Open Access Article
Open Access ArticleExploring the inhibitory potential of Nigella sativa against dengue virus NS2B/NS3 protease and NS5 polymerase using computational approaches†
Mamuna Mukhtar a,
Haris Ahmed Khan
a,
Haris Ahmed Khan ab and
Najam us Sahar Sadaf Zaidi
ab and
Najam us Sahar Sadaf Zaidi *a
*a
aAtta ur Rahman School of Applied Biosciences (ASAB), National University of Sciences and Technology (NUST), H-12, 44000, Islamabad, Pakistan. E-mail: sadafzaidi@asab.nust.edu.pk
bDepartment of Biotechnology, University of Mianwali, 42200, Punjab, Pakistan
First published on 16th June 2023
Abstract
Dengue fever, a highly infectious and rapidly spreading vector borne illness, is classified as a Neglected Tropical Disease (NTD) by WHO because they generally afflict the world's poor and historically have not received as much attention as other diseases. DENV NS2B/NS3 protease and NS5 polymerase are regarded as significant prospective therapeutic targets because of their critical involvement in the viral replication cycle. To date, no specific antiviral agents exist for dengue. The commonly used herbal plant Nigella sativa is known for its antibacterial, antiviral, anti-inflammatory, wound-healing, and dermatological properties. Nevertheless, not enough studies on the antiviral effects of Nigella sativa against DENV are reported. The current study used several prediction techniques to anticipate the oral bioavailability of substances, druglikeness, and non-toxic and non-mutagenic effects which could lead to the development of novel, safer medications. Therefore, the current study was conducted to explore the inhibitory potential of 18 phytochemicals from Nigella sativa against two important enzymes of dengue virus i.e., NS2B/NS3 and NS5. Promising results have been observed for NS2B/NS3 with Taraxerol (−9.1 kcal mol−1), isoquercetin (8.4 kcal mol−1), apigenin, and stigmasterol (−8.3 kcal mol−1). Similarly, NS5 has shown favorable outcomes with apigenin (−9.9 kcal mol−1), rutin (−9.3 kcal mol−1), nigellicine (−9.1 kcal mol−1), and stigmasterol (−8.8 kcal mol−1). MD simulations validated the structural flexibility of the NS2B/NS3-taraxerol and NS5-apigenin docking complexes based on an RMSF value below 5 Å. The study concluded that among the understudied phytocomponents of N. sativa, apigenin, nigellicine, nigellidine, dithymoquinone, taraxerol, campesterol, cycloeucalenol, stigmasterol and beta-sitosterol have been revealed as potential drug candidates, expected to show antiviral activity and promising drug likeliness. Phytochemicals on the short list may serve as inspiration for the creation of new drugs in the future. Further in vitro examination will assist in elucidating the molecular complexity of therapeutic and antiviral capabilities, opening several opportunities for researchers to identify novel medications throughout the drug development process.
Introduction
Dengue fever, a highly infectious viral and fast spreading vector-borne disease is responsible for more than 50 million dengue infections every year, and approximately 250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 people lose their lives to this terrible disease.1,2 The WHO classified dengue as a Neglected Tropical Disease (NTD), indicating that the disease is still largely uncontrolled globally, as these regions are likely to have more dengue cases.3 Dengue virus (DENV) belongs to the Flavivirus genus of family Flaviviridae and is mostly transmitted by Aedes mosquitoes, particularly Aedes aegypti. The four serotypes of dengue are responsible for an extensive spectrum of clinical signs and symptoms, from mild febrile illness to severe and fatal diseases like dengue shock syndrome (DSS) and dengue haemorrhagic fever (DHF).4–6 There are various challenges in creating effective medications against dengue fever, despite the large number of recent research efforts. Due to the fact that DENV exhibits antibody-dependent enhancement (ADE), which allows pre-existing antibodies to attach to DENV and increase the risk of dengue infection.7 Numerous vaccine candidates are undergoing preliminary clinical and preclinical research. The only approved vaccination at the moment is CYD-TDV (Dengvaxia®), however WHO advises using it for those between the ages of 9 and 45.8 Moreover, a re-infection with the same DENV serotype can occur after immunization due to genetic and antigenic variations9,10
000 people lose their lives to this terrible disease.1,2 The WHO classified dengue as a Neglected Tropical Disease (NTD), indicating that the disease is still largely uncontrolled globally, as these regions are likely to have more dengue cases.3 Dengue virus (DENV) belongs to the Flavivirus genus of family Flaviviridae and is mostly transmitted by Aedes mosquitoes, particularly Aedes aegypti. The four serotypes of dengue are responsible for an extensive spectrum of clinical signs and symptoms, from mild febrile illness to severe and fatal diseases like dengue shock syndrome (DSS) and dengue haemorrhagic fever (DHF).4–6 There are various challenges in creating effective medications against dengue fever, despite the large number of recent research efforts. Due to the fact that DENV exhibits antibody-dependent enhancement (ADE), which allows pre-existing antibodies to attach to DENV and increase the risk of dengue infection.7 Numerous vaccine candidates are undergoing preliminary clinical and preclinical research. The only approved vaccination at the moment is CYD-TDV (Dengvaxia®), however WHO advises using it for those between the ages of 9 and 45.8 Moreover, a re-infection with the same DENV serotype can occur after immunization due to genetic and antigenic variations9,10
DENV is a positive sense single-stranded RNA (+ssRNA) virus with an overall genome size of approximately 10.7 kb (i.e., 3391 amino acids) encoding a precursor polyprotein.11 The precursor polyprotein (5′-C-prME-NS1-NS2A-NS2B-NS3-NS4A-NS4BNS5-3′) is further cleaved to produce three structural proteins (capsid protein C, membrane protein prM and envelope protein E) and seven non-structural proteins (NS1, NS2a, NS2b, NS3, NS4a, NS4b and NS5).3,12 The genome of flaviviruses e.g., Dengue virus replicates by the RNA-dependent RNA polymerase (RdRp) domain of non-structural protein 5 (NS5). It also encodes a unique two-component NS2B/NS3 protease for its maturation and infectivity.13,14 Among non-structural proteins, NS3 and NS5 are known to perform a variety of enzymatic reactions. The NS5 protein is highly conserved between the four virus serotypes and is essential for virus replication and transcription. Since there is no enzyme with RdRp activity in humans, NS5 has been reported as an appropriate anti-dengue drug target.15,16 NS3 encodes the RNA helicase serine protease that cleaves the viral polypeptide into functional proteins, nucleocapsid triphosphatase (NTPase), and RNA triphosphatase (RTPase) activities. NS3 protease has apo behaviour and requires a small 14 kDa protein (i.e., NS2B protein), as a cofactor. The NS2B protein stabilizes the NS2B/NS3 complex and NS2B and NS3 perform proteolytic cleavage in the virus.17,18 Thus, these essential enzymatic activities of NS2B/NS3 and NS5 make them an attractive target for antiflaviviral drugs.
Along with the creation of several vaccine candidates, research is being done on antivirals that can stop the spread of viruses or stop illness. The first antiviral substance, acyclovir, entered clinical use in 1960, marking the field of antiviral medications' maturation. Gertrude B. Elion received the Nobel Prize in 1988 for the scientific discovery she made in 1959. Over time, many plants have been used to make rudimentary medicines that have been used to cure a variety of human ailments and illnesses.19 The antiviral activity of phytochemicals has been studied for burgeoning viral infections, and these natural antiviral compounds are the best replacements for antiviral medications or drugs due to their lower toxicity.20,21 Nigella sativa (N. sativa), a member of the family Ranunculaceae, is an annual flowering plant that grows in Mediterranean and Asian countries including Pakistan, India, Indonesia, Afghanistan, and Italy. It is commonly known as black seed, black cumin. In Pakistan it is called Kalonji while in China it is referred to as Hak Jung Chou.22,23 N. sativa has been used in the folk medicine systems such as Unani and Tibb, Ayurveda and Siddha for the treatment of different ailments.24 It is one of the most used herbal medicines throughout the world and pharmacological studies have confirmed the potential of N. sativa as an antidiabetic, anticancer, antioxidant, antimicrobial, hepatoprotective, neuroprotective, gastroprotective, spasmolytic, immunomodulator, analgesic, anti-inflammatory, and bronchodilator (Ahmad et al. 2021 (ref. 52)). Various chemical constituents of N. sativa have been identified, including dithymoquinone, -pinene, -pinene, thymoquinone, thymohydroquinone, thymol, nigellicine, carvacrol, p-cymene, nigellicine, nigllimine, nigellidine, alpha-hederin, and limonene. (Thakur et al. 2021;50 Kabir et al.76 2020).
Despite its clear severity and enormous morbidity, there is currently no direct-acting or host-directed antiviral therapy licensed to treat these illnesses, and the only approved vaccine used in certain countries is restricted to seropositive patients.25,26 A lot of attention has been paid to medicinal plants as a natural source of antiviral medications due to the poor clinical results and side effects of the dengue immunization program. For the treatment of viral infections, medicinal plants have long been thought of as the primary source of pharmacologically bioactive compounds. Additionally, the difficulties brought on by frequent virus mutations and the development of viral resistance to traditional antiviral therapies are igniting interest in natural substances as antiviral alternatives.27,28 The current study was conducted with the purpose of detecting the inhibitory potential of eighteen phytochemicals from Nigella sativa against two important enzymes of the dengue virus (i.e., NS2B/NS3 protease and NS5 polymerase). Molecular docking and MD simulation studies were conducted for ligand–protein complexes to determine the stability of docked structures and several pharmacokinetic analyses evaluated the drug-likeness and toxicity potential of the phytoligands for the evaluation of suitable leads to develop an anti-dengue drug.
Materials and methods
Ligand selection
For the present study, eighteen (18) bioactive components from seed oil and a seed source of black cumin (Nigella sativa) were selected from the literature. The selected phytochemicals have previously been studied for various having inhibitory potential against infectious diseases.23,29–31 3D structures of the selected phytochemicals were downloaded from PubChem in SDF formats and Biovia Discovery studio v2021 was used to convert them into PDB format. Ligands were prepared for docking by fixing charges and adding polar hydrogen before being saved in PDBQT format. The physicochemical properties and molecular formulas and their 3D structures of the selected phytochemicals are given in Table 1.Receptor preparation
The 3D crystal structures of two dengue virus proteins, i.e., NS2B/NNS3 protease (PDB ID: 2FOM) and NS5 polymerase (PDB ID: 4V0Q), available in the RCSB database, were downloaded from Protein Data Bank in PDB format. The downloaded files were opened in Discovery Studio to remove the original ligands and other small molecules attached to the protein structures. The dengue virus NS2BNS3 was originally bound with glycerol (GOL) whereas NS5 polymerase was complexed with S-adenosyl-L-homocysteine (SAH). The original ligand in NS5 and Curcumin, known to possess inhibitory potential against NS2BNS3 were used to serve as positive controls and their binding energies and interactions have been recorded for comparison. Both 3D protein structures (Fig. 1) were refined before docking analyses were performed. The refinement of the selected proteins was carried out by removing irrelevant ions, ligands (if any), and water molecules. Furthermore, polar hydrogen atoms and Kollman charges were added to the receptors and then saved in PDBQT format for docking.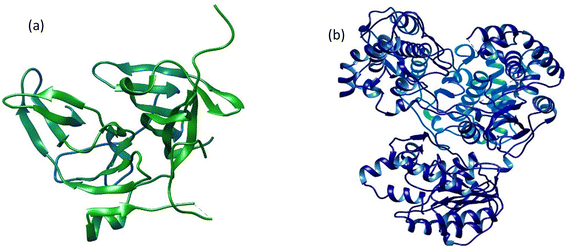 | ||
| Fig. 1 The three-dimensional crystal structures of two dengue virus proteins: (a) NS2B/NS3 protease and (b) NS5 polymerase. | ||
Prediction of activity spectra for substances (PASS) analysis
Prediction of Activity the Spectra for Substances (PASS) analysis program predicts the biological properties, pharmacological properties, drug-likeness, probable side effects, and mode of action of understudied phytoconstituents based on their structure–activity relationship with an established chemical entity. PASS analysis was carried out in this study through a variety of online and offline tools, mentioned below29,32Lipinski's rule of five
Lipinski's rule of five describes the molecular properties of drug compounds based on important pharmacokinetic parameters such as absorption, metabolism, distribution, and excretion. The rule is helpful in the design and development of drugs.33 The drug-likeness of the 18 phytochemicals of N. sativa included in the study was expected using the Lipinski rule of five. The online tool Molinspiration v2021.03 (“https://www.molinspiration.com/cgi-bin/properties”) was used to calculate the drug-likeness of phytochemicals and compared them with standard drugs. The drug-likeness was calculated using parameters such as the logarithm of the compound partition coefficient between n-octanol and water (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P 5), molecular weight (MW 500), number of hydrogen bond donors (NOHNH ≤ 5), hydrogen bond acceptor sites (NON ≤ 10), topological polar surface area (defined as a sum of surfaces of polar atoms in a molecule, TPSA ≤ 140 Å2), number of rotatable bonds (≤10). An orally active drug should not have more than one Lipinski violation to maintain its bioavailability.34
P 5), molecular weight (MW 500), number of hydrogen bond donors (NOHNH ≤ 5), hydrogen bond acceptor sites (NON ≤ 10), topological polar surface area (defined as a sum of surfaces of polar atoms in a molecule, TPSA ≤ 140 Å2), number of rotatable bonds (≤10). An orally active drug should not have more than one Lipinski violation to maintain its bioavailability.34
Bioactivity score prediction
Drug score values indicate the absolute potential of a prospective complex to be a drug candidate. A web-based tool, Molinspiration version 2021.03, predicted a bioactivity score of the phytoconstituents against the human receptors like G protein-coupled receptors (GPCRs), kinases, proteases, ion channels, enzymes, and nuclear receptors. If the bioactivity score is more than 0.0, then the compound is dynamic (active); if it is in the range between −5.0 and 0.0, then the complex is moderately active and if the bioactivity score is less than −5.0, then it is considered to be idle (inactive).35Toxicity potential study
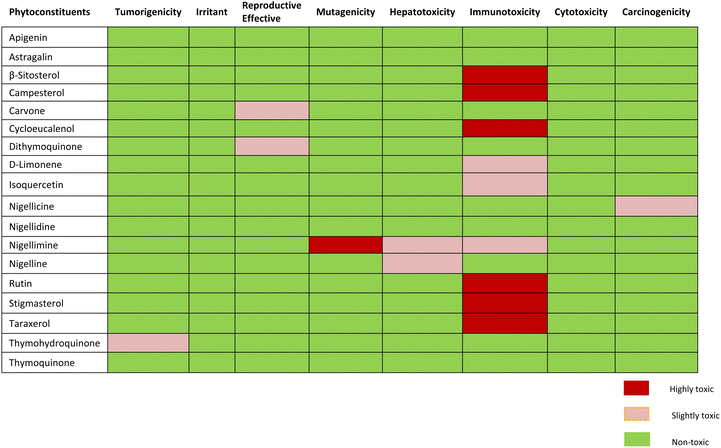
Toxicity risk assessment gives exploratory information on the potential side effects of phytoconstituents that can be employed in drug discovery and development. The Protox-II server and OSIRIS Data Warrior V5.2.1 software were used to observe drug-likeness and drug toxicity risk characteristics such as drug-likeness, tumorigenicity, carcinogenicity, immunotoxicity, hepatotoxicity, mutagenicity, reproductive and irritating effects.29,36 The understudy compounds have also been evaluated for prediction of their LD50 value and Drug toxicity class. The LD50 values for toxic dosages are frequently expressed in mg kg−1 body weight. The median lethal dosage, or LD50, is the quantity at which 50% of test subjects pass away after being exposed to a substance. According to the Globally Harmonized System (GHS) of categorization and labeling of substances, toxicity classes are established.37,38Pharmacokinetic property prediction
The absorption, distribution, metabolism, excretion, and toxicity (ADMET) properties of all phytoconstituents in this study were predicted using two online software, i.e., the Swiss-ADME software and ADMETlab. These programs examine a compound's important pharmacokinetic properties such as blood–brain barrier (BBB), distribution, GI absorption, metabolism as a P-glycoprotein (P-gp) substrate, cytochrome P450 s such as CYP1A2, CYP2C19, CYP2C9, CYP2D6, CYP3A4 inhibitor, and lipophilicity for plasma membrane absorption.39,40Docking
AutoDock Tools 1.5.6 was used to integrate phytoligands, which were considered “flexible,” into the protein targets that were considered “rigid.” AutoDock Vina is easy to use, has a fast processing speed, calculates the dimensions of grid boxes automatically, and conveniently estimates binding sites.41,42 In the present study, a grid box size of 40 × 40 × 40 (x, y, and z) and a grid spacing of 0.375 were used for both receptor proteins. The center of the grid for NS2B/NS3 protease was set at positions 0.456, −17.036, and 13.989 for x, y, and z, respectively, while for NS5 polymerase the centers (x, y, and z) were set at 24.892, 162.151, and 24.605, respectively. For each ligand, a configuration file containing grid box attributes was created and saved in.txt format. To begin the docking analysis, a set of codes (commands) was written in the command prompt to generate the output score.43 The results displayed the values of binding energy or the Gibbs free energy (G) in kcal mol−1, and the ligand with the maximum negative value was considered to have the highest binding affinity for a particular target protein. The results displayed the values of binding affinity and Gibbs free energy (G) in kcal mol−1, and the ligand with the maximum negative value was considered to have the highest binding affinity for a particular target protein. From the nine different conformations generated, the best-fit model (Model 1) was selected. The interaction profiles were displayed in 2D using the Discovery Studio v2021 and then evaluated.Molecular dynamic simulations
The investigation of the structural dynamics of the top docking complexes and the identification of motion were both carried out using iMODS. To determine deformability, B-factor, eigenvalue, variance, covariance map, and elastic network, normal mode analysis was performed on the docked complexes. The input PDB files were uploaded to iMODS with the default settings of 300 K constant temperature, 1 atm constant pressure, and 50 ns molecular dynamic simulation for each complex.44,45 The best docked structures were validated for their protein flexibility by performing molecular dynamic simulations using the CABS-flex 2.0 server and displayed as RMSF values (root mean square fluctuation). Simulations in CABS-flex were conducted using default parameters, i.e., protein rigidity: 1.0, number of cycles: 50, number of cycles between trajectories: 50, temperature range: 1.40, and a random number of generator seed of 5711. CABS-flex offers fast, high-resolution (10 ns) protein flexibility simulation at extremely reduced system constraints.46,47Results
Lipinski's rule of five
The physicochemical properties of eighteen phytoconstituents of N. sativa were assessed by PASS analysis by employing Lipinski's rule of five (Table 2). Three phytoconstituents (i.e., astragalin, isoquercetin and rutin) exhibited more than 1 Lipinski violation as they had more than five hydrogen bond donors (NOHNH) and more than ten hydrogen bond acceptors (NON) and their topological polar surface area (TPSA) was calculated to be greater than 140 Å2. Five compounds (i.e., β-sitosterol, campesterol, cycloeucalenol, stigmasterol and taraxerol) exhibited 1 violation as the value of clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P for these compounds was calculated to be greater than the standard value (i.e., ≤5). The remaining ten phytocompounds exhibited no violation. Since an ideal lead compound should not have more than one violation, fifteen compounds out of eighteen have met the criteria.
P for these compounds was calculated to be greater than the standard value (i.e., ≤5). The remaining ten phytocompounds exhibited no violation. Since an ideal lead compound should not have more than one violation, fifteen compounds out of eighteen have met the criteria.
| Phytoconstituents | clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P (≤5) P (≤5) |
MW (≤500) | NOHNH (≤5) | NON (≤10) | TPSA (≤140 Å2) | Number of rotatable bonds (≤10) | Lipinski's violations |
|---|---|---|---|---|---|---|---|
| Apigenin | 2.46 | 270.24 | 3 | 5 | 90.89 | 1 | 0 |
| Astragalin | 0.125 | 448.38 | 7 | 11 | 190.275 | 4 | 2 |
| β-Sitosterol | 8.62 | 414.718 | 1 | 1 | 20.228 | 6 | 1 |
| Campesterol | 8.305 | 400.691 | 1 | 1 | 20.228 | 5 | 1 |
| Carvone | 2.513 | 150.221 | 0 | 1 | 17.071 | 1 | 0 |
| Cycloeucalenol | 7.617 | 426.729 | 1 | 1 | 20.228 | 5 | 1 |
| Dithymoquinone | 1.703 | 328.408 | 0 | 4 | 68.284 | 2 | 0 |
| D-Limonene | 3.615 | 136.238 | 0 | 0 | 0 | 1 | 0 |
| Isoquercetin | −0.364 | 464.379 | 8 | 12 | 210.503 | 4 | 2 |
| Nigellicine | 1.652 | 246.266 | 1 | 5 | 64.238 | 1 | 0 |
| Nigellidine | 3.034 | 294.354 | 1 | 4 | 47.167 | 1 | 0 |
| Nigellimine | 1.894 | 203.241 | 0 | 3 | 31.36 | 2 | 0 |
| Nigelline | 2.107 | 195.218 | 1 | 4 | 47.566 | 4 | 0 |
| Rutin | −1.063 | 610.521 | 10 | 16 | 269.427 | 6 | 3 |
| Stigmasterol | 7.869 | 412.702 | 1 | 1 | 20.228 | 5 | 1 |
| Taraxerol | 8.023 | 426.729 | 1 | 1 | 1 | 1 | 1 |
| Thymohydroquinone | 3.258 | 166.22 | 2 | 2 | 40.456 | 1 | 0 |
| Thymoquinone | 1.90 | 164.20 | 0 | 2 | 34.14 | 1 | 0 |
Prediction of bioactivity score
The prediction of the bioactivity score revealed that Apigenin was biologically active as nuclear receptor ligand, kinase and enzyme inhibitor while remaining moderately active as GPCR ligand and ion channel modulator. β-Sitosterol, campesterol, taraxerol and cycloeucalenol have demonstrated significant activity as GPCR ligand, kinase inhibitor, ion channel modulator, nuclear receptor ligand, protease inhibitors and enzyme inhibitors as all their values were above the threshold level as described in materials and methods. Nigellidine was active as GPCR ligands, ion channel modulators, nuclear receptor ligands, and enzyme inhibitors while being moderately active as protease inhibitors. Dithymoquinone and stigmasterol were moderately active as ion channel modulator, kinase inhibitor and protease inhibitor and active as GPCR ligand, nuclear receptor ligand and enzyme inhibitor. Cycloeucalenol exhibited the highest value as protease inhibitor (i.e., 0.1) and as enzyme inhibitor (i.e., 0.6). Thymohydroquinone and thymoquinone were considered inactive because their maximum values were below the threshold level. The details of the bioactivity score prediction of all understudy phytoconstituents are given in Table 3.| Phytoconstituents | GPCR ligand | Ion channel modulator | Kinase inhibitor | Nuclear receptor ligand | Protease inhibitor | Enzyme inhibitor |
|---|---|---|---|---|---|---|
| a >0.00 active, −0.50 to 0.00 moderately active, <−0.50 inactive. | ||||||
| Apigenin | −0.07 | −0.09 | 0.18 | 0.34 | −0.25 | 0.26 |
| Astragalin | 0.06 | −0.05 | 0.10 | 0.20 | −0.05 | 0.41 |
| β-Sitosterol | 0.14 | 0.04 | −0.51 | 0.73 | 0.07 | 0.51 |
| Campesterol | 0.11 | 0.01 | −0.48 | 0.71 | 0.01 | 0.50 |
| Carvone | −1.23 | −0.30 | −2.51 | −0.54 | −1.21 | −0.45 |
| Cycloeucalenol | 0.14 | 0.14 | −0.37 | 0.92 | 0.10 | 0.61 |
| Dithymoquinone | 0.18 | −0.09 | −0.48 | 0.14 | −0.10 | 0.10 |
| D-Limonene | −0.91 | −0.27 | −2.01 | −0.34 | −1.38 | −0.21 |
| Isoquercetin | 0.06 | −0.04 | 0.13 | 0.20 | −0.06 | 0.42 |
| Nigellicine | −0.15 | 0.00 | −0.18 | −0.29 | −0.57 | 0.20 |
| Nigellidine | 0.07 | 0.01 | 0.29 | 0.10 | −0.32 | 0.19 |
| Nigellimine | 0.47 | −0.17 | −0.40 | −0.75 | −0.64 | −0.12 |
| Nigelline | −0.77 | −0.46 | −0.77 | −1.01 | −0.98 | −0.49 |
| Rutin | 0.05 | −0.52 | −0.14 | −0.23 | −0.07 | 0.12 |
| Stigmasterol | 0.12 | −0.08 | −0.48 | 0.74 | −0.02 | 0.53 |
| Taraxerol | 0.21 | 0.02 | −0.20 | 0.54 | 0.00 | 0.49 |
| Thymohydroquinone | −0.92 | −0.44 | −1.06 | −0.54 | −1.17 | −0.46 |
| Thymoquinone | −1.40 | −0.31 | −1.27 | −1.47 | −1.45 | −0.40 |
Pharmacokinetic property prediction
The ADMET analysis (Table 4) of understudied phytoconstituents suggested that apigenin, astragalin, isoquercetin, and rutin were unable to cross the blood–brain barrier, while all of the remaining selected phytoligands showed positive results, demonstrating that they can easily cross the BBB. Only β-sitosterol, stigmasterol, and nigellidine were found to be positive as permeability glycoprotein substrates (P-gp substrates), while the rest of the phytoconstituents were found to be negative. These results suggest that being non-Pgp substrates, the phytoconstituents will persist longer in the cells, leading to their increased pharmacokinetic efficacy. For incessant plasma concentrations and enhanced bioavailability, the understudy compounds were expected to inhibit the five classes of cytochromes P450, i.e., CYP1A2, CYP2C19, CYP2C9, CYP2D6, and CYP3A4. The results obtained confirmed CYP1A2 inhibition by apigenin, nigellidine, nigellimine, nigelline, and thymohydroquinone, the inhibition of CYP2D6 by apigenin, and the inhibition of nigellidine and CYP3A4 by apigenin alone. The rest of the compounds did not show inhibitory potential against cytochrome P450. The consensus lipophilicity (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P o/w) value was calculated to analyze the lipophilic potential of the phytoconstituents, resulting in the lipid-soluble nature of most of the compounds except astragalin, isoquercetin, and rutin. High human intestinal absorption (GI absorption) was observed for apigenin, carvone, dithymoquinone, nigellicine, nigellidine, nigellimine, nigelline, thymohydroquinone, and thymoquinone.
P o/w) value was calculated to analyze the lipophilic potential of the phytoconstituents, resulting in the lipid-soluble nature of most of the compounds except astragalin, isoquercetin, and rutin. High human intestinal absorption (GI absorption) was observed for apigenin, carvone, dithymoquinone, nigellicine, nigellidine, nigellimine, nigelline, thymohydroquinone, and thymoquinone.
| Phytoconstituents | CYP1A2 inhibition | CYP2C19 inhibition | CYP2C9 inhibition | CYP2D6 inhibition | CYP3A4 inhibition | P-gp substrate | BBB | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P o/w P o/w |
GI absorption |
|---|---|---|---|---|---|---|---|---|---|
| Apigenin | Yes | No | No | Yes | Yes | No | No | 2.11 | High |
| Astragalin | No | No | No | No | No | No | No | −0.09 | Low |
| β-Sitosterol | No | No | No | No | No | Yes | Yes | 8.025 | Low |
| Campesterol | No | No | No | No | No | No | Yes | 2.385 | Low |
| Carvone | No | No | No | No | No | No | Yes | 2.44 | High |
| Cycloeucalenol | No | No | No | No | No | No | Yes | 7.651 | Low |
| Dithymoquinone | No | No | No | No | No | No | Yes | 2.61 | High |
| D-Limonene | No | No | Yes | No | No | No | Yes | 3.37 | Low |
| Isoquercetin | No | No | No | No | No | No | No | −0.48 | Low |
| Nigellicine | No | No | No | No | No | No | Yes | 1.45 | High |
| Nigellidine | Yes | No | No | Yes | No | Yes | Yes | 2.82 | High |
| Nigellimine | Yes | No | No | No | No | No | Yes | 2.29 | High |
| Nigelline | Yes | No | No | No | No | No | Yes | 1.79 | High |
| Rutin | No | No | No | No | No | No | No | −0.038 | Low |
| Stigmasterol | No | No | No | No | No | Yes | Yes | 7.748 | Low |
| Taraxerol | No | No | No | No | No | No | Yes | 6.782 | Low |
| Thymohydroquinone | Yes | No | No | No | No | No | Yes | 2.39 | High |
| Thymoquinone | No | No | No | No | No | No | Yes | 1.85 | High |
Toxicity potential study
Assessment of expected drug-likeness and toxicity potential of selected phytochemicals by data warrior and protox-II server revealed that apigenin, astragalin, and thymoquinone were non-toxic. β-sitosterol, campesterol and cycloeucalenol, rutin, stigmasterol and taraxerol were appeared to have projected immunotoxin effects. Carvone and dithymoquinone were expected to have mild toxic effects on the reproductive system, while nigellicine appeared to be slightly carcinogenic. Nigelline and nigellimine have shown potential for causing hepatotoxicity whereas nigellimine is also positive for mild immunotoxicity and high mutagenicity. None of the phytoconstituents were found to be an irritant. The expected toxicity risk assessment of phytoconstituents of N. sativa is presented as in Table 5. For a compound to become a good drug candidate, its druglikeness score should be 1 or close to 1. If the drug score is zero or negative, then the understudy compound is not a good drug candidate. Nigellicine, nigellidine, apigenin, rutin, D-limonene, and stigmasterol have shown positive scores for drug-likeness. Computational calculations of predicted LD50 and toxicity class renders apigenin, astragalin, cycloeucalenol, dithymoquinone, D-limonene, isoquercetin and rutin to class-5 while β-sitosterol, campesterol, carvone, nigellicine and nigellidine, nigelline and stigmasterol to class-4. Nigellimine (pink highlight) was the only compound in class-III (i.e., toxic if swallowed) whereas taraxerol qualified to be sole potentially non-toxic lead compound having the highest LD50 value i.e., 7000 mg kg−1 (green highlight). The details of drug similarity and probable toxicity assessment of phytochemicals are given in Table 6.| Phytoconstituents | Druglikeness | Predicted LD50 | Predicted toxicity class |
|---|---|---|---|
| a Class I: fatal if swallowed (LD50 ≤ 5); Class II: fatal if swallowed (5 < LD50 ≤ 50). Class III: toxic if swallowed (50 < LD50 ≤ 300); Class IV: harmful if swallowed (300 < LD50 ≤ 2000). Class V: may be harmful if swallowed (2000 < LD50 ≤ 5000); Class VI: non-toxic (LD50 > 5000). | |||
| Apigenin | 0.28194 | 2500 mg kg−1 | Class V |
| Astragalin | −3.6679 | 5000 mg kg−1 | Class V |
| β-Sitosterol | −4.475 | 890 mg kg−1 | Class IV |
| Campesterol | −8.1908 | 890 mg kg−1 | Class IV |
| Carvone | −18.988 | 1640 mg kg−1 | Class IV |
| Cycloeucalenol | −7.6333 | 5000 mg kg−1 | Class V |
| Dithymoquinone | −21.855 | 2300 mg kg−1 | Class V |
| D-Limonene | 0.9165 | 4400 mg kg−1 | Class V |
| Isoquercetin | −3.6679 | 5000 mg kg−1 | Class V |
| Nigellicine | 2.1696 | 1300 mg kg−1 | Class IV |
| Nigellidine | 1.1929 | 1000 mg kg−1 | Class IV |
| Nigellimine | −1.7979 | 1300 mg kg−1 | Class III |
| Nigelline | −2.6175 | 1800 mg kg−1 | Class IV |
| Rutin | 1.9337 | 5000 mg kg−1 | Class V |
| Stigmasterol | 1.2217 | 890 mg kg−1 | Class IV |
| Taraxerol | −2.422 | 70000 mg kg−1 | Class VI |
| Thymohydroquinone | −2.3359 | 1000 mg kg−1 | Class IV |
| Thymoquinone | −1.1996 | 2400 mg kg−1 | Class V |
Docking analysis
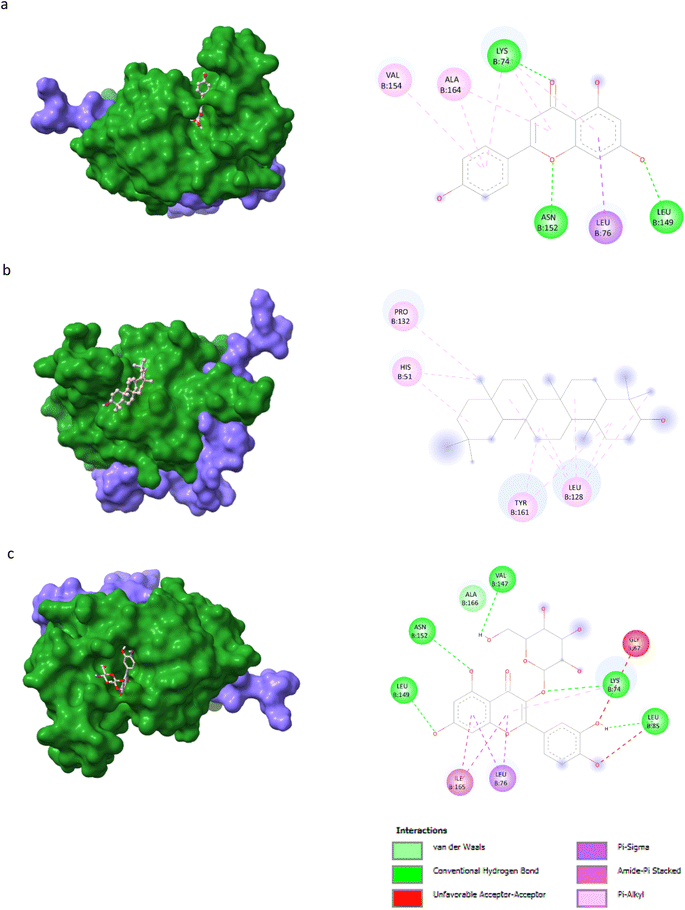
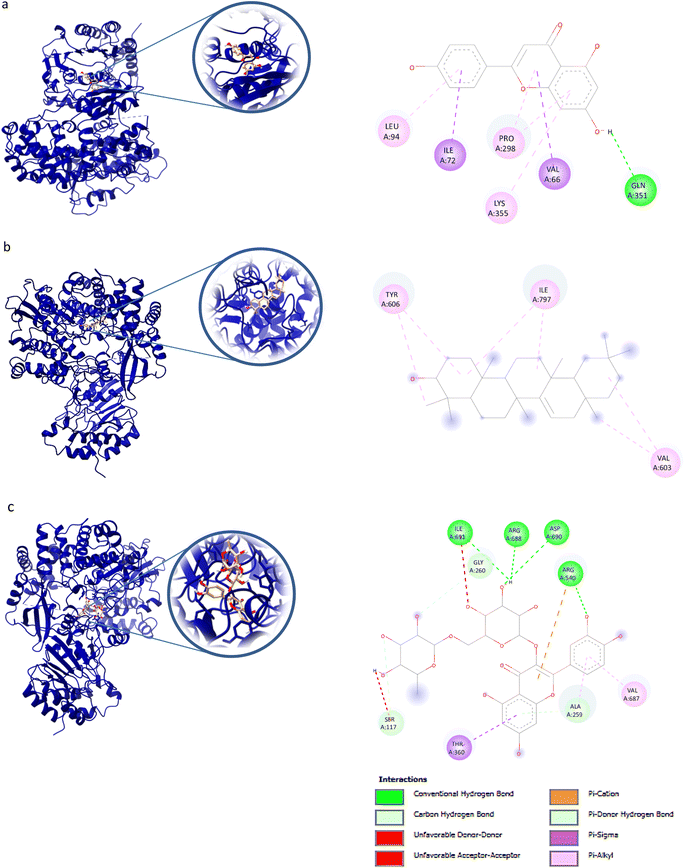
AutoDock Vina v1.5.6 was used to dock the target proteins with eighteen phytochemicals from Nigella sativa plants. The phytoligands showed different interactions at the binding pockets, displaying several binding affinities. The phytochemicals showed binding energies for NS2BNS3 ranging from −5.5 kcal mol−1 to −9.1 kcal mol−1. Taraxerol showed the highest binding affinity with lowest binding energy (i.e., −9.1 kcal mol−1), followed by Isoquercetin (i.e., −8.4 kcal mol−1), and the lowest binding affinity was recorded for D-limonene and nigelline (i.e., −5.5 kcal mol−1). The highest number of interacting residues were observed in the interaction of NS2BNS3 with astragalin, they have ten interacting amino acid residues (i.e., Asn167, Trp89, Gly87, Ala166, Val147, Glu88, Ile165, Asn152, Leu86, Lys74). Taraxerol, which has showed highest binding affinity containing 4 interacting amino acid residues i.e., Pro132, His51, Leu128, Tyr161. D-Limonene with least binding affinity showed only two interacting amino acid residues (i.e., Tyr161, Leu128). Docking interactions of selected phytochemicals from N. sativa with NS2BNS3 protease are shown in Table 7. The positive control (curcumin) has shown the binding affinity equals to Dithymoquinone i.e., −7.2 kcal mol−1 having two interacting residues i.e., Leu128, Pro132, Tyr161.The docking poses were viewed by ChimeraX whereas interacting amino acid residues were visualized by Discovery Studio. The top 3 docking poses for NS2BNS3 protease are shown in Fig. 2. The docking results of compounds against NS2BNS3 can be compared in following order: Taraxerol > Isoquercetin > Apigenin = β-Sitosterol = Cycloeucalenol = Stigmasterol > Rutin > Campesterol > Astragalin > Nigellicine > Nigellidine > Dithymoquinone > Nigellimine > Carvone = Thymohydroquinone = Thymoquinone > D-Limonene = Nigelline.| Ligand | Receptor proteins | Binding energies (kcal mol−1) | Interacting amino acid residues |
|---|---|---|---|
| Apigenin | NS2BNS3 | −8.3 | Val154, Ala164, Lys74, Asn152, Leu76, Leu149 |
| Astragalin | NS2BNS3 | −8.0 | Asn167, Trp89, Gly87, Ala166, Val147, Glu88, Ile165, Asn152, Leu86, Lys74 |
| β-Sitosterol | NS2BNS3 | −8.3 | His51, Phe130, Leu128 |
| Campesterol | NS2BNS3 | −8.1 | His51, Leu128, Phe130, Pro132 |
| Carvone | NS2BNS3 | −5.6 | Tyr150, Gly153, Pro132, Tyr161, Leu128 |
| Cycloeucalenol | NS2BNS3 | −8.3 | His51, Pro132, Leu128 |
| Dithymoquinone | NS2BNS3 | −7.2 | His51, Gly153, Pro132, Leu128 |
| D-Limonene | NS2BNS3 | −5.5 | Tyr161, Leu128 |
| Isoquercetin | NS2BNS3 | −8.4 | Leu149, Asn152, Ala166, Val147, Gly87, Lys74, Leu85, Leu76, Ile165 |
| Nigellicine | NS2BNS3 | −7.4 | Leu128, Pro132, Gly151, Tyr161 |
| Nigellidine | NS2BNS3 | −7.3 | Ala164, Leu76, Lys74, Asn167 |
| Nigellimine | NS2BNS3 | −6.2 | Leu76, Trp83, Leu85, Val147, Lys74, Asn152 |
| Nigelline | NS2BNS3 | −5.5 | Lys42, Val59, Val146, Lys145 |
| Rutin | NS2BNS3 | −8.2 | Leu128, Pro132, Gly151, Arg54, Trp50, Lys73, Asp75 |
| Stigmasterol | NS2BNS3 | −8.3 | His51, Leu128, Phe130, Pro132 |
| Taraxerol | NS2BNS3 | −9.1 | Pro132, His51, Leu128, Tyr161 |
| Thymohydroquinone | NS2BNS3 | −5.6 | Ala164, Val154, Ile123, Lys73 |
| Thymoquinone | NS2BNS3 | −5.6 | Asn152, Ile123, Ala164, Lys74, Val154 |
| Curcumin (positive control) | NS2BNS3 | −7.2 | Leu128, Pro132, Tyr161 |
For NS5 the phytochemicals showed binding energies ranging from −6.0 kcal mol−1 to −9.9 kcal mol−1 (Table 8). The highest binding affinity was recorded for apigenin with highest binding energy (i.e., −9.9 kcal mol−1), with rutin being the second highest (i.e., −9.3 kcal mol−1), while D-limonene bound with the lowest affinity (i.e., 6.0 kcal mol−1). Apigenin while having the highest binding affinity involved 06 interacting amino acid residues (i.e., Leu94, Ile72, Pro298, Lys355, Val66, Gln351). Rutin with second highest binding affinity involved Ile691, Gly260, Arg688, Asp690, Arg540, Val687, Ala259, and Thr360 as their 8 interacting amino acids. Highest number of interacting residues were observed in docking models of NS5-nigellicine (i.e., Arg352, Lys74, Arg352, Glu151, Val66, Leu94, Asn690, Ile165, Pro298) and NS5-nigellidine (i.e., Pro73, Lys96, Ile72, Leu94, Val66, Lys355, Pro298, Arg581, Glu296), both displayed 09 residues. The lowest i.e., 03 number of binding residues were involved in NS5-dithymoquinone (Val687, His52, Pro692) and NS5-taraxerol (Tyr606, Ile797, Val603). The positive control S-adenosyl-L-homocysteine (SAH) showed a binding affinity lesser than the majority of understudy phytocompounds. The binding energy −7.2 kcal mol−1 was higher than apigenin, astragalin, β-sitosterol, campesterol, cycloeucalenol, dithymoquinone, isoquercetin, nigellicine nigellidine, rutin, stigmasterol and taraxerol. The docking poses and interaction of amino acid residues was visualized by chimeraX and Discovery studio, respectively (the selected 3 docking poses for NS5 polymerase are shown in Fig. 3). 3D and 2D line models of remaining compounds are given in ESI Fig. S1.† The docking results of compounds can be summed up based on their binding affinities against NS5 in following order: Apigenin > Nigellicine > Rutin > Taraxerol > Stigmasterol > Isoquercetin > Astragalin = Campesterol > Nigellidine > β-Sitosterol = Cycloeucalenol > Dithymoquinone > Thymoquinone > Thymohydroquinone > Carvone > Nigelline > Nigellimine > D-Limonene.
| Ligand | Receptor proteins | Binding energies (kcal mol−1) | Interacting amino acid residues |
|---|---|---|---|
| Apigenin | NS5 | −9.9 | Leu94, Ile72, Pro298, Lys355, Val66, Gln351 |
| Astragalin | NS5 | −8.5 | Thr51, Ile691, Asp690, His52, Gln693, Val687, Ala259 |
| β-Sitosterol | NS5 | −8.0 | His52, Ile691, Ala535, Val687, Ala259, Tyr119 |
| Campesterol | NS5 | −8.5 | Tyr119, Ala535, His52, Val687, Ala259, Pro363, Tyr119 |
| Carvone | NS5 | −6.4 | Pro298, Val66, Ile72, Arg581, Pro582 |
| Cycloeucalenol | NS5 | −8.0 | Val603, Phe485, Ile797, Tyr606, Asp663 |
| Dithymoquinone | NS5 | −7.5 | Val687, His52, Pro692 |
| D-Limonene | NS5 | −6.0 | Pro298, Val66, Lys95, Pro73, Leu94, Ile72 |
| Isoquercetin | NS5 | −8.6 | His52, Thr51, Ile691, Asp690, Gln693, Val687, Ala259 |
| Nigellicine | NS5 | −9.1 | Arg352, Lys74, Arg352, Glu151, Val66, Leu94, Asn690, Ile165, Pro298 |
| Nigellidine | NS5 | −8.4 | Pro73, Lys96, Ile72, Leu94, Val66, Lys355, Pro298, Arg581, Glu296 |
| Nigellimine | NS5 | −6.1 | Lys300, Lys355, Tyr119, Thr360, His52, Ala259 |
| Nigelline | NS5 | −6.2 | Glu296, Pro298, Asn69, Lys355, Arg352, Val66 |
| Rutin | NS5 | −9.3 | Ile691, Gly260, Arg688, Asp690, Arg540, Val687, Ala259, Thr360 |
| Stigmasterol | NS5 | −8.8 | His52, Val687, Ala259, Tyr119, Pro363 |
| Taraxerol | NS5 | −9.0 | Tyr606, Ile797, Val603 |
| Thymohydroquinone | NS5 | −6.5 | Lys355, Lys95, Leu94, Pro298 |
| Thymoquinone | NS5 | −6.7 | Lys95, Leu94, Pro298, Lys355 |
| S-Adenosyl-L-homocysteine (SAH) (positive control) | NS5 | −7.2 | Glu356, Lys300, Val687, Ala259, Thr51, Ile691, His52, Ala535 |
MD simulations
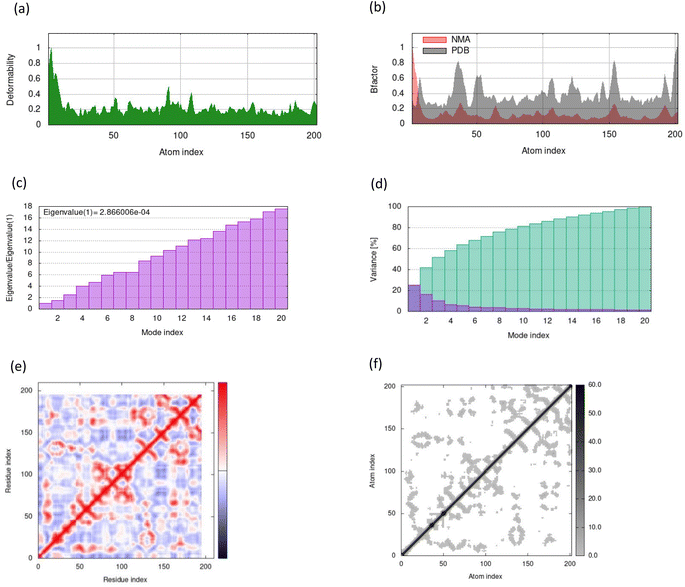
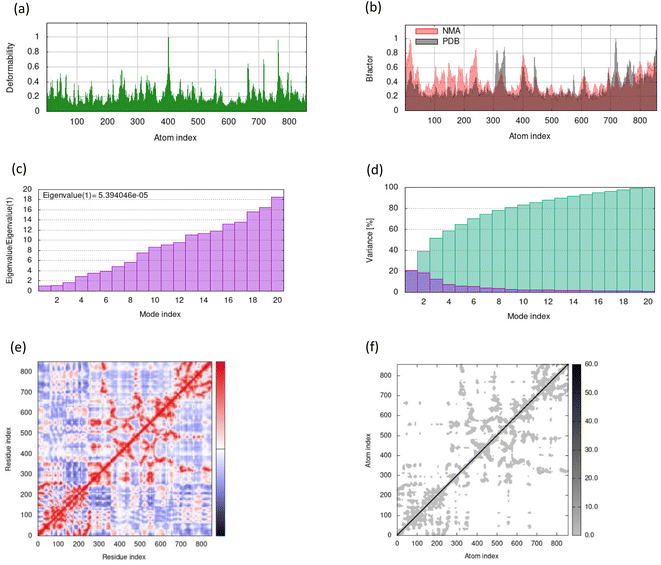
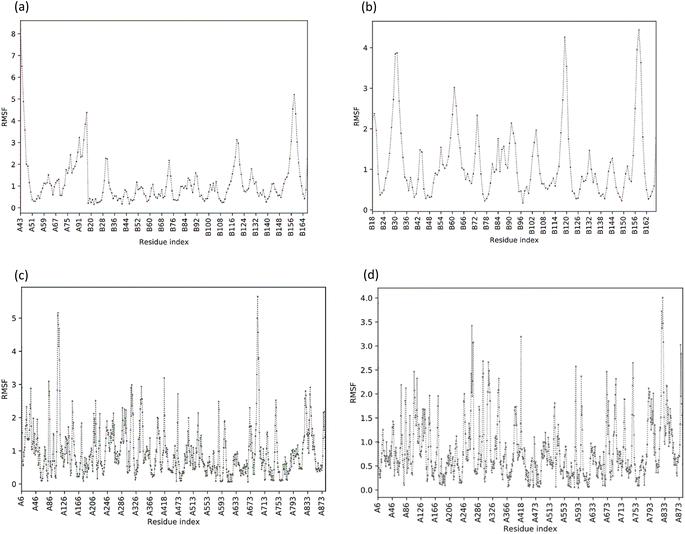
MD simulations in normal mode analysis have been performed for NS2BNS3 and NS5-apigenin based on their highest docking score. The highest peaks, which correspond to protein areas with high deformability, are shown in Fig. 4A and 5A. The normal mode analysis and the PDB field of the complexes are compared in the B-factor graphics. The B-factor column provides an averaged RMS since many PDB files of averaged NMR models lack B-factors (Fig. 4B and 5B). Every normal mode has an associated eigenvalue (Fig. 4C and 5C) and variance (Fig. 4D and 5D). The energy needed to distort the composite is correlated with the eigenvalue, and the smaller the value, the simpler the deformation. Correlations between residues in a complex are shown by the covariance matrix in Fig. 4E and 5E. The white hue denotes uncorrelated motion, whereas the red color depicts a respectable connection between residues. Anticorrelations are further illustrated by the blue color, the better the complex, the better the correlation. The relationships between the atoms are defined by the elastic maps of the complexes, where firmer regions are shown by darker gray areas and are illustrated in Fig. 5F and 6F.The structural flexibility of the NS2BNS3-taraxerol docking complex and the NS5-apigenin complex was evaluated by Cabs-flex 2.0. Our results indicated that the RMSF value of NS2BNS3 when bound with naturally existing ligan glycerol was as high as 08 Å and it reduced after binding with taraxerol, and the RMSF value of the complex was recorded below 4.5 Å (Fig. 6A and B). Similarly, after binding with apigenin, the RMSF value of the NS5 protein was reduced (4.0 Å) than the value while being complexed with SAH (Fig. 6C and D). Since the RMSF value recorded for selected complexes was closer to the ideal value, i.e., 3.8, the complexes were said to have stable interactions.
Discussion
Discovery and the development of novel drugs are expensive and time-consuming processes.39 A planned computational approach for rational drug design has been useful in assessing the potential of selected phytochemicals before wet-lab testing by predicting their drug likeness, oral bioavailability, efficacy, toxicity risk assessment, and so on.29,32 In silico screening and prediction of phytoconstituents with good pharmacodynamic and pharmacokinetic properties is time-saving and cost-effective. The current study attempted to investigate the antiviral potential of selected phytochemicals from N. sativa that have been previously documented to possess antiviral, antibacterial, and antifungal potentials. It is one of the most widely used herbal plants in the world. N. sativa oil was given to a 46 year-old HIV-positive patient in a documented clinical case and the findings of the investigation demonstrated full seroversion and rescue.48,49 HCV-RNA was undetectable in 195 HCV patients who received N. sativa and chloroquine combination treatment, according to an initial trial.50–53 The current study attempted to investigate the antiviral potential of selected phytochemicals such as apigenin,54 nigellicine, thymohydroquinone,55 nigellicine, dithymoquinone,55 nigllimine, and nigellidine, that have been tested in vitro to possess antiviral potentials.To compute the drug potential of understudy compounds, all 18 active phytoconstituents were tested using Lipinski's rule of five which envisages that strong absorption/permeation is expected when the MW < 500, clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P ≤ 5.0, there are ≤5 H-bond donors and ≤10 H-bond acceptors with no more than 1 Lipinski's violation otherwise its bioavailability is compromised.56,57 Out of 18 subjected phytoconstituents from N. sativa three compounds viz. astragalin, isoquercetin and rutin exhibited more than one Lipinski's violation and remaining fifteen phytocompounds qualified the criteria for being a good lead compound (Table 2). The phytocomponents were further evaluated for their drug-likeness capacity and to prevent inappropriate chemicals for future drug screening and to begin in vitro and in vivo evaluation, toxicity potential and bioavailability must be assessed.58 Apigenin, campesterol, cycloeucalenol, β-sitosterol, dithymoquinone, stigmasterol, taraxerol, nigellidine, and nigellicine have shown significant bioavailability (Table 3) and were found to have potential as significant protease and enzyme inhibitors.59–61 The majority of phytocomponents, including apigenin, campesterol, carvone, nigellicine, nigellidine, nigellimine, and nigelline, showed high human intestinal absorption (GI absorption), lipophilicity and blood–brain permeation, indicating high absorption and transport kinetics in the stomach and the ability to easily cross the BBB (Table 4). Toxicity risk assessment BBB (Table 5) through LD50 value and toxicity class BBB suggests that apigenin, astragalin, taraxerol and thymoquinone are expected to be safe as they exhibited non-mutagenic, non-tumorigenic, non-carcinogenic, non-irritant behaviour with no threats to reproductive systems. In compared to toxicity class 1–3, the LD50 values for oral acute toxicity of the majority of understudy phytocompounds fell into toxicity class 4 and 5, making them expectedly less dangerous. Only one compound was found in toxicity class 3 (i.e., nigellimine) whereas taraxerol secured place in class VI and was rendered as expectedly non-toxic (Table 6).
P ≤ 5.0, there are ≤5 H-bond donors and ≤10 H-bond acceptors with no more than 1 Lipinski's violation otherwise its bioavailability is compromised.56,57 Out of 18 subjected phytoconstituents from N. sativa three compounds viz. astragalin, isoquercetin and rutin exhibited more than one Lipinski's violation and remaining fifteen phytocompounds qualified the criteria for being a good lead compound (Table 2). The phytocomponents were further evaluated for their drug-likeness capacity and to prevent inappropriate chemicals for future drug screening and to begin in vitro and in vivo evaluation, toxicity potential and bioavailability must be assessed.58 Apigenin, campesterol, cycloeucalenol, β-sitosterol, dithymoquinone, stigmasterol, taraxerol, nigellidine, and nigellicine have shown significant bioavailability (Table 3) and were found to have potential as significant protease and enzyme inhibitors.59–61 The majority of phytocomponents, including apigenin, campesterol, carvone, nigellicine, nigellidine, nigellimine, and nigelline, showed high human intestinal absorption (GI absorption), lipophilicity and blood–brain permeation, indicating high absorption and transport kinetics in the stomach and the ability to easily cross the BBB (Table 4). Toxicity risk assessment BBB (Table 5) through LD50 value and toxicity class BBB suggests that apigenin, astragalin, taraxerol and thymoquinone are expected to be safe as they exhibited non-mutagenic, non-tumorigenic, non-carcinogenic, non-irritant behaviour with no threats to reproductive systems. In compared to toxicity class 1–3, the LD50 values for oral acute toxicity of the majority of understudy phytocompounds fell into toxicity class 4 and 5, making them expectedly less dangerous. Only one compound was found in toxicity class 3 (i.e., nigellimine) whereas taraxerol secured place in class VI and was rendered as expectedly non-toxic (Table 6).
The binding affinities of 18 selected phytochemicals from N. sativa have been studied against two target proteins of Dengue virus, i.e., NS2BNS3 protease and NS5 polymerase, using molecular, pharmacokinetic, and in silico approaches. A member of the S7 family of serine proteases, the DENV NS3 protease domain (NS3pro), which becomes fully active by interacting with the cofactor NS2B, facilitates polyprotein processing at certain sites. Due to its critical involvement in the viral replication cycle, NS2BNS3 protease is regarded as a significant prospective therapeutic target.62,63 NS5 is the most conserved and largest non-structural protein encoded by flaviviruses. Increased structural understanding and multifunctionality of the dengue NS5 protein will help accelerate drug development efforts targeting NS5 as an antiviral target.64,65 Improved in silico approaches, such as molecular docking, pharmacology networks, molecular dynamic simulations, and so on, now significantly contribute to the separation of prospective drugs.30
Molecular docking is a popular approach in computational drug discovery and development to anticipate the orientation of small molecules coupled to an enzyme or receptor binding pocket.66 Based on antiviral phytochemicals, the active site residues of the dengue NS2B/NS3 protease (Table 7) Lys73, Lys74, Leu76, Trp83, His51, Phe130, Leu128, Leu85, Glu88, Gly87, Ile165, Ala166, Ala164, Asn152, and Asn167 were observed to be involved in hydrophobic and hydrogen bond formation.12,67 Taraxerol (−9.1 kcal mol−1) interacted with Pro132, His51, Leu128, Tyr161, while astragalin showed binding affinity of −8.0 kcal mol−1 and interacted with residues Asn167, Trp89, Gly87, Ala166, Val147, Glu88, Ile165, Asn152, Leu86, Lys74. β-Sitosterol (−8.3 kcal mol−1), campesterol (8.1 kcal mol−1), and nigellidine (−7.3 kcal mol−1), each interacted actively with four amino acid residues of NS2B/NS3. For NS5 polymerase, apigenin (9.9 kcal mol−1) formed binding interactions with Leu94, Ile72, Pro298, Lys355, Val66, Gln351. Nigellicine (Arg352, Lys74, Arg352, Glu151, Val66, Leu94, Asn690, Ile165, Pro298) and nigellidine (Pro73, Lys96, Ile72, Leu94, Val66, Lys355, Pro298, Arg581, Glu296) interacted with nine residues in an active pocket with the binding affinity of −9.1 kcal mol−1 and −8.4 kcal mol−1, respectively (Table 8). Cycloeucalenol (−8.0 kcal mol−1) interacted with residues of Val603, Phe485, Ile797, Tyr606, Asp663 in the hydrophobic pocket. The NS5 residues Ser56, Gly81, Cys82, Arg84, Gly85, Thr104, Lys105, His110, Asp131, Val132, Asp146, and Gly148 are conserved in the binding pocket whereas the hydrophobic pocket contains Trp302, Phe354, Val358, Val577, Val579, Val603, and Gly599, Ala406, Ala407, Asn492, Glu507, Tyr606, and Ile797.68–70 An analysis of the interaction between dengue protease and ligands indicated significant contributions from van der Waals forces, conventional hydrogen bonding, carbon hydrogen bonding, alkyl interactions, and pi–alkyl interactions.71 The primary component ensuring the stability of the protein-ligand complex structure is hydrophobic interaction. However, hydrogen bonding has a less significant impact in the structural stability of the protein–ligand complexes. It is expected that existence of the hydrogen bond will matter to provide a better docking score because it has the highest negative weighted factor value among the other variables.42 MD simulations confirmed the structural flexibility of the NS2B/NS3-taraxerol docking complex and the NS5–apigenin complex, whose root mean square value fluctuated below the 5 Å value. RMSF values are utilized to calculate the atomic positional variation of each residue based on its C-alpha (Ca) atom.72,73 The complexes NS2BNS3-taraxerol and NS5-apigenin under iMODS investigation have a significant amount of deformability, according to the normal mode analysis. The low eigenvalue, covariance map, and elastic network of the complex under study are indications of the molecule's potential deformability and allowable dynamics.
The current study used several prediction techniques to anticipate the oral bioavailability of substances, which could lead to the development of novel, safer medications. In silico investigations can save a significant amount of cash and time before proceeding to the experimental stage. Dengue virus replication depends on two proteins, NS5 polymerase and NS2BNS3 main protease complexes,74 which are also the primary focus of antiviral drug research. A previous study66 has used the same target receptors to identify the potential inhibitors from Carica papaya and their results from molecular docking and ligand chemistry have been compared with our 18 understudied phytocomponents of N. sativa which suggests that apigenin, nigellicine, nigellidine, dithymoquinone, taraxerol, campesterol, cycloeucalenol, and beta-sitosterol have been revealed as the most potential lead compounds. According to another research, Nigella sativa demonstrated potent virucidal characteristics that inactivated encapsulated viruses from 2log10 (or 99%) to >4log10 (or 99.99%). The compound Apigenin was also shown to have antiviral, antibacterial, and antifungal effects.75
Conclusion
In conclusion, our research has identified a small number of possible phytochemicals with potential inhibitory effects against Dengue NS2BNS3 and NS5, which may be used for future in vitro and animal testing, formulation, and drug development. These phytoconstituents can be further investigated in vitro and in vivo as possible antiviral drugs for the treatment of dengue fever infections due to their expected bioavailability, druglikeness, and non-toxic and non-mutagenic effects.Author contributions
M. M. and H. A. K. conceptualized the study and performed the formal analyses. Validation was done by M. M., N. S. S. Z. and H. A. K. M. M. wrote the original draft. Original draft was reviewed and edited by N. S. S. Z. and H. A. K. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
We acknowledge the support of the Department of Industrial Biotechnology at Atta-ur-Rahman School of Applied Biosciences, NUST, for providing the research facilities and funding.References
- M. J. Sabir, N. B. S. Al-Saud and S. M. Hassan, Saudi J. Biol. Sci., 2021, 28, 5074–5080 CrossRef CAS PubMed.
- M. T. Qamar, U. A. Ashfaq, K. Tusleem, A. Mumtaz, Q. Tariq, A. Goheer and B. Ahmed, Pak. J. Pharm. Sci., 2017, 30, 2119–2137 Search PubMed.
- S. J. Thomas and T. P. Endy, Curr. Opin. Infect. Dis., 2011, 24, 442–450 CrossRef CAS PubMed.
- A. N. Anoopkumar and E. M. Aneesh, Environ. Dev. Sustain., 2021, 23, 11217–11239 CrossRef.
- S. Swaminathan and N. Khanna, Int. J. Infect. Dis., 2019, 84, S80–S86 CrossRef PubMed.
- C. P. Simmons and J. E. Farrar, N. Engl. J. Med., 2012, 366, 1423–1432 CrossRef CAS PubMed.
- A. Wilder-Smith, Bundesgesundheitsblatt - Gesundheitsforsch. - Gesundheitsschutz, 2020, 63, 40–44 CrossRef PubMed.
- M. Mukhtar, A. W. Wajeeha and N. Bibi, Int. J. Mol. Sci., 2022, 23, 13911 CrossRef CAS PubMed.
- J. Park, J. Kim and Y. S. Jang, J. Microbiol., 2022, 60, 247–254 CrossRef PubMed.
- C. J. Wollner, M. Richner, M. A. Hassert, A. K. Pinto, J. D. Brien and J. M. Richner, J. Virol., 2021, 95 Search PubMed.
- T. N. Adikari, N. Riaz, C. Sigera, P. Leung, B. M. Valencia, K. Barton, M. A. Smith, R. A. Bull, H. Li, F. Luciani, P. Weeratunga, T. L. Thein, V. W. X. Lim, Y. S. Leo, S. Rajapakse, K. Fink, A. R. Lloyd, D. Fernando and C. Rodrigo, Sci. Rep., 2020, 10, 18196 CrossRef CAS PubMed.
- R. A. D. Costa, J. Rocha, A. S. Pinheiro, A. Costa, E. Rocha, R. C. Silva, A. D. S. Goncalves, C. B. R. Santos and D. Brasil, Molecules, 2022, 27 CAS.
- L. Lim, M. Dang, A. Roy, J. Kang and J. Song, ACS Omega, 2020, 5, 25677–25686 CrossRef CAS PubMed.
- S. P. Lim, C. G. Noble, C. C. Seh, T. S. Soh, A. El Sahili, G. K. Chan, J. Lescar, R. Arora, T. Benson, S. Nilar, U. Manjunatha, K. F. Wan, H. Dong, X. Xie, P. Y. Shi and F. Yokokawa, PLoS Pathog., 2016, 12, e1005737 CrossRef PubMed.
- H. Shimizu, A. Saito, J. Mikuni, E. E. Nakayama, H. Koyama, T. Honma, M. Shirouzu, S. I. Sekine and T. Shioda, PLoS Negl. Trop. Dis., 2019, 13, e0007894 CrossRef PubMed.
- P. Coulerie, M. Nour, A. Maciuk, C. Eydoux, J. C. Guillemot, N. Lebouvier, E. Hnawia, K. Leblanc, G. Lewin, B. Canard and B. Figadere, Planta Med., 2013, 79, 1313–1318 CrossRef CAS PubMed.
- A. Wahaab, B. E. Mustafa, M. Hameed, N. J. Stevenson, M. N. Anwar, K. Liu, J. Wei, Y. Qiu and Z. J. V. Ma, Potential Role of Flavivirus NS2B-NS3 Proteases in Viral Pathogenesis and Anti-flavivirus Drug Discovery Employing Animal Cells and Models: A Review, 2021, vol. 14, p. 44 Search PubMed.
- N. Rasool, A. Ashraf, M. Waseem, W. Hussain and S. Mahmood, Turk. J. Biochem., 2019, 44, 261–277 CrossRef CAS.
- K. Mishra, N. Sharma, D. Diwaker, L. Ganju and S. Singh, J. Virol. Antivir. Res., 2013, 2, 2 Search PubMed.
- R. Ghildiyal, V. Prakash, V. Chaudhary, V. Gupta and R. Gabrani, in Plant-derived bioactives, Springer, 2020, pp. 279–295 Search PubMed.
- T. Rathinasabapathy, L. P. Sakthivel and S. Komarnytsky, J. Agric. Food Chem., 2022, 70, 2064–2076 CrossRef CAS PubMed.
- S. Desai, S. H. Saheb, K. K. Das and S. Haseena, Phytochemical Analysis of Nigella sativa and its Antidiabetic Effect, 2015 Search PubMed.
- S. L. Khan, F. A. Siddiqui, S. P. Jain and G. M. Sonwane, Coronaviruses, 2021, 2, 384–402 CrossRef CAS.
- A. Ahmad, A. Husain, M. Mujeeb, S. A. Khan, A. K. Najmi, N. A. Siddique, Z. A. Damanhouri and F. Anwar, Asian Pac. J. Trop. Biomed., 2013, 3, 337–352 CrossRef PubMed.
- J. Anjum, M. A. Sheikh, A. Tiwari, S. Sharma and B. Kumari, in Microbial and Biotechnological Interventions in Bioremediation and Phytoremediation, Springer, 2022, pp. 425–444 Search PubMed.
- A. McBride, P. Mehta, L. Rivino, A. V. Ramanan and S. J. Yacoub, Lancet Microbe, 2021, 2, e277 CrossRef CAS PubMed.
- Z. Hu, J. Lin, J. Chen, T. Cai, L. Xia, Y. Liu, X. Song and Z. J. He, Front. Pharmacol., 2021, 12, 1380 Search PubMed.
- R. Chakravarti, R. Singh, A. Ghosh, D. Dey, P. Sharma, R. Velayutham, S. Roy and D. J. Ghosh, RSC Adv., 2021, 11, 16711–16735 RSC.
- S. Siddiqui, S. Upadhyay, R. Ahmad, A. Gupta, A. Srivastava, A. Trivedi, I. Husain, B. Ahmad, M. Ahamed and M. A. Khan, J. Biomol. Struct. Dyn., 2022, 40, 3928–3948 CrossRef CAS PubMed.
- P. R. Ferdian, R. R. Elfirta, Q. Emilia and A. Z. N. Ikhwani, Ann. Bogor., 2021, 24 Search PubMed.
- R. S. Basurra, S. M. Wang and M. A. Alhoot, J. Pure Appl. Microbiol., 2021, 15, 29–41 CAS.
- R. Ahmad, PeerJ, 2019, 7, e6012 CrossRef PubMed.
- C. A. Lipinski, F. Lombardo, B. W. Dominy and P. J. Feeney, Adv. Drug Delivery Rev., 2012, 64, 4–17 CrossRef.
- P. Kulkarni, Y. Walunj and N. J. T. C. Dongare, Theoretical Validation of Medicinal Properties of Ocimum sanctum, 2021, p. 53 Search PubMed.
- D. Jindal and B. Rani, In Silico Studies of Phytoconstituents from Piper longum and Ocimum sanctum as ACE2 and TMRSS2 Inhibitors: Strategies to Combat COVID-19, 2022, pp. 1–18 Search PubMed.
- A. Agrwal, R. Saini, S. Bhandri, S. Verma, P. Srivastava and O. J. Prakash, Mater. Today: Proc., 2022, 67, 598–604 CAS.
- P. Banerjee, A. O. Eckert, A. K. Schrey and R. J. Preissner, Nucleic Acids Res., 2018, 46, W257–W263 CrossRef CAS PubMed.
- M. N. Drwal, P. Banerjee, M. Dunkel, M. R. Wettig and R. J. Preissner, Nucleic Acids Res., 2014, 42, W53–W58 CrossRef CAS PubMed.
- T. Khan, R. Ahmad, I. Azad, S. Raza, S. Joshi and A. R. Khan, Comput. Biol. Chem., 2018, 75, 178–195 CrossRef CAS PubMed.
- E. Berinyuy, J. Ibrahim and B. Alozieuwa, AROC in Pharmaceutical and Biotechnology, 2021, vol. 1, pp. 1–8 Search PubMed.
- D. Flamandita, K. Lischer, D. K. Pratami, R. Aditama and M. Sahlan, presented in part at the International Conference on Emerging Applications in Material Science and Technology: Iceamst 2020, 2020 Search PubMed.
- R. Novianty, S. Ananta and M. A. Karim, J. Phys. Conf., 2021, 1788 Search PubMed.
- V. Adrianne Dien-Yu, H. Siaw-San, C. Xavier Wezen and S. Edmund Ui-Hang, J. Appl. Biol. Biotechnol., 2022, 10(1), 38–44 Search PubMed.
- M. Ebrahimi and M. Alijanianzadeh, J. Mol. Divers., 2023, 1–24 Search PubMed.
- A. Ibrahim and E. U. Haq, Nat. Product Chem., 2023, 12, 97–108 Search PubMed.
- Y. Yuniwati, M. F. R. Syaban, S. G. Anoraga and F. L. Sabila, Acta. Inform. Med., 2022, 30, 91–95 CrossRef PubMed.
- R. Srivastava, Front. Chem., 2022, 10, 843642 CrossRef CAS PubMed.
- A. A. Onifade, A. P. Jewell and W. A. Adedeji, Afr. J. Tradit. Complement Altern. Med., 2013, 10, 332–335 Search PubMed.
- M. Riaz, M. Khan, R. Ahmad, L. H. AlLehaibi, N. Rahman and D. Deqiang, Bol. Latinoam. Caribe Plantas Med. Aromat., 2022, 21, 176–206 CrossRef CAS.
- S. Thakur, H. Kaurav and G. Chaudhary, Int. J. Res. Rev, 2021, 8, 342–357 CAS.
- M. Akram Khan and M. Afzal, Inflammopharmacology, 2016, 24, 67–79 CrossRef CAS PubMed.
- M. F. Ahmad, F. A. Ahmad, S. A. Ashraf, H. H. Saad, S. Wahab, M. I. Khan, M. Ali, S. Mohan, K. R. Hakeem and M. T. Athar, J. Herb. Med., 2021, 25, 100404 CrossRef PubMed.
- Z. Sheir, G. Badra and O. Salama, J. Liver, 2013, 2 Search PubMed.
- A. Hakobyan, E. Arabyan, A. Avetisyan, L. Abroyan, L. Hakobyan and H. Zakaryan, J. Arch. Virol., 2016, 161, 3445–3453 CrossRef CAS PubMed.
- E. R. Esharkawy, F. Almalki and T. B. Hadda, Bioorg. Chem., 2022, 120, 105587 CrossRef CAS PubMed.
- H. Tabassum and I. Z. Ahmad, Med. Chem., 2020, 16, 350–357 CrossRef CAS PubMed.
- Y. I. Shaikh, V. S. Shaikh, K. Ahmed, G. M. Nazeruddin and H. M. Pathan, The revelation of various compounds found in Nigella sativa L.(Black Cumin) and their possibility to inhibit COVID-19 infection based on the molecular docking and physical properties, 2020, vol. 11, pp. 31–35 Search PubMed.
- S. Riaz, M. A. Riaz, A. Zhanzhaksina, S. A. Raja and T. Javed, Silico Therapeutic and Pharmacological Evaluation of novel apolipoprotein E4 inhibitors against Alzheimer’s Disease, 2022, vol. 3, pp. 22–31 Search PubMed.
- I. Ghosh and P. J. Talukdar, World Sci. News, 2019, 124, 264–278 CAS.
- S. Bandyopadhyay, O. A. Abiodun, B. C. Ogboo, A. T. Kola-Mustapha, E. I. Attah, L. Edemhanria, A. Kumari, R. Jaganathan and N. S. Adelakun, J. Biomol. Struct., 2021, 1–17 Search PubMed.
- S. Hemaiswarya, P. K. Prabhakar and M. Doble, in Herb-Drug Combinations, Springer, 2022, pp. 235–244 Search PubMed.
- H. N. Saleem, F. Batool, H. J. Mansoor, S. Shahzad-ul-Hussan and M. Saeed, ACS Omega, 2019, 4, 1525–1533 CrossRef CAS.
- S. Bharadwaj, K. E. Lee, V. D. Dwivedi, U. Yadava, A. Panwar, S. J. Lucas, A. Pandey and S. G. Kang, Sci. Rep., 2019, 9, 19059 CrossRef CAS PubMed.
- J. O. Obi, H. Gutierrez-Barbosa, J. V. Chua and D. J. Deredge, Trop. Med. Infect. Dis., 2021, 6 Search PubMed.
- S. P. Lim, C. G. Noble and P. Y. Shi, Antiviral Res., 2015, 119, 57–67 CrossRef CAS PubMed.
- M. U. Farooq, B. Munir, S. Naeem, M. Yameen, S. Z. Iqbal, A. Ahmad, M. A. Mustaan, M. W. Noor, M. A. Nadeem and A. J. Ghaffar, Pak. J. Pharm. Sci., 2020, 33 Search PubMed.
- M. S. S. Shimu, S. Mahmud, T. E. Tallei, S. A. Sami, A. A. Adam, U. K. Acharjee, G. K. Paul, T. B. Emran, S. Zaman, M. S. Uddin, M. A. Saleh, S. Alshehri, M. M. Ghoneim, M. Alruwali, A. J. Obaidullah, N. R. Jui, J. Kim and B. Kim, Molecules, 2022, 27(3), 653 CrossRef CAS PubMed.
- C. N. Powers and W. N. Setzer, Comb. Chem. High Throughput Screening, 2016, 19, 516–536 CrossRef CAS PubMed.
- M. Tahir ul Qamar, A. Maryam, I. Muneer, F. Xing, U. A. Ashfaq, F. A. Khan, F. Anwar, M. H. Geesi, R. R. Khalid and S. A. Rauf, Sci. Rep., 2019, 9, 1–16 CrossRef CAS PubMed.
- K. Niranjan, M. Kumar and S. J. Bhatnagar, Indian J. Appl. Phys., 2017, 6, 114–118 CAS.
- A. A. Poola, P. S. Prabhu, T. K. Murthy, M. Murahari, S. Krishna, M. Samantaray and A. J. Ramaswamy, Front. Mol. Biosci., 2023, 10 Search PubMed.
- S. A. Mir, A. Firoz, M. Alaidarous, B. Alshehri, A. A. B. Dukhyil, S. Banawas, S. A. Alsagaby, W. Alturaiki, G. A. Bhat and F. J. Kashoo, Saudi J. Biol. Sci., 2022, 29, 394–401 CrossRef CAS PubMed.
- A. Nag, P. Verma, S. Paul and R. Kundu, Appl. Biochem. Biotechnol., 2022, 194, 4867–4891 CrossRef CAS PubMed.
- M. Hasan, M. M. Mia, S. U. Munna and M. M. H. Talha, Inform. Med. Unlocked, 2022, 30, 100932 CrossRef PubMed.
- S. Sharad and S. J. P. Kapur, Indian herb-derived phytoconstituent-based antiviral, antimicrobial and antifungal formulation: an oral rinse candidate for oral hygiene and the potential prevention of COVID-19 outbreaks, 2021, vol. 10, p. 1130 Search PubMed.
- Y. Kabir, Y. Akasaka-Hashimoto, K. Kubota and M. Komai, Heliyon, 2020, 6(10), e05343 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra02613b |
| This journal is © The Royal Society of Chemistry 2023 |