Open Access Article
Open Access ArticleBiotransformation of benzo[a]pyrene by Pannonibacter sp. JPA3 and the degradation mechanism through the initially oxidized benzo[a]pyrene-4,5-dihydrodiol to downstream metabolites†
Jingnan Jin *a,
Yahui Shia,
Baozhong Zhanga,
Dongjin Wana,
Qingye Zhangb and
Ying Lia
*a,
Yahui Shia,
Baozhong Zhanga,
Dongjin Wana,
Qingye Zhangb and
Ying Lia
aSchool of Environmental Engineering, Henan University of Technology, No. 100 Lianhua Street, High-Tech Industrial Development District, Zhengzhou, Henan 450001, China. E-mail: jingnanjin@haut.edu.cn; Fax: +86-371-67756982; Tel: +86-371-67756982
bCollege of Informatics, Huazhong Agricultural University, Wuhan 430070, China
First published on 21st June 2023
Abstract
Owing to its adverse effects on the environment and human health, benzo[a]pyrene (BaP) has attracted considerable attention and has been used as a model compound in ecotoxicology. In this study, Pannonibacter sp. JPA3 as a BaP-degrading strain was isolated from the production water of an oil well. The strain could remove 80% of BaP at an initial concentration of 100 mg L−1 after 35 d culture. The BaP-4,5-dihydrodiol, BaP-4,5-epoxide, 5-hydroxychrysene, and 2-hydroxy-1-naphthoic acid metabolites were identified in the biodegradation process. Simultaneously, the gene sequence coding for dioxygenase in the strain was amplified and a dioxygenase model was built by homology modeling. Combined with the identification of the metabolites, the interaction mechanism of BaP with dioxygenase was investigated using molecular docking. It was assumed that BaP was initially oxidized at the C4–C5 positions in the active cavity of dioxygenase. Moreover, a hypothesis for the progressive degradation mechanism of BaP by this strain was proposed via the identification of the downstream metabolites. In conclusion, our study provided an efficient BaP degrader and a comprehensive reference for the study of the degradation mechanism in terms of the degrading metabolites and theoretical research at the molecular level.
1. Introduction
Benzo[a]pyrene (BaP), a representative member of polycyclic aromatic hydrocarbons (PAHs) with five fused benzene rings, has raised extensive environmental concerns owing to its carcinogenic, teratogenic, and mutational properties.1–3 In general, BaP is derived from oil leak accidents and the incomplete combustion of coal, oil and gasoline. Nowadays, BaP is widely used as a model PAH in ecotoxicology research. Furthermore, considering its significant carcinogenicity to humans, the removal of BaP has become a research focus.4–6To date, many types of remediation technologies have been applied to eliminate PAHs from the environment, such as chemical oxidation, bioaccumulation and microbial degradation. Owing to its environment-friendly and cost-efficient characteristics, microbial degradation has been extensively studied.7,8 Simultaneously, the microbial degradation of BaP is complex in ecosystems on account of its low water solubility of 0.0023 mg L−1.9,10 Thus far, many kinds of microorganisms possessing PAH-catabolic enzymes have been screened by the microbial adaptation for the remediation of polluted environments, such as genera of Pseudomonas, Beijerinckia, Mycobacterium, Sphingomonas, Bacillus, and Stenotrophomonas.11–14
In fact, the biodegradation process for low-molecular-weight PAHs has been widely elucidated in detail. By contrast, the transformation pathway of high-molecular-weight PAHs is less understood.15 In the degradation process, the peripheral enzyme and fission enzyme in microorganisms work together to complete the microbial degradation process of PAHs. The former can identify and change PAHs into a degradable chemical structure, whereas latter guides the compounds into the conventional metabolism pathway.16,17 Dioxygenase plays a major role in the initial metabolic step of various PAHs by hydroxylating benzene rings.18 The dioxygenase system involving the aerobic degrading pathway is made up of a terminal oxygenase, ferredoxin and its reductase.19 Consequently, these three components of dioxygenase containing an α3β3 hexamer can be applied to perform a one-step oxidation process for PAHs.20 It introduces a molecule of O2 into the chemical structure of aromatic hydrocarbons and heterocycles, resulting in the formation of cis-dihydrodiol in the oxidation process.15,21
In this study, an efficient BaP-degrading strain from the production water of an oil well was isolated and applied in biodegradation experiments at different concentrations. The relative metabolites of BaP were detected for the analysis of the biodegradation mechanism. By sequencing the gene sequence coding for dioxygenase, the target enzyme was constructed utilizing homology-modeling technology. Subsequently, the interactions of BaP with the active cavity of the target enzyme were investigated at the molecular level via the molecular docking method. Combining the analysis of the metabolites and interaction mechanism, the process for the biodegradation of BaP was elucidated in the strain. Taken together, our study furnished an integrated view for understanding the degradation mechanism of BaP. Simultaneously, it also provided a basis for gene modification research to improve the biodegradation ability of BaP and other high-molecular-weight PAHs.
2. Material and methods
2.1 Chemicals
BaP and other PAHs (>98%) were purchased from Aladdin Biochemical Technology Company (Shanghai, China) and dissolved in dichloromethane with the final concentrations corresponding to the liquid culture systems. The oxygen-containing metabolites of BaP were prepared by chemical oxidation according to the method of Moody et al.22 Organic reagents of analytical grade were purchased from Rhawn Reagent Company (Shanghai, China).2.2 Isolation and identification of bacterium
The indigenous bacterium was acquired from the production water of an oil well in Dagang Oilfield, China. The enrichment culture technique was applied to the bacterial culture system containing 5 mL sample, 1.0 g L−1 BaP and 100 mL liquid mineral salt medium (MSM). The culture was performed at 28 °C and 150 rpm for 35 d. The MSM was adjusted to pH 7.0 for the culture experiments and its ingredients are described in Table S1.† In the process of bacterial domestication, the content of BaP was increased gradually from 1.0 to 3.0 g L−1 to screen the highly bioactive degrading bacteria by continuously expanding the selection pressure, respectively.The bacterial cells in the culture solution were isolated by the plate streak method on the solid MSM containing 1.6% agar. Briefly, the BaP solution dissolved in dichloromethane (1%, w/v) was coated evenly on the solid MSM. After the solvent was evaporated entirely, the bacterial cells were inoculated to the treated solid MSM medium. Based on this, a single bacterial strain was selected in terms of colony features, and this step was conducted three times to ensure bacterial purity. The 16S rDNA of the strain was amplified by the 27F and 1492R universal primers. The constitution of the amplification system and the amplification program were described in Tables S2 and S3,† respectively. The sequencing was executed by Majorbio Bio-pharm Technology Company (Shanghai, China). The strain was preserved at −80 °C for further study. The highly identical 16S rDNA sequences with the strain were searched by the BLAST program. The sequence alignment was carried out by ClustalX2. The phylogenetic relationship was analyzed by Mega5.05. The 16S rDNA sequence of the strain was submitted to GenBank with the accession number KT799678.
2.3 Test of biodegradation ability
The biodegradation experiment was performed using 1 mL bacterial suspension, 100 mL liquid MSM and different concentrations of BaP (100, 200, 400, and 800 mg L−1) in a 250 mL Erlenmeyer flask sealed with a sterile breathable film, respectively. The cells of the strain grown on BaP were collected by centrifugation. Subsequently, they were washed and resuspended in MSM to obtain a bacterial suspension with an optical density of 0.2. The measurements on all the culture samples were performed in triplicate for each analysis at 28 °C under agitation at 150 rpm. The autoclaved medium was regarded as the abiotic control. The growth activity of the strain was determined at 600 nm using a UV-vis absorption spectrophotometer (Shimadzu, Japan).To measure the remaining concentration of BaP, 5 mL culture solution was collected from the culture solution every 5 d and extracted with 15 mL ethyl acetate. The organic phase was isolated and dried using a column of anhydrous Na2SO4. Subsequently, it was concentrated using a rotary evaporator (Ruide Instruments, China) and held at 0.5 mL with n-hexane for the following analysis by GC-MS (Shimadzu, Japan). Finally, the target compounds were detected on an HP-5 capillary chromatogram column and the parameter settings are described in Table S4.†23 Statistical analysis was performed by ANOVA test on SPSS 26.
The comprehensive degradation ability of the strain for other substrates, including naphthalene (Nap), acenaphthylene (Ace), fluorene (Flu), phenanthrene (Phe), pyrene (Pyr) and benzo[k]fluoranthene (BF), was tested individually with an initial concentration of 100 mg L−1 under the same conditions as mentioned above. The extraction intervals for the other six PAH substrates were set at 15, 25, and 35 days, respectively. The other extraction and processing steps were the same as that for BaP.
2.4 Identification of metabolites
To obtain sufficient metabolites of BaP, an acidification step was added to the extraction process, as mentioned in Section 2.3. After the normal extraction operations, 0.1 mol L−1 HCl was used to acidify the remaining aqueous phase to pH 4.0. Then, it was extracted again with 15 mL ethyl acetate. Subsequently, the acidified and un-acidified organic phases were merged, dried and concentrated, as mentioned above for the subsequent determination by GC-MS.2.5 Amplification of dioxygenase gene
Two degenerate primers (sense primer: 5′-TGYCCNTAYCAYCARTGGACNTAYG-3′ and anti-sense primer: 5′-CRTANGTCCAYTGRTGRTANGGRCA-3′) were designed and applied to amplify the dioxygenase gene sequence in the strain. The system of gene amplification consisted of 1.00 μL each primer, 12.50 μL 2× Taq MasterMix, 0.50 μL genomic DNA and 10.00 μL ddH2O. The PCR procedure was executed as described in Table S5.† Finally, the gene sequence coding the alpha subunit of dioxygenase in this strain was amplified by the degenerate primers, which consisted of 1239 nucleotides and was stored in the GenBank database (accession no. OP821152).2.6 Homology modeling of target dioxygenase
The gene sequence coding for dioxygenase was translated to the protein sequence by DNAStar 5.0 for further research of homology modeling. The homology-modeling construction of the crystal structure of dioxygenase was performed on the SWISS-MODEL platform.24 The primary nucleotide sequence of the target enzyme was submitted to SWISS-MODEL to search for the template and build a three-dimensional model of the target enzyme based on the target–template alignment mode. Min et al.25 reported that the modeled structure would acquire a similar backbone construction under the condition of 30% identity. Moreover, a high-accuracy modeled structure will be obtained when the identity of the primary sequences of the template and target enzymes is greater than 50%. By sequence alignment, the crystal structure (ID: 3N9T, 2.0 Å resolution) of dioxygenase from the genus Pseudomonas was chosen as the template for the following homology-modeling study, which has an identity of 67.3% with dioxygenase in the strain. The PDB data of 3N9T displayed that the template enzyme is a polymer structure with two noncovalent Fe3+ ions. Finally, the structural quality of the dioxygenase model in the strain was verified by Ramachandran plots.2.7 Analysis of interaction mechanism
Based on the result of homology modeling, a modeled structure of dioxygenase was acquired and applied in the following study of molecular simulation employing the MOE program.26 The BaP structure in the ‘mol’ format was used as the docking ligand. The potential docking sites were searched based on the chemical structure of BaP. The Powell method and Tripos force field were used in the optimization process with 1000 maximum iterations. The FF99 and Gasteiger–Huckel charges were added in the docking process for the biopolymer and ligand, respectively. The interaction of the target enzyme and ligand was predicted by the molecular docking method with a proximity threshold value of 5.5. The polar hydrogens were added to the residues in the modeled target enzyme for the interaction between the ligand and dioxygenase. In conclusion, the interaction mechanism of dioxygenase and BaP was elucidated by combining the study of molecular interaction and the identification of the BaP metabolites.3. Results and discussion
3.1 Isolation and identification of bacterium
An effective BaP-degrading bacterium was isolated and identified by the enrichment culture and plate screening techniques. Its 16S rDNA sequence was made up of 1359 nucleotides. The phylogenetic analysis was performed and the result showed that the BaP-degrading strain possessed the closest genetic relationship with the bacterial genus Pannonibacter. This strain revealed the highest identity of 99.9% with Pannonibacter phragmitetus YS1 (MG735761), as shown in Fig. 1. It also shared an identity of 99.8% with Pannonibacter phragmitetus B16 (KT380507) and Pannonibacter phragmitetus L-s-R2A-19.4 (FR774557), respectively. Therefore, the strain was considered to be a member of the Pannonibacter genus and named Pannonibacter sp. JPA3. | ||
| Fig. 1 Phylogenetic relationship of Pannonibacter sp. JPA3 and the relevant strains of the Pannonibacter genus based on the neighbor-joining statistical method. | ||
To date, some types of BaP-degrading bacteria have been investigated, including the genera Mycobacterium, Hydrogenophaga, Sphingomonas and Pseudomonas.3,22,27 However, research on the biodegradation of BaP by the Pannonibacter genus is rare. Wang et al.28 briefly reported on the Pannonibacter phragmitetus strain CGMCC9175 with biodegradation ability for low-molecular-weight PAHs. Simultaneously, the application of the Pannonibacter genus mainly focused on the bacterial denitrification or degradation of fluorinated fungicides and asphaltene.29–31 Alternatively, in the present study, the isolated strain was applied to clarify its ability for the biodegradation of BaP and its interaction mechanism based on the modeled structure of dioxygenase.
3.2 Biodegradation of BaP
After a period of cultivation using BaP as the sole carbon source, the strain survived in microcosms from the production water of an oil well. The biotransformation of BaP by the strain was determined at different concentrations (100–800 mg L−1). As shown in Fig. 2A, 58% to 80% of BaP was removed after 35 d incubation. Simultaneously, the highest transformation percentage for BaP was observed at 100 mg L−1 initial concentration. With an increase in the concentration of BaP, the influence on the degradation percentage was low with a removal of 2% to 8% at the initial concentrations of 200 and 400 mg L−1. The rapid biotransformation between 10 and 25 d was associated with the corresponding increase in optical density, indicating a rapid increase in the cell concentration in the strain. The growth activity of the strain showed a decreasing trend in the slope after 25 days of incubation. When the concentration increased to 800 mg L−1, the degradation ability of the strain and its growth status were obviously difference from the other samples, and its degradation percentage decreased to 58% at the end of incubation. Taken together, we suppose that the biodegradation amount of BaP depended on the growth status of the bacterial cells. Similarly, the result also explained why BaP was degraded slower initially in the cultivation period.According to the report by Toyama et al.,32 the Mycobacterium strains derived from rhizosphere sediment could degrade 62% of BaP over 28 d. The strain Bacillus thuringiensis was used to investigate the degradation ability for 200 mg L−1 BaP, and eventually reached a degradation percentage of 80%, as described by Lu et al.13 Meanwhile, 78% of BaP was removed by a consortium of Bacillus, Paenibacillus, Serratia and Staphylococcus with the addition of succinate.33 The experimental concentrations of BaP in this study were higher than the natural environments at the μg L−1 level.3,34 Compared with other reported BaP-degrading bacteria, the strain showed efficient biodegradation ability for BaP at different test concentrations in this study.
3.3 Identification of metabolites
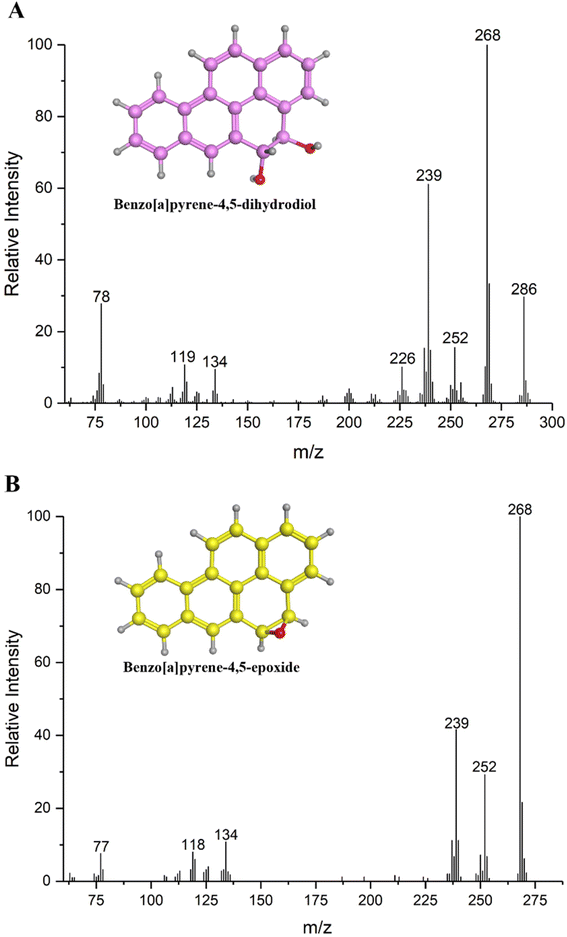
Four metabolites of BaP were detected during the biodegradation process by GC-MS. Metabolite I, with the chemical formula C20H14O2 and retention time of 23.62 min, gave a molecular ion peak (M+) at the m/z 286. Its base peak occurred at m/z 268 (M+ – 18, –H2O), and other fragment ions appeared at m/z 252 (M+ – 34, the loss of two hydroxyl groups), 239, 226, 134, 119 and 78. Accordingly, the key metabolite I was identified as BaP-4,5-dihydrodiol based on its retention time and mass spectrum data, as shown in Fig. 3A. To the best of our knowledge, to date, BaP-4,5-dihydrodiol as the BaP metabolite has not been reported in the genus Pannonibacter by researchers. Considering this, the strain could transform BaP into the dihydrodiol product at the C4 and C5 sites of the aromatic ring. Metabolite II has the chemical formula of C20H12O and was detected at 27.68 min (Fig. 3B). It exhibited a characteristic M+ peak at m/z 268 and fragment ions at m/z 252 [M+ – 16, –O], 239 [M+ – 29, –CHO], 134, 118 and 77. Consequently, metabolite II was identified as BaP-4,5-epoxide. Metabolite III was monitored at 19.87 min with the chemical formula of C18H12O. The mass spectrum data were made up of fragment ions including m/z 244, 215, 189, 122, 108 and 95. The fragment ion at m/z 244 indicated an M+ peak and the other ion at m/z 215 (M+ – 29) meant that an aldehyde group was lost from the BaP structure. According to the chromatographic retention time and mass spectrum data, metabolite III was identified as 5-hydroxychrysene. Meanwhile, the M+ peak of metabolite IV was discovered at m/z 188. The other ions were determined at m/z 170 (M+ – 18, –H2O), 144 (M+ – 44, –COO−) 126, 114, 88, 71 and 63, respectively. Simultaneously, metabolite IV possessed the chemical formula of C11H8O3 and retention time of 11.04 min. Therefore, it was identified as 2-hydroxy-1-naphthoic acid.In our study, metabolite I was detected in the BaP degradation process as the initial product. This proved that the biodegradation of BaP started at the C4–C5 positions, namely the K-region. Then, the 4,5-dihydrodiol metabolite was further degraded to the tetracyclic 5-hydroxychrysene. Seo et al.9 suggested that BaP was initially attacked by the oxidase at 4,5-, 7,8- and 9,10-sites. The 7,8- and 9,10-BaP-dihydrodiols could also be produced by the Beijerinckia sp. strain B1. Meanwhile, Mycobacterium sp. RJGII-135 could transform BaP into 7,8-dihydrodiol by ring oxidation. Subsequently, two ring-cleavage intermediates, 5-hydroxychrysene and 2-hydroxy-1-naphthoic acid, were detected as downstream metabolites.3 These results support our data, suggesting that the formation of BaP-4,5-dihydrodiol as the initial metabolite of BaP is the major step in the biodegradation of BaP by this strain.
3.4 Biodegradation of other substrates
In this study, six aromatic substrates were used to perform the biodegradation experiment to elucidate the comprehensive degradation ability of the JPA3 strain. This feature was necessary for the strain to ensure its biological activity in the complex polluted environment with multiple contaminants. As shown in Fig. 4, different letters indicate a significant difference (p < 0.05). For 2-ring Nap and 3-ring Ace and Phe, the strain could completely remove them at the end of culture, except for Flu, remaining with a percentage of 2%. Meanwhile, 97% of Pyr, 80% BaP and 73% of BF were degraded during the incubation period. Although each substrate had the same initial concentration of 100 mg L−1 at the beginning of the biodegradation, their residual amount gradually decreased as the cultivation time increased. Moreover, the degradation degree of each substrate gradually decreased with an increase in the aromatic ring number in each cultivation cycle. Consequently, the degradation ability of the JPA3 strain showed a significant difference (p < 0.05) between the degradation rate of each sample at 15, 25 and 35 d. Meanwhile, the experimental results proved that this strain possessed comprehensive biodegradation ability for different types of PAHs, favoring its future application in contaminated sites.3.5 Homology modeling of dioxygenase
To analyze the interaction mechanism of dioxygenase with BaP, a dioxygenase structure in the strain was constructed by homology modeling and verified by Ramachandran plots. As shown in Fig. S1,† the result showed that 94.5% of the amino acid residues were located in the favored region, and only 1.2% of residues were outliers. Therefore, the dioxygenase model in the strain was rational and effective in terms of the stereochemical structure, making it suitable for the following molecular docking research.3.6 Interaction analysis by molecular docking
The structure of dioxygenase in the strain was employed in the molecular docking process of BaP. The optimal docking conformations with a score of 4.15 were chosen by calculating the scoring functions. The combined conformation and site of BaP in the active site are displayed in Fig. 5A. The docking results showed that the ligands selected the binding cavity of the conserved α subunit in the dioxygenase surface. As shown in Fig. 5B, the interactions between BaP and the residues were composed of various intermolecular forces based on the chemical structural properties of BaP. Simultaneously, the primary interatomic distances were measured to illustrate the spatial conformations between them. The C5 atom on the BaP structure formed van der Waals (vdW) interaction forces with the Ile73 (2.6 Å) and Gln70 (1.9 Å) residues. In addition, vdW interaction forces were also found in C8–Leu74 and C9–Pro196. Also, the benzene ring on BaP and the terminal nitrogen atom of Arg69 formed an H–π bond. In the case of Gln70, one hydrogen atom and one nitrogen atom with high electronegativity formed an N–H covalent bond on the amino group. The formation of the covalent bond resulted in a strong bias of the shared electron pair towards the nitrogen atom between the nitrogen and hydrogen atoms. Consequently, the hydrogen atom became a bare proton with a positive charge and a tiny radius, which could attract the electronegative oxygen atoms into the active center of dioxygenase to participate in the oxidation process. Afterwards, BaP-4,5-dihydrodiol was produced by the biological oxidation process induced by dioxygenase. As shown in Fig. 6, the His204 and Phe220 residues could form π–π and π–H interactions with the aromatic ring. The vdW interactions were formed with the Phe348, Leu303, Asp201 and Met223 residues in the binding sites. | ||
| Fig. 6 Major binding forces between the initial metabolite BaP-4,5-dihydrodiol and the amino acid residues in the active site. | ||
Taken together, we considered that BaP was first oxidized at the C4–C5 sites when it entered the dioxygenase center. Moreover, the vdW interaction force played a leading role during the mutual recognition process between dioxygenase and BaP.35 Compared with BaP, the dihydrodiol metabolite could acquire a stronger binding force than before in the active cavity. The formation of intermolecular forces contributed to enhancing the binding potential between the active cavity and ligand. Meanwhile, the analysis of the key binding sites was also beneficial to the further study of gene modification to improve the biodegradation ability of this strain.
3.7 Analysis of degradation pathway
Based on the analysis of the metabolites and interaction mechanism, the biodegradation pathway for BaP by Pannonibacter sp. JPA3 was proposed (Fig. 7). The biodegradation of BaP initiated at the C4 and C5 sites on the chemical structure of BaP. Moreover, the dioxygenase induced two oxygen atoms in the C4–C5 positions to perform a double oxidation reaction. The discovery of the metabolite BaP-4,5-dihydrodiol verified the theoretical analysis results. According to the report by Moody et al.,22 BaP-4,5-epoxide is a precursor of 4,5-dihydrodiol, which is produced due to the oxidation action of cytochrome P450. This conclusion implies that another biodegradation pathway may exist in the strain, which is worth further research. According to the identification of the downstream metabolites, we assumed that the 4,5-dihydrodiol structure was further degraded by the ring cleavage mode in the K region. Then, the metabolites of 5-hydroxychrysene and 2-hydroxy-1-naphthoic acid were successively formed by the relative biochemical reactions in the strain. Eventually, a salicylic acid degrading pathway was speculated for the subsequent degradation.9 | ||
| Fig. 7 Proposed biodegradation pathway of benzo[a]pyrene in Pannonibacter sp. JPA3 based on the analysis of the ring-cleavage metabolites and molecular modeling. | ||
4. Conclusions
A BaP-degrading bacterium, Pannonibacter sp. JPA3, was screened and identified by the 16S rDNA sequence in this study. The amount of BaP gradually declined at the beginning of the biodegradation experiment. Finally, a degradation percentage of 80% was acquired at the end of the culture period in 100 mg L−1 BaP sample. A theoretical analysis was performed based on the homology modeling of the crystal structure of dioxygenase and molecular docking. Combined with the identification of the BaP-4,5-dihydrodiol metabolite, it was proven that the biological oxidation of BaP initially started at the C4–C5 positions. Then, 5-hydroxychrysene and 2-hydroxy-1-naphthoic acid were discovered in the liquid extraction solution as downstream metabolites. Finally, a salicylic acid degrading pathway was proposed for the further metabolism of the 2-hydroxy-1-naphthoic acid intermediate. Taken together, this study provides a comprehensive reference for the metabolic mechanism research of BaP in terms of the degrading metabolites and theoretical analysis at the molecular level.Author contributions
Jingnan Jin: conceptualization, formal analysis, funding acquisition, methodology, project administration, visualization, writing – original draft. Yahui Shi: investigation, data curation, formal analysis, writing – review & editing. Baozhong Zhang: resources, writing – review & editing. Dongjin Wan: resources, supervision. Qingye Zhang: software, visualization. Ying Li: resources, supervision.Conflicts of interest
The authors declare no conflict of interest, financial or otherwise.Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 41907302), the Fundamental Research Funds for the Henan Provincial Colleges and Universities in Henan University of Technology (No. 2018QNJH10), the Doctoral Scientific Research Start-up Foundation from Henan University of Technology (No. 2017BS027) and Training Program for Young Backbone Teachers of Henan University of Technology (No. 21420112).References
- P. Cheng, Z. Lin, X. Zhao, M. G. Waigi, G. K. Vasilyeva and Y. Gao, J. Hazard. Mater., 2022, 431, 128637 CrossRef CAS PubMed.
- K. Saravanakumar, S. Sivasantosh, A. Sathiyaseelan, A. Sankaranarayanan, K. V. Naveen, X. Zhang, M. Jamla, S. Vijayasarathy, V. V. Priya and D. MubarakAli, Environ. Pollut., 2022, 119207 CrossRef CAS PubMed.
- J. Schneider, R. J. Grosser, K. Jayasimhulu, W. L. Xue and D. Warshawsky, Appl. Environ. Microbiol., 1996, 62, 13–19 CrossRef CAS PubMed.
- J. Zhang, Y. He, X. Yan, C. Qu and P. Qi, Aquat. Toxicol., 2019, 214, 105239 CrossRef CAS PubMed.
- O. Kahla, S. M. B. Garali, F. Karray, M. B. Abdallah, N. Kallel, N. Mhiri, H. Zaghden, B. Barhoumi, O. Pringault and M. Quemeneur, Sci. Total Environ., 2021, 751, 141399 CrossRef CAS PubMed.
- K.-H. Kim, S. A. Jahan, E. Kabir and R. J. Brown, Environ. Int., 2013, 60, 71–80 CrossRef CAS PubMed.
- M. Marquès, M. Mari, C. Audí-Miró, J. Sierra, A. Soler, M. Nadal and J. L. Domingo, Chemosphere, 2016, 148, 495–503 CrossRef PubMed.
- J. Guo and X. Wen, Ecotoxicol. Environ. Saf., 2020, 207, 111292 CrossRef PubMed.
- J. S. Seo, Y. S. Keum and Q. X. Li, Int. J. Environ. Res. Public Health, 2009, 6, 278–309 CrossRef CAS PubMed.
- Y. Sun, Q. Zhou, X. Sheng, S. Li, Y. Tong, J. Guo, B. Zhou, J. Zhao, M. Liu and Z. Li, Chemosphere, 2021, 282, 131127 CrossRef CAS PubMed.
- Y. Zhong, T. Luan, L. Lin, H. Liu and N. F. Tam, Bioresour. Technol., 2011, 102, 2965–2972 CrossRef CAS PubMed.
- I. Ghosh, J. Jasmine and S. Mukherji, Bioresour. Technol., 2014, 166, 548–558 CrossRef CAS PubMed.
- Q. Y. Lu, K. Y. Chen, Y. Long, X. J. Liang, B. Y. He, L. H. Yu and J. S. Ye, J. Hazard. Mater., 2019, 366, 329–337 CrossRef CAS PubMed.
- A. Segura, V. Hernández-Sánchez, S. Marqués and L. Molina, Sci. Total Environ., 2017, 590, 381–393 CrossRef PubMed.
- J. Zeng, Q. Zhu, Y. Wu, H. Chen and X. Lin, Chemosphere, 2017, 185, 67–74 CrossRef CAS PubMed.
- S. Mishra, S. Singh and V. Pande, Bioresour. Technol., 2014, 164, 299–308 CrossRef CAS PubMed.
- J. M. DeBruyn, C. S. Chewning and G. S. Sayler, Environ. Sci. Technol., 2007, 41, 5426–5432 CrossRef CAS PubMed.
- A. Chemerys, E. Pelletier, C. Cruaud, F. Martin, F. Violet and Y. Jouanneau, Appl. Environ. Microbiol., 2014, 80, 6591–6600 CrossRef PubMed.
- A. Karlsson, J. V. Parales, R. E. Parales, D. T. Gibson, H. Eklund and S. Ramaswamy, Science, 2003, 299, 1039–1042 CrossRef CAS PubMed.
- C. E. Cerniglia, D. T. Gibson and R. H. Dodge, Appl. Environ. Microbiol., 1994, 60, 3931–3938 CrossRef CAS PubMed.
- P. Isaac, M. Lozada, H. M. Dionisi, M. C. Estévez and M. A. Ferrero, Int. Biodeterior. Biodegrad., 2015, 105, 1–6 CrossRef CAS.
- J. D. Moody, J. P. Freeman, P. P. Fu and C. E. Cerniglia, Appl. Environ. Microbiol., 2004, 70, 340–345 CrossRef CAS PubMed.
- J. N. Jin, J. Yao, W. J. Liu, Q. Y. Zhang and J. L. Liu, Environ. Sci. Pollut. Res., 2017, 24, 363–371 CrossRef CAS PubMed.
- A. Waterhouse, M. Bertoni, S. Bienert, G. Studer, G. Tauriello, R. Gumienny, F. T. Heer, T. A. P. de Beer, C. Rempfer and L. Bordoli, Nucleic Acids Res., 2018, 46, W296–W303 CrossRef CAS PubMed.
- J. Min, D. Lin, Q. Zhang, J. Zhang and Z. Yu, Eur. J. Med. Chem., 2012, 53, 150–158 CrossRef CAS PubMed.
- S. Vilar, G. Cozza and S. Moro, Curr. Top. Med. Chem., 2008, 8, 1555–1572 CrossRef CAS PubMed.
- Z. Yan, Y. Zhang, H. Wu, M. Yang, H. Zhang, Z. Hao and H. Jiang, RSC Adv., 2017, 7, 46690–46698 RSC.
- X. Wang, D. Jin, L. Zhou and Z. Zhang, Genome Announc., 2016, 4, e01675 Search PubMed.
- R. Koju, S. Miao, B. Liang, D. R. Joshi, Y. Bai, R. Liu and J. Qu, Chemosphere, 2020, 252, 126478 CrossRef CAS PubMed.
- W. F. Song, J. W. Wang, Y. C. Yan, L. Y. An, F. Zhang, L. Wang, Y. Xu, M. Z. Tian, Y. Nie and X. L. Wu, Int. Biodeterior. Biodegrad., 2018, 132, 18–29 CrossRef CAS.
- D. A. Alexandrino, A. P. Mucha, C. M. R. Almeida and M. F. Carvalho, J. Hazard. Mater., 2020, 394, 122545 CrossRef CAS PubMed.
- T. Toyama, T. Furukawa, N. Maeda, D. Lnoue, K. Sei, K. Mori, S. Kikuchi and M. Ike, Water Res., 2011, 45, 1629–1638 CrossRef CAS PubMed.
- R. Kotoky and P. Pandey, Chemosphere, 2020, 258, 127175 CrossRef CAS PubMed.
- J. S. Choi, S. H. Lim, S. R. Jung, L. P. Lingamdinne, J. R. Koduru, M. Y. Kwak, J. K. Yang, S. H. Kang and Y. Y. Chang, J. Environ. Manage., 2022, 317, 115403 CrossRef CAS PubMed.
- M. D. Wolfe and J. D. Lipscomb, J. Biol. Chem., 2003, 278, 829–835 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra01453c |
| This journal is © The Royal Society of Chemistry 2023 |