover, for K. pneumoniae, the 4-(furan-2-yl)-pyridine derivatives 2b and 6b exhibited MBC = 1.95 and 15.6 μg mL−1 higher than pyridine derivative 3b (MBC = 31.25 μg mL−1) and compared with Gentamicin (MBC = 62.5 μg mL−1). Similarly, the 4-(furan-2-yl)-pyridine derivatives 3b and 6b demonstrated MBC = 15.6 μg mL−1 compared to Gentamicin MBC = 31.25 μg mL−1 against MRSA strain. Moreover, compound 6b demonstrated slightly bactericidal resistance on P. aeruginosa with MBC value of 31.25 μg mL−1 with two folds higher than Gentamicin (MBC = 62.5 μg mL−1). Notably, for activity against yeast (Candida albicans), the 2-oxo-pyridine derivatives 2a, 2b, 3b, and Fluconazole displayed equipotent MFC value of 62.5 μg mL−1, while compound 6a showed remarkable MFC value of 15.6 μg mL−1 with four folds higher than Fluconazole. In contrast, compounds 3a and 6b exhibited fungicidal potency with MFC value of 125 μg mL−1 nearly half of Fluconazole. As well, all pyridine derivatives had fungicidal potency based on ratio (0.25–0.50).
Finally, the combination of pyridine with thiophene or furan at C4 and incorporation with benzylidene (containing electron donating group at ortho or para position) or 1,3-dioxo-1,3-dihydro-2H-indene moieties through imino group at N1 of 1,6-diaminopyridine enhancement the bactericidal and fungicidal activity and in some cases exhibited super-activity compared to Gentamicin and Fluconazole.
| Cpd no. | Biofilm inhibitory percentage (BI) at 1/8 MIC | |||
|---|---|---|---|---|
| MRSA | E. coli | P. aeruginosa | C. albicans | |
| a Gen. = Gentamicin; Dox. = Doxycycline; FCZ = Fluconazole. | ||||
| 2a | 27.6 ± 8.79 | 46.2 ± 0.95 | 1.6 ± 2.23 | 17.6 ± 0.92 |
| 2b | 64.7 ± 1.85 | 63.2 ± 0.28 | 3.1 ± 2.41 | 29.7 ± 1.50 |
| 3a | 61.5 ± 0.45 | 63.8 ± 0.45 | 0.70 ± 0.45 | 55.1 ± 2.77 |
| 3b | 59.9 ± 2.60 | 43.0 ± 0.90 | 10.1 ± 3.32 | 20.7 ± 0.85 |
| 6a | 55.5 ± 1.86 | 10.3 ± 2.36 | 26.5 ± 5.08 | 15.15 ± 5.26 |
| 6b | 61.0 ± 0.90 | 63.8 ± 1.60 | 45.4 ± 1.30 | 75.0 ± 0.51 |
| Gen. | 52.3 ± 1.65 | 45 ± 0.45 | 29.4 ± 1.53 | 35.9 ± 2.51 |
| Dox. | 70.8 ± 0.45 | 68.3 ± 0.79 | 47.1 ± 1.48 | — |
| Flu. | — | — | — | 57.6 ± 1.7 |
For MRSA, the most active derivatives 6a and 2b exhibited good biofilm inhibition (BI) percentage values ranging from 55.5% to 64.7%, compared with Gentamicin (BI = 52.3 ± 1.65%) and Doxycycline (BI = 70.8 ± 0.45%), while compound 2a showed lowest biofilm inhibitory percentage with value of 27.6%. Moreover, the biofilm inhibitory percentage of E. coli exhibited that compounds 2b, 3a, and 6b represented biofilm inhibition nearly 63% higher than Gentamicin (BI = 45 ± 0.45%), but still near to Doxycycline (BI ∼68.3%). In addition, 2-oxo-pyridine derivatives 2a and 3b demonstrated biofilm inhibitory percentage (BI ∼46.2 and 43.0%) very close to Gentamicin (BI = 45 ± 0.45%). In contrast, compound 6a showed the least biofilm inhibition of 10.3% against E. coli.
Regarding For P. aeruginosa biofilm activity, compound 6b showed the highest percentage of biofilm inhibition with 45.4% and very close to Doxycycline (BI = 47.1%) and displayed 1.5 folds higher than Gentamicin (BI = 29.4%), followed by 2-oxo-pyridine derivatives 6a and 3b with biofilm inhibition percentages of nearly 26.5 and 10.1%, respectively. However, compounds 2a, 2b, and 3a revealed a very low inhibition percentage (ranging from 0.7 to 3.1%). Moreover, the biofilm inhibitory percentage against C. albicans was tested. The results showed that 2-oxo-pyridines 6b donated the highest percentage of biofilm inhibition of 75% compared with Fluconazole (BI = 57.6%). At the same time, compound 3a revealed a biofilm inhibitory percentage of 55.1%, while compounds 2a, 3b, 2b, and 6a displayed a low biofilm inhibitory percentage (ranging from 15.15 to 29.7).
Finally, the most active derivatives showed good to potency biofilm inhibitory percentage at a low concentration from MIC value (1/8 MIC) compared to Gentamicin and Fluconazole. Moreover, compound 6b exhibited the most active member by broad-spectrum against biofilm inhibitory percentage against the three bacterial strains (MRSA, P. aeruginosa, and E. coli), and C. albicans as a fungal pathogen. Additionally, compounds 2b and 3a revealed significant biofilm inhibition with values near Gentamicin against MRSA, and E. coli. The biofilm inhibitory percentage against C. albicans was tested, and the results showed that 2-oxo-pyridines 6b and 3a donated a high percentage of biofilm inhibition of 75 and 55.1%, respectively, while compounds 2a, 3b, 2b, and 6a displayed the lower percentage of biofilm inhibition (ranging from 15.15 to 29.7).
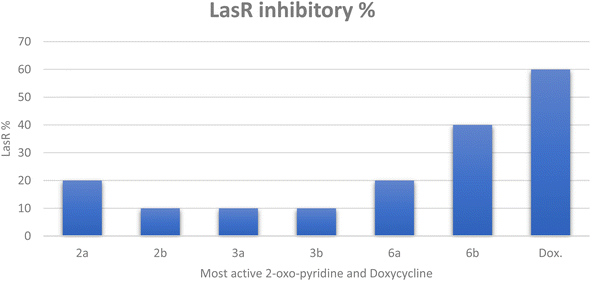 | ||
| Fig. 2 The inhibitory percentage of LasR gene expression when treated with most active 2-oxo-pyridine derivatives 2a, 2b, 3a, 3b, 6a, and 6b at 1/8 MICs. | ||
Finally, it can be concluded that the most active derivatives could downstream the production of LasR-regulated virulence factors. Our findings regarding P. aeruginosa, the model microorganism for QS studies, suggest that these derivatives act as anti-virulence and anti-quorum sensing.
The cytotoxic activity of the most active compounds on normal cell lines was evaluated to determine the safety of these derivatives. The IC50 values of the most active derivatives exhibited higher values ranging from 175.17 ± 3.49 to 344.27 ± 3.81 μg mL−1, indicating that these derivatives exhibited antimicrobial activity without harmful effects on normal cellular structure and functions (non-toxic). The cytotoxicity of the synthesized compounds was evaluated using MTT assay65,66 on Vero cells (normal cell line) and the results revealed excellent findings of most active compounds 2a, 2b, 3a, 3b, 6a, and 6b with IC50 values of 230.57 ± 6.52, 231.82 ± 2.21, 344.27 ± 3.81, 335.3 ± 7.95, 175.17 ± 3.49, 207.87 ± 5.14 μg mL−1 respectively (Tables 5, S4 and Fig. S1†).
| Cpd no. | The in vitro cytotoxic activity of the most active derivatives on Vero cell lines (IC50 ± SD) represented by μg mL−1 |
|---|---|
| 2a | 230.57 ± 6.52 |
| 2b | 231.82 ± 2.21 |
| 3a | 344.27 ± 3.81 |
| 3b | 335.3 ± 7.95 |
| 6a | 175.17 ± 3.49 |
| 6b | 207.87 ± 5.14 |
2.3. In silico studies
| Test items | Most active 2-oxo-pyridine derivatives 2a, 2b, 3a, 3b, 6a, 6b, and positive controls (Doxycycline and Gentamicin) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 2a | 2b | 3a | 3b | 6a | 6b | DOX | Gen | ||
| a M. soluble = moderately soluble; H. soluble = highly soluble; Dox. = Doxycycline; Gen. = Gentamicin. | |||||||||
| SwissADME | Molecular properties | ||||||||
| MLogP | 1.64 | 0.85 | 1.80 | 1.01 | 0.31 | −0.46 | −2.08 | −2.92 | |
| TPSA (Å2) | 145.43 | 130.33 | 156.43 | 141.33 | 170.34 | 155.24 | 181.62 | 199.73 | |
| M. wt. | 389.43 | 373.36 | 440.27 | 424.21 | 399.38 | 383.32 | 444.43 | 477.60 | |
| nHBA (NO) | 5 | 6 | 5 | 6 | 6 | 7 | 9 | 12 | |
| nHBD (OHNH) | 1 | 1 | 2 | 2 | 1 | 1 | 6 | 8 | |
| NRB | 5 | 5 | 3 | 3 | 2 | 2 | 2 | 7 | |
| Drug likeness and medicinal chemistry prediction | |||||||||
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S (ESOL) S (ESOL) |
−4.07 | −3.58 | −4.54 | −4.05 | −3.86 | −3.37 | −2.59 | 0.24 | |
| Solubility | M. soluble | Soluble | M. soluble | M. soluble | Soluble | Soluble | Soluble | H. soluble | |
| PAINS | 0 | 0 | 1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) N) |
1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) N) |
0 | 0 | 0 | 0 | |
| Lead-likeness | No (1) | No (1) | No (1) | No (1) | No (1) | No (1) | No (1) | No (1) | |
| Synthetic accessibility | 3.36 | 3.46 | 3.27 | 3.36 | 3.53 | 3.62 | 5.15 | 6.51 | |
| Bioavailability score | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 | 0.11 | 0.17 | |
| Lipinski rule (violation) | Yes (0) | Yes (0) | Yes (0) | Yes (0) | Yes (0) | Yes (0) | Yes (1) | No (2) | |
| Veber rule (violation) | No (1) | Yes (0) | No (1) | No (1) | No (1) | No (1) | Yes (0) | No (1) | |
All the 2-oxo-pyridine derivatives exhibited obeys to Lipinski rule of five without any violation. Lipinski rule considered important predictors for drug candidate to be oral bioavailability with molecular weight (M. wt.) less than 500 dalton, number of hydrogen bond acceptors (NO) less than or equal ten, number of hydrogen bond donor (OHNH) less than or equal to five, and finally, the partition coefficient MLogP ≤ 4.15. In addition, the Doxycycline showed number of hydrogen donors = 6 (i.e., higher than the standard rule), but belongs to Lipinski rule with one violation. On the other hand, Gentamicin doesn't fit the Lipinski rule due to two violations related to the number of hydrogen bond acceptors (nHBA (NO) = 12) and number of hydrogen bond donors (nHBD (OHNH) = 8). Moreover, all tested 2-oxo-pyridine derivatives 2a, 3a, 3b, 6a, 6b, and Gentamicin don't fit to Veber rule due to their topological polar surface area TPSA > 140 Å, except furano-pyridine 2b and Doxycycline Table 6.
Furthermore, in terms of drug likeness and medicinal chemistry predictions, three pyridine derivatives 2b, 6a, and 6b demonstrated solubility properties with log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S (ESOL) ranging from −0.46 to 0.85 compared to Doxycycline [log
S (ESOL) ranging from −0.46 to 0.85 compared to Doxycycline [log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S (ESOL) = −2.08] and Gentamicin [log
S (ESOL) = −2.08] and Gentamicin [log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S (ESOL) = 0.24]. In addition, the rest of derivatives 2a, 3a, and 3b exhibited moderate soluble with log
S (ESOL) = 0.24]. In addition, the rest of derivatives 2a, 3a, and 3b exhibited moderate soluble with log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S (ESOL) ranging from −4.54 to −4.05. Additionally, the values of synthetic accessibility indicated ease synthetic method with values ranging from 3.36 to 3.62 for most active derivatives compared with higher difficult synthetic accessibility to Doxycycline with values of 5.15 and Gentamicin (6.51). To our surprise, these derivatives exhibited an equal bioavailability score of 0.55, but still higher than positive controls. The drug likeness expression can be determined by how similar their properties are to those of existing drugs and all most active and positive controls (Doxycycline and Gentamicin) that showed violation in their molecular weight. The 6-amino-1-((5-bromo-2-hydroxybenzylidene)amino)-2-oxo-pyridine derivatives 3a and 3b showed the one Pan-assay interference compounds (PAINs) alarms due to imino group (C
S (ESOL) ranging from −4.54 to −4.05. Additionally, the values of synthetic accessibility indicated ease synthetic method with values ranging from 3.36 to 3.62 for most active derivatives compared with higher difficult synthetic accessibility to Doxycycline with values of 5.15 and Gentamicin (6.51). To our surprise, these derivatives exhibited an equal bioavailability score of 0.55, but still higher than positive controls. The drug likeness expression can be determined by how similar their properties are to those of existing drugs and all most active and positive controls (Doxycycline and Gentamicin) that showed violation in their molecular weight. The 6-amino-1-((5-bromo-2-hydroxybenzylidene)amino)-2-oxo-pyridine derivatives 3a and 3b showed the one Pan-assay interference compounds (PAINs) alarms due to imino group (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) in benzylidene moiety that has hydroxy group Table 6.
N) in benzylidene moiety that has hydroxy group Table 6.
The in silico toxicity predictions were also conducted for the 2-oxo-pyridine derivatives to determine their adverse effects and toxicity using Protox-II web tool as the same method described previously71,72 and the results listed in Table 7. The tested 2-oxo-pyridine derivatives belong to class four in toxicity classes according to globally harmonized system (GHS) that demonstrated LD50 from 600 mg kg−1 to 1300 mg kg−1 in comparison to Doxycycline (LD50 = 1007 mg kg−1) and Gentamicin (LD50 = 5000 mg kg−1 that belongs to class V). The tested pyridine derivatives displayed non-cytotoxic and non-immunotoxin compared with Doxycycline and Gentamicin that displayed immunotoxin behavior with probability 0.99 and 76, respectively. In addition, all derivatives showed activity to carcinogenic with probability ranging from 0.50–0.60, except for compound 3b and Doxycycline. For hepatotoxic, the 2-oxopyridine expected to cause hepatotoxic, except 6-amino-1-((4-ethoxybenzylidene)amino)-2-oxo-pyridine derivative 2a and 2b, as well as Gentamicin. Additionally, three pyridine derivatives 2a, 6a, and 6b increase rate of mutation, while the other derivatives 2b, 3a, 3b, and positive controls (Doxycycline and Gentamicin) exhibited inactive mutagenic potential.
| Oral toxicity prediction | Most active 2-oxo-pyridine derivatives 2a, 2b, 3a, 3b, 6a, 6b, and positive controls (Doxycycline and Gentamicin) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 2a | 2b | 3a | 3b | 6a | 6b | DOX | Gen | ||
| Oral toxicity prediction | |||||||||
| a DOX. = Doxycycline; Gen. = Gentamicin. | |||||||||
| ProTox-II prediction | LD50 mg kg−1 | 1000 | 1300 | 1000 | 600 | 600 | 600 | 1007 | 5000 |
| Toxicity class | IV | IV | IV | IV | IV | IV | IV | V | |
| Hepatotoxicity | Inactive | Inactive | Active | Active | Active | Active | Active | Inactive | |
| 0.55 | 0.52 | 0.62 | 0.67 | 0.52 | 0.56 | 0.54 | 0.83 | ||
| Carcinogenicity | Active | Active | Active | Inactive | Active | Active | Inactive | Inactive | |
| 0.55 | 0.57 | 0.50 | 0.50 | 0.58 | 0.60 | 0.77 | 0.74 | ||
| Immunotoxicity | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive | Active | Active | |
| 0.99 | 0.97 | 0.94 | 0.68 | 0.99 | 0.98 | 0.99 | 0.76 | ||
| Mutagenicity | Active | Inactive | Inactive | Inactive | Active | Active | Inactive | Inactive | |
| 0.51 | 0.51 | 0.53 | 0.57 | 0.51 | 0.50 | 0.95 | 0.77 | ||
| Cytotoxicity | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive | |
| 0.86 | 0.84 | 0.67 | 0.73 | 0.88 | 0.89 | 0.90 | 0.66 | ||
| Heat shock factor response element (HSE) | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive | |
| 0.92 | 0.91 | 0.92 | 0.91 | 0.93 | 0.92 | 0.98 | 0.98 | ||
| Mitochondrial membrane potential (MMP) | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive | |
| 0.64 | 0.67 | 0.59 | 0.62 | 0.53 | 0.55 | 0.96 | 0.84 | ||
| Phosphoprotein (tumor suppressor) p53 | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive | |
| 0.86 | 0.84 | 0.75 | 0.73 | 0.80 | 0.80 | 0.98 | 0.93 | ||
Finally, the most active derivatives revealed inactive to heat shock factor response, mitochondrial membrane potential, and phosphoprotein P53, i.e., these compounds displayed a safety profile when administered as a candidate drug, and therefore, it can be further studied in vivo in the future.
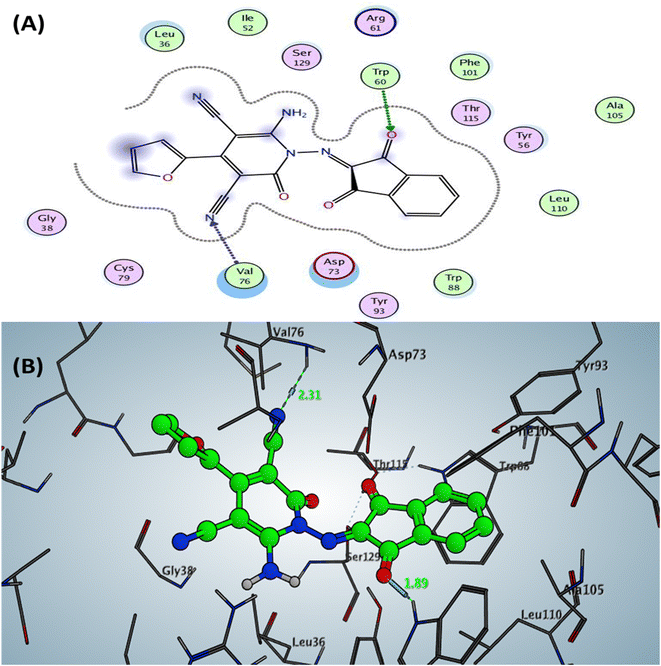 | ||
| Fig. 3 Docking pose and ligand interaction of compound 6b inside the active site of LasR (PDB: 6MVN) in P. aeruginosa (UCBPP-PA14) (A) 2D and (B) 3D structure. | ||
The Doxycycline as positive control revealed binding energy S = −6.36 kcal mol−1 (in vitro inhibitory percentage 60% at 1/8 MIC) with one backbone hydrogen bond donor between Leu39 and NH2 of amide group at ring A in tetracyclic structure with a distance of 2.41 and arene–H interaction between phenyl of phenol at ring D with the amino acid Tyr56 Fig. 4. In contrast, compound 6a demonstrated binding affinity S = −7.02 kcal mol−1 (in vitro inhibitory percentage 20% at 1/8 MIC) by forming two hydrogen bonds sidechain acceptors between the residue Arg61 and oxygen o carbonyl at C2 of pyridine nucleus and Trp60 with a nitrogen of nitrile group at C3 pf pyridine derivative with bond length 2.17 and 1.67 Å, respectively. In addition, Trp88 showed arene–arene interaction with the thiophene motif, as represented in Fig. 5.
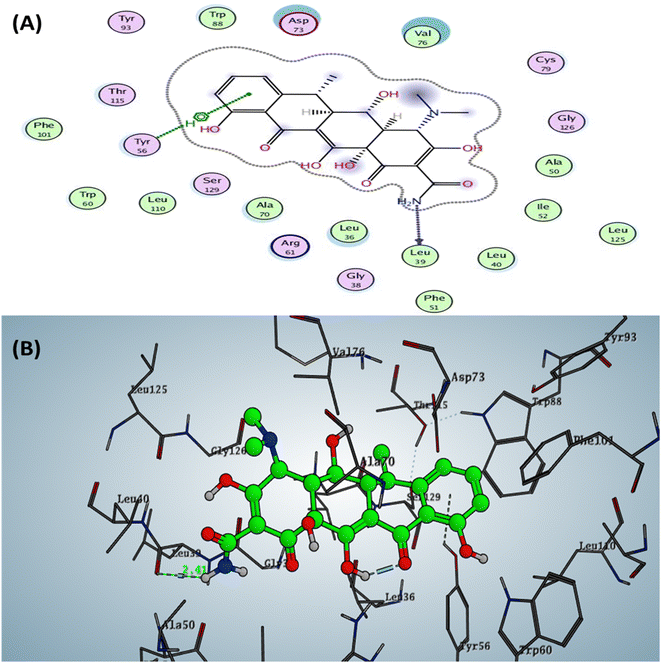 | ||
| Fig. 4 Docking pose and ligand interaction of Doxycycline inside the active site of LasR (PDB: 6MVN) in P. aeruginosa (UCBPP-PA14) (A) 2D and (B) 3D structure. | ||
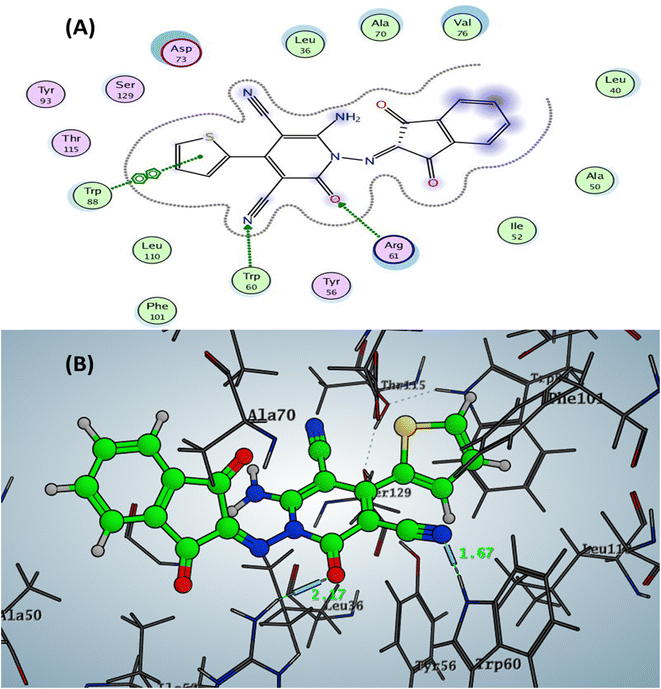 | ||
| Fig. 5 Docking pose and ligand interaction of compound 6a inside the active site of LasR (PDB: 6MVN) in P. aeruginosa (UCBPP-PA14) (A) 2D and (B) 3D structure. | ||
The designed 4-thiophenyl-2-oxo-pyridine derivative 2a and 3a revealed binding energy S = −6.74 and −6.57 kcal mol−1 through hydrogen bond sidechain acceptor between the residue Arg61. Firstly, for compound 2a, the arginine could form only one H-bond with the oxygen of the carbonyl group at C2 of the pyridine nucleus with a bond length of 2.45 Å. In contrast, compound 3a displayed two hydrogen bonds with a nitrogen of nitrile group with a bond length of 2.13 Å (with NH2 of arginine) and 2.49 Å (with nitrogen of imine of arginine). Moreover, the designed pyridine derivative 2b exhibited binding energy S = −6.51 kcal mol−1 (in vitro inhibitory percentage 10% at 1/8 MIC) with two hydrogen bonds sidechain acceptors between Arg61 and oxygen of carbonyl at C2 and nitrogen of nitrile at C3 on the pyridine nucleus with bond lengths of 2.31 and 2.63 Å. In addition, the amino group at C6 of pyridine could form a hydrogen bond donor with the sidechain amino acid The75 with a bond length of 2.06 Å. The docking complex of compound 3b demonstrated binding energy S = −6.61 kcal mol−1 (in vitro inhibitory percentage 10% at 1/8 MIC) through two hydrogen bonds with the sidechain residues. The first H-bond conducted between the Arg61 and the oxygen of carbonyl at C2 of pyridine with a bond length of 2.26 Å, while the other H-bond displayed with the hydroxyl group of benzylidene at N1 of pyridine with a bond length 2.04 Å Fig. 6 (all figures with complete size represented in ESI file Fig. S2–S17†).
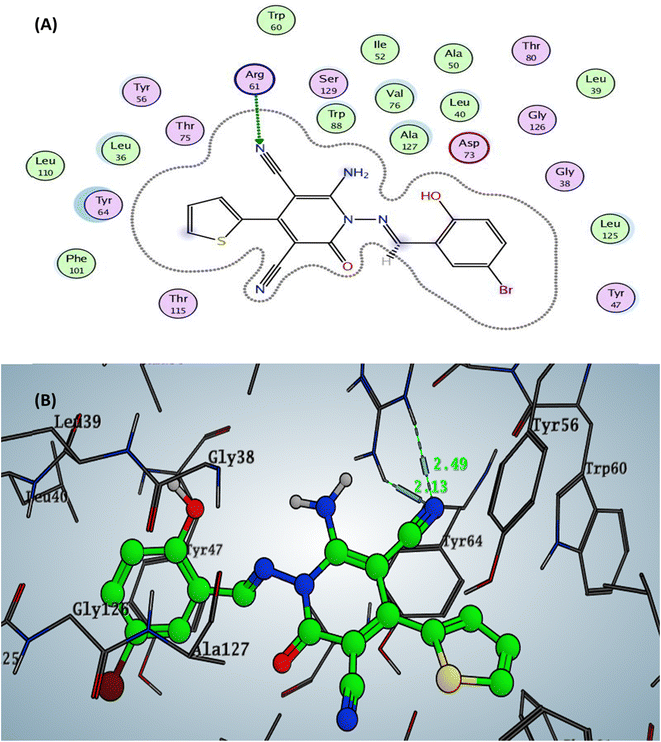 | ||
| Fig. 6 Docking pose and ligand interaction of compound 3a inside the active site of LasR (PDB: 6MVN) in P. aeruginosa (UCBPP-PA14) (A) 2D and (B) 3D structure. | ||
Finally, it can be concluded that the binding mode of furanone derivative and its binding energy as a co-crystallized ligand in the active site of the tested enzyme exhibited that our designed 2-oxo-pyridine nucleus act as QS inhibitors by downregulating LasR via H-bonds with the active site residues and hydrophobic interaction. Moreover, the carbonyl group at 2-oxo-pyridine derivative displayed H-bonds as the carbonyl of furanone.
3. Conclusion
This work introduced a new advancement in fighting P. aeruginosa by attenuating their virulence and disrupting their QS. A new series of 1,6-diamino-2-oxo-pyridine derivatives containing azomethine group at N1 of pyridine containing different fragment pharmacophores or tetracyclic rings containing pyridine were synthesized by green chemistry using stirring and CAN as a catalyst and reflux approaches. The in vitro antimicrobial activity was screened for synthesized pyridine derivatives with six promising candidates. The most active compounds 2a, 2b, 3a, 3b, 6a, and 6b showed significant and broad spectrum with MIC values of 3.9–500 μg mL−1 against bacterial strains compared to Gentamicin (MIC = 1.95–250 μg mL−1). Moreover, the MBC and MFC were determined, and their ratio with MIC revealed that these derivatives exhibited bactericidal and fungicidal potency. Anti-biofilm activity was evaluated against three bacteria (MRSA, E. coli, and P. aeruginosa) and C. albicans as fungal pathogen at concentrations 1/2, 1/4, and 1/8 MIC, and the results demonstrated strong reduction to biofilm formation. Furthermore, these six derivatives revealed down-regulation for the LasR gene as autoinducers for QS signals of P. aeruginosa with values ranging from 40 to 10% at low concentration (1/8 MIC) using the qPCR technique. The most active compound 6b reduced the expression of the lasR gene by 40% at 1/8 MIC compared to untreated P. aeruginosa. Subsequently, these derivatives exhibited IC50 values ranging from 175.17 ± 3.49 to 344.27 ± 3.81 μg mL−1 against Vero cells (normal cell lines), indicating non-toxic properties on normal cells. Additionally, the in silico ADMET properties, including molecular properties, drug-likeness, and some medicinal chemistry parameters, were predicted, and the tested pyridine derivatives possessed acceptable and good results as drug-like characteristics. The molecular docking simulation was determined for most active derivatives inside the LasR gene (PDB: 6MVN) to determine the binding mode interaction and binding energy that showed negative values ranging (−6.51 to −7.11 kcal mol−1) and, therefore, suggested that these derivatives can be used for further in vivo studies. Finally, we concluded that these novel compounds could provide additive value in fighting P. aeruginosa by attenuating their virulence and QS to overcome the continuous emergence of resistant strains and encourage us to test the efficacy of these compounds on other bacterial strains in the future.4. Experimental
4.1. Chemistry
4.2. Synthesis of target compounds
4.2.1.1 Method (a). To a solution of pyridine derivatives 1a,b (0.01 mmol) and the appropriate substituted aldehydes (0.01 mmol) or ketones (0.01 mmol) namely, 4-ethoxybenzaldehyde, 5-bromo-2-hydroxybenzaldehyde, 2-hydroxy-1-naphthaldehyde, and 9-anthraldehyde, ninhydrin, and isatin in dimethylformamide as a solvent catalyzed with acetic acid (1 mL). The solution mixture was heated under reflux for 3–6 h (monitoring by TLC). After the reaction is completed, the solution mixture was added to 20 mL cold ethanol, and the precipitated solid was filtered, washed with ethanol, dried, and crystallized from boiling with ethanol to yield the desired compounds.
4.2.1.2 Method (b). A mixture of pyridine derivatives 1a,b (0.01 mmol), substituted aldehydes (0.01 mmol) or ketones (0.01 mmol) (4-ethoxybenzaldehyde, 5-bromo-2-hydroxybenzaldehyde, 2-hydroxy-1-naphthaldehyde, and anthracene-9-carbaldehyde, ninhydrin, isatin), cerium ammonium nitrate (2 mol%) as Lewis acid catalyst, and acetonitrile (10 mL) as solvent were combined to a 100 mL rounded-flask and stirred at room temperature for 45–90 min (monitoring by TLC). After the reaction is completed, the solid product that precipitated was filtered out, washed with ethanol, dried, and crystallized from boiling with ethanol to produce the desired compounds with high yield.
4.2.1.2.1 6-Amino-1-((4-ethoxybenzylidene)amino)-2-oxo-4-(thiophen-2-yl)-1,2-dihydro-pyridine-3,5-dicarbonitrile (2a). M.p.: 215–217 °C; pale-yellow powder; yield 76% (89%); IR (KBr): νmax 3425, 3301 (NH2), 3080 (aromatic-H), 2981 (aliphatic-H), 2213 (CN), 1670 (CO), 1628 (CH = N); 1H NMR (δ, ppm) 1.36 (t, 3H,
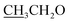 ), 4.15 (q, 2H,
), 4.15 (q, 2H, 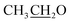 ), 7.10 (d, J = 8.8 Hz, 2H, Ar–H), 7.28 (t, J = 4.8 Hz, 1H, thiophene-H), 7.45 (d, J = 8.8 Hz, 1H, Ar–H), 7.56 (d, J = 3.6 Hz, 1H, thiophene-H), 7.96 (d, J = 8.8 Hz, 2H, Ar–H), 8.46 (s, 2H, NH2, D2O exchangeable), 8.79 (s, 1H, methylinic-H); 13CNMR (δ, ppm) 14.92
), 7.10 (d, J = 8.8 Hz, 2H, Ar–H), 7.28 (t, J = 4.8 Hz, 1H, thiophene-H), 7.45 (d, J = 8.8 Hz, 1H, Ar–H), 7.56 (d, J = 3.6 Hz, 1H, thiophene-H), 7.96 (d, J = 8.8 Hz, 2H, Ar–H), 8.46 (s, 2H, NH2, D2O exchangeable), 8.79 (s, 1H, methylinic-H); 13CNMR (δ, ppm) 14.92 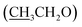 , 64.11
, 64.11 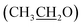 , 86.88 (
, 86.88 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –CN), 115.34 (CN), 115.17 (CN), 116.07, 124.54, 128.22, 128.87, 129.96, 130.79, 131.25, 132.33, 132.45, 133.80, 152.12 (Ar–Cs), 157.24 (CH = N), 159.68 (C–NH2), 164.02 (
–CN), 115.34 (CN), 115.17 (CN), 116.07, 124.54, 128.22, 128.87, 129.96, 130.79, 131.25, 132.33, 132.45, 133.80, 152.12 (Ar–Cs), 157.24 (CH = N), 159.68 (C–NH2), 164.02 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –OEt), 172.45 (C
–OEt), 172.45 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); anal calcd. For C20H15N5O2S (389.4): C, 61.68; H, 3.88; N, 17.98. Found: C, 67.61.52; H, 3.68; N, 17.81.
O); anal calcd. For C20H15N5O2S (389.4): C, 61.68; H, 3.88; N, 17.98. Found: C, 67.61.52; H, 3.68; N, 17.81.
4.2.1.2.2 6-Amino-1-((4-ethoxybenzylidene)amino)-2-oxo-4-(furan-2-yl)-1,2-dihydro-pyridine-3,5-dicarbonitrile (2b). M.p.: 226–228 °C; pale-yellow powder; yield 77% (89%); IR (KBr): νmax 3438, 3300 (NH2), 3119 (aromatic-H), 2981 (aliphatic-H), 2214 (CN), 1668 (CO), 1627 (CH = N). 1H NMR (δ, ppm) 1.35 (t, 3H,
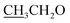 ), 4.13 (q, 2H,
), 4.13 (q, 2H, 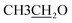 ), 6.81 (d, J = 3.6 Hz, 2H, furan-H), 7.07 (t, J = 5.6 Hz, 1H, furan-H), 7.31 (d, J = 8.8 Hz, 1H, Ar–H), 7.82 (d, J = 8.8 Hz 1H, Ar–H), 7.94 (d, J = 8.8 Hz, 2H, Ar–H), 8.34 (s, 2H, NH2, D2O exchangeable), 8.75 (s, 1H, methylinic-H); 13CNMR (δ, ppm) 14.99
), 6.81 (d, J = 3.6 Hz, 2H, furan-H), 7.07 (t, J = 5.6 Hz, 1H, furan-H), 7.31 (d, J = 8.8 Hz, 1H, Ar–H), 7.82 (d, J = 8.8 Hz 1H, Ar–H), 7.94 (d, J = 8.8 Hz, 2H, Ar–H), 8.34 (s, 2H, NH2, D2O exchangeable), 8.75 (s, 1H, methylinic-H); 13CNMR (δ, ppm) 14.99 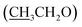 , 64.35
, 64.35 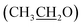 , 84.04 (
, 84.04 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –CN), 115.11, 115.28, (2CN), 116.26, 116.76, 117.58, 118.06, 124.35, 128.91, 130.02, 133.00, 145.22, 146.17, 147.51 (Ar–Cs), 159.64 (CH = N), 163.06 (C–NH2), 164.07 (C–OEt), 172.21 (C
–CN), 115.11, 115.28, (2CN), 116.26, 116.76, 117.58, 118.06, 124.35, 128.91, 130.02, 133.00, 145.22, 146.17, 147.51 (Ar–Cs), 159.64 (CH = N), 163.06 (C–NH2), 164.07 (C–OEt), 172.21 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); anal calcd. For C20H15N5O3 (373.37): C, 64.34; H, 4.05; N, 18.76. Found: C, 63.91.52; H, 3.98; N, 18.54.
O); anal calcd. For C20H15N5O3 (373.37): C, 64.34; H, 4.05; N, 18.76. Found: C, 63.91.52; H, 3.98; N, 18.54.
4.2.1.2.3 6-Amino-1-((5-bromo-2-hydroxybenzylidene)amino)-2-oxo-4-(thiophen-2-yl)-1,2-dihydropyridine-3,5-dicarbonitrile (3a). M.p.: 288–290 °C; brown powder; yield 80% (91%); IR (KBr): νmax 3305, 3210, 3102 (OH, NH2), 2967 (aromatic-H), 2927 (aliphatic-H), 2215 (CN), 1654 (CO), 1621 (CH = N). 1H NMR (δ, ppm) = 7.21 (t, 1H, thiophene-H), 7.31 (d, J = 3.0 Hz, 1H, thiophene-H), 7.34 (d, J = 7.1 Hz, 1H, Ar–H), 8.01 (d, J = 3.2 Hz, 1H, thiophene-H), 8.04 (d, J = 4.8 Hz, 1H, Ar–H), 8.07 (s, 1H, Ar–H), 8.33 (d, 2H, NH2, D2O exchangeable), 9.02 (s, 1H, methylinic-H), 10.56 (s, 1H, O–H, D2O exchangeable); 13CNMR (δ, ppm) 86.44 (
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –CN), 113.08, 114.77, 115.64 (2CN), 116.26, 116.74, 118.35, 119.72, 127.97, 145.23, 145.68, 146.68, 146.82, 147.00 (Ar–Cs), 157.51 (CH = N), 157.66 (C–NH2), 159.97 (C
–CN), 113.08, 114.77, 115.64 (2CN), 116.26, 116.74, 118.35, 119.72, 127.97, 145.23, 145.68, 146.68, 146.82, 147.00 (Ar–Cs), 157.51 (CH = N), 157.66 (C–NH2), 159.97 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 161.62 (C–OH); anal. calcd. For C18H10BrN5O2S (440.28): C, 50.96; H, 2.38; N, 16.51. Found: C, 50.52; H, 2.26; N, 16.22.
O), 161.62 (C–OH); anal. calcd. For C18H10BrN5O2S (440.28): C, 50.96; H, 2.38; N, 16.51. Found: C, 50.52; H, 2.26; N, 16.22.
4.2.1.2.4 6-Amino-1-((5-bromo-2-hydroxybenzylidene)amino)-2-oxo-4-(furan-2-yl)-1,2-dihydro-pyridine-3,5-dicarbonitrile (3b). M.p.: 246–248 °C; orange powder; yield 84% (90%); IR (KBr): νmax 3295, 3201 (NH2, OH), 2214 (CN), 1650 (CO), 1623 (CH = N). 1H NMR (δ, ppm) 7.27 (t, J = 4.4 Hz, 1H, furan-H), 7.54 (d, J = 3.2 Hz, 1H, furan-H), 7.58 (d, J = 7.2 Hz, 1H, Ar–H), 7.62 (d, J = 3.2 Hz, 1H, furan-H), 7.93 (d, J = 6.8 Hz, 1H, Ar–H), 8.28 (s, 1H, Ar–H), 8.47 (s, 2H, NH2, D2O exchangeable), 9.05 (s, 1H, methylinic-H), 10.86 (s, 1H, OH, D2O exchangeable); 13CNMR (δ, ppm) 83.10 (
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –CN), 115.28 (CN), 115.64 (CN), 116.74, 117.06, 120.01, 124.60, 132.32, 141.83, 145.14, 145.77, 146.05, 146.77, 146.98 (Ar–Cs), 157.74 (CH = N), 159.33 (C–NH2), 159.90 (C
–CN), 115.28 (CN), 115.64 (CN), 116.74, 117.06, 120.01, 124.60, 132.32, 141.83, 145.14, 145.77, 146.05, 146.77, 146.98 (Ar–Cs), 157.74 (CH = N), 159.33 (C–NH2), 159.90 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 161.52 (OH); anal. calcd. For C18H10BrN5O3 (424.21): C, 50.96; H, 2.38; N, 16.51. Found: C, 50.60; H, 2.26; N, 16.12.
O), 161.52 (OH); anal. calcd. For C18H10BrN5O3 (424.21): C, 50.96; H, 2.38; N, 16.51. Found: C, 50.60; H, 2.26; N, 16.12.
4.2.1.2.5 6-Amino-1-(((2-hydroxynaphthalen-1-yl)methylene)amino)-2-oxo-4-(thiophen-2-yl)-1,2-dihydropyridine-3,5-dicarbonitrile (4a). M.p.: 388–390 °C; brown red powder; yield 65% (88%); IR (KBr): νmax 3393, 3275, 3222 (NH2, OH), 3054 (aromatic-H), 3018 (aliphatic-H), 2213 (CN), 1655 (CO), 1631 (CH = N); 1H NMR (δ, ppm) 7.29 (t, J = 9.0 Hz, 1H, Ar–H), 7.57–7.64 (m, 4H, Ar–H), (d, J = 8.2 Hz, 2H, Ar–H), 7.94 (d, J = 8.0 Hz, 1H, Ar–H), 8.07 (d, J = 9.9 Hz, 2H, Ar–H), 8.25 (d, J = 9.6 Hz, 1H, Ar–H), 8.47 (d, 2H, NH2, D2O exchangeable), 9.09 (s, 1H, methylinic-H), 10.94 (s, 1H, OH, D2O exchangeable); 13CNMR (δ, ppm) 84.28 (
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –CN), 113.50, 115.07, 116.34 (CN), 116.94 (CN), 117.20, 118.86, 119.61, 128.43, 128.68, 128.80, 129.96, 130.23, 139.14, 139.24, 145.11, 145.65, 146.84 (Ar–Cs), 154.02 (CH = N), 155.07 (C–NH2), 157.55 (C–OH), 164.92 (C
–CN), 113.50, 115.07, 116.34 (CN), 116.94 (CN), 117.20, 118.86, 119.61, 128.43, 128.68, 128.80, 129.96, 130.23, 139.14, 139.24, 145.11, 145.65, 146.84 (Ar–Cs), 154.02 (CH = N), 155.07 (C–NH2), 157.55 (C–OH), 164.92 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); anal. calcd. For C22H13N5O2S (411.44): C, 64.22; H, 3.18; N, 17.02. Found: C, 63.88; H, 3.01; N, 16.78.
O); anal. calcd. For C22H13N5O2S (411.44): C, 64.22; H, 3.18; N, 17.02. Found: C, 63.88; H, 3.01; N, 16.78.
4.2.1.2.6 6-Amino-4-(furan-2-yl)-1-(((2-hydroxynaphthalen-1-yl)methylene)amino)-2-oxo-1,2-dihydropyridine-3,5-dicarbonitrile (4b). M.p.: 342–344 °C; gray powder; yield 67% (88%). IR (KBr): νmax 3385, 3266, 3115 (NH2, OH), 2926 (aliphatic-H), 2207 (CN), 1640 (CO), 1599 (CH = N); 1H NMR (δ, ppm) 7.27 (t, J = 8.0 Hz, 1H, Ar–H), 7.58 (d, J = 3.2 Hz, 2H, furan-H), 7.95 (d, J = 4.3 Hz, 2H, furan-H), 8.02 (d, J = 8.1 Hz, 1H, Ar–H), 8.07 (d, J = 9.9 Hz, 2H, Ar–H), 8.25 (d, J = 8.4 Hz, 1H, Ar–H), 8.53 (d, 2H, NH2, D2O exchangeable), 9.09 (s, 1H, methylinic-H), 11.49 (s, 1H, OH, D2O exchangeable). 13CNMR (δ, ppm) 82.86 (
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –CN), 113.96 (CN), 115.55 (CN), 116.32, 117.76, 118.86, 119.17, 119.87, 128.03, 128.53, 129.23, 129.63, 130.05, 139.11, 139.33, 145.03, 145.23, 146.69 (Ar–Cs), 153.84 (CH = N), 155.85 (C–NH2), 157.43 (C–OH), 162.74 (C
–CN), 113.96 (CN), 115.55 (CN), 116.32, 117.76, 118.86, 119.17, 119.87, 128.03, 128.53, 129.23, 129.63, 130.05, 139.11, 139.33, 145.03, 145.23, 146.69 (Ar–Cs), 153.84 (CH = N), 155.85 (C–NH2), 157.43 (C–OH), 162.74 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); anal. calcd. for C22H13N5O3 (395.38): C, 66.83; H, 3.31; N, 17.71. Found: C, 66.48; H, 3.11; N, 17.28.
O); anal. calcd. for C22H13N5O3 (395.38): C, 66.83; H, 3.31; N, 17.71. Found: C, 66.48; H, 3.11; N, 17.28.
4.2.1.2.7 6-Amino-1-((anthracen-9-ylmethylene)amino)-2-oxo-4-(thiophen-2-yl)-1,2-dihydro-pyridine-3,5-dicarbonitrile (5a). M.p.: 318–320 °C; brown powder; yield 73% (92%); IR (KBr): νmax 3372, 3322 (NH2), 3081 (aromatic-H), 2925 (aliphatic-H), 2212 (CN), 1691 (CO), 1647 (CH = N); 1H NMR (δ, ppm) 7.24 (t, J = 3.6 Hz, 1H, thiophene-H), 7.51 (d, J = 2.8 Hz, 2H, thiophene-H), 7.68 (t, J = 7.6 Hz, 2H, Ar–H), 7.83 (t, J = 6.8 Hz, 2H, Ar–H), 7.90 (d, J = 5.6 Hz, 2H, Ar–H), 8.27 (d, J = 8.1 Hz, 2H, Ar–H), 8.78 (s, 2H, NH2, D2O exchangeable), 8.92 (s, 1H, Ar–H), 9.04 (s, 1H, methylinic-H); 13CNMR (δ, ppm) 82.99 (
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –CN), 116.26 (CN), 116.74, (CN), 117.47, 118.34, 118.57, 120.02, 121.21, 121.71, 124.60, 126.62, 130.38, 134.10, 134.46, 135.03, 135.98, 139.10, 145.23, 145.68, 146.78, 146.83, 147.51, 151.27 (Ar–Cs), 157.66 (CH = N), 161.64 (C–NH2), 163.51(C
–CN), 116.26 (CN), 116.74, (CN), 117.47, 118.34, 118.57, 120.02, 121.21, 121.71, 124.60, 126.62, 130.38, 134.10, 134.46, 135.03, 135.98, 139.10, 145.23, 145.68, 146.78, 146.83, 147.51, 151.27 (Ar–Cs), 157.66 (CH = N), 161.64 (C–NH2), 163.51(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); anal. calcd. For C26H15N5OS (445.50): C, 70.10; H, 3.39; N, 15.72. Found: C, 69.92; H, 3.38; N, 15.61.
O); anal. calcd. For C26H15N5OS (445.50): C, 70.10; H, 3.39; N, 15.72. Found: C, 69.92; H, 3.38; N, 15.61.
4.2.1.2.8 6-Amino-1-((anthracen-9-ylmethylene)amino)-4-(furan-2-yl)-2-oxo-1,2-dihydropyridine-3,5-dicarbonitrile (5b). M.p.: 340–342 °C; brownish red powder; yield 69% (88%); IR (KBr): νmax 3381, 3271 (NH2), 3115 (aromatic-H), 2925 (aliphatic-H), 2206 (CN), 1670 (CO), 1640 (CH = N). 1H NMR (δ, ppm) 6.81 (m, 2H, Ar–H), 6.86 (t, J = 4.0 Hz, 1H, furan-H), 7.33 (d, J = 3.6 Hz, 2H, furan-H), 7.63 (t, J = 7.2 Hz, 2H, Ar–H), 8.06 (d, J = 6.6 Hz, 2H, Ar–H), 8.21 (d, J = 8.0 Hz, 1H, Ar–H), 8.40 (s, 2H, NH2, D2O exchangeable), 8.78 (d, J = 8.8 Hz, 2H, Ar–H), 8.92 (s, 1H, methylinic-H); 13CNMR (δ, ppm) 86.82 (
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –CN), 114.80 (CN), 116.77 (CN), 116.92, 118.57, 120.15, 124.60, 127.83, 128.35, 129.79, 131.40, 135.70, 137.70, 138.84, 139.53, 142.01, 145.17, 145.35, 145.54, 146.07, 146.49, 146.91, 147.16 (Ar–Cs), 155.93 (C–NH2), 157.60 (CH = N), 162.57 (C
–CN), 114.80 (CN), 116.77 (CN), 116.92, 118.57, 120.15, 124.60, 127.83, 128.35, 129.79, 131.40, 135.70, 137.70, 138.84, 139.53, 142.01, 145.17, 145.35, 145.54, 146.07, 146.49, 146.91, 147.16 (Ar–Cs), 155.93 (C–NH2), 157.60 (CH = N), 162.57 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); anal. calcd. For C26H15N5O2 (429.43): C, 72.72; H, 3.52; N, 16.31. Found: C, 72.43; H, 3.30; N, 15.97.
O); anal. calcd. For C26H15N5O2 (429.43): C, 72.72; H, 3.52; N, 16.31. Found: C, 72.43; H, 3.30; N, 15.97.
4.2.1.2.9 6-Amino-1-((1,3-dioxo-1,3-dihydro-2H-inden-2-ylidene)amino)-2-oxo-4-(thiophen-2-yl)-1,2-dihydropyridine-3,5-dicarbonitrile (6a). M.p.: 281–283 °C; black powder; yield 73% (89%); IR (KBr): νmax 3395, 3323 (NH2), 3092 (aromatic-H), 2211 (CN), 1658 (2CO), 1582 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1H NMR (δ, ppm) 7.43 (t, J = 4.0 Hz, 1H, thiophene-H), 7.82 (d, J = 4.0 Hz, 1H, thiophene-H), 8.12 (t, J = 8.0 Hz, 2H, Ar–H), 8.25 (d, J = 8.0 Hz, 2H, Ar–H), 8.37 (d, J = 4.0 Hz, 1H, thiophene-H), 8.78 (s, 2H, NH2, D2O exchangeable); 13CNMR (δ, ppm) 79.40 (
N); 1H NMR (δ, ppm) 7.43 (t, J = 4.0 Hz, 1H, thiophene-H), 7.82 (d, J = 4.0 Hz, 1H, thiophene-H), 8.12 (t, J = 8.0 Hz, 2H, Ar–H), 8.25 (d, J = 8.0 Hz, 2H, Ar–H), 8.37 (d, J = 4.0 Hz, 1H, thiophene-H), 8.78 (s, 2H, NH2, D2O exchangeable); 13CNMR (δ, ppm) 79.40 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –CN), 113.36 (CN), 113.90 (CN), 116.71, 118.95, 119.36, 122.28, 122.58, 124.41, 127.21, 127.86, 128.62, 134.95, 138.20, 138.71, 139.08 (Ar–Cs), 158.91 (C
–CN), 113.36 (CN), 113.90 (CN), 116.71, 118.95, 119.36, 122.28, 122.58, 124.41, 127.21, 127.86, 128.62, 134.95, 138.20, 138.71, 139.08 (Ar–Cs), 158.91 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 162.17 (C–NH2), 164.51 (CO), 168.51 (CO); anal. calcd. For C20H9N5O3S (399.38): C, 60.15; H, 2.27; N, 17.54. Found: C, 59.81.52; H, 2.11; N, 17.11.
N), 162.17 (C–NH2), 164.51 (CO), 168.51 (CO); anal. calcd. For C20H9N5O3S (399.38): C, 60.15; H, 2.27; N, 17.54. Found: C, 59.81.52; H, 2.11; N, 17.11.
4.2.1.2.10 6-Amino-1-((1,3-dioxo-1,3-dihydro-2H-inden-2-ylidene)amino)-4-(furan-2-yl)-2-oxo-1,2-dihydropyridine-3,5-dicarbonitrile (6b). M.p.: 305–307 °C; black powder; yield 66% (89%); IR (KBr): νmax 3374, 3313 (NH2), 3145 (aromatic-H), 2213 (CN), 1723, 1670 (2CO), 1650 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1H NMR (δ, ppm) 7.25 (t, J = 4.0 Hz, 1H, furan-H), 7.39 (t, J = 4.0 Hz, 1H, furan-H), 7.51 (d, J = 7.2 Hz, 1H, Ar–H), 7.81 (d, J = 3.1 Hz, 1H, furan-H), 7.91 (d, J = 7.7 Hz, 1H, Ar–H), 8.11 (d, J = 6.2 Hz, 1H, Ar–H), 8.34 (d, J = 7.8 Hz, 1H, Ar–H), 8.47 (s, 2H, NH2, D2O exchangeable); 13CNMR (δ, ppm) 81.56 (C–CN), 112.61 (CN), 114.81 (CN), 115.47, 119.89, 120.52, 123.70, 124.68, 127.49, 128.91, 129.48, 130.42, 137.50, 138.54, 139.54, 141.09 (Ar–Cs), 157.32 (C
N); 1H NMR (δ, ppm) 7.25 (t, J = 4.0 Hz, 1H, furan-H), 7.39 (t, J = 4.0 Hz, 1H, furan-H), 7.51 (d, J = 7.2 Hz, 1H, Ar–H), 7.81 (d, J = 3.1 Hz, 1H, furan-H), 7.91 (d, J = 7.7 Hz, 1H, Ar–H), 8.11 (d, J = 6.2 Hz, 1H, Ar–H), 8.34 (d, J = 7.8 Hz, 1H, Ar–H), 8.47 (s, 2H, NH2, D2O exchangeable); 13CNMR (δ, ppm) 81.56 (C–CN), 112.61 (CN), 114.81 (CN), 115.47, 119.89, 120.52, 123.70, 124.68, 127.49, 128.91, 129.48, 130.42, 137.50, 138.54, 139.54, 141.09 (Ar–Cs), 157.32 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 161.08 (C–NH2), 162.83 (CO), 166.95 (CO); anal. calcd. For C20H9N5O4 (383.32): C, 62.67; H, 2.37; N, 18.27. Found: C, 62.41.52; H, 2.06; N, 17.91.
N), 161.08 (C–NH2), 162.83 (CO), 166.95 (CO); anal. calcd. For C20H9N5O4 (383.32): C, 62.67; H, 2.37; N, 18.27. Found: C, 62.41.52; H, 2.06; N, 17.91.
4.2.1.2.11 6-Amino-2-oxo-1-((2-oxoindolin-3-ylidene)amino)-4-(thiophen-2-yl)-1,2-dihydropyridine-3,5-dicarbonitrile (8a). M.p.: 258–260 °C; brown powder; yield 76% (94%); IR (KBr): νmax 3389, 3286 (NH2, NH), 3115 (aromatic-H), 2208 (CN), 1729, 1637 (2CO), 1599 (CH = N); 1H NMR (δ, ppm) = 6.78 (t, J = 4.9 Hz, 1H, thiophene-H), 6.89 (d, J = 3.2 Hz, 1H, thiophene-H), 7.06 (t, J = 7.6 Hz, 1H, Ar–H), 7.33 (d, J = 3.6 Hz, 1H, thiophene-H), 7.48 (t, J = 7.2 Hz, 1H, Ar–H), 7.56 (d, J = 8.0 Hz, 2H, Ar–H), 8.4 (s, 2H, NH2, D2O exchangeable), 11.02 (s, 1H, NH-isatin, D2O exchangeable); 13CNMR (δ, ppm) 75.34 (C–CN), 109.15 (CN), 114.10 (CN), 114.28, 118.68, 119.01, 121.62, 122.87, 126.94, 129.71, 137.87, 142.63, 143.02, 145.54, 147.79 (Ar–Cs), 155.08 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 159.49 (NH2), 166.09 (CO), 172.12 (CO); anal. calcd. For C19H10N6O2S (386.39): C, 59.06; H, 2.61; N, 21.75. Found: C, 58.61.; H, 2.31; N, 21.21.
N), 159.49 (NH2), 166.09 (CO), 172.12 (CO); anal. calcd. For C19H10N6O2S (386.39): C, 59.06; H, 2.61; N, 21.75. Found: C, 58.61.; H, 2.31; N, 21.21.
4.2.1.2.12 6-Amino-4-(furan-2-yl)-2-oxo-1-((2-oxoindolin-3-ylidene)amino)-1,2-dihydropyridine-3,5-dicarbonitrile (8b). M.p.: 270–272 °C; brown powder; yield 71% (91%); IR (KBr): νmax 3374, 3313 (NH2, NH), 3045 (aromatic-H), 2213 (CN), 1723, 1670 (2CO), 1650 (CH = N); 1H NMR (δ, ppm) = 6.84 (t, J = 4.8 Hz, 1H, furan-H), 6.89 (d, J = 3.6 Hz, 1H, furan-H), 7.15 (t, J = 8.0 Hz, 1H, Ar–H), 7.38 (d, J = 4.8 Hz, 1H, furan-H), 7.47 (t, J = 7.2 Hz, 1H, Ar–H), 7.62 (d, J = 8.0 Hz, 2H, Ar–H), 8.06 (s, 2H, NH2, D2O exchangeable), 11.61 (s, 1H, NH-isatin, D2O exchangeable); 13C NMR (δ, ppm) 78.11(C–CN), 112.66 (CN), 114.28 (CN), 114.33, 117.15, 118.85, 120.16, 122.31, 125.31, 128.11, 136.87, 140.38, 141.11, 144.25, 147.10 (Ar–Cs), 151.55 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 158.28 (NH2), 164.82 (CO), 171.16 (CO); anal. calcd. For C19H10N6O3 (370.32): C, 61.62; H, 2.72; N, 22.69. Found: C, 61.41; H, 2.31; N, 22.43.
N), 158.28 (NH2), 164.82 (CO), 171.16 (CO); anal. calcd. For C19H10N6O3 (370.32): C, 61.62; H, 2.72; N, 22.69. Found: C, 61.41; H, 2.31; N, 22.43.
4.2.2.1 Method (a). To a solution of pyridine Schiff base derivatives 6a,b (0.01 mmol) or 8a,b (0.01 mmol) dimethylformamide (10 mL) as a solvent was heated under reflux for 17–20 h (monitoring by TLC). After the reaction is completed, the solution mixture was added to 20 mL cold ethanol, and the precipitated solid was filtered, washed with ethanol, dried, and crystallized from ethanol to yield the desired compounds.
4.2.2.2 Method (b). To a solution of pyridine Schiff base derivatives 6a,b (0.01 mmol) or 8a,b (0.01 mmol) and cerium ammonium nitrate (2 mol%), was dissolved in acetonitrile (5 mL), the solution was stirred at room temperature for 4–5 h (monitoring by TLC). After the reaction is completed, the solid product that precipitated was filtered out, washed with ethanol, dried, and crystallized from ethanol to produce the desired compounds with high yield.
4.2.2.3 Method (c). The 1,6-diamino-pyridine derivatives 1a,b (0.01 mol), and ninhydrin or isatin (1 mmol) were dissolved in DMF (10 mL), the solution was heated under reflux for 17–20 h (monitoring by TLC). After the reaction is completed, the mixture was added to 20 mL cold ethanol, and the precipitated solid was filtered, washed with ethanol, dried, and crystallized from ethanol.
4.2.2.4 Method (d). The 1,6-diamino-pyridine derivatives 1a,b (0.01 mol), ninhydrin or isatin (1 mmol), and cerium ammonium nitrate (2 mol%), were dissolved in acetonitrile (5 mL), the solution was stirred at room temperature for 4–5 h (monitoring by TLC). After the reaction is completed, the solid product that precipitated was filtered out, washed with ethanol, dried, and crystallized from suitable solvent to produce the desired compounds with high yield.
4.2.2.4.1 4,7-Dioxo-2-(thiophen-2-yl)-4,7dihydroindeno[1,2-e]pyrido[1,2-b][1,2,4]triazine-1,3-dicarbonitrile (7a). M.p.: 362–365 °C; brownish powder; yield 69% (91); IR (KBr): νmax 3092 (aromatic-H), 2212 (CN), 1740, 1683 (2CO), 1642 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1H NMR (δ, ppm) = 7.42 (t, J = 4.3 Hz, 1H, thiophene-H), 7.82 (d, J = 2.7 Hz, 2H, thiophene-H), 8.15–8.12 (m, 2H, Ar–H), 8.20 (d, J = 7.7 Hz, 1H, Ar–H), 8.36 (d, J = 6.8 Hz, 1H, Ar–H); 13CNMR (δ, ppm) 91.50, 99.17 (2
N); 1H NMR (δ, ppm) = 7.42 (t, J = 4.3 Hz, 1H, thiophene-H), 7.82 (d, J = 2.7 Hz, 2H, thiophene-H), 8.15–8.12 (m, 2H, Ar–H), 8.20 (d, J = 7.7 Hz, 1H, Ar–H), 8.36 (d, J = 6.8 Hz, 1H, Ar–H); 13CNMR (δ, ppm) 91.50, 99.17 (2![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –CN), 114.68, 115.67 (2CN), 125.70, 125.99, 128.78, 132.47, 133.16, 133.23, 137.16, 138.16, 138.56, 141.06, 148.28, 151.57, 153.09 (Ar–Cs), 156.20 (C
–CN), 114.68, 115.67 (2CN), 125.70, 125.99, 128.78, 132.47, 133.16, 133.23, 137.16, 138.16, 138.56, 141.06, 148.28, 151.57, 153.09 (Ar–Cs), 156.20 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 160.58 (CO), 183.62 (CO); anal. calcd. For C20H7N5O2S (381.37): C, 62.99; H, 1.85; N, 18.38. Found: C, 62.61.52; H, 1.78; N, 17.81.
N), 160.58 (CO), 183.62 (CO); anal. calcd. For C20H7N5O2S (381.37): C, 62.99; H, 1.85; N, 18.38. Found: C, 62.61.52; H, 1.78; N, 17.81.
4.2.2.4.2 4,7-Dioxo-2-(furan-2-yl)-4,7dihydroindeno[1,2-e]pyrido[1,2-b][1,2,4]triazine-1,3-dicarbonitrile (8b). M.p.: 352–354 °C; blue brown powder; yield 71% (84%); IR (KBr): νmax = 3092 (aromatic-H), 2211 (CN), 1647 (CO), 1607 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1H NMR (δ, ppm) 7.25 (t, J = 3.6 Hz, 1H, furan-H), 7.39 (t, J = 4.8 Hz, 1H, Ar–H), 7.51 (d, J = 3.2 Hz, 1H, furan-H), 7.80 (d, J = 4.0 Hz, 1H, furan-H), 7.71 (d, J = 5.6 Hz, 1H, Ar–H), 8.1 (t, J = 5.6 Hz, 1H, Ar–H) 8.21 (d, J = 8.4 Hz, 1H, Ar–H); 13CNMR (δ, ppm) 89.80, 97.93 (2
N); 1H NMR (δ, ppm) 7.25 (t, J = 3.6 Hz, 1H, furan-H), 7.39 (t, J = 4.8 Hz, 1H, Ar–H), 7.51 (d, J = 3.2 Hz, 1H, furan-H), 7.80 (d, J = 4.0 Hz, 1H, furan-H), 7.71 (d, J = 5.6 Hz, 1H, Ar–H), 8.1 (t, J = 5.6 Hz, 1H, Ar–H) 8.21 (d, J = 8.4 Hz, 1H, Ar–H); 13CNMR (δ, ppm) 89.80, 97.93 (2![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –CN), 113.48, 115.20 (2CN), 124.34, 125.34, 127.90, 131.95, 132.93, 135.57, 137.46, 138.33, 139.78, 141.66, 147.20, 149.37, 152.17 (Ar–Cs), 155.56 (C
–CN), 113.48, 115.20 (2CN), 124.34, 125.34, 127.90, 131.95, 132.93, 135.57, 137.46, 138.33, 139.78, 141.66, 147.20, 149.37, 152.17 (Ar–Cs), 155.56 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 158.47 (CO), 175.49 (CO); anal. calcd. For C20H7N5O3 (365.30): C, 65.76; H, 1.93; N, 19.17. Found: C, 65.61, H, 1.78; N, 18.81.
N), 158.47 (CO), 175.49 (CO); anal. calcd. For C20H7N5O3 (365.30): C, 65.76; H, 1.93; N, 19.17. Found: C, 65.61, H, 1.78; N, 18.81.
4.2.2.4.3 7-Oxo-9-(thiophen-2-yl)-7,12-dihydropyrido[1′,2′:2,3][1,2,4]triazino[5,6-b]indole-8,10-dicarbonitrile (9a). M.p.: 301–303 °C; brown powder; yield 65% (85%). IR (KBr): νmax = 3173 (NH), 3095 (aromatic-H), 2213 (CN), 1647 (CO), 1617 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1H NMR (δ, ppm) = 7.08 (t, J = 9.2 Hz, 1H, Ar–H), 7.33 (d, J = 3.6 Hz, 2H, thiophene-H), 7.48 (d, J = 7.2 Hz, 1H, Ar–H), 7.58 (t, J = 4.0 Hz, 1H, thiophene-H), 7.77 (d, J = 8.0 Hz, 1H, Ar–H), 7.88 (d, J = 8.0 Hz, 1H, Ar–H), 11.79 (s, 1H, NH-isatin, D2O exchangeable). 13CNMR (δ, ppm) = 85.48 (
N); 1H NMR (δ, ppm) = 7.08 (t, J = 9.2 Hz, 1H, Ar–H), 7.33 (d, J = 3.6 Hz, 2H, thiophene-H), 7.48 (d, J = 7.2 Hz, 1H, Ar–H), 7.58 (t, J = 4.0 Hz, 1H, thiophene-H), 7.77 (d, J = 8.0 Hz, 1H, Ar–H), 7.88 (d, J = 8.0 Hz, 1H, Ar–H), 11.79 (s, 1H, NH-isatin, D2O exchangeable). 13CNMR (δ, ppm) = 85.48 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –CN), 114.45, 115.21 (CN), 115.88 (CN), 117.16, 124.10, 126.62, 129.00, 131.19, 132.12, 135.70, 137.47, 139.08, 141.35, 144.29, 151.85 (Ar–Cs), 156.35, 163.69 (C
–CN), 114.45, 115.21 (CN), 115.88 (CN), 117.16, 124.10, 126.62, 129.00, 131.19, 132.12, 135.70, 137.47, 139.08, 141.35, 144.29, 151.85 (Ar–Cs), 156.35, 163.69 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 168.83 (CO); anal. calcd. For C19H8N6OS (368.37): C, 61.95; H, 2.19; N, 22.81. Found: C, 61.61.; H, 2.01; N, 22.41.
N), 168.83 (CO); anal. calcd. For C19H8N6OS (368.37): C, 61.95; H, 2.19; N, 22.81. Found: C, 61.61.; H, 2.01; N, 22.41.
4.2.2.4.4 9-(Furan-2-yl)-7-oxo-7,12-dihydropyrido[1′,2′:2,3][1,2,4]triazino[5,6-b]indole-8,10-dicarbonitrile (9b). M.p.: 310–312 °C; black brown powder; yield 61% (85%); IR (KBr): νmax = 3210 (NH), 2213 (CN), 1723, 1716 (2CO), 1621 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1H NMR (δ, ppm) 6.81 (t, J = 3.7 Hz, 1H, furan-H), 6.90 (d, J = 8.0 Hz, 1H, Ar–H), 7.11 (t, J = 8.0 Hz, 1H, Ar–H), 7.33 (d, J = 4.0 Hz, 2H, furan-H), 7.49 (d, J = 8.0 Hz, 1H, Ar–H), 7.58 (t, J = 6.8 Hz, 2H, Ar–H), 11.03 (s, 1H, NH, D2O exchangeable); 13CNMR (δ, ppm) = 83.53 (C–CN), 115.21 (CN), 115.88 (CN), 116.07, 116.41, 117.87, 125.56, 128.26, 129.55, 132.12, 135.70, 137.47, 140.28, 142.31, 148.18 (Ar–Cs), 154.41 (C
N); 1H NMR (δ, ppm) 6.81 (t, J = 3.7 Hz, 1H, furan-H), 6.90 (d, J = 8.0 Hz, 1H, Ar–H), 7.11 (t, J = 8.0 Hz, 1H, Ar–H), 7.33 (d, J = 4.0 Hz, 2H, furan-H), 7.49 (d, J = 8.0 Hz, 1H, Ar–H), 7.58 (t, J = 6.8 Hz, 2H, Ar–H), 11.03 (s, 1H, NH, D2O exchangeable); 13CNMR (δ, ppm) = 83.53 (C–CN), 115.21 (CN), 115.88 (CN), 116.07, 116.41, 117.87, 125.56, 128.26, 129.55, 132.12, 135.70, 137.47, 140.28, 142.31, 148.18 (Ar–Cs), 154.41 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 157.44 (C
N), 157.44 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 161.85 (C–NH), 165.94 (2CO); anal. calcd. For C19H8N6O2 (351.31): C, 64.77; H, 2.29; N, 23.85. Found: C, 64.41; H, 2.11; N, 22.99.
N), 161.85 (C–NH), 165.94 (2CO); anal. calcd. For C19H8N6O2 (351.31): C, 64.77; H, 2.29; N, 23.85. Found: C, 64.41; H, 2.11; N, 22.99.
4.3. Biological evaluation
| % inhibition = 1 − (OD sample/OD positive control) × 100 |
4.3.6.1 RNA extraction and conversion to DNA. Total RNA was isolated from P. aeruginosa (ATCC 2785) using the First High Pure RNA Isolation Kit (Cat. No. 11 828 665001, Roche Diagnostics GmbH, Germany) in accordance with the manufacturer's instructions. Using the QuantiTects Reverse Transcription Kit, one g of total RNA was reverse-transcribed into single-stranded complementary DNA (Qiagen, USA).80
4.3.6.2 Quantitative RT-PCR of lasR gene. RT-PCR was performed in two steps with a random primer hexamer, and any genomic DNA (gDNA) contamination was removed with gDNA wipeout buffer. Total cDNA (30 ng) was used as a template for amplification with the specific primers pair via Maxima SYBR Green/Fluorescein qPCR Master Mix. The forward and reverse gene-specific PCR primers for LasR were (5′-CTGTGGATGCTCAAGGACTAC-3′) and (5′-AACTGGTCTTGCCGATGG-3′), respectively. ropD was used as reference gene of primers' sequences of 5′-CGAACTGCTTGCCGACTT-3′ and 5′-GCGAGAGCCTCAAGGATAC-3′ for forward and reverse, respectively. Additionally, the sample was subjected to real-time PCR in duplicate and the mean values of the duplicates were used for subsequent analysis. Data was calculated using ΔΔCt method to estimate relative gene expression where fold change equal to (2−ΔΔct).80,81
4.4. The docking simulation
The docking simulation of the most active 2-oxo-pyridine derivatives was performed inside the active site of LasR protein. The crystal structure (PDB ID: 6MVN) involved two chains and our docking process was carried out using only one chain (chain A) and all water molecules and other ligand were deleted. The docking process was performed using Molecular Operating Environment (MOE) version 2014.0901.84–86 The co-crystallized 3-oxo-N-[(3S)-2-oxotetrahydrofuran-3-yl]decanamide was redocked inside the active site that generated under default parameters and exhibited binding energy S = −9.3504 kcal mol−1 and RMSD = 0.6986 Å, where the triangle matcher was selected as replacement and London dG as rescoring 1 with fifty retain and forcefield as refinement. The self-docking showed that co-crystallized ligand formed three hydrogen bond sidechain acceptors between Tyr56 and Ser129 with oxygen of carbonyl of amide group with bond length 1.68 and 1.97 Å and the third bond presented between the residue Trp60 with the oxygen of furanone derivatives with bond length of 2.19 Å. Additionally, the residue Asp73 bounded to NH of amide at C3 of furane derivative with hydrogen bond sidechain donor with bond length of 1.74 Å. Besides, the arene–H interaction was observed between the hydrogen of C5 on furane moiety and Trp60.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University for funding this work through a large group Research Project under grant number RGP2/50/44.References
- T. A. Hafiz, S. S. Alghamdi, M. A. Mubaraki, S. S. M. Alghamdi, A. Alothaybi, E. Aldawood and F. Alotaibi, J. Infect. Public Health, 2023, 16, 313–319 CrossRef PubMed.
- L. Yin, Y. Gou, Y. Dai, T. Wang, K. Gu, T. Tang, S. Hussain, X. Huang, C. He, X. Liang, G. Shu, F. Xu and P. Ouyang, Int. J. Mol. Sci., 2023, 24, 9288 CrossRef CAS PubMed.
- G.-F. Zhang, S. Zhang, B. Pan, X. Liu and L.-S. Feng, Eur. J. Med. Chem., 2018, 143, 710–723 CrossRef CAS PubMed.
- P. Dadgostar, Infect. Drug Resist., 2019, 3903–3910 CrossRef CAS PubMed.
- N. Nagasundaram, K. Padmasree, S. Santhosh, N. Vinoth, N. Sedhu and A. Lalitha, J. Mol. Struct., 2022, 1263, 133091 CrossRef CAS.
- E. R. A. Mahmoud, H. A. H. Ahmed, A. S. M. Abo-senna, O. K. M. Riad and M. M. A. A. – R. A.- Shadi, J. Radiat. Res. Appl. Sci., 2021, 14, 34–43 CAS.
- E. Leemans, K. V Mahasenan, M. Kumarasiri, E. Spink, D. Ding, P. I. O'Daniel, M. A. Boudreau, E. Lastochkin, S. A. Testero and T. Yamaguchi, Bioorg. Med. Chem. Lett., 2016, 26, 1011–1015 CrossRef CAS PubMed.
- A. Ragab, Y. A. Ammar, A. Ezzat, A. M. Mahmoud, M. Basseem, I. Mohamed, A. S. El-tabl and R. S. Farag, Comput. Biol. Med., 2022, 145, 105473 CrossRef CAS PubMed.
- A. Ezzat, M. B. I. Mohamed, A. M. Mahmoud, R. S. Farag, A. S. El-Tabl and A. Ragab, J. Mol. Struct., 2022, 1251, 132004 CrossRef CAS.
- P. A. Akinduti, O. W. George, H. U. Ohore, O. E. Ariyo, S. T. Popoola, A. I. Adeleye, K. S. Akinwande, J. O. Popoola, S. O. Rotimi, F. O. Olufemi, C. A. Omonhinmin and G. I. Olasehinde, Antibiotics, 2023, 12, 626 CrossRef CAS PubMed.
- R. Chen, G. Wang, Q. Wang, M. Zhang, Y. Wang, Z. Wan, Z. Si, Y. Bai, Z. Song, X. Lu and Y. Hao, Int. J. Microbiol. DOI:10.1007/s10123-023-00369-7.
- V. Zammuto, A. Spanò, E. Agostino, A. Macrì, C. De Pasquale, G. Ferlazzo, M. G. Rizzo, M. S. Nicolò, S. Guglielmino and C. Gugliandolo, Mar. Drugs, 2023, 21, 313 CrossRef CAS PubMed.
- A. Ragab, S. A. Fouad, Y. A. Ammar, D. S. Aboul-Magd and M. S. Abusaif, Antibiotics, 2023, 12, 128 CrossRef CAS PubMed.
- Y. A. Ammar, J. A. Micky, D. S. Aboul-Magd, S. M. A. Abd El-Hafez, S. A. Hessein, A. M. Ali and A. Ragab, Chem. Biol. Drug Des., 2023, 101, 245–270 CrossRef CAS PubMed.
- M. M. Abdelgalil, Y. A. Ammar, G. A. M. Elhag Ali, A. K. Ali and A. Ragab, J. Mol. Struct., 2023, 1274, 134443 CrossRef CAS.
- R. Rezende Mires de Carvalho, C. Silva Dias, L. Nogueira Paz, T. Melo de Lima Fires, C. Pereira Figueira, K. Araújo Damasceno and M. Hanzen Pinna, Heliyon, 2023, 9, e13802 CrossRef CAS PubMed.
- T. Yamaguchi, N. Higa, N. Okura, A. Matsumoto, I. Hermawan, T. Yamashiro, T. Suzuki and C. Toma, BMC Microbiol., 2018, 18, 64 CrossRef PubMed.
- A. A. N. Santos, C. P. Figueira, M. G. dos Reis, F. Costa and P. Ristow, Braz. J. Microbiol., 2015, 46, 1161–1164 CrossRef CAS PubMed.
- W. Mousa, N. Omar, O. K. M. Riad and M. E. A. Omran, Azhar International Journal of Pharmaceutical and Medical Sciences, 2022, 2, 94–104 CrossRef.
- P. Pachori, R. Gothalwal and P. Gandhi, Genes Dis., 2019, 6, 109–119 CrossRef PubMed.
- M. S. E. M. Badawy, O. K. M. Riad, F. A. Taher and S. A. Zaki, Int. J. Biol. Macromol., 2020, 149, 1109–1117 CrossRef CAS PubMed.
- E. P. Perikleous, D. Gkentzi, A. Bertzouanis, E. Paraskakis, A. Sovtic and S. Fouzas, Antibiotics, 2023, 12, 217 CrossRef CAS PubMed.
- A. Y. Alzahrani, Y. A. Ammar, M. Abu-Elghait, M. A. Salem, M. A. Assiri, T. E. Ali and A. Ragab, Bioorg. Chem., 2022, 119, 105571 CrossRef CAS.
- O. Kindler, O. Pulkkinen, A. G. Cherstvy and R. Metzler, Sci. Rep., 2019, 9, 12077 CrossRef.
- V. Bettenworth, B. Steinfeld, H. Duin, K. Petersen, W. R. Streit, I. Bischofs and A. Becker, J. Mol. Biol., 2019, 431, 4530–4546 CrossRef CAS PubMed.
- S. Mukherjee and B. L. Bassler, Nat. Rev. Microbiol., 2019, 17, 371–382 CrossRef CAS PubMed.
- S. S. Chourasiya, D. Kathuria, S. Singh, V. C. Sonawane, A. K. Chakraborti and P. V Bharatam, RSC Adv., 2015, 5, 80027–80038 RSC.
- S. Park, H.-S. Kim, K. Ok, Y. Kim, H.-D. Park and Y. Byun, Bioorg. Med. Chem. Lett., 2015, 25, 2913–2917 CrossRef CAS PubMed.
- S. Nizalapur, Ö. Kimyon, N. N. Biswas, C. R. Gardner, R. Griffith, S. A. Rice, M. Manefield, M. Willcox, D. S. Black and N. Kumar, Org. Biomol. Chem., 2016, 14, 680–693 RSC.
- M. E. Skindersoe, M. Alhede, R. Phipps, L. Yang, P. O. Jensen, T. B. Rasmussen, T. Bjarnsholt, T.-N. Tim, T. Tolker-Nielsen, N. Høiby and M. Givskov, Antimicrob. Agents Chemother., 2008, 52, 3648–3663 CrossRef CAS PubMed.
- M. Kurva, S. G. Pharande, A. Quezada-Soto and R. Gámez-Montaño, Tetrahedron Lett., 2018, 59, 1596–1599 CrossRef CAS.
- A. K. Gunthanakkala, M. S. Mangali, P. Venkatapuram and P. Adivireddy, J. Heterocycl. Chem., 2020, 57, 4164–4174 CrossRef CAS.
- M. A. Ismail, M. S. Abusaif, M. S. A. El-Gaby, Y. A. Ammar and A. Ragab, RSC Adv., 2023, 13, 12589–12608 RSC.
- R. Ayman, M. S. Abusaif, A. M. Radwan, A. M. Elmetwally and A. Ragab, Eur. J. Med. Chem., 2023, 249, 115138 CrossRef CAS PubMed.
- K. Harada, T. Ferdous, T. Harada, T. Takenawa and Y. Ueyama, Oncol. Lett., 2017, 14, 3349–3356 CrossRef PubMed.
- R. I. Sanchez, K. L. Fillgrove, K. L. Yee, Y. Liang, B. Lu, A. Tatavarti, R. Liu, M. S. Anderson, M. O. Behm and L. Fan, Xenobiotica, 2019, 49, 422–432 CrossRef CAS PubMed.
- T. S. U. Sihotang, A. D. W. Widodo and P. D. Endraswari, Ann. Med. Surg., 2022, 82, 104674 Search PubMed.
- F.-A. Khan, S. Yaqoob, S. Ali, N. Tanveer, Y. Wang, S. Ashraf, K. A. Hasan, S. A. M. Khalifa, Q. Shou, Z. Ul-Haq, Z.-H. Jiang and H. R. El-Seedi, Molecules, 2023, 28, 212 CrossRef CAS PubMed.
- A. Ragab, S. A. Fouad, O. A. A. Ali, E. M. Ahmed, A. M. Ali, A. A. Askar and Y. A. Ammar, Antibiotics, 2021, 10, 162 CrossRef CAS PubMed.
- A. K. Bass, E. Abdelhafez, M. El-Zoghbi, M. F. A. Mohamed, M. Badr and G. E.-D. A. A. Abuo-Rahma, J. Adv. Biomed. Pharm. Sci., 2021, 4, 81–86 Search PubMed.
- H. A. Soliman, A. H. Shamroukh, E. R. Kotb, K. Mahmoud, E. H. Anouar and M. I. Hegab, J. Mol. Struct., 2022, 1263, 133148 CrossRef CAS.
- A. M. Al-Etaibi and M. A. El-Apasery, Int. J. Environ. Res. Public Health, 2020, 17, 4714 CrossRef CAS PubMed.
- R. Alasbahi and M. Melzig, Sci. Pharm., 2008, 76, 471–484 CrossRef.
- A. M. Attia, A. I. Khodair, E. A. Gendy, M. A. El-Magd and Y. A. M. M. Elshaier, Lett. Drug Des. Discovery, 2020, 17, 124–137 CrossRef CAS.
- N. M. A. Gawad, H. H. Georgey, R. M. Youssef and N. A. El-Sayed, Eur. J. Med. Chem., 2010, 45, 6058–6067 CrossRef PubMed.
- N. Ryad, A.-S. My, M. M. Ismail and S. El Meligie, Chem. Pharm. Bull., 2018, 66, 939–952 CrossRef PubMed.
- L. K. A. Karem, F. Y. Waddai and N. H. Karam, J. Pharm. Sci. Res., 2018, 10, 1912–1917 Search PubMed.
- Y. Toubi, F. Abrigach, S. Radi, F. Souna, A. Hakkou, A. Alsayari, A. Bin Muhsinah and Y. N. Mabkhot, Molecules, 2019, 24, 3250 CrossRef PubMed.
- T. Tunc, A. B. Ortaakarsu, S. M. Hatipoglu, U. Kazancı, S. Karabocek, N. Karabocek, N. Dege and N. Karacan, J. Mol. Struct., 2022, 1254, 132299 CrossRef CAS.
- K.-C. Gan, K.-M. Sim, T.-M. Lim and K.-C. Teo, Lett. Org. Chem., 2020, 17, 191–198 CrossRef CAS.
- S. Javad, E. Samane, T. Raheleh and S. Hossein, Org. Prep. Proced. Int., 2019, 1–9 Search PubMed.
- M. Tahmasby, A. Darehkordi, M. Mohammadi and F. Nejadkhorasani, J. Mol. Struct., 2021, 1224, 129032 CrossRef CAS.
- A. Darehkordi, V. Salehi, F. Rahmani and M. Karimipour, Int. J. Biol. Macromol., 2018, 54, 554–558 CAS.
- A. A. Ali, H. Abd El-Wahab, M. S. Abusaif, A. Ragab, O. A. Abdel-Jaid, E. A. Eldeeb and Y. A. Ammar, Pigm. Resin Technol. DOI:10.1108/PRT-12-2022-0141.
- A. P. Zarecki, J. L. Kolanowski and W. T. Markiewicz, Molecules, 2020, 25, 1761 CrossRef CAS PubMed.
- M. Kalhor, S. Samiei and S. A. Mirshokraei, Green Chem. Lett. Rev., 2021, 14(3), 500–508 CrossRef CAS.
- E. A. Fayed, M. Mohsen, S. M. A. El-Gilil, D. S. Aboul-Magd and A. Ragab, J. Mol. Struct., 2022, 1262, 133028 CrossRef CAS.
- H. A. Mohamed, Y. A. Ammar, G. A. M. Elhagali, H. A. Eyada, D. S. Aboul-Magd and A. Ragab, J. Mol. Struct., 2023, 1287, 135671 CrossRef CAS.
- K. Zahmatkesh, K. A. Dilmaghani and Y. Sarveahrabi, Acta Chim. Slov., 2022, 69, 619–628 CrossRef CAS PubMed.
- J. Chadha, Ravi, J. Singh, S. Chhibber and K. Harjai, Front. Cell. Infect. Microbiol., 2022, 12, 899566 CrossRef CAS PubMed.
- F. M. Husain, I. Ahmad, M. Asif and Q. Tahseen, J. Biosci., 2013, 38, 835–844 CrossRef CAS PubMed.
- H. Ali Mohamed, Y. A. Ammar, G. A. M. Elhagali, H. A. Eyada, D. S. Aboul-Magd and A. Ragab, ACS Omega, 2022, 7, 4970–4990 CrossRef CAS PubMed.
- P. Awolade, N. Cele, N. Kerru and P. Singh, Mol. Diversity, 2021, 25, 2201–2218 CrossRef CAS PubMed.
- Y. A. Ammar, G. A. M. Elhagali, M. S. Abusaif, M. R. Selim, M. A. Zahran, T. Naser, A. B. M. Mehany and E. A. Fayed, Med. Chem. Res., 2021, 30, 1649–1668 CrossRef CAS.
- H. F. Rizk, M. A. El-Borai, A. Ragab, S. A. Ibrahim and M. E. Sadek, Polycyclic Aromat. Compd., 2023, 43, 500–522 CrossRef CAS.
- S. A. Ibrahim, A. Ragab and H. A. El-Ghamry, Appl. Organomet. Chem., 2022, 36, 1–17 Search PubMed.
- A. S. Hassan, N. M. Morsy, W. M. Aboulthana and A. Ragab, RSC Adv., 2023, 13, 9281–9303 RSC.
- R. R. Raslan, Y. A. Ammar, S. A. Fouad, S. A. Hessein, N. A. M. Shmiess and A. Ragab, RSC Adv., 2023, 13, 10440–10458 RSC.
- R. Ayman, A. M. Radwan, A. M. Elmetwally, Y. A. Ammar and A. Ragab, Arch. Pharm., 2022, e2200395 Search PubMed.
- E. A. Fayed, A. Ragab, R. R. Ezz Eldin, A. H. Bayoumi and Y. A. Ammar, Bioorg. Chem., 2021, 116, 105300 CrossRef CAS PubMed.
- A. S. Hassan, N. M. Morsy, W. M. Aboulthana and A. Ragab, Drug Dev. Res., 2023, 84, 3–24 CrossRef CAS PubMed.
- K. E. Saadon, N. M. H. Taha, N. A. Mahmoud, G. A. M. Elhagali and A. Ragab, J. Iran. Chem. Soc., 2022, 19, 3899–3917 CrossRef CAS.
- G. H. Elgemeie and S. H. Sayed, Synthesis, 2010, 37–41 Search PubMed.
- M. Balouiri, M. Sadiki and S. K. Ibnsouda, J. Pharm. Anal., 2016, 6, 71–79 CrossRef PubMed.
- Methods for Determining Bactericidal Activity of Antimicrobial Agents, Approved Guideline, CLSI document M26-A, Clinical and Laboratory Standards Institute, 950 West Valley Road, Suite 2500, Wayne, Pennsylvania 19087, USA Search PubMed.
- M. S. Kamel, M. O. Aboelez, M. R. Elnagar, E. K. Shokr, H. M. R. M. Selim, H. E. Abdel-Ghany, A. M. Drar, A. Belal, M. A. El Hamd and M. Abd El Aleem Ali Ali El-Remaily, ChemistrySelect, 2022, 7, e202203191 CrossRef CAS.
- F. Silva, S. Ferreira, J. A. Queiroz and F. C. Domingues, J. Med. Microbiol., 2011, 60(10), 1479–1486 CrossRef CAS PubMed.
- A. Carbone, S. Cascioferro, B. Parrino, D. Carbone, C. Pecoraro, D. Schillaci, M. G. Cusimano, G. Cirrincione and P. Diana, Molecules, 2021, 26(1), 81 CrossRef CAS PubMed.
- Y. Liu, L. Wu, J. Han, P. Dong, X. Luo, Y. Zhang and L. Zhu, Front. Microbiol., 2020, 11, 617473 CrossRef PubMed.
- M. M. Saleh, H. A. Abbas and M. M. Askoura, Microb. Pathog., 2019, 127, 31–38 CrossRef CAS PubMed.
- K. J. Livak and T. D. Schmittgen, Methods, 2001, 25, 402–408 CrossRef CAS PubMed.
- A. A. van de Loosdrecht, R. H. J. Beelen, G. J. Ossenkoppele, M. G. Broekhoven and M. M. A. C. Langenhuijsen, J. Immunol. Methods, 1994, 174, 311–320 CrossRef CAS PubMed.
- A. Ismail, A. S. Doghish, B. E. M. Elsadek, S. A. Salama and A. D. Mariee, Steroids, 2020, 160, 108656 CrossRef CAS PubMed.
- M. Eldeeb, E. F. Sanad, A. Ragab, Y. A. Ammar, K. Mahmoud, M. M. Ali and N. M. Hamdy, Biomedicines, 2022, 10, 722 CrossRef CAS PubMed.
- E. S. A. E. H. Khattab, A. Ragab, M. A. Abol-Ftouh and A. A. Elhenawy, J. Biomol. Struct. Dyn., 2022, 40, 1–19 CrossRef PubMed.
- S. A. El-Kalyoubi, A. Ragab, O. A. Abu Ali, Y. A. Ammar, M. G. Seadawy, A. Ahmed and E. A. Fayed, Pharmaceuticals, 2022, 15(3), 376 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra04230h |
| ‡ These authors contributed equally as first author. |
| This journal is © The Royal Society of Chemistry 2023 |