Open Access Article
Open Access ArticleA comprehensive review of bioleaching optimization by statistical approaches: recycling mechanisms, factors affecting, challenges, and sustainability
Tannaz Naseri a,
Vahid Beikia,
Seyyed Mohammad Mousavi
a,
Vahid Beikia,
Seyyed Mohammad Mousavi *ab and
Sebastien Farnaudc
*ab and
Sebastien Farnaudc
aBiotechnology Group, Chemical Engineering Department, Tarbiat Modares University, Tehran, Iran. E-mail: Mousavi_m@modares.ac.ir; Fax: +98-21-82884931; Tel: +98-21-82884917
bModares Environmental Research Institute, Tarbiat Modares University, Tehran, Iran
cCSELS, Faculty of Health & Life Sciences, Coventry University, Coventry, UK
First published on 7th August 2023
Abstract
A serious environmental problem is associated with the accumulation of solid waste on the Earth. Researchers are encouraged to find an efficient and sustainable method to recover highly profitable heavy metals and precious and base metals. Bioleaching is a green method of recovering valuable metals from solid waste. Optimizing the variables and conditions of the bioleaching process is crucial to achieving maximum metal recovery most cost-effectively. The conventional optimization method (one factor at a time) is well-studied. However, it has some drawbacks, such as the necessity of more experiments, the need to spend more time, and the inability to illuminate the synergistic effect of the variables. Optimization studies are increasingly utilizing response surface methodology (RSM) because it provides details about the interaction effects of variables with fewer experiments. This review discusses the application of RSM for bioleaching experiments from other solid wastes. It discusses the Central Composite and Box–Behnken designs as the most commonly used designs for optimizing bioleaching methods. The most influential factors for increasing the heavy metal recovery rate in applying RSM using the bioleaching process are recognized, and some suggestions are made for future research.
1. Introduction
A glance at the ever-growing industries and technologies reveals that waste containing heavy metals and organic compounds in the environment is increasing. These hazardous wastes may cause irreparable damage to humans and the environment.1 Heavy metals, in general, are metal elements with a density five times greater than water. Waste containing toxic metals usually contains arsenic as a metalloid, which is toxic even at low concentrations. In recent years, environmental contamination caused by these metals has raised concerns about public health and the environment. Although heavy metals are typically located in the Earth's crust, the industrial, domestic, and agricultural uses of these metals and alloys, as well as the recovery of these metals from mines, has led to a surge in the concentration of these metals in water bodies, soil, and air.2 Precious metals such as silver, gold, and platinum (PGMs), usually found in many wastes, are known for their high conductivity and chemical stability. Thus, the urge to recover these rare and precious metallic elements from solid waste has become more critical economically to the public health issues of heavy metals.3The traditional methods of metal recovery include pyrometallurgy and hydrometallurgy. Pyrometallurgical processes rely on thermal methods for metals to be recovered from wastes. Most thermal methods are prohibitive due to high energy consumption and hazardous gases (dioxin and furans) that result in environmental pollution. Hydrometallurgical processes use environmentally hazardous chemicals for metals leaching and generate a large amount of by-products, which requires further processing for disposal.4
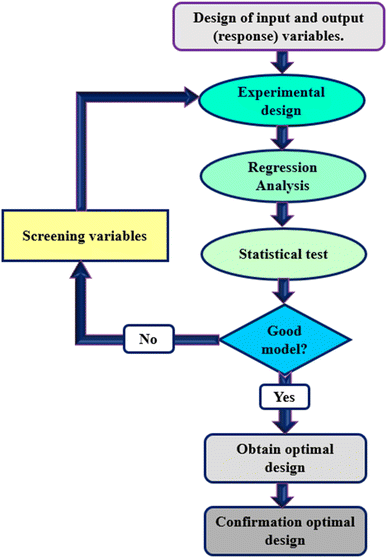
Many human diseases, including some forms of cancer, besides psychological and neurological disorders, behavioral disorders, organ disorders, and immunodeficiency, are linked to environmental toxicity exposure. Therefore, the control of these wastes is of significant importance. In addition, using environmentally friendly biotechnology approaches results in lower costs and higher efficiency, mainly when microorganisms are used to decrease the toxicity of these wastes.5 In the nineteenth century, bioleaching was known as a method for metal extraction from mines and ores. The bioleaching process (Fig. 1) is eco-friendly, with low energy consumption, low cost, and high efficiency and, therefore, the method of choice for heavy metals recovery. As a result, numerous major international companies are increasingly shifting to bioleaching as their preferred alternative to conventional methods.6
Bio-hydrometallurgy or bioleaching processes use the ability of a wide range of microorganisms, namely autotrophic and heterotrophic bacteria, and fungi, to remove the metal ions from solid wastes and transfer them into a solution.7 In this process, microorganisms can remove metal ions in two mechanisms, indirect and direct bioleaching. In the indirect mechanism, the bacteria have no physical contact with solid waste surfaces and will only indirectly dissolve solid waste through metabolite production. In the contact mechanism, bacteria can participate directly in the process. Thus, metals become soluble due to an electrochemical reaction.4 Autotrophic bacteria's growth depends on organic carbon as a carbon source. Instead, the carbon fixation from atmospheric CO2† promotes their growth. At the same time, O2 is a terminal electron acceptor for the respiratory chain in the metabolism of these bacteria, as illustrated in eqn (1).8
| O2 + 4H+ + 4e → 2H2O | (1) |
The most common species of autotrophic bacteria used in metal recovery from various wastes such as e-waste and industrial waste are Gram-negative bacteria Acidithiobacillus spp., which grow aerobically and include: Acidithiobacillus prosperus, Acidithiobacillus caldus, Acidithiobacillus thiooxidans (A. thiooxidans), Acidithiobacillus ferrooxidans (A. ferrooxidans), Leptosprillum ferrooxidans (L. ferrooxidans), Acidithiobacillus concretivorus, and Acidithiobacillus albertis.9 A. thiooxidans and A. ferrooxidans are exemplar species compatible with acidic medium (pH = 1–3), whereas other species from this family grow at higher pH. A. ferrooxidans as iron-oxidizing bacteria (IOB) and A. thiooxidans as sulfur-oxidizing bacteria (SOB) are prominent among bacterial species used for the recovery of metals.10
Heterotrophic bacteria and fungi require organic materials as the source of carbon. The role of these organic carbon sources is fundamental for these microorganisms' metabolisms, including organic acids (malic acid, oxalic acid, nitric acid, citric acid), which are secreted in the culture medium. The metals extraction from a solid matrix has been demonstrated with these microorganisms, often with their acidic and chelating features. Furthermore, because of the protein catabolism in these heterotrophic microorganisms, non-acidic complexes are also being produced, resulting in leaching systems of alkaline type.11,12 Several fungi such as Aspergillus niger (A. niger), Penicillium simplicissimum, Penicillium chrysogenum, and bacteria such as Gluconobacter oxydansseudomonas strains, Bacillus strains, and Chromobacterium violaceum have been used for recovering metals from various wastes.13
The bioleaching mechanism in fungi is linked with the production of the metabolites such as organic acids, which have a low molecular weight. Fungal bioleaching includes the following mechanisms: acidolysis, complexolysis, and redoxolysis. In the acidolysis mechanism, protons from producing organic acids solubilize metals, and the protonation of oxygen atoms coats the solid waste's surface. This mechanism is similar to acid leaching. Complexolysis mechanism, the organic acid-metal complex is produced by organic acid's carboxyl and hydroxyl group.14 In addition to fungi, cyanogenic bacteria have been shown to use this mechanism to recover valuable metals. Pseudomonas strains, Bacillus megaterium (B. megaterium), and Chromobacterium violaceum belong to this group of bacteria. Cyanogenic bacteria secrete cyanide as a secondary metabolite in their medium. They produce cyanide using HCN synthase and decarboxylated glycine at the end of the growth logarithmic phase of cyanogenic bacteria.42,43 As illustrated in eqn (2), cyanide can be secreted in two forms, including the hydrocyanic acid (HCN) and the cyanide anion (CN−), in a reversible reaction in equilibrium. Noteworthy is that due to the pKa of HCN, metal-cyanide complexes' generation occurs at high pH values.44
| HCN ↔ H+ + CN−, PK0a(HCN) = 9.21 ± 0.02 at 25 °C | (2) |
In the redoxolysis mechanism, metals mobilization from the solids waste results from oxidation–reduction reactions. This mechanism supplies the energy required for microbial growth by electron transfer.14 Table 1 shows the metals extracted from various solid wastes by different microorganisms.
| Microorganism | Solid wastes | Target metals | References |
|---|---|---|---|
| Autotrophic | |||
| A. thiooxidans | TPCBs | Cu and Au | 15 |
| LEDs | Cu, Ni, and Ga | 16 | |
| Tannery sludge | Cr | 17 | |
| Carbide slag | Zn, Ba, Ni and Li | 18 | |
| Refinery spent catalyst | Ni, V, Mo, and Al | 19 | |
| SCCs | Li, Co, Mn | 20 | |
| MPPCBs | Ni and Cd | 21 | |
| A. ferooxicidans | Mine tailings | Te | 22 |
| PCBs | Cu, Ni, and Fe | 23 | |
| PCBs | Cu and Ni | 24 | |
| WLED | Cu, Ni, and Ga | 25 | |
| SCCs | Li, Co, Mn | 26 | |
| LED | Cu, Ni, and Ga | 27 | |
| PCBs | Cu | 28 | |
| Low-grade ore | Cu | 29 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Hetrotrophic (fungi) | |||
| P. citrinum | LIBs | Li and Mn | 30–32 |
| Aspergillus niger | Zinc plant purification residue | Zn, Co, and Mn | 33 |
| Aspergillus niger | LCD | In | 34 |
| A. niger, Pseudomonas putida, Pseudomonas koreensis and P. bilaji | Iron rich laterite ore | Co and Ni | 35 |
| Aspergillus niger | Phosphorites | U, Sm, Th, and La | 36 |
| Aspergillus niger | WPCBs | Ni, Cu, and Zn | 37 |
| A. niger, P. simplicissimum | Vanadium-rich power plant residual ash | Ni, V | 38 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Hetrotrophic (bacteria) | |||
| Bacillus foraminis | AMOLED displays | Ag, Mo, and Cu | 39 |
| KB3B1 strain | Pyrolusite | Mn | 40 |
| Bacillus megaterium | Sulfide concentrate | Ni and Co | 41 |
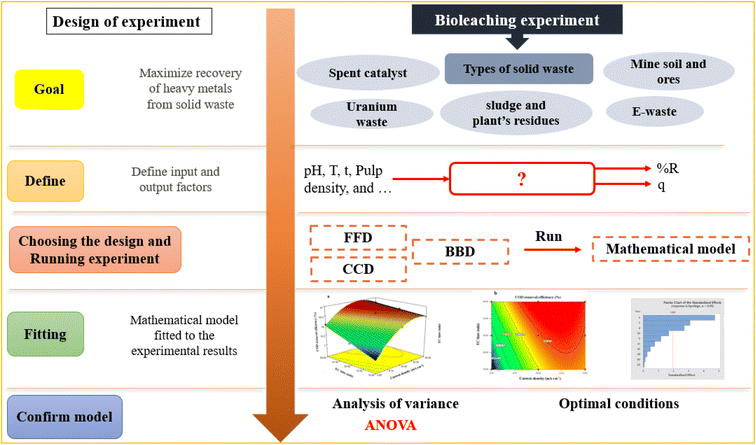
The percentage of metal bio-recovery is affected by different variables, including the substrates and their respective concentrations, O2 and CO2 concentrations, pH, temperature, inoculum density, the waste's particle size, solid-to-liquid ratio, bioleaching duration, and shaking speed.45 Investigating all the factors involved in the process requires many experiments, which is costly and time-consuming.46 In addition to each factor, their interaction highly affects the bioleaching rate, so even minor changes can be crucial for metal dissolution. Therefore, one of the most crucial challenges in using the bioleaching process to extract metals from solid wastes is the proper selection of experimental conditions obtained under the more widespread concept called test design of experiment (DOE).47 DOE is a systematic method to define the relationships between factors affecting a process and the responses. Since its development by Ronald Fisher in 1920, DOE has been increasingly used due to its numerous success in applications in increasing process efficiency and sustainability, lowering production costs, and improving our understanding of the input and output connection in the process.48
Without DOE, the experiments are carried out by a conventional one-factor-at-a-time optimization method. One factor is being analyzed in this procedure, while the others are kept constant. However, this approach is inefficient and misses possible interactions.49 DOE method is rapid, reliable, and identifies the interaction between parameters, although it reduces the total number of experiments, resulting in less material consumption and considerably lower laboratory work. Therefore, DOE aims to plan and execute experiments that may provide much information from the collected data in the smallest number of experimental runs.50 Using the DOE, a process can be modeled and optimized, so DOE is being used increasingly to evaluate the optimum response of heavy metals' recovery rates in some studies on solid waste bioleaching.51 In the past two decades, among many multivariate DOE-based designs, response surface methodology (RSM) has drawn extensive attention, especially in the design, optimization, and modeling of the bioleaching processes, but with very few thorough reviews available on applications of RSM in bioleaching processes. Therefore, in this paper, for the first time to the authors' knowledge, the fundamentals of RSM are examined, focusing on the applicability of this technique for the high recovery of heavy metals from various solid wastes.
2. RSM
Response Surface Methodology (RSM) is a method to optimize complex processes by applying multivariate approaches. RSM is a cost-effective and labor-saving method compared to other optimization methods, as it requires fewer experiments to assess multiple factors and their interactions. It is broadly applied in the biology and chemistry fields and the food industry and other areas.52 Using mathematical and statistical techniques, RSM analyses the relationship between multiple variables to obtain an optimal result when modeling and solving problems. In general, RSM is based on fitting the mathematical models (linear, square polynomial functions, and others) to the experimental results from the designed set of experiments and verifying the model obtained by the statistical methods.53 This methodology is advantageous to simultaneously examine several parameters at different levels by using a limited sequence of designed experiments and revealing the interactions between these parameters. Over the last decades, RSM was identified as the most effective and standard method for designing experiments among the various multivariate processes; this method focuses mainly on designing, modeling, and optimizing environmental and chemical investigations. Fig. 2 illustrates the total strategy in RSM.There are two types of significant designs in RSM, central composite design (CCD) and Box–Behnken design (BBD), which are applied to investigate the process variables at five and three levels, respectively.54 CCD and BBD are successful, broadly used, and reported in this literature for optimizing bioleaching processes. There are six main successive steps which are involved in the optimization and simulation of the bioleaching process: (1) the selection of independent factors which have a significant effect on the responses of the system through screening, (2) the choice of optimal experimental design, (3) running the experiments and obtaining the results, (4) model fitting to experimental data, (5) model confirmation using Analysis of Variance (ANOVA) and graphs, (6) determination of optimal conditions. Prior to explaining RSM and its application in the bioleaching process, it is better to explain the essential and common terms in this method. The following table explains these terms (Table 2).
| Main terms | Definition |
|---|---|
| Experimental design | A set of polynomial models which responses function can be approximated |
| Factors or independent variables | Experimental variables, namely pH, temperature, pulp density… that can change independently |
| Levels of a variable | Values of a variables, that is pH can be studied in different range (1–11) |
| Responses or dependent variables | Measure values from the experiment, such as heavy metals recovery rate in bioleaching process |
2.1. Screening of the independent variables
Screening the variables is the most crucial step in the bioleaching process that affects the recovery of metals. Therefore, it is required to select those variables with significant effects. Preliminary screening experiments, literature review, and consulting industries would give the researcher ideas to choose the influential factors. A screening design should determine which variables have a prominent interactive effect on the responses. If the factors and their domains are not selected correctly, the final results of this method will be unreliable. The metal removal using the bioleaching process is mainly affected by several environmental variables, namely initial pH, pulp density, bacteria growth temperature, substrate concentration, etc. Based on the selected solid waste and microorganisms, these factors could differ.2.2. Optimal experimental design
In the next step to evaluate the responses, the design of the experiment is accompanied by the selection of the points. Choosing an appropriate design method significantly impacts the creation of a response surface and the accuracy of the predicted model. RSM relies on the assertion that a response function can be approximated by using a polynomial model. Among the vast range of models used in the literature, first and second-order models are more frequently applied. The first-order model eqn (3) is provided as follows:
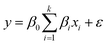 | (3) |
The first-order model comprises several elements: k signifies the number of variables, β0 denotes the constant term, βi represents the coefficients of the linear parameters, xi corresponds to the variables, and ε indicates the residual linked to the experiments. Consequently, the first-order model is unable to predict any curvature. To overcome this limitation, the second-order model is recommended. Two-level factorial designs are suitable for approximating first-order effects; however, they cannot predict higher-order effects. Therefore, a central point is used to evaluate curvature. To present the interaction between experimental variables, a second-order model eqn (4) should contain additional terms, as illustrated below:
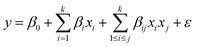 | (4) |
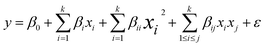 | (5) |
Also noteworthy is that the full quadratic polynomial eqn (6) is among the most used models to approximate the response. Ordinary least squares and second-order symmetrical designs are generally applied to estimate the coefficients.55,56
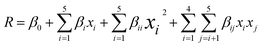 | (6) |
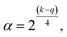 (3) all factors are studied in 5 levels (−α, −1, 0, +1, +α).55,56
(3) all factors are studied in 5 levels (−α, −1, 0, +1, +α).55,56Fig. 3(a) illustrates a CCD for two and three-variable optimizations. The factorial points are circular orbits, forming a circle and a sphere for two and three-variable optimizations. Additionally, for α > k1/2 the axial points are further from the center than the factorial points.
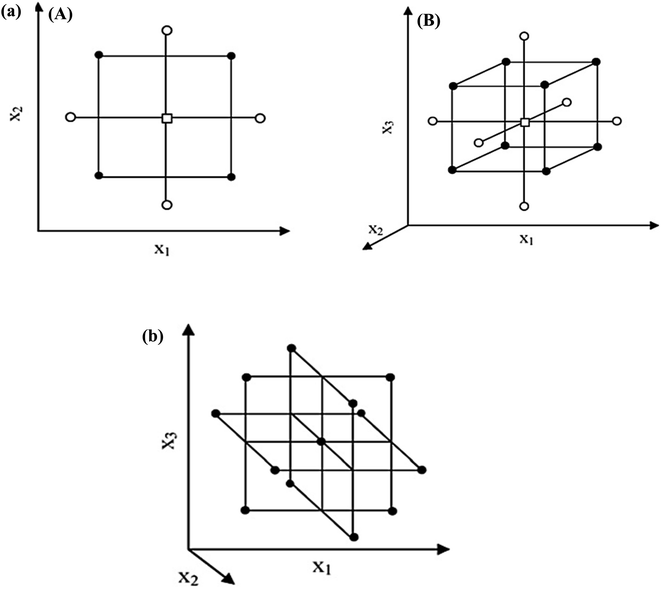 | ||
| Fig. 3 (a) CCD for the optimization of: (A) two variables (α = 1.41) and (B) three variables (α = 1.68), (b) BBD for the optimization of three variables.60 | ||
2.3. Running the experiments and obtaining the results
After determining the design method and establishing the values for the experiment design, the next step is to organize a set of experiments, especially those ordered using computer software. Then the obtained results would be analyzed in a mathematical model. In order to verify the entire procedure, this mathematical method must be fitted to the experimental results.2.4. Model fitting to experimental data
Two steps of codification and regression are essential to fit the selected mathematical model to experimental data. The first step includes transforming accurate obtained values to scaled dimensionless quantities because RSM mathematical models operate on coded inputs (−1, 0, +1) instead of actual values of factors. The weight of this step is due to equalizing the influence of each variable, making it possible to investigate values with different orders of magnitude. First, eqn (7) is used to convert actual values (zi) into coded values (xi).
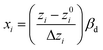 | (7) |
The second step, the regression step, using the coefficient of determination (R2), is a routine method for confirming an obtained model. However, the noteworthy fact is that a high R2 is not necessarily interpreted as a fit model. R2 is not a reliable value to indicate the bias of coefficient estimates. Moreover, it does not suggest the adequacy of a regression model. Hence, understanding the subject area associated with the residual plots improves the R2 value evaluation. It is also to be noted that the more the number of factors, the higher the R2 value falsely produced. This can occur during an over-fitting procedure, resulting from incorrectly incorporating random noise in the model's data.
The common strategy to overcome such obstacles and make reliable predictions is to report adjusted R2 and predicted R2 alongside the R2 and residual plots. Adjusted R2 compares the explanatory power of regression models that contain different numbers of factors, and it only increases if the new term improves the model more than it would be expected by chance. The new data shows a regression model ability of response prediction predicted R2. It is also known for preventing researchers from over-fitting a model since predicting random noise is impossible. Predicted R2 and adjusted R2 are of lesser values than the R2, although too many factors in a model can result in a wide gap between predicted R2 and R2. In conclusion, during the validation of a regression model, it is paramount to report the acquired R2, adjusted R2, and predicted R2.60,61
2.5. Model confirmation using ANOVA and graphs
Pertinent software should be used to obtain satisfactory results and responses in the study process. Sometimes the mathematical models obtained in step 4 do not describe the experimental domain studied, so it is necessary to use appropriate software that obtains the answer as three-dimensional surface and contour plots. These software outputs can help the researcher approximate the optimal response and conditions.ANOVA is a statistical model able to investigate the difference between groups. This method was invented by renowned biologist and statistician R. A. Fisher in 1925. His famous book “Statistical Methods for Research Workers” explored variance separation and helped form many statistical hypotheses. The basis of all of these methods was to divide the variance or data into several components. Nowadays, the use of ANOVA with this goal is hindering. In the simplest form, ANOVA can test the hypothesis of mean comparisons among several independent populations.
Another definition for ANOVA would be a set of mathematical functions and statistical methods employed to identify significant parameters in models with multiple parameters. With the help of ANOVA, we can identify the critical factors in the experiment and determine the model's accuracy.
To ensure that our model is suitable and consistent with the experimental results, it is essential to investigate some parameters and pathways. The F-tests and its “F-statistic” test statistic were named in honor of Ronald Fisher. The statistic F is the ratio of two variances. Variance is an index for measuring the dispersion rate, which shows how much data is dispersed from the mean. Larger values of variance indicate more excellent dispersion. The square of standard deviation is the variance. Using standard deviation rather than the variance in many sciences is more common because it coincides with the measured data. However, it shows the variance of payment in square units of measured data. However, in many real analyzes, the variance is used to perform calculations.62 The F-statistic is based on the ratio of the mean squares. While the term “mean squares” may be confusing, it is simply an approximation of population variance in which degrees of freedom (DOF) are used to calculate and estimate. However, by changing the variance containing the ratio, the F-test becomes very flexible. The F-test can be used in various situations with a variance ratio. Not surprisingly, the F-test can also assess variance equality.
The P-value is also an important parameter calculated based on the F-value. A p-value of less than 0.05 makes it statistically regarded as significant for each factor, and if it is larger, it is not statistically significant. The value of 0.05 is the default preset of p-value in most software. Suppose a parameter is not statistically significant but should be included in our model. In that case, the confidence level must be changed so as the confidence level decreases, our permissible p-value increases. The confidence level can be reduced to some extent, but this depends on the type of process. For instance, it may be acceptable to set the confidence level at 80% in one process but not in another. Usually, the confidence level can be reduced more easily in influential factors and controllable errors.57,60
2.6. Determination of optimal conditions
In the optimization technique, control variables in any method are adjusted to determine the proper factor levels, which lead to the most favorable results.63 Finding the optimum location of the model is easier with a graphical representation. It may be helpful to compare two types of graphs: the three-dimensional response surface (Fig. 4(a)) and the contours graph (Fig. 4(b)), which represents the surface projected onto a plane. Each contour determines the surface height. Responses in these graphs are expressed as functions of two factors. A simple visual inspection of the graph can determine whether the optimum value corresponds to a maximum or a minimum following the established optimization criterion.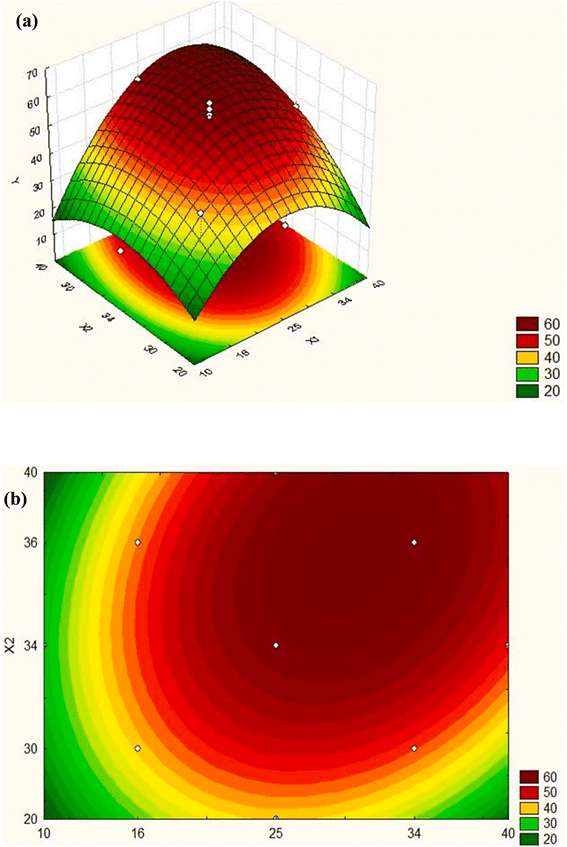 | ||
| Fig. 4 Response surface in the (a) three-dimensional space, (b) graph of contours. Y: response, X1 and X2: factors.57 | ||
For example, in a bioleaching process, optimizing factors (initial pH, waste concentration, sulfur concentration, and Fe2+ concentration) increase the bioleaching rate and enhance metals recovery.64 Setting each factor at its optimum value will be better with the software-predicted maximum value of metal removal efficiency, which is the main objective of a bioleaching process. Verifying RSM-suggested optimal conditions with statistical analysis and running confirmatory tests is paramount. This is achieved by comparing the modeled predictions with the real outcomes of experiments at optimal conditions. The developed model is cogent and insensitive to noises springing from factor-level alterations only if the experiment outcome complies with the predicted regression and ANOVA data values.
2.7. RSM limitations
RSM, as a tool, has been generally used for modeling and optimization of the bioleaching process. Although this method has many affirmative features, it has two significant limitations. First, the obtained model is accurate only within the experimental range, and extrapolation is not applicable. This issue stems from the fact that the standardized equations employed by RSM lack any methodological sense. RSM's first or significant limitation is that it is a ‘black box’ approach. In other words, the precision of the approximation cannot be estimated readily, or to say that estimating the size and order of the approximation errors is arduous. The second limitation is that RSM is a local analysis. The developed response surface is invalid for regions other than the studied ranges of factors.65 Fig. 5 briefly shows the steps for optimizing the bioleaching process of various solid wastes using RSM.3. Application of RSM in bioleaching processes
Bioleaching has the potential to recycle many types of solid wastes. This review presents the bioleaching process of different kinds of solid wastes that incorporate RSM with DOE tools for improving heavy metals removal efficiency in separate sections, including biorecovery from soils and ores, e-waste, spent catalysts, and sludge and plant's residues.3.1. Biorecovery from soil and ores
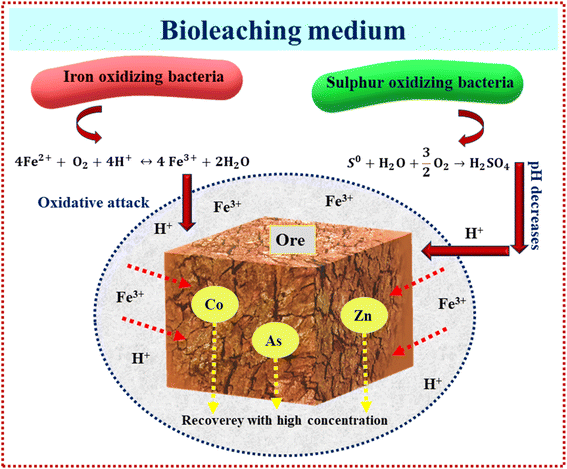
Bioleaching from ores and soil using RSM has been copiously studied in the literature as a green option preferred to traditional chemical leaching processes, as presented in Table 3. As shown in this table, most microorganisms are acidophilic species for the bioleaching of different ores, generating Fe3+ and H2SO4, hydrolysis of ferric iron in bioleaching medium releases protons, which together with H2SO4 contributes to suitable acid demand (eqn (8)–(12)).66 Fig. 6 illustrates the mechanism of acidophilic bacteria and their metabolites. Therefore, one of the main factors for these species is pH. Among 9 articles, 6 articles considered pH as a variable for optimization. According to Table 3, the optimum pH value is 2 or less, which leads to the proper oxidation of iron or sulfide ions and enhances the activities of microbes in the bioleaching process of aimed metals.66 In fungal species, carbon sources such as sucrose and glucose are the main factors affecting organic acid production and lead to better recovery of heavy metals;67 therefore, with optimization of this factor, metal's recovery as a response reaches its optimum rate.
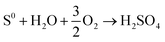 | (8) |
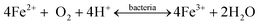 | (9) |
| Fe3+ + H2O → Fe(OH)2+ + H+ | (10) |
| Fe3+ + 2H2O → Fe(OH)2+ + 2H+ | (11) |
| Fe3+ + 3H2O → Fe(OH)3 + 3H+ | (12) |
| Microorganism | Type of waste | RSM | Optimization | Reference | ||||
|---|---|---|---|---|---|---|---|---|
| Method | Factors | Response(s) | Optimum condition | Optimum response | ||||
| Acidophilic bacterial | A. ferrooxidans, A. thiooxidans | Uranium-ore | CCD | pH, pulp density, initial Fe2+ | U% | pH = 1.58, pulp density = 10.11%, initial Fe2+ = 3.67 g L−1 | 93.71% | 73 |
| A. ferrooxidans, A. thiooxidans | Low-grade copper–molybdenum ore | Tween-20, initial Fe2+ | Cu%, Mo% | Tween-20 = 0.13 g L−1, initial Fe2+ = 6.75 g L−1 | 25%, 24% | 74 | ||
| A. ferrooxidans | Uranium-ore | CCD | Initial pH, aeration rates, inoculum, initial Fe2+ | U% | Initial pH = 1.45, aeration rate = 420 mL min−1, inoculum = 6% (v/v), initial Fe2 = 2.89 g L−1 | 92% | 75 | |
| A. ferrooxidans, A. thiooxidans | Low-grade sulfide ore | CCD | pH, pulp density, aeration rate | Cu% | pH = 1.5, pulp density = 10%, aeration rate = 140 rpm | 69.91% | 76 | |
| A. ferroxidans BYQ-12 | Traditionally purified realgar | BBD | Initial pH, Fe3+ conc., pulp density | As% | pH = 1.74, Fe3+ = 3.68 g L−1, pulp density = 0.95% (w/v) | 73.97% | 77 | |
| A. ferrooxidans | Sphalerite (zinc-ore) | CCD | pH, pulp density, initial Fe2+ conc., temp. | Zn conc. | pH = 1.94, pulp density = 4% w/v, initial Fe2+ conc. = 7.3 g L−1, temp. = 33.7 °C | 7.3 g L−1 | 78 | |
| A. ferrooxidans, A. thiooxidans, L. ferrooxidans | Molybdenite (molybdenum-ore) | CCD | Pulp density, pyrite, Ag ion conc. | Cu%, Mo% | Pulp density = 3%, pyrite = 5%, Ag ion conc. = 100 mg L−1 | 97%, 8.9% | 79 | |
| A. ferrooxidans, A. thiooxidans, L. ferrooxidans | Copper mine | CCD | pH, solid conc., inoculum | Cu%, Mo%, Re% | pH = 1.68, solid conc. = 0.95%, inoculum = 18.41% (v/v) | 64.72%, 2.76%, 31.3% | 80 | |
| M. cuprina Ar-4, M, prunae DSM10039, A. brierleyi DSM1561 | Chalcopyrite (Copper-ore) | BBD | Temp., pH, inoculum biomass | Cu conc. | Temp. = 66.1 °C, pH = 2, inoculum = 1.22 × 107 cells per mL | 3.93 g L−1 | 81 | |
| A. ferrooxidans | Uranium-ore | BBD | Particle size, aeration rates, irrigation rates | U% | Particle size = 5 mm, irrigation = 0.34 L m−2 min−1, aeration = 210 L m−2 min−1 | 63.85% | 82 | |
| A. ferrooxidans | Uranium-ore | BBD | Pulp density, aeration rate, agitation speed | U% | Pulp density = 5.8% w/v, aeration rate = 250 L h−1, agitation speed = 510 rpm | 95% | 83 | |
| Acidithiobacillus sp. | Uranium-ore | BBD | pH, temp., agitation rate inoculum, FeSO4·7H2O, (NH4)2SO4 | U% | pH = 1.96, temp. = 30.90 °C, agitation rate = 158 rpm, inoculum = 15.7%, FeSO4·7H2O = 13.83 g L−1, (NH4)2SO4 = 3.22 g L−1 | 83% | 84 | |
| A. ferrooxidans | Uranium-ore | CCD | Time, pH, pulp density | U% | Time = 48 h, pH = 2.0, pulp density = 5% | 100% | 85 | |
| Fungi | A. niger NCIM 548 | Lateritic chromite overburden | BBD | Temp., sucrose conc., fermentation days | Ni%, Co% | Temp. = 80 °C, sucrose conc. = 10%, fermentation days = 21 | 70.49%, 66.93% | 86 |
| A. niger | Uranium-ore | BBD | Initial pH, spore population sucrose conc. | V% | Initial pH = 3, spore population = 3 × 106 cells per mL, sucrose 1 conc. = 00 g L−1 | 44.8% | 87 | |
| Fungi and bacterial | Bacterial and fungal populations in the panchakavya | Mine soil | BBD | Time, panchakavya conc., agitation rate | Cu%, Pb% | Time = 54 h, panchakavya conc. = 10 mL, agitation rate = 120 rpm | 49%, 64% | 88 |
| Other species | Mesophilic bacteria | Low grade ore | CCD | Pulp density, pH, initial conc. of Fe2 | Cr% | Pulp density = 1.59%, pH = 2, initial conc. of Fe2 = 0 | 85.98% | 89 |
| Herbasprillum sp. GW103 | Mine soil | BBD | Cu% | Temp. = 30 °C, time = 60 h, CaCO3 = 1.75%, coconut oil cake = 3%, agitation rate = 140 rpm | 66% | 90 | ||
For instance, Sun et al. (2022) studied maximizing Ni, Cu, and Co leaching efficiency and minimizing the extraction of Mg and Fe ions from high-magnesium nickel sulfide ore using Acidithiobacillus ferrooxidans, Ferrimicrobium acidiphilum, and Leptospirillum ferrooxidans. Optimizing the particle size, acid addition, pulp density, and inoculation resulted in a recovery of 89.43% Ni, 36.78% Cu, 84.07% Co, 49.19% Mg, and 0.20 g L−1 of Fe was reached using CCD of RSM. Furthermore, the results showed that particle size is the most significant variable affecting Ni, Cu, and Co leaching efficiency. At the same time, acid addition is the most significant variable affecting Mg leaching efficiency.68
Tang et al. (2021) examined Acidithiobacillus caldus's potential for biodesulfurization of sulfide ore using a BBD-based RSM. The six studied parameters included temperature, particle size, shaking rate, inoculation, pH, and Tween80 concentration. At first, the Plackett–Burman screening design was used to identify which factors would be optimized to achieve the most effective result. As a result, particle size, shaking speed, and inoculation were selected as significant factors. Under optimum conditions of 120–140 mesh of particle size, 170 rpm of shaking speed, and 111 mL of inoculation size, desulfurization efficiency rose 8.1% over 5 days.69
Mo et al. (2019) investigated the effect of four significant parameters on U biorecovery: pH value, initial Fe2+, solid–liquid ratio, and inoculation percentage. CJ6-0, a consortium of microbial strains, was utilized consisting of Acidithiobacillus sp. (58.3%), Acidiphilium sp. (12.4%), Leptospirillum sp. (10.53%), and Ferrimicrobium sp. (9.15%). The interaction among selected variables was analyzed using the BBD, suggesting that the maximum U extracted was in the optimal conditions at 91.4%. Confirmation tests also revealed a 90.2% recovery, which was highly compliant with the predicted value.70 During the same year, column bioleaching of low-grade U-ore in lab-scale was also investigated by Jalali et al. (2019) using Acidithiobacillus Ferridurans SBU-SH2 KY497231, a newly isolated strain from a sulfur hot spring in Iran. The BBD is cheaper than CCD because it offers fewer design points with many factors. Experiments on samples provided from U-mines in Iran and the BBD design of RSM resulted in a maximum U extraction of 95.5%, considerably higher than the control solution without bacterial cells (11.33%). The optimization of conditions was limited to more effective parameters, namely pH, particle size, temperature, aeration, and irrigation rate. At the same time, a kinetic model was also offered for the rate of U recovery rate.71
Selvi and Aruliah (2018) evaluated the potential of an indigenous acidophilic bacterium characterized as Serratia marcescens SMAR1 towards the Zn removal from Zn spiked soil using the approach of bioleaching enhanced electrokinetic remediation (BEER) technology. It is known that electrokinetic remediation boasts high efficiency, low cost, and non-pollutant methods. Statistical optimization of Zn remediation process parameters, namely pH, contact time, and inoculum dosage, was investigated using RSM BBD. A maximum Zn reduction of 93.8% was achieved in the BEER process, compared to plain bioleaching (72.86%).72
3.2. Biorecovery from e-wastes
The bioleaching process has been broadly used in heavy metals removal from e-waste. Since many factors such as pH, substrate concentration, time, temperature, stirring speed, inoculum, pulp density, and agitation rate could affect the efficiency of the metals recovery rate, RSM can help optimize this process and maximize metals recovery. Table 4 summarizes some of the research implemented on e-waste using RSM.| Type of waste | Microorganism | RSM | Optimization | Reference | |||
|---|---|---|---|---|---|---|---|
| Method | Factors | Response(s) | Optimum condition | Optimum response | |||
| PCBs | A. thiooxidans | CCD | Biochar conc., pulp density | Cu%, Ni% | Biochar conc. = 1.6 g L−1, pulp density = 16 g L−1 | 36%, 64% | 24 |
| Waste computer motherboards | A. ferrooxidans, F. acidiphilum, L. ferriphilum, Thermoplasma acidophilum | CCD | Pulp density, initial pH, humic acid | Cu% | Pulp density = 1.35% (w/v), initial pH = 1.53, humic acid = 0.31 g L−1 | 100 | 97 |
| PCBs | A. ferrooxidans | BBD | Contact time, pulp density, particle size, temp. | Cu% | Contact time = 21 d, pulp density = 12 g L−1, particle size = 0.25–0.42 mm, temp. = 20 °C | 96.6% | 98 |
| LCD | A. thiooxidans | CCD | pH, sulfur conc., pulp density | In%, Sr% | pH = 2.6, sulfur conc. = 1.6% (w/v), pulp density = 8.6 g L−1 | 100%, 10% | 99 |
| PWBs | A. ferrooxidans, A. Thiooxidans | CCD | pH, sulfur conc., pulp density, initial FeSO4 conc. | Cu%, Zn%, Ni% | pH = 1.52, sulfur conc. = 6.75 g L−1, pulp density = 25 g L−1, initial FeSO4 conc. = 20 g L−1 | 94%, 92%, 96% | 100 |
| MPPCBs | B. megatherium | CCD | pH, pulp density, glycine conc | Au%, Cu% | pH = 10, pulp density = 9.13 g L−1, glycine conc. = 10 g L−1 | 72%, 65 g per tone MPPCBs | 101 |
| Zn–Mn batteries | A. ferrooxidans | CCD | pH, substrates conc., pulp density, temp. | Zn%, Mn% | pH = 1.9, substrate conc. = 28 g L−1, pulp density = 9.7%, temp. = 33 °C | 52% | 102 |
| A. Thiooxidans | pH = 1.8, substrate conc. = 29 g L−1, pulp density = 8%, temp. = 36.7 °C | 52.4% | |||||
| CPCBs | B. megatherium | CCD | pH, pulp density, particle mesh#, glycine conc | Au%, Cu% | pH = 10, pulp density = 2 g L−1, particle mesh# = 100, glycine conc. = 0.5 g L−1 | 36.81%, 13.26% | 103 |
| Ni–Cd and Ni–MH batteries | A. ferrooxidans | BBD | pH, initial Fe2+ conc. Powder size | Ni%, Cd%, Co% | pH = 1, initial Fe2+ conc. = 9.7 g L−1, powder size = 62 μm | 87%, 67%, 93.7% | 104 |
| PCBs | A. thiooxidans, A. ferrooxidans | CCD | pH, FeSO4·7H2O conc., sulfur conc. | Cu% | pH = 1.56, FeSO4·7H2O conc. = 16.88 g L−1, sulfur conc. = 5.44 g L−1 | 92.6% | 105 |
Nowadays, many electronic devices use printed circuit boards (PCBs). Hence, Trivedi et al. (2023) implemented the BBD of RSM for efficient enzymatic metal bioleaching from discarded cellphone PCBs using Aspergillus niger. They employed 27 runs to screen the factors that resulted in selecting glucose oxidase (GOx) concentration, Fe2+ concentration, pulp density, and shaking speed as influential factors. Then, they performed the BBD approach to optimize certain factors to maximize the extraction efficiency of Cu, Ni, Pb, and Zn. The optimal GOx concentration, Fe2+ concentration, pulp density, and shaking speed were 300 U L−1, 10 mM, 1 g L−1, and 335 rpm, respectively. At the suggested conditions, the extraction yield reached 100% Cu, 70% Ni, 40% Pb, and 100% Zn.91
Apart from PCBs, smartphone touch screens (SPTS) contain a significant amount of valuable metals, which can be used as secondary resources. In 2021, Pourhossein et al., used RSM to remove indium and strontium from organic light emitting diode type SPTS by adapting acidithiobacillus ferrooxidans. Designing the process for the CCD method, the authors considered ferrous sulfate concentration, pulp density, elemental sulfur, and initial pH as the most influential factors in maximizing the In and Sr recovery rates. RSM was used to optimize each parameter value for maximizing the In and Sr biorecovery processes. At optimal conditions of ferrous sulfate: 13.0 g L−1, pulp density 3 g L−1, initial sulfur concentration 5.6 (g L−1), and initial pH 1.1, In was recovered completely, but Sr was not appropriately recovered (only 5%).92
In another investigation, Arshadi et al. (2020) studied the RSM-optimized extraction of Cu and Ni from disposed of computer-printed circuit boards (CPCBs) utilizing Aspergillus niger. Optimization for maximizing the percentage of Cu and Ni recovery was done with pH of 5.15, pulp density of 10 g L−1, 1 × 107 spores of fungi, and 4.5 days for the time the powder was added, resulting in 97% of Cu and 74% of Ni. There was a claim that the time (day) the sample is added to the solution is essential to Aspergillus niger's ability to recover metals.93
Kumar et al. (2018) investigated the optimization of Au and Ag recovery using Pseudomonas balearica SAE1 from the computer-printed circuit boards (CPCB). To maximize Au and Ag recovery as the responses, they used Design-Expert Software to apply the CCD method with four factors: pulp density, temperature, initial pH, and glycine (as an additive) concentration. The optimal condition was found to be as follows: initial pH of 8.6, a temperature of 31.2 °C, pulp density of 5 g L−1, and glycine concentration of 6.8 g L−1. These values recovered 73.9% of Au and 41.6% of Ag. They also found that glycine concentration and pulp density noticeably influence the biorecovery rates of Au and Ag.94
Most portable devices enjoy a rechargeable battery, conventionally made using lithium. Over recent decades, the disposal of ever-increasing spent lithium-ion batteries (LIBs) integrated into laptops has raised serious environmental concerns. Researchers have recently focused on developing cost-cutting measures for metals recovery from e-waste. Therefore, Heydarian et al. (2018) conducted two-step research to introduce an effective and promising route for metal recovery using A. ferroxidans and A. thiooxidans. Evaluating the maximum value of metal recovery was done by optimizing more essential variables, namely initial pH, iron sulfate, and sulfur concentrations, using CCD. The maximum recovery of 99.2% Li, 50.4% Co, and 89.4% Ni was obtained. The results showed that the highest Li extraction could be reached at lower pH and higher sulfur concentration (Fig. 7). A toxicity assessment identified the bioleaching residual as a non-hazardous material and confirmed it was safe for disposal.95
 | ||
| Fig. 7 2D contour plots for Li dissolution.106 | ||
3.3. Bioleaching from spent catalysts
Malekian et al. (2019) recently used a spent de-coked catalyst from a refinery continuous catalytic reforming unit and investigated the bioleaching of platinum. Oxalic acid, a substance produced by A. niger, is known for its potential in bioleaching processes and is highly pH-dependent. Therefore, the oxalic acid concentration and the platinum recovery rates rose significantly by adjusting pH. Studying the more critical variables like the pulp density, temperature, and pH with the help of BBD led to a maximum Pt recovery of 37%. The undeniable role of oxalic acid was highlighted by evaluating the biorecovery rate of Pt in a blank medium in the absence of oxalic acid, demonstrating the biorecovery optimal conditions at just about 13%, which was far from the former conditions.96Shahrabi-Farahani et al. (2014) studied the bioleaching of molybdenum, Ni, and Al from hydrocracking spent catalyst in a slurry bubble bioreactor using A. thiooxidans. After adapting the bacteria to solid waste, bioleaching processes were evaluated with CCD to find the optimal condition. In order to achieve maximum biorecovery of three heavy metals in waste, three critical factors, such as the particle size of the solid waste, pulp density, and aeration rate, were selected. In this specific design, each variable was evaluated at five levels. The correlation between the recovery of heavy metals and the parameters was studied with a full quadratic model and two reduced cubic models. These correlations are presented as follows:
| Mo = 72.57 − 22.89A − 2.97B − 9.81C − 3.50AB − 2.75AC − 4.50BC − 8.02A2 − 6.25C2 − 14.53A2B + 13.56A2C + 19.14AB2 | (13) |
| Ni = 23.23 − 0.59A − 3.27B − 2.68C − 1.50AC − 2.50BC + 1.82B2 − 2.98A2B + 3.68A2C − 0.66AC2 | (14) |
| Al = 10.62 − 0.88A − 1.30B − 0.47C − 0.37AB + 0.13AC − 1.87BC − 0.52A2 + 1.07B2 + 0.71C2 | (15) |
A represents the particle size of solid waste, B represents the pulp density, and C represents the aeration rate. The optimal pulp density values, the solid waste particle size, and the aeration rate were 0.9% (w/v), 60.7 μm, and 209 mL min−1, respectively. At the optimal condition, maximum recovery of 87% Mo, 37% Ni, and 15% Al were reached after 7 days of the bioleaching process.107 The same year, Srichandan et al. (2014) studied Al, Ni, Mo, and V recovery from spent refinery catalysts using A. thiooxidans. In order to optimize the process, the CCD method was employed. Influential factors were initial pH (1.5–2.5), sulfur concentration (0.5–3 g L−1), and pulp density (1–10 g L−1). The most influential factor for Al, Mo, Ni, and V recovery was similar pH, considering the obtained models. After optimization and gaining optimal conditions as follows: pulp density of 1%, sulfur concentration 1.5% and pH 1.5, the recovery percentage for the target metals was: Ni 93%, Al 44%, Mo 34%, and vanadium 94%.107 Also, Motaghed et al. (2014) examined Pt and Rhenium biorecovery from the spent refinery catalyst by B. megaterium. They used the CCD method for the optimization of two influential factors (initial glycine concentration (0–15 g L−1) and pulp density (1–10%)). The analysis of variance results shows that the glycine concentration is more effective than pulp density in Re recovery. The optimal condition was reported as follows: initial glycine concentration = 12.8 g L−1 and pulp density = 4% (w/v). At these values, Pt and Re recoveries were 15.7% and 98%, respectively.109
Gholami et al. (2012) investigated Co, Mo, and Ni biorecovery using the fungus A. niger from spent catalysts. The following factors, pH (2.5–8.5), temperature (17.5–47.5 °C), inoculum percentage (0–12%), pulp density (0–4 g L−1), and rotation speed (100–160 rpm) were selected as the most effective in this process. Optimization was performed using the CCD method, and as a result, the optimal condition was reported as follows: pH = 5.0, temperature = 31.8 °C, pulp density = 2 g L−1, rotation speed = 115 rpm, and inoculum = 12%, the biorecovery percentage of Co, Mo, and Ni was 71%, 69%, and 46%, respectively.110
3.4. Biorecovery from sludge and plant's residues
Electroplating sludge contains valuable/toxic metals, so a green approach is needed to recover the valuable metals in it. Tian et al. (2022) recently studied Ni, Cu, Zn, and Cr recovery from electroplating sludge by sulfur-oxidizing strains. CCD method was applied to optimize four parameters of this process (pulp density (1–5%), temperature (15–55 °C), initial pH (0.6–1.6), and shaking speed (105–165 rpm)). Having these factors set at optimal values, Ni, Cu, Zn, and Cr biorecovery of 100%, 96.5%, 100%, and 76.1% were achieved, respectively. The values are presented: pulp density = 2%, temperature = 45 °C, initial pH = 0.8 and shaking speed = 150 rpm.116Barkusaraeya et al. (2021) investigated optimizing the biological leaching process for Zn extraction from paint sludge utilizing Acidithiobacillus thiooxidans. Using the CCD method in RSM, they considered temperature, shaking speed, pH, and particle size as the four factors with Zn recovery as the response. Optimization for maximal Zn removal was achieved with the temperature of 32 °C, speed shaking of 120 rpm, particle size of 1 mm, and pH of 4.2, resulting in 22% Zn removal efficiency.117
The simultaneous biological recovery of Cu, Cr, Zn, and Ni from sewage sludge was investigated by Li et al. (2018). Studying a mixed culture consisting of A. ferroxidans and A. thiooxidans, each bacterial culture was first examined in its pure form to enable comparisons. Influential variables included initial pH, solids concentration, ferrous, and sulfur ion concentration, which were studied and optimized via BBD of RSM in three different levels. Results indicated the mixed culture as the most influential culture in which a maximal extraction of 98.54% Cu, 57.99% Cr, 60.06% Ni, and 95.60% Zn was reached after setting each variable to their optimal value.64
Since biorecovery can be performed by fungi, Aspergillus sp. SMHS-3 isolate was introduced by Gholipour et al. (2018), which could oxidize sulfur media (toxic refinery spent sulfidic caustic). Although several sulfur-oxidizing fungi had been isolated during previous studies, it was claimed that this strain showed the highest activity. Sulfur decomposition rate optimization was determined based on the BBD of RSM by studying four impacting variables: pH, thiosulfate, sucrose, and Mo concentrations. Testing the selected optimum conditions evaluated by a 2-factor interaction (2FI) model ultimately revealed a 1.2-fold rise in the activity of sulfur conversion. The ANOVA of the fitted model also suggested a high significance in its prediction abilities with a P-value of less than 0.0001 (Table 5).108
| Source | Sum squares | df | Mean square | F-value | Prob > F |
|---|---|---|---|---|---|
| Model | 3193.62 | 10 | 319.36 | 90.69 | <0.0001 |
| A–A | 79.05 | 1 | 79.05 | 22.45 | 0.0002 |
| B–B | 415.36 | 1 | 415.36 | 117.95 | <0.0001 |
| C–C | 8.17 | 1 | 8.17 | 2.32 | 0.1462 |
| D–D | 353.17 | 1 | 353.17 | 100.29 | <0.0001 |
| AB | 148.84 | 1 | 148.84 | 42.27 | <0.0001 |
| AC | 24.50 | 1 | 24.50 | 6.96 | 0.0173 |
| AD | 1.10 | 1 | 1.10 | 0.31 | 0.5831 |
| BC | 1317.69 | 1 | 1317.69 | 374.19 | <0.0001 |
| BD | 453.69 | 1 | 453.69 | 128.84 | <0.0001 |
| CD | 392.04 | 1 | 392.04 | 111.33 | <0.0001 |
| Residual | 59.86 | 17 | 3.52 | ||
| Lack of fit | 59.60 | 14 | 4.26 | 47.74 | 0.0043 |
| Pure error | 0.27 | 3 | 0.089 | ||
| Corrected total | 3235.48 | 27 | |||
| Predicted R2 | 0.9450 | ||||
| Adjusted R2 | 0.9708 |
Ahmadi et al. (2017) studied Cu and Fe recovery from converter slags by A. ferrooxidans. A CCD method was applied to optimize the significant factors, including initial pH (1.5–3), initial Fe2+ concentration (0.5–9 g L−1), and pulp density (5–50 g L−1). Consequently, 95% Fe and 100% Cu was recovered at the optimal condition, which was presented as follows: initial pH = 1.8, pulp density = 1.4 g/100 mL, and initial Fe2+ = 7.3 g L−1.118 Table 6 indicates the research that has optimized the bioleaching processes from sludge and plant residues using RSM.
| Type of waste | Microorganism | RSM | Optimization | Reference | |||
|---|---|---|---|---|---|---|---|
| Method | Factors | Response(s) | Optimum condition | Optimum response | |||
| Waste sludge | S. acidophilus, At. Caldus, S. thermosulfidooxidans | CCD | Sludge solid content, sulfur conc. | Cu%, Zn%, Ni%, Pb% | Sludge solid content = 0.5%, sulfur conc. = 2.5% | 97%, 99%, 99%, 78% | 111 |
| Residues of Zn-plant | A. thiooxidans | CCD | pH, sulfur conc., pulp density | Zn% | pH = 3.3, sulfur conc. = 25.1 g L−1, pulp density = 21.5 g L−1 | 95% | 112 |
| Residues of Zn-plant | A. thiooxidans | CCD | pH, sulfur conc., pulp density | Zn% | pH = 3.3, sulfur conc. = 25.1 g L−1, pulp density = 21.5 g L−1 | 75% | 112 |
| Fuel-oil ash | A. thiooxidans | CCD | Pulp density, initial pH, sulfur conc. | V%, Ni%, Cu% | Pulp density = 1 g L−1, initial pH = 1, sulfur conc. = 9 g L−1 | 94.4%, 100%, 99.2% | 113 |
| Fuel-oil ash | A. ferroxidans | CCD | pH, initial Fe2+ conc., pulp density | V%, Ni%, Cu% | pH = 1.3, initial Fe2+ conc. = 2.6 g L−1, pulp density = 1% (w/v) | 74%, 95%, 88% | 114 |
| Dewatered metal-plating sludge | A. ferroxidans | CCD | pH, initial Fe2+ conc., pulp density | Cr%, Ni% | pH = 3, initial Fe2+ conc. = 1 g L−1, pulp density = 9 g L−1 | 55.6%, 58.2% | 109 |
| Chromate copper arsenate | Polyporales sp. KUC8959 | CCD | Culture filtrate conc., process time, temp. | Cu%, Cr%, As% | Culture filtrate conc. = 45.8%, process time = 20.6 h, temp. = 32.2 °C | 82.1%, 100%, 100% | 115 |
4. Discussion
RSM is recognized as a common and valuable method for designing and optimizing efficient bioleaching processes. In this review, 47 research articles on optimizing bioleaching processes have been analyzed, with a detailed description of how the RSM has helped the biorecovery of heavy metals from solid waste using microorganisms. Fig. 8 shows the features of the RSM reviewed in 47 bioleaching studies and illustrates the types of RSM design methods that have been used in these articles. As illustrated in Fig. 8(a), 65% of bioleaching articles have been using CCD, thanks to its features, such as precisely fitting a full quadratic and linear model by examining five levels of factors, considering extreme parameters combinations, and efficiency in the estimation of first- and second-order terms. Another popular method used in some of these articles is the BBD method, which, unlike the CCD, does not examine the borderline regions.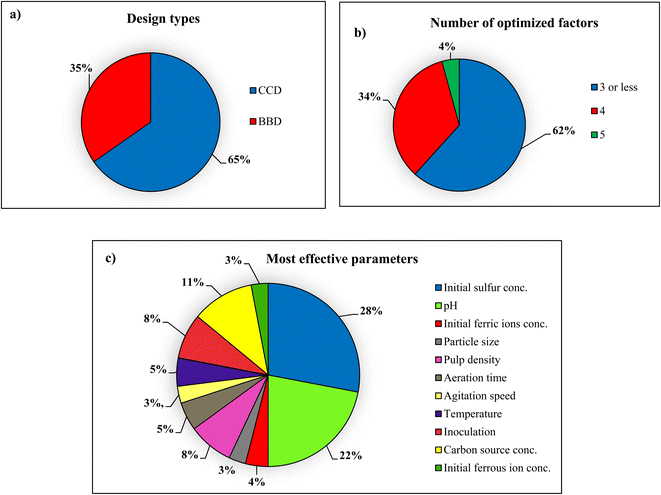 | ||
| Fig. 8 Charts of several RSM features utilized in the selected 47 papers that studied bioleaching (a) design types, (b) number of optimized factors, (c) most effective factors. | ||
As shown in Fig. 8(b), 62% of the articles reviewed in this study involve 3 or fewer influential factors to optimize the bioleaching process. Depending on the type of solid waste and the type of microorganism, and the mechanism of the bioleaching process, the number of factors was increased to 4 and over. Restricting the number of factors in RSM only limits the power of this method for designing and optimizing processes; the type and number of factors are central to the practical and maximal recovery of heavy metals in the biorecovery processes. Fig. 8(c) compares the most effectual factors among 64 models in 47 studied bioleaching papers, making them the most widely used factors in the bioleaching process, directly affecting heavy metal removal efficiency. These parameters' effectiveness was determined based on the coefficient of variables in the regression equation. Among them, pH and sulfur concentration have been considered the most critical factors; as mentioned before, high or very high pH is improper for the bioleaching process. Maintaining pH at low range, metal destruction occurs. Hence, pH is the most beneficial factor for optimizing in these papers. Also, sulfur concentration strongly affected the solubilization of heavy metals during the bioleaching process. It is worth noting that the concentration of added sulfur influences the acid production and pH variation in the bioleaching process.
5. Conclusion and future directions
Bioleaching is a green approach for heavy metal extraction. Depending on the type of process, process efficiency is affected by pulp density, particle size, pH, temperature, and inoculum density. To achieve the highest metal extraction rate, it is essential to identify the optimal number of factors to optimize the process. RSM offers a powerful and efficient way to explore complex interactions among multiple variables, identify key factors, and optimize process parameters to maximize desired outcomes. The value of RSM in bioleaching research lies in its ability to systematically investigate and optimize various factors that influence bioleaching efficiency and selectivity. By employing RSM, researchers can efficiently explore the interactive effects of multiple parameters, such as temperature, pH, particle size, microbial concentration, and nutrient availability. This approach enables the identification of optimal operating conditions, leading to enhanced leaching yields, reduced processing time, and improved resource utilization. Furthermore, RSM facilitates the identification and quantification of significant variables, enabling a deeper understanding of the underlying mechanisms and pathways involved in the bioleaching process. This knowledge is essential for the development of targeted strategies to overcome challenges and limitations, such as the presence of inhibitory substances, microbial competition, and the optimization of microbial consortia. Regarding sustainability, RSM can contribute to developing environmentally friendly and economically viable bioleaching processes. By optimizing process parameters through RSM, researchers can reduce resource consumption, minimize waste generation, and maximize metal recovery, thereby promoting the circular economy and minimizing the environmental impact of mining and metal extraction.Based on this review, researchers investigating the bioleaching process have widely used RSM with DOE tools to improve bioleaching efficiency. DOE tools such as CDD and BBD have been the most used to find the optimum conditions to remove heavy metals. According to our viewpoint, researchers must correctly choose factors and their levels to achieve success, which can be accomplished with screening design. BBD and CCD have fewer experimental runs over other designs and have proved time-saving approaches. This review shows that authors generated response surfaces using these models to obtain the reduced mathematical model to predict the response.
Many authors chose 3 or fewer factors, and the main factors applied for heavy metals removal were pH, pulp density, and inoculum size. Most of the microorganisms used in these studies are acidophilic species such as A. ferooxidans and A. thiooxidans. Therefore, pH is one of the main factors which affect Fe3+ and H2SO4 production and leads to better recovery of metals. Pulp density is another factor that is important to optimize and maximize the recovery of metal. In this review, most studies optimize pulp density with 0.2–9.7% (w/v) range for acidophilic microorganisms. Another fact to consider is that choosing the factors is varied based on the type of waste and microorganism. In some studies investigating the ore and soil microorganisms using an acidophilic microorganism, pH and pulp density were evaluated as the main factors. On the other hand, pH, nutrient/substrate concentration (Fe2+ and S0), and pulp density were considered essential factors for maximizing heavy metals removal from E-waste and sludge, and plant's residues. Since the structure of these wastes are more complex than ores and has a considerable percentage of heavy metals (high toxicity), high concentrations of metabolites in the bioleaching medium is essential.
For this reason, the production of metabolites is highly dependent on the nutrient/substrate concentration and pH, so optimization of these factors is vital in the high recovery of the target metal. In conclusion, the successes achieved and these studies demonstrate the benefits and the validity of using RSM as a method of DOE for the bioleaching of precious metals. The future research suggestions for the application of RSM in the bioleaching process are as follows:
(1) Integration of advanced optimization strategies: in addition to RSM, future research should explore the integration of other advanced optimization strategies, such as Dynamic Programming or Genetic Algorithms. These strategies can be applied to bioleaching experiments involving a large number of factors and can provide alternative approaches for experimental design, optimization, and decision-making.
(2) Multi-objective optimization: while RSM is effective in optimizing a single response variable, future studies should consider multi-objective optimization. This approach enables the simultaneous optimization of multiple responses, such as metal recovery, leaching rate, and energy consumption. By applying multi-objective optimization techniques, researchers can identify trade-offs and achieve a more comprehensive and sustainable bioleaching process.
(3) Advanced screening designs: alongside RSM, future research should employ advanced screening designs, such as Plackett–Burman or Taguchi orthogonal arrays, to effectively screen and select key factors and their appropriate levels. These designs allow for efficient identification of influential factors and reduction of experimental workload, thereby enhancing the success and efficiency of subsequent optimization studies.
(4) Comprehensive cost analysis: in addition to the technical aspects of bioleaching optimization, future studies should incorporate comprehensive cost analysis. This includes evaluating the economic feasibility, operational costs, and scalability of optimized bioleaching processes. Considering the cost implications will provide a more holistic understanding and support decision-making towards sustainable and commercially viable bioleaching operations.
(5) Scenario-based optimization: future research could explore multiple optimization scenarios to thoroughly explore different aspects and variables related to bioleaching processes. By considering diverse scenarios, such as variations in feedstock composition, leaching conditions, and recovery targets, researchers can gain a deeper understanding of process dynamics and broaden the applicability of optimization strategies.
Abbreviations
| ANOVA | Analysis of variance |
| BBD | Box–Behnken design |
| BEER | Bioleaching enhanced electrokinetic remediation |
| CCD | Central composite design |
| CPCB | Computer printed circuit boards |
| DOE | Design of experiment |
| DOF | Degrees of freedom |
| E-waste | Electronic waste |
| IOB | Iron-oxidizing bacteria |
| IT | Information technology |
| LCD | Liquid crystal display |
| LED | Light-emitting diode |
| LIB | lithium-ion battery |
| LSM | Least-squares method |
| MPPCB | Mobile phone printed circuit board |
| PCB | Printed circuit boards |
| PGM | Platinum group metals |
| PWB | Printed wire boards |
| RSM | Response surface methodology |
| SCC | Lithium coin cells |
| SOB | Sulfur-oxidizing bacteria |
| TPCBs | Telecommunication printed circuit boards |
| WLED | Waste light-emitting diodes |
| XRF | X-ray fluorescence spectrometry |
Author contributions
Tannaz Naseri: investigation, writing main parts of the original draft. Vahid Beygi: investigation, writing some parts of the original draft. Seyyed Mohammad Mousavi: project administration, funding acquisition, supervision, writing-review and editing. Sebastien Farnaud: review and language editing of the final version.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Tarbiat Modares University financially supported this study under grant number IG-39701.References
- M. T. Islam, N. Huda, A. Baumber, R. Shumon, A. Zaman, F. Ali, R. Hossain and V. Sahajwalla, J. Cleaner Prod., 2021, 316, 128297 CrossRef.
- M. L. Sall, A. K. D. Diaw, D. Gningue-Sall, S. Efremova Aaron and J.-J. Aaron, Environ. Sci. Pollut. Res., 2020, 27, 29927–29942 CrossRef CAS PubMed.
- A. Pathak, H. Al-Sheeha, R. Navvamani, R. Kothari, M. Marafi and M. S. Rana, Rev. Environ. Sci. Bio/Technol., 2022, 21, 1035–1059 CrossRef CAS.
- T. Naseri, F. Pourhossein, S. M. Mousavi, A. H. Kaksonen and K. Kuchta, Rev. Environ. Sci. Biotechnol., 2022, 21, 447–468 CrossRef CAS.
- T. Naseri, V. Beigi, A. Namdar, A. Keikavousi Behbahan and S. M. Mousavi, in Nano Technology for Battery Recycling, Remanufacturing, and Reusing, Elsevier, 2022, pp. 217–246 Search PubMed.
- F. Hosseinzadeh, S. O. Rastegar and M. Ashengroph, Process Biochem., 2021, 105, 1–7 CrossRef CAS.
- F. Pourhossein and S. M. Mousavi, Waste Manage., 2018, 79, 98–108 CrossRef CAS PubMed.
- M. E. Hoque and O. J. Philip, Mater. Sci. Eng., C, 2011, 31, 57–66 CrossRef CAS.
- B. K. Biswal, U. U. Jadhav, M. Madhaiyan, L. Ji, E.-H. Yang and B. Cao, ACS Sustainable Chem. Eng., 2018, 6, 12343–12352 CrossRef CAS.
- S. Ilyas and J. Lee, ChemBioEng Rev., 2014, 1, 148–169 CrossRef.
- T. Gu, S. O. Rastegar, S. M. Mousavi, M. Li and M. Zhou, Bioresour. Technol., 2018, 261, 428–440 CrossRef CAS PubMed.
- F. Anjum, M. Shahid and A. Akcil, Hydrometallurgy, 2012, 117–118, 1–12 CrossRef CAS.
- I. Asghari and S. M. Mousavi, Rev. Environ. Sci. Bio/Technol., 2014, 13, 139–161 CrossRef CAS.
- S. Mohanty, S. Ghosh, B. Bal and A. P. Das, Rev. Environ. Sci. Bio/Technol., 2018, 17, 791–811 CrossRef CAS.
- F. Pourhossein and S. M. Mousavi, Resour., Conserv. Recycl., 2022, 187, 106599 CrossRef CAS.
- F. Pourhossein, S. M. Mousavi and F. Beolchini, Resour., Conserv. Recycl., 2022, 182, 106306 CrossRef CAS.
- J. Yao, M. Wang, L. Wang, M. Gou, J. Zeng and Y.-Q. Tang, Environ. Sci. Pollut. Res., 2022, 29, 48509–48521 CrossRef CAS PubMed.
- M. Pirsaheb, S. Zadsar, S. O. Rastegar, T. Gu and H. Hossini, Environ. Technol. Innovation, 2021, 22, 101480 CrossRef CAS.
- A. Pathak, H. Srichandan and D. J. Kim, J. Environ. Manage., 2019, 242, 372–383 CrossRef CAS PubMed.
- T. Naseri, N. Bahaloo-Horeh and S. M. Mousavi, J. Environ. Manage., 2919, 235, 357–367 CrossRef PubMed.
- M. Arshadi and S. M. Mousavi, Sep. Purif. Technol., 2015, 147, 210–219 CrossRef CAS.
- Y. Zhan, X. Shen, M. Chen, K. Yang and H. Xie, Lett. Appl. Microbiol., 2022, 75, 1076–1083 CrossRef CAS PubMed.
- M. Arshadi, F. Pourhossein, S. M. Mousavi and S. Yaghmaei, Sep. Purif. Technol., 2021, 272, 118701 CrossRef CAS.
- S. Kadivar, F. Pourhossein and S. M. Mousavi, J. Environ. Manage., 2021, 280, 111642 CrossRef CAS PubMed.
- F. Pourhossein and S. M. Mousavi, J. Hazard. Mater., 2019, 378, 120648 CrossRef CAS PubMed.
- T. Naseri, N. Bahaloo-Horeh and S. M. Mousavi, J. Cleaner Prod., 2019, 220, 483–492 CrossRef CAS.
- F. Pourhossein and S. M. Mousavi, Waste Manage., 2018, 79, 98–108 CrossRef CAS PubMed.
- W. Gu, J. Bai, B. Dong, X. Zhuang, J. Zhao, C. Zhang, J. Wang and K. Shih, Hydrometallurgy, 2017, 171, 172–178 CrossRef CAS.
- M. L. M. Rodrigues, K. C. S. Lopes, H. C. Leôncio, L. A. M. Silva and V. A. Leão, Chem. Eng. J., 2016, 284, 1279–1286 CrossRef CAS.
- T. Naseri, S. M. Mousavi, A. Liese and K. Kuchta, J. Environ. Manage., 2023, 343, 118197 CrossRef CAS PubMed.
- T. Naseri, S. M. Mousavi and K. Kuchta, Waste Manage., 2023, 157, 47–59 CrossRef CAS PubMed.
- T. Naseri and S. M. Mousavi, Int. J. Biol. Macromol., 2022, 209, 1133–1143 CrossRef CAS PubMed.
- F. Faraji, R. Golmohammadzadeh, H. Sharifidarabad and F. Rashchi, Int. J. Environ. Sci. Technol., 2023, 20, 8785–8798 CrossRef CAS.
- J. Cui, N. Zhu, F. Mao, P. Wu and Z. Dang, Sci. Total Environ., 2021, 790, 148151 CrossRef CAS PubMed.
- M. H. Nasab, M. Noaparast, M. Abdollahi, H. Amoozegar and M. Ali, Hydrometallurgy, 2020, 193, 105309 CrossRef.
- Y. Osman, A. Gebreil, A. M. Mowafy, T. I. Anan and S. M. Hamed, World J. Microbiol. Biotechnol., 2019, 35, 93 CrossRef PubMed.
- F. Faraji, R. Golmohammadzadeh, F. Rashchi and N. Alimardani, J. Environ. Manage., 2018, 217, 775–787 CrossRef CAS PubMed.
- P. Rasoulnia and S. M. Mousavi, RSC Adv., 2016, 6, 9139–9151 RSC.
- M. Golzar-Ahmadi and S. M. Mousavi, Waste Manage., 2021, 131, 226–236 CrossRef CAS PubMed.
- D. A. Prasidya, W. Wilopo, I. W. Warmada and E. Retnaningrum, Biodiversitas, 2019, 20(7) DOI:10.13057/biodiv/d200716.
- X. Cui, Q. Gu, X. Liu, J. Wen, A. Lu, H. Ding, F. Yang, H. Shang, B. Wu, M. Zhang and X. Wang, Miner. Metall. Process., 2018, 35, 221–229 Search PubMed.
- F. Vakilchap, S. M. Mousavi, M. Baniasadi and S. Farnaud, Rev. Environ. Sci. Bio/Technol., 2020, 19, 509–530 CrossRef CAS.
- V. Beiki, S. M. Mousavi and T. Naseri, J. Environ. Manage., 2023, 344, 118399 CrossRef CAS PubMed.
- M. Arshadi and S. M. Mousavi, Bioresour. Technol., 2014, 174, 233–242 CrossRef CAS PubMed.
- I. Asghari and S. M. Mousavi, Rev. Environ. Sci. Bio/Technol., 2014, 13, 139–161 CrossRef CAS.
- S. O. Rastegar, S. M. Mousavi, M. Rezaei and S. A. Shojaosadati, J. Ind. Eng. Chem., 2014, 20, 3096–3101 CrossRef CAS.
- R. E. Kempson and R. Mead, Statistician, 1990, 39, 91 CrossRef.
- T. J. Xu and Y. P. Ting, Enzyme Microb. Technol., 2004, 35, 444–454 CrossRef CAS.
- G. A. Lujan-Moreno, P. R. Howard, O. G. Rojas and D. C. Montgomery, Expert Syst. Appl., 2018, 109, 195–205 CrossRef.
- O. A. Mohamed, S. H. Masood and J. L. Bhowmik, Adv. Manuf., 2015, 3, 42–53 CrossRef CAS.
- F. Gerayeli, F. Ghojavand, S. M. Mousavi, S. Yaghmaei and F. Amiri, Sep. Purif. Technol., 2013, 118, 151–161 CrossRef CAS.
- Y. Zhu, J. Yu, C. Jiao, J. Tong, L. Zhang, Y. Chang, W. Sun, Q. Jin and Y. Cai, Heliyon, 2019, 5, e02374 CrossRef CAS PubMed.
- P. Van Aken, R. Van den Broeck, J. Degrève and R. Dewil, J. Cleaner Prod., 2017, 161, 1432–1441 CrossRef CAS.
- N. Manojkumar, C. Muthukumaran and G. Sharmila, J. King Saud Univ., Eng. Sci., 2022, 34(3), 198–208 Search PubMed.
- J. M. Donohue, E. C. Houck and R. H. Myers, Oper. Res., 1993, 41, 880–902 CrossRef.
- R. H. Myers, A. I. Khuri and W. H. Carter, Technometrics, 1989, 31, 137–157 Search PubMed.
- L. M. S. Pereira, T. M. Milan and D. R. Tapia-Blácido, Biomass Bioenergy, 2021, 151, 106166 CrossRef CAS.
- N.-K. Nguyen and J. J. Borkowski, J. Stat. Plan. Inference, 2008, 138, 294–305 CrossRef.
- S. Karimifard and M. R. Alavi Moghaddam, Sci. Total Environ., 2018, 640–641, 772–797 CrossRef CAS PubMed.
- M. A. Bezerra, R. E. Santelli, E. P. Oliveira, L. S. Villar and L. A. Escaleira, Talanta, 2008, 76, 965–977 CrossRef CAS PubMed.
- T. Lundstedt, E. Seifert, L. Abramo, B. Thelin, Å. Nyström, J. Pettersen and R. Bergman, Chemom. Intell. Lab. Syst., 1998, 42, 3–40 CrossRef CAS.
- L. St»hle and S. Wold, Chemom. Intell. Lab. Syst., 1989, 6, 259–272 CrossRef.
- V. A. Sakkas, M. A. Islam, C. Stalikas and T. A. Albanis, J. Hazard. Mater., 2010, 175, 33–44 CrossRef CAS PubMed.
- H. Li, M. Ye, L. Zheng, Y. Xu, S. Sun, Q. Du, Y. Zhong, S. Ye and D. Zhang, Water Sci. Technol., 2018, 2017, 390–403 CrossRef PubMed.
- D. C. Cox and P. Baybutt, Risk Anal., 1981, 1, 251–258 CrossRef.
- A. H. Kaksonen, A.-M. Lakaniemi and O. H. Tuovinen, J. Cleaner Prod., 2020, 264, 121586 CrossRef CAS.
- P. Rasoulnia, R. Barthen, K. Valtonen and A.-M. Lakaniemi, Waste Biomass Valorization, 2021, 12, 5545–5559 CrossRef CAS.
- J. zhi Sun, B. Wu, B. wei Chen and J. kang Wen, J. Cent. South Univ., 2022, 29, 1488–1499 CrossRef.
- J. Tang, Y. Feng, Z. Wu, S. Zhang, E. K. Sarkodie, H. Jin, R. Yuan, W. Pan and H. Liu, Minerals, 2021, 11, 20–30 Search PubMed.
- X. Mo, X. Li and J. Wen, J. Radioanal. Nucl. Chem., 2019, 321, 579–590 CrossRef CAS.
- F. Jalali, J. Fakhari and A. Zolfaghari, Hydrometallurgy, 2019, 185, 194–203 CrossRef CAS.
- A. Selvi and R. Aruliah, Chemosphere, 2018, 207, 753–763 CrossRef CAS PubMed.
- C. Liu, B. Liao, S. Nie, X. Wang, Z. Sun, J. Wang and P. Ke, J. Radioanal. Nucl. Chem., 2021, 329, 1045–1060 CrossRef CAS.
- W. Liu, Z. Liu and C. Liu, Russ. J. Non-Ferr. Met., 2021, 62, 382–389 CrossRef.
- H. Z. Tavakoli, M. Abdollahy, S. J. Ahmadi and A. K. Darban, Trans. Nonferrous Met. Soc. China, 2017, 27, 2691–2703 CrossRef CAS.
- M. Naderi, S. Z. Shafaie, M. Karamoozian and S. Gharanjik, J. Min. Environ., 2017, 8, 523–537 Search PubMed.
- L. Yan, H. Hu, S. Zhang, P. Chen, W. Wang and H. Li, Electron. J. Biotechnol., 2017, 25, 50–57 CrossRef.
- D. F. Haghshenas, B. Bonakdarpour, E. K. Alamdari and B. Nasernejad, Hydrometallurgy, 2012, 111, 22–28 CrossRef.
- H. Abdollahi, S. Z. Shafaei, M. Noaparast and Z. Manafi, Int. J. Min. Geo-Eng., 2017, 51, 151–159 Search PubMed.
- H. Abdollahi, S. Z. Shafaei, M. Noaparast, Z. Manafi and N. Aslan, Trans. Nonferrous Met. Soc. China, 2013, 23, 219–230 CrossRef CAS.
- J. L. Song, S. J. Liu, C. Jiang, M. Resources and C. Academy, 2015, 1130, pp. 338–341.
- H. Z. Tavakoli, M. Abdollahy, S. J. Ahmadi and A. K. Darban, Russ. J. Non-Ferr. Met., 2017, 58, 188–199 CrossRef.
- M. Eisapour, A. Keshtkar, M. A. Moosavian and A. Rashidi, Ann. Nucl. Energy, 2013, 54, 245–250 CrossRef CAS.
- F. Fatemi, M. Arabieh and S. Jahani, Radiochim. Acta, 2016, 104, 239–246 CrossRef CAS.
- A. Rashidi, R. Roosta-Azad and S. J. Safdari, J. Radioanal. Nucl. Chem., 2014, 301, 341–350 CrossRef CAS.
- S. Biswas and K. Bhattacharjee, Sep. Purif. Technol., 2014, 135, 100–109 CrossRef CAS.
- J. Safdari, R. Roostaazad and A. Rashidi, Ann. Nucl. Energy, 2013, 56, 48–52 CrossRef.
- L. Praburaman, J.-H. Park, M. Govarthanan, T. Selvankumar, S.-G. Oh, J.-S. Jang, M. Cho, S. Kamala-Kannan and B.-T. Oh, Chemosphere, 2015, 138, 127–132 CrossRef CAS PubMed.
- M. Y. Moghaddam, S. Z. Shafaei, M. Noaparast, F. D. Ardejani, H. Abdollahi, M. Ranjbar, M. Schaffie and Z. Manafi, Trans. Nonferrous Met. Soc. China, 2015, 25, 4126–4143 CrossRef CAS.
- M. Govarthanan, G.-W. Lee, J.-H. Park, J. S. Kim, S.-S. Lim, S.-K. Seo, M. Cho, H. Myung, S. Kamala-Kannan and B.-T. Oh, Chemosphere, 2014, 109, 42–48 CrossRef CAS PubMed.
- A. Trivedi and S. Hait, J. Environ. Manage., 2023, 326, 116797 CrossRef CAS PubMed.
- F. Pourhossein, O. Rezaei, S. M. Mousavi and F. Beolchini, J. Environ. Health Sci. Eng., 2021, 19, 893–906 CrossRef CAS PubMed.
- M. Arshadi, A. Esmaeili and S. Yaghmaei, Hydrometallurgy, 2020, 197, 105464 CrossRef CAS.
- A. Kumar, H. S. Saini and S. Kumar, 3 Biotech., 2018, 8, 100 CrossRef CAS PubMed.
- A. Heydarian, S. M. Mousavi, F. Vakilchap and M. Baniasadi, J. Power Sources, 2018, 378, 19–30 CrossRef CAS.
- H. Malekian, M. Salehi and D. Biria, Waste Manage., 2019, 85, 264–271 CrossRef CAS PubMed.
- Q. Zhao, L. Tong, A. R. Kamali, W. Sand and H. Yang, Hydrometallurgy, 2020, 197, 105437 CrossRef CAS.
- M. P. Murugesan, K. Kannan and T. Selvaganapathy, Mater. Today Proc., 2019, 26, 2720–2728 CrossRef.
- M. J. Jowkar, N. Bahaloo-Horeh, S. M. Mousavi and F. Pourhossein, J. Cleaner Prod., 2018, 180, 417–429 CrossRef CAS.
- M. Mostafavi, F. Rashchi, S. Beikzadeh Noei and N. Mostoufi, in Solid State Phenomena, Trans Tech Publ, 2017, vol. 262, pp. 692–695 Search PubMed.
- M. Arshadi, S. M. Mousavi and P. Rasoulnia, Waste Manag., 2016, 57, 158–167 CrossRef CAS PubMed.
- Z. Niu, Q. Huang, B. Xin, C. Qi, J. Hu, S. Chen and Y. Li, J. Chem. Technol. Biotechnol., 2016, 91(3), 608–617 CrossRef CAS.
- M. Arshadi and S. M. Mousavi, Bioresour. Technol., 2015, 175, 315–324 CrossRef CAS PubMed.
- M. I. Bajestani, S. M. Mousavi and S. A. Shojaosadati, Sep. Purif. Technol., 2014, 132, 309–316 CrossRef.
- G. Liang, J. Tang, W. Liu and Q. Zhou, J. Hazard. Mater., 2013, 250, 238–245 CrossRef PubMed.
- A. Heydarian, S. M. Mousavi, F. Vakilchap and M. Baniasadi, J. Power Sources, 2018, 378, 19–30 CrossRef CAS.
- M. Shahrabi-Farahani, S. Yaghmaei, S. M. Mousavi and F. Amiri, Sep. Purif. Technol., 2014, 132, 41–49 CrossRef CAS.
- S. Gholipour, P. Mehrkesh, E. Azin, H. Nouri, A. A. Rouhollahi and H. Moghimi, J. Environ. Chem. Eng., 2018, 6, 2762–2767 CrossRef CAS.
- M. Motaghed, S. M. Mousavi, S. O. Rastegar and S. A. Shojaosadati, Bioresour. Technol., 2014, 171, 401–409 CrossRef CAS PubMed.
- R. Mafi Gholami, S. M. Mousavi and S. M. Borghei, J. Ind. Eng. Chem., 2012, 18, 218–224 CrossRef.
- S.-Y. Chen and W.-H. Chen, J. Environ. Sci. Health, Part A: Toxic/Hazard. Subst. Environ. Eng., 2013, 48, 1094–1104 CrossRef CAS PubMed.
- M. Sethurajan, P. N. L. Lens, E. R. Rene, J. van de Vossenberg, D. Huguenot, H. A. Horn, L. H. A. Figueiredo and E. D. van Hullebusch, J. Chem. Technol. Biotechnol., 2017, 92, 512–521 CrossRef CAS.
- S. O. Rastegar, S. M. Mousavi, S. A. Shojaosadati and T. Gu, RSC Adv., 2016, 6, 21756–21764 RSC.
- S. O. Rastegar, S. M. Mousavi and S. A. Shojaosadati, RSC Adv., 2015, 5, 41088–41097 RSC.
- Y.-S. Choi, J.-J. Kim, M.-J. Kim, J. Son and G.-H. Kim, J. Environ. Manage., 2013, 121, 6–12 CrossRef CAS PubMed.
- B. Tian, Y. Cui, Z. Qin, L. Wen, Z. Li and H. Chu, J. Environ. Manage., 2022, 312, 114927 CrossRef CAS PubMed.
- F. Honarjooy Barkusaraey, R. Mafigholami, M. Faezi Ghasemi and G. Khayati, J. Environ. Sci. Health, Part A: Toxic/Hazard. Subst. Environ. Eng., 2021, 56, 1243–1252 CrossRef CAS PubMed.
- S. Ahmadi, M. Vafaie Sefti, M. M. Shadman, Z. Azimi Dijvejin and H. Hosseini, J. Adv. Environ. Health Res., 2017, 5, 154–162 CAS.
Footnote |
| † Carbon dioxide. |
| This journal is © The Royal Society of Chemistry 2023 |