Open Access Article
Open Access ArticleMass spectrometry-based assays for assessing replicative bypass and repair of DNA alkylation in cells
Jiaxian Li
,
Zhihai Hu
,
Dandan Liu
* and
Pengcheng Wang
 *
*
Institute of Surface Analysis and Chemical Biology, University of Jinan, Jinan, Shandong 250022, P. R. China. E-mail: 202111100006@stu.ujn.edu.cn; ila_wangpc@ujn.edu.cn
First published on 22nd May 2023
Abstract
Endogenous metabolism and environmental exposure can give rise to DNA alkylation, which can elicit deleterious biological consequences. In the search for reliable and quantitative analytical methods to elucidate the impact of DNA alkylation on the flow of genetic information, mass spectrometry (MS) has attracted increasing attention, owing to its unambiguous determination of molecular mass. The MS-based assays obviate conventional colony-picking methods and Sanger sequencing procedures, and retained the high sensitivity of postlabeling methods. With the help of the CRISPR/Cas9 gene editing method, MS-based assays showed high potential in studying individual functions of repair proteins and translesion synthesis (TLS) polymerases in DNA replication. In this mini-review, we have summarized the development of MS-based competitive and replicative adduct bypass (CRAB) assays and their recent applications in assessing the impact of alkylation on DNA replication. With further development of MS instruments for high resolving power and high throughput, these assays should be generally applicable and efficient in quantitative measurement of the biological consequences and repair of other DNA lesions.
1. Introduction
DNA is intrinsically unstable and its integrity in cells is constantly challenged by various endogenous and exogenous chemicals, resulting in a plethora of DNA lesions.1–3 For instance, it can undergo spontaneous deamination and depurination under physiological conditions. In addition, DNA, as a poly-nucleophile, can react with many electrophile species, leading to covalently bound adducts.3,4 Among them, DNA alkylation constitutes a major type of DNA damage due to the ubiquitous presence of alkylating agents in the environment and within cells.5 If not repaired, the alkylated DNA may perturb the efficiency and fidelity of DNA replication and transcription, thereby compromising the flow of genetic information and conferring adverse human health consequences.5,6 Indeed, it has been demonstrated that DNA alkylation can be cytotoxic, teratogenic, and carcinogenic.7To counteract the DNA alkylation and reduce its subsequent deleterious effects, cells have equipped diverse cellular repair pathways to collectively modulate alkylation, such as direct removal, base-excision repair (BER), nucleotide-excision repair (NER), mismatch repair (MMR), homologous recombination (HR), and non-homologous end joining (NHEJ).1,2,4,5 In addition, the translesion DNA synthesis (TLS) pathway utilizes low-fidelity DNA polymerases to bypass lesions in case the replication machinery is stalled by alkylation blockage.8,9 However, thus far, the roles of specific TLS polymerase in mutagenesis still largely remain elusive. Understanding the replicative bypass of DNA alkylation and involvement of repair proteins as well as TLS polymerases necessitates the investigation of how this type of lesions perturbs the flow of genetic information in cells under various gene manipulation.
The synthesis and construction of site-specifically modified genomes facilitate pinpointing mutation spectra of specific lesions with accuracy and certainty.10,11 Conventionally, extensive colony picking and Sanger sequencing procedures are required to assess the biological consequences of lesions and elucidate the roles of DNA polymerases and repair proteins.10,12–14 However, the relatively small sample size may result in some mutations of low frequencies being overlooked.15 Recent years witnessed the advancement of mass spectrometry (MS) in investigating the occurrence and biological consequences of DNA lesions.16–18 Liquid chromatography-tandem mass spectrometry (LC-MS/MS) has been successfully employed as an efficient approach in assessing how DNA lesions compromise DNA replication in vitro and in vivo, which has been reviewed by others.17–19 Here, we place emphasis on recent discoveries and compare the replicative bypass of different types of alkyl lesions in cells.
2. MS-based assay for replication study in cells
With the availability of site-specifically modified oligodeoxyribonucleotides (ODN), Essigmann and co-workers introduced a lesion bypass and mutagenesis assay to assess how DNA replication is compromised by certain lesions.20,21 In that assay, the bypass efficiency and mutation frequency of a defined lesion can be quantitatively determined by restriction digestion and postlabeling. This assay is then further developed by Wang's lab and the resultant competitive and replicative adduct bypass (CRAB) assay has been employed to evaluate the genetic perturbations caused by DNA lesions during replication in E. coli as well as in mammalian cells.22–24As shown in Fig. 1a, in E. coli cells, the experimental procedure begins with the synthesis and construction of lesion-containing and lesion-free M13 phage genomes in addition to a competitor M13 genome which possesses three additional nucleotides. As transfection efficiency for each independent experiment is not consistent, the competitor genome is premixed with a lesion-containing or lesion-free genome and serves as an internal standard in the calculation of bypass efficiencies with the assumption that the bypass efficiency of the corresponding control (lesion-free) M13 genome is 100%. After co-transfection and replication in E. coli, the progeny M13 genomes are harvested and the region of interest containing either the initial lesion site or the lesion-free control, or the competitor is amplified by PCR. The resultant PCR products are digested by appropriate restriction enzymes and subjected to LC-MS/MS analysis (Fig. 1b). In addition, with postlabeling, the digested products can also be analyzed by polyacrylamide gel electrophoresis (PAGE). Furthermore, by switching the order of restriction enzyme addition, the lesion-situated strand or its complementary strand can be selectively labeled and four potential types of replication products can be resolved from each other with two electrophoreses (Fig. 1c).15,25–27 Note that TLS polymerases in E. coli cells can be induced due to SOS response, which is named after the distress signal “SOS” in the Morse alphabet.28 Thus, to investigate the roles of specific TLS polymerases, the E. coli cells are generally treated with UV irradiation to initiate the SOS response.28
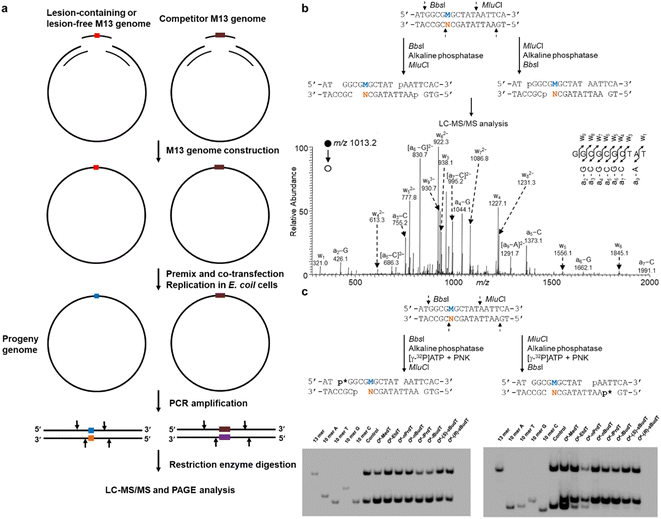 | ||
| Fig. 1 (a) Schematic illustration of experimental procedures of CRAB in E. coli cells. (b) Sequential restriction enzyme digestion for MS-based replication assay in E. coli cells (top) and a representative MS spectrum (bottom) showing the T → C mutation elicited by O4-methylthymidine (O4-MedT). (c) Sequential restriction enzyme digestion for the selective labeling of the strand initially bearing the lesion or the complementary strand, and the representative images of PAGE gel. The ‘M’ labeled in blue and ‘N’ labeled in orange indicate the nucleotide at the lesion-situated stand and lesion-complementary strand after replication, respectively. Reproduced from ref. 25 with permission from Oxford, copyright 2015. | ||
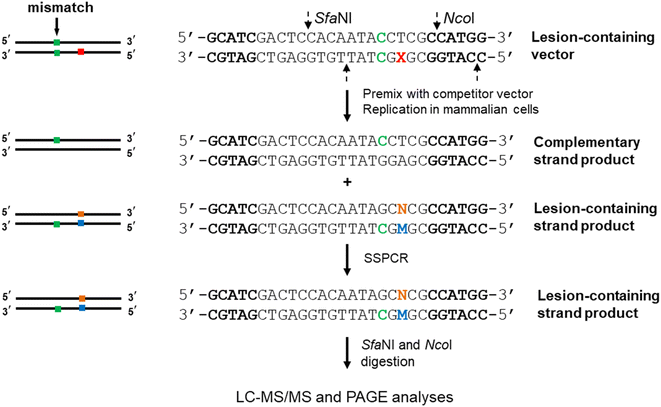
The CRAB assay also extended to mammalian cells by changing the single-stranded phage plasmid to a double-stranded shuttle vector that is capable of propagating in mammalian cells (cf. Fig. 2). Similarly, lesion-containing/lesion-free and competitor plasmids are constructed first via a gapped vector-based strategy. As the lesion may exert an impediment to replication, the non-lesion strand in the double-stranded plasmid usually is preferentially replicated, rendering it difficult to accurately determine the mutation frequencies and bypass efficiencies. As a result, a mismatch is introduced into the double-stranded plasmid containing the lesion site, enabling the independent assessment of the products arising from the replication of the lesion-bearing and lesion-free complementary strands. The progeny genomes of the plasmid are isolated from host cells, and the residual unreplicated plasmid is removed by DpnI digestion. The progeny plasmids are then amplified by PCR. The region of interest in the progeny plasmids is amplified with strand-specific PCR (SSPCR) so that the strand complementary to the lesion-containing can be selectively amplified. The resulting PCR products are restriction digested and subjected to LC-MS/MS and PAGE analyses.29–33 A summary of experimental procedures of CRAB in E. coli and mammalian cells are shown in Table 1.
| E. coli cells | (1) The lesion-containing, lesion-free, and competitor M13 genomes are constructed by incorporation of specific ODNs into the EcoRI-linearized M13 genome with two scaffolds |
| (2) The scaffolds and unligated M13 genomes are degraded with T4 DNA polymerase | |
| (3) The lesion-containing or lesion-free M13 genome is premixed with the competitor genome | |
| (4) The competent E. coli cells are transfected with the premixed M13 genomes | |
| (5) The transfected E. coli cells are cultured in Lysogeny Broth (LB) media | |
| (6) The culture media are centrifuged, and the cell pellets are harvested | |
| (7) The progeny M13 genomes are extracted | |
| (8) The region of interest in the purified single-stranded M13 genome is PCR amplified using two primers | |
| (9) The PCR products are digested with sequential restriction enzymes | |
| (10) The resultant digestion mixture is subjected to LC-MS analysis and postlabeling for PAGE | |
| Mammalian cells | (1) The double-stranded shuttle vector is nicked with Nt.BstNBI to produce a gapped vector |
| (2) The gap is filled with lesion-containing, lesion-free, or competitor ODN and ligated with T4 DNA ligase | |
| (3) The supercoiled plasmid is purified by agarose gel electrophoresis | |
| (4) The lesion-containing or lesion-free plasmid is premixed with the competitor plasmid | |
| (5) The overnight-cultured cells are transfected with the premixed plasmids | |
| (6) The transfected cells are harvested after 24 h, and the progenies of the plasmid are isolated | |
| (7) The residual unreplicated plasmid is removed by DpnI digestion | |
| (8) The region of interest in the progeny genomes is amplified with SSPCR | |
| (9) The PCR products are digested with sequential restriction enzymes | |
| (10) The resultant digestion mixture is subjected to LC-MS analysis and postlabeling for PAGE |
3. Applications of MS-based replication assays
In the past few years, a number of discoveries have been made using the abovementioned strategy to assess the effect of DNA alkylation on replication in cells.25–27,29–36 Here, we discuss the effects of alkyl lesions on replication in E. coli and mammalian cells and compare the difference between the major groove and minor groove lesions. In addition, alkylation on the phosphate backbone will also be described.3.1 Alkyl lesions in the major groove
The O4-alkylthymidine (O4-alkyldT) and O6-alkyl-2′-deoxyguanosine (O6-alkyldG) lesions represent two types of alkyl lesions on pyrimidine and purine bases. Systematic experiments have been performed to assess how the size of the alkyl group, from methyl to butyl, at the O4 position of thymine and O6 position of guanine (cf. Fig. 3) affects the efficiency and fidelity of DNA replication in E. coli cells.25,26 It is found that O4-alkyldT lesions direct exclusively misincorporation of 2′-deoxyguanosine opposite the lesion site,25 while O6-alkyldG lesions elicit only G → A mutation,26 which can be attributed to their distinct base-pairing properties. In addition, both types of lesions are not strong impediments to DNA replication in E. coli cells and the SOS-induced polymerases play redundant roles in bypassing these lesions, except for O4-sBudT, which exhibits moderate blockage to DNA replication and requires polymerase (Pol) V to facilitate efficient bypass.25,26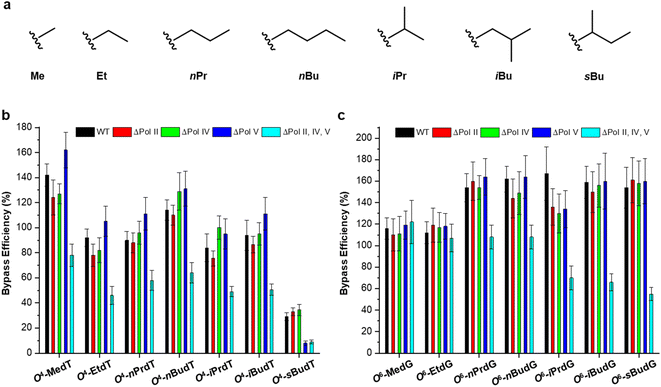 | ||
| Fig. 3 Chemical structures of the examined alkyl groups (a) and replicative bypass efficiencies of O2-alkyldT and O4-alkyldG lesions (b and c) in E. coli cells. Reproduced from ref. 25 with permission from Oxford, copyright 2015 and ref. 26 with permission from Elsevier, copyright 2018. | ||
Direct removal by O6-alkylguanine DNA alkyltransferase (MGMT) is an important repair pathway for O4-alkyldT and O6-alkyldG lesions.5 In E. coli cells, two types of MGMT are encoded, i.e., Ogt and Ada.37 Thus, their impact on the repair of these two types of lesions was also evaluated, and found that Ogt but not Ada is involved in repairing O4-alkyldT lesions25 while both of them can repair O6-alkyldG lesions with Ogt more efficiently than Ada.26
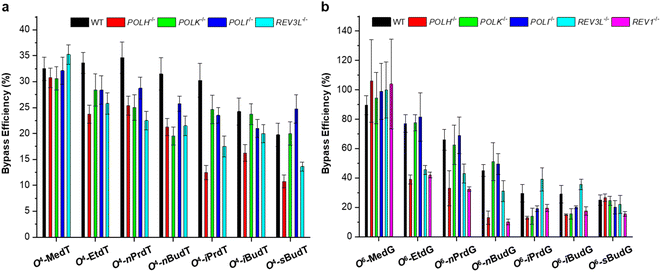
O4-alkyldT and O6-alkyldG lesions also elicit exclusively T → C and G → A mutation in mammalian cells, respectively. However, different from E. coli cells, these two types of lesions exert some blockage to DNA replication (cf. Fig. 4).29,30 It is shown that O4-alkyldT lesions are moderately blocking DNA replication, with bypass efficiencies ranging from 20% to 33%. For O6-alkyldG lesions, they also show moderate impediment to DNA replication with the exception of O6-MedG. The DNA adduct, 4-(3-pyridyl)-4-oxobut-1-yl (POB), was also studied, which is generated by tobacco-specific nitrosamine.31 It was found that while O4-POBdT still directs only T → C mutation, O6-POBdG elicited primarily the G → A transition (∼75%) together with a low frequency of the G → T transversion (∼3%). In addition, O6-POBdG exerts a higher blockage effect on DNA replication compared to O4-POBdT.31
 | ||
| Fig. 4 Replicative bypass efficiencies of O4-alkyldT (a) and O6-alkyl-dG lesions (b) in mammalian cells. Reproduced from ref. 29 with permission from Oxford, copyright 2016 and ref. 30 with permission from Elsevier, copyright 2019. | ||
The roles of repair proteins and TLS polymerases in replicative bypass and mutagenesis were also investigated in mammalian cells. For O4-alkyldT lesions, deficiency in Pol η or Pol ζ, but not Pol κ or Pol ι, leads to pronounced drops in bypass efficiencies for all the O4-alkyldT lesions. In addition, depletion of Pol η, Pol κ, or Pol ι does not significantly alter the T → C mutation for any of the O4-alkyldT lesions, with the exception that loss of Pol η results in significant decreases in T → C mutation for O4-EtdT and O4-nPrdT. Moreover, the depletion of Pol ζ results in significant drops in T → C mutation frequencies for all O4-alkyldT lesions except O4-MedT and O4-nBudT. For O6-alkyldG lesions, depletion of REV1 significantly decreases the bypass efficiency of all lesions except for O6-MedG. In addition, individual ablation of Pol η or Pol ζ confers a pronounced reduction in bypass efficiency for all straight-chain lesions. In terms of replication fidelity, Pol η, and Pol ζ are involved in the error-prone bypass of the straight-chain lesions, whereas Pol κ favors the accurate bypass of the branched-chain lesions. Moreover, it was found that MGMT is effective in removing the smaller alkyl groups from the O6 position of guanine, whereas repair of the branched-chain lesions relies on NER. Interestingly, the roles of TLS polymerases involved in replicative bypassing of these two types of lesions are similar to other major groove lesions, such as N7-guanine and N6-adenine cross-links.38,39 Several reports indicated that Pol η is required for efficient bypass of DNA adducts in the major groove.29,30,38,39
3.2 Alkyl lesions in the minor groove
Two representative minor-groove alkyl lesions were systematically investigated using the CRAB assay, i.e., O2-alkylthymidine (O2-alkyldT) and N2-alkyl-2′-deoxyguanosine (N2-alkyldG).27,32,33 These two types of lesions direct promiscuous mutations, but the results are not consistent in E. coli and in mammalian cells. In E. coli cells, all four nucleotides can be inserted opposite the O2-alkyldT lesion site,40 while T → C mutation is absent in mammalian cells.32 For N2-alkyldG, it is nonmutagenic in E. coli cells,27 while it directs G → A and G → C mutations in mammalian cells.33As shown in Fig. 5, both lesions are strong impediments to DNA replication in mammalian cells,32,33 which is consistent with the results obtained in E. coli cells.27,40 In addition, the bypass efficiencies decrease with the increase in the size of the alkyl group. The involvement of TLS polymerases was also evaluated by conducting experiments in isogenic cells engineered with CRISPR/Cas9 genome editing method. It was found that the replicative bypass of O2-alkyldT lesions requires Pol η and Pol ζ, which resulted in higher mutation frequencies. In contrast, Rev1, Pol ι, and Pol κ are involved in bypassing N2-alkyldG lesions, and ablation of these three polymerases elicits substantial frequencies of G → A transition and G → T transversion. Interestingly, further depletion of Pol ζ in Pol κ- or Pol ι-deficient cells elevated G → A and G → T mutations but decreased bypass efficiencies. Different from major-groove lesions, the roles of TLS polymerases are lesion-specific in bypassing adducts in minor groove.
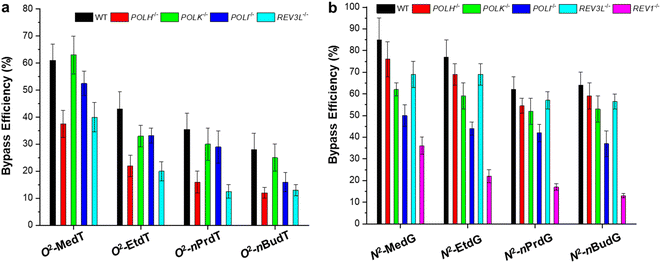 | ||
| Fig. 5 Replicative bypass efficiencies of O2-alkyldT (a) and N2-alkyl-dG lesions (b) in mammalian cells. Reproduced from ref. 32 with permission from Elsevier, copyright 2018 and ref. 33 with permission from American Chemical Society, copyright 2019. | ||
3.3 Alkyl lesions on the phosphate backbone
Apart from alkylation on nucleobases, the oxygen atoms of the internucleotide phosphate group can also be alkylated by alkylating agents and give rise to phosphotriester adducts (PTEs),41 which are known to be persistent in mammalian cells.42 As the phosphotriester contains a chiral phosphorus center, two configurations (Sp or Rp) can be formed depending on the non-carbon-bound oxygen atom that is alkylated (cf. Fig. 6).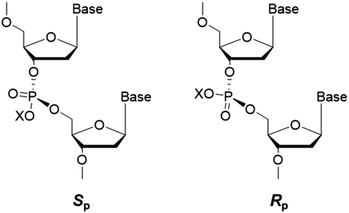 | ||
| Fig. 6 Sp and Rp diastereomers of alkyl phosphotriester residues in DNA. The ‘X’ indicates the substituted alkyl group. | ||
Using the CRAB assay, Wu et al. found that the Sp diastereomer of the alkyl-PTE lesions could be efficiently bypassed in E. coli cells, while the Rp counterparts moderately impede DNA replication.34 In addition, the bypass of the alkyl-PTE lesions does not require any of the three TLS polymerases (Pol II, Pol IV, and Pol V) and is not modulated by SOS induction. It was found that at the flanking TT dinucleotide site, Sp-Me-PTE induces TT → GT and TT → GC mutations, and the induction of these mutations requires Ada protein. Additionally, the mutation frequencies are not affected by individual depletion of TLS polymerases, though simultaneous ablation of all three polymerases results in a slight decrease in TT → GT mutation.34
The replication bypass and fidelity of alkyl-PTE lesions were also investigated at different flanking dinucleotide sites, i.e., XT and TX (X = A, C, or G).35 It was shown that DNA replication was highly efficient, and the replication products contain 85–90% AT and 5–10% TG for Sp-Me-PTE in the sequence contexts of 5′-XT-3′, largely independent of the flanking base. In addition, the Rp diastereomer of Me-PTEs at XT sites and both diastereomers of Me-PTEs at TX sites exhibited error-free replicative bypass. Moreover, both diastereomers of nBu-PTEs at TX and XT sites are non-mutagenic.35 In another report, both diastereomers of POB-PTEs exhibit low blocking effects on DNA replication with bypass efficiencies higher than 80%, and neither diastereomer is mutagenic.36
4. Conclusions and perspectives
The advances in MS instrumentation have rendered it a viable and efficient tool for elucidating the biological consequences of DNA adducts.16,18,19,43 In this review, we summarized the development of MS-based strategies for assessing replicative bypass and mutagenesis of alkylated DNA lesions, and recent findings in E. coli and mammalian cells. These MS-based assays demonstrate comparable sensitivity to restriction endonuclease and postlabeling (REAP) assay and are highly reproducible that a mutation frequency of around 0.5% can be reliably determined with a standard deviation being ∼0.1%.20,21,24,44 As all these assays use the entire progeny genomes and obviate the tedious colony counting, their quantitative results would be more reliable. The better reliability has been manifested by the low G → T transversion conferred by O6-POBdG which was detected using MS-based assays but absent in the conventional method.15 It should be noted that PAGE (cf. Fig. 1c), though can distinguish the four potential types of replication products as aforementioned, requires a specific sequence of lesion-containing strands. Hence, the PAGE analysis lacks the ability to determine the involvement of flanking base. However, these MS-based assays still cannot fully assess the impact of sequence context on DNA replication, as the pool of ODN sequences is confined by the restriction sites recognized by enzymes. Another limitation of MS-based assays is that these assays generally cannot provide kinetic information about nucleotide insertion and the interaction between DNA and proteins. Therefore, methods, such as surface plasmon resonance45 and biolayer interferometry,46 that can reveal the binding affinity and kinetics between lesions and repair proteins/polymerases would complement MS-based assays.Although DNA alkylation is generally considered deleterious, alkylating agents are a class of widely used drugs in chemotherapy.5,47 Understanding how alkylating agents exert their therapeutic effects and how cells respond to chemotherapeutic alkylating agents would be important for drug discovery and development.5 In addition, it will provide insight into the proper use of chemotherapeutic drugs. All these necessitate elucidation of the roles of repair proteins and TLS polymerases in coping with alkyl DNA lesions. MS displays high potential in these applications, owing to its unambiguous quantification. Moreover, by utilizing the CRISPR/Cas9 genome editing method,48–50 manipulating single and even multiple genes involved in DNA replicative bypass becomes more facile and proficient, enabling the investigation of individual or synergistic roles of repair proteins and TLS polymerases. Furthermore, MS-based assays have also been adopted in the evaluation of the impact of DNA lesions on transcription, rendering them more versatile in analytical method development.51–54
Owing to the importance of understanding the biological consequence of DNA lesions, the development, and applications of lesion bypass and mutagenesis assays have attracted numerous attention.10,55 Unfortunately, the CRAB assays described here are of relatively low throughput and the analysis costs hours even days to complete. In addition, it cannot provide genome-wide information on DNA adducts. Thus, many other biological assays for the determination of DNA lesion bypass and mutation have been developed, such as chromatin immunoprecipitation (ChIP)-based assay56 and next-generation sequencing (NGS)-based assay.44 For instance, the involvement of polymerases ν and θ in the replicative bypass of 8-oxo-7,8-dihydroguanine was assessed with high-throughput, using the construction of a shuttle vector-based NGS assay.57 However, MS-based assays can still be a complementary tool to pinpoint the exact type of mutations including frameshift mutations.43,58 As the mutagenesis caused by environmental exposure will be more complicated in the human body,55,59 MS with high resolving power is indispensable in quantitative and/or qualitative measurement. Therefore, combining MS and other analytical methods would be an effective way to circumvent the above-mentioned limitations. In addition, improvement of sample preparation techniques, development of programmable software, and application of high-resolution MS (e.g., Orbitrap and time-of-flight) would be of top priority. With the development of online separation and automatic injection, we envision that high throughput and unattended analysis can be achieved in the future.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank the financial support from the Natural Science Foundation of China (No. 21906065), and the Shandong Provincial Grant for the Talent-Leading Teams.References
- N. Chatterjee and G. C. Walker, Environ. Mol. Mutagen., 2017, 58, 235–263 CrossRef CAS PubMed.
- E. C. Friedberg, G. C. Walker, W. Siede, R. D. Wood, R. A. Schultz and T. Ellenberger, DNA Repair and Mutagenesis, ASM Press, Washington, D.C., 2005 Search PubMed.
- K. S. Gates, Chem. Res. Toxicol., 2009, 22, 1747–1760 Search PubMed.
- N. C. Bauer, A. H. Corbett and P. W. Doetsch, Nucleic Acids Res., 2015, 43, 10083–10101 CAS.
- D. Fu, J. A. Calvo and L. D. Samson, Nat. Rev. Cancer, 2012, 12, 104–120 CrossRef CAS PubMed.
- Y. Yu, P. Wang, Y. Cui and Y. Wang, Anal. Chem., 2018, 90, 556–576 CrossRef CAS PubMed.
- L. P. Bignold, Anticancer Res., 2006, 26, 1327–1336 CAS.
- X. Ma, T.-S. Tang and C. Guo, Environ. Mol. Mutagen., 2020, 61, 680–692 CrossRef CAS PubMed.
- S. Fujii and R. P. Fuchs, Microbiol. Mol. Biol. Rev., 2020, 84, e00002–e00020 CrossRef PubMed.
- A. K. Basu and J. M. Essigmann, Chem. Res. Toxicol., 2022, 35, 1655–1675 Search PubMed.
- H. O. Smith, C. A. Hutchison III, C. Pfannkoch and J. C. Venter, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 15440–15445 CrossRef CAS PubMed.
- J. C. Delaney and J. M. Essigmann, Chem. Res. Toxicol., 2008, 21, 232–252 Search PubMed.
- O. Ziv, N. Diamant, S. Shachar, A. Hendel and Z. Livneh, Methods Mol. Biol., 2012, 920, 529–542 CrossRef CAS PubMed.
- A. L. Livingston, V. L. O'Shea, T. Kim, E. T. Kool and S. S. David, Nat. Chem. Biol., 2008, 4, 51–58 CrossRef CAS PubMed.
- P. Wang, J. Leng and Y. Wang, J. Biol. Chem., 2019, 294, 3899–3908 CrossRef CAS PubMed.
- S. Liu and Y. Wang, Chem. Soc. Rev., 2015, 44, 7829–7854 RSC.
- C. You and Y. Wang, Acc. Chem. Res., 2016, 49, 205–213 CrossRef CAS PubMed.
- M. Dizdaroglu, E. Coskun and P. Jaruga, Free Radical Res., 2015, 49, 525–548 CrossRef CAS PubMed.
- J. Guo and R. J. Turesky, High-Throughput, 2019, 8, 13 CrossRef CAS PubMed.
- J. C. Delaney, L. Smeester, C. Wong, L. E. Frick, K. Taghizadeh, J. S. Wishnok, C. L. Drennan, L. D. Samson and J. M. Essigmann, Nat. Struct. Mol. Biol., 2005, 12, 855–860 CrossRef CAS PubMed.
- J. C. Delaney and J. M. Essigmann, Methods Enzymol., 2006, 408, 1–15 CAS.
- H. Hong, H. Cao and Y. Wang, Nucleic Acids Res., 2007, 35, 7118–7127 CrossRef CAS PubMed.
- B. Yuan, T. R. O'Connor and Y. Wang, ACS Chem. Biol., 2010, 5, 1021–1027 CrossRef CAS PubMed.
- B. Yuan, H. Cao, Y. Jiang, H. Hong and Y. Wang, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 8679–8684 CrossRef CAS PubMed.
- P. Wang, N. J. Amato, Q. Zhai and Y. Wang, Nucleic Acids Res., 2015, 43, 10795–10803 CrossRef CAS PubMed.
- P. Wang and Y. Wang, J. Biol. Chem., 2018, 293, 15033–15042 CrossRef CAS PubMed.
- Y. Wang, J. Wu, J. Wu and Y. Wang, Chem. Res. Toxicol., 2021, 34, 695–698 Search PubMed.
- C. Janion, Int. J. Biol. Sci., 2008, 4, 338–344 Search PubMed.
- J. Wu, L. Li, P. Wang, C. You, N. L. Williams and Y. Wang, Nucleic Acids Res., 2016, 44, 9256–9265 CAS.
- H. Du, P. Wang, L. Li and Y. Wang, J. Biol. Chem., 2019, 294, 11144–11153 CrossRef CAS PubMed.
- H. Du, J. Leng, P. Wang, L. Li and Y. Wang, J. Biol. Chem., 2018, 293, 11100–11108 CrossRef CAS PubMed.
- J. Wu, P. Wang, L. Li, C. You and Y. Wang, J. Biol. Chem., 2018, 293, 8638–8644 CrossRef CAS PubMed.
- J. Wu, H. Du, L. Li, N. E. Price, X. Liu and Y. Wang, ACS Chem. Biol., 2019, 14, 1708–1716 CrossRef CAS PubMed.
- J. Wu, P. Wang and Y. Wang, Nucleic Acids Res., 2018, 46, 4013–4021 CrossRef CAS PubMed.
- J. Wu, J. Yuan, N. E. Price and Y. Wang, J. Biol. Chem., 2020, 295, 8775–8783 CrossRef CAS PubMed.
- J. Wu and Y. Wang, Chem. Res. Toxicol., 2020, 33, 308–311 Search PubMed.
- M. C. Wilkinson, P. M. Potter, L. Cawkwell, P. Georgiadis, D. Patel, P. F. Swann and G. P. Margison, Nucleic Acids Res., 1989, 17, 8475–8484 CrossRef CAS PubMed.
- S. Wickramaratne, S. Ji, S. Mukherjee, Y. Su, M. G. Pence, L. Lior-Hoffmann, I. Fu, S. Broyde, F. P. Guengerich, M. Distefano, O. D. Scharer, Y. Y. Sham and N. Tretyakova, J. Biol. Chem., 2016, 291, 23589–23603 CrossRef CAS PubMed.
- P. P. Ghodke, G. Gonzalez-Vasquez, H. Wang, K. M. Johnson, C. A. Sedgeman and F. P. Guengerich, J. Biol. Chem., 2021, 296, 100444 CrossRef CAS PubMed.
- Q. Zhai, P. Wang, Q. Cai and Y. Wang, Nucleic Acids Res., 2014, 42, 10529–10537 CrossRef CAS PubMed.
- G. D. D. Jones, R. C. Le Pla and P. B. Farmer, Mutagenesis, 2010, 25, 3–16 CrossRef CAS PubMed.
- L. Den Engelse, G. J. Menkveld, R. J. De Brij and A. D. Tates, Carcinogenesis, 1986, 7, 393–403 CrossRef CAS PubMed.
- J. R. Edwards, H. Ruparel and J. Ju, Mutat. Res., 2005, 573, 3–12 CrossRef CAS PubMed.
- S.-c. Chang, B. I. Fedeles, J. Wu, J. C. Delaney, D. Li, L. Zhao, P. P. Christov, E. Yau, V. Singh, M. Jost, C. L. Drennan, L. J. Marnett, C. J. Rizzo, S. S. Levine, F. P. Guengerich and J. M. Essigmann, Nucleic Acids Res., 2015, 43, 5489–5500 CrossRef CAS PubMed.
- P. G. Stockley and B. Persson, Methods Mol. Biol., 2009, 543, 653–669 CrossRef CAS PubMed.
- J. K. Barrows and M. W. Van Dyke, PLoS One, 2022, 17, e0263322 CrossRef CAS PubMed.
- Y. Peng and H. Pei, J. Zhejiang Univ., Sci., B, 2021, 22, 47–62 CrossRef CAS PubMed.
- Y. Ma, L. Zhang and X. Huang, FEBS J., 2014, 281, 5186–5193 CrossRef CAS PubMed.
- D. Gupta, O. Bhattacharjee, D. Mandal, M. K. Sen, D. Dey, A. Dasgupta, T. A. Kazi, R. Gupta, S. Sinharoy, K. Acharya, D. Chattopadhyay, V. Ravichandiran, S. Roy and D. Ghosh, Life Sci., 2019, 232, 116636 CrossRef CAS PubMed.
- P. D. Hsu, E. S. Lander and F. Zhang, Cell, 2014, 157, 1262–1278 CrossRef CAS PubMed.
- C. You, X. Dai, B. Yuan, J. Wang, J. Wang, P. J. Brooks, L. J. Niedernhofer and Y. Wang, Nat. Chem. Biol., 2012, 8, 817–822 CrossRef CAS PubMed.
- X. He, P. Wang and Y. Wang, Anal. Chem., 2021, 93, 1161–1169 CrossRef CAS PubMed.
- Y. Tan, S. Guo, J. Wu, H. Du, L. Li, C. You and Y. Wang, J. Am. Chem. Soc., 2021, 143, 16197–16205 CrossRef CAS PubMed.
- Y. Tan, J. Wu, G. Clabaugh, L. Li, H. Du and Y. Wang, DNA, 2022, 2, 221–230 CrossRef PubMed.
- D. H. Phillips, DNA Repair, 2018, 71, 6–11 CrossRef CAS PubMed.
- D. Wu, A. Banerjee, S. Cai, N. Li, C. Han, X. Bai, J. Zhang and Q.-E. Wang, DNA Repair, 2021, 108, 103230 CrossRef CAS PubMed.
- X. Zheng, D. Chen, Y. Zhao, X. Dai and C. You, Anal. Chem., 2022, 94, 11627–11632 CrossRef CAS PubMed.
- P. Kowalczyk, J. M. Ciesla, M. Komisarski, J. T. Kusmierek and B. Tudek, Mutat. Res., 2004, 550, 33–48 CrossRef CAS PubMed.
- N. Saini and D. A. Gordenin, Environ. Mol. Mutagen., 2018, 59, 672–686 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |