DOI:
10.1039/D3RA00153A
(Paper)
RSC Adv., 2023,
13, 9418-9427
Theoretical and solid-state structures of three new macroacyclic Schiff base complexes and the investigation of their anticancer, antioxidant and antibacterial properties†
Received
8th January 2023
, Accepted 8th March 2023
First published on
22nd March 2023
Abstract
Three symmetrical macroacyclic Schiff base complexes were prepared from a ligand derived from the condensation reaction of 1,4-bis(2-aminophenyl)piperazine and salicylaldehyde (L) with Cu(II), Ni(II) and Co(II) perchlorates. The ligand and the Schiff base complexes were characterized by elemental analyses, UV-vis, FT-IR and mass spectrometry, with the structure of [CoL] being determined by a single crystal X-ray structural analysis. In this complex, the cobalt is in a distorted trigonal prismatic coordination environment, surrounded by the six donor atoms of the deprotonated hexadentate ligand. In order to compare the experimental and theoretical data and determine structure parameters in all complexes, Density Functional Theory (DFT) calculations at the B3lyp and BP86 levels with Def2-TZVP basis set have been carried out. NBO and QTAIM analyses have been used to describe the nature of M–O and M–N bonding in these complexes. The synthesized complexes were screened for their antioxidant activities using the DPPH free radical scavenging assay while their bactericidal activity was evaluated by the paper disc diffusion method against both Gram-negative and Gram-positive bacteria, revealing strong antioxidant activities and moderate effectiveness against all bacteria tested. The cytotoxicity of the metal complexes was also investigated against AGS (gastric) and A549 (lung) adenocarcinoma cells.
1. Introduction
Schiff-base complexes can be considered as the one of the most significant stereochemical models in transition metal coordination chemistry due to the fact that they are structurally diverse and are easily prepared.1,2 They have a very significant role in the development of coordination chemistry which has resulted in a wide variety of research ranging from physicochemical and biochemical studies to purely synthetic studies.3 There has been much research into metal complexes with chelate ligands with the aim of mimicking the reactions found in biochemical systems, including the redox functions of different metalloenzymes and the coordination and reactivity of dioxygen. Metalloenzymes can have specific functions, such as redox4–9 and structural and catalytic roles10–14 and accordingly there has been intensive research into metal complexes that can mimic the active sites of metalloproteins. In order for the metal to have a similar coordination environment to those found in proteins, many model metal complexes have been obtained using sterically bulky polydentate ligands. 2-Hydroxy Schiff base ligands and their complexes derived from the reaction of derivatives of salicylaldehyde with amines have been extensively studied in great detail for their various crystallographic, structural and magnetic features.15,16 Schiff bases and their metal complexes have been studied for their important bio-medicinal properties including antibacterial, fungicidal, antioxidant, anticancer, and herbicidal activities.17,18 The origin of some pathophysiological abnormalities in the human body, including cancers, is free radicals (˙OH− and ˙O2−). Antioxidants can be used to protect the human body from such abnormalities.19,20 Thus, it is important to produce potential drugs with both strong antioxidant and strong anticancer properties to protect the body from the various types of cancers. In recent years our research group has reported on the synthesis and biological properties of a number of macrocyclic and macro-acyclic Schiff base ligands incorporating the piperazine group, together with their metal complexes.20–25 Of particular interest has been those hexadentate ligands which have a potentially N4O2 donor set.26 In the current study, we have extended this work by the synthesis of a new hexadentate Schiff base ligand from the condensation of 1,4-bis(2-aminophenyl)piperazine with salicylaldehyde and prepared the Co(II), Ni(II) and Cu(II) complexes. The cytotoxicity of the complexes were then investigated against AGS (gastric) and A549 (lung) adenocarcinoma cells, their antioxidant activity was studied by the in vitro DPPH free radical scavenging method while their antibacterial activity was measured against a variety of bacterial strains. To enable a comparison between experimental and theoretical data and determine structure parameters Density functional theory (DFT) calculations been utilized while NBO and QTAIM analyses have been used to describe the nature of M–O and M–N (M = Co, Ni and Cu) bonding in these complexes.
2. Experimental
2.1. Materials
Salicylaldehyde, 1-fluoro-2-nitrobenzene, piperazine, metal salts and solvents were commercial products from Merck and used without further purification.
Caution! Perchlorate salts are potentially explosive. Only small amounts of materials should be prepared, with caution, and handled in small quantities.
2.2. Physical measurements
Infrared spectra were collected using KBr pellets on a BIO-RAD FTS-40A spectrophotometer (4000–400 cm−1). CHN analyses were carried out using a PerkinElmer, CHNS/O elemental analyzer model 2400 series 2. 1H and 13C NMR spectra were taken in CDCl3 on a Bruker Avance 400 MHz spectrometer using Si(CH3)4 as an internal standard. Electron impact mass spectra were performed on an Agilent technologies (HP) 5973 mass (EI at 70 eV) spectrometer.
2.3. X-ray crystal structure determination
Orange single crystals of formula [C30H26N4O2Co] were crystallized by vapour diffusion of ether into methanol. Data were collected at 100 K at the MX2 beamline at the Australian Synchrotron, fitted with a silicon double crystal monochromator and Dectris Eiger 16M detector, the wavelength being tuned to approximate Mo-Kα radiation (λ = 0.710872 Å).27 Data reduction was performed with XDS28 using multi-scan absorption corrections. Using Olex2,29 the structure was solved with the ShelXT30 structure solution program using Intrinsic Phasing and refined with the ShelXL31 refinement package using Least Squares minimization on F2, using all data. All non‐hydrogen atoms were refined with anisotropic displacement parameters, while all hydrogen atoms were placed at geometrical estimates and refined using the riding model. The maximum and minimum electron density peaks were 0.43 and −0.59 e Å−3. Crystallographic data are given in Table 1. Selected bond lengths and angles are given in Table 2.
Table 1 Crystal data and structure refinement for the complex [CoL]
| CCDC number |
2109477 |
| Empirical formula |
C30H26CoN4O2 |
| Formula weight |
533.48 |
| Temperature/K |
100.0(2) |
| Crystal system |
Monoclinic |
| Space group |
C2/c |
|
a/Å |
19.669(4) |
|
b/Å |
7.3570(15) |
|
c/Å |
17.509(4) |
|
α/° |
90 |
|
β/° |
104.87(3) |
|
γ/° |
90 |
| Volume/Å3 |
2448.8(9) |
|
Z
|
4 |
|
ρ
calc g/cm3 |
1.447 |
|
µ/mm |
0.737 |
|
F(000) |
1108.0 |
| Crystal size/mm3 |
0.1 × 0.05 × 0.015 |
| Radiation |
Synchrotron (λ = 0.71087) |
| 2θ range for data collection/° |
5.564 to 64.148 |
| Index ranges |
−27 ≤ h ≤ 27, −10 ≤ k ≤ 10, −26 ≤ l ≤ 25 |
| Reflections collected |
43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 102 102 |
| Independent reflections |
3866 [Rint = 0.0358, Rsigma = 0.0164] |
| Data/restraints/parameters |
3866/0/168 |
| Goodness-of-fit on F2 |
1.156 |
| Final R indexes [I >= 2σ(I)] |
R
1 = 0.0315, wR2 = 0.0961 |
| Final R indexes [all data] |
R
1 = 0.0321, wR2 = 0.0966 |
| Largest diff. peak/hole/e Å−3 |
0.43/−0.59 |
Table 2 Bond lengths (Å) and selected angles (°) for [CoL]a
| Atom–atom |
Length/Å |
|
Symmetry code i: −x, y, ½ − z.
|
| Co1–O1 |
1.9769(9) |
| Co1–N1 |
2.1228(11) |
| Co1N2 |
2.2874(11) |
| Atom–atom–atom |
Angle/° |
| O1–Co1–O1i |
127.42(6) |
| O1–Co1–N1 |
86.82(5) |
| O1–Co1–N1i |
87.74(4) |
| O1i–Co1–N2 |
93.69(4) |
| O1–Co1–N2 |
133.26(4) |
| N1i–Co1–N1 |
167.70(6) |
| N1–Co1–N2 |
72.03(4) |
| N1–Co1–N2i |
119.33(4) |
| N2–Co1–N2i |
64.35(5) |
2.4. Computational methods
The geometries of all complexes studied here, were fully optimized in the gas phase at the B3lyp32 and BP86 (ref. 33,34) levels of theory with Def2-TZVP basis set35 using the Gaussian 09 package.36 Vibrational frequency analysis, computed at the same levels of theory, show that the optimized structures of all complexes are at the stationary points without any imaginary frequency. The crystal structure coordinates of the Co(II) complex [CoL] were used as the basis for the DFT calculations for the three [ML] (M = Co, Ni and Cu) complexes. It should be noted that the Co(II) complex is in the high-spin state and the Ni(II) complex is in the low-spin state (magnetic measurements are included in result and discussion) were studied using density functional methods. The nature of coordinated bonds has been investigated using Atoms-In-Molecules (AIM)37 at the BP86/TZVP level of theory. The topological parameters of the QTAIM analysis were calculated using the AIM2000 program.38 Also, Natural Bond Orbital (NBO) analysis was carried out with the internal module Gaussian 09 at the BP86/TZVP level of theory.39
2.5. Cytotoxicity in vitro
Gastric Cell Line (AGS; IBRC C10071) and lung cell line (A549; IBRC C10080) were purchased from the Iranian Biological Resource Center (IBRC; Tehran, Iran). DMEM: Ham's F12, used in culturing the lung cancer cell line and Gastric cell line, was supplemented with L-glutamine, 10% Fetal Bovine Serum (FBS), 100 units mL−1 penicillin and 100 µg mL−1 streptomycin. The cells were kept in a humidified atmosphere at 37 °C with a concentration of 5% CO2. Standard solutions were made by dissolving the compounds in DMSO to give a concentration of 20 mM L−1. These solutions were then diluted to give concentrations of 1.56, 3.13, 6.25, 12.50, 25 and 50 mM L−1 which were subsequently added to the culture medium. Assessment of cell viability was performed using the modified cell viability (MTT) method in which the reduction of MTT (3-[4,5-dimethylthiazolyl]-2,5-diphenyltetrazolium bromide) was detected. In each well of a 96-well plate 25 was allocated 5 × 104 cells with 100 µL RPMI medium supplemented with 10% FBS. After allowing 24 hours for cell adhesion, diluted solutions of the tested compounds (0 to 50 µM) were added. In order to prevent the toxicity of the solvent, 0.5% (v/v) was chosen as the final DMSO concentration. After a further 12 h 10 µL MTT was added followed by incubating the plates at 37 °C for an additional 4 h. The formazan blue that was subsequently formed was dissolved in DMSO (100 µL) and the optical density (OD) of the solution in each well was measured at 490 nm using an ELISA plate reader to determine the cell survival curves. Each test was carried out in triplicate and the mean value was used to determine the IC50 value – the concentration of the tested compound which inhibits the growth of the cells by 50%.
2.6. Antioxidant property
The electron donation abilities of the complexes were evaluated by bleaching of the purple-colored chloroform solution of DPPH (2,2-diphenyl-1-picrylhydrazyl).9 Briefly, 2.5 mL of different concentrations of each sample (0.2–1 mg mL−1 in methanol) were added to 1 mL of 0.3 mM DPPH solution and the mixture incubated for 30 min at room temperature in the dark. The reduction of free radicals was measured by reading the absorbance at 517 nm. Ascorbic acid and quercetin, standard antioxidants, were used as standards. The percentage inhibition of the production of free radicals by DPPH (I%) was calculated using the following formula:
| I (%) = [1 − (As − Ab)/Ac] × 100 |
where, As is the absorbance of the reaction mixture containing 2.5 mL of sample + 1 mL of DPPH, Ab is the absorbance of the reaction mixture containing 2.5 mL of sample + 1 mL methanol, and Ac is the absorbance of the control sample containing 1 mL of DPPH + 2.5 mL methanol. The IC50 value, defined as the concentration of the sample leading to 50% reduction of the initial DPPH concentration, was calculated from the linear regression plot of the concentration of the test sample against the mean percentage of the antioxidant activity.40,42,43
2.7. Antibacterial activity
The antibacterial activity of synthesized compounds was studied against two Gram-positive (Staphylococcus aureus, and Bacillus subtilis) and two Gram-negative bacteria (Escherichia coli, and Klebsiella oxytoca). Each of the complexes was dissolved in methanol to give a concentration of 1 mg mL−1 which was then filtered through a 0.45 µm Millipore. Each solution was then added to a suspension of 1.5 × 108 bacteria mL−1 (10 mL) which were then spread out on the agar medium. Vancomycin, tetracycline, ampicillin, and penicillin were also applied as antibiotic standards. The volume of swelling from the edge and the diameter of the inhibition region of each sample was given in mm.
2.8. Synthesis
2.8.1 Ligand synthes'is (H2L).
1,4-Bis(2-aminophenyl)piperazine (A) was synthesized according to the literature procedure.41 A solution of salicylaldehyde (1 mmol, 0.15 g) in absolute ethanol (30 mL) was added drop wise to a solution of A (0.5 mmol, 0.13 g) in absolute ethanol (20 mL) and the mixture was gently refluxed and stirred overnight. The resulting product was then filtered off, washed with ethanol and dried in vacuum (Scheme 1).
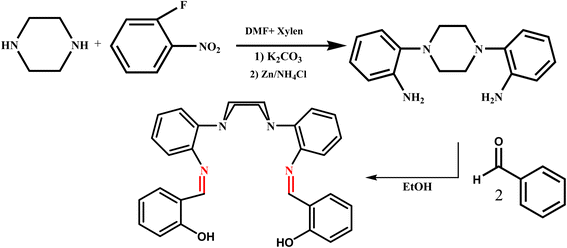 |
| | Scheme 1 The processes for the synthesis of ligand. | |
Yield: 0.19 g (84%). Anal. Calc. for C30H28N4O2 (MW: 476.57): C, 75.61; H, 5.92; N, 11.76 Found: C, 75.52; H, 6.08; N, 11.64. EI-MS (m/z): 476.4 IR (KBr, cm−1): 3230 ν (OH), 1618 ν (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). 1H NMR (CDCl3–d6, ppm) δH = 2.55–2.86 (s, H), 6.58–7.88(Haromatic), 8.62 (Himini), 13.43 (OH). 1C NMR(CDCl3, ppm): δc: 55.8–57.8(Cpy), 115.8–149.8(Caromatic), 160.6(Cimini), 161.4(C–OH).
N). 1H NMR (CDCl3–d6, ppm) δH = 2.55–2.86 (s, H), 6.58–7.88(Haromatic), 8.62 (Himini), 13.43 (OH). 1C NMR(CDCl3, ppm): δc: 55.8–57.8(Cpy), 115.8–149.8(Caromatic), 160.6(Cimini), 161.4(C–OH).
2.8.2 Synthesis of complexes.
General procedure: in order to prepare the metal complexes, the ligand (L) (1 mmol), dissolved in 25 mL of methanol, was added to a solution of M(ClO4)2·6H2O (1 mmol) (M: Ni, Co, Cu) dissolved in 20 mL of methanol. The solution was then refluxed for 24 h, after which triethylamine (3 drops) was added and the reaction mixture was allowed to cool to room temperature. The precipitate was filtered off, washed with excess of ethanol and then dried in vacuum. The products were characterized as the pure compounds (Scheme 2).
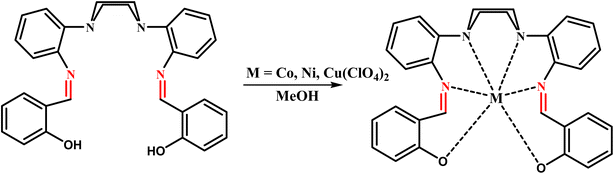 |
| | Scheme 2 The processes for the synthesis of macrocyclic complexes. | |
[CoL]: orange single crystals. Yield: 0.45 g (76%). Anal. Calc. for C30H26CoN4O2 (MW: 533.49): C, 67.54; H, 4.91; N, 10.50. Found: C, 67.47; H, 4.75; N, 10.32. EI-MS (m/z): 533IR (KBr, cm−1): 1611 ν (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N).
N).
[NiL]: green powder. Yield: 0.5 g (82.3%). M. p. 315 °C. Anal. Calc. for C30H26NiN4O2 (MW: 533.25): C, 67.57; H, 4.91; N, 10.51. Found: C, 67.49; H, 4.83; 10.42%. IR (cm−1, KBr): 1610 (s, m C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). MS (EI): (m/z) = 533.19.
N). MS (EI): (m/z) = 533.19.
[CuL]: green powder. Yield: 0.49 g (87%). Anal. Calc. for C30H26CuN4O2 (MW: 537.14): C, 66.96; H, 4.87; N, 10.41. Found: C, 66.79; H, 4.71; N, 10.32. EI-MS (m/z): 537.09 IR (KBr, cm−1): 1606 ν (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N).
N).
3. Result and discussion
We prepared the hexadentate Schiff base ligand with a N4O2 donor set by condensation of 1,4-bis(2-aminophenyl)piperazine, with 2-hydroxy-benzaldehyde in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio. Elemental analysis, IR, mass, 1H and 13C NMR spectroscopy were used to characterize the formation of the ligand (L). The infrared spectrum shows a strong absorption band at ∼1618 cm−1, assigned to the C
2 ratio. Elemental analysis, IR, mass, 1H and 13C NMR spectroscopy were used to characterize the formation of the ligand (L). The infrared spectrum shows a strong absorption band at ∼1618 cm−1, assigned to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching vibration, and with the absence of bands due to C
N stretching vibration, and with the absence of bands due to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and NH2 stretching vibrations, indicates the formation of the Schiff-base ligand. The mass spectrum of the ligand shows the expected peak at 476.4 m/z. The Ni(II), Co(II) and Cu(II) complexes were prepared by direct reaction of the ligand and metal salts in a 1
O and NH2 stretching vibrations, indicates the formation of the Schiff-base ligand. The mass spectrum of the ligand shows the expected peak at 476.4 m/z. The Ni(II), Co(II) and Cu(II) complexes were prepared by direct reaction of the ligand and metal salts in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio in methanol and characterized by mass and IR spectroscopy and by elemental analysis. In the IR spectra the imine stretching bands found for the Ni(II), Co(II) and Cu(II) complexes, 1610 cm−1, 1611 cm−1 and 1606 cm−1, respectively, were shifted to lower frequency compared to the free ligand (1618 cm−1) consistent with the coordination of the imine nitrogen to the metal ions. Furthermore, there no absorption bands in the 3400–3200 cm−1 region, due O–H stretching bands, confirming that the deprotonation of the phenol had occurred. Considering the great similarity of the IR spectra of the complexes, it can be concluded that their structure is similar. Also, the calculated spectra of the ligand and complexes were almost identical with the experimental spectra (Fig. S1†–S8). The UV-visible absorption spectra of the ligand and its complexes were measured in 10−4 M DMSO solutions at room temperature. The intense bands 266 nm of the ligand, may be assigned to π → π* transitions of the azomethine. The other peak which has appeared in the 310 nm region of the ligand spectrum is attributed to n → π* transition. In the spectra of the related complexes, the bands due to the ligand (π → π* and n → π* transitions) are still pronounced with a small shift in position with respect to the corresponding bands in the free ligand. In addition, a charge transfer band at 400–437 nm has been also observed in the electronic spectra of the ligand and complexes. The band at 483–494 nm in electronic spectra of the complexes can be attributed to MLCT (Fig. S9–S12†).
1 ratio in methanol and characterized by mass and IR spectroscopy and by elemental analysis. In the IR spectra the imine stretching bands found for the Ni(II), Co(II) and Cu(II) complexes, 1610 cm−1, 1611 cm−1 and 1606 cm−1, respectively, were shifted to lower frequency compared to the free ligand (1618 cm−1) consistent with the coordination of the imine nitrogen to the metal ions. Furthermore, there no absorption bands in the 3400–3200 cm−1 region, due O–H stretching bands, confirming that the deprotonation of the phenol had occurred. Considering the great similarity of the IR spectra of the complexes, it can be concluded that their structure is similar. Also, the calculated spectra of the ligand and complexes were almost identical with the experimental spectra (Fig. S1†–S8). The UV-visible absorption spectra of the ligand and its complexes were measured in 10−4 M DMSO solutions at room temperature. The intense bands 266 nm of the ligand, may be assigned to π → π* transitions of the azomethine. The other peak which has appeared in the 310 nm region of the ligand spectrum is attributed to n → π* transition. In the spectra of the related complexes, the bands due to the ligand (π → π* and n → π* transitions) are still pronounced with a small shift in position with respect to the corresponding bands in the free ligand. In addition, a charge transfer band at 400–437 nm has been also observed in the electronic spectra of the ligand and complexes. The band at 483–494 nm in electronic spectra of the complexes can be attributed to MLCT (Fig. S9–S12†).
The magnetic susceptibility of the complexes was measured at room temperature. Co-L effective magnetic moment value (µeff = 4.23 B.M.) is typical of high-spin d7 systems with three unpaired electrons. The Ni–L complex is a low spin (with d8 configuration) paramagnetic moiety because its magnetic moment is (µeff = 2.6 B.M.).
In the 1H NMR spectrum of the ligand (L) exhibits peaks for the aromatic hydrogen atoms, in the range of δ 6.55–7.88, and peaks for the aliphatic hydrogen atoms in the range of δ 2.55–2.86 ppm. The signal exhibited at 8.62 ppm in the spectra has been assigned to the imine protons and the signal at 13.43 ppm has been assigned to the OH protons (Fig. S13†). 13C NMR spectrum of the ligand (L) shows peaks ranging from δ 115.8 to 149.80 for the carbons of the aromatic ring and peaks in the range of δ 55.8 to 58.8 for the aliphatic carbons. The signal at 160.6 ppm is assigned to the carbon of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N group while the signal at 161.4 ppm is assigned to the carbon attached to the OH group (Fig. S14†).
N group while the signal at 161.4 ppm is assigned to the carbon attached to the OH group (Fig. S14†).
3.1. X-ray structure
The structure of [CoL] is almost identical to those found for [2,2'-{(piperazine-1,4-diyl)bis[(2,1-phenylene)(azanylylidene)methanylylidene]}bis(6-methoxyphenolato)]-cobalt(II) [CoL1],44 and [2,2'-{(piperazine-1,4-diyl)bis[(2,1-phenylene)(azanylylidene)methanylylidene]}bis(4-nitrophenolato)]-cobalt(II) [CoL2]45 cobalt complexes of the hexadentate ligand that have either methoxy groups or nitro groups, respectively, attached to the phenol rings. In the present compound the cobalt atom lies on a 2-fold axis, so that the asymmetric unit comprises half the molecule. As with [CoL1] and [CoL2], the cobalt atom is in a distorted trigonal prismatic coordination environment (Fig. 1 and 2), surrounded by the six donor atoms of the deprotonated hexadentate ligand. The absence of a counter ion and the values of the Co–O and Co–N bond lengths are consistent with the oxidation state being Co(II). The atoms N1, N2 and O1i and O1, N1i and N2i (symmetry code i: −x, y, ½ − z) form the two triangular faces, which are almost coplanar, the dihedral angle being 6.81(8)°, while the angles between the rectangular faces are 56.21(4)°, 61.90(3)° and 61.90(3)°. Continuous Shape Measure (CShM) calculations, based on a trigonal prismatic geometry,47 give a value of 2.536 (compared to 2.099 for [CoL1] and 1.249 for [CoL2]). The Co–O distances are 1.9769(9) Å, while the Co–Npiperazine distances, 2.2874(11) Å, are longer than the Co–Nimine distances of 2.1228(11) Å, similar to that observed for [CoL1]. Trigonal prismatic coordination for metal complexes is rare,48 a search of the Cambridge Structural Database (CSD Version 5.43 June 2022 Update)49 found only 11 cobalt(II) structures with a N2O4 trigonal prismatic coordination environment, other than [CoL1] and [CoL2], (out of a total of 151 trigonal prismatic Co complexes), only one of which, ((2,2'-((piperazine-1,4-diyl)bis(propane-3,1-diyl(nitrilo)methylylidene))bis(6-methoxyphenolato))-cobalt(II)),50 had a N3O donor set in each of the rectangular faces, as found here for the present complex. The cobalt atom lies in the plane of the atoms of the two (aminomethyl)phenol groups, while the 5-membered metallocyclic rings are in an envelope conformation with the Co atom lying out of the plane of the other 4 atoms. The dihedral angle between the two (aminomethyl)phenol groups is 59.71(3)° while the dihedral angle between the diaminophenyl rings is 72.34(5)°. There are no hydrogen bonds in the structure.
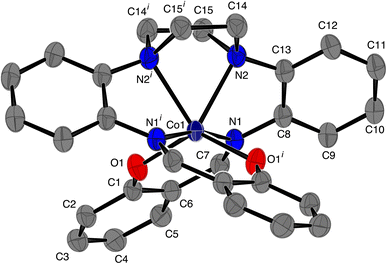 |
| | Fig. 1 ORTEP46 diagram of [CoL], ellipsoids are drawn at the 50% probability level. Hydrogen atoms have been omitted for clarity; symmetry code i: −x, y, ½ − z. | |
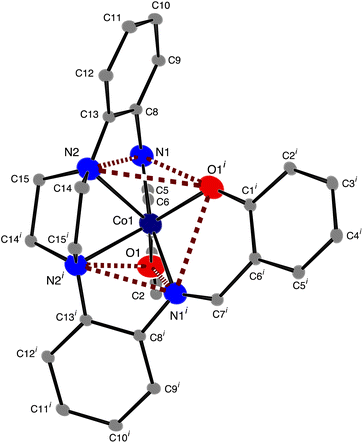 |
| | Fig. 2 ORTEP46 diagram of [CoL], ellipsoids are drawn at the 50% probability level for Co, O and N atoms and at the 10% probability level for carbon atoms. Hydrogen atoms have been omitted for clarity; symmetry code i: −x, y, ½ − z. | |
3.2. Theoretical studies
The optimized structures of the ML (M = Co, Ni and Cu) complexes, at the B3lyp and BP86 levels with the Def2-TZVP basis set, are shown in Fig. 3; the starting point for the calculations were the crystal structure coordinates of the Co(II) complex. The calculations show that the optimized structure of the CoL complex is similar to its X-ray structure, in which the two oxygen atoms and four nitrogen atoms of the macroacyclic ligand are coordinated to the Co atom in a distorted trigonal-prismatic environment. Selected calculated bond lengths and bond angles of optimized structures of ML (M = Co, Ni and Cu) complexes and corresponding experimental values obtained from the X-ray crystal are reported in Table 3. A comparison between experimental and calculated bond lengths values for CoL complex show that there is a good agreement between the experimental and calculated values using both B3lyp and BP86, but that BP86 shows slightly better agreement than B3lyp (Table 3). For example, as can be seen in Table 1, the calculated bond length values using BP86 for the Co←O and Co←N2 bonds are about 0.031 Å and 0.065 Å greater than experimental values, respectively, while the Co←N1 bond length is similar to the experimental value. For B3lyp the values for the Co←O, Co←N2 and Co←N1 bond lengths are about 0.031 Å, 0.096 Å and 0.039 Å greater than experimental values, respectively. A comparison of the experimental and calculated bond angles for CoL show small differences. For example, the calculated values of the bond angles differ from the corresponding experimental values by around 0.09°–3.5° for BP86 and around 0.48°–7.7° for B3lyp. A can also be seen in Table 3, changing the M atom from Co(II) to Ni(II) and Cu(II), leads to a decrease in the values of the M←O and M←N1 bond lengths and an increase in the values of the M←N2 bond length. Therefore, values of the M←N2 bond lengths in the Ni(II) and Cu(II) complexes are very large and thus outside the range of the usual M←N bonds, which shows that these complexes can have a square planar geometries. The QTAIM and NBO analyses were also used to characterize the type and nature of M←O and M←N bonds in the ML (M = Co, Ni and Cu) complexes. The results of AIM and NBO analyses were summarized in Table 4. The NBO results for all complexes studied here, demonstrate that the M (M = Co, Ni and Cu) atom has a positive charge which is an acceptor atom and two oxygen atoms and four nitrogen atoms of the macroacyclic ligand as donor atoms have negative charges. As can be seen, the amount of the charge transfer from the ligand to metal ions are about 1.12 e, 1.40 e and 1.11 e in the CoL, NiL and CuL complexes, respectively. These results also show that in the above complexes, maximum charge transfer occurs between M and L atoms in M←O and M←N1 bonds and minimum charge transfer occurs in M←N2 bond. The Wiberg bond indices (WBIs) for are also reported in Table 4. The largest values of WBI are attributed to M←O and M←N1 bonds and the smallest ones are attributed to M←N2 bonds. Also, the AIM analysis was used to investigate the bond critical points (BCPs) at M←O and M←N bonds bond for above complexes. The topological properties calculated at bond critical points for M←O and M←N bonds are listed in Table 4. The AIM results show that there is a good agreement between the amounts of electron density ρ(rc) of M←O and M←N bonds with calculated WBI in above complexes. Also, the amounts of ∇2ρ(rc) and −Gc/Vc for the M←O and M←N bonds show that the nature of these bonds is partly covalent (of ∇2ρ(rc) > 0 and −Gc/Vc < 1).
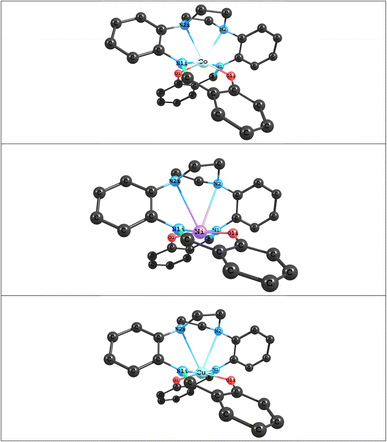 |
| | Fig. 3 Optimized structures of ML (M = Co, Ni and Cu) complexes at the BP86 levels with Def2-TZVP basis set. | |
Table 3 The experimental and calculated important bond lengths (Å) and bond angles (°) of ML (M = Co, Ni and Cu) complexes
| M–L bond |
CoL |
NiL |
CuL |
| Bond lengths |
Bond lengths |
Bond lengths |
| Experimental |
B3LYP/TZVP |
BP86/TZVP |
B3LYP/TZVP |
BP86/TZVP |
B3LYP/TZVP |
BP86/TZVP |
| M–O1 |
1.9769 |
2.00763 |
2.00825 |
1.87339 |
1.87510 |
1.94832 |
1.96688 |
| M–N1 |
2.1228 |
2.16204 |
2.11821 |
1.94185 |
1.91793 |
2.03740 |
2.01885 |
| M–N2 |
2.2874 |
2.38339 |
2.35270 |
2.97098 |
2.98149 |
2.958 |
2.89248 |
| M–L bond |
CoL |
NiL |
CuL |
| Bond angles |
Bond angles |
Bond angles |
| Experimental |
B3LYP/TZVP |
BP86/TZVP |
B3LYP/TZVP |
BP86/TZVP |
B3LYP/TZVP |
BP86/TZVP |
| O1–M1–O1i |
127.42 |
135.147 |
130.977 |
168.430 |
168.009 |
152.680 |
151.724 |
| O1–M1–N1 |
86.82 |
85.549 |
86.915 |
91.586 |
92.398 |
90.435 |
91.010 |
| O1–M1–N1i |
87.74 |
90.091 |
87.946 |
89.067 |
88.273 |
91.468 |
90.969 |
| O1i–M1–N2 |
93.69 |
92.706 |
92.888 |
81.305 |
80.885 |
88.123 |
88.143 |
| O1–M1–N2 |
133.26 |
127.102 |
130.894 |
109.444 |
110.287 |
117.172 |
118.100 |
| N1i–M1–N1 |
167.70 |
168.564 |
167.594 |
173.510 |
173.582 |
171.939 |
171.887 |
| N1–M1–N2 |
72.03 |
70.901 |
71.816 |
65.534 |
65.822 |
65.047 |
65.266 |
| N1–M1–N2i |
119.33 |
119.806 |
119.715 |
108.185 |
107.978 |
107.197 |
106.940 |
| N2–M1–N2i |
64.35 |
63.034 |
63.644 |
51.338 |
51.314 |
52.331 |
52.248 |
Table 4 Natural charges of M, N and O atoms, Wiberg bond indices (WBI) and the topological properties calculated at bond critical points of M–L bonds in ML (M = Co, Ni and Cu) complexes at the BP86/def2-TZVP level of theory
| M⋯L bond |
M⋯L bond length |
NBO |
QTAIM |
| qM |
qL |
WBI |
ρ(rc) |
∇2ρ(rc) |
−Gc/Vc |
| Co⋯O1 |
2.00825 |
0.87993 |
−0.61831 |
0.3509 |
0.0769 |
0.3702 |
−0.8759 |
| Co⋯N1 |
2.11821 |
−0.44971 |
0.2775 |
0.0679 |
0.2716 |
−0.8558 |
| Co⋯N2 |
2.35270 |
−0.39086 |
0.1427 |
0.0436 |
0.1468 |
−0.8744 |
| Ni⋯O1 |
1.87510 |
0.59715 |
−0.53645 |
0.5119 |
0.1023 |
0.5205 |
−0.8639 |
| Ni⋯N1 |
1.91793 |
−0.39071 |
0.4710 |
0.1040 |
0.4284 |
−0.8224 |
| Ni⋯N2 |
2.98149 |
−0.39311 |
0.0192 |
0.0152 |
0.0380 |
−0.9250 |
| Cu⋯O1 |
1.96688 |
0.89204 |
−0.60467 |
0.3310 |
0.0834 |
0.3834 |
−0.8498 |
| Cu⋯N1 |
2.01885 |
−0.45849 |
0.2942 |
0.0836 |
0.3089 |
−0.8151 |
| Cu⋯N2 |
2.89248 |
−0.39400 |
0.0283 |
0.0165 |
0.0419 |
−0.9490 |
3.3. Cytotoxicity in vitro
MTT assay was used to analyze the cytotoxicity of the synthesized compounds against lung (A549) and gastric (AGS) cell lines. The reduction of the yellow MTT by the mitochondrial dehydrogenase's enzymatic activity was followed by analyzing the cell viability. Fig. 4 illustrates the percentage of the cell viability of the comparative cytotoxicity patterns The IC50 values of the complexes, the amount of material that kills 50% of the cells, was measured from the dose-survival curves for the growth inhibition of the two cell lines (Table 5). A comparison of the IC50 values shows that AGS cells (average IC50 value; 6.48 µM) are more chemosensitive than the A549 cells (average IC50 value; 25.46 µM) and that Cu–L has the highest efficiency in stopping the growth of both cell lines. In addition, while the IC50 values of all three compounds against AGS cells are similar, the cytotoxic potency of CuL against the A549 cell line is much greater than found for the other two complexes. Accordingly, all three complexes may warrant further investigation as therapeutic agents against AGS cell lines, while it would appear that only CuL may warrant further investigation against A549 cell lines. In this regard, as lung cancer is the leading cause of death through the world, causing 1.6 million deaths with ∼1.8 million new cases each year, new therapeutic agents against lung cancer are needed.51,52 These values are much lower than the values found for [CoL2] (73), [NiL2] (67), and [CuL2] (67),45 indicating that the absence of nitro groups in the present ligand has significantly enhanced the cytotoxic potency.
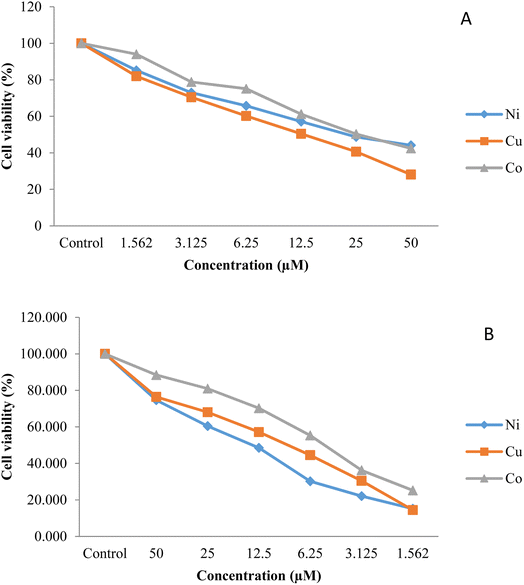 |
| | Fig. 4 Graphical representation of the cytotoxicity of the synthesized compounds against (A) lung (A549) and (B) gastric (AGS) cancer cell lines obtained by MTT assay. | |
Table 5 Comparison of cytotoxicity (IC50 values based on µM)
| Structure |
AGS |
A549 |
| Ni-L |
7.29 ± 0.24 |
29.32 ± 0.6 |
| Cu-L |
5.01 ± 0.19 |
13.86 ± 0.91 |
| Co-L |
7.14 ± 0.21 |
33.21 ± 1.02 |
3.4. Antioxidant property
The antioxidant proficiency of the synthesized compounds was studied by carrying out the DPPH˙ (2,2-diphenyl-2-picryl-hydrazyl) free radical scavenging technique and the results were reported in terms of radical scavenging percentage and IC50 values. Table 6 shows the antioxidant activity of all complexes along with standards, ascorbic acid, and quercetin. It can be seen that the compounds displayed moderate to significant antioxidant activity ranging from 36.6 to 79.7% with IC50 values of 0.33 to 0.56 mg mL−1 which were less than the standards ascorbic acid and quercetin. The effectiveness of the samples as DPPH radical scavengers were in the order: Cu (IC50 = 0.33 mg mL−1) > Ni (IC50 = 0.39 mg mL−1) > Co (IC50 = 0.56 mg mL−1) indicating that the presence of Cu in Schiff base complexes can be effective in producing antioxidant activity in lower concentrations. These IC50 values are, on average, higher than the values found for [CoL2] (0.20), [NiL2] (0.40), and [CuL2] (0.19),45 indicating that the absence of nitro groups in the present ligand has significantly reduced the antioxidant activity of the complexes.
Table 6 The Antioxidant activity of the synthesized compounds and two standards (ascorbic acid and quercetin)
| Sample |
Concentration (mg mL−1) |
Average |
IC50 |
| 0.2 |
0.4 |
0.6 |
0.8 |
1 |
| Ni |
39.57 |
51.01 |
56.89 |
69.34 |
78.69 |
59.10 |
0.39 |
| Co |
36.62 |
46.49 |
53.79 |
60.84 |
69.31 |
53.41 |
0.56 |
| Cu |
49.65 |
61.43 |
66.95 |
74.01 |
79.70 |
66.35 |
0.33 |
| Ascorbic acid |
74.24 |
75.91 |
79.25 |
81.25 |
84.55 |
79.04 |
0.13 |
| Quercetin |
75.26 |
81.36 |
85.75 |
88.44 |
89.29 |
84.02 |
0.13 |
3.5. Antibacterial activity
Antibacterial activity studies of the complexes were carried out by the in vitro disc diffusion method. All compounds were screened against two Gram-positive bacterial strains Staphylococcus aureus (S. aureus), Bacillus subtilis (B. subtilis), and two Gram-negative bacteria strain Escherichia coli (E. coli), and Klebsiella oxytoca (K. oxytoca). The antibacterial activities of the synthesized compounds, presented in Table 7, are greater against the Gram-negative bacteria than towards the Gram-positive ones. This behavior can be attributed to the structures of bacteria cell wall which results in different rates of diffusion of the compounds into the cells. A large part of cell walls in the Gram-positive bacteria consists of thick layers of peptidoglycan, which is a more rigid structure than the Gram-negative bacteria which has a much thinner layer of peptidoglycan. Based on the obtained results, the Cu-complex (with average inhibition zone: 24.8 mm) has the most effect on both Gram-negative and -positive bacteria. B. subtilis (+) showed the most resistance, and K. oxytoca (−) was the most sensitive bacteria to the synthesized compounds. Compared to the reference antibiotics the antibacterial activities of synthesized compounds show significant inhibitory potency against the tested bacteria and that the Cu complex has shown greater antibacterial activity than the other complexes.
Table 7 Antibacterial activity of synthesized compounds and four antibiotics against selected bacterial strains
| Sample |
Inhibition zone (mm) |
|
B. subtilis
|
S. aureus
|
E. coli
|
K. oxytoca
|
| Ni |
17 |
19.3 |
22.5 |
24.3 |
| Co |
14.5 |
17 |
19 |
22.5 |
| Cu |
22 |
23.8 |
25.8 |
27.5 |
| Penicillin |
13 |
13 |
18 |
15 |
| Ampicillin |
14 |
16 |
12 |
14 |
| Vancomycine |
17 |
15 |
23 |
21 |
| Tetracycline |
21 |
22 |
27 |
23 |
| Methanol |
Na |
Na |
Na |
Na |
4. Conclusions
We have prepared a new hexadentate N4O2 Schiff-base ligand (L) derived from the condensation of 1,4-bis(2-aminophenyl)piperazine with 2-hydroxy-benzaldehyde and prepared the Ni(II), Cu(II) and Co(II) complexes the direct reaction of the ligand with the metal perchlorates. The ligand and complexes were characterized by IR and Mass spectroscopy, elemental analysis and by the determination of the single crystal X-ray structure of [CoL]. In this complex, the cobalt is in a distorted trigonal prismatic coordination environment, surrounded by the six donor atoms of the deprotonated hexadentate ligand, similar to the other complexes with the ligands containing methoxy or nitro substitutents on the phenol rings.44,45 The geometries of [ML] (M = Co, Ni and Cu) complexes were fully optimized using the B3lyp and BP86 levels of theory with def2-TZVP basis set. The results show that the CoL complex studied here has the distorted trigonal-prismatic geometry, similar to X-ray crystal structure of its complex, and Ni(II) and Cu(II) complexes can have the square planar geometries. Therefore there are possible structures of square planar for Ni(II) and Cu(II) complexes. The nature of the M←O and M←N bonds in the ML (M = Co, Ni and Cu) complexes were determined by QTAIM and NBO analyses. According to the NBO analysis, the maximum values of the Wiberg bond indices and charge transfer are attributed to the M←O and M←N1 bonds in NiL complex. The topological properties that were calculated confirm the covalent character of the M←O and M←N bonds in the [ML] complexes. (CuL) illustrated its greater toxicity properties than other complexes. The antibacterial studies have shown that the complex [CuL] has the greatest activity, on both on Gram-negative and Gram-positive bacteria, with E. coli (−) being the most sensitive bacteria to [CuL] while S. aureus (+) was the most resistant bacteria. The antioxidant activity of the synthesized compounds showed that, at high concentrations, the complex [CuL], had a similar scavenging activity to quercetin. Also [CuL] illustrated its greater toxicity properties than other complexes. The absence of nitro groups in the present ligand, compared to the complexes with the ligand containing nitro groups,45 has enhanced the cytotoxic potency of the complexes but lowered the antioxidant activity.
Conflicts of interest
There are no conflicts to declare.
Acknowledgements
We are grateful to the Faculty of Chemistry of Bu Ali Sina University, for financial support. This research was undertaken, in part, using the MX2 beamline at the Australian Synchrotron, part of ANSTO, Victoria, Australia, and made use of the Australian Cancer Research Foundation (ACRF) detector.
References
- M. Rezaeivala and H. Keypour, Coord. Chem. Rev., 2014, 280, 203–253 CrossRef CAS.
- B. K. Panda and A. Chakravorty, J. Organomet. Chem., 2005, 690, 3169–3175 CrossRef CAS.
- K. R. Krishnapriya and M. Kandaswamy, Polyhedron, 2005, 24, 113–120 CrossRef CAS.
- K. N. Kumar and R. Ramesh, Polyhedron, 1999, 18, 1561–1568 CrossRef.
-
J. M. Lehn, Supramolecular Chemistry Concepts and Perspectives, Wiley-VCH, Weinheim, Germany, 1st edn, 1995 Search PubMed.
- B. Belghoul, I. Maier, A. Weiterlich, A. Toutianoush, A. Raman Rabindranath and B. Tieke, Langmuir, 2007, 23, 5062–5069 CrossRef CAS PubMed.
- B. K. Tripuramallu, R. Kishore and S. K. Das, Polyhedron, 2010, 29, 2985–2990 CrossRef CAS.
- R. E. Mewis and S. J. Archibald, Coord. Chem. Rev., 2010, 254, 1686–1712 CrossRef CAS.
- C. X. Ding, J. Ni, Y. H. Yang, S. W. Ng, B. W. Wang and Y. S. Xie, CrystEngComm, 2012, 14, 7312–7319 RSC.
- L. D. S. Yadav and S. Singh, Indian J. Chem., Sect. B: Org. Chem. Incl. Med. Chem., 2001, 40, 440–442 Search PubMed.
- H. Zaiyin, L. Yi, Q. Songsheng and F. Ying, Thermochim. Acta, 1998, 320, 121–126 CrossRef.
- Z. Guo, R. Xing, S. Liu, Z. Zhong, X. Ji, L. Wang and P. Li, Carbohydr. Res., 2007, 342, 1329–1332 CrossRef CAS PubMed.
- Z. H. A. El-Wahab and M. R. El-Sarrag, Spectrochim. Acta, Part A, 2004, 60, 271–277 CrossRef PubMed.
- M. Wang, L. F. Wang, Y. Z. Li, Q. X. Li, Z. D. Xu and D. M. Qu, Transition Met. Chem., 2001, 26, 307–310 CrossRef CAS.
- A. Th. Chaviara, P. J. Cox, K. H. Repana, R. M. Papi, K. T. Papazisis, D. Zambouli, A. H. Kortsaris, D. A. Kyriakidis and C. A. Bolos, J. Inorg. Biochem., 2004, 98, 271–1283 CrossRef PubMed.
- M. M. Omar, G. G. Mohamed and A. A. Ibrahim, Spectrochim. Acta, Part A, 2009, 73, 358–369 CrossRef CAS PubMed.
- K. Shanker, R. Rohini, V. Ravinder, P. M. Reddy and Y. Ho, Spectrochim. Acta, Part A, 2009, 73, 205–211 CrossRef PubMed.
- X. Qiao, Z.-Y. Ma, C.-Z. Xie, F. Xue, Y.-M. Zhang, J.-Y. Xu, Z.-Y. Qiang, D.-S. Lou, G.-J. Chen and S.-P. Yan, J. Inorg. Biochem., 2011, 5, 728–737 CrossRef PubMed.
- C. X. Ding, C. Gao, S. W. Ng, B. W. Wang and Y. S. Xie, Chem. – Eur. J., 2013, 19, 9961–9972 CrossRef CAS PubMed.
- A. Pal, B. Biswas, S. K. Mondal, C.-H. Lin and R. Ghosh, Polyhedron, 2012, 31, 671–675 CrossRef CAS.
- K. Rissanen, J. Huuskonen and A. Koskinen, J. Chem. Soc., Chem. Commun., 1993, 771–772 RSC.
- H. Keypour, M. Rezaeivala, L. Valencia and P. Perez-Lourido, Polyhedron, 2008, 27, 3172–3176 CrossRef CAS.
- H. Keypour, M. Rezaeivala, L. Valencia, S. Salehzadeh, P. Perez-Lourido and H. Khavasi, Polyhedron, 2009, 28, 3533–3541 CrossRef CAS.
- H. Keypour, M. Rezaeivala, L. Valencia, S. Salehzadeh, P. Perez-Lourido and A. H. Mahmoudkhani, Polyhedron, 2009, 28, 3415–3418 CrossRef CAS.
- H. Keypour, M. Rezaeivala, L. Valencia and P. Perez-Lourido, Polyhedron, 2009, 28, 4096–4100 CrossRef CAS.
- H. Keypour, N. Rahpeyma, N. Arzhangi, M. Rezaeivala, Y. Elerman, O. Buyukgungor and L. Valencia, Polyhedron, 2010, 29, 144–1148 CrossRef.
- D. Aragão, J. Aishima, H. Cherukuvada, R. Clarken, M. Clift, N. P. Cowieson, D. J. Ericsson, C. L. Gee, S. Macedo, N. Mudie, S. Panjikar, J. R. Price, A. Riboldi-Tunnicliffe, R. Rostan, R. Williamson and T. T. Caradoc-Davies, J. Synchrotron Radiat., 2018, 25, 885–891 CrossRef PubMed.
- W. Kabsch, Acta Crystallogr., 2010, D66, 125–132 CrossRef PubMed.
- O. V. Dolomanov, L. J. Bourhis, R. J. Gildea, J. A. K. Howard and H. Puschmann, J. Appl. Crystallogr., 2009, 42, 339–341 CrossRef CAS.
- G. M. Sheldrick, Acta Crystallogr., 2015, A71, 3–8 CrossRef PubMed.
- G. M. Sheldrick, Acta Crystallogr., 2015, C71, 3–8 CrossRef PubMed.
- A. D. Becke, J. Chem. Phys., 1993, 98, 5648–5652 CrossRef CAS.
- A. D. Becke, Phys. Rev. A, 1988, 38, 3098–3100 CrossRef CAS PubMed.
- J. P. Perdew, Phys. Rev. B: Condens. Matter Mater. Phys., 1986, 33, 8822–8824 CrossRef PubMed.
- F. Weigend and R. Ahlrichs, Phys. Chem. Chem. Phys., 2005, 7, 3297–3305 RSC.
-
M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery Jr, J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, T. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, O. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski and D. J. Fox, GAUSSIAN 09, Revision D.01, Gaussian, Inc., Wallingford, CT, 2013 Search PubMed.
-
R. F. W. Bader, Atoms in Molecules: A Quantum Theory, Oxford University Press, Oxford, 1990 Search PubMed.
-
R. F. W. Bader, AIM2000 program package, Ver. 2.0, McMaster University, Hamilton, Canada, 2000 Search PubMed.
-
E. D. Gladdening, A. E. Reed, J. A. Carpenter and F. Weinhold, NBO Version 3.1 Search PubMed.
- L. L. Mensor, F. S. Menezes, G. G. Leitão, A. S. Reis, T. C. dos Santos, C. S. Coube and S. G. Leitão, Phytother. Res., 2001, 15(2), 127–130 CrossRef CAS PubMed.
-
M. B. Hursthouse, M. E. Light and I. A. Fallis, CSD Communication, Private Communication 218006, 2003 Search PubMed.
- A. J. Hataway and D. E. Underhill, J. Chem. Soc., 1961, 3091 RSC.
- W. J. Geary, Coord. Chem. Rev., 1971, 7, 81 CrossRef CAS.
- M. M. Keypour, F. Forouzandeh, R. Azadbakht, J. Khanabadi and M. A. Moghadam, J. Mol. Struct., 2021, 1232, 130024 CrossRef CAS.
- A. Tafazzoli, H. Keypour, S. H. M. Farida, Z. Ahmadvand and R. W. Gable, J. Mol. Struct., 2023, 1276, 134770 CrossRef CAS.
-
C. K. Johnson and M. N. Burnett, ORTEPIII. Report ORNL-6895, Oak Ridge National Laboratory, Tennessee, USA, 1996 Search PubMed.
-
M. Llunell, D. Casanova, J. Cirera, J. Bofill, P. Alemany, S. Alvarez, M. Pinsky and D. Avnir, SHAPE v. 2.1. Program for the Calculation of Continuous Shape Measures of Polygonal and Polyhedral Molecular Fragments, University of Barcelona, Barcelona, Spain, 2013 Search PubMed.
- E. Cremades, J. Echeverría and S. Alvarez, Chem.–Eur. J., 2010, 34, 10380 CrossRef PubMed.
- C. R. Groom, I. J. Bruno, M. P. Lightfoot and S. C. Ward, Acta Crystallogr., 2016, B72, 171–179 Search PubMed.
- Y. Peng, T. Bodenstein, K. Fink, V. Mereacre, C. E. Anson and A. K. Powell, Phys. Chem. Chem. Phys., 2016, 18, 30135 RSC.
- H. Keypour, M. Mahmoudabadi, A. Shooshtari, M. Bayat, F. Mohsenzadeh and R. W. Gable, J. Mol. Struct., 2018, 1155, 196 CrossRef CAS.
- H. Keypour, M. T. Rezaei, M. Jamshidi, S. H. Moazzami Farida and R. Karamian, Inorg. Chem. Commun., 2021, 125C, 108443 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 2109477. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d3ra00153a |
|
| This journal is © The Royal Society of Chemistry 2023 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article *a,
Saadat
Hajari
a,
Fereshteh
yaghoobi
b,
Seyed Hamed
Moazzami Farida
c,
Masoud
Saadati
*a,
Saadat
Hajari
a,
Fereshteh
yaghoobi
b,
Seyed Hamed
Moazzami Farida
c,
Masoud
Saadati
 c and
Robert William
Gable
c and
Robert William
Gable
 d
d
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 102
102![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). 1H NMR (CDCl3–d6, ppm) δH = 2.55–2.86 (s, H), 6.58–7.88(Haromatic), 8.62 (Himini), 13.43 (OH). 1C NMR(CDCl3, ppm): δc: 55.8–57.8(Cpy), 115.8–149.8(Caromatic), 160.6(Cimini), 161.4(C–OH).
N). 1H NMR (CDCl3–d6, ppm) δH = 2.55–2.86 (s, H), 6.58–7.88(Haromatic), 8.62 (Himini), 13.43 (OH). 1C NMR(CDCl3, ppm): δc: 55.8–57.8(Cpy), 115.8–149.8(Caromatic), 160.6(Cimini), 161.4(C–OH).![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N).
N).![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). MS (EI): (m/z) = 533.19.
N). MS (EI): (m/z) = 533.19.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N).
N).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio. Elemental analysis, IR, mass, 1H and 13C NMR spectroscopy were used to characterize the formation of the ligand (L). The infrared spectrum shows a strong absorption band at ∼1618 cm−1, assigned to the C
2 ratio. Elemental analysis, IR, mass, 1H and 13C NMR spectroscopy were used to characterize the formation of the ligand (L). The infrared spectrum shows a strong absorption band at ∼1618 cm−1, assigned to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching vibration, and with the absence of bands due to C
N stretching vibration, and with the absence of bands due to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and NH2 stretching vibrations, indicates the formation of the Schiff-base ligand. The mass spectrum of the ligand shows the expected peak at 476.4 m/z. The Ni(II), Co(II) and Cu(II) complexes were prepared by direct reaction of the ligand and metal salts in a 1
O and NH2 stretching vibrations, indicates the formation of the Schiff-base ligand. The mass spectrum of the ligand shows the expected peak at 476.4 m/z. The Ni(II), Co(II) and Cu(II) complexes were prepared by direct reaction of the ligand and metal salts in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio in methanol and characterized by mass and IR spectroscopy and by elemental analysis. In the IR spectra the imine stretching bands found for the Ni(II), Co(II) and Cu(II) complexes, 1610 cm−1, 1611 cm−1 and 1606 cm−1, respectively, were shifted to lower frequency compared to the free ligand (1618 cm−1) consistent with the coordination of the imine nitrogen to the metal ions. Furthermore, there no absorption bands in the 3400–3200 cm−1 region, due O–H stretching bands, confirming that the deprotonation of the phenol had occurred. Considering the great similarity of the IR spectra of the complexes, it can be concluded that their structure is similar. Also, the calculated spectra of the ligand and complexes were almost identical with the experimental spectra (Fig. S1†–S8). The UV-visible absorption spectra of the ligand and its complexes were measured in 10−4 M DMSO solutions at room temperature. The intense bands 266 nm of the ligand, may be assigned to π → π* transitions of the azomethine. The other peak which has appeared in the 310 nm region of the ligand spectrum is attributed to n → π* transition. In the spectra of the related complexes, the bands due to the ligand (π → π* and n → π* transitions) are still pronounced with a small shift in position with respect to the corresponding bands in the free ligand. In addition, a charge transfer band at 400–437 nm has been also observed in the electronic spectra of the ligand and complexes. The band at 483–494 nm in electronic spectra of the complexes can be attributed to MLCT (Fig. S9–S12†).
1 ratio in methanol and characterized by mass and IR spectroscopy and by elemental analysis. In the IR spectra the imine stretching bands found for the Ni(II), Co(II) and Cu(II) complexes, 1610 cm−1, 1611 cm−1 and 1606 cm−1, respectively, were shifted to lower frequency compared to the free ligand (1618 cm−1) consistent with the coordination of the imine nitrogen to the metal ions. Furthermore, there no absorption bands in the 3400–3200 cm−1 region, due O–H stretching bands, confirming that the deprotonation of the phenol had occurred. Considering the great similarity of the IR spectra of the complexes, it can be concluded that their structure is similar. Also, the calculated spectra of the ligand and complexes were almost identical with the experimental spectra (Fig. S1†–S8). The UV-visible absorption spectra of the ligand and its complexes were measured in 10−4 M DMSO solutions at room temperature. The intense bands 266 nm of the ligand, may be assigned to π → π* transitions of the azomethine. The other peak which has appeared in the 310 nm region of the ligand spectrum is attributed to n → π* transition. In the spectra of the related complexes, the bands due to the ligand (π → π* and n → π* transitions) are still pronounced with a small shift in position with respect to the corresponding bands in the free ligand. In addition, a charge transfer band at 400–437 nm has been also observed in the electronic spectra of the ligand and complexes. The band at 483–494 nm in electronic spectra of the complexes can be attributed to MLCT (Fig. S9–S12†).
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N group while the signal at 161.4 ppm is assigned to the carbon attached to the OH group (Fig. S14†).
N group while the signal at 161.4 ppm is assigned to the carbon attached to the OH group (Fig. S14†).





