Open Access Article
Open Access ArticleEfficient enzymatic synthesis of chiral 2,3-dihydro-1,4-benzodioxane motif using engineered Candida antarctica lipase B†
Zhiyun Wu *,
Weifeng Shi,
Ming Jin and
Wei Zhou
*,
Weifeng Shi,
Ming Jin and
Wei Zhou
Department of Clinical Laboratory, The Third Affiliated Hospital of Soochow University, Changzhou, Jiangsu 213003, China. E-mail: zhiyunwu@126.com
First published on 21st June 2023
Abstract
Chiral motifs of 2,3-dihydro-1,4 benzodioxane are extensively utilized in diverse medicinal substances and bioactive natural compounds, exhibiting significant biological activities. Notable examples of such therapeutic agents include prosympal, dibozane, piperoxan, and doxazosin. In this work, using 1,4-benzodioxane-2-carboxylic acid methyl ester as the substrate, after screening 38 CALB covariant residues, we found that mutants A225F and A225F/T103A can catalyze the kinetic resolution of the substrate. The effect of temperature, cosolvent, and cosolvent concentration on kinetic resolution was investigated, revealing that the best results were achieved at 30 °C with 20% n-butanol as a cosolvent, resulting in an optimal resolution (e.e.s 97%, E = 278) at 50 mM substrate concentration. Structure analysis showed that mutation sites 225 and 103 are not among the sites that interact directly with the substrate, which means that covariant amino acids that interact remotely with the substrate also regulate enzyme catalysis. This research may provide us with a new strategy for enzyme evolution.
1 Introduction
The chiral motif of 2,3-dihydro-1,4 benzodioxane is widely used in various biologically active natural products and therapeutic agents with crucial biological activity.1–4 It has been proven that the absolute configuration of the C2 stereo centroid of benzodioxane greatly influences its biological activity.5 Several 1,4-benzodioxanes have displayed intriguing characteristics as α- or β-blocking agents, exhibiting antihypertensive properties.6 Additionally, some of these compounds have shown affinities towards serotonin receptors involved in schizophrenia, headaches, and nervous breakdowns, such as the antihypertensive drug (R)-doxazosin,7 selective α2C adrenergic receptor antagonist,8 the antidepressant MKC-242,9 the potent α1D-adrenergic antagonist WB4101,10 the 5-HT1A receptor agonist BSF-190555,11 and the natural product flavolignan silybin, found in Silybum marianum Gaertn, recognized for its antihepatotoxic activities12 (Fig. 1).Numerous reports on the preparation of chiral 2,3-dihydro-1,4 benzodioxane motifs signify the significance of this fundamental unit. Several approaches have been developed in recent decades to construct critical chiral 2,3-dihydro-1,4-benzodioxane frameworks.13–15 Most synthetic methods are chemical syntheses involving the building blocks in chiral pools or crystallization of chiral amino diastereomers. For example, Buchwald and colleagues prepared chiral 1,4-benzodiazepines and their derivatives by palladium-catalyzed intramolecular etherification of halogenated aromatic hydrocarbons with chiral alcohol groups.9 Palladium-catalyzed asymmetric intramolecular O-arylation coupling reactions were developed by Cai and his team using asymmetric demethylation.16 These reactions resulted in the production of chiral 1,4-benzodiazepines with enantioselectivity ranging from moderate to excellent. Meanwhile, Zhang and his team accomplished the asymmetric hydrogenation of 1,4-benzodioxane compounds using Ir catalysis, which led to the formation of chiral 1,4-benzodioxane compounds.17 Yin optimized the Ir-catalyzed asymmetric hydrogenation process, achieving a production of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ton, which was then utilized to establish more efficient synthetic pathways for a number of crucial biologically active compounds.18
000 ton, which was then utilized to establish more efficient synthetic pathways for a number of crucial biologically active compounds.18
Nevertheless, the majority of the reported approaches for synthesis encounter the issue of either low yield or low enantioselectivity. In certain instances, multiple crystallization steps may be necessary to attain a high level of enantiomer purity. Moreover, chemical synthesis methods are environmentally unfriendly, and the subsequent separation costs are too high. Currently, biocatalysis is becoming an increasingly helpful tool for organic chemists to synthesize biologically active natural and non-natural products.19,20 Hence, the development of a biocatalytic system that is both highly efficient and enantioselective is imperative for the construction of chiral 2,3-dihydro-1,4 benzodioxane motifs and their derivatives. Lipases (EC 3.1.1.3) are enzymes that facilitate the hydrolysis of carboxylic acid esters in aqueous environments, as well as esterification or transesterification reactions in organic solvents.19 Candida antarctica lipase B (CALB; Novozyme 435), one of the most widely utilized biocatalysts in both industry and academia, remains a prominent choice due to its availability in large quantities for industrial expression.21,22
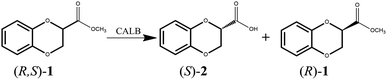
With the aim of developing a productive method for kinetic resolution, we screened a covariant amino acid site-saturated mutant library that was constructed previously23 and obtained mutants that could catalyze enantioselective hydrolysis of 1,4-benzodioxane-2-carboxylic acid methyl ester (R, S)-1 to synthesize optically pure 1,4-benzodioxane molecules. The specific reaction diagram is shown in Fig. 2.
2 Experiment
2.1 Strains, plasmids, and chemicals
The CALB gene was artificially synthesized by GenScript Crop (Nanjing, China). The constructed plasmid of all mutants come from our previous work.23 Escherichia coli Rosetta (DE3) competent cells was acquired from Vazyme (Nanjing, China). The transformed E. coli was regularly cultured for 12 h at 37 °C in LB broth comprising tryptone (1% w/v), sodium chloride (1% w/v), and yeast extract (0.5% w/v) or on LB agar plates supplemented with 100 μg mL−1 ampicillin. All substrates were obtained from Sigma (Sigma-Aldrich).2.2 Recombinant protein expression and purification
According to the CALB gene sequence, amplification of the CALB gene with NcoI and NotI sites was carried out using the forward primer 5′-GCGCCATGGCCTTACCTAGTGGTTCCGAC-3′ and the reverse primer 5′-GCTGCGGCCGCTCAAGGAGTAACAATTCCTGAAC-3′. Polymerase chain reaction (PCR) amplification was performed with ProofastTM (ATG Biotechnology Co., Ltd) Super Fidelity polymerase and the following temperature program: denaturation at 98 °C for 3 minutes; followed by 30 cycles of amplification (15 seconds at 98 °C, 15 seconds at 55 °C, 2 minutes at 72 °C); and a final extension at 72 °C for 10 minutes, followed by a hold at 10 °C. After digestion with NcoI and XhoI, the PCR product was cloned into the vector pET-22b (pET22b-CALB). Subsequently, pET22b-WT was electroporated into E. coli Rosetta (DE3). The recombinant plasmid pET22b-CALB was confirmed by DNA sequencing. E. coli Rosetta (DE3)/pET22b-CALB, harboring the CALB gene, was cultured in 100 mL LB medium supplemented with 100 mg L−1 ampicillin and 20 mg L−1 chloramphenicol at 37 °C on a rotary shaker at 180 rpm for 2–3 h. Upon reaching an OD600 of 0.6–0.8, a final concentration of 0.1 mM IPTG (isopropyl beta-D-thiogalactopyranoside) was added to induce CALB expression at 22 °C and 180 rpm for 12 h. Cells were harvested by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min, and CALB protein expression was confirmed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). The recombinant protein was expressed and purified using a previously described method.24
000 rpm for 10 min, and CALB protein expression was confirmed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). The recombinant protein was expressed and purified using a previously described method.24
2.3 Lipase-catalyzed hydrolysis of (±)-methyl 1,4-benzodioxan-2-carboxylate
To determine the mutants that could catalyze racemic methyl 1,4-benzodioxan-2-carboxylate, 5 μL of substrate solution (5 mg) was added to a 5 mL reaction system (4 mL PBS buffer and 1 mL n-butanol). After the substrate was fully dissolved, different CALB mutants (1 mg) were added and placed in a 30 °C shaker at 220 rpm for 1 h. Once a specific level of conversion was achieved, as determined by chiral high-performance liquid chromatography (HPLC), the reaction was stopped by the addition of MTBE (methyl tertiary butyl ether) followed by centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm to eliminate the enzyme and particles in suspension. The resulting clear solution was separated by decantation, and the centrifuged residue was then extracted with 5 mL ethyl acetate separately. The organic layers were combined and placed in a rotary evaporator to evaporate and dry. Then, 200 μL of mobile phase was added to dissolve, the sample was centrifuged at 12
000 rpm to eliminate the enzyme and particles in suspension. The resulting clear solution was separated by decantation, and the centrifuged residue was then extracted with 5 mL ethyl acetate separately. The organic layers were combined and placed in a rotary evaporator to evaporate and dry. Then, 200 μL of mobile phase was added to dissolve, the sample was centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min, and the supernatant was taken and filtered with a 0.45 μm nylon filter head for HPLC detection.
000 rpm for 5 min, and the supernatant was taken and filtered with a 0.45 μm nylon filter head for HPLC detection.
2.4 HPLC methods
The quantities of substrates in the reaction mixture and their conversion were assessed by analysing the relative area percentages (APs) of the HPLC peaks without applying any correction. HPLC was used to monitor the hydrolysis of the substrate (R, S)-methyl 1,4-benzodioxan-2-carboxylate using a Daicel CHIRALCEL OJ-H column (5 μm, 4.6 mm × 250 mm) at 30 °C with UV detection at 220 nm. The flow rate was 1 mL min−1 with solvent (n-hexane/isopropanol (90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, V/V)). The retention time for the two enantiomers of (R, S)-1 was 25.2 min and 27.1 min, respectively (Fig. S1†). In order to quantify the substrate (R, S)-1, a standard curve was established for this compound. Standard curves were generated by plotting the peak area from high-performance liquid chromatography (HPLC) against the substrate concentration for authentic standards. These curves were then utilized to determine the potencies of different batches of (R, S)-1. The solvent used for the HPLC analysis was a mixture of hexane and isopropyl alcohol (90
10, V/V)). The retention time for the two enantiomers of (R, S)-1 was 25.2 min and 27.1 min, respectively (Fig. S1†). In order to quantify the substrate (R, S)-1, a standard curve was established for this compound. Standard curves were generated by plotting the peak area from high-performance liquid chromatography (HPLC) against the substrate concentration for authentic standards. These curves were then utilized to determine the potencies of different batches of (R, S)-1. The solvent used for the HPLC analysis was a mixture of hexane and isopropyl alcohol (90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10), and standard samples with concentrations of 2.5, 5, 10, 25, 50, and 100 mM substrate were employed. Three parallels were made for each concentration, and HPLC analysis of a total of 18 samples was performed. Then, the standard curve was generated with the concentration as the abscissa and the peak area as the ordinate. The results are shown in Fig. S2.† Using the standard curve formula, the substrate concentration can be calculated based on the peak area.
10), and standard samples with concentrations of 2.5, 5, 10, 25, 50, and 100 mM substrate were employed. Three parallels were made for each concentration, and HPLC analysis of a total of 18 samples was performed. Then, the standard curve was generated with the concentration as the abscissa and the peak area as the ordinate. The results are shown in Fig. S2.† Using the standard curve formula, the substrate concentration can be calculated based on the peak area.
For the resolution evaluation of chiral drug intermediates, both yield and enantiomer purity should be considered. Chemical yield is expressed by conversion rate Conv., and enantiomeric purity is expressed by enantiomeric excess (e.e.s). Enantioselectivity (E) is a comprehensive evaluation of enzyme-catalyzed reactions, representing the efficiency of catalyst resolution. The higher the E value, the higher the enantiomer selectivity of the enzyme. The calculation formula is as follows:
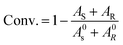 | (1) |
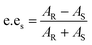 | (2) |
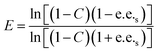 | (3) |
A0s, A0R—the concentrations of the two enantiomers of 1,4-benzodioxane-2-carboxylic acid methyl ester (R, S)-1 before the reaction; As, AR—the concentration of the two enantiomers of the substrate that have not been resolved after the reaction.
2.5 Optimization of CALB catalyzed hydrolysis of (R, S)-1 on a small scale: effects of cosolvent, cosolvent concentration and, temperature
The effects of cosolvent, cosolvent concentration, and temperature on enzyme activities were assessed by measuring the initial rates of hydrolysis of (R, S)-1 under different reaction conditions. To investigate the effect of cosolvent on the CALB-catalyzed kinetic resolution of (R, S)-1, cosolvents (20%), including DMSO, acetonitrile, isopropyl alcohol, isopropyl ether, acetic acid, ethyl ester, and toluene, were used. After confirming that n-butanol was the best cosolvent, we further explored the influence of n-butanol content on the reaction.Concentrations of n-butanol of 0%, 1%, 5%, 20%, 50%, 80%, and 100% were selected. The optimum temperature for wild-type CALB and its variants was determined using standard assay conditions at different temperatures ranging from 15 °C to 40 °C in 5 °C increments. The reaction samples were diluted with methyl tert-butyl (MTBE) and detected by HPLC.
2.6 Time course
We determined 20% n-butanol as an organic solvent in the two-phase reaction system and then made time progression curves for the catalytic substrates of wild-type CALB, mutant A225F, and A225F/T103A. In a 10 mL reaction system (8 mL PBS phosphoric acid buffer and 2 mL n-butanol), 100 μL substrate solution and 2 mg enzyme were added for the reaction, and 500 μL reaction solution was removed every 20 min into a 2 mL EP tube. The reaction was stopped by adding 1 mL MTBE, and the extraction was performed 3 times. The extraction liquid was dried in a rotary evaporator, and then 200 μL of mobile phase was added to dissolve it. The sample was centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min, and the supernatant was collected and filtered through a 0.45 μm nylon filter head. HPLC was used to determine the resolution effect at different reaction time.
000 rpm for 5 min, and the supernatant was collected and filtered through a 0.45 μm nylon filter head. HPLC was used to determine the resolution effect at different reaction time.
2.7 Structure analysis
The crystal structure of CALB (PDB ID: 1TCA)25 was used for substrate analogue (R-1) docking analysis. The interactions between CALB mutants and the ligand were obtained by conducting molecular docking using AutoDock Vina (http://vina.scripps.edu/).26 Molecular Operating Environment (MOE) was used to predict the 3D structures of the ligand bound to the protein. After multiple independent screenings, the possible small molecule conformation was obtained, which provides a basis for subsequent dynamics simulation. The representative conformation after docking was used as the initial conformation for further minimization and MD simulation. MD simulations were performed using the GPU accelerated code (pmemd) of the Amber 20 (ref. 27) and AmberTools 18 packages.28 The partial charge of the ligand was set to coincide with the electrostatic potential generated at HF/6-31 G(d) levels using the RESP (suppressed electrostatic potential) mode. The solvent box of the system was constructed using Packmol, and the solvent was a 20% n-butanol solution. The particle mesh Ewald (PME) method was employed to treat long-range electrostatic interactions.29 The addition of Na+ ions is intended to neutralize the system. AMBER ff14SB and General AMBER force field (GAFF) were applied to the residue and ligand, respectively. To generate the substrate's force field, the antechamber was utilized, while the SHAKE algorithm constrained hydrogen-containing bonds.30 The system underwent a 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-step steepest descent minimization process and was then heated and briefly equilibrated in the NVT ensemble at 303 K for 30 ps. Three independent trajectories of 50 ns each were simulated. All the figures of the molecular model were created using PyMol 2.5 (http://www.pymol.org).
000-step steepest descent minimization process and was then heated and briefly equilibrated in the NVT ensemble at 303 K for 30 ps. Three independent trajectories of 50 ns each were simulated. All the figures of the molecular model were created using PyMol 2.5 (http://www.pymol.org).
3 Results and discussion
3.1 Identification of the mutant
The search for a biocatalyst with high enantioselectivity and activity is a crucial step in the practical application of enzymatic catalysis. Instead of using a raditional substrate-guided screening strategy,31,32 we screened 38 covariant functional amino acid sites that were distributed inside the fundamental structure of the protein obtained from previous work.23 Although wild-type CALB and many mutants showed hydrolysing activity, only mutants A225F and A225F/T103A exhibited enantioselectivity toward (R, S)-1, as shown in Fig. S3.† After 30 min reaction, the mutants A225F and A225F/T103A almost completely hydrolysed one of the substrates with one configuration (S type) but not the other. However, after 30 min of reaction, the wild-type CALB had a large amount of residues in both substrates with two configurations. Therefore, the mutants A225F and A225F/T103A were identified to catalyze the resolution of methyl 1,4-benzodioxan-2-carboxylate.3.2 Effect of cosolvent
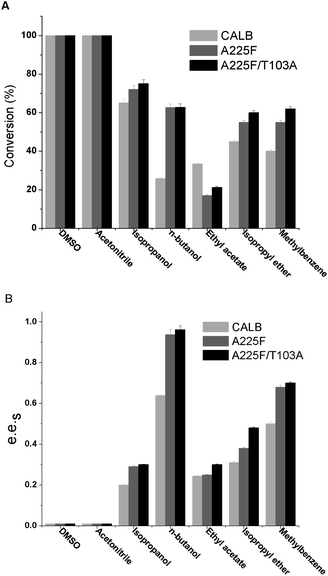
After determining the mutants, we explored the influence of cosolvents on the reaction, and 20% of various polar and nonpolar solvents were added to the reaction system. The results are shown in Fig. 3, and it can be found that n-butanol as a cosolvent is conducive to enzyme catalysis of 1,4-benzodioxane-2-carboxylic acid methyl ester, and the e.e.s of both mutant A225F and A225F/T103A are above 0.9. In polar solvents DMSO and acetonitrile, the reaction effect is very similar to that in the water phase, and the conversion rate reaches nearly 100%. However, the enzyme has no selectivity for substrates. In the nonpolar solvents isopropyl ether and toluene, although the enzyme has partial selectivity for the substrate, the resolution effect is not as good as that of n-butanol. Among the above seven cosolvents, the n-butanol reaction system is the best. Even if the mechanism underlying the influence of cosolvents on the enzyme enantioselectivity remains unclear, it is very important to screen suitable co-solvents in chiral resolution experiments. The incorporation of a cosolvent can exert a favorable impact on both the reaction equilibrium33 and kinetics34 as well as enhancing the enzyme's stability against denaturation.35 The cosolvent of n-butanol could have a greater attraction than water, allowing for a strong interaction with lipase and the potential to alter its three-dimensional structure during the preparation stage as possible explanation of the phenomenon.3.3 Effect of n-butanol concentration
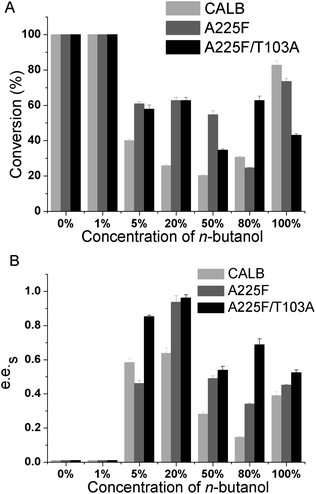
After determining n-butanol as the best cosolvent in the reaction system, we further explored the influence of different concentrations of n-butanol on the reaction. The resolution effects of wild-type, mutant A225F and A225F/T103A on the substrate were investigated under 0%, 1%, 5%, 20%, 50%, 80%, and 100% n-butanol contents. The results are shown in Fig. 4, and with increasing n-butanol content, the enzyme catalysis effect was improved, and the best selectivity was achieved at 20%. The e.e.s value of mutant A225F/T103A reached 0.95. When the concentration of n-butanol continued to increase, the selectivity of the enzyme to the reaction decreased. The results showed that the resolution effect of the reaction was good in the two-phase system, and the optimal concentration of n-butanol was 20%.3.4 Effect of temperature
In enzymatic catalysis, temperature is very important because it can influence enzyme activity, specificity, stability, and reaction equilibrium.36 To investigate the influence of the temperature on the resolution of (R, S)-1 catalyzed by CALB in 20% n-butanol, the reaction temperature was varied from 15 °C to 40 °C. As shown in Fig. 5, with increasing temperature, the conversion rate continues to increase, and the e.e.s value reaches the highest value of 0.97 at 30 °C. This may be due to the decrease in solution viscosity at higher temperatures, which reduces mass transfer limitations. However, as the temperature rises further, the e.e.s value decreases. This may be because high temperature would render the enzyme inactive. Therefore, we set the optimal reaction temperature at 30 °C.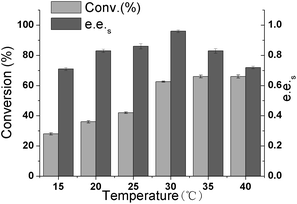 | ||
| Fig. 5 Effect of temperature on the A225F/T103A mutant catalysis of 1,4-benzodioxane-2-carboxylic acid methyl ester. | ||
3.5 Time course
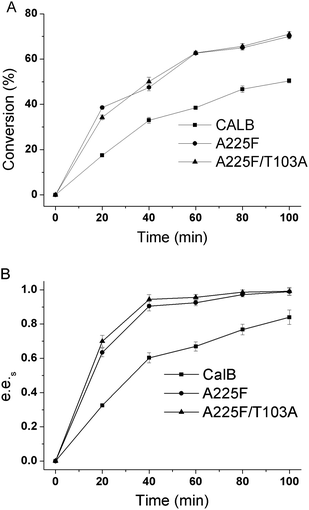
In a 10 mL reaction system with substrate concentration of 50 mM, reaction temperature of 30 °C and n-butanol content of 20%, we made a time progression curve for the kinetic resolution of substrates by wild-type CALB, mutant A225F, and A225F/T103A. As shown in Fig. 6, the conversion rate and e.e.s value increased with time, and the mutant had a good resolution effect on the substrate after 40 min of reaction.The conversion rate of A225F/T103A was 50%, e.e.s was 0.97, and the E value reached 278. At this time, the conversion rate of wild-type CALB was only 30%, and the e.e.s value was only 0.6. At 100 min, the conversion rate of the reaction catalyzed by wild-type CALB reached 50%, and the e.e.s value was only 0.83. Therefore, we can confirm that the mutant A225F/T103A has good resolution on the substrate after 40 min of reaction under this condition.
3.6 Structural analysis of the evolution
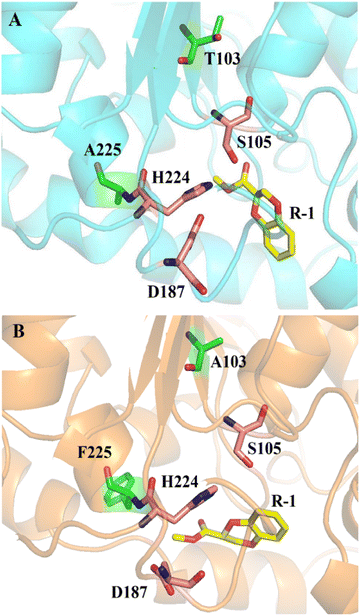
The experimental results showed that the wild type could catalyze (R, S)-1 without selectivity, while the mutant could not catalyze the R-1 substrate, making it have high enantioselectivity to (R, S)-1. Using R-1 as substrate, we performed docking computations and 50 ns molecular dynamics simulations with wild-type CALB and mutant A225F/T103A, which provided structural information related to the enhanced enantioselectivity in mutant A225F/T103A. The binding poses of R-1 in the active site pockets of CALB and A225F/T103A are shown in Fig. 7. However, the docking results showed that sites 225 and 103 are far from the substrate and will not interact directly with the substrate. This indicates that covariant amino acids might remotely affect enzyme activity and enantioselectivity, which means that key residues distant from the catalytic site are also important for enzyme catalysis.37,38 The site H224 is near 225, and we supposed that maybe the mutation at site 225 resulted in a change in the catalytic triad (S105-D187-H224) conformation (Fig. S4†), which made the R-type substrate unable to pass through the channel, thus increasing enantioselectivity. Additionally, we performed calculations to determine the specific activity of both CALB and mutants towards the substrate (R, S)-1. Surprisingly, we discovered that the activity of the mutants was comparable to that of the wild-type CALB (Table S1†). Therefore, we were able to enhance enantioselectivity without compromising enzyme activity. Unlike the traditional rational design, the new SCA algorithm, SCA. SIM predicted 38 covariant residues in Candida antarctica lipase B (CALB) that are not confined to the substrate binding pocket. The amino acid mutations far from the active center provide us with a new idea for protein evolution, and this also reaffirms that covariant amino acids can be used for enzyme engineering research.39,404 Conclusions
In conclusion, A225F and A225F/T103A have been identified as efficient mutants for the enantioselective hydrolysis of 1,4-benzodioxane-2-carboxylic acid methyl ester. The effect of temperature, cosolvent, and cosolvent concentration on kinetic resolution was investigated, revealing that the best results were achieved at 30 °C with 20% n-butanol as a cosolvent, resulting in an optimal resolution (e.e.s 97%, E = 278) at 50 mM of substrate concentration. Molecular docking suggested that the mutations are far from the active center, which gives us a new insight for protein evolution. Our study provides a new environmentally friendly and promising biosynthesis method for chiral 2,3-dihydro-1,4 benzodioxane motifs.Author contribution
Zhiyun Wu conducted the experiments, analysed the results, and wrote the paper. Weifeng Shi, Wei Zhou and Ming Jin completed the article review and contributed to the preparation of the figures. All authors analysed the results and approved the final version of the manuscript.Conflicts of interest
The authors declare no competing interests.Acknowledgements
This work was supported by grants from Jiangsu Innovative and Entrepreneurial Talent Programme (JSSCBS20211594) and the Changzhou Science and Technology Project (Applied Based Research, No. CJ20220102).References
- S. Wang, Y. Chen, S. Zhao, X. Xu, X. Liu, B. F. Liu and G. Zhang, Bioorg. Med. Chem. Lett., 2014, 24, 1766–1770 CrossRef CAS PubMed.
- O. Cruz-Lopez, M. C Nunez, A. Conejo-Garcia, M. Kimatrai and J. M Campos, Curr. Org. Chem., 2011, 15, 869–887 CrossRef CAS.
- C. Bolchi, F. Bavo, R. Appiani, G. Roda and M. Pallavicini, Eur. J. Med. Chem., 2020, 200, 112419 CrossRef CAS PubMed.
- A. Sen and S. H. Chikkali, Org. Biomol. Chem., 2021, 19, 9095–9137 RSC.
- W. Quaglia, M. Pigini, M. Giannella and C. Melchiorre, J. Med. Chem., 1990, 33, 2946–2948 CrossRef CAS PubMed.
- R. R. Ruffolo, Jr., W. Bondinell and J. Paul Hieble, J. Med. Chem., 1995, 38, 3681–3716 CrossRef PubMed.
- S. Ma, L. Ren, D. Zhao, Z. Zhu, M. Wang, H. Lu and L. Duan, Acta Pharmacol. Sin., 2006, 27, 1423–1430 CrossRef CAS PubMed.
- S. D. Patel, W. M. Habeski, H. Min, J. Zhang, R. Roof, B. Snyder, G. Bora, B. Campbell, C. Li and D. Hidayetoglu, Bioorg. Med. Chem. Lett., 2008, 18, 5689–5693 CrossRef CAS PubMed.
- S. Kuwabe, K. E. Torraca and S. L. Buchwald, J. Am. Chem. Soc., 2001, 123, 12202–12206 CrossRef CAS PubMed.
- L. Fumagalli, M. Pallavicini, R. Budriesi, C. Bolchi and M. Canovi, J. Med. Chem., 2013, 56, 6402–6412 CrossRef CAS PubMed.
- A. M. Birch, P. A. Bradley, J. C. Gill, F. Kerrigan and P. L. Needham, J. Med. Chem., 1999, 42, 3342–3355 CrossRef CAS PubMed.
- N. R. Farnsworth and A. S. Bingel, Problems and Prospects of Discovering New Drugs from Higher Plants by Pharmacological Screening, in New Natural Products and Plant Drugs with Pharmacological, Biological or Therapeutical Activity, Proceedings in Life Sciences, ed. H. Wagner and P. Wolff, Springer, Berlin, Heidelberg, 1977, DOI:10.1007/978-3-642-66682-7_1.
- A. Rouf, M. A. Aga, B. Kumar and S. C. Taneja, Tetrahedron Lett., 2013, 54, 6420–6422 CrossRef CAS.
- C. Bolchi, M. Pallavicini, L. Fumagalli, N. Marchini, B. Moroni, C. Rusconi and E. Valoti, Tetrahedron: Asymmetry, 2005, 16, 1639–1643 CrossRef CAS.
- V. Straniero, G. Lodigiani, L. Suigo and E. Valoti, Chirality, 2022, 34, 1053–1064 CrossRef CAS PubMed.
- W. Yang, J. Yan, Y. Long, S. Zhang, J. Liu, Y. Zeng and Q. Cai, Org. Lett., 2013, 15, 6022–6025 CrossRef CAS PubMed.
- Y. Wang, J. Xia, G. Yang and W. Zhang, Tetrahedron, 2018, 74, 477–482 CrossRef CAS.
- X. Yin, Y. Huang, Z. Chen, Y. Hu, L. Tao, Q. Zhao, X.-Q. Dong and X. Zhang, Org. Lett., 2018, 20, 4173–4177 CrossRef CAS PubMed.
- T. Matsuda, Future directions in biocatalysis, Elsevier, 2017 Search PubMed.
- S. Wu, R. Snajdrova, J. C. Moore, K. Baldenius and U. T. Bornscheuer, Angew. Chem., 2020, 60, 88–119 CrossRef PubMed.
- D. Sharma, B. Sharma and A. Shukla, Biotechnology, 2011, 10, 23–40 CrossRef CAS.
- P. Chandra, R. Singh and P. K. Arora, Microb. Cell Fact., 2020, 19, 169 CrossRef CAS PubMed.
- Z. Wu, H. Liu, L. Xu, H. F. Chen and Y. Feng, FASEB J., 2020, 34, 1983–1995 CrossRef CAS PubMed.
- M. W. Larsen, U. T. Bornscheuer and K. Hult, Protein Expression Purif., 2008, 62, 90–97 CrossRef CAS PubMed.
- J. Uppenberg, M. T. Hansen, S. Patkar and T. A. Jones, Structure, 1994, 2, 293–308 CrossRef CAS PubMed.
- J. Eberhardt, D. Santos-Martins, A. F. Tillack and S. Forli, J. Chem. Inf. Model., 2021, 61, 3891–3898 CrossRef CAS PubMed.
- R. Salomon-Ferrer, D. A. Case and R. C. Walker, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2013, 3, 198–210 CAS.
- D. A. Case, T. E. Cheatham III, T. Darden, H. Gohlke, R. Luo, K. M. Merz Jr, A. Onufriev, C. Simmerling, B. Wang and R. J. Woods, J. Comput. Chem., 2005, 26, 1668–1688 CrossRef CAS PubMed.
- T. Darden, D. York and L. Pedersen, J. Chem. Phys., 1993, 98, 10089–10092 CrossRef CAS.
- J.-P. Ryckaert, G. Ciccotti and H. J. Berendsen, J. Comput. Phys., 1977, 23, 327–341 CrossRef CAS.
- Y. Zhang, B. Xia, Y. Li, X. Lin and Q. Wu, Biomacromolecules, 2021, 22, 918–926 CrossRef CAS PubMed.
- J. Qiao, D. Yang, Y. Feng, W. Wei, X. Liu, Y. Zhang, J. Zheng and X. Ying, RSC Adv., 2023, 13, 10468–10475 RSC.
- N. I. Ruzich and A. S. Bassi, Can. J. Chem. Eng., 2010, 88, 277–282 CrossRef CAS.
- J. Wang, G.-X. Sun, L. Yu, F.-A. Wu and X.-J. Guo, Bioresour. Technol., 2013, 128, 156–163 CrossRef CAS PubMed.
- T. Arakawa and S. Timasheff, Biophys. J., 1985, 47, 411–414 CrossRef CAS PubMed.
- Z. Xu, Y.-K. Cen, S.-P. Zou, Y.-P. Xue and Y.-G. Zheng, Crit. Rev. Biotechnol., 2020, 40, 83–98 CrossRef CAS PubMed.
- D. Kim, M. H. Noh, M. Park, I. Kim, H. Ahn, D.-y. Ye, G. Y. Jung and S. Kim, Metab. Eng., 2022, 74, 49–60 CrossRef CAS PubMed.
- N. Halabi, O. Rivoire, S. Leibler and R. Ranganathan, Cell, 2009, 138, 774–786 CrossRef CAS PubMed.
- J. Xie, W. Zhang, X. Zhu, M. Deng and L. Lai, Elife, 2023, 12, e81850 CrossRef PubMed.
- Y. Liu, Z. Yan, X. Lu, D. Xiao and H. Jiang, Sci. Rep., 2016, 6, 24117 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra02623j |
| This journal is © The Royal Society of Chemistry 2023 |