Open Access Article
Open Access ArticleA near infrared fluorescent probe for rapid sensing of peroxynitrite in living cells and breast cancer mice†
Zixiang Xua,
Zhencai Xu *b and
Dong Zhang*b
*b and
Dong Zhang*b
aDepartment of Oncology, Affiliated Hospital of Nantong University, Medical School of Nantong University, Nantong 226001, China
bGuanyun People's Hospital, Lianyungang, Jiangsu 222000, China. E-mail: xu2454287273@163.com; zhangd123456@sina.com
First published on 14th March 2023
Abstract
Peroxynitrite (ONOO−) plays an essential role in numerous physiological and pathological processes owing to its strong oxidation and nitrification. Many studies have shown that ONOO− abnormalities are associated with inflammatory diseases, even cancer, such as arthritis, hepatitis, pneumonia, and breast cancer. Thus, developing a trustworthy technology to monitor ONOO− levels is critical in inflammatory or cancer illnesses. Herein, an ultrafast near-infrared (NIR) fluorescent probe (Cy–OH–ONOO) is proposed to detect ONOO− within 30 s. The probe's borate moiety is oxidized and separated from Cy–OH–ONOO, releasing a NIR fluorescence signal after interacting with ONOO− under physiological circumstances. In addition, the probe displays good selectivity and sensitivity towards ONOO− compared to other related biological species. Moreover, it is applied to the image and detects the level fluctuation of ONOO− in living cells and breast cancer mice based on excellent features with high biocompatibility and low toxicity of the developed probe. Therefore, Cy–OH–ONOO could serve as a powerful imaging tool to understand the correlation of ONOO− with inflammatory or breast cancer pathophysiological processes and to assess ONOO− levels in cellular oxidative stress.
1. Introduction
Cell proliferation, apoptosis, and differentiation are regulated by reactive nitrogen species (RNS), which are intracellular signaling molecules that play a part in two important processes: pathological and physiological.1–3 During the invasion of foreign substances, it is possible that their abnormal levels could lead to the development of various ailments, such as inflammation, cardiovascular diseases, neurological diseases, and even cancer.4,5 Peroxynitrite (ONOO−), a critical RNS, is generated by the intracellular diffusion of superoxide anion (O2˙−) and nitric oxide (NO).6–8 Because of its powerful oxidation and nitration, it may react with most compounds in cells, such as proteins, DNA, RNA, and lipids. Furthermore, it has been identified as a key molecule in the process of signal transduction.9,10 Unfortunately, the overgeneration of ONOO− may interfere with the physiological activities of cells, thereby causing damage to human tissues and organs.11–13 Several recent studies have shown that ONOO− is closely related to inflammatory and cancer diseases. For example, breast cancer is now one of the most common types of invasive cancers and the second leading cause of cancer-related deaths in women worldwide, and its incidence and mortality rates are rapidly increasing.14,15 Developing an effective technique for identifying the ONOO− in living creatures is essential to appreciate its involvement in breast cancer and enhance the understanding of disease onset and progression.Recently, various analysis methods have been developed to trace ONOO−, including the UV-vis technique, electrochemical analysis, ELISA, and electron spin resonance.16–18 However, despite these methods being developed to detect ONOO−, there is still a challenge because its half-lives are relatively short, and its levels are low in the biology system. It is necessary to develop a novel technique for monitoring the fluctuations of ONOO− to better understand the regulatory mechanisms of ONOO− in the human body. With its non-invasiveness, simple operation, real-time responsiveness, and spatiotemporal resolution, the fluorescence probe and imaging method has become a popular tool for visualizing biological species.19–24 Several fluorescent probes have been constructed for the detection of ONOO−. This probe design is primarily due to the cleavage of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, oxidation of boronate, hydrazides, N-dearylation, and others.25–30 Compared to the UV or visible region, near-infrared (NIR) fluorescent probes have unique advantages, which include less light damage, less background fluorescence, and deeper tissue penetration.31–33 Consequently, it is critical to build a NIR fluorescent probe that is eventually proficient in detecting ONOO− in a fast and precise manner in living cells.
C, oxidation of boronate, hydrazides, N-dearylation, and others.25–30 Compared to the UV or visible region, near-infrared (NIR) fluorescent probes have unique advantages, which include less light damage, less background fluorescence, and deeper tissue penetration.31–33 Consequently, it is critical to build a NIR fluorescent probe that is eventually proficient in detecting ONOO− in a fast and precise manner in living cells.
We reasonably design a NIR fluorescent probe (Cy–OH–ONOO) to detect quickly and selectively intracellular ONOO−. The probe comprises a hemicyanine dye (Cy-OH) and a phenylboronic acid ester derivative with a particular recognition moiety for ONOO−. The developed probe exhibits high specificity and sensitivity. In the presence of ONOO−, the probe promptly cleaves the boronic ester group and emits a NIR fluorescence signal (705 nm) via the intramolecular charge transfer (ICT) mechanism. Because of the probe's minimal biotoxicity and high biocompatibility, it is being engaged for the detection of the sensitivity of intracellular ONOO− under oxidative stress. This probe reveals great potential for imaging ONOO− in biological systems, which is beneficial for the diagnosis of breast cancer diseases.
2. Experimentation portion
2.1 Reagents and apparatus
Aladdin provided 1,2-myristate-1,3-acetate (PMA), lipopolysaccharide (LPS), and 4-bromomethylphenylboronic acid pinacol ester (Shanghai, China). All the other compounds were obtained from Maclean and were not further purified. UV-2910 and F-7000 were used to determine the spectra of the UV-vis and fluorescence predominantly, respectively. A Bruker nuclear magnetic resonance spectrometer was used to perform 1H and 13C NMR. Olympus FV1000 was used to capture the fluorescence images.2.2 Synthesis of compound 2
9 mL of DMF and 50 mL of CH2Cl2 were added to a three-necked flask and swirled at 0 °C for 10 min. Then, 10 mL of phosphorus tribromide (PBr3) was added dropwise to the mixture. After stirring for about 1 h, 4 mL cyclohexanone was added dropwise and stirred vigorously overnight (12 h) at room temperature. The pH was adjusted to neutral, extracted with CH2Cl2, and subjected to the following step without further purification (Scheme S1†).2.3 Synthesis of compound 3
Compound 2 (1.89 g, 10 mmol), Cs2CO3 (6.5 g, 2 mmol), and 2-hydroxy-4-methoxybenzaldehyde (1.52 g, 10 mmol) were put in a round bottom flask. This mixture was dissolved in 30 mL of dry DMF and stirred at room temperature (25 °C) for 18 h. Water was used to quench the reaction; then, the mixture was extracted using CH2Cl2. Anhydrous Na2SO4 was carried out by drying the organic phase. The crude product was chromatographed on a silica column using petroleum ether and CH2Cl2 (3/1, v/v) to obtain yellow solids. 1H NMR (500 MHz, CDCl3), δ (ppm): 10.31 (s, 1H), 7.11 (d, 1H), 6.82–6.48 (m, 3H), 3.85 (s, 3H), 2.57–2.51 (m, 2H), 2.42 (t, 2H), 1.69 (m, 2H).2.4 Synthesis of compound 4
After dissolving compound 3 (1.21 g, 5 mmol) in 20 mL dry CH2Cl2, BBr3 (1.5 mL, 15 mmol) was added dropwise to the solution at 0 °C, and the mixture was allowed to react at room temperature for one night. It was put into ice water, and the pH was adjusted to neutral. The removal of the solvent under lower pressure after extraction with CH2Cl2. The crude product was chromatographed on a silica column using CH2Cl2/CH3OH (100/1, v/v) to obtain compound 4. 1H NMR (500 MHz, d6-DMSO) δ (ppm): 10.28 (s, 1H), 10.18 (s, 1H), 7.18 (d, 1H), 6.95 (s, 1H), 6.71 (s, 1H), 6.63 (d, 1H), 2.56–2.51 (d, 2H), 2.29 (t, 2H), 1.69–1.61 (m, 2H).2.5 Synthesis of compound 5
The following compounds were dissolved in CH3CN: compound 4 (0.228 g, 1 mmol); K2CO3 (0.331 g, 2.4 mmol); and 4-(bromomethyl)benzeneboronic acid (0.325 g, 1.1 mmol). After stirring and refluxing for 12 h, the solvent was removed. The acquired crude product was purified by chromatographing on a silica column with CH2Cl2/CH3OH (30/1, v/v) to obtain yellow solids. 1H NMR (500 MHz, CDCl3), δ (ppm): 10.32 (s, 1H), 7.86 (d, J = 8.0 Hz, 2H), 7.45 (d, J = 7.9 Hz, 2H), 7.09 (d, J = 8.5 Hz, 1H), 6.75 (s, 2H), 6.66 (s, 1H), 5.14 (s, 2H), 2.62–2.55 (m, 2H), 2.46 (t, J = 6.0 Hz, 2H), 1.74 (dd, J = 12.3, 6.1 Hz, 2H), 1.37 (s, 12H). 13C NMR (125 MHz, CDCl3) δ(ppm): 187.7, 160.4, 153.3, 139.3, 135.1, 127.5, 126.5, 114.9, 112.6, 111.5, 101.6, 83.9, 70.2, 30.0, 24.9, 21.5, 20.4.2.6 Synthesis of the Cy–OH–ONOO compound
10 mL of anhydrous ethanol, piperidine (0.1 mL), compound 5 (0.444 g, 1 mmol), and compound 6 (0.315 g, 1 mmol) were added into a three-necked bottom flask. The mixture was stirred and refluxed under N2 for 12 h. After being cooled to room temperature, the solvent was removed under reduced pressure. The obtained crude product was performed on silica column chromatography with CH2Cl2/CH3OH (50/1, v/v) to produce yellow solids. 1H NMR (500 MHz, CDCl3) δ 8.63 (d, J = 14.7 Hz, 1H), 7.90 (s, 2H), 7.56–7.48 (m, 4H), 7.46–7.42 (m, 2H), 7.37 (s, 1H), 7.22 (s, 1H), 6.96 (d, J = 8.6 Hz, 1H), 6.88 (s, 1H), 6.62 (d, J = 14.9 Hz, 1H), 5.27 (s, 2H), 4.64 (dd, J = 14.3, 7.1 Hz, 2H), 2.81 (dd, J = 52.9, 5.6 Hz, 4H), 1.97 (s, 2H), 1.80 (s, 6H), 1.56 (s, 3H), 1.36 (s, 12H). 13C NMR (125 MHz, CDCl3) δ (ppm) 152.0, 146.6, 145.4, 138.2, 135.1, 134.9, 127.7, 127.6, 126.6, 126.3, 126.1, 121.4, 117.6, 104.4, 92.1, 83.8, 83.6, 65.3, 44.7, 44.2, 36.3, 32.6, 28.7, 28.0, 24.9, 24.4, 10.8. HR-MS: calcd for C40H45BNO4+, 613.3460; found [M + H]+, 614.3444.2.7 A combination of cell culture and confocal microscopy
The two cells (RAW 264.7 and HepG2 cells) were grown in DMEM supplemented with 1% antibiotics (streptomycin/penicillin) and 10% FBS at 37 °C in a 5% CO2 environment with a 95% humidified atmosphere. The cytotoxicity of the Cy–OH–ONOO probe was investigated using the CCK-8 experiment. The cells were cultured in 96-well plates (5000–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well) for 24 h. Next, the different densities of the probe were added. The cells were rinsed with DMEM media after 24 h of incubation, and then 10 μL CCK-8 was added. After the incubation period (4 h), the optical density was measured at 450 nm (Tecan, Austria). A fluorescent image was received at Olympus FV1000 and fluorescence collection windows in the range of 690–750 nm. All the experiments were performed in compliance with relevant laws or guidelines and followed institutional guidelines. Furthermore, all animal procedures were performed following the Guidelines for Care and Use of Laboratory Animals of Guanyun People's Hospital, and the experiments were approved by the Animal Ethics Committee of the Guanyun People's Hospital Animal Ethics Committee.
000 cells per well) for 24 h. Next, the different densities of the probe were added. The cells were rinsed with DMEM media after 24 h of incubation, and then 10 μL CCK-8 was added. After the incubation period (4 h), the optical density was measured at 450 nm (Tecan, Austria). A fluorescent image was received at Olympus FV1000 and fluorescence collection windows in the range of 690–750 nm. All the experiments were performed in compliance with relevant laws or guidelines and followed institutional guidelines. Furthermore, all animal procedures were performed following the Guidelines for Care and Use of Laboratory Animals of Guanyun People's Hospital, and the experiments were approved by the Animal Ethics Committee of the Guanyun People's Hospital Animal Ethics Committee.
3. Results and discussion
3.1 Synthesis design of Cy–OH–ONOO
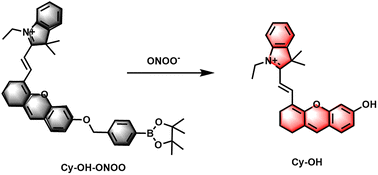
The molecular formula and the proposed reaction mechanism of the probe Cy–OH–ONOO toward ONOO− are explained in Scheme 1. In this design, hemicyanine dye was introduced as NIR fluorophores owing to its great biocompatibility and spectral properties. Recent studies have confirmed that phenylboronic acid and its esters can respond quickly to ONOO− and are superior to H2O2; thus, they can be good candidate reaction sites to achieve ONOO− specific response.34–36 The pinacol ester of 4-(bromomethyl)benzene-boronic acid was chosen as an ONOO− responsive unit. In contrast, ONOO− can transform phenylboronate into phenol, followed by 1,6-self-elimination of the phenol derivative, releasing the fluorophore Cy–OH–ONOO, which leads to the turn-on of the fluorescent signal. The ESI† shows the detailed synthesis technique as well as 1H NMR, 13C NMR, and HRMS characterizations of Cy–OH–ONOO.3.2 Cy–OH–ONOO response to ONOO−
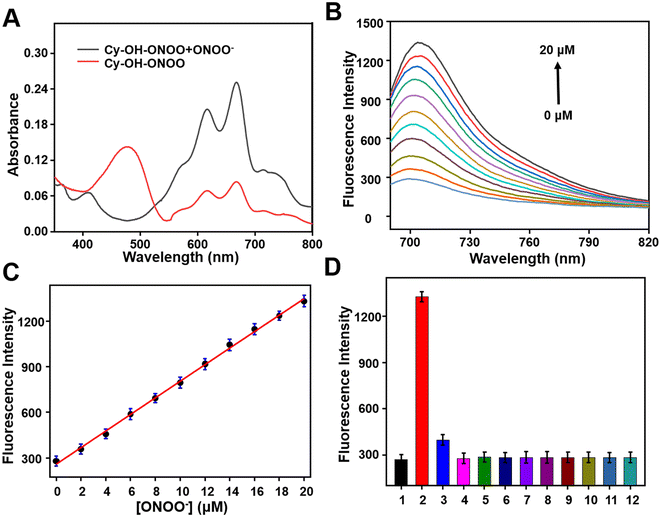
The spectrum characteristics of the compound Cy–OH–ONOO were investigated in a PBS buffer solution with 5% ethanol (EtOH/PBS = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 19, v/v, pH = 7.4). As illustrated in Fig. 1A, the Cy–OH–ONOO probe had maximum absorbance centered at 480 nm and a weak double peak (610 and 670 nm). After the addition of ONOO−, there was a strong double peak around 610 and 670 nm, and the absorption peak at 480 nm disappeared (Fig. 1A). Then, the fluorescent spectrum of Cy–OH–ONOO to ONOO− was investigated as well. Initially, the Cy–OH–ONOO probe displayed a negligible fluorescence emission peak. After being treated with ONOO−, Cy–OH–ONOO exhibited an obvious fluorescence emission around 705 nm, which should be attributed to the leaving of p-boratebenzyl in ONOO− and in turn led to the onset of the fluorescent signal. Moreover, the fluorescence intensity of Cy–OH–ONOO with varied ONOO− content was further evaluated (Fig. 1B). There was a satisfying linear relationship between fluorescence intensity and ONOO− level in the range of 0–20 μM (Fig. 1C). The regression equation was determined as F705 nm = 54.5294 [ONOO−] + 259.4823, with a satisfactory linear relationship (R2 = 0.9874) between fluorescence intensity and ONOO− level in the range of 0–20 μM (Fig. 1C). The detection limit (LOD) was calculated as 56 nM using a standard method of 3σ/k, demonstrating that Cy–OH–ONOO can identify ONOO− in the presence of other compounds.
19, v/v, pH = 7.4). As illustrated in Fig. 1A, the Cy–OH–ONOO probe had maximum absorbance centered at 480 nm and a weak double peak (610 and 670 nm). After the addition of ONOO−, there was a strong double peak around 610 and 670 nm, and the absorption peak at 480 nm disappeared (Fig. 1A). Then, the fluorescent spectrum of Cy–OH–ONOO to ONOO− was investigated as well. Initially, the Cy–OH–ONOO probe displayed a negligible fluorescence emission peak. After being treated with ONOO−, Cy–OH–ONOO exhibited an obvious fluorescence emission around 705 nm, which should be attributed to the leaving of p-boratebenzyl in ONOO− and in turn led to the onset of the fluorescent signal. Moreover, the fluorescence intensity of Cy–OH–ONOO with varied ONOO− content was further evaluated (Fig. 1B). There was a satisfying linear relationship between fluorescence intensity and ONOO− level in the range of 0–20 μM (Fig. 1C). The regression equation was determined as F705 nm = 54.5294 [ONOO−] + 259.4823, with a satisfactory linear relationship (R2 = 0.9874) between fluorescence intensity and ONOO− level in the range of 0–20 μM (Fig. 1C). The detection limit (LOD) was calculated as 56 nM using a standard method of 3σ/k, demonstrating that Cy–OH–ONOO can identify ONOO− in the presence of other compounds.
3.3 Selectivity
Whether the substance can be specifically detected is the criterion for evaluating the pros and cons of the probe. The selectivity monitoring of the probe Cy–OH–ONOO on ONOO− was evaluated in PBS buffer (EtOH/PBS = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 19, v/v, pH = 7.4). As depicted in Fig. 1D, an obvious NIR fluorescence enhancement was found after the addition of ONOO− (20 μM). It was previously reported that H2O2 can react with phenylboronate; however, it did not cause strong fluorescence fluctuations. Subsequently, other reactive species (HOCl, ·OH, HNO, NO, O2˙−, NO2− and BuOO·), common anions or cations (Na+, K+, Ca2+, HCO3−, Br− and HS−) and amino acid (glutamic acid (Glu), glutathione (GSH), tryptophane (Trp), serine (Ser), cysteine (Cys), methionine (Met), aspartic acid (Asp), homocysteine (Hcy), threonine (Thr) and lysine (Lys)) was inspected to see if they could cause changes in the probe's fluorescence signal (Fig. 1D and S1†). As expected, inert fluorescence was observed, further indicating that the Cy–OH–ONOO probe can specifically respond to ONOO− over other species.
19, v/v, pH = 7.4). As depicted in Fig. 1D, an obvious NIR fluorescence enhancement was found after the addition of ONOO− (20 μM). It was previously reported that H2O2 can react with phenylboronate; however, it did not cause strong fluorescence fluctuations. Subsequently, other reactive species (HOCl, ·OH, HNO, NO, O2˙−, NO2− and BuOO·), common anions or cations (Na+, K+, Ca2+, HCO3−, Br− and HS−) and amino acid (glutamic acid (Glu), glutathione (GSH), tryptophane (Trp), serine (Ser), cysteine (Cys), methionine (Met), aspartic acid (Asp), homocysteine (Hcy), threonine (Thr) and lysine (Lys)) was inspected to see if they could cause changes in the probe's fluorescence signal (Fig. 1D and S1†). As expected, inert fluorescence was observed, further indicating that the Cy–OH–ONOO probe can specifically respond to ONOO− over other species.
3.4 The pH effect and response speed
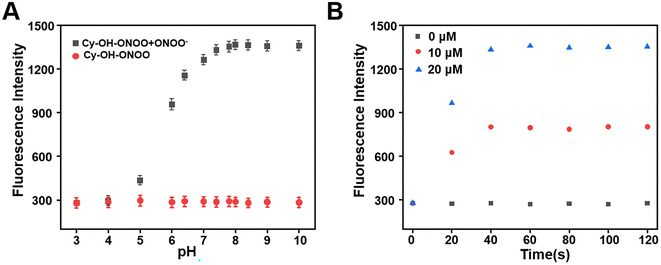
The effect of pH on Cy–OH–ONOO was investigated in the absence or presence of ONOO−. As we expected, the fluorescence intensity of Cy–OH–ONOO was passive and remained constant in the range of 3–10 pH in the absence of ONOO−, which suggested that pH could not cause fluctuations in the fluorescence intensity of the probe. However, the fluorescence intensity of the Cy–OH–ONOO probe increased rapidly and stabilized in physiological pH in the presence of ONOO− (Fig. 2A). However, the response time of Cy–OH–ONOO to ONOO− was examined. As shown in Fig. 2B, the fluorescence intensity of Cy–OH–ONOO eventually reached a plateau within 30 s at variable ONOO− concentrations (e.g., 0, 10, and 20 μM). The findings revealed that the probe can detect ONOO− levels in cells in real time. These results revealed that the Cy–OH–ONOO probe was useful for monitoring trace ONOO− variations in physiological conditions based on these findings.3.5 Appearance of ONOO− in living cells
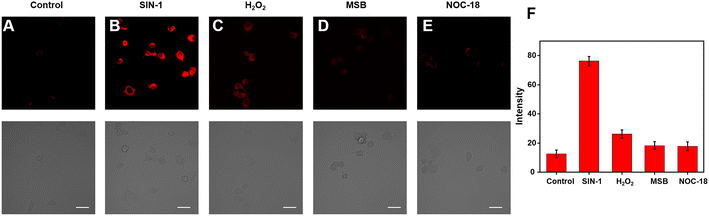
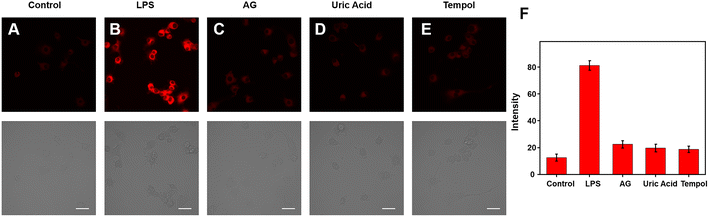
Because of its excellent spectrum properties, we investigated the ability of Cy–OH–ONOO to detect ONOO− in living cells. The Cells Counting Kit-8 (CCK-8) test was carried out to determine the cytotoxicity and biocompatibility of Cy–OH–ONOO prior to cell imaging. HepG2 and RAW 264.7 cells were selected as discussed subjects. The vitality of these cells was high when they were treated with various concentrations of Cy–OH–ONOO, demonstrating that the probe's cytotoxicity was little in this setting (Fig. S3†). It also suggested that Cy–OH–ONOO has a lot of potential for biological imaging applications. Therefore, we investigated the ability of Cy–OH–ONOO to image exogenous ONOO− in living cells. As shown in Fig. 3A, the RAW 264.7 cells displayed weak fluorescence signals when treated with Cy–OH–ONOO for 30 min. Subsequently, the RAW 264.7 cell line was incubated with 3-morpholino-sydnonimine (SIN-1, an ONOO− donor) for 60 min and then stained by Cy–OH–ONOO before imaging (Fig. 3B). There is an apparent fluorescence after the RAW 264.7 cells were treated with SIN-1, revealing that Cy–OH–ONOO was able to detect ONOO− in living cells. However, the cells were treated with 100 μM H2O2 and then stained with Cy–OH–ONOO; a negative fluorescent signal was obtained. Following that, the cells were treated with MSB (an O2˙− donor) or NOC-18 (an NO donor). In neither set of cells, there was no significant increase in the fluorescence signal, showing that these chemicals cannot produce changes in the fluorescence intensity of the probes. These findings suggest that this probe might be utilized to detect ONOO− but not other active species in cells.The potential application of Cy–OH–ONOO for imaging endogenous ONOO− was further investigated in RAW 264.7 cells. RAW 264.7 cells produced elevated levels of inducible nitric oxide synthase (iNOS) when stimulated by lipopolysaccharide (LPS) and PMA through a cellular inflammation response.37–39 Only one probe was used to stain the RAW 264.7 cell, and we discovered an inert fluorescent signal (Fig. 4A). A clear fluorescence enhancement signal was obtained after the cells were preincubated with LPS/PMA, indicating that intracellular ONOO− concentrations were increased in response to oxidative stress activation (Fig. 4B). In the next attempt, a nitric oxide synthase (NOS) inhibitor AG was utilized to verify that the fluctuations in fluorescence signals were caused by endogenous ONOO−, not other substances. As expected, we observed a decrease in fluorescence after the cells had been pre-incubated with LPS/PMA and then with AG for 30 min, revealing that the change in fluorescence signal was indeed from ONOO− rather than other biomolecular (Fig. 4C). Similarly, uric acid (an ONOO− scavenger) and superoxide scavenger 2,2,6,6-tetramethylpiperidine-N-oxyl (Tempol) was also employed to investigate whether the fluorescent signal was generated by the reaction of the probe with ONOO−. A weaker fluorescence phenomenon was found, indicating that the generation of ONOO− was inhibited (Fig. 4D and E). Moreover, it is visually declared that only the presence of ONOO− can cause fluctuations in the fluorescence signal. The above imaging results proved that Cy–OH–ONOO can respond to exogenous and endogenous conditions concerning the ONOO− state in the system of biology.
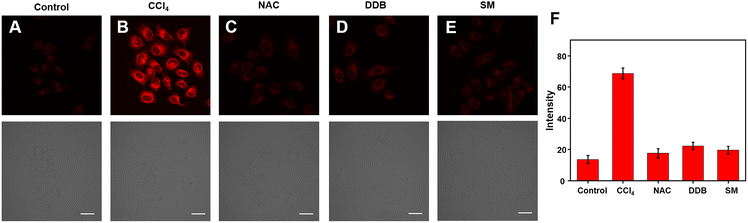
The Cy–OH–ONOO probe was used to scan exogenous and endogenous ONOO−. We applied it to detect ONOO− in CCl4-induced toxic hepatitis. Once CCl4 enters the cell, 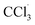 and CCl3OO· are produced, thereby tracking the influx of cell membrane and Ca2+.40 This process results in oxidative damage to hepatocytes and intracellular oxidative stress, which in turn leads to toxic hepatitis. As depicted in Fig. 5A, HepG2 cells treated with Cy–OH–ONOO showed weak fluorescence. When HepG2 cells were pretreated with CCl4 before being treated with Cy–OH–ONOO, a significant fluorescence signal was found. The levels of ONOO− were higher in CCl4-induced toxic hepatitis according to these findings, suggesting that ONOO− could serve as a biomarker in inflammatory diseases (Fig. 5B). Subsequently, the cells were treated with hepatoprotective medicine, including N-acetylcysteine (NAC), silymarin (SM), and bifendatatum (DDB). We found a negligible fluorescence signal (Fig. 5C–E). These findings showed that the Cy–OH–ONOO probe was an effective tool for detecting ONOO− levels in CCl4-induced toxic hepatitis cells.
and CCl3OO· are produced, thereby tracking the influx of cell membrane and Ca2+.40 This process results in oxidative damage to hepatocytes and intracellular oxidative stress, which in turn leads to toxic hepatitis. As depicted in Fig. 5A, HepG2 cells treated with Cy–OH–ONOO showed weak fluorescence. When HepG2 cells were pretreated with CCl4 before being treated with Cy–OH–ONOO, a significant fluorescence signal was found. The levels of ONOO− were higher in CCl4-induced toxic hepatitis according to these findings, suggesting that ONOO− could serve as a biomarker in inflammatory diseases (Fig. 5B). Subsequently, the cells were treated with hepatoprotective medicine, including N-acetylcysteine (NAC), silymarin (SM), and bifendatatum (DDB). We found a negligible fluorescence signal (Fig. 5C–E). These findings showed that the Cy–OH–ONOO probe was an effective tool for detecting ONOO− levels in CCl4-induced toxic hepatitis cells.
3.6 Bioimaging applications of Cy–OH–ONOO in zebrafish
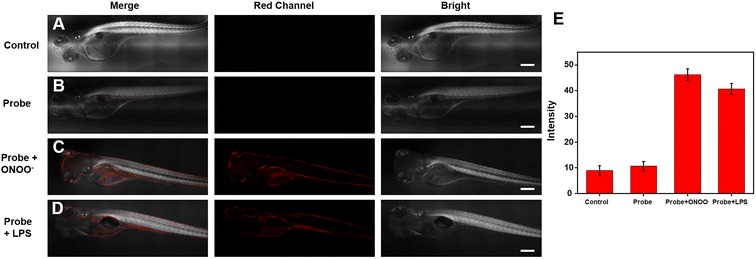
Considering that the efficiency with ONOO− can be imaged and detected in living cells, we wondered whether the Cy–OH–ONOO probe could be used to scan ONOO− in zebrafish. The zebrafish were photographed immediately without any processing as a control group; substantially, no fluorescence signal phenomenon was observed (Fig. 6A). Next, the zebrafish were stained by the probe for 30 min. As we expected, weak red fluorescence was found, indicating that the probe could easily enter zebrafish (Fig. 6B). However, ONOO− was added to these zebrafish, which exhibited stronger red fluorescence (Fig. 6C). It was discovered that the Cy–OH–ONOO probe could detect exogenous ONOO− in zebrafish. To determine the utility of Cy–OH–ONOO in imaging endogenous ONOO−, the zebrafish were preconditioned with LPS and PMA to promote the production of ONOO− (Fig. 6D). After the zebrafish were treated with the probe, an obvious fluorescence enhancement signal was obtained. These experiments demonstrated that the Cy–OH–ONOO probe could act as an effective tool to image and detect exogenous/endogenous ONOO− in vivo.3.7 Bioimaging applications of Cy–OH–ONOO in mice breasts
We attempted to apply the probe to breast cancer tumor imaging in vivo. Here, we implanted 4T1 cells in situ in mice to form breast cancer tumors. We used a syringe to inject the probe in situ into the skin near the breast cancer tumor site in mice. As a control group, we also injected the same dose of Cy–OH–ONOO into the upper left side of the tumor site. As shown in Fig. 7, after injection of the probe solution, the tumor site exhibited stronger fluorescence compared to the non-tumor site. This indicates that the probe had good tumor imaging performance, which further proves that it has a strong ability to identify tumors. These experiments demonstrated that the Cy–OH–ONOO probe could act as an effective tool to image and detect exogenous/endogenous ONOO− in vivo.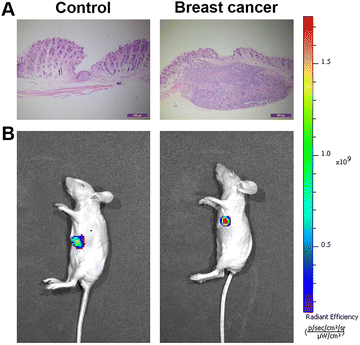 | ||
| Fig. 7 Fluorescent imaging of ONOO− in mice. (A) H&E analysis images. (B) Fluorescent imaging of ONOO− in normal mice and breast mice. | ||
4. Conclusion
In conclusion, we developed a Cy–OH–ONOO NIR fluorescent probe, which may be employed to track and image intracellular dynamic ONOO− contents. Under physiological conditions, the probe demonstrated exceptional specificity and sensitivity to ONOO− in various analyses. Furthermore, the aspects that are valuable for low biocompatibility cytotoxicity enabled the Cy–OH–ONOO probe to image ONOO− in complex biological conditions. In addition, Cy–OH–ONOO was employed to identify ONOO− upregulation in CCl4-induced toxic hepatitis and LPS stimulation in living cells. Moreover, the probe was successfully applied to detect ONOO− levels in mice with breast cancer, which helps to further investigate the relationship between ONOO− and breast cancer. Therefore, these favorable properties of Cy–OH–ONOO make it promising for bioimaging applications, especially for studying the relationship between ONOO− and inflammatory or cancer diseases.Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.Author contributions
Zixiang Xu designed and synthesized and purified the compound, in addition, performed data analysis and wrote the manuscript. Zhencai Xu and Dong Zhang contributed to the idea of the study, revised the manuscript, and approved the final version. All authors contributed to the article and approved the submitted version.Conflicts of interest
The authors declare that no conflicts of interest exist.References
- Y. Hu, Y. Wang, X. Wen, Y. Pan, X. Cheng, R. An, G. Gao, H. Y. Chen and D. Ye, Research, 2020, 4087069 CAS.
- C. Hwang, A. J. Sinskey and H. F. Lodish, Science, 1992, 257, 1496–1501 CrossRef CAS PubMed.
- X.-X. Chen, L.-Y. Niu, N. Shao and Q.-Z. Yang, Anal. Chem., 2019, 91, 4301–4306 CrossRef CAS PubMed.
- D. Wolf and K. Ley, Circ. Res., 2019, 124, 315–327 CrossRef CAS PubMed.
- P. Pacher, J. S. Beckman and L. Liaudet, Physiol. Rev., 2007, 87, 315–424 CrossRef CAS PubMed.
- S. Goldstein, J. Lind and G. Merényi, Chem. Rev., 2005, 105, 2457–2470 CrossRef CAS PubMed.
- R. Radi, J. Biol. Chem., 2013, 288, 26464–26472 CrossRef CAS PubMed.
- C. Szabó, H. Ischiropoulos and R. Radi, Nat. Rev. Drug Discovery, 2007, 5, 662–680 CrossRef.
- D. Gius and D. R. Spitz, Antioxid. Redox Signaling, 2006, 8, 1249–1252 CrossRef CAS.
- G. Ferrer-Sueta and R. Radi, ACS Chem. Biol., 2009, 4, 161–177 CrossRef CAS PubMed.
- S. B. Digerness, K. D. Harris and J. W. Kirklin, Free Radical Biol. Med., 1999, 27, 1386–1392 CrossRef CAS PubMed.
- Y. Li, X. Xie, X. Yang, M. Li, X. Jiao, Y. Sun, X. Wang and B. Tang, Chem. Sci., 2017, 8, 4006–4011 RSC.
- X. Z. Luo, Z. Y. Cheng, R. Wang and F. B. Yu, Anal. Chem., 2021, 93, 2490–2499 CrossRef CAS.
- C. Tang, H. Tong, B. Liu, X. Wang, Y. Jin, E. Tian and F. Wang, Anal. Chem., 2022, 94, 14012–14020 CrossRef CAS PubMed.
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, Ca-Cancer J. Clin., 2021, 71, 209–249 CrossRef.
- C. Amatore, S. Arbault, D. Bruce, P. de Oliveira, M. Erard and M. Vuillaume, Chem. - Eur. J., 2001, 7, 4171–4179 CrossRef CAS.
- Y. Chen, C. Chen, W. Chen, J. L. Zweier, O. Augusto, R. Radi and R. P. Mason, J. Biol. Chem., 2004, 279, 18054–18062 CrossRef CAS PubMed.
- A. Daiber, M. Oelze, M. August, M. Wendt, K. Sydow, H. Wieboldt, A. L. Kleschyov and T. Munzel, Free Radical Res., 2004, 38, 259–269 CrossRef CAS.
- Z. Tian, X. Tian, L. Feng, Y. Tian, X. Huo, B. Zhang, S. Deng, X. Ma and J. Cui, J. Mater. Chem. B, 2019, 7, 4983–4989 RSC.
- H. Kobayashi, M. Ogawa, R. Alford, P. L. Choyke and Y. Urano, Chem. Rev., 2010, 110, 2620–2640 CrossRef CAS PubMed.
- J. Lu, L. Ji and Y. Yu, RSC Adv., 2021, 11, 35093–35098 RSC.
- R. Kumar, W. S. Shin, K. Sunwoo, W. Y. Kim, S. Koo, S. Bhuniya and J. S. Kim, Chem. Soc. Rev., 2015, 44, 6670–6683 RSC.
- J. Li, L. Chen, Q. Wang, H. Liu, X. Hu, L. Yuan and X. Zhang, Anal. Chem., 2018, 90, 4167–4173 CrossRef CAS PubMed.
- X. Chen, F. Wang, J. Y. Hyun, T. Wei, J. Qiang, X. Ren, I. Shin and J. Yoon, Chem. Soc. Rev., 2016, 45, 2976–3016 RSC.
- Z. Chen, W. Ren, Q. Wright and H. Ai, J. Am. Chem. Soc., 2013, 135, 14940–14943 CrossRef CAS.
- W. Qu, C. Niu, X. Zhang, W. Chen, F. Yu, H. Liu, X. Zhang and S. Wang, Talanta, 2019, 197, 431–435 CrossRef CAS.
- C. Zhao, J. An, L. Zhou, Q. Fei, F. Wang, J. Tan, B. Shi, R. Wang, Z. Guo and W.-H. Zhu, Chem. Commun., 2016, 52, 2075–2078 RSC.
- D. Liu, S. Feng and G. Feng, Sens. Actuators, B, 2018, 269, 15–21 CrossRef CAS.
- D. Yang, H. L. Wang, Z. N. Sun, N. W. Chung and J. G. Shen, J. Am. Chem. Soc., 2006, 128, 6004–6005 CrossRef CAS.
- Z. Lei, C. Sun, P. Pei, S. Wang, D. Li, X. Zhang and F. Zhang, Angew. Chem., Int. Ed., 2019, 58, 8166–8171 CrossRef CAS.
- X. Luo, R. Wang, C. Lv, G. Chen, J. You and F. Yu, Anal. Chem., 2020, 92, 1589–1597 CrossRef CAS PubMed.
- S. Y. Liu, H. Xiong, J. Q. Yang, S. H. Yang, Y. F. Li, W. C. Yang and G. F. Yang, ACS Sens., 2018, 3, 2118–2128 CrossRef CAS PubMed.
- L. Yuan, W. Lin, K. Zheng, L. He and W. Huang, Chem. Soc. Rev., 2013, 42, 622–661 RSC.
- L. C. Murfin, M. Weber, S. J. Park, W. T. Kim, C. M. Lopez-Alled, C. L. McMullin, F. Pradaux-Caggiano, C. L. Lyall, G. KociokKöhn, J. Wenk, S. D. Bull, J. Yoon, H. M. Kim, T. D. James and S. E. Lewis, J. Am. Chem. Soc., 2019, 141, 19389–19396 CrossRef CAS PubMed.
- D. Li, S. Wang, Z. Lei, C. Sun, A. M. El-Toni, M. S. Alhoshan, Y. Fan and F. Zhang, Anal. Chem., 2019, 91, 4771–4779 CrossRef CAS PubMed.
- Z. Wang, W. Wang, P. Wang, X. Song, Z. Mao and Z. Liu, Anal. Chem., 2021, 93, 3035–3041 CrossRef CAS PubMed.
- R. B. Lorsbach, W. J. Murphy, C. J. Lowenstein, S. H. Snyder and S. W. Russell, J. Biol. Chem., 1993, 268, 1908–1913 CrossRef CAS.
- X. Sun, Q. Xu, G. Kim, S. E. Flower, J. P. Lowe, J. Yoon, J. S. Fossey, X. Qian, S. D. Bull and T. D. James, Chem. Sci., 2014, 5, 3368–3373 RSC.
- A. C. Sedgwick, X. Sun, G. Kim, J. Yoon, S. D. Bul and T. D. James, Chem. Commun., 2016, 52, 12350–12352 RSC.
- Y. Tian, D. Y. Zhou, W. L. Jiang, Z. P. She, Y. Li and C. Y. Li, Talanta, 2021, 223, 121720 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Additionally, the fluorescence spectra and pictures and structural characterizations of the probe. See DOI: https://doi.org/10.1039/d3ra01024d |
| This journal is © The Royal Society of Chemistry 2023 |