Open Access Article
Open Access ArticleMagnetic dummy template molecularly imprinted particles functionalized with dendritic nanoclusters for selective enrichment and determination of 4-methylnitrosamino-1-(3-pyridyl)-1-butanone (NNK) in tobacco products
Yani Zhenga,
Yin Daia,
Junqiang Hongb,
Huizhu Fana,
Qing Zhangac,
Wei Jiangd,
Wei Xua,
Jianwen Feia and
Junli Hong *a
*a
aSchool of Pharmacy, Nanjing Medical University, Nanjing, Jiangsu 211166, China. E-mail: junlihong@njmu.edu.cn; Fax: +86 25 86868476; Tel: +86 25 86868476
bDepartment of Radiotherapy, Fujian Medical University Affiliated Xiamen Humanity Hospital, Xiamen, Fujian 361000, China
cAnhui Province Key Laboratory of Research & Development of Chinese Medicine, Hefei 230012, China
dYunnan Key Laboratory of Tobacco Chemistry, Research and Development Center of China Tobacco Yunnan Industrial Co. Ltd, Kunming, Yunnan 650231, China
First published on 5th May 2023
Abstract
The compound 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), one of the tobacco specific nitrosamines (TSNAs), is widely recognized as a major carcinogen found in tobacco products, environmental tobacco smoke and wastewater. Thus, a selective enrichment and sensitive detection method for monitoring the risk of NNK exposure is highly desirable. In this study, a magnetic molecularly imprinted polymer (MMIP) functionalized with dendritic nanoclusters was synthesized to selectively recognize NNK via the dummy template imprinting strategy, aiming to avoid residual template leakage and increase the imprinting efficiency. The nanocomposites were characterized by scanning electron microscopy (SEM), transmission electron microscopy (TEM), energy dispersive spectroscopy (EDS), X-ray diffraction (XRD), Fourier transform infrared (FTIR) spectroscopy, as well as vibrating sample magnetometry (VSM) and nitrogen adsorption/desorption analysis. The resulting MMIPs exhibited high adsorption capacity, fast binding kinetics and good selectivity for trace amounts of NNK. A rapid, low cost and efficient method for detecting NNK in tobacco products was established using magnetic dispersive solid-phase extraction coupled with HPLC-DAD with a good linear range of 0.1–250 μg mL−1. The limit of detection (LOD) and limit of quantification (LOQ) of NNK were 13.5 and 25.0 ng mL−1, respectively. The average recoveries were 87.8–97.3% with RSDs lower than 3%. The results confirmed that the MMIPs could be used as an excellent selective adsorbent for NNK, with potential applications in the pretreatment of tobacco products.
1 Introduction
Tobacco-specific nitrosamines (TSNAs) are strong carcinogens commonly found in tobacco products and cigarette smoke. TSNAs are N-nitroso-derivatives of pyridine-alkaloids, including 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), N′-nitrosonornicotine (NNN), N′-nitrosoanabasine (NAB) and N′-nitrosoanatabine (NAT). During air curing of fresh green tobacco, fermentation, storage, and tobacco combustion, nicotine and minor tobacco alkaloids can also be converted into TSNAs through nitrosation. NNK has been shown to increase the risk of several malignancies, including lung, esophageal, liver and pancreatic cancer, even at low levels.1–3 NNK is classified as a confirmed human carcinogen (group 1) by the International Agency for Research on Cancer (IARC) with a No Significant Risk Level (NSRL) of 14 ng per day.4 Thus, it is necessary to monitor NNK in the complex tobacco matrix at trace levels.A number of analytical techniques for the determination of NNK have been developed, including GC, HPLC-MS/MS, GC/MS and thermal energy analysis.5–7 However, because of the low concentration of NNK in tobacco products and the interference of complex matrix components, the detection of NNK is a major analytical challenge. In order to effectively solve this problem, several pretreatment methods for the extraction of NNK have been established, using adsorbents such as mesoporous silica, metal organic frameworks8 and zeolite.9 Although these materials generally achieve the purpose of adsorption using a deformable molecular sieve, they have shortcomings such as no specific chemical recognition sites, low rebinding capacity, large consumption of organic solvents and inevitably adsorption of tobacco aroma substances, which limit the application scope. Therefore, a highly selective, rapid and environmentally friendly method for the enrichment and determination of NNK in tobacco products is urgently needed.
Molecular imprinted polymers (MIPs) are artificial locks with specific cavities, which can be used as analyte-recognizing parts for sensors and in some cases, instead of natural affinity compounds such as receptors or antibodies.10 Nowadays, it have attracted great interest, due to the advantages of sensitivity, good adsorption, desired surface properties and high separation efficiency. For application aspect, MIPs are widely used in the molecularly imprinted polymers (MIPs) based affinity sensors, and MIPs are especially suitable for sample pretreatment or chromatography as adsorbent or stationary phase.11,12 In recent years, surface molecular imprinted technology has attracted increasing attention due to advantages such as high utilization ratio of binding sites, fast mass transfer, and high surface-to-volume ratio.13 The carrier material of surface molecularly imprinted polymer directly affects its adsorption characteristics. Up to now, the MIPs for some molecules, such as toxic, expensive, unstable and poorly soluble targets, limited the wide application of MIPs. Furthermore, template leakage will severely deteriorating the accuracy and precision of the trace analysis. Pseudo template, a compound which has similar spatial structure with the target molecule, is an effective substitute to solve these problems. This structural analogue used during MIPs design and production could generate imprinting sites that are the same or similar to those produced by target molecule, and exhibited selectivity towards the target analyte that is similar to the imprinting sites formed by the target molecule. Hence, the accuracy of trace analysis may improve. Polyamide amine (PAMAM) dendrimer is a highly branched three-dimensional macromolecule which has superior features including abundant terminal amine groups, high internal porosity, as well as good structural homogeneity, biocompatibility and chemical stability.14,15 In addition, magnetic separation is a rapid and convenient method for controlling adsorption and desorption process of target compounds from complicated matrices by a magnet without additional centrifugation or filtration. More importantly, this magnetic separation strategy achieves the purification of the analysis system, improves the enrichment ability and extraction power, and overcomes the shortcomings of traditional pretreatment methods that may have poor recovery rates and low adsorbent reusability.16 Thus, the grafting of dendritic nanoclusters onto magnetic nanoparticles may improve the adsorption capacity, binding kinetics and separation efficiencies of surface molecularly imprinted polymers.
In this work, we prepared high performance NNK magnetic surface molecular imprinting polymers (NNK-MMIPs) functionalized with dendritic nanoclusters. The aim of this study was to develop multifunction NNK-MMIPs through a simple synthetic method for selective separation and enrichment of NNK in tobacco products. Considering the high toxicity of NNK, the dummy template molecular imprinting technique was used to avoid template leaking and reduce the interference in trace detection. The morphology and composition of NNK-MMIPs were characterized using scanning electron microscopy (SEM), FTIR spectroscopy and X-ray diffraction (XRD) and so on. Finally, NNK-MMIPs were successfully applied for the selective recognition, enrichment and trace detection of NNK coupled with HPLC-DAD in tobacco products, and encouraging results were obtained.
2 Materials and methods
2.1 Materials and reagents
FeCl3·6H2O, FeCl2·4H2O and tetraethyl orthosilicate (TEOS) were obtained from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). NH3·H2O and glutaraldehyde (GA, 50% in water) were purchased from Macklin Reagent Co., Ltd. (Shanghai, China). Methanol and ethanol were obtained from Jiangsu Hanbon Sci. & Tech. Co., Ltd. γ-Aminopropyl triethoxysilane (KH550) was obtained from Diamond Advanced Material of Chemical Inc. Methacrylic acid (MAA) was obtained from Shanghai Lingfeng Chemical Reagent Co., Ltd. Polyamidoamine dendrimer (PAMAM) (ethylenediamine core, generation 3, 20 wt% in methanol), 4-(N-nitrosomethylamino)-1-(3-pyridyl)butan-1-one (NNK) and ethylene glycol dimethacrylate (EGDMA) were purchased from Sigma-Aldrich. Nicotinylmethylamide (NMAM), 3-methyl-pyridine and nicotinamide were purchased from Aladdin Reagent Co., Ltd. (Shanghai, China). Furthermore, 2,2′-azobisisobutyronitrile (AIBN) was obtained from Shanghai NO. 4 Reagent & H.V. Chemical Co., Ltd. Deionized water was used throughout the experiments. All other chemicals were analytical grade and obtained commercially.2.2 Apparatus
HPLC was performed with a Shimadzu (Japan) system using an LC-20AT pump, SPD-M20A, DAD and LabSolutions chromatographic workstation. Other instruments included a MilliQ® (Millipore Co. Milford, MA, USA) water purification system, Shimadzu (Japan) AUW220D electronic balance (accuracy: 0.01 mg), KQ5200 Ultrasonic Cleaner (Kunshan Ultrasonic Instrument Co., Ltd., China), DZG-6020 vacuum driving oven (Shanghai Samsung Laboratory Equipment Co., Ltd., China) and IKA®C-MAG HS 7 and IKA®-Werke GmbH & Co. KG temperature magnetic mixer (IKA® Processing Equipment, Germany). Transmission electron microscopy (TEM) and high-resolution transmission electron microscopy (HRTEM) graph was obtained by JEM-2100E transmission electron microscope (JEOL, Japan). Scanning electron microscopy (SEM) graph was obtained by HITACHI SU8010. The phase structure of the as-prepared samples was characterized by X-ray power diffraction (XRD) with Bruker D8 ADVANCE. The Fourier Infrared spectra was measured by Thermo Nicolet IS5. The BET record was gained from Micromeritics ASAP2020/ASAP2460. The VSM data was obtained from Quantum Design's MPMS SQUID-VSM.2.3 Preparation of magnetic nanoparticles functionalized with dendritic nanoclusters
Fe3O4 nanoparticles were synthesized referring to previous articles published by our laboratory.17,18 In a preparative process, 0.1 mol FeCl2·4H2O and 0.2 mol FeCl3·6H2O in 80 mL deionized water were heated to 80 °C under nitrogen flow with stirring. Then, aqueous ammonia was dripped slowly until the pH of the reaction solution reached 10, after which it was allowed to react for 30 min. The black products obtained by magnetic separation were cleaned with ethanol followed by deionized water and dried under vacuum. Then, Fe3O4 nanoparticles were sequentially modified with TEOS and APTES to introduce amino groups. Typically, Fe3O4@SiO2 nanoparticles were synthesized using sol–gel method, 0.15 g of Fe3O4 nanoparticles were dispersed in 120 mL of ethanol, and the mixture was homogenized by ultrasonication for 15 min prior to the addition of 1 mL NH3·H2O. After being stirred vigorously for 45 min at 30 °C, 1.5 mL TEOS combing 30 mL ethanol were dripped into the solution. After 45 min, 0.3 mL APTES was added and allowed to react for another 12 h to obtain the aminated Fe3O4@SiO2.Subsequently, Fe3O4@SiO2@PAMAM nanoparticles were synthesized by Schiff base reaction, 50 mg of aminated Fe3O4@SiO2 were dissolved in 7.5 mL water, after which 2.5 mL of glutaraldehyde solution (20 wt%) were added to the solution and slowly shaken for 6 h at room temperature. After rinsing three times with 20 mM PBS (pH 7.4) to remove the unreacted glutaraldehyde, 2 mL of PAMAM dendrimers (0.1 wt%) were added to the aldehyde-functionalized Fe3O4@SiO2. The mixture was gently shaken overnight at 37 °C and then washed with PBS (pH 7.4) three times and dried.
2.4 Preparation of dummy template NNK-MMIPs
In a technological process, molecularly imprinted polymers were synthesized by free radical polymerization. A total of 50 mg of the modified magnetic nanoparticles and 39 mg nicotinylmethylamide and MAA (0.78 mmol) were dispersed in 50 mL of methanol–water (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v), and incubated at room temperature for 1 h in order to occur prepolymerization. Afterwards, 3.12 mmol EGDMA and 30 mg AIBN were added as cross-linker and initiator, respectively. The mixture was stirred for 24 h at 50 °C under nitrogen gas. After the reaction, the product was separated with a magnet. Then, the template molecule was eluted by washing with methanol–acetic acid (9
1, v/v), and incubated at room temperature for 1 h in order to occur prepolymerization. Afterwards, 3.12 mmol EGDMA and 30 mg AIBN were added as cross-linker and initiator, respectively. The mixture was stirred for 24 h at 50 °C under nitrogen gas. After the reaction, the product was separated with a magnet. Then, the template molecule was eluted by washing with methanol–acetic acid (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) 3 times, and NNK-MMIPs were successfully obtained. The NNK-MNIPs were prepared in the same manner in the absence of templates.
1, v/v) 3 times, and NNK-MMIPs were successfully obtained. The NNK-MNIPs were prepared in the same manner in the absence of templates.
2.5 Adsorption experiments
The adsorption experiments were carried out as follows: 20 mg of NNK-MMIPs and NNK-MNIPs were added into the methanol–water (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) solution containing NNK standard at various concentrations (2.5–80 mM). After shaking for 2 h at 25 °C, the materials were separated by applying an external magnetic field, then washed with methanol (5 mL, 2 times) and desorption solution (10 mL, methanol–acetic acid, 9
1, v/v) solution containing NNK standard at various concentrations (2.5–80 mM). After shaking for 2 h at 25 °C, the materials were separated by applying an external magnetic field, then washed with methanol (5 mL, 2 times) and desorption solution (10 mL, methanol–acetic acid, 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v), respectively. Then, the desorption solution was dried under a nitrogen stream. The obtained residue was redissolved in the mobile phase, and measured by HPLC-DAD. The dynamic adsorption was tested with the NNK standard solution (50 mM in methanol).
1, v/v), respectively. Then, the desorption solution was dried under a nitrogen stream. The obtained residue was redissolved in the mobile phase, and measured by HPLC-DAD. The dynamic adsorption was tested with the NNK standard solution (50 mM in methanol).
2.6 Selective adsorption
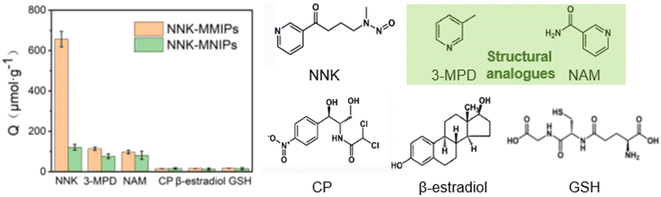
The selectivity of NNK-MMIPs and NNK-MNIPs was assessed using NNK as target compound, 3-methyl-pyridine and nicotinamide as structural analogues, chloramphenicol, β-estradiol and glutathione as reference substances for comparison at a concentration of 10 mM. The remaining steps were the same as those listed in the adsorption experiment.2.7 Chromatographic conditions
The chromatographic separation was performed on a Diamonsil® C18 column (5 μm, 150 mm × 4.6 mm). The mobile phase consisted of mobile phase A (10 mM ammonium acetate in water) and mobile phase B (10 mM ammonium acetate in methanol). The step gradient was as follows: 0–4.5 min 12% B; 4.5–4.6 min linear increase to 60% B; 4.6–9.0 min linear increase to 95% B; 9.0–10.0 min decreased to 12% B. The detection wavelength was 230 nm and the injection volume was 10 μL.2.8 Analysis of tobacco samples
Briefly, 10 g dried tobacco (purchasing from a supermarket) was burned and the mainstream smoke was collected using Cambridge filter pads. Each pad with trapped smoke particulates was sonicated in 10 mL of ammonium acetate (0.1 mM) for 30 min. The extraction solution was then filtered through a 0.45 μm filtering membrane, dried under a nitrogen stream and redissolved in 10 mL methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (v/v = 9
water (v/v = 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1).
1).
Then, 20 mg of NNK-MMIPs were dispersed in 5 mL of the sample solution and shaken at room temperature for 30 min. Then, the NNK-MMIPs were separated by applying an external magnetic field, washed with methanol and eluted with 10 mL desorption solution (methanol–acetic acid, 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) by ultrasonic treatment for 20 min. The supernatant was collected and dried under a nitrogen stream, after which the residues were redissolved in 200 μL of the mobile phase for further HPLC-DAD analysis.
1, v/v) by ultrasonic treatment for 20 min. The supernatant was collected and dried under a nitrogen stream, after which the residues were redissolved in 200 μL of the mobile phase for further HPLC-DAD analysis.
3 Results and discussion
3.1 Principle for NNK enrichment and detection using NNK-MMIPs
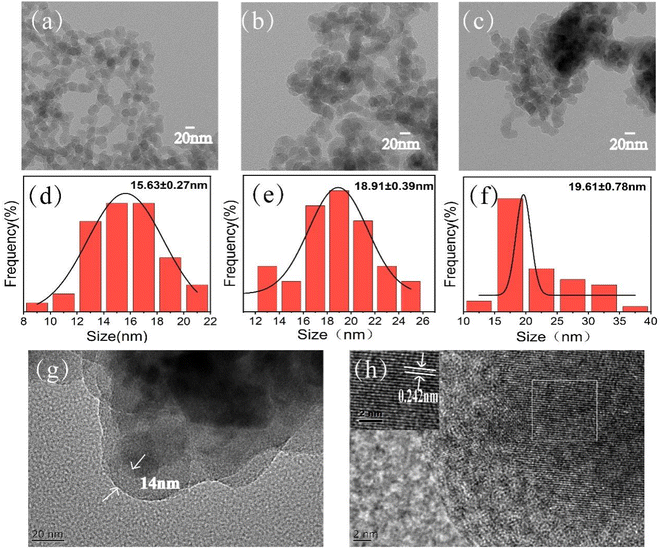
The molecularly imprinted polymer specific for NNK was synthesized at the surface of ∼15 nm magnetic silica nanoparticles functionalized with dendritic modifications using the dummy template strategy (Scheme 1). First, dendritic nanoclusters were successfully assembled at the surface of the magnetic nanoparticles through simple free-radical polymerization, followed by further copolymerization between methacrylic acid (MAA), ethylene glycol dimethacrylate (EGDMA) and 2,2′-azobisisobutyronitrile (AIBN) in the presence of templates. After elution of the templates, a large number of tailor-made cavities on the surface of magnetic silica nanoparticles (NNK-MMIPs) were formed. NNK-MMIPs with this prearranged structure and specific molecular recognition ability were successfully applied for the separation and enrichment of analytes coupled with HPLC-DAD detection.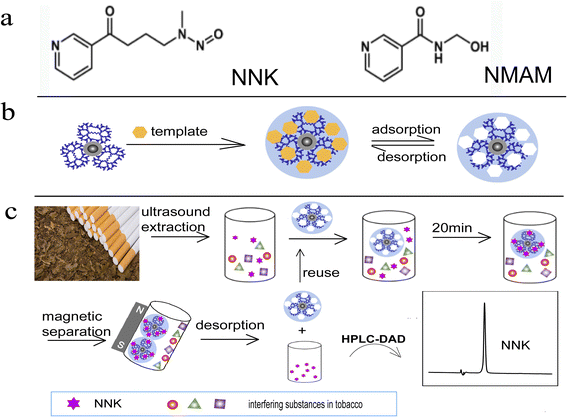 | ||
| Scheme 1 Schematic illustration (a) template comparison; (b) synthesis of NNK-MMIPs; (c) application of NNK-MMIPs for NNK detection in real samples. | ||
The protruding dendritic nanostructures and open cavities on the surface of molecularly imprinted polymers make a great contribution to their fast binding-kinetics, high adsorption capacity, more effective imprinting sites and good selectivity for the target species. Moreover, in order to avoid the leakage of a trace amount of the target NNK, which may have potential genotoxicity and carcinogenicity, a structural analogue of NNK, nicotinylmethylamide, was used as the dummy template. As for the chemical structure, nicotinylmethylamide and NNK both have a 3-pyridine carbon group structure, and the chemical structural terminus of nicotinylmethylamide has the similar number of hydrogen bond donor acceptors as the nitrosamine group of NNK, resulting the generation of similar imprinting sites, since the template and functional monomer in the present molecularly imprinted polymer were bonded by noncovalent hydrogen bond. What's more, nicotinylmethylamide is an easily obtainable, low-cost, and highly soluble compound, is used for cholagogic and anti-inflammatory purposes, with much lower toxicity compared to NNK because of the lack of a nitroso group.
Here, the dummy molecularly imprinted polymer was used to avoid sample contamination or false positive readings, thereby increasing the accuracy and precision of the analysis. Furthermore, because of the good magnetic properties, NNK-MMIPs can avoid tedious filtration and centrifugation steps to achieve rapid and efficient separation using an external magnetic field. Based on these improvements, we used NNK-MMIPs as dispersive solid-phase extraction (DSPE) materials coupled with HPLC-DAD for the selective monitoring of trace NNK in tobacco product samples.
3.2 Characterization
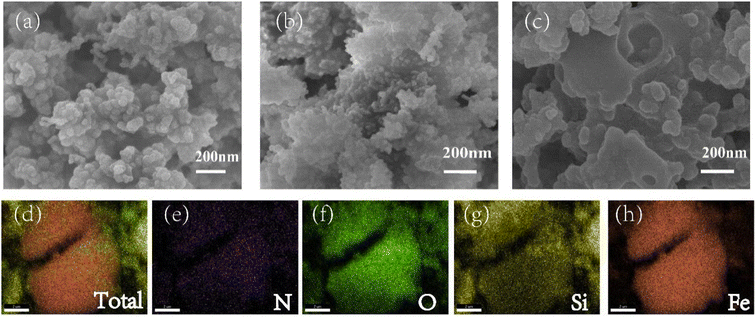
More details were revealed by SEM. Compared with Fe3O4@SiO2 (Fig. 2a), Fe3O4@SiO2@PAMAM (Fig. 2b) showed a more uneven and rough surface. After the synthesis of imprinted polymers (Fig. 2c), the surface became relatively smooth and large holes appeared, which were attributed to the imprinted cavities. Moreover, energy dispersive spectroscopy (EDS) and elemental mapping analysis showed the distribution of N, O, Si, Fe in the NNK-MMIPs (Fig. 2d–h). These results further confirmed the successful grafting of dendritic nanoclusters onto the nanomaterial.
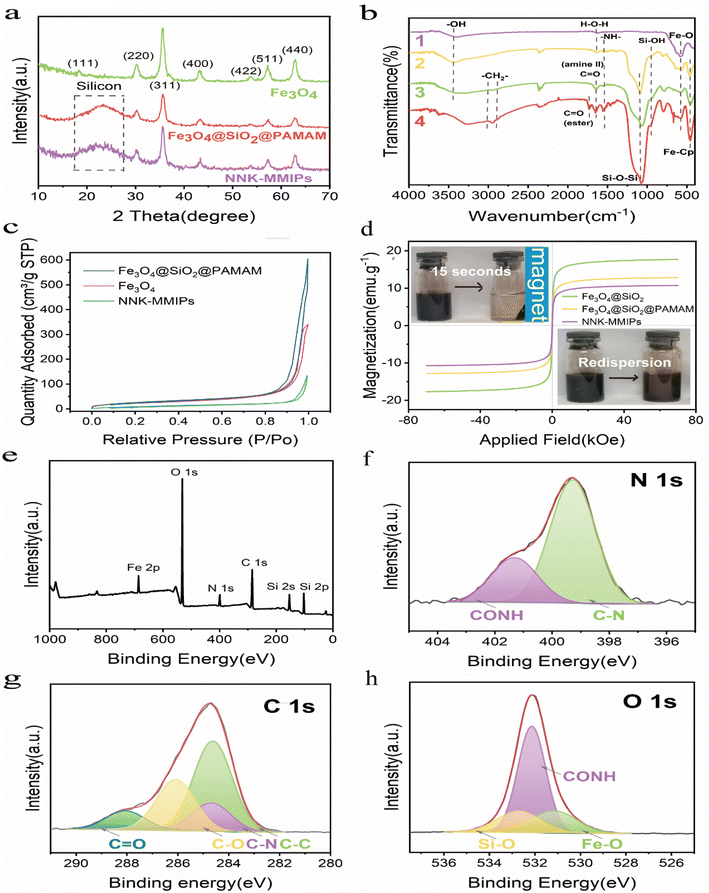
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O from secondary amine groups, respectively. In the NNK-MMIPs samples (curve 4), the characteristic peak at 1724 cm−1 was ascribed to the stretching vibration of C
O from secondary amine groups, respectively. In the NNK-MMIPs samples (curve 4), the characteristic peak at 1724 cm−1 was ascribed to the stretching vibration of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups in the crosslinking agent (EGDMA), which confirmed that the imprinted polymers are successfully synthesized.24,25 Thus, the data confirmed the successful grafting of dendritic nanoclusters and introduction of the molecular imprinting layer.
O groups in the crosslinking agent (EGDMA), which confirmed that the imprinted polymers are successfully synthesized.24,25 Thus, the data confirmed the successful grafting of dendritic nanoclusters and introduction of the molecular imprinting layer.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C–O, C–N and C–C, respectively (Fig. 3g). The O 1s high resolution spectrum displayed peaks at 532.78, 532.18 and 531.08 eV were assigned to Si–O, CONH and Fe–O bonds, respectively (Fig. 3h). The above results further confirmed the successful grafting of dendritic nanoclusters on the surface of Fe3O4 NPs, which was consistent with the EDS elemental mapping.
O, C–O, C–N and C–C, respectively (Fig. 3g). The O 1s high resolution spectrum displayed peaks at 532.78, 532.18 and 531.08 eV were assigned to Si–O, CONH and Fe–O bonds, respectively (Fig. 3h). The above results further confirmed the successful grafting of dendritic nanoclusters on the surface of Fe3O4 NPs, which was consistent with the EDS elemental mapping.3.3 Evaluation of the absorption characteristics of NNK-MMIPs
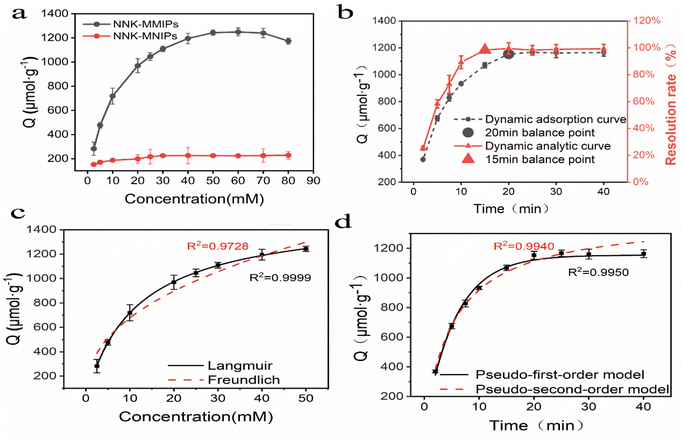
As shown in Fig. 4c, the binding properties of isotherms could be estimated by application of the Langmuir isotherm model with a high R2 value (0.99995). This suggested that the adsorption occurred at specific homogeneous sites within the adsorbent, forming a monolayer on the surface of the sorbent.
3.4 Method validation
Under the optimized conditions of HPLC coupled with NNK-MMIPs as SPE polymer materials, good linearity was achieved in the range of 0.1–250 μg mL−1 for NNK and the regression was Y = 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 639X − 68
639X − 68![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 384 with a correlation of 0.9993. The limit of detection (LOD, S/N = 3) and limit of quantification (LOQ, S/N = 10) was 13.5 ng mL−1 and 25.0 ng mL−1 for NNK, respectively.
384 with a correlation of 0.9993. The limit of detection (LOD, S/N = 3) and limit of quantification (LOQ, S/N = 10) was 13.5 ng mL−1 and 25.0 ng mL−1 for NNK, respectively.
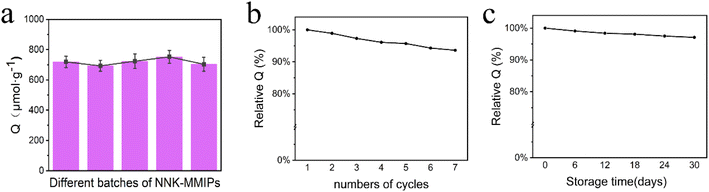
The precision and accuracy were assessed by analyzing tobacco samples spiked with NNK at three concentration levels. As shown in Table 1, the average recoveries were in the range of 87.8–97.3%, with RSD in the range of 1.53–2.71%. All these data confirmed that we developed an accurate, reliable and practical method for the separation and determination of trace levels of NNK in complex tobacco samples.
| Sample | Measured (ng g−1) | Spiked (ng g−1) | Found (ng g−1) | Recovery (%) | RSD (%) |
|---|---|---|---|---|---|
| S1 | 8.32 | 6.60 | 14.74 | 97.3 | 2.37 |
| 8.50 | 16.25 | 93.3 | 1.83 | ||
| 9.90 | 17.69 | 94.6 | 1.53 | ||
| S2 | 7.16 | 5.70 | 12.47 | 93.2 | 2.71 |
| 7.20 | 13.72 | 91.1 | 1.68 | ||
| 8.60 | 14.71 | 87.8 | 1.53 |
3.5 Sample analysis
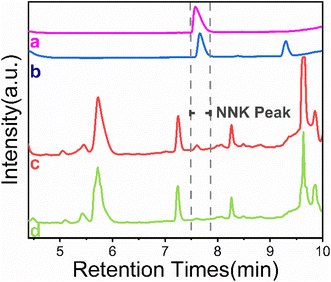
For final validation, the proposed method was applied successfully to the analysis of NNK in real samples. Fig. 7 shows that NNK in the complex samples could not be detected by HPLC without enrichment (curve c). Compared with the direct HPLC analysis, NNK was effectively enriched and the interference was removed after extraction with NNK-MMIPs (curves b and d). As can be seen in the chromatograms, the concentration of NNK was high enough to be quantitatively analyzed (curve b). The contents of NNK was found to be 7.16 and 8.32 ng g−1, respectively (Table 1). These results compared favorably with other published methods, as shown in Table 2. The characteristics of our method proved that, due to the selective binding sites at the surface of NNK-MMIPs, the extraction procedure can reach the equilibrium quickly. Moreover, it does not require special instrumentation, consumes much less toxic organic solvents, and has a good clean-up and concentration effect for the analysis of NNK in tobacco samples.| Methods | Materials | Target | Sample type | LOD | Linear range | Time (min) | Ref. |
|---|---|---|---|---|---|---|---|
| Online SPE LC-MS/MS | C18 | NNAL | Plasma | 0.09 pg mL−1 | 0.5–100 pg mL−1 | 10 | 27 |
| UPLC-MS/MS | C18 | NNK | Tissue | 0.005 ng mL−1 | 0.04–1.25 ng mL−1 | 3 | 28 |
| CSEI-sweeping-MEKC | Silica capillary column | NNK | Urine | 16 ng mL−1 | 25–500 ng mL−1 | 9 | 29 |
| SPME-HPLC | PPY-coated capillary | NNK | Cell cultures | 63 ng mL−1 | 20–250 ng mL−1 | 19 | 30 |
| HPLC | NNK-MMIPs | NNK | Tobacco | 13.5 ng mL−1 | 0.1–250 μg mL−1 | 10 | This work |
4 Conclusions
An efficient and simple method using magnetic dummy template molecularly imprinted particles functionalized with dendritic nanoclusters coupled with HPLC was developed for the enrichment and separation of NNK from complex tobacco samples. Firstly, the highly branched dendritic nanoclusters possessed plentiful imprinted sites to increase the specific surface area, resulting in high adsorption capacity, fast adsorption kinetics and great selectivity. Secondly, the dummy template was sufficiently similar to the target and could avoid the leakage of trace amounts of NNK, which is potentially genotoxic and carcinogenic. The dummy template strategy can be used to accurately determine trace analytes even if some of the template was not completely removed from the NNK-MMIPs completely. Thirdly, the magnetic separation was more efficient, fast and economical by avoiding additional centrifugation or filtration. The strategy is novel, inexpensive, and easy to operate without requiring complicated instruments such as LC-MS/MS.The combination of NNK-MMIPs with HPLC was successfully applied to determine trace amounts of NNK in tobacco samples. The results confirmed satisfactory separation, with high recovery, precision and sensitivity. This research strategy has great potential for monitoring trace amounts of other compounds in complex samples.
In the future, we believe that sensitive detection of microorganisms and biological macromolecules such as nucleotides and proteins in complex samples can be achieved by using molecularly imprinted polymers. It is promising to construct MIP-based sensors for multi responsive patterns or biomedical implantability through the design and development of molecularly imprinted polymers with different properties. Moreover, molecular imprinting technology is expected to be combined with other technologies to achieve automated online analysis.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Medical Research Foundation of Jiangsu Commission of Health (No. Z2020009), the Foundation of Anhui Province Key Laboratory of Research & Development of Chinese Medicine (AKLPDCM202301), the Open Project of Yunnan Key Laboratory of Tobacco Chemistry (No. 11700167) and Chinese Postdoctoral Science Foundation (No. 2017M621789).References
- S. Sarlak, C. Lalou, A. C. B. Sant'Anna-Silva, W. Mafhouf, M. De Luise, B. Rousseau, J. Izotte, S. Claverol, D. Lacombe, E. Nikitopoulou, M. Yang, M. Oliveira, C. Frezza, G. Gasparre, H. R. Rezvani, N. D. Amoedo and R. Rossignol, Antioxid. Redox Signaling, 2022, 36, 525–549 CrossRef CAS PubMed.
- Z. Zhuang, J. Li, G. Sun, X. Cui, N. Zhang, L. Zhao, P. K. S. Chan and R. Zhong, Chem. Res. Toxicol., 2020, 33, 470–481 Search PubMed.
- Y. Wang, L. Shi, J. Li, H. Wang and H. Yang, Toxicol. In Vitro, 2020, 63, 104740 CrossRef CAS PubMed.
- X. Tang, N. Benowitz, L. Gundel, B. Hang, C. M. Havel, E. Hoh, P. Jacob III, J. H. Mao, M. Martins-Green, G. E. Matt, P. J. E. Quintana, M. L. Russell, A. Sarker, S. F. Schick, A. M. Snijders and H. Destaillats, Environ. Sci. Technol., 2022, 56, 12506–12516 CrossRef CAS PubMed.
- S. H. Edwards, L. M. Rossiter, K. M. Taylor, M. R. Holman, L. Zhang, Y. S. Ding and C. H. Watson, Chem. Res. Toxicol., 2017, 30, 540–551 Search PubMed.
- H. Wang, X. Li, J. Guo, B. Peng, H. Cui, K. Liu, S. Wang, Y. Qin, P. Sun, L. Zhao, F. Xie and H. Liu, Inhalation Toxicol., 2016, 28, 89–94 Search PubMed.
- K. A. Shah and H. T. Karnes, Crit. Rev. Toxicol., 2010, 40, 305–327 CrossRef CAS PubMed.
- W. B. Zhu, L. Gao, Y. K. Zhang, Q. Zhang, Y. Hong, W. J. Shen, Y. Wang and J. H. Zhu, Microporous Mesoporous Mater., 2022, 333, 111730 CrossRef CAS.
- C. L. Shi, X. D. Sun, Y. H. Gao, S. J. Zheng, S. H. Li, J. Yang, Y. Z. Wang, J. W. Xiong, Y. Shen, Y. Wang and J. H. Zhu, J. Hazard. Mater., 2019, 365, 196–204 CrossRef CAS PubMed.
- S. Ramanavicius and A. Ramanavicius, Adv. Colloid Interface Sci., 2022, 305, 102693 CrossRef CAS PubMed.
- S. Ramanavicius, U. Samukaite-Bubniene, V. Ratautaite, M. Bechelany and A. Ramanavicius, J. Pharm. Biomed. Anal., 2022, 215, 114739 CrossRef CAS PubMed.
- S. Ramanavicius, A. Jagminas and A. Ramanavicius, Polymers, 2021, 13, 974 CrossRef CAS PubMed.
- S. Shi, D. Fan, H. Xiang and H. Li, Food Chem., 2017, 237, 198–204 CrossRef CAS PubMed.
- Z. Guo, C. M. Pedersen, P. Wang, M. Ma, Y. Zhao, Y. Qiao and Y. Wang, J. Agric. Food Chem., 2021, 69, 5105–5112 CrossRef CAS PubMed.
- F. Avila-Salas, R. I. Gonzalez, P. L. Rios, I. Araya-Duran and M. B. Camarada, J. Chem. Inf. Model., 2020, 60, 2966–2976 CrossRef CAS PubMed.
- Y. Huang, W. Zhang, M. Bai and X. Huang, Chem. Eng. J., 2020, 380, 122392 CrossRef CAS.
- C. Zhu, W. Zhu, L. Xu and X. Zhou, Anal. Chim. Acta, 2019, 1047, 21–27 CrossRef CAS PubMed.
- Q. Han, X. Shen, W. Zhu, C. Zhu, X. Zhou and H. Jiang, Biosens. Bioelectron., 2016, 79, 180–186 CrossRef CAS PubMed.
- Y. Gao, Y. Chen, M. Li, L. Jia, L. Zhang and J. Zhu, Sens. Actuators, B, 2021, 329, 129137 CrossRef CAS.
- R. Dawn, M. Zzaman, F. Faizal, C. Kiran, A. Kumari, R. Shahid, C. Panatarani, I. M. Joni, V. K. Verma, S. K. Sahoo, K. Amemiya and V. R. Singh, Braz. J. Phys., 2022, 52, 99 CrossRef CAS.
- H. Kiziltaş, T. Tekin and D. Tekin, J. Environ. Chem. Eng., 2020, 8, 104160 CrossRef.
- S. Kutluay, S. Horoz, Ö. Şahin, A. Ekinci and M. Ş. Ece, Int. J. Energy Res., 2021, 45, 20176–20185 CrossRef CAS.
- M. Ş. Ece, A. Ekinci, S. Kutluay, Ö. Şahin and S. Horoz, J. Mater. Sci.: Mater. Electron., 2021, 32, 18192–18204 CrossRef CAS.
- K. Prabakaran, P. J. Jandas, J. Luo, C. Fu and Q. Wei, Colloids Surf., A, 2021, 611, 125859 CrossRef CAS.
- Z. Wu, W. Liu, S. Zhang, Z. Peng, Y. Dong, Z. Huang, M. Zhong, Y. Ye, X. Su and Y. Liang, RSC Adv., 2022, 12, 20785–20791 RSC.
- M. M. Rahman, A. Z. Shafiullah, A. Pal, M. A. Islam, I. Jahan and B. B. Saha, Energies, 2021, 14, 7478 CrossRef CAS.
- J. Zhang, X. Liu, B. Shi, Z. Yang, Y. Luo, T. Xu, D. Liu, C. Jiang, G. Du, N. Lu, C. Zhang, Y. Ma, R. Bai and J. Zhou, Environ. Res., 2022, 214, 113811 CrossRef CAS PubMed.
- T. Meikopoulos, O. Begou, T. Panagoulis, E. Kontogiannidou, D. G. Fatouros, J. H. Miller, G. Theodoridis and H. Gika, Anal. Bioanal. Chem., 2022, 414, 7865–7875 CrossRef CAS PubMed.
- Y. Yang, H. Nie, C. Li, Y. Bai, N. Li, J. Liao and H. Liu, Talanta, 2010, 82, 1797–1801 CrossRef CAS PubMed.
- W. M. Mullett, K. Levsen, J. Borlak, J. Wu and J. Pawliszyn, Anal. Chem., 2002, 74, 1695–1701 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |