Open Access Article
Open Access ArticleReconfigurable DNA triplex structure for pH responsive logic gates†
Mingxuan Qi‡
a,
Peijun Shi‡b,
Xiaokang Zhang b,
Shuang Cuib,
Yuan Liu
b,
Shuang Cuib,
Yuan Liu b,
Shihua Zhoua and
Qiang Zhang
b,
Shihua Zhoua and
Qiang Zhang *a
*a
aKey Laboratory of Advanced Design and Intelligent Computing, Ministry of Education, School of Software Engineering, Dalian University, Dalian 116622, China. E-mail: zhangq@dlu.edu.cn
bSchool of Computer Science and Technology, Dalian University of Technology, Dalian 116024, China
First published on 28th March 2023
Abstract
The DNA triplex is a special DNA structure often used as a logic gate substrate due to its high stability, programmability, and pH responsiveness. However, multiple triplex structures with different C−G−C+ proportions must be introduced into existing triplex logic gates due to the numerous logic calculations involved. This requirement complicates circuit design and results in many reaction by-products, greatly restricting the construction of large-scale logic circuits. Thus, we designed a new reconfigurable DNA triplex structure (RDTS) and constructed the pH-responsive logic gates through its conformational change that uses two types of logic calculations, ‘AND’ and ‘OR’. The use of these logic calculations necessitates fewer substrates, further enhancing the extensibility of the logic circuit. This result is expected to promote the development of the triplex in molecular computing and facilitate the completion of large-scale computing networks.
1 Introduction
In recent years, DNA nanotechnology1 has developed rapidly and become the most dynamic frontier discipline in molecular biology. It has been widely used in fields of DNA computing,2–4 nanomachines,5–7 biosensing8,9 and so on. Among them, DNA computing is a new computing model based on the principle of biochemical reaction, which has the characteristics of high parallelism, mass storage capacity, and extremely low energy consumption etc. It shows great advantages in optimizing storage methods10–12 and solving non-traditional computing problems.13–15 The DNA logic gate, which has functions like logical analysis, judgment and multi-input recognition, is regarded as the basis of DNA computing.16–18 It has shown great potential practicability in the fields of analytical detection and disease diagnosis.19–22 The existing logic gates usually use DNA duplex as the substrate and DNA strand displacement as the tool,23–25 combine with materials like proteins,26–28 small molecules,29,30 ions31,32 and so on to build logic circuits with logic calculations and signal amplification functions, and then promote the completion of large-scale computing networks.33,34 Among them, it is worth noting that DNA strand displacement is an important brick for the construction of logic gates. DNA strand displacement is a kind of molecular dynamics process which indicates the hybridized DNA duplex is replaced by another DNA strand through partial or complete base pairing.In addition, special DNA structures such as G-quadruplexes,35,36 i-motif37,38 and triplex39,40 can also be used as substrates of logic gates. Among them, DNA triplex is a structure formed through the connection between the Hoogsteen interactions and a cytosine-rich triplex forming oligonucleotide (TFO) on the basis of DNA duplex.41 In the nearly acidic case, triplex forms C−G−C+ hydrogen bond through cytosine protonation. Due to the strong dependence of the hydrogen bond on H+ in the solution, the triplex has unique pH responsiveness. pH has a significant impact on life activities, and the change of H+ concentration is an important reference index for pathological monitoring and environmental detection.42–45 Therefore, it is significant to construct molecular devices such as logic circuits,40,46 biosensors47–49 and nanoswitches50–52 that can rapidly respond to pH changes.
Above all, researchers have constructed a variety of different C−G−C+ triplex structures based on the pH responsiveness of the triplex. It was combined with programmable activated toehold53 to construct the detachable logic circuit for self-assembly of spherical nucleic acids, with optochemical materials54 to construct the switching circuit for logic calculation and probability calculation and with micro-gels55 to complete the multi-mode chemical reaction circuit for controlling the swelling of soft matter. These logic circuits improve the response ability to external pH changes and expand the applicability of the circuit by using the triplex structure. However, in order to complete diverse logical calculations, a great deal of different C−G−C+ ratio of triplex structures usually need to be introduced, which increases the difficulty of circuit design. And due to the high similarity of the triplex sequence in the reactions, it is easy to produce many kinds of by-products. These problems make the construction of large-scale logic circuit faces great challenges.56,57
Therefore, we design a new reconfigurable DNA triplex structure (RDTS), which uses pH to control conformational change and complete two logic gates named as ‘AND’ and ‘OR’. Firstly, we verified the sequence specificity and pH responsiveness of triplex. It is proved that triplex can quickly respond to external pH changes. Secondly, we constructed a reconfigurable triplex structure in which cytosine protonated and TFO adhered closely to it under acid condition. Cytosine deprotonation and TFO separation in the structure were performed under alkaline conditions, and RDTS formation was confirmed by a step-by-step assembly. Finally, we construct a pH responsive logic gate based on RDTS. A stable triplex structure is formed which appears as ‘AND’ gate at pH 5 and triplex can change its conformation and perform as ‘OR’ gate at pH 8. The triplex structure we designed has the characteristics of reconfigurability, high stability and programmability, and the logic gates based on it can produce different logical operations under two kinds of pH. The logic gates can use fewer substrates to complete two kinds of logic calculations which increases the extensibility and flexibility of the logic circuit. It is expected to build large-scale logic circuits responding to different pH ranges and provide more ideas for promoting the research of DNA triplex in the fields of DNA computing and biosensing.
2 Experimental section
2.1 Materials
All DNA sequences were purchased from Sangon Biotech (Shanghai) Co, Ltd. The unmodified DNA strand was purified by PAGE, and the DNA strand modified by fluorescence group (Cy5) and quenched group (BHQ-2) was purified by HPLC. Dissolve all DNA strands in ultra-pure water as a reserve solution. Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific Inc, Waltham, MA, USA) was used for quantification, and the absorption intensity was measured at λ = 260 nm. Other chemicals are analytically grade and do not require further purification for use.2.2 Annealing
All DNA structures were dispersed in a 20 μL 1× TAE/Mg2+ buffer (pH 5, 12.5 mM Mg2+) at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio under acidic conditions. Under alkaline conditions, they were also dispersed in 20 μL 1× TAE/Mg2+ buffer (pH 8, 12.5 mM Mg2+) at 1
1 molar ratio under acidic conditions. Under alkaline conditions, they were also dispersed in 20 μL 1× TAE/Mg2+ buffer (pH 8, 12.5 mM Mg2+) at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio. These structures were annealed by polymerase chain reaction (PCR) thermal cycling machine (95 °C for 5 min; 65 °C for 30 min; 50 °C for 30 min; 37 °C for 30 min; 25 °C for 30 min). During pH adjustment, the buffer was adjusted to pH 5 using acetic acid and to pH 8 using NaOH.
1 molar ratio. These structures were annealed by polymerase chain reaction (PCR) thermal cycling machine (95 °C for 5 min; 65 °C for 30 min; 50 °C for 30 min; 37 °C for 30 min; 25 °C for 30 min). During pH adjustment, the buffer was adjusted to pH 5 using acetic acid and to pH 8 using NaOH.
In the reconfigurable DNA triplex structure assembly module, after annealing the RDTS, the input and substrate were mixed at a molar ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and reacted at room temperature for 30 min. In the pH responsive logic gates module, after annealing of state 1 or state 2, the input (IN1, IN2) and the substrate were mixed at a molar ratio of 1.2
1 and reacted at room temperature for 30 min. In the pH responsive logic gates module, after annealing of state 1 or state 2, the input (IN1, IN2) and the substrate were mixed at a molar ratio of 1.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 under both acid–base conditions, and the reaction was carried out at room temperature for 30 min.
1 under both acid–base conditions, and the reaction was carried out at room temperature for 30 min.
2.3 Native PAGE characterization
DNA strands (3 μM) were dispersed in 20 μL 1× TAE/Mg2+ buffer (pH 5, 12.5 mM Mg2+) at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio and mixed with 50% glycerol under acidic conditions. The samples run on 12% PAGE at a constant voltage of 95 V for 240 min at 4 °C. DNA strands (1.5 μM) were dispersed in 20 μL 1× TAE/Mg2+ buffer (pH 8, 12.5 mM Mg2+) at 1
1 molar ratio and mixed with 50% glycerol under acidic conditions. The samples run on 12% PAGE at a constant voltage of 95 V for 240 min at 4 °C. DNA strands (1.5 μM) were dispersed in 20 μL 1× TAE/Mg2+ buffer (pH 8, 12.5 mM Mg2+) at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio and mixed with 50% glycerol under alkaline conditions. The samples run on 12% PAGE at a constant voltage of 85 V for 210 min at 25 °C. After the polyacrylamide gel electrophoresis was completed, the gel was placed in stains-all staining solution for 1 hour for staining. After fading by light, Canon scanner was used to image the gel.
1 molar ratio and mixed with 50% glycerol under alkaline conditions. The samples run on 12% PAGE at a constant voltage of 85 V for 210 min at 25 °C. After the polyacrylamide gel electrophoresis was completed, the gel was placed in stains-all staining solution for 1 hour for staining. After fading by light, Canon scanner was used to image the gel.
It's worth noting that the native PAGE run at 4 °C for acidic conditions and 25 °C for alkaline conditions. The DNA triplex is stable under acidic conditions. And the electrophoresis apparatus generates extra heat during polyacrylamide gel electrophoresis. These heat causes the temperature of the electrophoretic solution to rise, and higher temperature will greatly affect the stability of the triplex, which can affect our PAGE analysis results under acidic conditions.58 Therefore, we can resist the heat from the electrophoresis apparatus by adjusting the reaction temperature to 4 °C and make the PAGE analysis results under acidic conditions more worthy of analysis. But the heat generated by the machine has little effect the stability of the DNA duplex and strands under alkaline conditions. The experimental temperature can be set to 25 °C under alkaline conditions.
2.4 Fluorescence kinetic measurements and normalization method
Fluorescence kinetic measurements were performed using TECAN Spark 10 M Multifunctional microplate detector. In both acidic and alkaline conditions, 0.1 μM DNA strands were dispersed into 50 μL 1× TAE/Mg2+ buffer (pH 5, 12.5 mM Mg2+) or 50 μL 1× TAE/Mg2+ buffer (pH 8, 12.5 mM Mg2+) at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio, respectively. In corresponding fitted pH-dependent curve, 0.3 μM DNA strands were used. All reactions were performed at 25 °C. All fluorescence measurements were shown as mean and standard deviation (n = 3) to ensure the certainty of reactions.
1 molar ratio, respectively. In corresponding fitted pH-dependent curve, 0.3 μM DNA strands were used. All reactions were performed at 25 °C. All fluorescence measurements were shown as mean and standard deviation (n = 3) to ensure the certainty of reactions.
In the fluorescence normalization method, the minimum value of the reaction is normalized to 0, and the maximum value is normalized to 1, which represents the fluorescence curve with the logic output of 1 (ΔF/MaxΔF). It should be specifically pointed out that we set the threshold for the pH responsive logic gate at 0.4. We set the threshold as 0.4, when the logic gate output whose normalized fluorescence value is lower than 0.4, its logic output means 0; otherwise, it means 1. To ensure that the half-completion time of each curve is constant, it is required to normalize each curve with logic output of 1 separately for the OR gate because there are multiple reactions with logic output of 1. Next, we need to explain how to select the curve as normalization. The curve of an OR gate that has a logic output of 0 is considerably different from the curve that has a logic output value of 1. During the experiment, there was almost no increase in its fluorescence. So we don't select the curve that has a logic output of 0 for normalization. We cannot utilize the curve that has the maximum value of three curves whose logic output are 1 for normalization. Herein, we select the curve whose three logic outputs are 1 in OR gate and whose lower output value for normalization. The curve we chose for normalization makes a fair comparison between the 1 curve and the 0 curve.
3 Results and discussion
3.1 Verification of DNA triplex properties
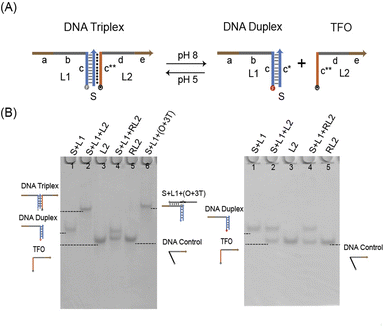
Verifying the sequence specificity of TFO is an important prerequisite for DNA triplex design. DNA triplex usually consists of cytosine-rich TFO and a DNA duplex by Hoogsteen interactions (Fig. 1A). And strands of random sequences cannot interact with duplex (Fig. S1†). Therefore, we need to prove the sequence specificity. We tested whether strand RL2 (DNA Control, 39 nt) whose domain c** contains random sequences and strand L2 (TFO, 39 nt) whose domain c** contains cytosine-rich sequences could create DNA triplex with DNA duplex. According to the result of PAGE experiment, under acidic conditions (Fig. 1B, left), TFO can form DNA triplex (lane 2) with DNA duplex that parallel to the contrast lane (lane 6). DNA control and DNA duplex are divided into two clear bands (lane 4) which shows that the strand of random sequence cannot form the triplex. This property was further verified by fluorescence kinetic experiments (Fig. S2†). As a result, we have confirmed the triplex sequence specificity.The verification of the pH responsiveness of triplex is a key part in the design of triplex structure. The cytosine in the DNA triplex will be protonated in the near acidic situation, and then generate the C−G−C+ hydrogen bond between the DNA duplex and the TFO, which is assembled into the DNA triplex, was the result. So the pH responsiveness of DNA triplex is caused by the dependence of the C−G−C+ structure hydrogen bond on H+ in solution. Therefore, the higher the C−G−C+ hydrogen bond content of triplex, the stronger the dependence of its structure on H+. In this research, the DNA triplex containing 40% C−G−C+ was used, which formed triplex at pH 5 and dissociated at pH 8. We verified through PAGE that under pH 5 (Fig. 1B, left), the duplex can be bonded with L2 to form DNA triplex. However, at pH 8 (Fig. 1B, right), the duplex failed to form triplex with either L2 (lane 2) or RL2 (lane 4).
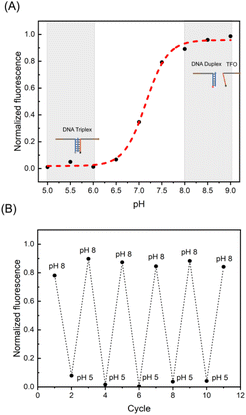
In addition, we have demonstrated the pH responsiveness of the triplex by cyclic pH-jump experiments (Fig. 2A) and corresponding fitted pH-dependent curve (Fig. 2B). In corresponding pH-dependent curve demonstrated the progressive separation of DNA triplex can be by gradually increasing the pH value of the solution. And in cyclic pH-jump experiment, adding 0.88 M acetic acid to pH 5, and then adding 0.9 M NaOH to pH 8 after 30 min. Five such pH jumps were made (the trend diagram of triplex with pH in Fig S3†).
The results are in line with expectations. DNA duplex and TFO can form Hoogsteen hydrogen bonds under acidic conditions and fluorescence and quencher will quickly pull into the Fluorescence Resonance Energy Transfer (FRET) that leads to a decline in fluorescence value. DNA triplex is separated and FRET cannot be generated at pH 8, so the fluorescence increases. Finally, we experimentally demonstrated the pH responsiveness and sequence specificity of DNA triplex and determined the C−G−C+ ratio and pH response range of RDTS based on these properties.
3.2 Assembly of reconfigurable DNA triplex structures
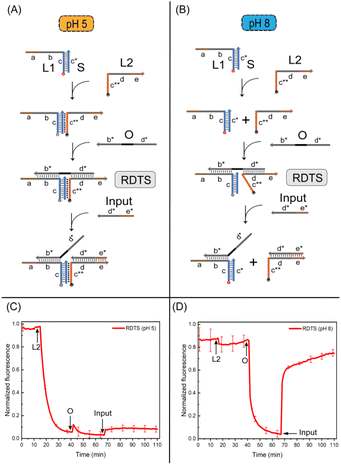
The assembly of reconfigurable DNA triplex structures are central to building the pH responsive logic gates. Here, strand O and DNA triplex were connected by Watson-Crick hydrogen bond to construct the reconfigurable triplex structure. First, we designed the assembly method of RDTS under both acidic and alkaline conditions. Because of strand O's presence, the structure was relatively stable at both pH values. It was different that after input (23 nt) is added, cytosine protonation occurs between strand L2 and duplex under acidic conditions (Fig. 3A), but strand L2 will not separate from RDTS. In the alkaline case (Fig. 3B), cytosine in RDTS is deprotonated. Strand L2 cannot be closely connected to RDTS and forms duplex with strand input, then RDTS is detached. The RDTS is reconfigurable because its transformation of conformation after pH changes. We then verified the assembly of RDTS by polyacrylamide gel at different pH values. The strand O was added into RDTS respectively at pH 5 (Fig. S4†) and pH 8 (Fig. S5†). It can be observed that the corresponding lanes formed large structures under both conditions, strand O was successfully attached to DNA triplex. So the assembly of RDTS was realized. Strand input needs to be added to verify the assembly of the RDTS.It can be observed that the corresponding lane had a larger structure at pH 5, indicating that input combines with RDTS. There were two lanes observed which were parallel to the two contrast lanes set, proving that the conformation of the structure changed at this time and the structure separated when responding to the same input.
The results were also verified by fluorescence kinetic measurements. Whether in acidic or alkaline conditions (as shown in Fig. 3C and D), the fluorescence value decreased after the addition of L2 to DNA duplex. However, the fluorescence value fluctuated and recovered immediately at pH 5 after strand O was added. The fluorescence value will decrease significantly at pH 8. When strand O was attached to L2 and duplex, it can pull the distance between them and the fluorescence value will also decrease. Finally, we added two times the concentration of strand input in two cases respectively. It can be observed that RDTS is stable in the acid case and L2 will not break out of the structure. So the fluorescence value does not change much. RDTS underwent a conformational transformation under alkaline conditions, L2 and IN2 formed duplex which fluorescence value would increase rapidly. The reconfigurable DNA triplex structure was successfully established in response to pH changes. This phenomenon also inspired the construction of the following pH responsive logic gates.
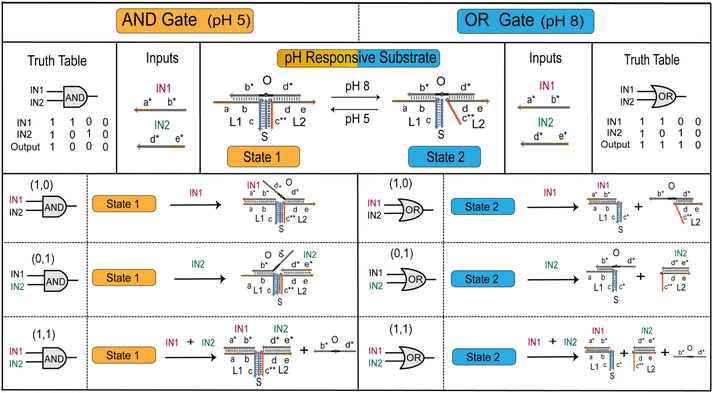
3.3 Construction of pH responsive logic gates
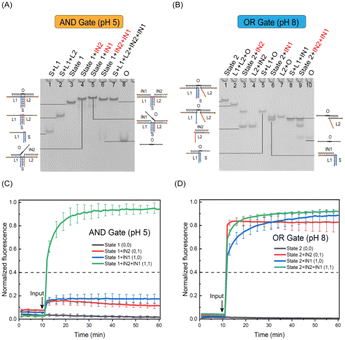
RDTS has stable structure under acid and is not affected by input. For basic conformational conversion, input can separate the structure. We designed the pH responsive logic gates that complete two kinds of logic calculations through one kind of structure (Fig. 4). First, the structural state of RDTS under acid and alkaline conditions was set as state 1 and state 2 respectively. After adding strand IN1 in the alkaline case, both cases will undergo strand displacement reaction with strand O. After the reaction is completed, IN1 will be attached to strand L1. However, the difference is that strand O is not completely separated from state 1 after bonding in acid, the quenching group is not far away from the two fluorescence groups, so the fluorescence value in the solution will not change when IN1 is input alone. But, due to the absence of C−G−C+ hydrogen bond in state 2 under alkaline conditions, once IN1 is fully bonded to strand L1, strand O and L2 will be detached from state 2, making fluorescence Cy5 modified on L1 far away from quenching group BHQ-2 modified on strand O which leads to the increase of fluorescence value. This change of fluorescence is also shown on the side of IN2 input alone, after IN2 input, the fluorescence value remains unchanged in the acid condition. While in the alkaline condition, L2 will form duplex with IN2 and break away from the original structure, and the fluorescence value increases.However, when IN1 and IN2 are input at the same time in two cases, both will undergo strand displacement reaction with strand O which make the strand O originally attached to the original structure out of state 1 or state 2, the quenching group BHQ-2 will be completely far away from the fluorescence group Cy5 modified in strand L1 and strand L2, and the fluorescence value will rise. The above changes in fluorescence values with the introduction of input conform to the ‘AND’ gate in the case of acid, and to the ‘OR’ gate in the case of alkaline.
Then, we verified the structures involved in the logic gate in the two cases by PAGE experiment. First, IN1, IN2, IN1 + IN2 were mixed with state 1 or state 2 at room temperature for 0.5 h. Through the experimental results, it was observed that under the acid condition (Fig. 5A), when IN1 and IN2 were respectively inputted, a larger structure than state 1 was generated, and no loss of strand O was caused. In addition to a larger structure than state 1, strand O replaced by the reaction can also be observed in the IN1 + IN2 strip. These two products can correspond to the contrast lanes. It has been verified that after IN1 and IN2 are input at the same time, strand O can shed the DNA triplex structure. As for the verification of the structure under alkaline conditions, it also conforms to the experimental expectation (Fig. 5B), and the structures generated by the reaction can all be in line with the contrast lanes. In summary, we have realized the verification of the structures involved in the pH responsive logic gate.
Finally, we further verified the calculations of pH responsive logic gate through the fluorescence kinetic experiment. It shows that under acidic conditions (Fig. 5C), after adding 1.2 times of IN1, IN2 and IN1 + IN2 to state 1, two fluorescence curves of input IN1 and IN2 can be observed without obvious fluorescence rise effect. The normalized fluorescence value is lower than the threshold value 0.4, the output was 0 at this time. Due to the high stability of the structure under acidic conditions, there is a certain Seesaw reaction17 when strand IN2 replaces strand O, and O will stick back, so the fluorescence value will be slightly reduced. After inputting the fluorescence curve of IN1 + IN2, a relatively obvious fluorescence rising effect can be seen, which is higher than the threshold value, the output is ‘1’. These conform to the logic calculations of ‘AND’ gate. Meanwhile, added 1.2 times of IN1, IN2 and IN1 + IN2 to state 2 respectively under alkaline conditions (Fig. 5D). And it can be observed that the fluorescence curves all increase significantly, they are all greater than the threshold value which logic output means ‘1’. While the fluorescence value of substrate without input is unchanged which logic output is ‘0’. These conform to logic expression of ‘OR’ gate. We also switch between states 1 and 2, the logic calculations are consistent with the change which further demonstrates the flexibility of pH responsive logic gates (Fig. S6 and S7†). The results show that RDTS can be transformed into state 1 and state 2 with the change of external pH and two logical expressions of ‘AND’ and ‘OR’ can be generated. These can expand the application scenario of the logic circuit based on DNA triplex and realize more complex logic operations.
In conclusion, the pH responsive logic gate designed by us presents as ‘AND’ gate at pH 5 and ‘OR’ gate at pH 8 which has been verified by PAGE and fluorescence kinetic experiment. The construction of pH responsive logic gate provides more possibilities for the achievement of pH responsive large logic circuits.
4 Conclusions
Herein, we have designed the reconfigurable DNA triplex structure and determined the pH response range of RDTS and composition of C−G−C+ hydrogen bond by testing the pH responsiveness and sequence specificity of triplex. Using the conformational transformation of RDTS in both pH 5 and pH 8 and adding the strands input to it can produce different fluorescence effects, and this phenomenon can identify the reconfigurability of RDTS. Finally, through the conversion of RDTS controlled by pH, the pH responsive logic gate presented as ‘AND’ gate at pH 5 and ‘OR’ gate at pH 8 was constructed to make the application of RDTS in the field of logic circuits come true. Different from general triplex logic circuits, reconfigurable DNA triplex logic gates can accomplish two kinds of logic calculations and the same logic expression with fewer substrates. In the construction of large-scale logic circuits, the design rules can be further simplified and the generation of by-products can be reduced.Besides, the RDTS we designed is universality which can be combined with antibodies to construct a nanodevice for information exchange between proteins and with microgel to control the swelling behavior of microgel. It can also anchor on the surface of cell membrane to construct biosensors for monitoring cell physiological status. Therefore, not only can the triplex we designed apply to complete pH responsive logic gate, but have a wide application future in the fields of drug delivery and biosensing.
Author contributions
Mingxuan Qi and Peijun Shi, conceptualization, methodology and validation. Xiaokang Zhang and Shuang Cui, formal analysis. Peijun Shi, investigation. Yuan Liu, data curation. Mingxuan Qi, writing—original draft. Peijun Shi and Shihua Zhou, writing—review & editing. Mingxuan Qi, visualization. Shihua Zhou, supervision. Shihua Zhou and Qiang Zhang, project administration. Qiang Zhang, funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is supported by 111 Project (No. D23006), by the National Natural Science Foundation of China (No. 62272079, 61972266), Liaoning Revitalization Talents Program (No. XLYC2008017), Natural Science Foundation of Liaoning Province (No. 2021-MS-344, 2021-KF-11-03, 2022-KF-12-14), the Postgraduate Education Reform Project of Liaoning Province (No. LNYJG2022493), the Dalian Outstanding Young Science and Technology Talent Support Program (No. 2022RJ08).Notes and references
- M. Madsen and K. V. Gothelf, Chem. Rev., 2019, 119, 6384–6458 CrossRef CAS PubMed.
- Y. K. Li, J. Q. Deng, Z. W. Han, C. Liu, F. Tian, R. Xu, D. Han, S. H. Zhang and J. S. Sun, J. Am. Chem. Soc., 2021, 143, 1290–1295 CrossRef CAS PubMed.
- Y. F. Pei, T. Y. Bian, Y. L. Liu, Y. Liu, Y. J. Xie and J. Song, Nano Lett., 2022, 22, 3003–3010 CrossRef CAS PubMed.
- K. Matange, J. M. Tuck and A. J. Keung, Nat. Commun., 2021, 12, 1358 CrossRef CAS PubMed.
- K. Leung, K. Chakraborty, A. Saminathan and Y. Krishnan, Nat. Nanotechnol., 2019, 14, 176–183 CrossRef CAS PubMed.
- Y. Zhang, W. W. Chen, Y. Y. Fang, X. B. Zhang, Y. Liu and H. X. Ju, J. Am. Chem. Soc., 2021, 143, 15233–15242 CrossRef CAS PubMed.
- P. Yang, R. X. Zhou, C. P. Kong, L. Fan, C. Dong, J. B. Chen, X. D. Hou and F. Li, ACS Nano, 2021, 15, 16870–16877 CrossRef CAS PubMed.
- K. Wang, H. H. Wang, C. H. Li, Z. N. Yin, R. J. Xiao, Q. Z. Li, Y. Xiang, W. Wang, J. Huang, L. Chen, P. P. Fang and K. W. Liang, Sci. Adv., 2021, 7, eabe3516 CrossRef CAS PubMed.
- E. Y. Dai, L. Han, J. Liu, Y. C. Xie, H. J. Zeh, R. Kang, L. L. Bai and D. L. Tang, Nat. Commun., 2020, 11, 6339 CrossRef CAS PubMed.
- Q. Yin, Y. Zheng, B. Wang and Q. Zhang, IEEE/ACM Trans. Comput. Biol. Bioinf., 2021, 19, 3384–3394 Search PubMed.
- P. H. Wang, Z. N. Mu, L. J. Sun, S. Q. Si and B. Wang, Front. Bioeng. Biotechnol., 2022, 10, 916615 CrossRef PubMed.
- D. Zhu, Z. Huang, S. Liao, C. Zhou, S. Yan and G. Chen, IEEE Trans. Nanobioscience, 2022 DOI:10.1109/TNB.2022.3220795.
- X. Li, P. F. Han, W. Q. Chen, C. N. Gao, S. Wang, T. Song, M. Y. Niu and A. Rodriguez-Paton, Briefings Bioinf., 2023, 24, bbac524 CrossRef PubMed.
- B. Cao, P. Shi, Y. Zheng and Q. Zhang, Comput. Biol. Med., 2022, 151, 106269 CrossRef CAS PubMed.
- L. J. Sun, B. Cao, Y. Liu, P. J. Shi, Y. F. Zheng, B. Wang and Q. Zhang, J. Phys. Chem. B, 2022, 126, 8708–8719 CrossRef CAS PubMed.
- F. Wang, H. Lv, Q. Li, J. Li, X. L. Zhang, J. Y. Shi, L. H. Wang and C. H. Fan, Nat. Commun., 2020, 11, 121 CrossRef CAS PubMed.
- L. Qian and E. Winfree, Science, 2011, 332, 1196–1201 CrossRef CAS PubMed.
- J. Chen, S. N. Fu, C. Y. Zhang, H. Y. Liu and X. Su, Small, 2022, 18, 2108008 CrossRef CAS PubMed.
- Y. Du, P. Peng and T. Li, ACS Nano, 2019, 13, 5778–5784 CrossRef CAS PubMed.
- C. H. Ouyang, S. B. Zhang, C. Xue, X. Yu, H. Xu, Z. M. Wang, Y. Lu and Z. S. Wu, J. Am. Chem. Soc., 2020, 142, 1265–1277 CrossRef CAS PubMed.
- E. Amalfitano and K. Pardee, Nat. Chem. Biol., 2022, 18, 356–358 CrossRef CAS PubMed.
- Y. Q. Yu, Q. Q. Guo, W. L. Jiang, H. Zhang and C. X. Cai, Anal. Chem., 2021, 93, 11298–11304 CrossRef CAS PubMed.
- C. Xing, Z. Y. Chen, J. D. Dai, J. Zhou, L. P. Wang, K. L. Zhang, X. F. Yin, C. H. Lu and H. H. Yang, ACS Appl. Mater. Interfaces, 2020, 12, 6336–6342 CrossRef CAS PubMed.
- J. B. Zhu, F. Boskovic, B. N. T. Nguyen, J. R. Nitschke and U. F. Keyser, Nano Lett., 2021, 21, 1368–1374 CrossRef CAS PubMed.
- T. C. Xie, Y. H. Deng, J. R. Zhang, Z. Zhang, Z. Hu and T. B. Wu, Nucleic Acids Res., 2022, 50, 8431–8440 CrossRef CAS PubMed.
- T. Q. Song, A. Eshra, S. L. Shah, H. Bui, D. Fu, M. Yang, R. Mokhtar and J. Reif, Nat. Nanotechnol., 2019, 14, 1075–1081 CrossRef CAS PubMed.
- X. D. Zheng, J. Yang, C. J. Zhou, C. Zhang, Q. Zhang and X. P. Wei, Nucleic Acids Res., 2019, 47, 1097–1109 CrossRef CAS PubMed.
- X. K. Zhang, Q. Zhang, Y. Liu, B. Wang and S. H. Zhou, Comput. Struct. Biotechnol. J., 2020, 18, 2107–2116 CrossRef CAS PubMed.
- A. Prokup, J. Hemphill and A. Deiters, J. Am. Chem. Soc., 2012, 134, 3810–3815 CrossRef CAS PubMed.
- Y. S. Mi, J. Zhao, H. Q. Chu, Z. X. Li, M. M. Yu and L. L. Li, Anal. Chem., 2021, 93, 2500–2509 CrossRef CAS PubMed.
- J. Zhang, H. Q. Fu and X. Chu, Anal. Chem., 2021, 93, 14675–14684 CrossRef CAS PubMed.
- G. Shi, C. Yan and J. H. Chen, Anal. Chem., 2021, 93, 3273–3279 CrossRef CAS PubMed.
- C. J. Liu, Y. Liu, E. Q. Zhu, Q. Zhang, X. P. Wei and B. Wang, Nucleic Acids Res., 2020, 48, 10691–10701 CrossRef CAS PubMed.
- Z. X. Zhou, Y. Ouyang, J. B. Wang and I. Willner, J. Am. Chem. Soc., 2021, 143, 5071–5079 CrossRef CAS PubMed.
- D. Chen, S. Yang, H. Y. Han, L. B. Song, D. Huang, X. Lin, X. P. Xu and Q. F. Yang, Chem.–Eur. J., 2021, 27, 11627–11632 CrossRef CAS PubMed.
- S. Wang, J. Sun, J. H. Zhao, S. S. Lu and X. R. Yang, Anal. Chem., 2018, 90, 3437–3442 CrossRef CAS PubMed.
- H. D. Huang, Z. Z. Guo, C. J. Zhang, C. Cui, T. Fu, Q. L. Liu and W. H. Tan, ACS Appl. Mater. Interfaces, 2021, 13, 30397–30403 CrossRef CAS PubMed.
- X. D. Lin, Y. Q. Liu, J. K. Deng, Y. L. Lyu, P. C. Qian, Y. F. Lia and S. Wang, Chem. Sci., 2018, 9, 1774–1781 RSC.
- T. Li, D. Ackermann, A. M. Hall and M. Famulok, J. Am. Chem. Soc., 2012, 134, 3508–3516 CrossRef CAS PubMed.
- J. Zheng, Q. W. Wang, L. Shi, P. Peng, L. L. Shi and T. Li, Angew. Chem., Int. Ed., 2021, 60, 20858–20864 CrossRef CAS PubMed.
- Y. W. Hu, A. Cecconello, A. Idili, F. Ricci and I. Willner, Angew. Chem., Int. Ed., 2017, 56, 15210–15233 CrossRef CAS PubMed.
- Y. Yang and Z. D. Deng, Appl. Phys. Rev., 2019, 6, 011309 Search PubMed.
- F. Wang, R. R. Chang, R. R. Ma, H. W. Qiu and Y. Q. Tian, ACS Sustain. Chem. Eng., 2021, 9, 10142–10153 CrossRef CAS.
- D. Wencel, A. Kaworek, T. Abel, V. Efremov, A. Bradford, D. Carthy, G. Coady, R. C. N. McMorrow and C. McDonagh, Small, 2018, 14, 1803627 CrossRef PubMed.
- L. Liu, Y. You, K. Zhou, B. Y. Guo, Z. Cao, Y. L. Zhao and H. C. Wu, Angew. Chem., Int. Ed., 2019, 58, 14929–14934 CrossRef CAS PubMed.
- H. J. Qi, S. Z. Yue, S. Bi, C. F. Ding and W. L. Song, Chem. Commun., 2018, 54, 8498–8501 RSC.
- L. Liu, C. X. Dou, J. W. Liu, X. N. Wang, Z. M. Ying and J. H. Jiang, Anal. Chem., 2018, 90, 11198–11202 CrossRef CAS PubMed.
- X. X. Chen, T. S. Chen, L. J. Ren, G. F. Chen, X. H. Gao, G. X. Li and X. L. Zhu, ACS Nano, 2019, 13, 7333–7344 CrossRef CAS PubMed.
- F. Chen, Q. J. Lu, L. N. Huang, B. W. Liu, M. L. Liu, Y. Y. Zhang and J. W. Liu, Angew. Chem., Int. Ed., 2021, 60, 5453–5458 CrossRef CAS PubMed.
- A. Idili, A. Vallee-Belisle and F. Ricci, J. Am. Chem. Soc., 2014, 136, 5836–5839 CrossRef CAS PubMed.
- Y. X. Hu, Z. Y. Wang, Z. K. Chen and L. Q. Pan, Nucleic Acids Res., 2020, 48, 8591–8600 CrossRef CAS PubMed.
- N. Farag, R. Mattossovich, R. Merlo, L. Nierzwicki, G. Palermo, A. Porchetta, G. Perugino and F. Ricci, Angew. Chem., Int. Ed., 2021, 60, 7283–7289 CrossRef CAS PubMed.
- Y. J. Guo, D. B. Yao, B. Zheng, X. B. Sun, X. Zhou, B. Wei, S. Y. Xiao, M. He, C. X. Li and H. J. Liang, ACS Nano, 2020, 14, 8317–8327 CrossRef CAS PubMed.
- X. W. Xiong, M. S. Xiao, W. Lai, L. Li, C. H. Fan and H. Pei, Angew. Chem., Int. Ed., 2021, 60, 3397–3401 CrossRef CAS PubMed.
- Q. Tang, W. Lai, P. P. Wang, X. W. Xiong, M. S. Xiao, L. Li, C. H. Fan and H. Pei, Angew. Chem., Int. Ed., 2021, 60, 15013–15019 CrossRef CAS PubMed.
- C. Y. Zhou, H. M. Geng, P. F. Wang and C. L. Guo, Small, 2019, 15, 1903489 CrossRef CAS PubMed.
- H. M. Su, J. L. Xu, Q. Wang, F. Wang and X. Zhou, Nat. Commun., 2019, 10, 5390 CrossRef PubMed.
- I. M. A. del Mundo, E. J. Cho, K. N. Dalby and K. M. Vasquez, Nucleic Acids Res., 2019, 47, e73 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra00536d |
| ‡ These authors contribute equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |