Open Access Article
Open Access Articleα-Alkylidene δ-lactones inhibit quorum sensing phenotypes in Chromobacterium strain CV026 showing interaction with the CviR receptor†
Fernanda Faveroab,
Terezinha Alves Tolentinoa,
Vinicius Fernandesc,
Werner Treptow*c,
Alex Leite Pereira*b and
Angelo Henrique Lira Machado *a
*a
aInstituto de Química, Universidade de Brasília, Campus Universitário Darcy Ribeiro, Asa Norte, Brasília, DF 70910‐900, Brazil. E-mail: nagelo@unb.br
bCampus of Ceilândia, University of Brasília, Centro Metropolitano, Conjunto A, Ceilândia Sul, Brasília, DF 72220-275, Brazil. E-mail: alexpereira@unb.br
cLaboratório de Biologia Teórica e Computacional, Departamento de Biologia Celular, Universidade de Brasília, Campus Universitário Darcy Ribeiro, Asa Norte, Brasília, DF 70910-900, Brazil. E-mail: treptow@unb.br
First published on 15th June 2023
Abstract
Disruption of bacterial quorum sensing (QS) is presented as a promising strategy to overcome clinically relevant and phytopathogenic bacteria. This work presents α-alkylidene δ-lactones as new chemical scaffolds that inhibit the biosynthesis of violacein in the biosensor strain Chromobacterium CV026. Three molecules displayed higher than 50% violacein reduction when tested at concentrations lower than 625 µM. The most active α-alkylidene δ-lactone inhibited the hydrolysis of chitin concomitantly with the inhibition of violacein production in CV026, suggesting the disruption of its QS machinery. Further, RT-qPCR and competition experiments showed this molecule to be a transcriptional inhibitor of the QS-regulated operon vioABCDE. Docking calculations suggested a good correlation between binding affinity energies and inhibition effects, with all molecules positioned within the CviR autoinducer-binding domain (AIBD). The most active lactone yielded the best binding affinity energy, most probably due to its unprecedented binding with the AIBD. Our results show α-alkylidene δ-lactones as promising chemical scaffolds for the development of new QS inhibitors affecting LuxR/LuxI-systems.
Introduction
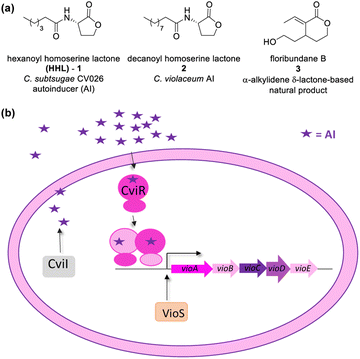
Lactones display several biological functions such as antimicrobial, antifungal, antiparasitic, anti-inflammatory, and cytotoxic, and others have been attributed to them.1–6 They are usually secondary metabolites and can play several roles in cellular physiology, including cell–cell signalling such as Quorum Sensing (QS).17 Bacteria produce chemical signals, called autoinducers (AI), to scan their population growth.8–10 At a threshold concentration, an AI enables bacteria to synchronize changes in gene expression on a population scale, activating or repressing genes that benefit the survival of the community.Several important biological processes have been described as being regulated by QS, including bioluminescence, virulence, and the synthesis of toxins and antibiotics. It is important to note that QS-controlled genes are essential for surveillance, survival, and adaptation to environmental changes.7,11 Gram-negative bacteria use N-acyl homoserine γ-lactones (AHL) like 1 and 2 as autoinducers (Fig. 1a).8 The AHL are synthesized by LuxI and recognized by LuxR-type proteins that act as transcription factors controlling QS genes. Homologs of the LuxI–LuxR QS module occur in a variety of Gram-negative bacteria, including clinically relevant species (such as Pseudomonas aeruginosa, Burkholderia cepacia, Aeromonas hydrophila, Chromobacterium violaceum, Yersinia spp.) as well as in the phytopathogen Agrobacterium tumefaciens.8,11
Resistance to antibiotics is a challenge for public health. Approaches based on the inhibition of QS are promising therapies to control bacterial infections.12–16 QS inhibitors (QSI) are supposed to cause a lower evolutionary pressure than antibiotics, leading to slower emergence of resistance.17 QS inhibition can occur in three ways: the degradation of AI, the interruption of autoinducer biosynthesis, and the use of antagonists of the receptor proteins.18
In the literature, the chemical structure of AIs has been extensively used as the chemical scaffold for the synthesis of antagonists or LuxR-type proteins.8,19 Natural products, such as flavonoids, flavones, and furanones, have already proven their effectiveness as QS inhibitors.20–22
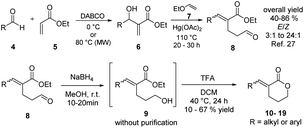
CV026 is a model strain in QSI studies. It is known as an AHL-deficient mutant of Chromobacterium strain ATCC 31532 that has deleted the luxI-type gene cviI, and the vioS, the repressor gene of the CviI/R system.23,24 CV026 only produces violacein in response to an externally supplied hexanoyl-L-homoserine lactone (HHL). Chromobacterium strains ATCC 31532 and CV026 were recently reclassified as Chromobacterium subtsugae.25
As in C. violaceum, QS in CV026 controls biofilm formation, chitinase enzymes, and violacein biosynthesis. A gene cluster found in an 8-kb DNA locus encodes the proteins of CV026 related to violacein biosynthesis as a single operon (vioABCDE). The CviR-trigged expression of vioABCDE produces large and polycistronic RNA transcripts that contiguously harbour the coding information for translation of the proteins involved in violacein biosynthesis (VioA, VioB, VioC, VioD, and VioE).26,27 The production of this purple pigment is an easily quantifiable QS-regulated function.23 The CviR of CV026 recognizes HHL (1) as an autoinducer, and that of C. violaceum recognizes decanoyl homoserine lactone (2) (Fig. 1a). The CviR–AI complex is an X-shaped homodimer that functions as a transcriptional activator controlling the expression of target genes.13
CV026 has been used in a screening assay to detect QS modulators that display an inhibitory effect on C. violaceum and Pseudomonas aeruginosa biofilms.21,28,29 Arena and co-workers studied the effect of seven coumarin skeletons as inhibitors of biofilm and QS mechanisms in C. violaceum and P. aeruginosa.30
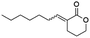
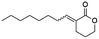
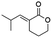
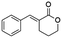
Blackwell and co-workers reviewed several compounds capable of modulating the QS in P. aeruginosa.19 5-Membered lactones are widely studied because they mimic molecular motifs held by the cognate AI. However, 6-membered lactones have been little explored as QS modulators. The α-alkylidene δ-lactones (3, Fig. 1a) are members of this chemical group that show significant biological activities.31 The comparison of the α-alkylidene δ-lactone chemical structure with that of the AHLs allows one to highlight that the conformational restriction imposed by the alkylidene moiety can mimic the effect of the chirality at Cα and the exocyclic planar amide on positioning the side chain of the acyl moiety of the AHL within the autoinducer-binding domain (AIBD) of CviR. To the best of our knowledge, the impact of this structural modification on the modulation of Gram-negative QS has not been evaluated so far. This work studied the effects of α-alkylidene δ-lactones against the QS of C. subtsugae strain CV026. Furthermore, this work probed the production of violacein, and the hydrolysis of chitin, well-known phenotypes of this QS system.32 RT-qPCR and competitive assays were conducted to assess the impact of α-alkylidene δ-lactones on the expression of the CviR-controlled vioABCDE operon. Molecular docking calculations were conducted to evaluate the binding affinity of these lactones to CviR, and to probe their structure–activity relationships (SAR) as QSI.
Results
Synthesis of the α-alkylidene δ-lactones
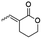
A representative library of α-alkylidene δ-lactones (10–19) was synthesized in good overall yields from the aldehydes 8 by their chemoselective reduction with sodium borohydride in methanol, followed by acidic cyclization without further purification (Scheme 1). The aldehydes 8 were obtained in a stereoselective way by a previously reported route of synthesis, starting from Morita–Baylis–Hillman adducts prepared with readily available aldehydes 4 and ethyl acrylate 5.33Synthesis of chlorolactone (20)
Chlorolactone (20) was synthesized by a synthetic route based on that proposed by Bassler and co-workers, as shown in the ESI (Scheme S1†).34 This molecule is a known inhibitor of the CV026 QS and was used as a positive inhibition control to validate our biological evaluation.13Biological evaluation
| % (inhibition) = (µODtestsolvent − µODtestmolecule) × 100%/µODtestsolvent | (1) |
Table 1 shows the violacein inhibition results achieved by δ-lactones. Some of them had their best percentages of inhibition at higher tested concentrations, but for comparison reasons here we show inhibition results at 625 µM. The complete set of inhibition results is found in the ESI (Fig. S1 to S15).†
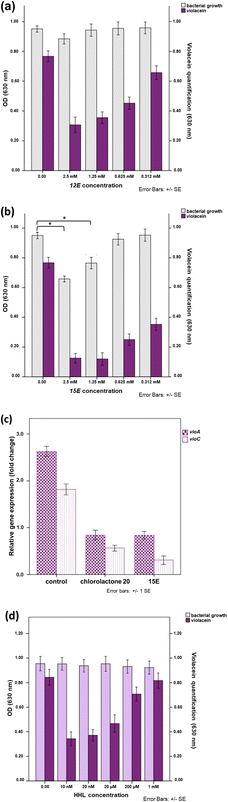
Aliphatic lactones with longer chains delivered better inhibition values when compared to the short ones. 12E (with 52% inhibition at a concentration of 1.25 mM, Fig. 2a) and 13E (with 42% inhibition at a concentration of 1.25 mm, Fig. S7†). For the aromatic ones, the molecule 15E showed inhibition of 75% and 85% at concentrations of 312 µM and 625 µM, respectively (Table 1 and Fig. 2b), achieving the best result among all the α-alkylidene δ-lactones. It is worth mentioning that the 2.5 and 1.25 mM concentrations were not considered, as they showed a decrease in microbial growth. In the validation test with 20, inhibition of the violacein biosynthesis by 95% was observed at a concentration of 32 µM (Fig. S15†). This result agreed with the previous one reported by Bassler for the inhibition of HHL-producing C. subtsugae strain CV31532, which showed IC50 = 2.6 µM.34
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 times the natural activation dose of the HHL.13 These results suggested that autoinducer 1 and lactone 15E act as competitive antagonists.
000 times the natural activation dose of the HHL.13 These results suggested that autoinducer 1 and lactone 15E act as competitive antagonists.| Molecule | Binding affinity (kcal mol−1) | Hydrophilic interaction (H-bond)a | Hydrophobic interactions | logP |
|---|---|---|---|---|
a Amino acid residues highlighted are non-coincidental to CviR![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HHL (2) complex.b Non-conventional H-bond. HHL (2) complex.b Non-conventional H-bond. |
||||
| HHL (1) | −7.0 | Tyr80, Trp84, Tyr88, Asp97, Ser155 | — | 0.952 |
| Positive inhibition control (20) | −8.8 | Tyr80, Trp84, Tyr88, Asp97, Ser155 | Tyr88 | 1.474 |
| Negative inhibition control (21) | 0.0 | No interactions within the AIBD | No interactions within the AIBD | 4.787 |
| 10E | −6.2 | Trp84, Asp97, Ser155 | — | 1.665 |
| 10Z | −6.4 | Trp84, Asp97, Ser155 | — | 1.665 |
| 11E | −6.5 | Tyr80, Trp84, Tyr88, Asp97, Ser155 | — | 2.110 |
| 11Z | −6.3 | Tyr80, Trp84, Tyr88, Asp97, Ser155 | — | 2.110 |
| 12E | −7.0 | Tyr80, Trp84, Ser155 | — | 3.444 |
| 12Z | −7.4 | Tyr80, Trp84, Asp97 | — | 3.888 |
| 13E | −7.4 | Tyr80, Trp84, Asp97 | — | 4.333 |
| 13Z | −7.5 | Tyr80, Trp84, Asp97 | — | 4.333 |
| 14E | −6.5 | Tyr80, Asp97, Ser155, Trp111, Thr140 | — | 2.397 |
| 15E | −8.4 | Tyr80, Tyr88, Asp97, Ser155 | Trp111 | 2.882 |
| 16E | −8.3 | Tyr88, Asp97, Ser155, Trp111 | Tyr88 | 3.486 |
| 17E | −8.1 | Tyr88, Asp97, Ser155, Leu85b, Trp111 | Tyr88 | 3.651 |
| 18E | −8.5 | Trp84, Asp97, Asn77 | — | 2.526 |
| 19E | −8.0 | Trp84, Asp97, Asn77 | — | 2.505 |
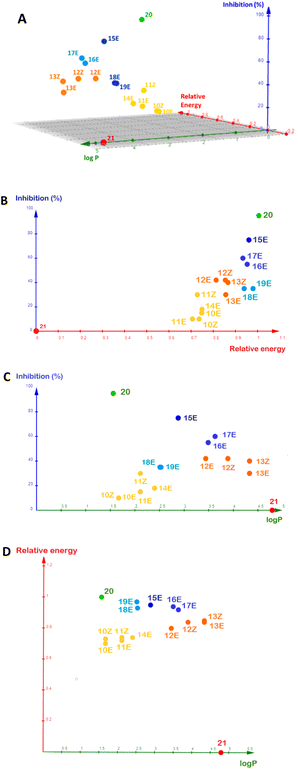
To summarize this information, we plotted logP, inhibition percentage, and normalized docking energy on a single 3D graphic (Fig. 5). The docking energies were normalized by the lowest value computed, which corresponds to positive inhibition control 20. For comparison purposes, we chose a molecule from Fallarero's work (21, Fig. S32†) as a negative inhibition control.21 It did not affect violacein biosynthesis at 400 µM and its calculated logP is 4.787.
Discussion
The α-alkylidene δ-lactones here investigated were tested for their ability to interfere with violacein biosynthesis, with all of them displaying an inhibitory effect around 625 µM. The biosynthesis of violacein is a well-known QS-regulated phenotype in strain CV026 and can be described as a two-step event: (a) the transcription of the vioABCDE genes allowing the production of the enzymes responsible for violacein biosynthesis; and (b) the transformation of tryptophan into violacein accomplished by the vioABCDE metabolic pathway.26 Starting from this premise, the inhibition of violacein biosynthesis observed for the α-alkylidene δ-lactones could come independently from (1) the disruption of the transcriptional event mediated by CviR or (2) the antagonist action of these lactones in one of the biosynthetic steps catalysed by vioABCDE enzymes.The chemical structure of the α-alkylidene δ-lactones is similar to the one of AHL 1, the cognate autoinducer of the CV026 QS. They have a hydrophilic lactone attached at Cα to a lipophilic hydrocarbon side chain, and this chemical similarity prompted us to suggest that the inhibitory profile observed for the α-alkylidene δ-lactones could be due to the competitive antagonist binding of these lactones to the CviR AIBD, disrupting the QS in CV026.
Lactone 15E showed the best inhibition of violacein biosynthesis in CV026, 85% at concentrations of 625 µM, and was selected for further investigation of the origin of the α-alkylidene δ-lactone's inhibitory activity here observed. In a competitive binding assay with 15E and cognate autoinducer 1, a complete restoration of violacein production was achieved as the autoinducer concentration reached similar values to those used for 15E. Additionally, RT-qPCR experiments with 15E showed a decrease in the expression of vioABCDE RNA transcripts. These results characterize it as a QSI that works at the transcriptional level.
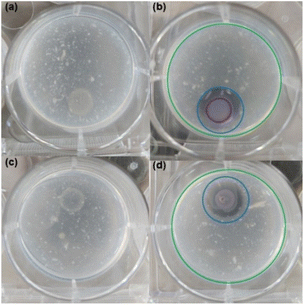
The α-alkylidene δ-lactone 15E also inhibited hydrolysis of chitin, a well-known QS-regulated phenotype in strain CV026, concomitantly with the suppression of violacein biosynthesis.16 This result was observed similarly to that for QS inhibitor 20, endorsing the initial hypothesis that, at the transcriptional level, α-alkylidene δ-lactones can inhibit the AHL-based QS in CV026.
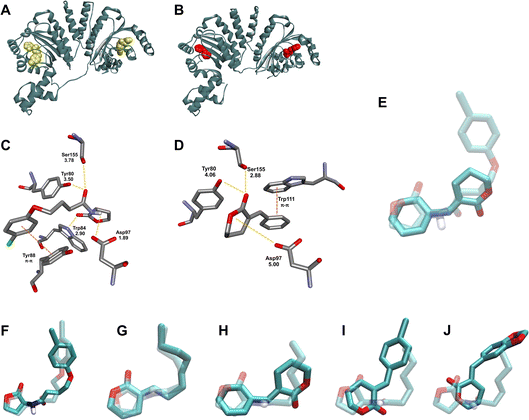
The blind molecular docking studies conducted with the CviR crystal structure (PDB ID 3QP5) were used here to evaluate if and how these new QSI α-alkylidene δ-lactones interact with CviR. All tested δ-lactones showed interaction with the AIBD of CviR as their most stable pose (Fig. S18–S31†). The molecular docking energies computed for the δ-lactones ranged from −6.2 kcal mol−1 to −8.5 kcal mol−1 (Table 2), with a similar magnitude to that computed for cognate AI 1, (−7.0 kcal mol−1) and positive inhibition control 20 (−8.8 kcal mol−1). Negative control 21 could not be found in the AIBD, and relative energy equal to zero was assigned to it (Fig. S32†).
In addition to the similar docking energies shown by 20 and 15E (respectively, −8.8 kcal mol−1 vs. −8.4 kcal mol−1), these QS inhibitors share four hydrophilic interactions in the CviR AIBD. Docking information, together with RT-qPCR results and the competition assay, endorse the hypothesis that the observed QS inhibition caused by 15E occurs due to the competition between this δ-lactone and cognate autoinducer 1 to bind CviR, inhibiting the expression of the vioABCDE operon at the transcriptional level in CV026.
The logP of the α-alkylidene δ-lactones was calculated (Table 2), and all values were lower than 5, suggesting their ability to diffuse across the bacterial membrane.36 However, the logP could not be assigned as a good descriptor for the analysis of SAR regarding the inhibition measured for these δ-lactones (Fig. 5d). The aliphatic lactones showed logP ranging from 1.6–4.3, and those with the best inhibition – namely 12E, 12Z, 13E, and 13Z – had the logP > 3.000, far from the positive inhibitory control 20 (logP = 1.474). A different trend was observed for the aromatic δ-lactones. Their logP varied over a much smaller range, between 2.501 and 3.651, when compared to the aliphatic ones. These values were closer to the logP for positive inhibitory control 20, suggesting that the presence of the phenyl group, substituted or not, improves the inhibitory activity.
To evaluate the correlation between the interaction energies obtained by docking and the per cent inhibition, these energies were normalized by the one computed for complex CviR![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20. In contrast to the logP, relative interaction energy and QS inhibition percentage had a good correlation, as can be seen in Fig. 5b.
20. In contrast to the logP, relative interaction energy and QS inhibition percentage had a good correlation, as can be seen in Fig. 5b.
Hughson, Bassler and co-workers introduced a seminal discussion concerning the CviR conformational changes imposed by the acyl chains of AHL-based ligands.13 Long-chain AHLs showed competitive inhibitory activity on this enzyme. To better understand this fact at the molecular level, they analysed the X-ray crystal structure of the CviR complexed with different AHLs, including 1, 2, AHL with C8 acyl side chain, and aromatic analogue 20. Those that showed inhibitory activity had the acyl side chain oriented toward Met 89, resulting in conformational changes for CviR that prevented interaction with the DNA, thereby inhibiting the transcription of vioABCDE. Our results corroborate this discussion, because the α-alkylidene δ-lactones with longer aliphatic chains yielded better inhibition results for those with alkyl ones (10–13), in addition to better relative interaction energies (Fig. 5c). These findings prompted us to suggest that longer aliphatic side chains, in addition to the increase in the permeability of the lactones through the cell envelope and their cellular concentration, optimize the interaction with the AIBD to promote the conformational changes in CviR needed to prevent its interaction with DNA, in a way similar to that proposed by Hughson, Bassler and co-workers for the AHL inhibitors (Fig. 4g).
The behaviour of the docking results for the aromatic δ-lactones in the AIBD was less convergent than that for those with alkyl side chains. Compounds 15E–18E were positioned with their lactone moiety in the same AIBD region where the homoserine lactone of the AHLs would be, with the benzylidene group oriented toward the same AIBD region occupied by the aryl group of 20. This pose suggests the same π–π stacking interaction with Trp88 observed for this known inhibitor. Despite this similarity between these four δ-lactones, the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O for 17E and 18E was pointed toward the Trp84 residue of CviR (Fig. 4J and S30,† respectively), in contrast to 15E and 16E, which had their carbonyl oriented to the opposite direction, making a hydrogen bond with Tyr88 (Fig. 4I and, S28,† respectively). These differences did not impose significant changes on their docking energy, but they did seem to affect their inhibitory activity on violacein biosynthesis.
O for 17E and 18E was pointed toward the Trp84 residue of CviR (Fig. 4J and S30,† respectively), in contrast to 15E and 16E, which had their carbonyl oriented to the opposite direction, making a hydrogen bond with Tyr88 (Fig. 4I and, S28,† respectively). These differences did not impose significant changes on their docking energy, but they did seem to affect their inhibitory activity on violacein biosynthesis.
A particular difference in the binding pose for α-benzylidene δ-lactone 15E is noteworthy. Its lactone ring was found positioned at the same AIBD region occupied by the AHL's acyl chain, and its aromatic moiety was oriented toward the same AIBD region occupied by the AHL's homoserine lactone (Fig. 4H and S27†). This unprecedented docking pose positioned the phenyl ring of the δ-lactone 15E parallel to the Trp111 residue, suggesting π–π stacking, which, to the best of our knowledge, has never been reported before for lactone-based CviR ligands.
A comparison of the inhibition measured for the α-benzylidene δ-lactones, their relative docking energies, and logP (Fig. 5) endorses the binding pose of 15E at the AIBD as the basis for its inhibitory effect.
Conclusion
A representative library of compounds belonging to α-alkylidene δ-lactones was evaluated for their inhibitory activity for violacein production in a C. subtsugae CV026 bioassay. Out of the 14 compounds, two (12E and 15E) proved to be the most promising ones. The competitive binding assay between 15E and cognate autoinducer 1 showed a dose-dependent pattern, while violacein production was gradually restored by increasing the concentration of autoinducer 1. Lactone 15E also inhibited the chitin hydrolysis concomitantly with violacein production in a C. subtsugae CV026 bioassay; both are well-known QS-mediated phenotypes, suggesting that this α-alkylidene δ-lactone is working at a transcriptional level, disrupting the CV026 QS machinery. RT-qPCR experiments with 15E showed a decrease in the expression of vioABCDE RNA transcripts assessed via two target loci (vioA and vioC with 68% and 80% decrease, respectively). These results characterize 15E as a QSI that works at the transcriptional level in CV026. Docking studies for the α-alkylidene δ-lactones with CviR suggested that molecular interactions of the lactone moiety with AIBD can mimic the interactions of lactone 1. The aromatic lactones have π–π stacking interaction that improves their binding with the CviR protein. The most active compound, 15E, showed the best binding affinity by docking studies, most probably due to its unprecedented binding positioning with the AIBD. The original hypothesis that motivated the evaluation of α-alkylidene δ-lactones as modulators of the QS in CV026 was based on the structural similarity between them and the cognate AI, suggesting that α-alkylidene δ-lactones could mimic the AI pose within the CviR AIBD. Surprisingly, blind docking calculations for CviR![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15E complex yielded the ligand positioned within the CviR AIBD but inverted with respect to the cognate AI, with the phenyl group of 15E fitting the AI's lactone pocket, and the lactone ring of 15E filling the AI's acyl chain pocket. Although this pose seems divergent from our original hypothesis, lactone 15E presents interactions with the same amino acid residues that are functionally important for QS inhibition, in agreement with the inhibition mechanism proposed by Hughson and Bassler for Chromobacterium spp.13 Taken together, these docking calculations suggest that the shortening of the spacer between the lactone and the aryl ring still allows 15E-like molecules to block the CviR AIBD by a molecular binding mechanism distinct from classic AHL-based inhibitors such as the inhibitor 20 proposed by Bassler. This study shows that the α-alkylidene δ-lactone is a new chemical scaffold with the potential to act as a QSI in Gram-negative LuxR/LuxI-type QS systems, and further studies are ongoing in our group to explore them in searching for novel optimized QSIs.
15E complex yielded the ligand positioned within the CviR AIBD but inverted with respect to the cognate AI, with the phenyl group of 15E fitting the AI's lactone pocket, and the lactone ring of 15E filling the AI's acyl chain pocket. Although this pose seems divergent from our original hypothesis, lactone 15E presents interactions with the same amino acid residues that are functionally important for QS inhibition, in agreement with the inhibition mechanism proposed by Hughson and Bassler for Chromobacterium spp.13 Taken together, these docking calculations suggest that the shortening of the spacer between the lactone and the aryl ring still allows 15E-like molecules to block the CviR AIBD by a molecular binding mechanism distinct from classic AHL-based inhibitors such as the inhibitor 20 proposed by Bassler. This study shows that the α-alkylidene δ-lactone is a new chemical scaffold with the potential to act as a QSI in Gram-negative LuxR/LuxI-type QS systems, and further studies are ongoing in our group to explore them in searching for novel optimized QSIs.
Experimental section
Chemistry
Solvents obtained from commercial sources were treated, before being used, according to procedures previously described in the literature. Anhydrous DMF was purchased from Sigma-Aldrich in a sure-seal flask. Nuclear magnetic resonance (NMR) experiments were performed on the following devices: Varian Mercury Plus (300 MHz for 1H and 75 MHz for 13C) 7.04 T and Bruker Ascend (600 MHz for 1H 150 and MHz for 13C) 14.1 T. chemical shifts (δ) were expressed in parts per million (ppm). Coupling constants are expressed in Hertz (Hz). The analysed samples were dissolved in deuterated chloroform (CDCl3). For 1H NMR spectra, TMS tetramethylsilane (0.00 ppm) was used as an internal reference, and for 13C NMR spectra, deuterated chloroform (77.0 ppm). The spectra were processed using the MestreNova 6.0 program.QSI synthesis
The α-alkylidene δ-lactones of this study were synthesized as described by Machado and co-workers.33Ethyl (E/Z)-2-ethylidene-5-oxopentanoate (8; R = Me mixture of isomers). Physical state: light yellow oil. Yield: 86% (E/Z 3.0
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0). 1H NMR (600 MHz, CDCl3, 25 °C) δ E 9.78 (t, J = 1.5 Hz, 1H), Z 9.76 (t, J = 1.3 Hz, 1H), E 6.92 (q, J = 7.2 Hz, 1H), Z 6.11 (q, J = 7.2H, 1H), E 4.24–4.17 (m, 2H), E/Z 2.65–2.56 (m, 4H), Z 1.99 (d, J = 7.2 Hz, 3H), E 1.84 (d, J = 7.2 Hz, 3H), E/Z 1.33–1.27 (m, 3H). 13C NMR (75 MHz, CDCl3, 25 °C) δ E 201.7, 167.2, 138.6, 131.4, 60.5, 43.0, 14.2. Z 201.7, 167.4, 138.6, 130.9, 60.2, 43.4, Z 14.3, E/Z 19.2. MS (70 eV) m/z: (M)+ 170, 41(100) IR (ATR): 2984, 1715, 1448, 1379, 1263, 1182. HRMS (ESI-TOF): [M + Na]+ calcd for C9H14NaO3 193.0835, found 193.0831.
1.0). 1H NMR (600 MHz, CDCl3, 25 °C) δ E 9.78 (t, J = 1.5 Hz, 1H), Z 9.76 (t, J = 1.3 Hz, 1H), E 6.92 (q, J = 7.2 Hz, 1H), Z 6.11 (q, J = 7.2H, 1H), E 4.24–4.17 (m, 2H), E/Z 2.65–2.56 (m, 4H), Z 1.99 (d, J = 7.2 Hz, 3H), E 1.84 (d, J = 7.2 Hz, 3H), E/Z 1.33–1.27 (m, 3H). 13C NMR (75 MHz, CDCl3, 25 °C) δ E 201.7, 167.2, 138.6, 131.4, 60.5, 43.0, 14.2. Z 201.7, 167.4, 138.6, 130.9, 60.2, 43.4, Z 14.3, E/Z 19.2. MS (70 eV) m/z: (M)+ 170, 41(100) IR (ATR): 2984, 1715, 1448, 1379, 1263, 1182. HRMS (ESI-TOF): [M + Na]+ calcd for C9H14NaO3 193.0835, found 193.0831.
Ethyl (E/Z)-2-(3-oxopropyl)pent-2-enoate (8; R = Et mixture of isomers). Physical state: colorless oil. Yield: 67% (E/Z 3.0
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0). 1H NMR (600 MHz, CDCl3, 25 °C) δ E 9.78 (t, J = 1.4 Hz, 1H), Z 9.77 (t, J = 1.4 Hz, 1H), E 6.81 (t, J = 7.6 Hz, 1H), Z 5.97 (t, J = 7.4 Hz, 1H), E/Z 4.24–4.17 (m, 2H), E/Z 2.66–2.55 (m, 2H), E/Z 2.26–2.19 (m, 2H), E/Z 1.33–1.28 (m, 3H), E/Z 1.08–1.00 (M, 3H). 13C NMR (75 MHz, CDCl3, 25 °C) δ E 201.7, 145.4, 129.8, 60.5, 43.3, 21.8, 13.2. Z 201.7, 145.6, 129.7, 60.5, 43.4, 21.9, 13.2, E/Z 167.4, E/Z 19.4. IR (KBr): 2976; 2729; 1709; 1644; 1461; 1372; 859. HRMS (ESI-TOF): [M + H]+ calcd for C10H17O3 185.1172, found 185.1180.
1.0). 1H NMR (600 MHz, CDCl3, 25 °C) δ E 9.78 (t, J = 1.4 Hz, 1H), Z 9.77 (t, J = 1.4 Hz, 1H), E 6.81 (t, J = 7.6 Hz, 1H), Z 5.97 (t, J = 7.4 Hz, 1H), E/Z 4.24–4.17 (m, 2H), E/Z 2.66–2.55 (m, 2H), E/Z 2.26–2.19 (m, 2H), E/Z 1.33–1.28 (m, 3H), E/Z 1.08–1.00 (M, 3H). 13C NMR (75 MHz, CDCl3, 25 °C) δ E 201.7, 145.4, 129.8, 60.5, 43.3, 21.8, 13.2. Z 201.7, 145.6, 129.7, 60.5, 43.4, 21.9, 13.2, E/Z 167.4, E/Z 19.4. IR (KBr): 2976; 2729; 1709; 1644; 1461; 1372; 859. HRMS (ESI-TOF): [M + H]+ calcd for C10H17O3 185.1172, found 185.1180.
Ethyl (E/Z)-4-methyl-2-(3-oxopropyl)pent-2-enoate (8; R = iPro mixture of isomers). Physical state: colorless oil. Yield: 67% (E/Z 7.1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0). 1H NMR (600 MHz, CDCl3, 25 °C) δ E 9.74 (t, J = 1.4 Hz, 1H), Z 9.72 (t, J = 1.5 Hz, 1H), E 6.57 (d, J = 10.2 Hz, 1H), Z 5.69 (dd, J = 9.8 Hz, 1H), E/Z 4.19–4.12 (m, 2H), E/Z 2.66–2.50 (m, 5H), E/Z 1.28–1.18 (m, 3H), E 0.99 (d, J = 6.6 Hz, 6H), Z 0.94 (d, J = 6.6 Hz, 6H). 13C NMR (151 MHz, CDCl3, 25 °C) δ E 201.7, 167.6, 128.1, 60.6, 43.7, 22.2, 14.2, E/Z 150.4, E/Z 27.9, E/Z 19.6, Z 201.6, 167.5, 127.8, 176.5, 60.3, 43.5, 22.2, 14.2. MS (70 eV) m/z: 198 (M+), 81 (100). IR (ATR): 2964, 2869, 1708, 1645, 1448, 1313, 1256. HRMS (ESI-TOF): [M + H]+ calcd for C11H19O3 199.1329, found 199.1337.
1.0). 1H NMR (600 MHz, CDCl3, 25 °C) δ E 9.74 (t, J = 1.4 Hz, 1H), Z 9.72 (t, J = 1.5 Hz, 1H), E 6.57 (d, J = 10.2 Hz, 1H), Z 5.69 (dd, J = 9.8 Hz, 1H), E/Z 4.19–4.12 (m, 2H), E/Z 2.66–2.50 (m, 5H), E/Z 1.28–1.18 (m, 3H), E 0.99 (d, J = 6.6 Hz, 6H), Z 0.94 (d, J = 6.6 Hz, 6H). 13C NMR (151 MHz, CDCl3, 25 °C) δ E 201.7, 167.6, 128.1, 60.6, 43.7, 22.2, 14.2, E/Z 150.4, E/Z 27.9, E/Z 19.6, Z 201.6, 167.5, 127.8, 176.5, 60.3, 43.5, 22.2, 14.2. MS (70 eV) m/z: 198 (M+), 81 (100). IR (ATR): 2964, 2869, 1708, 1645, 1448, 1313, 1256. HRMS (ESI-TOF): [M + H]+ calcd for C11H19O3 199.1329, found 199.1337.
Ethyl (E/Z)-2-(3-oxopropyl)non-2-enoate (8; R = Hex mixture of isomers). Physical state: colorless oil. Yield: 80% (E/Z 4.0
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0). 1H NMR (600 MHz, CDCl3, 25 °C) δ E 9.78 (t, J = 1.4 Hz, 1H), Z 9.77 (t, J = 1.4 Hz, 1H), E 6.82 (t, J = 7.6 Hz, 1H), Z 5.99 (t, J = 7.4 Hz, 1H), E/Z 4.23–4.17 (m, 2H), E/Z, 2.64–2.55 (m, 4H), E/Z 2.22–2.17 (m, 2H), E/Z 1.48–1.21 (m, 11H), E/Z 0.90–0.86 (m, 3H). 13C NMR (151 MHz, CDCl3, 25 °C) δ E 201.6, 167.3, 144.2, 130.2, 60.5, 43.3, 31.7, 29.3, 29.0, 28.60, 22.6, 14.0, E/Z 19.5, E/Z 14.2, Z 201.7, 167.3, 144.3, 129.8, 60.2, 43.5, 31.7, 29.3, 28.9, 28.5, 22.5, 14.0. MS (70 eV) m/z: 240 (M+), 137 (100). IR (ATR): 2925, 2865, 2858, 1708, 1456, 1363, 1270, 1183. HRMS (ESI-TOF): [M + H]+ calcd for C14H25O3 241.1798, found 241.1800.
1.0). 1H NMR (600 MHz, CDCl3, 25 °C) δ E 9.78 (t, J = 1.4 Hz, 1H), Z 9.77 (t, J = 1.4 Hz, 1H), E 6.82 (t, J = 7.6 Hz, 1H), Z 5.99 (t, J = 7.4 Hz, 1H), E/Z 4.23–4.17 (m, 2H), E/Z, 2.64–2.55 (m, 4H), E/Z 2.22–2.17 (m, 2H), E/Z 1.48–1.21 (m, 11H), E/Z 0.90–0.86 (m, 3H). 13C NMR (151 MHz, CDCl3, 25 °C) δ E 201.6, 167.3, 144.2, 130.2, 60.5, 43.3, 31.7, 29.3, 29.0, 28.60, 22.6, 14.0, E/Z 19.5, E/Z 14.2, Z 201.7, 167.3, 144.3, 129.8, 60.2, 43.5, 31.7, 29.3, 28.9, 28.5, 22.5, 14.0. MS (70 eV) m/z: 240 (M+), 137 (100). IR (ATR): 2925, 2865, 2858, 1708, 1456, 1363, 1270, 1183. HRMS (ESI-TOF): [M + H]+ calcd for C14H25O3 241.1798, found 241.1800.
Ethyl (E/Z)-2-(3-oxopropyl)dec-2-enoate (8; R = Hept mixture of isomers). Physical state: colorless oil. Yield: 96% (E/Z 3.5
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0). 1H NMR (600 MHz, CDCl3, 25 °C) δ E 9.77 (t, J = 1.4 Hz, 1H), Z 9.75 (m, 1H), E 6.80 (t, J = 7.6 Hz, 1H), Z 5.97 (t, J = 7.4 Hz, 1H), E/Z 4.24–4.11 (m, 2H), E/Z 2.65–2.50 (m, 4H), Z 2.42 (q, J = 7.3 Hz, 2H), E 2.15 (q, J = 7.3 Hz, 2H), E/Z 1.48–1.10 (m, 10H), E/Z 0.95–0.75 (m, 6H). 13C NMR (151 MHz, CDCl3, 25 °C) δ Z 201.8, E 201.7, Z 167.5, E 167.4, Z 144.4, E 144.2, E 130.3, Z 129.8, E 60.5, Z 60.2, Z 43.5, E 43.4, Z 31.8, E 31.7, Z 29.7, E 29.4, Z 29.3, E 29.1, Z 28.8, E 28.7, E 28.6(3), Z 28.5(8), Z 27.4, E 22.6, Z 22.1, E 19.6, E/Z 14.3, E/Z 14.1. MS (70 eV) m/z: 254 (M+), 137 (100). IR (ATR): 2959, 2932, 2851, 1717, 1643, 1456, 1368, 1256, 1188. HRMS (ESI-TOF): [M + Na]+ calcd for C15H27O3 255.1955, found 255.1948.
1.0). 1H NMR (600 MHz, CDCl3, 25 °C) δ E 9.77 (t, J = 1.4 Hz, 1H), Z 9.75 (m, 1H), E 6.80 (t, J = 7.6 Hz, 1H), Z 5.97 (t, J = 7.4 Hz, 1H), E/Z 4.24–4.11 (m, 2H), E/Z 2.65–2.50 (m, 4H), Z 2.42 (q, J = 7.3 Hz, 2H), E 2.15 (q, J = 7.3 Hz, 2H), E/Z 1.48–1.10 (m, 10H), E/Z 0.95–0.75 (m, 6H). 13C NMR (151 MHz, CDCl3, 25 °C) δ Z 201.8, E 201.7, Z 167.5, E 167.4, Z 144.4, E 144.2, E 130.3, Z 129.8, E 60.5, Z 60.2, Z 43.5, E 43.4, Z 31.8, E 31.7, Z 29.7, E 29.4, Z 29.3, E 29.1, Z 28.8, E 28.7, E 28.6(3), Z 28.5(8), Z 27.4, E 22.6, Z 22.1, E 19.6, E/Z 14.3, E/Z 14.1. MS (70 eV) m/z: 254 (M+), 137 (100). IR (ATR): 2959, 2932, 2851, 1717, 1643, 1456, 1368, 1256, 1188. HRMS (ESI-TOF): [M + Na]+ calcd for C15H27O3 255.1955, found 255.1948.
Ethyl (E)-2-benzylidene-5-oxopentanoate (8; R = Ph). Physical state: light green oil. Yield: 67% (E/Z 24.0
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0). 1H NMR (600 MHz, CDCl3, 25 °C) δ 9.74 (t, J = 1.4 Hz, 1H), 7.72 (s, 1H), 7.38–7.25 (m, 5H), 4.25 (q, J = 7.1 Hz, 2H), 2.85–2.81 (m, 2H), 2.67–2.63 (m, 2H), 1.32 (t, J = 7.1 Hz, 3H). 13C NMR (151 MHz, CDCl3, 25 °C) δ 201.3, 167.7, 140.3, 135.3, 131.4, 129.0, 128.7, 128.6, 61.0, 43.2, 20.3, 14.3. MS (70 eV) m/z: 232 (M+), 129 (100). IR (ATR): 2979, 2738, 1701, 1627, 1488, 1365. HRMS (ESI-TOF): [M + Na]+ calcd for C14H16NaO3 255.0992, found 255.0990.
1.0). 1H NMR (600 MHz, CDCl3, 25 °C) δ 9.74 (t, J = 1.4 Hz, 1H), 7.72 (s, 1H), 7.38–7.25 (m, 5H), 4.25 (q, J = 7.1 Hz, 2H), 2.85–2.81 (m, 2H), 2.67–2.63 (m, 2H), 1.32 (t, J = 7.1 Hz, 3H). 13C NMR (151 MHz, CDCl3, 25 °C) δ 201.3, 167.7, 140.3, 135.3, 131.4, 129.0, 128.7, 128.6, 61.0, 43.2, 20.3, 14.3. MS (70 eV) m/z: 232 (M+), 129 (100). IR (ATR): 2979, 2738, 1701, 1627, 1488, 1365. HRMS (ESI-TOF): [M + Na]+ calcd for C14H16NaO3 255.0992, found 255.0990.
Ethyl (E)-2-(4-bromobenzylidene-5-oxopentanoate) (8; R = 4-Br-Ph). Physical state: colorless oil. Yield: 40% (E/Z 17.0
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0). 1H NMR (600 MHz, CDCl3, 25 °C) δ 9.77 (t, J = 1.3 Hz, 1H), 7.65 (s, 1H), 7.54–7.51 (m, 2H), 7.21–7.17 (m, 2H), 4.28 (q, J = 7.1 Hz, 2H), 2.85–2.81 (m, 2H), 2.70–2.66 (m, 2H), 1.35 (t, J = 7.1 Hz, 3H). 13C NMR (151 MHz, CDCl3, 25 °C) δ 201.0, 167.5, 138.9, 134.1, 132.1, 130.5, 122.9, 61.2, 43.0, 20.2, 14.3. MS (70 eV) m/z: 310 (M+), 129(100). IR (ATR): 2979, 1702, 1673, 1585, 1481, 1369. HRMS (ESI-TOF): [M + H]+ calcd for C14H16BrO3 311.0277, found 311.0287.
1.0). 1H NMR (600 MHz, CDCl3, 25 °C) δ 9.77 (t, J = 1.3 Hz, 1H), 7.65 (s, 1H), 7.54–7.51 (m, 2H), 7.21–7.17 (m, 2H), 4.28 (q, J = 7.1 Hz, 2H), 2.85–2.81 (m, 2H), 2.70–2.66 (m, 2H), 1.35 (t, J = 7.1 Hz, 3H). 13C NMR (151 MHz, CDCl3, 25 °C) δ 201.0, 167.5, 138.9, 134.1, 132.1, 130.5, 122.9, 61.2, 43.0, 20.2, 14.3. MS (70 eV) m/z: 310 (M+), 129(100). IR (ATR): 2979, 1702, 1673, 1585, 1481, 1369. HRMS (ESI-TOF): [M + H]+ calcd for C14H16BrO3 311.0277, found 311.0287.
Ethyl (E)-2-(4-chlorobenzylidene-5-oxopentanoate) (8; R = 4-Cl-Ph). Physical state: colorless oil. Yield: 73% (E/Z 18.0
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0). 1H NMR (300 MHz, CDCl3, 25 °C) δ 9.78 (t, J = 1.3 Hz, 1H), 7.68 (s, 1H), 7.40–7.23 (m, 5H), 4.28 (q, J = 7.1 Hz, 2H), 2.87–2.78 (m, 2H), 2.73–2.65 (m, 2H), 1.35 (t, J = 7.1 Hz, 3H). 13C NMR (151 MHz, CDCl3, 25 °C) δ 201.2, 167.3, 139.1, 134.6, 133.4, 133.7, 132.1, 130.4, 128.9, 61.3, 43.0, 20.4, 14.4. MS (70 eV) m/z: 266 (M+), 129 (100) IR (ATR): 2984, 2724, 1702, 1635,1592, 1492, 1256, 1166. HRMS (ESI-TOF): [M + H]+ calcd for C14H16ClO3 267.0782, found 267.0786.
1.0). 1H NMR (300 MHz, CDCl3, 25 °C) δ 9.78 (t, J = 1.3 Hz, 1H), 7.68 (s, 1H), 7.40–7.23 (m, 5H), 4.28 (q, J = 7.1 Hz, 2H), 2.87–2.78 (m, 2H), 2.73–2.65 (m, 2H), 1.35 (t, J = 7.1 Hz, 3H). 13C NMR (151 MHz, CDCl3, 25 °C) δ 201.2, 167.3, 139.1, 134.6, 133.4, 133.7, 132.1, 130.4, 128.9, 61.3, 43.0, 20.4, 14.4. MS (70 eV) m/z: 266 (M+), 129 (100) IR (ATR): 2984, 2724, 1702, 1635,1592, 1492, 1256, 1166. HRMS (ESI-TOF): [M + H]+ calcd for C14H16ClO3 267.0782, found 267.0786.
Ethyl (E)-2-(4-nitrobenzylidene-5-oxopentanoate) (8; R = 4-NO2-Ph). Physical state: yellow oil. Yield: 71% (E/Z 6.0
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0). 1H NMR (600 MHz, CDCl3, 25 °C) δ Z 9.80 (s, 1H), E 9.73 (s, 1H), E 8.23 (d, J = 8.7 Hz, 2H), Z 8.13 (d, J = 8.7 Hz, 2H), E 7.71 (s, 1H), E 7.45 (d, J = 8.8 Hz, 2H), Z 7.34 (d, J = 8.8 Hz, 2H), Z 6.76 (s, 1H), E 4.27 (q, J = 7.1 Hz, 2H), Z 4.10 (q, J = 7.1 Hz, 2H), E/Z 2.81–2.76 (m, 2H), E/Z 2.76–2.71 (m, 4H), E 2.68 (t, J = 7.6 Hz, 2H), Z 1.33 (t, J = 7.1 Hz, 3H), Z 1.08 (t, J = 7.1 Hz, 3H). 13C NMR (151 MHz, CDCl3, 25 °C) δ E 200.4, E/Z 166.9, E/Z 147.5, E/Z 141.9, E/Z 137.6, E 129.7, E 123.9, E 61.5, E 42.9, E/Z 27.8, E/Z 14.3, Z 200.4, Z 167.9, Z 129.0, Z 123.4, Z 61.2, 42.4, E/Z 20.3. HRMS (ESI-TOF): [M + H]+ calcd for C14H16NO5 278.1023, found 278.1042.
1.0). 1H NMR (600 MHz, CDCl3, 25 °C) δ Z 9.80 (s, 1H), E 9.73 (s, 1H), E 8.23 (d, J = 8.7 Hz, 2H), Z 8.13 (d, J = 8.7 Hz, 2H), E 7.71 (s, 1H), E 7.45 (d, J = 8.8 Hz, 2H), Z 7.34 (d, J = 8.8 Hz, 2H), Z 6.76 (s, 1H), E 4.27 (q, J = 7.1 Hz, 2H), Z 4.10 (q, J = 7.1 Hz, 2H), E/Z 2.81–2.76 (m, 2H), E/Z 2.76–2.71 (m, 4H), E 2.68 (t, J = 7.6 Hz, 2H), Z 1.33 (t, J = 7.1 Hz, 3H), Z 1.08 (t, J = 7.1 Hz, 3H). 13C NMR (151 MHz, CDCl3, 25 °C) δ E 200.4, E/Z 166.9, E/Z 147.5, E/Z 141.9, E/Z 137.6, E 129.7, E 123.9, E 61.5, E 42.9, E/Z 27.8, E/Z 14.3, Z 200.4, Z 167.9, Z 129.0, Z 123.4, Z 61.2, 42.4, E/Z 20.3. HRMS (ESI-TOF): [M + H]+ calcd for C14H16NO5 278.1023, found 278.1042.
Ethyl (E)-2-(benzo[d][1,3]dioxol-5-ylmethylene)-5-oxopentanoate (8; R = 3,4-(O-CH2-O)-Ph). Physical state: yellow oil. Yield: 75% (E/Z 16.0
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0). 1H NMR (600 MHz, CDCl3, 25 °C) δ 9.76 (t, J = 1.4 Hz, 1H), 7.63 (s, 1H), 6.82–6.78 (m, 4H), 5.96 (s, 2H); 4.23 (q, J = 7.1 Hz, 2H), 2.89–2.81 (m, 2H), 2.67–2.63 (m, 2H), 1.30 (t, J = 7.1 Hz, 3H). 13C NMR (151 MHz, CDCl3, 25 °C) δ 201.2, 168.1, 148.0, 139.9, 130.1, 129.2, 124.0, 109.0, 108.6, 101.4, 61.0, 43.1, 20.3, 13.9. MS (70 eV) m/z: 276 (M+), 115 (100). IR (ATR): 2978, 2896, 1702, 1623, 1494, 1446, 1365. HRMS (ESI-TOF): [M + H]+ calcd for C15H17O5 277.1071, found 277.1071.
1.0). 1H NMR (600 MHz, CDCl3, 25 °C) δ 9.76 (t, J = 1.4 Hz, 1H), 7.63 (s, 1H), 6.82–6.78 (m, 4H), 5.96 (s, 2H); 4.23 (q, J = 7.1 Hz, 2H), 2.89–2.81 (m, 2H), 2.67–2.63 (m, 2H), 1.30 (t, J = 7.1 Hz, 3H). 13C NMR (151 MHz, CDCl3, 25 °C) δ 201.2, 168.1, 148.0, 139.9, 130.1, 129.2, 124.0, 109.0, 108.6, 101.4, 61.0, 43.1, 20.3, 13.9. MS (70 eV) m/z: 276 (M+), 115 (100). IR (ATR): 2978, 2896, 1702, 1623, 1494, 1446, 1365. HRMS (ESI-TOF): [M + H]+ calcd for C15H17O5 277.1071, found 277.1071.
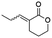
(E)-3-Ethylidenetetrahydro-2H-pyran-2-one (10E). CAS No [61203-10-9] physical state: yellow oil. Yield: 38%. 1H NMR (600 MHz, CDCl3, 25 °C) δ 7.14 (qt, J = 7.2, 2.4 Hz, 1H), 4.32–4.29 (m, 2H), 2.54–2.50 (m, 2H), 1.94 (dt, J = 11.1, 6.5 Hz, 2H), 1.79 (dt, J = 7.2, 1.4 Hz, 3H). 13C NMR (151 MHz, CDCl3, 25 °C) δ 166.7, 141.3, 126.5, 68.3, 23.6, 22.5, 14.2. MS (70 eV) m/z: 126 (M+), 67 (100). IR (ATR): 2947, 1710, 1636, 1396, 1251, 1141, 1072. HRMS (ESI-TOF): [M + H]+ calcd for C7H11O2 127.0754, found 127.0750.
(Z)-3-Ethylidenetetrahydro-2H-pyran-2-one (10Z). CAS No [61203-11-0] physical state: yellow oil. Yield: 18%. 1H NMR (300 MHz, CDCl3, 25 °C) δ 6.18 (qt, J = 7.2, 1.8 Hz, 1H), 4.30–4.24 (m, 2H), 2.61–2.52 (m, 2H), 2.10 (dt, J = 7.3, 1.8 Hz, 3H), 1.96–1.88 (m, 2H). 13C NMR (75 MHz, CDCl3, 25 °C) δ 165.9, 142.6, 125.7, 68.6, 29.1, 23.2, 16.2. MS (70 eV) m/z: 126 (M+), 67 (100). IR (ATR): 2941, 1710, 1630, 1440, 1394, 1325, 1251, 1141, 1078. HRMS (ESI-TOF): [M + H]+ calcd for C7H11O2 127.0754, found 127.0752.
(E)-3-Propylidenetetrahydro-2H-pyran-2-one (11E). CAS No [98011-26-8] physical state: yellow oil. Yield: 67%. 1H NMR (600 MHz, CDCl3, 25 °C) δ 7.06–7.01 (m, 1H), 4.33–4.29 (m, 2H), 2.54–2.51 (m, 2H), 2.20–2.14 (m, 2H), 1.96–1.92 (m, 2H), 1.09–1.07 (m, 3H). 13C NMR (75 MHz, CDCl3, 25 °C) δ 166.8, 147.8, 125.0, 68.6, 23.7, 23.0, 21.6, 12.7. MS (70 eV) m/z: 140 (M+), 67 (100). IR (ATR): 2970, 2866, 1710, 1636, 1383, 1302, 1244, 1153, 1089. HRMS (ESI-TOF): [M + H]+ calcd for C8H13O2 141.0910, found 141.0920.
(Z)-3-Propylidenetetrahydro-2H-pyran-2-one (11Z). CAS No [98011-27-9] physical state: yellow oil. Yield: 10%. 1H NMR (300 MHz, CDCl3, 25 °C) δ 6.02 (tt, J = 7.2, 1.7 Hz, 1H), 4.29–4.23 (m, 2H), 2.60–2.50 (m, 4H), 1.97–1.86 (m, 2H), 1.03 (t, J = 7.5 Hz, 3H). 13C NMR (75 MHz, CDCl3, 25 °C) δ 165.8, 149.6, 124.1, 68.6, 29.3, 23.4, 23.2, 13.6. MS (70 eV) m/z: 140 (M+), 67 (100). IR (ATR): 2964, 2878, 1705, 1630, 1389, 1239, 1141, 1066. HRMS (ESI-TOF): [M + H]+ calcd for C8H13O2 141.0910, found 141.0919.
(E)-3-Heptylidenetetrahydro-2H-pyran-2-one (12E). CAS No [68872-81-1] physical state: yellow oil. Yield: 50%. 1H NMR (600 MHz, CDCl3, 25 °C) δ 7.06 (tt, J = 7.5, 2.4 Hz, 1H), 4.32–4.29 (m, 2H), 2.52 (t, J = 6.6 Hz, 2H), 2.14 (q, J = 7.4 Hz, 2H), 1.95–1.91 (m, 2H), 1.49–1.43 (m, 2H), 1.36–1.23 (m, 6H), 0.89 (t, J = 6.9 Hz, 3H). 13C NMR (75 MHz, CDCl3, 25 °C) δ 166.8, 146.8, 125.3, 68.5, 31.6, 29.0, 28.3, 28.1, 23.6, 22.6, 22.5, 14.0. MS (70 eV) m/z: 196 (M+), 113 (100). IR (ATR): 2935, 2854, 1716, 1624, 1394, 1256, 1170. HRMS (ESI-TOF): [M + H]+ calcd for C12H21O2 197.1542, found 197.1529.
(Z)-3-Heptylidenetetrahydro-2H-pyran-2-one (12Z). CAS No [68872-79-7] physical state: yellow oil. Yield: 10%. 1H NMR (600 MHz, CDCl3, 25 °C) δ 6.06 (tt, J = 7.2, 1.7 Hz, 1H), 4.30–4.28 (m, 2H), 2.63–2.57 (m, 4H), 1.96–1.93 (m, 2H), 1.33–1.30 (m, 6H), 0.90 (t, J = 7.0 Hz, 3H). 13C NMR (75 MHz, CDCl3, 25 °C) δ 165.9, 148.4, 124.5, 68.8, 31.6, 29.0, 28.3, 28.1, 23.4, 22.6, 22.5, 14.0. MS (70 eV) m/z: 196 (M+), 139 (100). IR (ATR): 2965, 2936, 2876, 1715, 1635, 1396, 1236, 1139. HRMS (ESI-TOF): [M + H]+ calcd for C12H21O2 197.1542, found 197.1530.
(E)-3-Octylidenetetrahydro-2H-pyran-2-one (13E) new compound. Physical state: yellow oil. Yield: 54%. 1H NMR (600 MHz, CDCl3, 25 °C) δ 7.06 (tt, J = 7.5, 2.3 Hz, 1H), 4.31–4.26 (m, 2H), 2.52 (m, 2H), 2.14 (q, J = 7.5 Hz, 2H), 1.94–1.93 (m, 2H), 1.31–1.27 (m, 11H), 0.88 (t, J = 7.0 Hz, 3H). 13C NMR (151 MHz, CDCl3, 25 °C) δ 166.7, 146.8, 125.3, 68.5, 31.7, 29.4, 29.1, 28.3, 28.1, 23.6, 22.7, 22.6, 14.1. MS (70 eV) m/z: 210 (M+), 113 (100). IR (ATR): 2912, 2861, 1716, 1630, 1457, 1389, 1262, 1164, 1106, 1078. HRMS (ESI-TOF): [M + H]+ calcd for C13H23O2 211.1698, found 211.1690.
(Z)-3-Octylidenetetrahydro-2H-pyran-2-one (13Z) new compound. Physical state: yellow oil. Yield: 26%. 1H NMR (600 MHz, CDCl3, 25 °C) δ 6.04 (tt, J = 7.2, 1.7 Hz, 1H), 4.28–4.25 (m, 2H), 2.64–2.53 (m, 4H), 1.96–1.88 (m, 2H), 1.46–1.38 (m, 2H), 1.35–1.21 (m, 9H), 0.88 (t, J = 7.1 Hz, 3H). 13C NMR (151 MHz, CDCl3, 25 °C) δ 166.3, 148.5, 124.7, 68.8, 31.8, 29.8, 29.4, 29.3, 29.3, 29.1, 23.4, 22.7, 14.1. MS (70 eV) m/z: 210 (M+), 139 (100). IR (ATR): 2923, 2861, 1716, 1630, 1394, 1233, 1135. HRMS (ESI-TOF): [M + H]+ calcd for C13H23O2 211.1698, found 211.1690.
(E)-3-(2-Methylpropylidene)tetrahydro-2H-pyran-2-one (14E) new compound. Physical state: yellow oil. Yield: 18%. 1H NMR (600 MHz, CDCl3, 25 °C) δ 6.89 (dt, J = 10.1, 2.3 Hz, 1H), 4.34–4.30 (m, 2H), 2.65–2.55 (m, 1H), 2.56 (td, J = 6.7, 2.3 Hz, 2H), 1.97–1.91 (m, 2H), 1.04 (d, J = 6.7 Hz, 6H). 13C NMR (75 MHz, CDCl3, 25 °C) δ 166.9, 152.4, 123.3, 68.4, 27.5, 23.3, 22.7, 21.4. MS (70 eV) m/z: 154 (M+), 81 (100). IR (ATR): 2958, 2861, 1710, 1624, 1394, 1313, 1256, 1135. HRMS (ESI-TOF): [M + H]+ calcd for C9H15O2 155.1072, found 155.1075.
(E)-3-Benzylidenetetrahydro-2H-pyran-2-one (15E). CAS No [98011-25-7] physical state: yellow solid. Yield: 47%. 1H NMR (600 MHz, CDCl3, 25 °C) δ 7.93 (t, J = 2.3 Hz, 1H), 7.46–7.34 (m, 6H), 4.42–4.38 (m, 2H), 2.89 (dt, J = 6.59, 2.47 Hz, 2H), 2.00–1.95 (m, 2H). 13C NMR (151 MHz, CDCl3, 25 °C) δ 167.1, 141.5, 135.2, 130.3, 129.2, 128.7, 125.8, 68.8, 26.1, 23.2. MS (70 eV) m/z: 188 (M+), 115 (100). IR (ATR): 2977, 1700, 1623, 1483, 1439, 1395, 1264, 1169, 1111. HRMS (ESI-TOF): [M + H]+ calcd for C12H13O2 189.0916, found 189.0906.
(E)-3-(4-Chlorobenzylidene)tetrahydro-2H-pyran-2-one (16E). CAS No [2409927-23-5] physical state: yellow solid. Yield: 45%. 1H NMR (600 MHz, CDCl3, 25 °C) δ 7.86 (s, 1H), 7.42–7.35 (m, 4H), 4.56–4.31 (m, 2H), 2.85 (td, J = 6.7, 2.2 Hz, 1H), 2.03–1.77 (m, 1H). 13C NMR (151 MHz, CDCl3, 25 °C) δ 166.6, 140.3, 135.1, 133.3, 131.6, 128.9, 126.3, 68.6, 25.8, 23.1. MS (70 eV) m/z: 188 (M+), 115(100). IR (ATR): 2999, 2964, 2888, 1699, 1619, 1590, 1480, 1394, 1256, 1164, 822. HRMS (ESI-TOF): [M + H]+ calcd for C12H12ClO2 223.0526, found 223.0533.
(E)-3-(4-Bromobenzylidene)tetrahydro-2H-pyran-2-one (17E). CAS No [2639400-91-0] physical state: yellow solid. Yield: 52%. 1H NMR (600 MHz, CDCl3, 25 °C) δ 7.84 (t, J = 2.2 Hz, 1H), 7.57–7.54 (m, 2H), 7.32–7.28 (m, 2H), 4.43–4.39 (m, 1H), 2.84 (td, J = 6.6, 2.4 Hz, 1H), 2.03–1.96 (m, 1H). 13C NMR (151 MHz, CDCl3, 25 °C) δ 166.7, 140.1, 133.9, 131.8, 131.6, 126.5, 123.4, 68.7, 26.0, 22.9. MS (70 eV) m/z: 265 (M+), 267 ((M+2)+), 128 (100). IR (ATR): 2992, 2964, 2935, 2889, 2854, 1705, 1612, 1585, 1481, 1262, 1175. HRMS (ESI-TOF): [M + H]+ calcd for C12H1279BrO2 267.0021, found 266.9992.
(E)-3-(Benzo[d][1,3]dioxol-5-ylmethylene)tetrahydro-2H-pyran-2-one (18E). CAS No [906423-89-0] physical state: yellow solid. Yield: 42%. 1H NMR (600 MHz, CDCl3, 25 °C) δ 7.83 (t, J = 2.1 Hz, 1H), 7.12–6.94 (m, 2H), 6.87 (d, J = 8.0 Hz, 1H), 6.03 (s, 2H), 4.47–4.34 (m, 2H), 2.87 (td, J = 6.7, 2.2 Hz, 2H), 2.09–1.94 (m, 2H). 13C NMR (151 MHz, CDCl3, 25 °C) δ 167.2, 148.5, 147.9, 141.4, 129.2, 126.0, 123.8, 109.8, 108.6, 101.5, 68.5, 26.1, 23.0. MS (70 eV) m/z: 232 (M+), 148 (100). IR (ATR): 2970, 1681, 1590, 1509, 1452, 1273, 1239, 1164. HRMS (ESI-TOF): [M + H]+ calcd for C13H13O4 233.0808, found 233.0811.
(E)-3-(4-Nitrobenzylidene)tetrahydro-2H-pyran-2-one (19E). CAS No [1334334-84-7] physical state: yellow solid. Yield: 20%. 1H NMR (300 MHz, CDCl3, 25 °C) δ 8.32–8.23 (m, 1H), 7.93 (t, J = 2.4 Hz, 1H), 7.61–7.54 (m, 2H), 4.49–4.40 (m, 2H), 2.87 (td, J = 6.6, 2.5 Hz, 2H), 2.05–1.97 (m, 2H). 13C NMR (75 MHz, CDCl3, 25 °C) δ 165.9, 141.2, 138.8, 130.6, 129.5, 123.7, 68.6, 25.83, 23.0. MS (70 eV) m/z: 232 (M+), 115 (100). IR (ATR): 2970, 1699, 1515, 1331, 1164, 1112, 1066. HRMS (ESI-TOF): [M + H]+ calcd for C12H12NO4 234.0766, found 234.0779.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v), under stirring for 18 hours at room temperature. Then, the reaction was acidified to pH = 1 with 1 M HCl, and a white solid was filtered through a Büchner funnel, yielding 1.7 g (7.9 mmol, 86% yield) of a solid characterized as S4.† A solution of 0.5 g of S4† (2 mmol) with 0.4 g of 1-ethyl-3-(3-dimethyl aminopropyl) carbodiimide hydrochloride (EDC·HCl, 2 mmol) in 20 mL of dry dichloromethane was prepared in a 50 mL flask under inter atmosphere. The reaction was stirred for 5 minutes at 0 °C. To this mixture, 2.3 mmol of lactone and 1.3 mL of Et3N (9.3 mmol) were added. The reaction was left under stirring for an additional 1 hour at 0 °C and 18 hours at room temperature. The reaction was then washed with 5 × 20 mL of HCl (1 M), and 5 × 20 mL of a saturated solution of NaHCO3. The organic phase was dried with Na2SO4 and the solvent was removed on a rotary evaporator. Purification was done on a silica gel column (dichloromethane/ethyl acetate 2
1 v/v), under stirring for 18 hours at room temperature. Then, the reaction was acidified to pH = 1 with 1 M HCl, and a white solid was filtered through a Büchner funnel, yielding 1.7 g (7.9 mmol, 86% yield) of a solid characterized as S4.† A solution of 0.5 g of S4† (2 mmol) with 0.4 g of 1-ethyl-3-(3-dimethyl aminopropyl) carbodiimide hydrochloride (EDC·HCl, 2 mmol) in 20 mL of dry dichloromethane was prepared in a 50 mL flask under inter atmosphere. The reaction was stirred for 5 minutes at 0 °C. To this mixture, 2.3 mmol of lactone and 1.3 mL of Et3N (9.3 mmol) were added. The reaction was left under stirring for an additional 1 hour at 0 °C and 18 hours at room temperature. The reaction was then washed with 5 × 20 mL of HCl (1 M), and 5 × 20 mL of a saturated solution of NaHCO3. The organic phase was dried with Na2SO4 and the solvent was removed on a rotary evaporator. Purification was done on a silica gel column (dichloromethane/ethyl acetate 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5). The fractions were collected, and the solvent was removed on a rotary evaporator. Then, the material was submitted to a high vacuum to remove any residual solvent, yielding 0.41 mg (1.4 mmol, 61% yield) of a solid characterized as a known inhibitor, 20.
0.5). The fractions were collected, and the solvent was removed on a rotary evaporator. Then, the material was submitted to a high vacuum to remove any residual solvent, yielding 0.41 mg (1.4 mmol, 61% yield) of a solid characterized as a known inhibitor, 20.
(S)-4-(4-Chlorophenoxy)-N-(2-oxotetrahydrofuran-3-yl)butanamide (20). CAS No [1453857-28-7] 1H RMN (600 MHz, CDCl3, 25 °C): δ 7.24–7.20 (m, 2H), 6.84–6.79 (m, 2H), 6.23 (d, J = 5.4 Hz, 1H), 4.56 (ddd, J = 11.6, 8.6, 6.1 Hz, 1H), 4.48–4.44 (m, 1H), 4.28 (ddd, J = 11.2, 9.3, 5.9 Hz, 1H), 3.98 (td, J = 6.1, 1.5 Hz, 2H), 2.84–2.77 (m, 1H), 2.47 (dd, J = 10.8, 4.1 Hz, 2H), 2.13 (ddd, J = 13.3, 7.2, 6.2 Hz, 3H). 13C RMN (151 MHz, CDCl3, 25 °C): δ 175.4, 172.9, 157.3, 129.3, 125.7, 115.7, 67.0, 66.0, 49.2, 32.3, 30.4, 24.8. EM (70 eV) m/z: 170 (100). IR (ATR) cm−1: 3291, 1767, 1643, 1540, 1493, 1243, 1189, 660.
Bacterial strains and growth conditions
C. subtsugae strain CV026 (AHL-deficient mutant of C. subtsugae strain ATCC 31532 – genotype cviI![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mini-Tn5, Kmr, AHL−) was kept under cryopreservation (−80 °C) in LB medium supplemented with 50% glycerol. CV026 was recovered with kanamycin (50 µg mL−1) and cultured in LB broth (pH 7.2) under stirring at 200 rpm for 24 hours at 28 °C. To obtain isolated colonies, LB agar supplemented with HHL (2) (4 nM) was streaked with 10 µL of bacterial culture and incubated at 28 °C for 48 h. Isolated CV026 colonies were surveyed for the steady production of violacein.
mini-Tn5, Kmr, AHL−) was kept under cryopreservation (−80 °C) in LB medium supplemented with 50% glycerol. CV026 was recovered with kanamycin (50 µg mL−1) and cultured in LB broth (pH 7.2) under stirring at 200 rpm for 24 hours at 28 °C. To obtain isolated colonies, LB agar supplemented with HHL (2) (4 nM) was streaked with 10 µL of bacterial culture and incubated at 28 °C for 48 h. Isolated CV026 colonies were surveyed for the steady production of violacein.
Violacein quantification assay
Isolated colonies of CV026 (LB agar – 48 hours at 28 °C) were used to make a bacterial suspension in LB medium adjusted to 2.0 McFarland scale. The bacterial suspension was used to inoculate the test medium in a proportion of 10% v/v. The test medium was then supplemented with the cognate autoinducer (N-hexanoyl-L-homoserine 96% lactone, Sigma-Aldrich), at a final concentration of 10 µM. A 1.95 mL aliquot of this culture was transferred to a conical falcon tube (v = 15 mL) and supplemented with 50 µL of the tested molecules in an acetone solution of 25%, in different final concentrations (2.5 mM, 1.25 mM, 625 µM and 312 µM). The assays were incubated under agitation at 200 rpm at 30 °C for 24 hours and then were put in a kiln to completely evaporate the liquid medium. Violacein was solubilized from dried bacterial pellets with 1 mL of DMSO. After 24 h of agitation, the absorbance of soluble violacein was measured at 630 nm using a microplate reader (Biotek – ELx808™).The same experimental conditions were repeated, in parallel, in the absence of the cognate auto-inducer (“blank”) for evaluating the bacterial growth by measuring turbidity (630 nm) after 24 hours under agitation at 200 rpm and 30 °C in a microplate reader (Biotek – ELx808™).
Competitive assay
Competition assays for the CviR binding site were performed following the same set-up as the violacein quantification assay, but adopting different concentrations of the cognate AI, 1, (10 nm to 1 mM) and a fixed concentration of lactone 20 (312 µM).Chitinase assay on agar plates
The protocol established by Chernin et al. for testing chitin hydrolysis in Chromobacterium CV026 was conducted with minor modifications. Isolated colonies of CV026 (LB agar – 48 hours at 28 °C) were used to make a bacterial suspension in NaCl 0.9% (w/w) medium adjusted to 2.0 McFarland scale. A 10 µL aliquot of this suspension was placed onto plates with a semi-minimal medium consisting of 10% LB (v/v), 0.2% (w/v) colloidal chitin32 and supplemented with HHL at a final concentration of 10 µM (positive control of chitinase hydrolysis – PC). A plate without HHL was made for the negative control of chitinase hydrolysis (NC). 20 and 15E were evaluated in chitinase hydrolysis assay. In the test medium, the respective molecules were added at the final concentrations of 32 µM for 20 and 312 µM for 15E. The plates were incubated at 30 °C for 72 hours until zones of clearing of the chitin could be seen around the colonies.RT-qPCR assays
Total RNA was extracted from CV026 cells in the same condition as in the violacein quantification assay using the ReliaPrep™ RNA Miniprep System (Promega) as indicated by the manufacturer's instructions. Primers for vioA, vioC, and gyrB were designed using the web-based tool Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast) (Table 3).37| Primer | Sequence |
|---|---|
| vioA-F | ATCCGGAAATCCAGAGCTTC |
| vioA-R | ACTTGTCGCCCTTGAAGTAG |
| vioC-F | GCCTTTTTCGACCGTTACTTC |
| vioC-R | CTATGCATGTAGCGGGTGTA |
| gyrB-F | TCACCATCAATCCGGACAAC |
| gyrB-R | TCTTGTTGCGGTTCATGTATTC |
Total RNA was used as a template for the reverse transcription reaction using the GoTaq® 1-Step RT-qPCR System (Promega). The RT-qPCR was performed in one step according to the manufacturer's protocol. The reaction mix (20 µL) was made up as follows: 10 µL GoTaq® qPCR Master Mix, 2.5 µL forward and reverse primers (150 nM), 0.4 µL GoScript™ RT Mix for 1-Step RT-qPCR and 4 µL of the RNA template (20 ng). RT-qPCR was performed using Applied Biosystems Quant Studio™ 3 Real-Time PCR System (Applied Biosystems Inc., CA, USA). The reaction conditions are as follows: reverse transcriptase reaction at 37 °C for 15 minutes, RT enzyme inactivation at 95 °C for 10 minutes, followed by 40 cycles of denaturation at 95 °C for 10 s, annealing at 54 °C for 30 s and extension at 72 °C for 30 s. The relative mRNA expression of vioA and vioC was calculated using the ΔΔCt method. gyrB was used as an internal reference gene for normalization. These experiments were independently conducted 3 times.
Evaluation of the antagonistic activity of compounds 9–21 through molecular docking analysis
In this study, AutoDock Vina 1.1.2 and VMD 1.9.3 (ref. 35 and 38) programs were used for molecular docking and visualization. Molecular docking was used for the prediction of the interaction between the possible QSI and CviR. The structures of the molecules were optimized using density functional theory at B3LYP/6-311G (d,p) level in Gaussian 6.06.39 The enzyme structure of CviR (PDB ID 3QP5) was obtained from the RCSB protein data bank (https://www.rcsb.org/) and the molecular docking studies were carried out using the dimer structure. The grid box center was set at the centre of the dimer (x = 39, y = −0.15, z = 15.71), and the dimensions of the box were set at 60 Å × 90 Å × 100 Å. The docking results are a list of the free energy of binding (ΔG) of the protein and molecules binding poses. This can be considered as a measure of a predicted binding affinity in kcal mol−1. Last, the complex structures were evaluated using VMD 1.9.3 program, and the distance between the molecules and the amino acids was measured.Redocking was used to ensure that this computational tool was able to mimic the interactions between ligand and protein. The overlap between the chlorolactone pose from the X-ray crystal structure and the docking one (Fig. S17†) is considered good for determining docking as an accurate computational predictor of interactions between ligand and protein. Molecule 21 had no interaction within the binding site, as seen in Fig. S32,† even when the grid box was reduced to the AIBD size.
It is important to mention that we were careful to use the correct pdb for the strain tested. The 3QP5 pdb file is the crystallographic structure of the X-ray of the CviR complex with the Chlorolactone 20 (C. subtsugae ATCC 31532/CV026).
LogP
Chemicalize was used for the prediction of logP. https://chemicalize.com/developed by ChemAxon (https://www.chemaxon.com).Author contributions
W. T., A. L. P. and A. H. L. M. designed the research. F. F. and T. A. T. performed the synthetic chemistry research. F. F. performed the biological assays, and the computational biology research. F. F., T. A. T., V. F., W. T., A. L. P. and A. H. L. M. analysed the data. F. F., A. L. P. and A. H. L. M. wrote the paper. T. A. T., V. F., W. T., A. L. P. and A. H. L. M. revised the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brazil (CAPES) – Finance Code 001, Conselho Nacional de Desenvolvimento Científico e Tecnológico – Brazil (CNPq) – 477418/2013-9, Fundação de Apoio à Pesquisa do Distrito Federal – Brazil (FAPDF) 193.001.049/2015 and 0193.001.560/2017, Decanato de Pesquisa e Pós-graduação – Brazil (DPG/UnB) – DPG 0004/2021.Notes and references
- H. M. C. Ferraz, F. I. Bombonato, M. K. Sano and L. S. Longo, Quim. Nova, 2008, 31, 885 CrossRef CAS
.
- S. L. Robinson, J. K. Christenson and L. P. Wackett, Nat. Prod. Rep., 2019, 36, 458 RSC
.
- A. Albrecht, J. F. Koszuk, J. Modranka, M. Różalski, U. Krajewska, A. Janecka, K. Studzian and T. Janecki, Bioorg. Med. Chem., 2008, 16, 4872 CrossRef CAS PubMed
.
- T. Janecki, Natural Lactones and Lactams: Synthesis, Occurrence and Biological Activity, Wiley‐VCH, Weinheim, 2014 Search PubMed
.
- S. Mukherjee and A. T. Biju, Chem.–Asian J., 2018, 13, 2333 CrossRef CAS PubMed
.
- D. M. Souza, L. L. Machado and A. H. L Machado, Tetrahedron Lett., 2019, 60, 1811–1813 CrossRef CAS
.
- M. Whiteley, S. Diggle and E. Greenberg, Nature, 2017, 551, 313–320 CrossRef CAS PubMed
.
- A. Camilli and B. B. Bassler, Science, 2006, 311, 1113–1116 CrossRef CAS PubMed
.
- M. Thoendel, J. S. Kavanaugh, C. E. Flack and A. R. Horswill, Chem. Rev., 2011, 111, 117–151 CrossRef CAS PubMed
.
- X. Huang, O. P. Duddy, J. E. Silpe, J. E. Paczkowski, J. Cong, B. R. Henke and B. L. Bassler, J. Biol. Chem., 2020, 295, 291 CrossRef PubMed
.
- A. G. Palmer, A. C. Senechal, T. C. Haire, N. P. Mehta, S. D. Valiquette and H. E. Backwell, ACS Chem. Biol., 2018, 13, 31115–31122 CrossRef PubMed
.
- X. Zhao, Z. Yu and T. Ding, Microorganisms, 2020, 8, 425 CrossRef CAS PubMed
.
- G. Chen, L. R. Swem, D. L. Swem, D. L. Stauff, C. T. O'Loughlin, P. D. Jeffrey, B. L. Bassler and F. M. Hughson, Mol. Cell, 2011, 42, 199 CrossRef CAS PubMed
.
- S. Haque, D. K. Yadav, S. C. Bisht, N. Yadav, V. Singh, K. K. Dubey, A. Jawed, M. Wahid and S. A. Dar, J. Chemother., 2019, 31, 161 CrossRef CAS PubMed
.
- L. Lu, M. Li, G. Yi, L. Liao, Q. Cheng, J. Zhu, B. Zhang, Y. Wang, Y. Chen and M. Zeng, J. Pharm. Anal., 2021, 12, 1 Search PubMed
.
- B. Rémy, S. Mion, L. Plener, M. Elias, E. Chabrière and D. Daudé, Front. Pharmacol., 2018, 9, 203 CrossRef PubMed
.
- E. Shaw and W. M. Wuest, RSC Med. Chem., 2020, 11, 358 RSC
.
- P. Piewngam, J. Chiou, P. Chatterjee and M. Otto, Expert Rev. Anti-Infect. Ther., 2020, 18, 499 CrossRef CAS PubMed
.
- M. E. Mattmann and H. E. Blackwell, J. Org. Chem., 2010, 75, 6737 CrossRef CAS PubMed
.
- T. Zang, B. W. K. Lee, L. M. Cannon, K. A. Ritter, S. Dai, D. Ren, T. K. Wood and Z. S. Zhou, Bioorg. Med. Chem. Lett., 2009, 19, 6200 CrossRef CAS PubMed
.
- S. Manner and A. Fallarero, Int. J. Mol. Sci., 2018, 19, 1346 CrossRef PubMed
.
- O. M. Vandeputte, M. Kiendrebeogo, T. Rasamiravaka, C. Stévigny, P. Duez, S. Rajaonson, B. Diallo, A. Mol, M. Baucher and M. el Jaziri, Microbiology, 2011, 157, 2120 CrossRef CAS PubMed
.
- K. H. McClean, M. K. Winson, L. Fish, A. Taylor, S. R. Chhabra, M. Camara, M. Daykin, J. H. Lamb, S. Swift, B. W. Bycroft, G. S. A. B. Stewart and P. Williams, Microbiology, 1997, 143, 3703 CrossRef CAS PubMed
.
- G. Devescovi, M. Kojic, S. Covaceuszach, M. Cámara, P. Williams, I. Bertani, S. Subramoni and V. Venturi, Front. Microbiol., 2017, 8, 349 CrossRef PubMed
.
- A. M. Harrison and S. D. Soby, AMB Express, 2020, 10, 202 CrossRef CAS PubMed
.
- T. Hoshino, Appl. Microbiol. Biotechnol., 2011, 91, 1463–1475 CrossRef CAS PubMed
.
- N. Durán, G. Z. Justo, M. Durán, M. Brocchi, L. Cordi, L. Tasic, G. R. Castro and G. Nakazato, Biotechnol. Adv., 2016, 34, 1030 CrossRef PubMed
.
- A. Chang, S. Sun, L. Li, X. Dai, H. Li, Q. He and H. Zhu, Bioorg. Chem., 2019, 91, 103140 CrossRef CAS PubMed
.
- X. Zhuang, A. Zhang and W. Chu, Int. Microbiol., 2020, 23, 215 CrossRef CAS PubMed
.
- R. E. D'Almeida, R. D. I. Molina, C. M. Viola, M. C. Luciardi, C. Nieto Peñalver, A. Bardón and M. E. Arena, Bioorg. Chem., 2017, 73, 37 CrossRef PubMed
.
- A. Albrecht, Ł. Albrecht and T. Janecki, Eur. J. Org. Chem., 2011, 2747 CrossRef CAS
.
- L. S. Chernin, M. K. Winson, J. M. Thompson, S. Haran, B. W. Byceoft, I. Chet, P. Willians and G. S. A. B. Stewart, J. Bacteriol., 1998, 180, 4435–4441 CrossRef CAS PubMed
.
- V. S. Silva, T. A. Tolentino, T. C. A. F. Rodrigues, F. F. M. Santos, D. F. S. Machado, W. A. Silva, H. C. B. De Oliveira and A. H. L. Machado, Org. Biomol. Chem., 2019, 17, 4498 RSC
.
- L. R. Swem, D. L. Swem, C. T. O'Loughlin, R. Gatmaitan, B. Zhao, S. M. Ulrich and B. L. Bassler, Mol. Cell, 2009, 35, 143 CrossRef CAS PubMed
.
- O. Trott and A. J. Olson, J. Comput. Chem., 2010, 31, 455 CAS
.
- C. Vraka, L. Nics, K. H. Wagner, M. Hacker, W. Wadsak and M. Mitterhauser, Nucl. Med. Biol., 2017, 50, 1 CrossRef CAS PubMed
.
- J. Ye, G. Coulouris, I. Zaretskaya, I. Cutcutache, S. Rozen and T. L. Madden, BMC Bioinf., 2012, 13, 134 CrossRef CAS PubMed
.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14, 33 CrossRef CAS PubMed
.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji, X. Li, M. Caricato, A. V. Marenich, J. Bloino, B. G. Janesko, R. Gomperts, B. Mennucci, H. P. Hratchian, J. V. Ortiz, A. F. Izmaylov, J. L. Sonnenberg, D. Williams-Young, F. Ding, F. Lipparini, F. Egidi, J. Goings, B. Peng, A. Petrone, T. Henderson, D. Ranasinghe, V. G. Zakrzewski, J. Gao, N. Rega, G. Zheng, W. Liang, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, K. Throssell, J. A. MontgomeryJr, J. E. Peralta, F. Ogliaro, M. J. Bearpark, J. J. Heyd, E. N. Brothers, K. N. Kudin, V. N. Staroverov, T. A. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. P. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, J. M. Millam, M. Klene, C. Adamo, R. Cammi, J. W. Ochterski, R. L. Martin, K. Morokuma, O. Farkas, J. B. Foresman and D. J. Fox, Gaussian 16, Revision C.01, Gaussian, Inc., Wallingford CT, 2016 Search PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra01975f |
| This journal is © The Royal Society of Chemistry 2023 |