Open Access Article
Open Access ArticleEcotoxicological significance of bio-corona formation on micro/nanoplastics in aquatic organisms
Camil Rex Ma,
Abhrajit Debroya,
M. Joyce Nirmala b and
Amitava Mukherjee
b and
Amitava Mukherjee *a
*a
aCentre for Nanobiotechnology, Vellore Institute of Technology, Vellore 632014, India. E-mail: amitav@vit.ac.in; amit.mookerjea@gmail.com
bDepartment of Chemical Engineering, Indian Institute of Technology Madras, Chennai 600036, India
First published on 28th July 2023
Abstract
The unsustainable manufacturing, utilization and inadequate handling of plastics have led to a surge in global plastic pollution. In recent times, there has been increasing concern about the plausible hazards associated with exposure to micro/nanoplastics (M/NPs). As aquatic systems are considered to be the likely sink for M/NPs, it is crucial to comprehend their environmental behavior. The bioavailability, toxicity and fate of M/NPs in the environment are predominantly dictated by their surface characteristics. In the aquatic environment, M/NPs are prone to be internalized by aquatic organisms. This may facilitate their interaction with a diverse array of biomolecules within the organism, resulting in the formation of a biocorona (BC). The development of BC causes modifications in the physicochemical attributes of the M/NPs including changes to their size, stability, surface charge and other properties. This review details the concept of BC formation and its underlying mechanism. It provides insight on the analytical techniques employed for characterizing BC formation and addresses the associated challenges. Further, the eco-toxicological implications of M/NPs and the role of BC in modifying their potential toxicity on aquatic organisms is specified. The impact of BC formation on the fate and transport of M/NPs is discussed. A concise outlook on the future perspectives is also presented.
1. Introduction
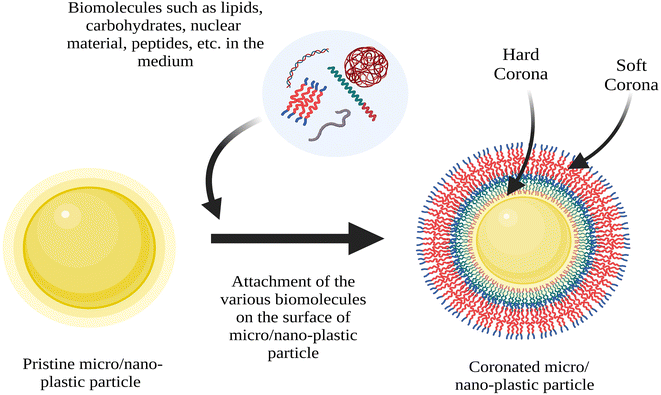
Plastics are organic polymers characterized by their substantial molecular mass and synthesized through the polymerization of monomers derived from petroleum, natural gas or coal.1,2 Plastics find extensive application in daily life owing to their cost-effectiveness, resilience and lightweight nature.3,4 In 2019, worldwide plastic production surged to 368 million metric tons (Mt) and it is projected to undergo a twofold increase within a span of 20 years.5,6 Current estimates indicate that the accumulation of plastic waste in the environment is projected to surpass 250 million tons by the year 2025.7 According to estimates, only 9% of the plastic waste produced globally has undergone recycling, with 12% being incinerated, resulting in a substantial 79% being accumulated in the environment.8 The COVID-19 pandemic has exacerbated the generation and mishandling of plastic waste.9–12 Upon introduction into water systems, a substantial proportion of plastic debris undergoes fragmentation or degradation. Consequently, this results in the formation of plastic fragments known as mesoplastics (>5 mm and <25 mm), and further degradation leads to the generation of microplastics (<5 mm) or nanoplastics (<1 mm or 1000 nm).6,13 This phenomenon carries inherent environmental risks that have captured global attention.14 In recent years, there has been a progressive increase in the abundance of micro- and nanoplastics (M/NPs) within aquatic environments.15 Primary M/NPs refer to the particles deliberately manufactured with multiple sizes, surface charge and incorporated into various products.16,17 On the other hand, secondary M/NPs are generated through physical or chemical fragmentation of larger plastic particles within the environment, as well as through biological processes such as digestion by the macroinvertebrates.18,19 In addition, M/NPs present in the aquatic ecosystems could interact with a wide range of organic compounds, pollutants and other environmental components through adsorption and desorption processes.20,21 These interactions have significant implications on the bioavailability, degradation, toxicity fate and transport of both MPs/NPs and the associated compounds.22–24 The interactions between M/NPs and various environmental components have gained substantial attention, and thus, intensive research is carried out in this area. The interactions between M/NPs and environmental components are affected by a range of intrinsic factors, which include the properties of M/NPs and environmental components, and extrinsic factors such as pH, temperature and ionic strength.25,26 Besides, the hydrophobic surface and the specific surface area of M/NPs enable them to competitively adsorb organic biomolecules.14 The term “corona” refers to the substances adsorbed on the fine particulates, and it is comprised of both “hard” (hardcorona) and “soft” (soft corona) layer.27,28 The hard corona is typically composed of a monolayer of tightly bound molecules that forms within the first few minutes of the particle's interaction with its surrounding media. The soft corona, on the other hand, consists of loosely bound molecules that cover the hard corona. Unlike the stable hard corona, the components of the soft corona are unstable and can quickly exchange with the surrounding environment.28Natural organic matter (NOM) can undergo adsorption onto M/NPs and results in the formation of a biomolecular-coated layer referred to as the ecocorona (EC).25 NOM is an intricate blend of organic compounds encompassing humic and fulvic acids, proteins, lipids, carbohydrates, and various other organic substances.29 Similarly, when M/NPs come into contact with biological fluids or biological systems, a layer of biomolecules forms on their surface, which is commonly referred to as the biocorona (BC).30 Typically, BC comprises diverse biomolecules such as proteins, lipids, nucleic acids and carbohydrates. Further, BC formation has been observed to exert notable influence on the transport, uptake, distribution, biotransformation and toxicity of M/NPs.28,30,31 Based on our comprehensive literature survey, we found that there is only one review article by Cao et al. that extensively examines the formation of BC formation pertaining to M/NPs.28 Despite the profusion of scientific literature discussing corona formation on nanoparticles, there remains a paucity of review articles that specifically addresses BC formation in the context of M/NPs. Thus, we have endeavored to bridge the knowledge gap by elucidating the mechanisms underlying the formation of BC and explicating its importance in the ecotoxicity, fate and transport of M/NPs. Further, a succinct overview of the analytical techniques employed to characterize BC formation on M/NPs is provided, while shedding light on the challenges encountered in implementing these methods.
2. Ecotoxicological implications of M/NPs on aquatic organisms
Plastic pollution in freshwater and marine habitats has emerged as a serious matter of concern throughout the world.32 M/NPs present in the aquatic environment have the ability to interact with various organic compounds such as drugs, pesticides and other pollutants by means of adsorption and desorption.33 Such interactions have a profound impact on the environmental dynamics, bioavailability, degradation, transport and toxicity of M/NPs.34–37 The diverse adverse effects such as oxidative damage, retorted growth, organelle damage, early mortality, decreased food uptake, inflammation, immune malfunctions and aberrant behavior are the major detrimental effects of M/NPs on the aquatic biota.38–432.1 Freshwater systems
Globally, several studies have documented the occurrence of M/NPs in the freshwater systems such as surface waters, rivers, lakes, rivers sediments, reservoirs and drinking water.44–48 This abundance and bioavailability of M/NPs serve as a major threat to the freshwater biota.49,50 For instance, several detrimental effects of M/NPs have been observed in the toxicological investigations on zebrafish (Danio rerio).51 Studies have unveiled that zebrafish exposed to M/NPs exhibited diminished food intake, disruption in the intestinal functioning, villi disintegration, excessive ROS production, inflammatory responses and modification in gills and intestinal mucosa.52–56 Recently, Hansjosten et al. (2022) reported that polystyrene nanoplastics (PS-NPs) of 31.2 mg L−1 inhibited the hatching of zebrafish embryo.57 Exposure of PS-NPs with fathead minnow (Pimephale spromelas) showed that the PS-NPs affected their innate immune response, enhanced oxidative burst and activated the neutrophil function.58 Similarly, PS-NPs of 0.1 μm inhibited the brain acetylcholinesterase (AchE) activity in red tilapia (Oreochromis niloticus).59 Common goby (Pomatoschistus microps) juveniles have exhibited less predatory ability and efficiency on exposure to PE for 24 h.60 Similarly, 10 days exposure of polyethylene (PE)-MPs to rice midge (Chironomus tepperi) larvae significantly decreased the survival rate and diminished the adult emergence rate. Furthermore, it was reported that the uptake of 1 μm PE MPs triggered the inhibition of locomotion in Daphnia magna.61 Additionally, combination toxicity studies on cadmium MPs revealed that the exposure to MPs has elevated the ROS generation and neurotoxicity in the Asian clam (Corbicula fluminea).622.2 Marine systems
Freshwater and terrestrial habitats are considered to be the sources and pathways through which M/NPs are transported to the marine systems.60,63 Several reports highlighted the presence of M/NPs in oceans, marine culture zones, fish farms, salts from oceans and even in the commercial salts.64–67 Studies have shown that the M/NPs are frequently ingested by marine organisms and lead to various detrimental effects. For instance, the interaction of MPs with gilthead seabream (Sparus aurata) has significantly enhanced the catalase and glutathione reductase (GRd) activity.68 Mediterranean mussel (Mytilus galloprovincialis) interaction with PE and PS-MPs resulted in the MPs accumulation in their gills, hemolymph and significant inflammatory responses.69 Similarly, blue mussel (Mytilus edulis) on exposure to PE-MPs showed a denotable histological change and significant inflammatory responses.37 In addition, it was reported that exposure to PE-MPs (11–13 μm) might lead to a significant change in the acetylcholinesterase activity of Scrobicularia plana (clam) that pave the way for neurotoxity.70 It was reported that scallop (Pecten maximus) uptakes PS-NPs and get accumulated in its digestive tract and tubules followed by deposition in muscles, kidneys and gonads.71 The toxicological investigation of carboxylated nanoplastics (COOH–NPs) on the larvae of marine rotifer (Brachionus plicatilis) for 48 h showed a significant lethal effect.72 However, aminated nanoplastics (NH2–NPs) showed a negligible lethality as they got aggregated at the microscale level. Studies on the NPs uptake behavior of Artemia franciscana reveals the presence of NPs in their gut and on the mandible, tail and appendages.73 The results revealed the fecal release of ingested COOH–NPs (5–100 mg L−1), though, the complete excretion was not observed.74The previous reports clearly demonstrate that the characteristics of microplastics, including their type, size, surface charges, and interactions with other organic and inorganic compounds, play a critical role in determining the extent of toxicity imposed upon freshwater and marine organisms. Notably, the interaction between biological molecules and M/NPs would lead to significant variations to the properties of M/NPs. These insights underscore the imperative of comprehensively understanding the intricate dynamics of M/NPs and their interactions in order to assess the implications they might have on the aquatic biota.
3. Biocorona formation on M/NPs
An ecologically relevant encapsulation of particles like plastics, MPs or NPs or non-plastics by various biomolecules such as polysaccharides, proteins, etc. is often referred to as an “environmental- or ecocorona”.31,75 M/NPs in the aquatic environment can easily interact with the bio-molecules such as nucleic acid and proteins secreted by the aquatic organisms.76,77 The particle surface will undergo substantial changes owing to these interactions and this alters the biological reactivity of the particles altogether.78,79 Sometimes, the particles can be internalized by the organisms, and they can interact with the biomolecules in the intracellular environment leading to formation of BC.80–82 Following their release into the environment, various bio-organics may compete with one another to adsorb the hydrophobic surface of M/NPs.83 BC is also formed over the M/NPs through EPS from the exo-proteome (proteins released as a result of signaling and habitat adaptation) or generated through the metabolic activity of aquatic organisms (DNA, carbohydrate, protein and so on).84 The formation of EC essentially involved natural/dissolved organic matter (NOM/DOM), pesticides, persistent organic pollutants (POPs), metabolite adsorption, and on the other hand, BC is all about proteins, metabolites, lipids and polysaccharides.30 Researchers have documented the EC formation on NPs within the culture medium of Daphnia magna. Additionally, they have observed the formation of BC within the organism's physiological fluid after homogenization.85,86To form BC (Fig. 1), M/NPs interact with the biomolecules through hydrophobic and electrostatic modes.87,88 Due to a variety of nanomaterial (NM) usage, particularly in the field of biomedicine, the formation of BC on the surface of NMs has lately received substantial attention.89,90 The BC is dynamic in nature, since, the protein adsorbs and desorbs from NM surfaces with time.78,91 As a result, the BC develops two distinct layers: (1) the hard corona, and (2) the soft corona.92 On the other hand, hard corona, experiences a significant number of conformational changes, sluggish exchange rates, lengthy retention times and high binding affinities.93 Due to differences in the forces involved and the chemical composition, the proteins in the hard and soft corona layers would interact differently in the aquatic environment.94–97
Bio-membranes like vesicles, engaged in various physiological functions such as storage of carbon, protection of tissues and resistance to external chemicals, must have lipids as the necessary component.98 The biological role and structure of lipid are presumably less well understood than proteins. In biological systems, there are several lipid derivatives that differ in their hydrophobic tails and hydrophilic heads, including waxes, triacylglycerides, phospholipids, sphingolipids and glycolipids.99 Lipids, particularly unsaturated lipids, may be oxidized during their interaction with M/NPs, leading to the production of –O–, –OH, –OOH, and hypochlorite moieties on hydrophobic tails of lipid molecules. These modified lipids frequently participate in a variety of biological activities. For instance, iron-dependent programmed cell death has recently been linked to ferroptosis, which is activated by lipid peroxides as an essential danger signal.100,101
The surface properties of the pristine or as formed M/NPs, including surface charge and functionalization, play a determinant role in their biological interactions.102 Presently, reactive functional groups such as sulfonic, carboxylic, phenoxy hydroxyl, alcoholic hydroxyl, carbonyl, hydroxide and amine groups103 are noted in the environmental polymers. In addition, multiple chemical groups might attach on the surface of M/NPs during the migration and degradation in the environment. The presence of various surface chemical groups largely dictates the fate and toxicity of M/NPs in the environment. In biological systems, corona formation on M/NPs surface might result in three types of adverse effects, i.e., antagonism, synergism and independent action.104
Various intermolecular interactions (Fig. 2) that influences the adsorption of biomolecule on the surface of M/NPs may include ionic (electrostatic) interaction (attractive and repulsive), van der Waals forces, hydrogen bonding, hydrophobic interactions, hydration forces and acid–base interactions. The key driving forces such as hydrophobic and ionic interactions, combined with an entropy gain originate from the release of small molecules (e.g., water and counter-ions), from the interface between the biomolecules and the surface. Hence, the size of the biomolecules will largely influence its ability to adsorb onto surfaces.105
From the preceding discussion it is evident that the interaction(s) between various biomolecules and M/NPs is influenced by properties of biomolecules, and physico-chemical characteristics of the M/NPs. This will in turn determine the types of forces involved in the interactions in forming multiple layers of surface bound biomolecules. This will critically influence the fate and biological reactivity of the M/NPs.
4. Ecological implications of BC
The contamination of M/NPs poses a significant environmental crisis on a global scale.6,106 The adherence of various biomolecules to the surface of M/NPs results in the formation of BC, which alters the surface properties of the M/NPs.6 The formation of BC is closely tied to the fate and toxicological consequences of M/NPs.43,107 The aquatic ecosystem is highly vulnerable to the risks associated with M/NPs pollution, with BC formation playing a crucial role in determining the toxicological effects on aquatic organisms (Fig. 3). In our study, we have emphasized the toxicological implications of M/NPs on aquatic organisms, specifically zoobenthos, zooplankton and nekton. The biomolecules present in these organisms are instrumental in the formation of BC on M/NPs.4.1 Effect of BC formation on the eco-toxicity of M/NPs
It is apparent from the preceding studies that the formation of the BC on M/NPs can elicit either a decrease or an increase in the toxic potential of these particles. The extent of such alterations is primarily determined by the specific biomolecules involved in the corona formation, in addition to the inherent physiochemical properties of the M/NPs. This observation highlights the critical influence of both biomolecular interactions and the intrinsic characteristics of MPs in shaping their overall toxicological impact.
4.2 Impact of BC on fate and transport of M/NPs
The fate and transport of M/NPs in the aquatic systems are highly influenced by the corona formation on M/NPs. For instance, the impact of proteins (BSA and LYZ) on the transportation and retention of NPs was studied in order to gain insight into the corona formation on NPs. The electrostatic charge of proteins, along with salinity levels and the size of the NPs, play a significant role in NP transportation. BSA effectively dispersed NPs of sizes 200 nm and 500 nm by enhancing steric repulsion forces, thereby, greatly improving their transportability. Conversely, the formation of a LYZ corona resulted in a substantial NP aggregation and hindered the transport of 200 nm NPs. Interestingly, LYZ exhibited a minimal binding affinity for the surfaces of 500 nm and 1000 nm NPs, suggesting a limited impact on the transportation of larger-sized NPs. However, when seawater salinity decreased from 35 to 3.5 PSU, LYZ was found to induce aggregation of 500 nm NPs, thereby, impeding their transportability.114 The impact of BC on the transportation and deposition of microplastics (MPs) in quartz sand was investigated under NaCl solutions with ionic strengths of 5 mM and 25 mM. COOH–MPs and NH2–MPs were utilized as MPs with negative and positive surface charges respectively. BSA and trypsin (TRY) were selected as proteins, each possessing distinct electrical charges. The presence of these two proteins had contrasting effects on the transport of COOH–MPs in quartz sand. It was found that negatively charged BSA facilitated the transport of COOH–MPs in quartz sand, while positively charged trypsin hindered COOH–MPs transport. Interestingly, both types of proteins enhanced the transport of NH2–MPs in quartz sand, regardless of their opposite effects on COOH–MPs. The improved transport of COOH–MPs in suspensions containing BSA was attributed to the steric interaction caused by the adsorption of BSA corona onto the surface of COOH–MPs as well as the repulsive effects, resulting from the presence of BSA in the solutions. On the other hand, the reduced movement of COOH–MPs in suspensions with trypsin was due to the larger size and decreased electrostatic repulsion of COOH–MPs resulting from trypsin adsorption onto their surface as well as the additional deposition sites, created by trypsin adsorption onto the quartz sand. The enhanced electrostatic repulsion resulting from BSA adsorption onto NH2–MPs surfaces led to increased transport of NH2–MPs when accompanied by BSA in the solutions.115 In a recent study conducted by Li et al. (2021), it was observed that BSA exhibited diverse effects on the aggregation kinetics of different surface-coated NH2–NPs under environmental conditions. The surface functional groups of PS-NPs played a crucial role in determining the amount of protein adsorption and the electrostatic destabilization or steric stabilization of PS-NPs. Due to the distinct surface characteristics of PS-NPs and the complex interactions between PS-NPs and BSA, a low concentration of BSA promoted the aggregation of negatively charged PS-NPs and PS-bare aggregates, while retarding the aggregation of positively charged NH2–NPs. The formation of a protein corona provided significant stability to the bare and negatively charged PS-NPs, indicating their high mobility in surface waters.96 Similarly, the effects of BSA (negatively charged) and bovine trypsin (TRY) (positively charged) on the aggregation of PS-NPs were investigated using time-resolved dynamic light scattering. The PS-NPs remained stable in the presence of BSA at moderately high protein concentrations (>10 mg L−1) but exhibited rapid aggregation in the presence of TRY. These findings clearly demonstrate the significant influence of electrostatic charge of proteins and their concentration on the aggregation behavior of NPs.The aggregation kinetics of PS-NPs in the aquatic environment were examined using five different proteins, namely BSA, bovine hemoglobin (BH), human serum albumin (HSA), bovine casein (BS) and collagen type I (Col I). The experiments were conducted under natural water conditions, with variations in pH, protein concentration and ionic strength to mimic real-world scenarios. The findings revealed that the structure of the proteins, their electrostatic characteristics, and the chemical composition of the solution played significant roles in PSNP–protein interactions, leading to distinct aggregation patterns in NaCl and CaCl2 solutions. In the presence of BSA, BS and Col I, PS-NPs remained stable in NaCl solution due to steric hindrance and electrostatic repulsion. However, at a concentration of 300 mM NaCl, BH destabilized PS-NPs, resulting in an aggregation rate of 1.71 nm s−1. On the other hand, in CaCl2 solutions with a concentration below 20 mM, HSA, BH and Col I destabilized PS-NPs through mechanisms such as steric hindrance, cation bridging and compression of the double layer. In contrast, BS stabilized PS-NPs by precipitating Ca2+ ions reduced the screening effect of charges. The quantity of proteins and the pH of the solution influenced the formation of the PC, protein structure and surface charge, and all that impacted the stability of PS-NPs.116
The aforementioned studies provide valuable insights into how BC formation and water chemistry affect the fate and transport of M/NPs in aquatic systems. One of the critical factors in the fate of bio-coronated M/NPs is the surface charge of the attached biomolecules. The surface properties of these biomolecules influence the overall charge of the M/NPs, affecting its stability, aggregation, and interaction with other particles or surfaces. For instance, if the biomolecules confer a positive charge to the M/NPs, they may have a higher tendency to repel each other, leading to reduced aggregation and improved dispersion in the water column. Apart from the impact of the surface charge of the attached biomolecules, the surface functionalization of M/NPs also plays a significant role in their fate and transport. Similarly, particle size is another crucial aspect influencing the fate and transport of bio-coronated M/NPs. Smaller nanoparticles generally have a larger surface area relative to their volume, leading to increased interactions with biomolecules and the surrounding environment. The nature of the surrounding medium, including pH, chemical composition of the water and the presence of other dissolved substances, also has a profound impact on the behavior of bio-coronated M/NPs. Understanding the fate and transport of bio-coronated M/NPs is a complex and multifaceted process due to the intricate interplay of all these factors. A comprehensive grasp of these process is required for evaluating the environmental implications of BC.
5. Potential methods to characterize biocorona formation
The accurate quantification and identification of the composition of BC are crucial for comprehending the ecological implications of M/NPs. This knowledge is pivotal as the composition and quantity of the BC directly influences the properties of M/NPs. Various analytical techniques have been employed to characterize the composition of BC formed on diverse nanomaterials. In many instances, a combination of multiple detection methods has proven to be the most efficacious approach. Generally, two primary approaches, namely ex situ and in situ techniques/methods, are utilized for the analysis of BC formation on M/NPs.5.1 In situ techniques/approaches
In situ approach assists in assessing the BC while the nanomaterial is disseminated in the physiological environment.117 Numerous researchers have engaged in in situ measurements to gain a more comprehensive understanding of the formation of the soft corona and the protein corona.118 These investigations have enabled them to obtain reliable information on the interactions between NM–protein and protein–protein within the chosen medium, without the need for purification.119 Initially, in situ approaches were proposed for characterizing corona formation, which has been supported by material characterization techniques. Such techniques include Fourier transform infrared spectroscopy (FTIS), Raman spectroscopy (RS), time of flight secondary ion mass spectrometry (TOF-SIMS), ultraviolet-visible spectroscopy (UV-vis), atomic force microscopy (AFM), etc.,28,120–122 However, the majority of these methodologies are inadequate for identifying all the diverse molecular constituents present in coronas. The dispersion of M/NPs is also a critical factor in determining the composition of the adsorbed corona. For instance, the formation of aggregates is a common occurrence among M/NPs, particularly, under high salinity conditions.112,123 The formation of corona on these agglomerated M/NPs will vary from the widely dispersed M/NPs.123 Hence, efforts directed at comprehending the process of BC formation on M/NPs should involve a comprehensive assessment of M/NPs, encompassing an in-depth analysis of their colloidal dynamics.5.2 Ex situ techniques/approaches
The ex situ approach proves valuable in evaluating the BC by isolating the nanomaterial–corona complex from its physiological milieu.117,145 The nanomaterial could be separated through exposure to extreme temperatures, high salinity, enzymatic treatment and detergents. Isolation can be complete or partial, depending on the strength of the interaction between macro-biomolecules and the nanomaterial, followed by, subsequent analyses.145 This strategy yields highly effective results for quantitative corona structure analysis. While the ex situ approach is commonly employed for protein corona (PC) analysis and has provided valuable insights into this phenomenon, it does not provide definitive information regarding the protein adsorption process or precise details about the soft corona.117,119| Type of M/NPs | Size | Method of BC isolation and measurement | BC composition | Refs. |
|---|---|---|---|---|
| a Acronyms: CD: circular dichroism; CLSM: confocal laser scanning microscopy; ELISA: enzyme-linked immunosorbent assay; FM: fluorescence microscope; FS: fluorescent spectroscopy; LSCM: laser scanning confocal microscope; MS: mass spectrometry; nano-HPLC-ESI-MS/MS: nano-high performance liquid chromatography-electrospray ionisation tandem mass spectrometry; NDS: nanodrop spectroscopy; PQ kit: protein quantification kit. | ||||
| PS M/NPs | 55–1000 nm | Centrifugation, ELISA, CLSM, FS, NDS and PQ kit | Proteins, DNA and carbohydrates | 159 |
| PS-NPs | 300 nm | Centrifugation, TEM, SDS-PAGE, nano-HPLC-Q exactive MS, BCA assay, and western blotting | Proteins | 160 |
| PS-NPs (NH2 and COOH) | 90–110 nm | Centrifugation, SDS PAGE, SA and MS | Proteins | 161 |
| PS-NPs (COOH) | 60 nm | Centrifugation, SEM and TEM | Proteins and carbohydrates | 162 |
| PS-NPs | 23 nm | Centrifugation, SEC, FM, DLS and PQ kit | Uronic acid, and proteins | 31 |
| PS-NPs (NH2) | 100–120 nm | Centrifugation, bradford assay, SDS-PAGE, gel staining, nano-LC-MS/MS, ELISA and LSCM | Proteins | 163 |
| PS-NPs (NH2) | 50 nm | Centrifugation, SDS PAGE and nano-HPLC-ESI-MS/MS | Proteins (32 kDa) | 164 |
| PS-NPs (NH2 COOH and plain) | 50–150 nm | SEM, FS, CD spectra, UV-vis spectra FT-IR and EDS | Proteins | 96 |
| PS M/NPs | 50–500 nm | Centrifugation, CLSM and FS | Glycoprotein biopolymer | 165 |
| PS-NPs (COOH) | 60 nm | Centrifugation, TEM, SDS-PAGE, FS, and SEC | Proteins | 162 |
| PS-NPs | 20/100 nm | DLS, FS, and CD spectra | Proteins (BSA and TRY) | 166 |
| PS-NPs (NH2) | 50 nm | DLS, TEM, centrifugation, SDS-PAGE gel staining and nano-HPLC-ESI-MS/MS | Proteins | 113 |
5.3 Limitations and challenges in characterizations
Although various analytical techniques are employed in BC analysis, several challenges are associated with these processes. Centrifugation, a crucial pre-treatment process, presents several obstacles in corona isolation. One major challenge is the potential for false positives. During centrifugation, proteins or protein complexes that did not originally bind to the nanomaterial as well as proteins, and that only bind to the proteins attached to the nanomaterial but not the nanomaterial itself, may sediment along with the nanomaterial and its corona, leading to false positive results. Additionally, false negatives could occur due to protein dissociation from the nanomaterial–corona complex induced by the centrifugal forces.146 Moreover, the centrifugation approach fails to separate the soft corona because the protein–protein interaction in this structure are too weak to withstand the centrifugal forces.147 In the case of SEC, selectivity decreases when applied to analytes with extremely high molar mass, and there is a possibility for analytes to interact with the stationary phase.146 DLS, UV-vis, and fluorescent microscopy are essential methods for evaluating the interaction between biomolecules and M/NPs.167 However, these techniques have limitations in terms of qualification and sensitivity. While DLS has traditionally been employed to study protein-nanomaterial interactions, a critical limitation arises from the presence of unbound proteins that could interfere with the scattering signal.119 Consequently, sample purification is necessary, which results in the loss of the soft corona that cannot be detected. In the case of fluorescent microscopy, a significant challenge is the need for fluorescence labeling that adds complexity to the experimental processes. TEM, SEM and AFM are indispensable microscopic techniques in the study of coronas. However, these approaches have limitations in terms of low particle population, qualification and quantification. TEM requires sample preparation that could potentially affect the morphology of M/NP-corona complexes. Additionally, the counterstaining becomes necessary due to the inadequate contrast resulting from the smaller size of the nanomaterial and thin protein layer.168 AFM allows for 3D imaging but suffers from poor efficiency.169 On the other hand, energy dispersive X-ray (EDX) analysis exhibits high efficiency but is not suitable for detecting organic compounds. RS provides the advantage of analyzing both organic and inorganic compounds but it has limitation in terms of low spatial resolution.164 FTIR has limitations in detecting the inorganic compounds and lacks sensitivity and accuracy.146In contrast, MS-based approaches are highly effective analytical techniques due to their high sensitivity, resolution, reproducibility, accuracy, quantification and selectivity.170–172 However, TOF-SIMS is not suitable for analyzing unknown materials and performs poorly in quantification assessments.173 GC-MS excels in the analysis and identification of trace amounts of organic molecules with high sensitivity and repeatability, but it cannot analyze thermally labile compounds.28 LC-MS offers better throughput, high sensitivity, greater precision compared to PAGE and reduced user bias.145 However, this approach involves multiple steps in the sample pre-treatment process. While ICP-MS analysis could detect low levels of metal elements (0.3 ppb), they would only represent the average metal amounts of the overall particle population.28 Moreover, it falls short in characterizing the molecular structure of organic coronas. CE-MS has limitations in terms of loading capacity and the direct analysis of high molecular weight proteins.174
6. Conclusion and future prospects
This review provides a concise overview of scientific knowledge pertaining to the formation of BC on the M/NPs, analytical techniques for characterizing BC, the eco-toxicity of M/NPs and the ecological implications of BC formation in the aquatic systems. The abundant presence of M/NPs in aquatic environments enables their interaction with various biomolecules leading to the BC formation. The resulting BC alters the size, stability, surface charge, and other physical and chemical properties of M/NPs, which in turn, affect their bioavailability, eco-toxicity and fate, ultimately resulting in diverse ecological effects. The scientific evidence clearly indicates that the BC formation can have both non-toxic and toxic impacts on the aquatic organisms.Characterizing the composition of BC poses challenges, as it requires a combination of multiple analytical techniques and the development of new characterization technologies. In particular, detecting BC formation in the biological system in situ remains a hurdle. The utilization of radiological or fluorescent labeling strategies could aid in the detection of BC in the biological systems. The formation of BC is significantly influenced by environmental factors such as pH, ionic strength, temperature and conductivity. Therefore, scientific investigations are needed to understand the contributions of these factors to the BC formation. Most of the research conducted on BC has utilized commercial M/NPs, model proteins or other biomaterials in the laboratory settings. To gain a comprehensive understanding of the interaction between BC and M/NPs in the aquatic environments, field research is highly recommended. The toxicological experiments involving bio-coronated M/NPs and aquatic organisms should be conducted under ecologically relevant conditions, as recommended by Organisation for Economic Cooperation and Development (OECD) guidelines. While protein corona studies have been predominant in aquatic biota, further investigations on other types of coronas such as lipids, nucleic acids and other types of BC would enhance our knowledge of their ecological impacts.
Conflicts of interest
There are no conflicts to declare.References
- K. K. Khoaele, O. J. Gbadeyan, V. Chunilall and B. Sithole, Sustainability, 2023, 15, 5233 CrossRef CAS.
- N. R. Lakkimsetty, B. Al Rahbi, S. Karunya, G. Kavitha and S. Gundu, in AIP Conference Proceedings, AIP Publishing LLC, 2023, vol. 2690, p. 20051 Search PubMed.
- N. P. Ivleva, A. C. Wiesheu and R. Niessner, Angew. Chem., Int. Ed., 2017, 56, 1720–1739 CrossRef CAS.
- K. Kruse, K. Knickmeier, D. Brennecke, B. Unger and U. Siebert, in Marine Mammals: A Deep Dive into the World of Science, Springer International Publishing Cham, 2023, pp. 49–62 Search PubMed.
- L. Lebreton and A. Andrady, Palgrave Commun., 2019, 5, 1–11 CrossRef.
- T. R. Walker and L. Fequet, TrAC, Trends Anal. Chem., 2023, 116984 CrossRef CAS.
- J. R. Jambeck, R. Geyer, C. Wilcox, T. R. Siegler, M. Perryman, A. Andrady, R. Narayan and K. L. Law, Science, 2015, 347, 768–771 CrossRef CAS PubMed.
- R. Geyer, J. R. Jambeck and K. L. Law, Sci. Adv., 2017, 3, e1700782 CrossRef PubMed.
- M. Shams, I. Alam and M. S. Mahbub, Environ. Adv., 2021, 5, 100119 CrossRef CAS PubMed.
- M. Hasan, A. R. M. T. Islam, M. M. M. F. Jion, M. N. Rahman, S. D. Peu, A. Das, A. B. M. M. Bari, M. S. Islam, S. C. Pal, A. Islam, T. R. Choudhury, M. R. J. Rakib, A. M. Idris and G. Malafaia, Sci. Total Environ., 2023, 164164 CrossRef CAS PubMed.
- T. K. Dey, M. Rasel, T. Roy, M. E. Uddin, B. K. Pramanik and M. Jamal, Sci. Total Environ., 2023, 867, 161390 CrossRef CAS.
- A. Li, H. Cui, Y. Sheng, J. Qiao, X. Li and H. Huang, J. Environ. Chem. Eng., 2023, 110092 CrossRef CAS.
- N. B. Hartmann, T. Hüffer, R. C. Thompson, M. Hassellöv, A. Verschoor, A. E. Daugaard, S. Rist, T. Karlsson, N. Brennholt, M. Cole, M. P. Herrling, M. C. Hess, N. P. Ivleva, A. L. Lusher and M. Wagner, Environ. Sci. Technol., 2019, 53, 1039–1047 CrossRef CAS PubMed.
- P. Zhang, Y. Liu, L. Zhang, M. Xu, L. Gao and B. Zhao, Ecotoxicol. Environ. Saf., 2022, 243, 113997 CrossRef CAS PubMed.
- V. Kumar, E. Singh, S. Singh, A. Pandey and P. C. Bhargava, Chem. Eng. J., 2023, 459, 141568 CrossRef CAS.
- P. Fabbri and M. Messori, in Modification of Polymer properties, ed. C. F. Jasso-Gastinel and J. M. Kenny, William Andrew Publishing, 2017, pp. 109–130 Search PubMed.
- J.-L. Xu, X. Lin, J. J. Wang and A. A. Gowen, Sci. Total Environ., 2022, 851, 158111 CrossRef CAS PubMed.
- Y. Song, R. Qiu, J. Hu, X. Li, X. Zhang, Y. Chen, W. M. Wu and D. He, Sci. Total Environ., 2020, 746, 141289 CrossRef CAS PubMed.
- C. C. Rodrigues, R. F. Salla and T. L. Rocha, J. Hazard. Mater., 2023, 444, 130382 CrossRef CAS PubMed.
- Y. Yu, W. Y. Mo and T. Luukkonen, Sci. Total Environ., 2021, 797, 149140 CrossRef CAS PubMed.
- S. Liu, H. Ding, Y. Song, Y. Xue, M. Bi, M. Wu, C. Zhao, M. Wang, J. Shi and H. Deng, J. Hazard. Mater., 2023, 450, 131089 CrossRef CAS PubMed.
- H. Qu, R. Ma, H. Barrett, B. Wang, J. Han, F. Wang, P. Chen, W. Wang, G. Peng and G. Yu, Environ. Int., 2020, 136, 105480 CrossRef CAS PubMed.
- A. M. de Souza, A. L. Santos, D. S. Araújo, R. R. de B. Magalhães and T. L. Rocha, J. Hazard. Mater. Adv., 2022, 6, 100068 CrossRef.
- T. Wang, L. Wang, Q. Chen, N. Kalogerakis, R. Ji and Y. Ma, Sci. Total Environ., 2020, 748, 142427 CrossRef CAS PubMed.
- S. Kihara, S. Ghosh, D. R. McDougall, A. E. Whitten, J. P. Mata, I. Köper and D. J. McGillivray, Biointerphases, 2020, 15, 51002 CrossRef CAS.
- I. Ali, X. Tan, J. Li, C. Peng, I. Naz, Z. Duan and Y. Ruan, J. Clean. Prod., 2022, 376, 134314 CrossRef CAS.
- I. Lynch and K. A. Dawson, Nano Today, 2008, 3, 40–47 CrossRef CAS.
- J. Cao, Q. Yang, J. Jiang, T. Dalu, A. Kadushkin, J. Singh, R. Fakhrullin, F. Wang, X. Cai and R. Li, Part. Fibre Toxicol., 2022, 19, 55 CrossRef.
- W. T. Cooper, J. C. Chanton, J. D'Andrilli, S. B. Hodgkins, D. C. Podgorski, A. C. Stenson, M. M. Tfaily and R. M. Wilson, Mass Spectrom. Rev., 2022, 41, 215–239 CrossRef CAS PubMed.
- K. E. Wheeler, A. J. Chetwynd, K. M. Fahy, B. S. Hong, J. A. Tochihuitl, L. A. Foster and I. Lynch, Nat. Nanotechnol., 2021, 16, 617–629 CrossRef CAS PubMed.
- M. Junaid and J. Wang, Water Res., 2021, 201, 117319 CrossRef CAS PubMed.
- G. Lamichhane, A. Acharya, R. Marahatha, B. Modi, R. Paudel, A. Adhikari, B. K. Raut, S. Aryal and N. Parajuli, Int. J. Environ. Sci. Technol., 2023, 20, 4673–4694 CrossRef CAS.
- F. Avazzadeh Samani and L. Meunier, J. Environ. Sci. Health, Part A: Toxic/Hazard. Subst. Environ. Eng., 2023, 58, 222–235 CrossRef CAS.
- S. L. Wong, B. B. Nyakuma, K. Y. Wong, C. T. Lee, T. H. Lee and C. H. Lee, Mar. Pollut. Bull., 2020, 158, 111432 CrossRef CAS PubMed.
- Q. Liu, Y. Chen, Z. Chen, F. Yang, Y. Xie and W. Yao, Sci. Total Environ., 2022, 851, 157991 CrossRef CAS PubMed.
- M. Vighi, J. Bayo, F. Fernández-Piñas, J. Gago, M. Gómez, J. Hernández-Borges, A. Herrera, J. Landaburu, S. Muniategui-Lorenzo and A.-R. Muñoz, Rev. Environ. Contam. Toxicol., 2021, 257, 163–218 CAS.
- S. Sangkham, O. Faikhaw, N. Munkong, P. Sakunkoo, C. Arunlertaree, M. Chavali, M. Mousazadeh and A. Tiwari, Mar. Pollut. Bull., 2022, 181, 113832 CrossRef CAS.
- N. U. Benson, O. D. Agboola, O. H. Fred-Ahmadu, G. E. De-la-Torre, A. Oluwalana and A. B. Williams, Front. Mar. Sci., 2022, 9, 291 Search PubMed.
- M. C. Guerrera, M. Aragona, C. Porcino, F. Fazio, R. Laurà, M. Levanti, G. Montalbano, G. Germanà, F. Abbate and A. Germanà, Appl. Sci., 2021, 11, 5768 CrossRef CAS.
- D. Gao, X. Liu, M. Junaid, H. Liao, G. Chen, Y. Wu and J. Wang, Sci. Total Environ., 2022, 155620 CrossRef CAS.
- V. Thiagarajan, S. A. Alex, R. Seenivasan, N. Chandrasekaran and A. Mukherjee, Sci. Total Environ., 2021, 784, 147262 CrossRef CAS PubMed.
- L. Natarajan, D. Soupam, S. Dey, N. Chandrasekaran, R. Kundu, S. Paul and A. Mukherjee, Toxicol. Rep., 2022, 9, 1953–1961 CrossRef CAS PubMed.
- H. Gong, R. Li, F. Li, X. Guo, L. Xu, L. Gan, M. Yan and J. Wang, J. Hazard. Mater., 2023, 443, 130266 CrossRef CAS PubMed.
- J. Shi, Y. Dong, Y. Shi, T. Yin, W. He, T. An, Y. Tang, X. Hou, S. Chong and D. Chen, Environ. Res., 2022, 210, 112855 CrossRef CAS PubMed.
- K. Neelavannan, I. S. Sen, A. M. Lone and K. Gopinath, Chemosphere, 2022, 290, 133354 CrossRef CAS PubMed.
- C. Yuan, H. Almuhtaram, M. J. McKie and R. C. Andrews, Chemosphere, 2022, 286, 131881 CrossRef CAS PubMed.
- A. R. de Carvalho, L. Riem-Galliano, A. Ter Halle and J. Cucherousset, Environ. Pollut., 2022, 309, 119760 CrossRef CAS PubMed.
- X. Cai, H. Chen, B. Huang and J. Lu, Ecotoxicol. Environ. Saf., 2022, 232, 113254 CrossRef CAS.
- W. Li, X. Li, J. Tong, W. Xiong, Z. Zhu, X. Gao, S. Li, M. Jia, Z. Yang and J. Liang, Sci. Total Environ., 2023, 856, 159030 CrossRef CAS.
- A. Al Mamun, T. A. E. Prasetya, I. R. Dewi and M. Ahmad, Sci. Total Environ., 2023, 858, 159834 CrossRef PubMed.
- H. Çelebi, T. Bahadır, İ. Şimşek and Ş. Tulun, Environ. Sci. Proc., 2023, 25, 69 Search PubMed.
- Y. Lu, Y. Zhang, Y. Deng, W. Jiang, Y. Zhao, J. Geng, L. Ding and H. Ren, Environ. Sci. Technol., 2016, 50, 4054–4060 CrossRef CAS PubMed.
- Q. Chen, M. Gundlach, S. Yang, J. Jiang, M. Velki, D. Yin and H. Hollert, Sci. Total Environ., 2017, 584, 1022–1031 CrossRef PubMed.
- L. Lei, S. Wu, S. Lu, M. Liu, Y. Song, Z. Fu, H. Shi, K. M. Raley-Susman and D. He, Sci. Total Environ., 2018, 619, 1–8 CrossRef PubMed.
- G. Limonta, A. Mancia, A. Benkhalqui, C. Bertolucci, L. Abelli, M. C. Fossi and C. Panti, Sci. Rep., 2019, 9, 1–11 CrossRef CAS PubMed.
- D. de Mello Pereira, S. C. Mazon, E. J. Mendes, R. Brunetto, B. Ozelame, F. S. Zembruski, A. L. F. Dalcin, I. B. Marsaro, G. P. Aguiar and J. A. Lutinski, J. Toxicol. Environ. Heal. Part A, 2023, 1–14 Search PubMed.
- I. Hansjosten, M. Takamiya, J. Rapp, L. Reiner, S. Fritsch-Decker, D. Mattern, S. Andraschko, C. Anders, G. Pace and T. Dickmeis, Environ. Sci. Nano, 2022, 9, 375–392 RSC.
- A. Greven, T. Merk, F. Karagöz, K. Mohr, M. Klapper, B. Jovanović and D. Palić, Environ. Toxicol. Chem., 2016, 35, 3093–3100 CrossRef CAS PubMed.
- J. Ding, S. Zhang, R. M. Razanajatovo, H. Zou and W. Zhu, Environ. Pollut., 2018, 238, 1–9 CrossRef CAS PubMed.
- S.-A. Strungaru, R. Jijie, M. Nicoara, G. Plavan and C. Faggio, TrAC, Trends Anal. Chem., 2019, 110, 116–128 CrossRef CAS.
- S. Rehse, W. Kloas and C. Zarfl, Chemosphere, 2016, 153, 91–99 CrossRef CAS PubMed.
- S. Parra, S. Varandas, D. Santos, L. Félix, L. Fernandes, E. Cabecinha, J. Gago and S. M. Monteiro, Water, 2021, 13, 394 CrossRef CAS.
- J. K. H. Wong, K. K. Lee, K. H. D. Tang and P.-S. Yap, Sci. Total Environ., 2020, 719, 137512 CrossRef CAS.
- Z. Feng, T. Zhang, Y. Li, X. He, R. Wang, J. Xu and G. Gao, Sci. Total Environ., 2019, 696, 133948 CrossRef CAS PubMed.
- S. Gündoğdu, Food Addit. Contam.: Part A, 2018, 35, 1006–1014 CrossRef.
- Q. Liu, Z. Chen, Y. Chen, F. Yang, W. Yao and Y. Xie, J. Agric. Food Chem., 2021, 69, 10450–10468 CrossRef CAS PubMed.
- A.-N. Mubin, S. Arefin, M. S. Mia, A. R. M. T. Islam, A. B. M. M. Bari, M. S. Islam, M. M. Ali, M. A. B. Siddique, M. S. Rahman and V. Senapathi, Sci. Total Environ., 2023, 164224 CrossRef CAS.
- X. Capó, J. J. Company, C. Alomar, M. Compa, A. Sureda, A. Grau, B. Hansjosten, J. Lopez-Vazquez, J. B. Quintana and R. Rodil, Sci. Total Environ., 2021, 767, 144976 CrossRef.
- C. G. Avio, S. Gorbi, M. Milan, M. Benedetti, D. Fattorini, G. d'Errico, M. Pauletto, L. Bargelloni and F. Regoli, Environ. Pollut., 2015, 198, 211–222 CrossRef CAS PubMed.
- S. O'Donovan, N. C. Mestre, S. Abel, T. G. Fonseca, C. C. Carteny, B. Cormier, S. H. Keiter and M. J. Bebianno, Front. Mar. Sci., 2018, 5, 143 CrossRef.
- M. Al-Sid-Cheikh, S. J. Rowland, K. Stevenson, C. Rouleau, T. B. Henry and R. C. Thompson, Environ. Sci. Technol., 2018, 52, 14480–14486 CrossRef CAS PubMed.
- L. Manfra, A. Rotini, E. Bergami, G. Grassi, C. Faleri and I. Corsi, Ecotoxicol. Environ. Saf., 2017, 145, 557–563 CrossRef CAS PubMed.
- M. Sendra, E. Sparaventi, J. Blasco, I. Moreno-Garrido and C. V. M. Araujo, Ecotoxicol. Environ. Saf., 2020, 188, 109853 CrossRef PubMed.
- E. Bergami, E. Bocci, M. L. Vannuccini, M. Monopoli, A. Salvati, K. A. Dawson and I. Corsi, Ecotoxicol. Environ. Saf., 2016, 123, 18–25 CrossRef CAS PubMed.
- D. Chakraborty, S. Giri, L. Natarajan, N. Chandrasekaran and A. Mukherjee, J. Indian Inst. Sci., 2022, 102, 621–637 CrossRef.
- H. Hadiyanto, A. Khoironi, I. Dianratri, S. Suherman, F. Muhammad and S. Vaidyanathan, Heliyon, 2021, 7, e07676 CrossRef CAS PubMed.
- J. Windheim, L. Colombo, N. C. Battajni, L. Russo, A. Cagnotto, L. Diomede, P. Bigini, E. Vismara, F. Fiumara and S. Gabbrielli, Int. J. Mol. Sci., 2022, 23, 10329 CrossRef CAS PubMed.
- P. Del Pino, B. Pelaz, Q. Zhang, P. Maffre, G. U. Nienhaus and W. J. Parak, Mater. Horiz., 2014, 1, 301–313 RSC.
- P. C. Ke, S. Lin, W. J. Parak, T. P. Davis and F. Caruso, ACS Nano, 2017, 11, 11773–11776 CrossRef CAS PubMed.
- D. Magrì, M. Veronesi, P. Sánchez-Moreno, V. Tolardo, T. Bandiera, P. P. Pompa, A. Athanassiou and D. Fragouli, Environ. Pollut., 2021, 271, 116262 CrossRef.
- O. Hollóczki and S. Gehrke, ChemPhysChem, 2020, 21, 9–12 CrossRef PubMed.
- P. M. Gopinath, V. Saranya, S. Vijayakumar, M. Mythili Meera, S. Ruprekha, R. Kunal, A. Pranay, J. Thomas, A. Mukherjee and N. Chandrasekaran, Sci. Rep., 2019, 9, 8860 CrossRef.
- L. Zeng, J. Gao, Y. Liu, J. Gao, L. Yao, X. Yang, X. Liu, B. He, L. Hu and J. Shi, TrAC, Trends Anal. Chem., 2019, 118, 303–314 CrossRef CAS.
- F. Nasser and I. Lynch, J. Proteomics, 2016, 137, 45–51 CrossRef CAS PubMed.
- J. Armengaud, J. A. Christie-Oleza, G. Clair, V. Malard and C. Duport, Expert Rev. Proteomics, 2012, 9, 561–575 CrossRef CAS PubMed.
- O. O. Fadare, B. Wan, K. Liu, Y. Yang, L. Zhao and L.-H. Guo, Environ. Sci. Technol., 2020, 54, 8001–8009 CrossRef CAS PubMed.
- L. Canesi, T. Balbi, R. Fabbri, A. Salis, G. Damonte, M. Volland and J. Blasco, NanoImpact, 2017, 8, 89–98 CrossRef.
- G. V Lowry, K. B. Gregory, S. C. Apte and J. R. Lead, 2012.
- D. Docter, D. Westmeier, M. Markiewicz, S. Stolte, S. K. Knauer and R. H. Stauber, Chem. Soc. Rev., 2015, 44, 6094–6121 RSC.
- J. Kuruvilla, A. P. Farinha, N. Bayat and S. Cristobal, Nanoscale Horiz., 2017, 2, 55–64 RSC.
- C. D. Walkey and W. C. W. Chan, Chem. Soc. Rev., 2012, 41, 2780–2799 RSC.
- G. Pulido-Reyes, F. Leganes, F. Fernández-Piñas and R. Rosal, Environ. Toxicol. Chem., 2017, 36, 3181–3193 CrossRef CAS PubMed.
- S. Yang, Y. Liu, Y. Wang and A. Cao, Small, 2013, 9, 1635–1653 CrossRef CAS.
- F. Barbero, L. Russo, M. Vitali, J. Piella, I. Salvo, M. L. Borrajo, M. Busquets-Fité, R. Grandori, N. G. Bastús and E. Casals, in Seminars in Immunology, Elsevier, 2017, vol. 34, pp. 52–60 Search PubMed.
- C. Li, Y. Ma, X. Liu, R. Huang, R. Su, W. Qi, J. Che and Z. He, Ecotoxicol. Environ. Saf., 2021, 214, 112115 CrossRef CAS PubMed.
- X. Li, E. He, K. Jiang, W. J. G. M. Peijnenburg and H. Qiu, Water Res., 2021, 190, 116742 CrossRef CAS PubMed.
- T. Cedervall, I. Lynch, S. Lindman, T. Berggård, E. Thulin, H. Nilsson, K. A. Dawson and S. Linse, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2050–2055 CrossRef CAS.
- G. Van Meer, D. R. Voelker and G. W. Feigenson, Nat. Rev. Mol. Cell Biol., 2008, 9, 112–124 CrossRef CAS.
- S. K. Tayebati, Molecules, 2018, 23, 2257 CrossRef PubMed.
- S. Xu, H. Zheng, R. Ma, D. Wu, Y. Pan, C. Yin, M. Gao, W. Wang, W. Li and S. Liu, Nat. Commun., 2020, 11, 3484 CrossRef CAS.
- H. Zheng, J. Jiang, S. Xu, W. Liu, Q. Xie, X. Cai, J. Zhang, S. Liu and R. Li, Nanoscale, 2021, 13, 2266–2285 RSC.
- T. S. Galloway, M. Cole and C. Lewis, Nat. Ecol. Evol., 2017, 1, 116 CrossRef PubMed.
- B. S. S. Ramasamy and S. Palanisamy, Environ. Sci. Pollut. Res., 2021, 28, 43258–43273 CrossRef CAS PubMed.
- Y. Cao, M. Zhao, X. Ma, Y. Song, S. Zuo, H. Li and W. Deng, Sci. Total Environ., 2021, 788, 147620 CrossRef CAS PubMed.
- N. Mei, J. Hedberg, I. Odnevall Wallinder and E. Blomberg, ACS Omega, 2019, 4, 21778–21791 CrossRef CAS PubMed.
- B. Chen, Z. Zhang, T. Wang, H. Hu, G. Qin, T. Lu, W. Hong, J. Hu, J. Penuelas and H. Qian, J. Hazard. Mater., 2023, 451, 131198 CrossRef CAS.
- M. Junaid, J. A. Siddiqui, S. Liu, R. Lan, Z. Abbas, G. Chen and J. Wang, Sci. Total Environ., 2023, 163679 CrossRef CAS PubMed.
- F. Manzi, P. Schlösser, A. Owczarz and J. Wolinska, Philos. Trans. R. Soc., B, 2023, 378, 20220013 CrossRef CAS.
- H. Luo, Q. Du, Z. Zhong, Y. Xu and J. Peng, Sci. Total Environ., 2022, 851, 157948 CrossRef CAS PubMed.
- L. Canesi, C. Ciacci, E. Bergami, M. P. Monopoli, K. A. Dawson, S. Papa, B. Canonico and I. Corsi, Mar. Environ. Res., 2015, 111, 34–40 CrossRef CAS PubMed.
- J. Jiménez-Lamana, L. Marigliano, J. Allouche, B. Grassl, J. Szpunar and S. Reynaud, Anal. Chem., 2020, 92, 11664–11672 CrossRef PubMed.
- F. Akhatova, I. Ishmukhametov, G. Fakhrullina and R. Fakhrullin, Int. J. Mol. Sci., 2022, 23, 806 CrossRef CAS.
- L. F. Marques-Santos, G. Grassi, E. Bergami, C. Faleri, T. Balbi, A. Salis, G. Damonte, L. Canesi and I. Corsi, Nanotoxicology, 2018, 12, 847–867 CrossRef CAS PubMed.
- Z. Dong, Y. Hou, W. Han, M. Liu, J. Wang and Y. Qiu, Water Res., 2020, 182, 115978 CrossRef CAS.
- H. Rong, L. He, M. Li, M. Zhang, K. Yi, P. Han and M. Tong, Sci. Total Environ., 2021, 756, 143837 CrossRef CAS PubMed.
- Z. Huang, C. Chen, Y. Liu, S. Liu, D. Zeng, C. Yang, W. Huang and Z. Dang, Water Res., 2022, 219, 118522 CrossRef CAS PubMed.
- F. Pederzoli, G. Tosi, M. A. Vandelli, D. Belletti, F. Forni and B. Ruozi, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2017, 9, e1467 Search PubMed.
- F. Traldi, P. Liu, I. Albino, L. Ferreira, A. Zarbakhsh and M. Resmini, Int. J. Mol. Sci., 2023, 24, 2810 CrossRef CAS PubMed.
- R. García-Álvarez and M. Vallet-Regí, Nanomaterials, 2021, 11, 888 CrossRef PubMed.
- N. Jin, Y. Song, R. Ma, J. Li, G. Li and D. Zhang, Anal. Chim. Acta, 2022, 1197, 339519 CrossRef CAS.
- Y. Chen, D. Wen, J. Pei, Y. Fei, D. Ouyang, H. Zhang and Y. Luo, Curr. Opin. Environ. Sci. Health., 2020, 18, 14–19 CrossRef.
- C. Du, J. Wu, J. Gong, H. Liang and Z. Li, Surf. Interface Anal., 2020, 52, 293–300 CrossRef CAS.
- C. González-Fernández, K. Tallec, N. Le Goïc, C. Lambert, P. Soudant, A. Huvet, M. Suquet, M. Berchel and I. Paul-Pont, Chemosphere, 2018, 208, 764–772 CrossRef PubMed.
- R. Bilardo, F. Traldi, A. Vdovchenko and M. Resmini, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2022, 14, e1788 CAS.
- J. Wu, R. Jiang, W. Lin and G. Ouyang, Environ. Pollut., 2019, 245, 836–843 CrossRef CAS.
- Y. Wang and Y. Ni, Talanta, 2014, 119, 320–330 CrossRef CAS.
- T. Zhang, G. Zhu, B. Lu, Z. Qian and Q. Peng, Med. Res. Rev., 2021, 41, 1835–1850 CrossRef CAS.
- M. P. Monopoli, D. Walczyk, A. Campbell, G. Elia, I. Lynch, F. Baldelli Bombelli and K. A. Dawson, J. Am. Chem. Soc., 2011, 133, 2525–2534 CrossRef CAS.
- G. Sarau, L. Kling, B. E. Oßmann, A.-K. Unger, F. Vogler and S. H. Christiansen, Appl. Spectrosc., 2020, 74, 1155–1160 CrossRef CAS.
- W. J. Shim, S. H. Hong and S. E. Eo, Anal. Methods, 2017, 9, 1384–1391 RSC.
- S. Mariano, S. Tacconi, M. Fidaleo, M. Rossi and L. Dini, Front. Toxicol., 2021, 3, 636640 CrossRef PubMed.
- P. Rivera-Gil, D. Jimenez De Aberasturi, V. Wulf, B. Pelaz, P. Del Pino, Y. Zhao, J. M. De La Fuente, I. Ruiz De Larramendi, T. Rojo and X.-J. Liang, Acc. Chem. Res., 2013, 46, 743–749 CrossRef CAS PubMed.
- G. Li, Z. Yang, Z. Pei, Y. Li, R. Yang, Y. Liang, Q. Zhang and G. Jiang, Talanta, 2022, 249, 123701 CrossRef CAS PubMed.
- W. Fu, J. Min, W. Jiang, Y. Li and W. Zhang, Sci. Total Environ., 2020, 721, 137561 CrossRef CAS PubMed.
- T. Kopac, Int. J. Biol. Macromol., 2021, 169, 290–301 CrossRef CAS PubMed.
- C. Gruian, E. Vanea, S. Simon and V. Simon, Biochim. Biophys. Acta, Proteins Proteomics, 2012, 1824, 873–881 CrossRef CAS PubMed.
- E. Vanea and V. Simon, Appl. Surf. Sci., 2013, 280, 144–150 CrossRef CAS.
- P.-L. Latreille, M. Le Goas, S. Salimi, J. Robert, G. De Crescenzo, D. C. Boffito, V. A. Martinez, P. Hildgen and X. Banquy, ACS Nano, 2022, 16, 1689–1707 CrossRef CAS PubMed.
- H. Tiernan, B. Byrne and S. G. Kazarian, Spectrochim. Acta, Part A, 2020, 241, 118636 CrossRef CAS PubMed.
- Y. Tang, Y. Zhuang, S. Zhang, Z. J. Smith, Y. Li, X. Mu, M. Li, C. He, X. Zheng and F. Pan, ACS Cent. Sci., 2021, 7, 768–780 CrossRef CAS PubMed.
- J. De Meutter and E. Goormaghtigh, Anal. Chem., 2021, 93, 13441–13449 CrossRef CAS PubMed.
- T. Pazderka and V. Kopecký Jr, Spectrochim. Acta, Part A, 2017, 185, 207–216 CrossRef CAS PubMed.
- J.-L. Xu, K. V. Thomas, Z. Luo and A. A. Gowen, TrAC, Trends Anal. Chem., 2019, 119, 115629 CrossRef CAS.
- T. K. Dey, M. E. Uddin and M. Jamal, Environ. Sci. Pollut. Res., 2021, 28, 16925–16947 CrossRef CAS PubMed.
- R. Fakhrullin, L. Nigamatzyanova and G. Fakhrullina, Sci. Total Environ., 2021, 772, 145478 CrossRef CAS PubMed.
- H. Cai, E. G. Xu, F. Du, R. Li, J. Liu and H. Shi, Chem. Eng. J., 2021, 410, 128208 CrossRef CAS.
- L. Böhmert, L. Voß, V. Stock, A. Braeuning, A. Lampen and H. Sieg, Nanoscale Adv., 2020, 2, 563–582 RSC.
- C. Weber, S. Morsbach and K. Landfester, Angew. Chem., Int. Ed., 2019, 58, 12787–12794 CrossRef CAS PubMed.
- J. C. Prata, J. P. da Costa, A. V. Girão, I. Lopes, A. C. Duarte and T. Rocha-Santos, Sci. Total Environ., 2019, 686, 131–139 CrossRef CAS.
- R. R. Hurley, A. L. Lusher, M. Olsen and L. Nizzetto, Environ. Sci. Technol., 2018, 52, 7409–7417 CrossRef CAS PubMed.
- Y. T. Ho, B. Poinard, E. L. L. Yeo and J. C. Y. Kah, Analyst, 2015, 140, 1026–1036 RSC.
- P. Khramtsov, T. Kalashnikova, M. Bochkova, M. Kropaneva, V. Timganova, S. Zamorina and M. Rayev, Int. J. Pharm., 2021, 599, 120422 CrossRef CAS PubMed.
- Z. Zhu, J. J. Lu and S. Liu, Anal. Chim. Acta, 2012, 709, 21–31 CrossRef CAS.
- C. Vidaurre-Agut, E. Rivero-Buceta, E. Romaní-Cubells, A. M. Clemments, C. D. Vera-Donoso, C. C. Landry and P. Botella, ACS Omega, 2019, 4, 8852–8861 CrossRef CAS PubMed.
- E. Rodríguez-Suárez and A. D. Whetton, Mass Spectrom. Rev., 2013, 32, 1–26 CrossRef PubMed.
- I. Ali, Q. Cheng, T. Ding, Q. Yiguang, Z. Yuechao, H. Sun, C. Peng, I. Naz, J. Li and J. Liu, J. Clean. Prod., 2021, 313, 127863 CrossRef CAS.
- R. Qiao, K. Lu, Y. Deng, H. Ren and Y. Zhang, Sci. Total Environ., 2019, 682, 128–137 CrossRef CAS PubMed.
- N. Fernández-Iglesias and J. Bettmer, Nanoscale, 2015, 7, 14324–14331 RSC.
- L. M. Hernandez, N. Yousefi and N. Tufenkji, Environ. Sci. Technol. Lett., 2017, 4, 280–285 CrossRef CAS.
- R.-F. Shiu, C. I. Vazquez, C.-Y. Chiang, M.-H. Chiu, C.-S. Chen, C.-W. Ni, G.-C. Gong, A. Quigg, P. H. Santschi and W.-C. Chin, Sci. Total Environ., 2020, 748, 141469 CrossRef CAS PubMed.
- Y. Tan, X. Zhu, D. Wu, E. Song and Y. Song, Environ. Sci. Technol., 2020, 54, 11485–11493 CrossRef CAS PubMed.
- G. Grassi, E. Gabellieri, P. Cioni, E. Paccagnini, C. Faleri, P. Lupetti, I. Corsi and E. Morelli, Sci. Total Environ., 2020, 725, 138457 CrossRef CAS PubMed.
- O. O. Fadare, B. Wan, L.-H. Guo, Y. Xin, W. Qin and Y. Yang, Environ. Sci. Nano, 2019, 6, 1466–1477 RSC.
- L. Canesi, C. Ciacci, R. Fabbri, T. Balbi, A. Salis, G. Damonte, K. Cortese, V. Caratto, M. P. Monopoli and K. Dawson, Environ. Res., 2016, 150, 73–81 CrossRef CAS PubMed.
- S. Summers, T. Henry and T. Gutierrez, Mar. Pollut. Bull., 2018, 130, 258–267 CrossRef CAS PubMed.
- X. Li, E. He, B. Xia, Y. Liu, P. Zhang, X. Cao, L. Zhao, X. Xu and H. Qiu, Environ. Sci. Nano, 2021, 8, 1560–1570 RSC.
- R. R. Samal, H. S. Navani, S. Saha, B. Kisan and U. Subudhi, J. Hazard. Mater., 2023, 131496 CrossRef CAS.
- M. Mahmoudi, A. M. Abdelmonem, S. Behzadi, J. H. Clement, S. Dutz, M. R. Ejtehadi, R. Hartmann, K. Kantner, U. Linne and P. Maffre, ACS Nano, 2013, 7, 6555–6562 CrossRef CAS PubMed.
- H. Zhang, J. Huang, Y. Wang, R. Liu, X. Huai, J. Jiang and C. Anfuso, Opt. Commun., 2018, 406, 3–17 CrossRef CAS.
- M. Magro, M. Zaccarin, G. Miotto, L. Da Dalt, D. Baratella, P. Fariselli, G. Gabai and F. Vianello, Anal. Bioanal. Chem., 2018, 410, 2949–2959 CrossRef CAS PubMed.
- X.-X. Zhou, S. He, Y. Gao, Z.-C. Li, H.-Y. Chi, C.-J. Li, D.-J. Wang and B. Yan, Anal. Chem., 2021, 93, 6698–6705 CrossRef CAS PubMed.
- J. Kruszewska, J. Zajda and M. Matczuk, Talanta, 2021, 226, 122153 CrossRef CAS.
- D. Heller-Krippendorf, L. Veith, R. ter Veen, D. Breitenstein, E. Tallarek, B. Hagenhoff and C. Engelhard, Surf. Interface Anal., 2019, 51, 1078–1092 CrossRef CAS.
- F. P. Gomes and J. R. Yates III, Mass Spectrom. Rev., 2019, 38, 445–460 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |