DOI:
10.1039/D3RA01378B
(Paper)
RSC Adv., 2023,
13, 15182-15189
Preparation and in vivo imaging of NIR-emissive carbonized polymer dots derived from biomass olive leaves with a quantum yield of 71.4%†
Received
1st March 2023
, Accepted 27th April 2023
First published on 18th May 2023
Abstract
The conversion of biomass materials into high value-added chemicals is receiving more and more attention. Herein, biomass olive leaves are converted into carbonized polymer dots (CPDs) through a simple hydrothermal reaction. The CPDs show near infrared light emission properties, and the absolute quantum yield reaches a record breaking value of 71.4% under the excitation wavelength of 413 nm. Detailed characterization determines that CPDs only contain three elements: carbon, hydrogen and oxygen, which is very different from most carbon dots which contain nitrogen atoms. Subsequently, NIR fluorescence imaging both in vitro and in vivo is performed to test their feasibility as fluorescence probes. The metabolic pathways of CPDs in the living body are inferred by studying the bio-distribution of CPDs in major organs. Their outstanding advantage is expected to further broaden the application field of this material.
Introduction
Biomass sources have aroused great interest due to their advantages of being inexpensive, easy to obtain, green, abundant and eco-friendly.1–4 Converting biomass materials into valuable products such as fuels and nanomaterials is critical to sustainability development.5–10 Due to the multiple applications of carbon-based nanomaterials in optoelectronics, photocatalysis, light-emitting devices, and solar cells, therefore, the conversion of biomass materials into nanocarbon materials (nanotubes, graphene, fullerenes, etc.) is a promising way.11–14 Particularly, owing to the advantages of abundant low-cost sources, excellent biocompatibility, high quantum yield (QY), and low cytotoxicity, carbonized polymer dots (CPDs) have attracted immense attention in recent years.15–20 It is widely believed that the CPD is a transition material between polymer and fully carbonized CDs due to it possessing many functional groups and polymer chains on the surface.21 Various carbon sources including small molecules, polymers and biomass materials are used to prepare CPDs via hydrothermal carbonization, microwave and pyrolysis method.22 However, the literature survey found that the fluorescence emission wavelength of most CPDs derived from biomass is in the blue and green spectral regions.23–25 Few attempts succeed in NIR emissive CPDs that direct derives from the biomass raw materials. It has been confirmed that the imaging in the NIR region can greatly improve the contrast and resolution of the results since autofluorescence and light scattering from living organisms are minimized.26–29 Therefore, the directly converting of biomass raw materials into NIR emissive CPDs will greatly broad the scope of application of biomass materials. In this work, a novel type of NIR emission CPDs with maximum emission wavelength of 683 nm is prepared by a one-step hydrothermal method using olive leaf as the precursor. The NIR fluorescence of CPDs can be excited under wide wavelength ranging from ultraviolet to red. More importantly, the absolute quantum yield (AQY) of the CPDs is as high as 74% under the excitation wavelength at 413 nm. Additionally, a narrow full width at half maximum with about 25 nm is detected, suggesting superior property of the novel CPDs. In vitro and vivo imaging of the CPDs is carried out based on the carefully structure research. The results demonstrate that the CPDs can serve as outstanding NIR fluorescent probe with excellent biocompatibility and long retention time. Finally, the possible metabolic pathways of CPDs are drawn up based on imaging experiments of major organs.
Experiment
Materials and methods
Olive leaves were presented by Longnan Xiangyu Olive Development Co., Ltd. All reagents were commercially obtained and used without further purification.
Synthesis of RCPDs
The dried olive leaves are ground and passed through a 200-mesh sieve. Then, 1.0 g powder was added in 20 mL of acetone solution, stirred well. The solution was transferred to a poly (tetrafluoroethylene) Teflon-lined autoclave (50 mL) and heated in an oven at 120 °C for 5 h. The resulting mixture was centrifuged (8000 r min−1, 10 min) after the reactor was cooled to room temperature. The dark-green supernatant solution was collected. And through 0.22 μm polyethersulfone membrane was used to remove large particles. A green powder sample was obtained after drying the solvent with nitrogen, named RCPDs.
Characterization
A nicolet iS50 FTIR spectrophotometer is used for the Fourier transformed infrared (FTIR) analysis in the range of 400 to 4000 cm−1 with the resolution of 4 cm−1. A Shimadzu 1780 UV-vis spectrophotometer was used to obtain the UV-vis absorption spectra. Fluorescence spectroscopy was performed with a Shimadzu RF-5301 PC spectrophotometer. The absolute quantum yield was measured on FLS 920 with an integrating sphere (Edinburgh). The 1H, 13C and HQSC NMR spectra were performed with a Bruker AVANCE NMR spectrometer (600 MHz) using DMSO-d6 as the solvent. The morphologies are characterized using an 6701F field emission scanning electron microscopy (SEM) and JEM-2100F (TEM) (HITACHI, Tokyo, Japan). The chemical composition and distribution of the products are obtained by energy dispersive X-ray spectroscopy (EDS). XPS was investigated by using ESCALAB 250 spectrometer with a mono X-ray source Al Kα excitation (1486.6 eV). Dynamic light scattering (DLS) was performed using a Zetasizer Nano-ZS (Malvern Instruments). The thermogravimetric analysis (TGA) was performed on a PerkinElmer TGA 7 thermogravimetric analyser under a nitrogen atmosphere from room temperature to 700 °C with a heating rate of 10 °C min−1. The PL and Raman spectrum (Raman) was recorded on a Horiva (Lab Ram HR-800) spectrometer. The contact angle was measured with a Ramé-Hart 200-F1 standard goniometer at ambient temperature.
Cell cytotoxicity and imaging assay
Cancer cell line HUVEC and MCF-7 were obtained from Northwest Normal University. They were grown in DMEM supplemented with 10% (v/v) fetal bovine serum, 100 U per mL penicillin and 100 μg per mL streptomycin. All the cells were grown in an incubator with the environmental conditions of 5% CO2 at 37 °C. The cell cytotoxicity was valued by the cell counting Kit-8 Assay (CCK-8). In brief, 100 μL cells (5 × 104 mL) were seeded in 96-well plates and cultivated in incubator for 24 h. RCPDs with different concentrations were added into the 96-well plates when the cells were under the logarithmic growth phase. After incubating with materials for 24 h, supernatant in wells were removed and replaced by culture medium contained with 10% CCK-8 solution. After 1 h reaction in the incubator, optical density (O.D) of the well plate was detected by microplate reader. The DMSO was used as control. Cell viability of cancer cells were calculated according to the OD value of experimental samples versus that of control groups. For the cell imaging, the cells were inoculated 6-well plates with the concentration of 5000 cells per mL and cultured in incubator with 5% CO2 at 37 °C. The RCPDs with final concentration of 25 and 100 μg mL−1 were added into the culture system. The supernatant medium was removed and cells were washed with phosphate buffer solution for three times after 24 h incubation. Cell imaging pictures could be taken by the MF52-N fluorescence microscope.
In vivo toxicity and fluorescence imaging
Healthy male Kunming mice (20 ± 2 g) were purchased from experimental animal centre of Lanzhou University and randomly divided into two groups (n = 3) and were given of RCPDs one time via tail vein (20 μL injection volume, 10 mg kg−1), control mice received DMSO. All animals were sacrificed after 7 days. Major organs (heart, liver, spleen, lung, and kidneys) were removed, washed with PBS and fixed in 4% formaldehyde, embedded in paraffin, sectioned, and then stained with hematoxylin and eosin for histological analysis. Mice were intravenously injected the RCPDs solution (10 mg kg−1) and fluorescence imaging was conducted at 0.5, 1, 2, 4, 6 and 10 h post-injection. In vitro and vivo fluorescence imaging experiments of major organs were conducted in ultra-sensitive fluorescence imaging system for small animals (VISQUE In vivo Smart-LF, viewers).
Results and discussion
Fabrication and characterization
Many biomass materials are employed to fabricate the carbon quantum dots. The NIR emissive carbonized polymer dots (RCPDs) are prepared using the olive leaf as resources via a solvothermal method at 120, 150 and 180 °C for 5 h in acetone solution. The detail prepared process is depicted in the Scheme 1. The polymer properties of the samples are first characterized by using 1H and 13C NMR in d6-DMSO (Fig. S1†). Hydrogen atoms with various structures are contained in the RCPDs samples. The hydrogen with chemical shifts between 0 and 2 ppm is in the CH, CH2 and CH3 groups that are directly linked to carbon atoms.30 Lots of hydrogens that are directly attached to heteroatoms (such as oxygen) and indirectly attached to the aromatic ring can be detected at chemical shifts between 2.0 and 5.0 ppm. A small number of peaks found between chemical shifts 5.0 and 8.0 ppm are attributed to hydrogen directly attached to the double bond and the aromatic ring.31 A pair of peaks of the chemical shift at 8.75 ppm may be attributed to the phenolic hydroxyl group in catechol. The exceedingly small peaks between the chemical shifts of 9.0 and 13.0 ppm may be associated with active hydrogen groups such as carboxyl groups in the structure.32 We are surprised to find that almost all the linkage types of hydrogen atoms mentioned in organic chemistry are included in the structure of RCPDs. As the reaction temperature rises from 120 to 180 °C, except for the increase in the peak intensity at the chemical shifts of 2.2 and 2.5 ppm, the peak intensities of the other types of hydrogen are significantly reduced, which is due to the fact that high-temperature oxidation and condensation increase the number of sp2 hybridized carbon atoms in the sample, which in turn increases the number of hydrogen atoms on the sp2 carbon. The dense signal appears at the chemical shift of 10–75 ppm in the 13C NMR spectrum, which is due to the presence of a large number of aliphatic (sp3) carbon atoms in the structure. The signal is significantly reduced between the chemical shifts of 75–90 ppm, which should be attributed to carbon atoms that are directly connected to heteroatom such as oxygen. The signals in the range of 100–185 ppm indicate the presence of sp2 carbon atoms. Interesting, the signal of carboxyl groups is observed at 210 ppm. With the increase of reaction temperature from 120 to 180 °C, the intensity of the carbon peak between 10–20 ppm and 80–180 ppm decreases obviously, while that between 20 and 80 ppm increases. This is because the graphitization degree of the sample increases with the increase of temperature. In addition, the peak at 210 ppm increases with reaction temperature, suggesting the number of oxygen-containing functional groups also increases. The prepared RCPDs contain various structures of carbon and hydrogen elements, which are rarely seen in previous literature. This may be related to the diversity of active components in olive leaves. The direct connection relationship between carbon and hydrogen in RCPDs-120 is explored using two-dimensional nuclear magnetic resonance (HSQC) technique (Fig. S2†). This assigned to the RCPDs possess a polymer/carbon hybrid structure rather than the structure of carbon main body.33 The signal of the carbon directly connected to a hydrogen with a chemical shift between 0 and 3 ppm appears at a chemical shift between 10 and 55 ppm, corresponding to region A on the spectrum. This indicates that there is abundant saturated alkane moiety in RCPDs. The peaks of hydrogen and carbon atoms in unsaturated hydrocarbon structures are between 3–4.5 and 55–100 ppm, respectively, which are located in region B of Fig. S2b.† The signal of both hydrogen and carbon region B is less than that of in region A, indicating the content of unsaturated hydrocarbons is less than that of saturated hydrocarbons. The correlation aromatics signal is mainly contained in region C, where the chemical shifts of hydrogen and carbon spectra are 2.5–5 and 50–90 ppm, respectively. However, no carbon atoms were found directly linked to hydrogen atoms with chemical shifts between 8 and 13 ppm, suggesting that these hydrogen atoms should be combined with heteroatoms such as oxygen. The SEM and TEM of the RCPDs are depicted in the Fig. 1. Many spherical and ellipsoidal nanoparticles can be observed from low resolution images for the RCPDs-120 sample (Fig. 1a). The particle size is relatively uniform, and the particles are independent of each other without crosslinking and agglomeration. It can be found from the high-resolution photos that the surface of the particles is relatively smooth and has no porous features (Fig. 1b). Nanoparticles are about 30–50 nm in size (calculated by 200). When the preparation temperature is raised to 150 °C, there are almost no single particles in the sample and a large number of nano-sized particles are observed with crosslinked network structures, although the particle size did not change much compared with the RCPDs-120. High-resolution SEM images show that the particles have lost their spherical structure and become completely random particles. At the same time, the cross-linking of particles is not uniform, and multiple cross-linking particles tend to form clusters (Fig. 1e and f). As the temperature rises from 120 to 150 °C, the powder of the sample also turns into oily in appearance. As the temperature rises further to 180 °C, the cross-linked network structure appears in a wider visual field. And the random nanoparticles stuck more tightly to each other, creating structures like grape clusters. The high-resolution images show a significant reduction in the size of the nanoparticles and the polydispersity has increased further, which may be ascribed to the complex polymerization process and inexact reaction conditions. The highly crosslinked network structure is generated by the process of dehydration and carbonization. The morphology structure of RCPDs is next investigated via the TEM (Fig. 1c, g and k). Several uniform dispersion regular spherical nanoparticles can be seen without apparent aggregation and crosslinked network structure. The diameters are around 50–70 nm for the RCPDs-120. Moreover, with the increase of temperature, the spherical structure of the sample gradually loses its regularity, and wrinkles and large aggregates appear which is caused by the cross-linking network structure in the sample. The high-resolution TEM (Fig. 1d, h and l) further confirmed almost all the nanoparticles in the three samples have no lattice fringes, indicating the amorphous characteristics of as-prepared RCPDs. This was confirmed by SAED patterns of the three samples (Fig. S3†). Their show broad diffraction halos and a faint larger one, indicating that the as-prepared RCPDs are amorphous in structure. This may be related to the complicated formation process of the RCPDs. The EDS and elemental mapping of three samples revealed the existence of C and O elements (Fig. S4†). The contents of C and O elements are about 70.83 and 29.17 wt% in the RCPDs-120, respectively. The content of the both elements in the RCPDs-150 increases to the 73.53% and 26.47%. The value further increases to be 79.51% and 20.49% as the temperature up to 180 °C. The increase of carbon content and the decrease of oxygen content indicate that the carbonization degree of samples increases with the reaction temperature. Curiously, no heteroatoms content is detected in the three samples prepared at different temperatures. There are few natural biomass materials that can directly prepare CPDs without heteroatoms.34 In this study, the CPDs prepared from olive leaves did not contain any heteroatoms except oxygen atoms, and showed high purity, which provided a good opportunity for precise control of heteroatoms in CPDs. The C, H and O components are uniformly distributed on the three nanoparticles from the mapping. The chemical structure changes as the temperature increases are quantitate analysed used X-ray photoelectron spectroscopy (XPS) (Fig. 2). The survey spectrum of the three samples is presented in Fig. 2a–c. It is clearly found that no other elements except carbon and oxygen have been detected in both RCPDs-120 and 150 sample. Negligible 2.35% nitrogen content is detected in RCPDs-180 samples, which may be caused by nitrogen doping from the air atmosphere during the preparation process. The quantitative analysis results are inserted in the corresponding survey spectrum. With the increase of reaction temperature, the carbon content decreased and the oxygen content increased in the three samples. This differs from the results of EDS, which may be due to the deeper penetration of EDS tests, where the detected content is the average for the whole block, while XPS can only detect the depth of 5–10 nm on the sample surface, and the measured content is the composition of the sample surface. Moreover, the C/O ratio calculated from the XPS results fell from 3.4 to 2.6 as the temperature increased from 120 to 180 °C, which may be caused by the continuous oxidation of samples. The
high-resolution C 1s spectrum (Fig. 2d) revealed five different types of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, C–C, C–H, C–O, COOH and C
C, C–C, C–H, C–O, COOH and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group in the RCPDs-120, the corresponding bonding energy is located at 281.95, 282.53, 282.86, 283.56, 284.58 and 286.34 eV.35 However, as the temperature rises to 150 °C, the C 1s in the form of carboxyl group is almost completely lost, which is caused by the oxidation of samples at high temperature. For the RCPDs-180, the existence form of C 1s is basically the same as the samples at RCPDs-150. The relative content of chemical bonds of RCPD in XPS spectra is calculated by using the proportion of each integral area in the fitted spectrum (Table S1†). The carbonization degree, as an important parameter to evaluate the polymer/carbon hybrid structure of CPDs, has an important effect on the properties of CPDs. The higher carbonization temperatures resulted in increasing content of C–C/C
O group in the RCPDs-120, the corresponding bonding energy is located at 281.95, 282.53, 282.86, 283.56, 284.58 and 286.34 eV.35 However, as the temperature rises to 150 °C, the C 1s in the form of carboxyl group is almost completely lost, which is caused by the oxidation of samples at high temperature. For the RCPDs-180, the existence form of C 1s is basically the same as the samples at RCPDs-150. The relative content of chemical bonds of RCPD in XPS spectra is calculated by using the proportion of each integral area in the fitted spectrum (Table S1†). The carbonization degree, as an important parameter to evaluate the polymer/carbon hybrid structure of CPDs, has an important effect on the properties of CPDs. The higher carbonization temperatures resulted in increasing content of C–C/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, which revealed the carbonization degree really increased. The O 1s XPS spectrum displayed two peaks at 530.4 eV and 532.5 eV is related to C–O and C
C, which revealed the carbonization degree really increased. The O 1s XPS spectrum displayed two peaks at 530.4 eV and 532.5 eV is related to C–O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group in all samples (Fig. 2g–i). The obvious content increase and decrease in C–O and C
O group in all samples (Fig. 2g–i). The obvious content increase and decrease in C–O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O is found as the temperature increases. The above structure clearly reflects that the oxygen-containing functional groups decrease with increasing temperature. The FTIR spectroscopy of RCPDs is shown in Fig. S5a.† The stretching vibrations of –OH appear around 3430 cm−1. The absorption bonds located at 2890 and 2980 cm−1 are attributed to the stretching vibrations model of –CH2– and –CH3 group.36 There are characteristic peaks of C
O is found as the temperature increases. The above structure clearly reflects that the oxygen-containing functional groups decrease with increasing temperature. The FTIR spectroscopy of RCPDs is shown in Fig. S5a.† The stretching vibrations of –OH appear around 3430 cm−1. The absorption bonds located at 2890 and 2980 cm−1 are attributed to the stretching vibrations model of –CH2– and –CH3 group.36 There are characteristic peaks of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C (1608, 1510, and 1445 cm−1) and carbonyl C
C (1608, 1510, and 1445 cm−1) and carbonyl C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (1630 cm−1) in the RCPDs. Stretching vibrations of C–C and C–O localized at about 1330, 1424 and 1150 cm−1, respectively.37 It can be found that the red fluorescent RCPDs carry a variety of chemical functional groups including hydroxyl, carbonyl, and double bonds. The characteristic peaks of almost all functional groups decrease obviously with the increase of temperature, which is due to the continuous reduction of the types of functional groups caused by oxidation and dehydration. The thermal stability and components of the as-prepared RCPDs is also tested via the thermogravimetric analysis (TGA, Fig. S5b†). The physically absorbed water on the surface of RCPDs is removed firstly when the temperature below 150 °C, and the corresponding weight loss is about 5%. Approximately 84.03, 83.88 and 85.83% of the mass loss occurred at temperatures ranging from 200 °C to 550 °C for the three samples, which can be attributed to the combustion of the RCPDs sample. A large exothermic peak on the DTG curve is found. This similar with as the TGA process of typical hydrocarbon polymers such as polyethylene, which may be due to the existence of similar chains structures in RCPDs.38 The XRD patterns of the RCPDs are shown in Fig. S5c.† It can be found that two broad peaks cantered at 15.46° and 20.29° are observed in three samples, indicative of a characteristic of weak crystalline.39 This is completely different from the number of lattices spacing for graphene and GQDs, in which the lattice spacing value is 0.21 nm for (100) facet spacing or 0.34 nm for (002) facet. Both peaks become sharp with the increase of temperature, indicating that the crystallinity increases as the temperature. The highly disordered carbon atoms in the RCPDs may be related to the abundance of oxygen-containing functional groups and the absence of nitrogen-containing functional groups in the quantum dot. The average size of the RCPDs nanoparticles is also estimated to be 42, 34 and 32 nm using the Scherer's formula by full width at half maximum of the peak at the 2θ of 15.46°.40 The average particle size is tested by dynamic light scattering (DLS) in the DMSO since the solubility of RCPD in water is not good (Fig. S5d†). The image suggested that the value of D10, D50 and D90 is around 17.01 nm, 37.23 nm and 49.11 nm for the RCPDs-120, respectively. This means that the average size is less than 17.01 nm, 27.23 nm and 49.11 nm particles account for 10%, 50% and 90% of all particles. The average size and PDI of RCPDs-120 in DMSO solution are 42.28 ± 12.5 nm and 0.212, respectively. The particle size curves of the both RCPDs-150 and 180 samples in DMSO completely coincide. The curve of RCPDs-150 is overlaid by RCPDs-180. The average particle size is about 35.01 ± 8.25 nm. A single peak appears in all three samples, this indicates that all RCPDs basically exist in the monodisperse form in DMSO. The curves of RCPDs-120 are more consistent with normal distribution than that of 150 and 180, indicating that the particle size distribution is relatively uniform. The wettability and Raman characteristics were studied using RCPDs-120 as the representative (Fig. S6a and b†). The RCPDs-120 exhibits an excellent water wettability property. A nearly zero water contact angle can be found when a droplet is placed on the surface of RCPDs-120. This may be due to a large density of oxygen containing functional groups in the RCPDs are polar and capable of hydrogen bonding with water molecules. The Raman spectrum of the RCPDs-120 is also tested. Unfortunately, even though the 783 nm laser is selected for excitation, no obvious D or G bands are detected in the Raman spectra. This may be caused by the fact that the Raman signal is masked by the strong fluorescence signal and the low content of carbon lattice structure in the sample.41 The photoluminescence properties of RCPDs-120 are also investigated in the solid state at room temperature (Fig. S6c†). The RCPDs-120 film on glass substrate is excited at 532 nm. The excitation power is kept below ∼1 mW with a 3 mm beam diameter to avoid heating of the substrate. An emission peak maximum at 683 nm can be observed and the curve shows excellent symmetry. The PL spectra of RCPDs-120 in the solid state is nearly identical with that of in the DMSO solution, indicates a lack of interparticle electronic coupling.42 This is likely due to oxygen-containing functional group passivating the surface of the RCPDs. The UV absorption spectrum of the RCPDs in the DMSO with concentration of 20 μg mL−1 is exhibited in the Fig. S6d.† A series of absorption peaks located at 420 nm, 450 nm, 505 nm, 540 nm, 620 nm and 675 nm is detected in three samples, suggesting the RCPDs sample possessed a very broad absorption from UV to deep red-light region. The UV-vis absorption intensity decreases obviously with the increase of preparation temperature under same concentration. The digital photograph is displayed in the Fig. S6d.† The clear light-green apparent colour transmits into strong deep red fluorescence under UV light excitation. The excitation-independent emission spectrum of the three RCPDs samples in the DMSO is shown in Fig. S6a, d and g.† It can be found that the emission peak cantered at around 685 nm and accompanied by a narrow full width at half maximum around 24 nm for all samples. As the excitation wavelength increases from 360 nm to 430 nm, the fluorescence emission intensity increases first and then decreases, which reaches the peak when the excitation wavelength is 420 nm. Interesting, a very weak shoulder emission peak appears near 720 nm, which assigned to the existence of the vibration fine structure.43 The concentration-dependent fluorescence emission intensity is shown in Fig. S7b, e and h.† With the decrease of RCPDs concentration, the intensity is also dropped. However, it should be noted that the maximum emission wavelength and full width at half maximum did not change with vary of concentration. The fluorescence emission spectra of the RCPDs sample in a binary solvent mixture of H2O and DMSO with different water fiction are shown in Fig. S7c, f and i.† When the percentage of water is up to 50 vol%, the fluorescence of the sample is almost completely quenched for three samples. With the increase of water content, a large number of RCPD particles agglomerate together to form aggregates with large particle sizes, leading to fluorescence quenching, which is called aggregation induced quenching (ACQ) effect.16 The absolute fluorescence quantum yield of the RCPDs is determined in the DMSO solution. The excitation and emission range are 398–428 nm and 650–780 nm (Table S1†). The QY reached 71.4%, 28.25% and 12.93% under 414 nm excitation for the RCPDs-120, 150 and 180 samples. The value of 71.4% is the highest QY for deep red/NIR fluorescent CDs under the excitation 414 nm wavelengths ever reported, to the best of our knowledge, which may be due to the prohibition of energy dissipation through nonradioactive channels.44 Moreover, the aggregation caused quenching reduces due to π–π stacking interactions after formation
of nanodots. The fluorescence emission of the RCPDs-120 is further confirmed with CIE1931 and CIE1976 coordinates.45 Fig. S8† shows the colour coordinates of the red samples in the CIE tristimulus chromaticity diagram. The value of x, y and u′, v′ for the RCPDs-120 sample is (0.73121, 0.26879) and (0.61408, 0.50789), respectively. This further proves the excellent deep red fluorescent luminescence properties of the implemented RCPDs-120, which gives the immense potential as a fluorescence visualization biosensing platform in biological systems.
O (1630 cm−1) in the RCPDs. Stretching vibrations of C–C and C–O localized at about 1330, 1424 and 1150 cm−1, respectively.37 It can be found that the red fluorescent RCPDs carry a variety of chemical functional groups including hydroxyl, carbonyl, and double bonds. The characteristic peaks of almost all functional groups decrease obviously with the increase of temperature, which is due to the continuous reduction of the types of functional groups caused by oxidation and dehydration. The thermal stability and components of the as-prepared RCPDs is also tested via the thermogravimetric analysis (TGA, Fig. S5b†). The physically absorbed water on the surface of RCPDs is removed firstly when the temperature below 150 °C, and the corresponding weight loss is about 5%. Approximately 84.03, 83.88 and 85.83% of the mass loss occurred at temperatures ranging from 200 °C to 550 °C for the three samples, which can be attributed to the combustion of the RCPDs sample. A large exothermic peak on the DTG curve is found. This similar with as the TGA process of typical hydrocarbon polymers such as polyethylene, which may be due to the existence of similar chains structures in RCPDs.38 The XRD patterns of the RCPDs are shown in Fig. S5c.† It can be found that two broad peaks cantered at 15.46° and 20.29° are observed in three samples, indicative of a characteristic of weak crystalline.39 This is completely different from the number of lattices spacing for graphene and GQDs, in which the lattice spacing value is 0.21 nm for (100) facet spacing or 0.34 nm for (002) facet. Both peaks become sharp with the increase of temperature, indicating that the crystallinity increases as the temperature. The highly disordered carbon atoms in the RCPDs may be related to the abundance of oxygen-containing functional groups and the absence of nitrogen-containing functional groups in the quantum dot. The average size of the RCPDs nanoparticles is also estimated to be 42, 34 and 32 nm using the Scherer's formula by full width at half maximum of the peak at the 2θ of 15.46°.40 The average particle size is tested by dynamic light scattering (DLS) in the DMSO since the solubility of RCPD in water is not good (Fig. S5d†). The image suggested that the value of D10, D50 and D90 is around 17.01 nm, 37.23 nm and 49.11 nm for the RCPDs-120, respectively. This means that the average size is less than 17.01 nm, 27.23 nm and 49.11 nm particles account for 10%, 50% and 90% of all particles. The average size and PDI of RCPDs-120 in DMSO solution are 42.28 ± 12.5 nm and 0.212, respectively. The particle size curves of the both RCPDs-150 and 180 samples in DMSO completely coincide. The curve of RCPDs-150 is overlaid by RCPDs-180. The average particle size is about 35.01 ± 8.25 nm. A single peak appears in all three samples, this indicates that all RCPDs basically exist in the monodisperse form in DMSO. The curves of RCPDs-120 are more consistent with normal distribution than that of 150 and 180, indicating that the particle size distribution is relatively uniform. The wettability and Raman characteristics were studied using RCPDs-120 as the representative (Fig. S6a and b†). The RCPDs-120 exhibits an excellent water wettability property. A nearly zero water contact angle can be found when a droplet is placed on the surface of RCPDs-120. This may be due to a large density of oxygen containing functional groups in the RCPDs are polar and capable of hydrogen bonding with water molecules. The Raman spectrum of the RCPDs-120 is also tested. Unfortunately, even though the 783 nm laser is selected for excitation, no obvious D or G bands are detected in the Raman spectra. This may be caused by the fact that the Raman signal is masked by the strong fluorescence signal and the low content of carbon lattice structure in the sample.41 The photoluminescence properties of RCPDs-120 are also investigated in the solid state at room temperature (Fig. S6c†). The RCPDs-120 film on glass substrate is excited at 532 nm. The excitation power is kept below ∼1 mW with a 3 mm beam diameter to avoid heating of the substrate. An emission peak maximum at 683 nm can be observed and the curve shows excellent symmetry. The PL spectra of RCPDs-120 in the solid state is nearly identical with that of in the DMSO solution, indicates a lack of interparticle electronic coupling.42 This is likely due to oxygen-containing functional group passivating the surface of the RCPDs. The UV absorption spectrum of the RCPDs in the DMSO with concentration of 20 μg mL−1 is exhibited in the Fig. S6d.† A series of absorption peaks located at 420 nm, 450 nm, 505 nm, 540 nm, 620 nm and 675 nm is detected in three samples, suggesting the RCPDs sample possessed a very broad absorption from UV to deep red-light region. The UV-vis absorption intensity decreases obviously with the increase of preparation temperature under same concentration. The digital photograph is displayed in the Fig. S6d.† The clear light-green apparent colour transmits into strong deep red fluorescence under UV light excitation. The excitation-independent emission spectrum of the three RCPDs samples in the DMSO is shown in Fig. S6a, d and g.† It can be found that the emission peak cantered at around 685 nm and accompanied by a narrow full width at half maximum around 24 nm for all samples. As the excitation wavelength increases from 360 nm to 430 nm, the fluorescence emission intensity increases first and then decreases, which reaches the peak when the excitation wavelength is 420 nm. Interesting, a very weak shoulder emission peak appears near 720 nm, which assigned to the existence of the vibration fine structure.43 The concentration-dependent fluorescence emission intensity is shown in Fig. S7b, e and h.† With the decrease of RCPDs concentration, the intensity is also dropped. However, it should be noted that the maximum emission wavelength and full width at half maximum did not change with vary of concentration. The fluorescence emission spectra of the RCPDs sample in a binary solvent mixture of H2O and DMSO with different water fiction are shown in Fig. S7c, f and i.† When the percentage of water is up to 50 vol%, the fluorescence of the sample is almost completely quenched for three samples. With the increase of water content, a large number of RCPD particles agglomerate together to form aggregates with large particle sizes, leading to fluorescence quenching, which is called aggregation induced quenching (ACQ) effect.16 The absolute fluorescence quantum yield of the RCPDs is determined in the DMSO solution. The excitation and emission range are 398–428 nm and 650–780 nm (Table S1†). The QY reached 71.4%, 28.25% and 12.93% under 414 nm excitation for the RCPDs-120, 150 and 180 samples. The value of 71.4% is the highest QY for deep red/NIR fluorescent CDs under the excitation 414 nm wavelengths ever reported, to the best of our knowledge, which may be due to the prohibition of energy dissipation through nonradioactive channels.44 Moreover, the aggregation caused quenching reduces due to π–π stacking interactions after formation
of nanodots. The fluorescence emission of the RCPDs-120 is further confirmed with CIE1931 and CIE1976 coordinates.45 Fig. S8† shows the colour coordinates of the red samples in the CIE tristimulus chromaticity diagram. The value of x, y and u′, v′ for the RCPDs-120 sample is (0.73121, 0.26879) and (0.61408, 0.50789), respectively. This further proves the excellent deep red fluorescent luminescence properties of the implemented RCPDs-120, which gives the immense potential as a fluorescence visualization biosensing platform in biological systems.
 |
| | Scheme 1 Schematic illustration of the preparation of RCPDs. | |
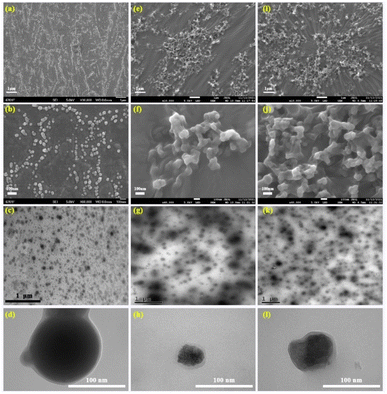 |
| | Fig. 1 The topology of RCPDs-120 (a, b: SEM and c, d: TEM images), RCPDs-150 (e, f: SEM and g, h: TEM) and RCPDs-180 (i, j: SEM and k, l: TEM). | |
 |
| | Fig. 2 XPS spectra of RCPDs-120 (a, d and g), RCPDs-150 (b, e and h) and RCPDs-180 (c, f and i). | |
Toxicity and imaging
Using RCPDs-120 as a representative, the application of the prepared CPDs in biological imaging was researched. The proliferating cells (MCF-7) and epithelial cells (HUVEC) are selected as tumour cells and normal cells, respectively, and the cytotoxicity of RCPDs is studied by MTT method. The result is summarized in Fig. S9.† A significant proliferation promotion effect can be found for both cells at concentrations of 200 μg mL−1 after 24 h incubation. The promotion effect of proliferation gradually decreased as the concentration decreased to 25 μg mL−1. Moreover, no significant inhibitory effect is observed of both cells, confirming the good biocompatibility and low cytotoxicity of RCPDs. This may be related to the fact that RCPDs contains only three elements of carbon, hydrogen, and oxygen, which are similar with the effect of sugar in cell culture medium. The in vitro fluorescence imaging is carried out by MCF-7 cells. The strong red fluorescence is clearly observed under blue (405 nm) excitation as shown in Fig. 3, suggesting the RCPDs can be rapidly uptake by cancer cells and displays great potential for bioimaging applications.46–48 Interesting, additionally, it can be found that the RCPD can distributed into both cytoplasm and cytoplasts in the MCF-7 cells. This intuitively reflects that RCPD has no effect on the functions of various organelles, such as DNA replication and nutrient transport, which further confirms its low toxicity and good biocompatibility. The histological examination of organs is also performed to assess the side effects such as tissue damage, inflammation, and lesions from the RCPDs at dose of 10 mg kg−1 by H&E staining method. The major organs (brain, heart, liver, lung, and kidney) are collected from the mice at 7 days after intravenous administration. It can be found that no major organ (brain, heart, liver, lung, testicle, and kidney) is abnormal, and no apparent histopathological abnormalities, degenerations, or lesions are observed (Fig. S10†). This suggests that the RCPD have no obvious toxicity to the mice as a bio-imaging agent, which may be ascribed to the optimal characteristics of RCPDs such as chemical structures, fractions, sizes, metabolism, and bio-distribution. Subsequently, the tube fluorescence imaging of RCPD solutions is tested at different excitation wavelengths, and the results are shown in Fig. S11.† The free DMSO is used as control. The excitation and emission wavelength parameters of different fluorescence imaging is also summarized in Fig. S10.† It can be found that all the four excitation wavelengths can excite RCPDs solution, showing its wide fluorescence excitation performance. Meanwhile, the luminance of RCPDs fluorescence imaging decreases with the increase of excitation wavelength. It should be note that pure DMSO can be excited by blue light at wavelengths 390–490 nm. The quantitative fluorescence intensity of the RCPDs solution in different excitation wavelengths is shown in Fig. S12.† The signal reaches the maximum under 620–650 nm excitation. Based on the above analysis, the in vivo imaging performance of RCPDs in mice is evaluated with 690–740 nm emission under 620–640 nm excitation. In vivo fluorescence images are collected at time points of 0.5, 1.0, 2.0, 4.0, 6.0 and 10.0 h after the intravenous administration of RCPDs (10 mg kg−1). The images are depicted in Fig. 4. The clear fluorescence signals are observed in the body of the mice at 0.5 h post-injection time points. Although the fluorescence signal attenuates significantly over time, it should be noted that even after 10 hours of injection, the fluorescence signal in the small animal is still obvious, indicating that the RCPD has a long residence time in the body. The fluorescence imaging experiments of major organs (brain, heart, lung, liver, spleen, kidney and intestine) are carried out at different time points after injection in order to better explore the distribution of RCPDs in vivo. The results are summarized in Fig. 4A. It can be found that the fluorescence signals mainly accumulated in live, stomach, kidney and intestine with negligible distribution in brain and lung. No fluorescent signal is observed in the heart, spleen and testes, indicating that RCPD did not accumulate in these sites. Meanwhile, the fluorescence intensity of these organs decreased with time. Fig. 4B depicted that the fluorescence signal intensity in the kidney, liver and intestine organs decreased as time going by. The signal in the intestine reaches its highest value at 0.5 hour after injection, while the signal in the liver and kidney reaches its peak at 4 hours. Therefore, it can be anticipant that RCPDs can be excreted either through the kidney system or through metabolized in the liver into the intestines. It has two metabolic pathways. The above results indicated that the RCPDs are capable of long-term in vivo imaging.49
 |
| | Fig. 3 Fluorescence images of HeLa cells incubated with 50 and 100 μg mL−1 RCPDs-120 for 24 h. | |
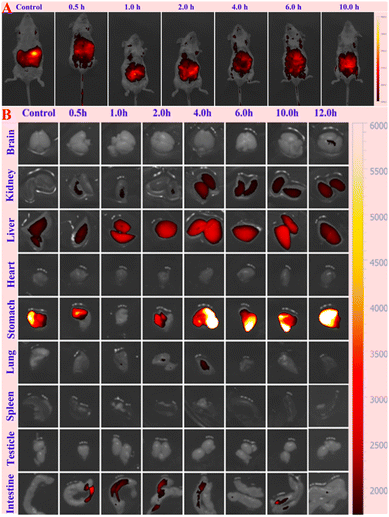 |
| | Fig. 4 In vivo (A) and ex vivo (B) imaging of mice with intravenous injection of the RCPDs at different time points. | |
Conclusion
In summary, the novel NIR fluorescence emissive carbonized polymer dots are designed and successful prepared using olive leaf as raw materials through a solvothermal method without further purification. The deep red emissive RCPDs with the FWHM of 25 nm possess the QY as high as 71.4% under 414 nm excitation. NMR characterizations recognize that the RCPDs contain many alkyls group. The results of XPS and EDS identify the absence of N elements in the RCPDs. Detail characterizations confirms that the RCPDs is a unique agglomerate formed by the stacking of O heterocyclic and aromatic rings, which is proved to govern the PL centre. The in vitro investigations confirmed the RCPDs showed good biocompatibility and low toxicity. The in vivo study indicates that RCPDs exhibits long-term residence time without systemic toxicity. The metabolic pathway of RCPDs is mainly excreted by the kidney and hepatobiliary system.
Conflicts of interest
The authors declare no competing financial interest.
Acknowledgements
The authors acknowledge the support of Natural Science Foundation of Gansu Province (No: 20JR10RA511, 2019B-084) and the Dongguan Science and technology of social development Program under Grant number 202050715001823.
References
- J. K. Saha and A. Dutta, Waste Biomass Valorization, 2022, 13, 1385–1429 CrossRef PubMed.
- M. A. Krishnan, K. Yadav, P. Roach and V. Chelvam, Biomater. Sci., 2021, 6, 2295–2312 RSC.
- Z. Wang, W. W. Wang, P. Z. Wang, X. J. Song, Z. Q. Mao and Z. H. Liu, Anal. Chem., 2021, 93, 3035–3041 CrossRef CAS PubMed.
- T. Li, D. D. Zhi, Z. H. Guo, J. Z. Li, Y. Chen and F. B. Meng, Green Chem., 2022, 24, 647–674 RSC.
- C. P. Ge, F. G. Shen, Y. Y. Yin, K. W. Chang, X. Zhang, P. X. Zhou, J. M. Li, Y. X. Liu and C. B. Lu, Talanta, 2021, 223, 121758 CrossRef CAS PubMed.
- B. Shi, N. Ren, L. Y. Gu, G. Xu, R. C. Wang, T. L. Zhu, Y. Zhu, C. H. Fan, C. C. Zhao and H. Tian, Angew. Chem., Int. Ed., 2019, 58, 16826–16830 CrossRef CAS PubMed.
- H. D. Li, D. Kim, Q. C. Yao, H. Y. Ge, J. Chung, J. L. Fan, J. Y. Wang, X. J. Peng and J. Yoon, Angew. Chem., Int. Ed., 2020, 133, 17408–17429 CrossRef.
- M. A. Miller and R. Weissleder, Nat. Rev. Cancer, 2017, 17, 399–414 CrossRef CAS PubMed.
- E. E. Hoover and J. A. Squier, Nat. Photonics, 2013, 7, 93–101 CrossRef CAS PubMed.
- W. R. Legant, L. Shao, J. B. Grimm, T. A. Brown, D. E. Milkie, B. B. Avants, L. D. Lavis and E. Betzig, Nat. Methods, 2016, 13, 359–365 CrossRef PubMed.
- L. R. Amjith and B. Bavanish, Chemosphere, 2022, 293, 133579 CrossRef CAS PubMed.
- B. Del Rosal and A. Benayas, Small Methods, 2018, 2, 1800075–1800091 CrossRef.
- L. L. Wang, W. Du, Z. J. Hu, K. Uvdal, L. Li and W. Huang, Angew. Chem., Int, Ed., 2019, 58, 14026–14043 CrossRef CAS PubMed.
- C. X. Yan, Y. T. Zhang and Z. Q. Guo, Coord. Chem. Rev., 2021, 427, 213556–213567 CrossRef CAS.
- C. Kang, S. Tao, F. Yang and B. Yang, Aggregate, 2022, 3, e169 CrossRef CAS.
- Y. B. Song, S. J. Zhu, J. R. Shao and B. Yang, J. Polym. Sci., Part A-1: Polym. Chem., 2017, 55, 610–615 CrossRef CAS.
- C. L. Xia, S. J. Zhu, T. L. Feng, M. X. Yang and B. Yang, Adv. Sci., 2019, 6, 1901316–1901338 CrossRef CAS PubMed.
- S. J. Zhu, Y. B. Song, X. H. Zhao, J. R. Shao, J. H. Zhang and B. Yang, Nano Res., 2015, 8, 355–381 CrossRef CAS.
- Q. Zeng, T. Feng, S. Tao, S. Zhu and B. Yang, Light: Sci. Appl., 2021, 10, 142 CrossRef CAS PubMed.
- S. Y. Tao, T. L. Feng, C. Y. Zheng, S. J. Zhu and B. Yang, J. Phys. Chem. Lett., 2019, 10, 5182–5188 CrossRef CAS PubMed.
- S. Y. Tao, S. J. Zhu, T. L. Feng, C. L. Xia, Y. B. Song and B. Yang, Mater. Today Chem., 2017, 6, 13–25 CrossRef.
- K. Jiang, L. Zhang, J. F. Lu, C. X. Xu, C. Z. Cai and H. W. Lin, Angew. Chem., Int. Ed., 2016, 128, 7384 CrossRef.
- W. X. Meng, X. Bai, B. Y. Wang, Z. Y. Liu, S. Y. Lu and B. Yang, Energy Environ. Mater., 2019, 2, 19–39 Search PubMed.
- W. Yang, T. Shimanouchi and Y. Kimura, ACS Sustainable Chem. Eng., 2015, 3, 591–598 CrossRef CAS.
- Z. F. Wang, J. F. Yu, X. Zhang, N. Li, B. Liu, Y. Y. Li, Y. H. Wang, W. X. Wang, Y. Z. Li, L. C. Zhang, S. Dissanayake, S. L. Suib and L. Y. Sun, ACS Appl. Mater. Interfaces, 2016, 8, 1434–1439 CrossRef CAS PubMed.
- S. S. Jing, Y. S. Zhao, R. C. Sun, L. X. Zhong and X. W. Peng, ACS Sustainable Chem. Eng., 2019, 7, 7833–7843 CrossRef CAS.
- H. E. Karahan, M. D. Ji, J. L. Pinilla, X. X. Han, A. Mohamed, L. Wang, Y. L. Wang, S. L. Zhai, A. Montoya, H. Beyenal and Y. Chen, J. Mater. Chem. B, 2020, 8, 9668–9678 RSC.
- Y. H. Liu, C. Zhu, Y. Gao, L. Yang, J. Y. Xu, X. T. Zhang, C. Lu, Y. F. Wang and Y. Z. Zhu, Appl. Surf. Sci., 2020, 510, 145437–145447 CrossRef CAS.
- J. J. Liu, Y. J. Geng, D. W. Li, H. Yao, Z. P. Huo, Y. F. Li, K. Zhang, S. J. Zhu, H. T. Wei, W. Q. Xu, J. L. Jiang and B. Yang, Adv. Mater., 2020, 32, 1906641–1906649 CrossRef CAS PubMed.
- S. T. Zhang, L. Yao, Q. M. Peng, W. J. Li, Y. Y. Pan, R. Xiao, Y. Gao, C. Gu, Z. M. Wang, P. Lu, F. Li, S. J. Su, B. Yang and Y. G. Ma, Adv. Funct. Mater., 2015, 25, 1755–1762 CrossRef CAS.
- W. C. Chen, Y. Yuan, S. F. Ni, Q. X. Tong, F. L. Wong and C. S. Lee, Chem. Sci., 2017, 8, 3599–3608 RSC.
- Z. H. Zhang, Y. M. Zhang, Y. J. Li, Y. F. Zhang, L. H. Qi, Q. Y. Yang, T. B. Wei, H. Yao and Q. Lin, J. Lumin., 2020, 227, 117546–117552 CrossRef CAS.
- W. J. Sun, L. Luo, Y. S. Feng, Y. W. Qiu, C. R. Shi, S. S. Meng, X. Y. Chen and H. M. Chen, Adv. Mater., 2020, 32, 2000377–2000384 CrossRef CAS PubMed.
- S. J. Chen, H. Wang, Y. N. Hong and B. Z. Tang, Mater. Horiz., 2016, 3, 283–293 RSC.
- Y. Jiao, Y. F. Gao, Y. T. Meng, W. J. Lu, Y. Liu, H. Han, S. M. Shuang, L. Li and C. Dong, ACS Appl. Mater. Interfaces, 2019, 11, 16822–16829 CrossRef CAS PubMed.
- X. H. Gao, H. X. Gao, T. H. Zhou and G. Liu, Sol. Energy Mater. Sol. Cells, 2018, 176, 93–99 CrossRef CAS.
- L. L. Pan, S. Sun, L. Zhang, K. Jiang and H. W. Lin, Nanoscale, 2016, 8, 17350–17356 RSC.
- Q. Xu, P. Pu, J. G. Zhao, C. B. Dong, C. Gao, Y. S. Chen, J. R. Chen, Y. Liu and H. J. Zhou, J. Mater. Chem. A, 2015, 3, 542–546 RSC.
- F. De Santis and R. Pantani, J. Therm. Anal. Calorim., 2013, 112, 1481–1488 CrossRef.
- M. Z. Kassaee, E. Motamedi and M. Majdi, Chem. Eng. J., 2011, 172, 540–549 CrossRef CAS.
- S. N. Qu, X. Y. Wang, Q. P. Lu, X. Y. Liu and L. J. Wang, Angew. Chem., Int. Ed., 2012, 51, 12215–12218 CrossRef CAS PubMed.
- H. Peng and J. Travas-Sejdic, Chem. Mater., 2009, 21, 5563–5565 CrossRef CAS.
- Y. W. Zheng, D. Yang, X. Wu, H. R. Yan, Y. C. Zhao, B. Feng, K. Duan, J. Weng and J. X. Wang, RSC Adv., 2015, 5, 90245–90254 RSC.
- F. Zhang, X. T. Feng, Y. Zhang, L. P. Yan, Y. Z. Yang and X. G. Liu, Nanoscale, 2016, 8, 8618–8632 RSC.
- S. J. Zhu, Q. N. Meng, L. Wang, J. H. Zhang, Y. B. Song, H. Jin, K. Zhang, H. Sun, H. Y. Wang and B. Yang, Angew. Chem., Int. Ed., 2013, 52, 3953–3957 CrossRef CAS PubMed.
- O. H. C. Cheng, T. Qiao, M. Sheldon and D. H. Son, Nanoscale, 2020, 12, 13113–13118 RSC.
- Z. F. Wang, F. L. Yuan, X. H. Li, Y. C. Li, H. Z. Zhong, L. Z. Fan and S. H. Yang, Adv. Mater., 2017, 29, 1702910–1702916 CrossRef PubMed.
- J. J. Liu, D. W. Li, K. Zhang, M. X. Yang, H. C. Sun and B. Yang, Small, 2018, 14, 1703919–1703928 CrossRef PubMed.
- J. Miller, P. Mulligan and C. E. Johnson, Powder Technol., 2020, 375, 28–32 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: UV-vis absorption spectrum and fluorescence emission spectrum of RCPDs at different concentration, excitation wavelength and water content; the absolute PLQY of RCPDs in DMSO solution under the excitation and emission wavelength of 414 nm and 680 nm; IR spectrum, TGA curves, XRD pattern and Raman spectrum, PL spectrum; 1H, 13C and 1H-13C HSQC spectrum; the size distribution histogram; the EDS curve and elements mapping; DLS size distribution and contact angle image; cell viability of HUVEC cells and MCF-7 cells after incubation with various concentrations of RCPDs for 24 h; histological evaluation of RCPDs toxicity in vivo; tube fluorescence images of RCPDs; the signal intensity in the tube and major organs in mice at different time points; the relative content of chemical bonds of RCPDs in XPS spectra. See DOI: https://doi.org/10.1039/d3ra01378b |
| ‡ Equal contribution: Zhiqiang Zhao, Qin Luo and Shengjing Chu contributed equally to this work. |
|
| This journal is © The Royal Society of Chemistry 2023 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article b,
Jijun Xu*d,
Weibing Xu
b,
Jijun Xu*d,
Weibing Xu *c and
Muhua Yi*e
*c and
Muhua Yi*e
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, C–C, C–H, C–O, COOH and C
C, C–C, C–H, C–O, COOH and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group in the RCPDs-120, the corresponding bonding energy is located at 281.95, 282.53, 282.86, 283.56, 284.58 and 286.34 eV.35 However, as the temperature rises to 150 °C, the C 1s in the form of carboxyl group is almost completely lost, which is caused by the oxidation of samples at high temperature. For the RCPDs-180, the existence form of C 1s is basically the same as the samples at RCPDs-150. The relative content of chemical bonds of RCPD in XPS spectra is calculated by using the proportion of each integral area in the fitted spectrum (Table S1†). The carbonization degree, as an important parameter to evaluate the polymer/carbon hybrid structure of CPDs, has an important effect on the properties of CPDs. The higher carbonization temperatures resulted in increasing content of C–C/C
O group in the RCPDs-120, the corresponding bonding energy is located at 281.95, 282.53, 282.86, 283.56, 284.58 and 286.34 eV.35 However, as the temperature rises to 150 °C, the C 1s in the form of carboxyl group is almost completely lost, which is caused by the oxidation of samples at high temperature. For the RCPDs-180, the existence form of C 1s is basically the same as the samples at RCPDs-150. The relative content of chemical bonds of RCPD in XPS spectra is calculated by using the proportion of each integral area in the fitted spectrum (Table S1†). The carbonization degree, as an important parameter to evaluate the polymer/carbon hybrid structure of CPDs, has an important effect on the properties of CPDs. The higher carbonization temperatures resulted in increasing content of C–C/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, which revealed the carbonization degree really increased. The O 1s XPS spectrum displayed two peaks at 530.4 eV and 532.5 eV is related to C–O and C
C, which revealed the carbonization degree really increased. The O 1s XPS spectrum displayed two peaks at 530.4 eV and 532.5 eV is related to C–O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group in all samples (Fig. 2g–i). The obvious content increase and decrease in C–O and C
O group in all samples (Fig. 2g–i). The obvious content increase and decrease in C–O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O is found as the temperature increases. The above structure clearly reflects that the oxygen-containing functional groups decrease with increasing temperature. The FTIR spectroscopy of RCPDs is shown in Fig. S5a.† The stretching vibrations of –OH appear around 3430 cm−1. The absorption bonds located at 2890 and 2980 cm−1 are attributed to the stretching vibrations model of –CH2– and –CH3 group.36 There are characteristic peaks of C
O is found as the temperature increases. The above structure clearly reflects that the oxygen-containing functional groups decrease with increasing temperature. The FTIR spectroscopy of RCPDs is shown in Fig. S5a.† The stretching vibrations of –OH appear around 3430 cm−1. The absorption bonds located at 2890 and 2980 cm−1 are attributed to the stretching vibrations model of –CH2– and –CH3 group.36 There are characteristic peaks of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C (1608, 1510, and 1445 cm−1) and carbonyl C
C (1608, 1510, and 1445 cm−1) and carbonyl C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (1630 cm−1) in the RCPDs. Stretching vibrations of C–C and C–O localized at about 1330, 1424 and 1150 cm−1, respectively.37 It can be found that the red fluorescent RCPDs carry a variety of chemical functional groups including hydroxyl, carbonyl, and double bonds. The characteristic peaks of almost all functional groups decrease obviously with the increase of temperature, which is due to the continuous reduction of the types of functional groups caused by oxidation and dehydration. The thermal stability and components of the as-prepared RCPDs is also tested via the thermogravimetric analysis (TGA, Fig. S5b†). The physically absorbed water on the surface of RCPDs is removed firstly when the temperature below 150 °C, and the corresponding weight loss is about 5%. Approximately 84.03, 83.88 and 85.83% of the mass loss occurred at temperatures ranging from 200 °C to 550 °C for the three samples, which can be attributed to the combustion of the RCPDs sample. A large exothermic peak on the DTG curve is found. This similar with as the TGA process of typical hydrocarbon polymers such as polyethylene, which may be due to the existence of similar chains structures in RCPDs.38 The XRD patterns of the RCPDs are shown in Fig. S5c.† It can be found that two broad peaks cantered at 15.46° and 20.29° are observed in three samples, indicative of a characteristic of weak crystalline.39 This is completely different from the number of lattices spacing for graphene and GQDs, in which the lattice spacing value is 0.21 nm for (100) facet spacing or 0.34 nm for (002) facet. Both peaks become sharp with the increase of temperature, indicating that the crystallinity increases as the temperature. The highly disordered carbon atoms in the RCPDs may be related to the abundance of oxygen-containing functional groups and the absence of nitrogen-containing functional groups in the quantum dot. The average size of the RCPDs nanoparticles is also estimated to be 42, 34 and 32 nm using the Scherer's formula by full width at half maximum of the peak at the 2θ of 15.46°.40 The average particle size is tested by dynamic light scattering (DLS) in the DMSO since the solubility of RCPD in water is not good (Fig. S5d†). The image suggested that the value of D10, D50 and D90 is around 17.01 nm, 37.23 nm and 49.11 nm for the RCPDs-120, respectively. This means that the average size is less than 17.01 nm, 27.23 nm and 49.11 nm particles account for 10%, 50% and 90% of all particles. The average size and PDI of RCPDs-120 in DMSO solution are 42.28 ± 12.5 nm and 0.212, respectively. The particle size curves of the both RCPDs-150 and 180 samples in DMSO completely coincide. The curve of RCPDs-150 is overlaid by RCPDs-180. The average particle size is about 35.01 ± 8.25 nm. A single peak appears in all three samples, this indicates that all RCPDs basically exist in the monodisperse form in DMSO. The curves of RCPDs-120 are more consistent with normal distribution than that of 150 and 180, indicating that the particle size distribution is relatively uniform. The wettability and Raman characteristics were studied using RCPDs-120 as the representative (Fig. S6a and b†). The RCPDs-120 exhibits an excellent water wettability property. A nearly zero water contact angle can be found when a droplet is placed on the surface of RCPDs-120. This may be due to a large density of oxygen containing functional groups in the RCPDs are polar and capable of hydrogen bonding with water molecules. The Raman spectrum of the RCPDs-120 is also tested. Unfortunately, even though the 783 nm laser is selected for excitation, no obvious D or G bands are detected in the Raman spectra. This may be caused by the fact that the Raman signal is masked by the strong fluorescence signal and the low content of carbon lattice structure in the sample.41 The photoluminescence properties of RCPDs-120 are also investigated in the solid state at room temperature (Fig. S6c†). The RCPDs-120 film on glass substrate is excited at 532 nm. The excitation power is kept below ∼1 mW with a 3 mm beam diameter to avoid heating of the substrate. An emission peak maximum at 683 nm can be observed and the curve shows excellent symmetry. The PL spectra of RCPDs-120 in the solid state is nearly identical with that of in the DMSO solution, indicates a lack of interparticle electronic coupling.42 This is likely due to oxygen-containing functional group passivating the surface of the RCPDs. The UV absorption spectrum of the RCPDs in the DMSO with concentration of 20 μg mL−1 is exhibited in the Fig. S6d.† A series of absorption peaks located at 420 nm, 450 nm, 505 nm, 540 nm, 620 nm and 675 nm is detected in three samples, suggesting the RCPDs sample possessed a very broad absorption from UV to deep red-light region. The UV-vis absorption intensity decreases obviously with the increase of preparation temperature under same concentration. The digital photograph is displayed in the Fig. S6d.† The clear light-green apparent colour transmits into strong deep red fluorescence under UV light excitation. The excitation-independent emission spectrum of the three RCPDs samples in the DMSO is shown in Fig. S6a, d and g.† It can be found that the emission peak cantered at around 685 nm and accompanied by a narrow full width at half maximum around 24 nm for all samples. As the excitation wavelength increases from 360 nm to 430 nm, the fluorescence emission intensity increases first and then decreases, which reaches the peak when the excitation wavelength is 420 nm. Interesting, a very weak shoulder emission peak appears near 720 nm, which assigned to the existence of the vibration fine structure.43 The concentration-dependent fluorescence emission intensity is shown in Fig. S7b, e and h.† With the decrease of RCPDs concentration, the intensity is also dropped. However, it should be noted that the maximum emission wavelength and full width at half maximum did not change with vary of concentration. The fluorescence emission spectra of the RCPDs sample in a binary solvent mixture of H2O and DMSO with different water fiction are shown in Fig. S7c, f and i.† When the percentage of water is up to 50 vol%, the fluorescence of the sample is almost completely quenched for three samples. With the increase of water content, a large number of RCPD particles agglomerate together to form aggregates with large particle sizes, leading to fluorescence quenching, which is called aggregation induced quenching (ACQ) effect.16 The absolute fluorescence quantum yield of the RCPDs is determined in the DMSO solution. The excitation and emission range are 398–428 nm and 650–780 nm (Table S1†). The QY reached 71.4%, 28.25% and 12.93% under 414 nm excitation for the RCPDs-120, 150 and 180 samples. The value of 71.4% is the highest QY for deep red/NIR fluorescent CDs under the excitation 414 nm wavelengths ever reported, to the best of our knowledge, which may be due to the prohibition of energy dissipation through nonradioactive channels.44 Moreover, the aggregation caused quenching reduces due to π–π stacking interactions after formation
of nanodots. The fluorescence emission of the RCPDs-120 is further confirmed with CIE1931 and CIE1976 coordinates.45 Fig. S8† shows the colour coordinates of the red samples in the CIE tristimulus chromaticity diagram. The value of x, y and u′, v′ for the RCPDs-120 sample is (0.73121, 0.26879) and (0.61408, 0.50789), respectively. This further proves the excellent deep red fluorescent luminescence properties of the implemented RCPDs-120, which gives the immense potential as a fluorescence visualization biosensing platform in biological systems.
O (1630 cm−1) in the RCPDs. Stretching vibrations of C–C and C–O localized at about 1330, 1424 and 1150 cm−1, respectively.37 It can be found that the red fluorescent RCPDs carry a variety of chemical functional groups including hydroxyl, carbonyl, and double bonds. The characteristic peaks of almost all functional groups decrease obviously with the increase of temperature, which is due to the continuous reduction of the types of functional groups caused by oxidation and dehydration. The thermal stability and components of the as-prepared RCPDs is also tested via the thermogravimetric analysis (TGA, Fig. S5b†). The physically absorbed water on the surface of RCPDs is removed firstly when the temperature below 150 °C, and the corresponding weight loss is about 5%. Approximately 84.03, 83.88 and 85.83% of the mass loss occurred at temperatures ranging from 200 °C to 550 °C for the three samples, which can be attributed to the combustion of the RCPDs sample. A large exothermic peak on the DTG curve is found. This similar with as the TGA process of typical hydrocarbon polymers such as polyethylene, which may be due to the existence of similar chains structures in RCPDs.38 The XRD patterns of the RCPDs are shown in Fig. S5c.† It can be found that two broad peaks cantered at 15.46° and 20.29° are observed in three samples, indicative of a characteristic of weak crystalline.39 This is completely different from the number of lattices spacing for graphene and GQDs, in which the lattice spacing value is 0.21 nm for (100) facet spacing or 0.34 nm for (002) facet. Both peaks become sharp with the increase of temperature, indicating that the crystallinity increases as the temperature. The highly disordered carbon atoms in the RCPDs may be related to the abundance of oxygen-containing functional groups and the absence of nitrogen-containing functional groups in the quantum dot. The average size of the RCPDs nanoparticles is also estimated to be 42, 34 and 32 nm using the Scherer's formula by full width at half maximum of the peak at the 2θ of 15.46°.40 The average particle size is tested by dynamic light scattering (DLS) in the DMSO since the solubility of RCPD in water is not good (Fig. S5d†). The image suggested that the value of D10, D50 and D90 is around 17.01 nm, 37.23 nm and 49.11 nm for the RCPDs-120, respectively. This means that the average size is less than 17.01 nm, 27.23 nm and 49.11 nm particles account for 10%, 50% and 90% of all particles. The average size and PDI of RCPDs-120 in DMSO solution are 42.28 ± 12.5 nm and 0.212, respectively. The particle size curves of the both RCPDs-150 and 180 samples in DMSO completely coincide. The curve of RCPDs-150 is overlaid by RCPDs-180. The average particle size is about 35.01 ± 8.25 nm. A single peak appears in all three samples, this indicates that all RCPDs basically exist in the monodisperse form in DMSO. The curves of RCPDs-120 are more consistent with normal distribution than that of 150 and 180, indicating that the particle size distribution is relatively uniform. The wettability and Raman characteristics were studied using RCPDs-120 as the representative (Fig. S6a and b†). The RCPDs-120 exhibits an excellent water wettability property. A nearly zero water contact angle can be found when a droplet is placed on the surface of RCPDs-120. This may be due to a large density of oxygen containing functional groups in the RCPDs are polar and capable of hydrogen bonding with water molecules. The Raman spectrum of the RCPDs-120 is also tested. Unfortunately, even though the 783 nm laser is selected for excitation, no obvious D or G bands are detected in the Raman spectra. This may be caused by the fact that the Raman signal is masked by the strong fluorescence signal and the low content of carbon lattice structure in the sample.41 The photoluminescence properties of RCPDs-120 are also investigated in the solid state at room temperature (Fig. S6c†). The RCPDs-120 film on glass substrate is excited at 532 nm. The excitation power is kept below ∼1 mW with a 3 mm beam diameter to avoid heating of the substrate. An emission peak maximum at 683 nm can be observed and the curve shows excellent symmetry. The PL spectra of RCPDs-120 in the solid state is nearly identical with that of in the DMSO solution, indicates a lack of interparticle electronic coupling.42 This is likely due to oxygen-containing functional group passivating the surface of the RCPDs. The UV absorption spectrum of the RCPDs in the DMSO with concentration of 20 μg mL−1 is exhibited in the Fig. S6d.† A series of absorption peaks located at 420 nm, 450 nm, 505 nm, 540 nm, 620 nm and 675 nm is detected in three samples, suggesting the RCPDs sample possessed a very broad absorption from UV to deep red-light region. The UV-vis absorption intensity decreases obviously with the increase of preparation temperature under same concentration. The digital photograph is displayed in the Fig. S6d.† The clear light-green apparent colour transmits into strong deep red fluorescence under UV light excitation. The excitation-independent emission spectrum of the three RCPDs samples in the DMSO is shown in Fig. S6a, d and g.† It can be found that the emission peak cantered at around 685 nm and accompanied by a narrow full width at half maximum around 24 nm for all samples. As the excitation wavelength increases from 360 nm to 430 nm, the fluorescence emission intensity increases first and then decreases, which reaches the peak when the excitation wavelength is 420 nm. Interesting, a very weak shoulder emission peak appears near 720 nm, which assigned to the existence of the vibration fine structure.43 The concentration-dependent fluorescence emission intensity is shown in Fig. S7b, e and h.† With the decrease of RCPDs concentration, the intensity is also dropped. However, it should be noted that the maximum emission wavelength and full width at half maximum did not change with vary of concentration. The fluorescence emission spectra of the RCPDs sample in a binary solvent mixture of H2O and DMSO with different water fiction are shown in Fig. S7c, f and i.† When the percentage of water is up to 50 vol%, the fluorescence of the sample is almost completely quenched for three samples. With the increase of water content, a large number of RCPD particles agglomerate together to form aggregates with large particle sizes, leading to fluorescence quenching, which is called aggregation induced quenching (ACQ) effect.16 The absolute fluorescence quantum yield of the RCPDs is determined in the DMSO solution. The excitation and emission range are 398–428 nm and 650–780 nm (Table S1†). The QY reached 71.4%, 28.25% and 12.93% under 414 nm excitation for the RCPDs-120, 150 and 180 samples. The value of 71.4% is the highest QY for deep red/NIR fluorescent CDs under the excitation 414 nm wavelengths ever reported, to the best of our knowledge, which may be due to the prohibition of energy dissipation through nonradioactive channels.44 Moreover, the aggregation caused quenching reduces due to π–π stacking interactions after formation
of nanodots. The fluorescence emission of the RCPDs-120 is further confirmed with CIE1931 and CIE1976 coordinates.45 Fig. S8† shows the colour coordinates of the red samples in the CIE tristimulus chromaticity diagram. The value of x, y and u′, v′ for the RCPDs-120 sample is (0.73121, 0.26879) and (0.61408, 0.50789), respectively. This further proves the excellent deep red fluorescent luminescence properties of the implemented RCPDs-120, which gives the immense potential as a fluorescence visualization biosensing platform in biological systems.





