Open Access Article
Open Access ArticleAza-Michael promoted glycoconjugation of PETIM dendrimers and selectivity in mycobacterial growth inhibitions†
Biswajit Sarkara,
Avisek Mahapa*bc,
Kalyan Deya,
Rakshit Manhasc,
Dipankar Chatterji*b and
Narayanaswamy Jayaraman *a
*a
aDepartment of Organic Chemistry, Indian Institute of Science, Bangalore 560 012, India. E-mail: jayaraman@iisc.ac.in
bMolecular Biophysics Unit, Indian Institute of Science, Bangalore 560 012, India. E-mail: dipankar@iisc.ac.in
cInfectious Disease Department, CSIR-Indian Institute of Integrative Medicine, Jammu-180001, India. E-mail: avisek.mahapa@gmail.com
First published on 3rd February 2023
Abstract
The benign nature of aza-Michael addition reaction in aqueous solutions is demonstrated herein to conduct a direct glycoconjugation of amine-terminated poly(ether imine) (PETIM) dendrimers. Zero to three generations of dendrimers, possessing up to 16 amine functionalities at their peripheries, undergo aza-Michael reaction with unsaturated sugar vinyl sulfoxide in aq. MeOH solutions and afford the corresponding dendrimers modified with multiple glycosyl moieties at the periphery. First order kinetics of the glycoconjugation is monitored at varying temperatures and the rate constants are observed to be 60–508 s−1, for zero and first generation dendrimers. The antibacterial effects of amine-terminated dendrimers and the corresponding glycoconjugates are studied across Gram-positive, Gram-negative and acid-fast bacteria. Among the species, M. smegmatis and M. tuberculosis showed the greatest growth inhibition effect at micromolar concentrations, for the native amine-terminated and the corresponding glycoconjugated dendrimers. Quantitative assays are performed to adjudge the inhibition efficacies of dendrimers and the glycoconjugates. Selectivity to inhibit M. smegmatis and M. tuberculosis growth, and minimal effects on other bacterial species by dendrimers and glycoconjugates are emphasized.
Introduction
Many biological processes depend on the post-translational modifications of proteins and lipids with carbohydrate moieties.1–3 Cell surface modifications,4 assembly of N-glycoproteins,5 evolution of glycoarrays,6 and development of drugs and vaccines are enabled by glycoconjugations with appropriate sugar moieties.7–11 As a result, robust conjugation methodologies have been developed, which include amidations, thiol–ene, urea formations, Michael addition, click reactions and more.12–21 In these instances, a reactive moiety is tethered to the sugar derivative so as to enable glycoconjugation with the chosen receptors. We demonstrated recently a glycoconjugation methodology which relies on the aza-Michael addition reaction of a number of oxygen, nitrogen, carbon and sulfur nucleophiles with a sugar vinyl sulfoxide under aqueous conditions.22 The reactions are conducted under aqueous milieu, as a result, glycoconjugation on to proteins becomes feasible. The side chain amine functionality of lysine residues in proteins reacts with sugar vinyl sulfoxide in such glycoconjugations. In advancing further, the utility of the methodology for glycoconjugation of dendritic macromolecules is undertaken herein.Synthesis of monodispersed dendritic macromolecules requires tight control over their molecular weights and the number of chain ends.23–25 Interfacing dendrimers with diverse biological, chemical and materials entities is one of the major advancements in contemporary macromolecular research.26–29 Carbohydrate-binding proteins, namely, lectins and antibodies are the first point of attachment of carbohydrate ligands on to biologically-relevant receptors.30 Following these clues, intense efforts were undertaken in the glycoconjugation of dendrimers. In this context, multitude of developments also include exploiting efficacies of glycodendrimers to study multivalent carbohydrate–protein interactions, inhibitions of enzyme functions, in gene and drug delivery applications.31,32
An area of intense investigations relates to the role of the cationic dendrimers as a new class of antibacterial agents.33–45 When a microbial infection is treated with traditional antibiotics at sub-lethal concentrations or for a long time, it inversely induces resistance to the pathogens, prompting efforts to mitigate the multidrug-resistant strains. Extensively drug-resistant M. tuberculosis, Methicillin-resistant S. aureus, beta-lactamase-producing E. coli and P. aeruginosa are prominent among multidrug-resistant (MDR) strains, associated with healthcare infections. Effective antibacterial regimens become an important query, in efforts to address such disease-causing bacterial infections to hosts.
Studies of poly(lysine),43,44 poly(amido amine),45 poly(propylene imine),39,40 poly(phosphane)35 and carbosilane dendrimers38 show that the antibacterial properties of these dendrimers arise due to the cationic sites present within the structure. The antibacterial efficacies of such dendrimer also enhance further upon conjugation with small molecule inhibitors. The series of poly(ether imine) (PETIM) dendrimers were assessed earlier for their cytotoxicity and biological effects, in mammalian cell cultures, from which non-toxicity profiles were observed for the dendrimers possessing hydroxyl functionalities at their peripheries.46–49 This series of dendrimers also possesses varied functional moieties, namely, amines and carboxylic acids at the peripheries. The amine functionalities afford cationic sites to the dendrimer, suitable to interfere with the cell wall structures and functions. Specific relevance herein is the membrane cell wall biosynthesis in a bacterium. Controlling the biosynthesis of cell wall components assumes a greater significance to mitigate bacterial pathogenicity to host cells. The presence of multiple chain ends in dendritic macromolecules attracted efforts to identify potent antimicrobial properties, either in the native or chain end-functionalized forms.50 The clustering of functional groups present at the peripheries of dendrimers leads to specific and non-specific interactions of the designed dendrimer agents with the microbial targets. The dendrimers with quaternary ammonium groups are effective against Gram-negative E. coli and Gram-positive S. aureus strains. Concerning dendrimers functionalized with sugars, it was demonstrated early by Magnusson and co-workers that glycoconjugation of dendrimers with di- to tetravalent galabiosides led to more effective hemagglutination inhibition of S. suis than the monomer galabioside.51 PAMAM dendrimers are by far the most studied and are known to engage in membrane disruption against Gram-positive and Gram-negative pathogens against opportunistic pathogens.52 An increasing interest in the antimicrobial properties of dendrimers arises in order to develop more effective dendrimer-based antimicrobial agents against MDR strains.
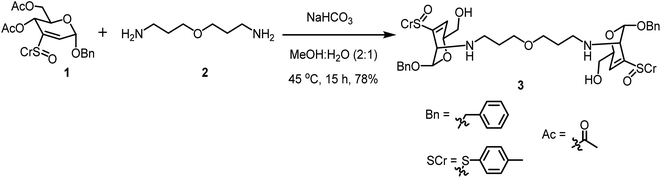
We report herein a glycoconjugation method, where the modified sugar moieties are covalently-linked to amine functionalities present at the peripheries of zero to three generations of PETIM dendrimers. Following synthesis, the native, amine-terminated dendrimers and the corresponding glycoconjugates are studied in order to evaluate their efficacies on the growth of varied planktonic bacterial strains. Details are presented herein.
Results
Synthesis
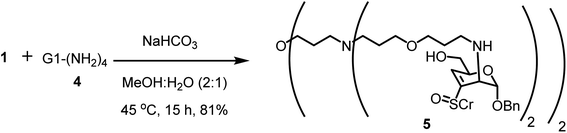
One-step glycoconjugation of amine-terminated PETIM dendrimers53 in an aqueous medium, namely, aza-Michael addition, is described herein. Zero, first, second and third generation dendrimers possess 2, 4, 8 and 16 peripheral amine moieties, respectively, and the amine functionality acts as the aza-Michael donor. Benzyl 4,6-di-O-acetyl-2,3-dideoxy-3-(p-tolylsulfinyl)-α-D-erythro-hex-2-enopyranoside (1), possessing 2,3-unsaturated vinyl sulfoxide functionality acted as the Michael acceptor. The reaction of sugar vinyl sulfoxide 1 (ref. 54) with the bis-amine 2 (ref. 55) is shown in Scheme 1. The reaction was conducted in an aqueous solution, under basic condition (pH ∼ 7.5–8) and the reaction afforded the bis-conjugated product 3. Three molar equivalents of sulfoxide 1 were used to one molar equivalent of acceptor 2 and the reaction occurring at 45 °C was identified to be optimal for the glycoconjugation. Insolubility of 1 in water necessitated MeOH for solubilization and the reaction was conducted at 45 °C. Following column purification of the reaction mixture, bis-glycoconjugate 3 was secured in a good yield. As observed earlier, an addition–elimination occurred to afford the 3,4-unsaturated vinyl sulfoxide. The basic pH of the reaction also led to hydrolysis of the acetate moiety, leading to product 3 with a primary hydroxyl group. The addition occurred from the axial face to afford the manno-configured product, as observed generally for the primary amine additions.54 Olefinic proton at C-2 carbon of 1 at 6.80 ppm disappeared and the olefinic proton at C-4 of 3 appeared at 6.59 ppm. Further, the anomeric H-1 in 1 appearing at 5.4 ppm up-field shifted to 5.10 ppm in 3, the JH-1–H-2 was 2.6 Hz, implying a trans-diequatorial disposition of H-1–H-2 protons in 3. The dendrimer methylene protons in 3 appeared at 3.32, 2.44 and 1.55 ppm and shifted up-field by ∼0.2–0.3 ppm after conjugation with sulfoxide 1. The anomeric resonance shifted down-field to 94.2 ppm for product 3, whereas that for vinyl sulfoxide 1 appeared at 93.7 ppm and 93.5 ppm. The methylene carbon adjacent to amine moiety in 3 appeared at 54.7 ppm, that for bis-amine 1 appeared at 47.5 ppm. The mass spectrum of 3 showed molecular ion peak as [M + H+] species, appearing at m/z 845.3532 (calcd mass m/z 845.3505).First generation PETIM dendrimer 4 (ref. 53) with 4 peripheral amine moieties was subjected to glycoconjugation with vinyl sulfoxide 1 (Scheme 2). The reaction was conducted with one-and-half molar equivalent of 1 per amine moiety in tetra-amine 4. Following reaction over 15 h, the solution was evaporated, suspended in water and triturated with EtOAc several times in order to remove excess of vinyl sulfoxide. The product thus obtained was analysed to determine the extent of conjugation of sugar moiety with the dendrimer 5. MALDI-TOF mass analysis revealed the molecular ion peak as [M + 2H+] adduct at m/z 2018.380 (calcd m/z 2018.9737), corresponding to complete conjugation at all the available primary amine moieties (Fig. S8†). In addition, multiple peaks due to the presence of acetate moiety at C-6 carbon were also seen. Further the conjugation was ascertained by comparison of the resonances of aromatic moiety with methylene moiety adjacent to the amine functionality in the 1H NMR spectrum of 5. The NMR spectra were recorded in CD3OD. The resonances appeared broad and in sets of resonances, presumably due to sulfoxide chirality and the resulting diastereomeric nature of the product. The methylene moiety adjacent to primary amine in 4 appeared 2.79 ppm. Whereas in the glycoconjugated product 5, methylene moieties adjacent to the amine sites appeared at 2.67–3.00 ppm. The H-4 olefinic proton and the H-1 proton of 5 were found to resonate at 6.54 and 5.13 ppm, respectively, in 1H NMR spectra. The peak corresponding to anomeric carbon resonated at 95.3 ppm in 13C NMR spectra. These newly observed peaks in 1H and 13C NMR spectroscopy ascertained the formation of conjugated dendrimer 5.
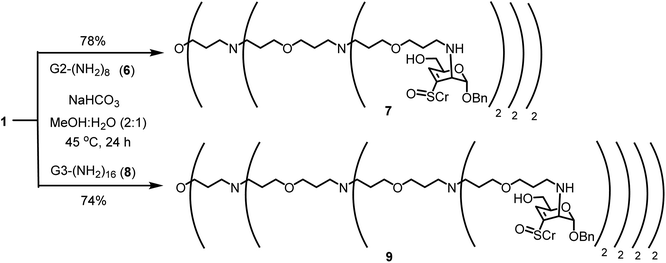
Glycoconjugations of G2-(NH2)8 and G3-(NH2)16,53 possessing 8 and 16 amine moieties, respectively, were conducted (Scheme 3). The reactions were conducted in aq. MeOH for 24 h at 45 °C, in both the reactions. Removal of solvents and trituration with EtOAc afforded glycoconjugates 7 and 9, in 78% and 74%, respectively.
NMR spectra of these conjugated products 7 and 9 were recorded in CD3OD and the resonances appeared broad. Major observations were that: (i) the acetate moiety at C-6 position was susceptible to hydrolysis; (ii) with an exponential increase in the molecular weights of glycoconjugates 7 and 9 without acetate moiety, 4362.19 and 9052.65 g mol−1, respectively, gradual line-broadening and reduced signal-to-noise ratio of the peaks occurred in 1H and 13C NMR spectra and (iii) mass spectra of these derivatives could not be secured by ESI-MS and MALDI-TOF mass spectrometries. Upon conjugation, the H-4, H-1 protons in 7 and 9 appeared at 6.59, 5.30 and 6.60, 5.19 ppm in 1H NMR spectra, respectively. Whereas, the anomeric carbon appeared at 93.9 and 95.4 ppm in 13C NMR spectra, respectively. For these higher generations of dendrimers, the extent of conjugation was an average, calculated from the integral values of 1H NMR spectra. In the case of second-generation amine 6, the methylene moieties adjacent to amines appeared at 2.94 and 2.60 ppm, as apparent triplets. For the conjugation product 7, these and tolyl-CH3 protons appeared 2.21–2.85 ppm, as broad resonances. The aryl moieties of sugar component appeared at 7.38–7.48 ppm and 7.68–7.73 ppm. Integration of these aryl moiety protons with that of the methylene moieties adjacent to amine sites and tolyl-CH3 protons enabled to adjudge the extent of conjugation. In this manner, a conjugation of 7 out of 8 sugar moieties was adjudged for the complete structure as given for 7 in Scheme 3. A similar analysis of the 1H NMR spectrum of product 9 was also undertaken. In third-generation dendrimer 8, methylene protons adjacent to amine sites appeared at 3.03 and 2.54 ppm, as broad peaks. A comparison of the integrations of these and tolyl-CH3 protons, 2.19–2.71 ppm and the aromatic moieties, 7.25–7.42 and 7.66–7.74, in 9 suggested a conjugation of 14 sugar moieties for the complete structure as given in Scheme 3. This incomplete conjugation also appeared to cause reduced resolutions of the NMR spectra in these glycoconjugates.
Determination of the kinetic constants
The kinetics of the conjugation was monitored, by 1H NMR spectroscopy, in a mixture of CD3OD and D2O. Disappearance of the methylene protons adjacent to primary amine of dendrimer scaffold was chosen to monitor the conjugation, occurring at 25, 35 and 45 °C, over 7 h. The conjugation led to a reduction in the intensity of this methylene resonance at ∼2.67 ppm in bis-amine 2, adjacent to primary amine moiety (–CH2NH2). Conjugation of G1 generation dendrimer also led to a reduction of the intensity of resonance at ∼2.56 ppm, corresponding to –CH2NH2 moiety. Due to overlapping resonances, kinetics of the conjugations in the case of G2 and G3 generation could not be conducted. 1H NMR data were collected for up to 7 h and the time course analysis was fit to a first-order kinetic equation and the rate constants of conjugation were calculated (Table 1). Rate of the reaction increased approximately twice for each 10 °C raise in the temperature. The rate constant was found to be higher for the first (4) generation dendrimer than bis-amine 2 (Table 1).| Dendrimer generation | Kinetic constant (s−1) | ||
|---|---|---|---|
| 25 °C | 35 °C | 45 °C | |
| G0 (2) | 61.2 | 158.4 | 284.4 |
| G1 (4) | 144.1 | 295.2 | 507.6 |
Bacterial growth assays
In the initial screening, the antibacterial efficacies of amine-terminated dendrimers and the corresponding glycoconjugates (25 μg mL−1) were evaluated on the growth of E. coli DH5α (Gram-negative), P. aeruginosa (Schroeter) Migula 27853 (Gram-negative), S. aureus Rosenbach 25923 (Gram-positive) and M. smegmatis mc2 155 (acid fast). Kanamycin (Kan) was used as positive antibacterial control for the assays. The bacterial growth profiles are shown in Fig. 1, the corresponding growth inhibition percentages in the presence and absence of dendrimers are given in Table 2. The untreated cells were considered as negative control (NC). The growth assays yielded a generation time of ∼40 min, 90 min, 51 min. and 4.8 h for E. coli, P. aeruginosa, S. aureus and M. smegmatis, respectively, without the presence of the dendrimer agents.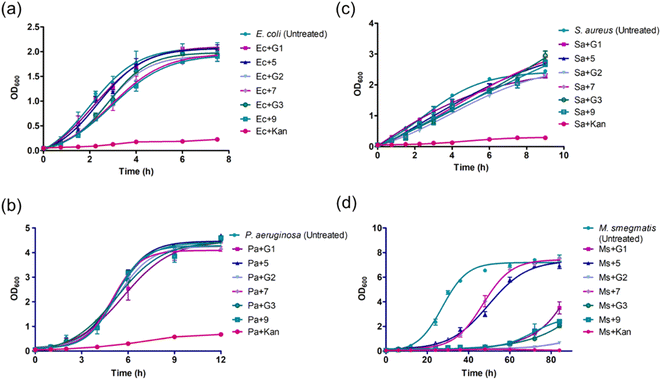 | ||
| Fig. 1 Bacterial growth observed in the presence of different native and glycoconjugated dendrimer generations: (a) E. coli DH5α; (b) P. aeruginosa; (c) S. aureus and (d) M. smegmatis mc2 155. | ||
| M. smegmatis | E. coli | P. aeruginosa | S. aureus | |
|---|---|---|---|---|
| Untreated | 100.0 | 100.0 | 100.0 | 100.0 |
| G-1 | 8.8 ± 1.5 | 97.1 ± 3.4 | 91.0 ± 1.9 | 91.9 ± 2.7 |
| 5 | 56.8 ± 0.23 | 95.3 ± 3.4 | 99.5 ± 5.6 | 88.1 ± 4.4 |
| G-2 | 4.0 ± 0.08 | 85.1 ± 2.9 | 94.5 ± 2.8 | 67.2 ± 4.7 |
| 7 | 62.0 ± 2.7 | 80.0 ± 3.4 | 95.5 ± 0.2 | 82.8 ± 3.6 |
| G-3 | 7.8 ± 0.24 | 87.0 ± 1.1 | 95.3 ± 0.5 | 81.8 ± 2.4 |
| 9 | 9.4 ± 2.9 | 78.0 ± 3.3 | 95.7 ± 0.07 | 81.1 ± 0.7 |
As seen in Fig. 1a and b, the growth profiles of E. coli and P. aeruginosa encountered little or no inhibition of the bacterial growth in the presence of dendrimers, the maximum inhibition percentages were 22% and 6% for the conjugate 9, respectively. The native un-conjugated G-2 dendrimer showed 32% growth inhibition of Gram-positive S. aureus (Fig. 1c). Further 10–12% growth inhibitions were observed in the presence of remaining native and glycoconjugated dendrimers, in comparison to untreated control. Among the strains, M. smegmatis was observed to undergo a significant growth reduction. G-1: 91.2%; 5: 43.2%; G-2: 43.8%; 7: 38%; G-3: 92.2% and 9: 90.6% (Fig. 1d), as compared to the untreated control. Also, the doubling times of M. smegmatis cells increased significantly: G-1: 12.5 h; 5: 2.1 h; G-2: 17.7 h; 7: 3.7 h; G-3: 31 h and 9: 7.4 h (Fig. 1d).
The above observations reveal that the dendrimer agents are effective to inhibit the growth of mycobacterium strains and, to some extent, Gram-positive S. aureus. The native and glycoconjugated dendrimers did not act on Gram-negative E. coli or P. aeruginosa strains, even when cationic amine sites constitute the dendrimers. A defining feature of the Gram-positive and Gram-negative strains is the altered cell wall structure compositional features. We presume that the cell surface-active dendrimer agents might differentiate these features, leading to the most effects of the dendrimer agents on acid-fast M. smegmatis, followed by Gram-positive S. aureus and minimal effects on the Gram-negative P. aeruginosa and E. coli strains.
MIC values determination for M. smegmatis and M. tuberculosis
Due to the selectivity of the dendrimers towards mycobacterial growth inhibition, further evaluation of the MIC value was conducted on M. smegmatis and, importantly, M. tuberculosis H37Rv cells and the results are presented in Table 3. The MIC values found for M. smegmatis were 50 μg mL−1 for G1, G2 and G3 generations of amine terminated PETIM dendrimers. Whereas, the same for M. tuberculosis H37Rv cells were 100 μg mL−1, 100 μg mL−1 and 200 μg mL−1, respectively. The glycoconjugated dendrimer generations G1 and G2, the MIC values were found 100 μg mL−1 and 200 μg mL−1, in each case for M. smegmatis and M. tuberculosis H37Rv, respectively. These values reiterated that the amine terminated dendrimers are relatively more efficient in inhibiting mycobacterial growth than the glycoconjugated dendrimers. When MIC of M. smegmatis was evaluated against third generation glycoconjugate 9, twice the concentration as that of the lower generation dendrimer glycoconjugates, implying that lower generation dendrimer glycoconjugates surpass inhibition efficiencies as compared to the high generation glycoconjugates. Bacterial strains S. aureus, E. coli and P. aeruginosa did not show growth inhibitions in the presence of the dendrimers and the corresponding glycoconjugates.| Compound | M. smegmatis MIC value (μg mL−1) | M. tuberculosis MIC value (μg mL−1) |
|---|---|---|
| a NA: MIC value is not within the limit. | ||
| G1 | 50 | 100 |
| 5 | 100 | 200 |
| G-2 | 50 | 100 |
| 7 | 100 | 200 |
| G3 | 50 | 200 |
| 9 | 200 | NA |
Discussion
Dendrimers with cationic sites at the peripheries are known to act as bactericidal agents, as a result of ionic interactions with anionic bacterial cell membranes, that lead to penetration and permeability of the exogeneous dendrimers into the cellular plasma environment and the damage of the intracellular components.45,50 Carbohydrate-functionalized dendrimers were studied, specifically as anti-adhesive agents that block the binding of the host cell surface glycans with lectins present at the outer membranes of a bacterium.56,57 Among the most studied are the polycationic poly(amido amine) (PAMAM) dendrimers that show anti-bacterial effects on Gram-positive and Gram-negative bacteria, in low μg mL−1 concentrations. Such bactericidal effects of the amine-terminated PAMAM dendrimers were off-set when the peripheral functionalities were modified with poly(ethylene glycol) moieties.58 Similar bactericidal effects were established to poly(propylene imine) (PPI) and carbosilane dendrimers, possessing amine functionalities at their peripheries.59,60 Further, the antibacterial efficacies of maltose sugar-functionalized fourth generation PPI dendrimers were demonstrated in low micromolar concentrations against Gram-positive and Gram-negative bacteria.39,40 Bare PPI dendrimer, without the sugar functionalization, was found to be more cytotoxic than the sugar functionalized PPI dendrimer. Apoptosis, coupled with oxidative stress, of cells was found to be major causes for the antibacterial activity. The maltose-functionalized fourth generation PPI dendrimer was found to disrupt outer membrane of bacterial cells, thereby permitting muralytic enzyme, endolysin, to penetrate and degrade peptidoglycans, leading to lysis of bacterial cells.40One of the well-studied polycationic macromolecules are the polylysine glycoconjugates that act on both the Gram-positive and Gram-negative bacterial cell membranes and rupture membrane structures, leading to cytoplasm damage and bacterial cell lysis.43 The selectivity of the cell lysis by the polylysine glycoconjugate was found to be about half that of the native polylysine, even when the microbial targeting was superior with the polymer glycoconjugate. Reduction in the cationic sites density in the polymer glycoconjugate was proposed to cause reduced cell lysis, related with the increased minimum inhibitory concentration required to affect the bacterial growth. Important outcomes of our present study are that (i) the native dendrimers are more effective to inhibit the growth Gram-positive and acid-fast bacteria and very minimal or none of the inhibition effect on Gram-negative bacteria; (ii) the native dendrimers with dense cationic sites are far more mycobacterial growth inhibitors than the corresponding glycoconjugates and (iii) the higher generation native dendrimers are far more effective inhibitors than the glycoconjugates. The pattern reiterates that amine-rich dendrimers are by-far the efficient inhibitors and the corresponding glycoconjugations compensate the cationic sites induced growth inhibitions and reduce the inhibitory efficacies.
The selectivity to inhibit the growth of mycobacterial strains M. smegmatis and M. tuberculosis is revealing. We presume that the cell surface composition of various components is likely the origin of this selectivity. The bacterial outer layer is a complex, multi-layered arrangement of biological macromolecules. The organization and the components of the bacterial cell wall differ across bacterial species. The cell wall structures of E. coli and P. aeruginosa, used as representatives from Gram-negative bacteria, Gram-positive S. aureus, M. tuberculosis and acid-fast M. smegmatis are categorized broadly.61,62 The Gram-negative bacterial cell wall consists of layers of the inner membrane, the peptidoglycan and the outer membrane. The outer membrane is a bilayer of lipid, mainly composed of lipopolysaccharide. The Gram-positive bacteria lacks such an outer membrane. Repeating N-acetyl glucosamine and N-acetyl muramic acid disaccharide, along with cross-linked pentapeptide side chains, constituted the peptidoglycan layer. This peptidoglycan layer in Gram-positive bacteria is more dense than Gram-negative bacteria. Further, the Gram-positive bacteria possesses long anionic polymers called teichoic acids. On the other hand, the mycobacterial cell wall is different, called myco-membrane, composed of mycolic acids (MA). This outer membrane is critical for mycobacterial physiology and protects mycobacteria from different stress. This lipid layer also provides high resistance to many broad spectra of antibiotics. The cell wall is orders of magnitude less permeable, than the most resistant Gram-negative bacteria E. coli and P. aeruginosa. The lipids content in the mycobacterial cell wall is 60%, which is threefold higher than most Gram-negative bacteria. The mycolic acids in the mycobacterial cell wall are covalently linked with cell wall polysaccharides, as in lipomannan and lipoarabinomannan.63 This unique architecture and higher acidic moiety in mycobacteria provide negative charge to the mycobacterium cell wall compared. We presume that the un-modified cationic dendrimers interact efficiently with the anionic mycobacterial cell wall, when compared to other bacterial strains. This interaction at the mycobacterial cell wall plays an important role in the mycobacterial growth inhibition by dendrimers.
Conclusion
A benign glycoconjugation of amine-rich PETIM dendrimers of 0–3 generations, possessing 2–16 amine functionalities at the peripheries, is conducted with sugar moieties in aqueous MeOH solutions, by an aza-Michael addition reaction of sugar vinyl sulfoxide. The conjugation is observed to be faster in the case of the first generation dendrimer, than zero generation bis-amine derivative. The glycoconjugated dendrimers were studied for their antibacterial properties against Gram-positive, Gram-negative and acid-fast bacteria. Among the bacteria, the growths of M. smegmatis and M. tuberculosis are inhibited profoundly by the native, unmodified and glycoconjugated dendrimers, whereas Gram-negative bacterial strains are not affected. Results of this study show that the dendrimers inhibit the mycobacterial growth specifically and such a specificity opens up leads for the development of target-driven dendrimer-based anti-mycobacterial agents.Experimental section
G0-Glycoconjugate (3)
A mixture of PETIM G0-(NH2)2 2 (4 mg, 0.03 mmol) and sugar vinyl sulfoxide 1 (41 mg, 0.09 mmol) in MeOH/H2O (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (1.5 mL) was added with NaHCO3 (2 mg) and stirred at 45 °C for 15 h. The crude reaction mixture was evaporated and purified using column chromatography (SiO2, 100–200 mesh) (20% MeOH/CHCl3) to afford compound 3, as a gum (20 mg, 78%). Rf = 0.2 (20% MeOH/CHCl3); 1H NMR (400 MHz, CDCl3): δ 7.63–7.59 (m, 4H), 7.36–7.22 (m, 14H), 6.68 (d, J = 16 Hz, 0.5H), 6.59 (app. s, 1.5H), 5.15–5.10 (m, 2H), 4.87–4.81 (m, 2H), 4.63–4.51 (m, 4H), 3.85–3.75 (m, 4H), 3.34–3.32 (m, 4H), 2.92 (s, 2H), 2.45–2.41 (m, 4H), 2.39 (s, 6H), 2.11–2.09 (m, 4H), 1.57–1.52 (m, 4H); 13C NMR (CDCl3, 100 MHz) δ 143.8, 141.9, 139.5, 137.3, 130.2, 130.1, 129.9, 128.7, 128.6, 128.3, 128.2, 128.1, 128.0, 126.7, 125.8, 94.25, 71.2, 69.9, 69.1, 64.6, 54.73, 44.3, 30.5, 29.8, 21.6 and 14.2. ESI-MS m/z: [M + Na]+ calcd For C46H56N2O9S2H, 845.3505; found: 845.3532.
1) (1.5 mL) was added with NaHCO3 (2 mg) and stirred at 45 °C for 15 h. The crude reaction mixture was evaporated and purified using column chromatography (SiO2, 100–200 mesh) (20% MeOH/CHCl3) to afford compound 3, as a gum (20 mg, 78%). Rf = 0.2 (20% MeOH/CHCl3); 1H NMR (400 MHz, CDCl3): δ 7.63–7.59 (m, 4H), 7.36–7.22 (m, 14H), 6.68 (d, J = 16 Hz, 0.5H), 6.59 (app. s, 1.5H), 5.15–5.10 (m, 2H), 4.87–4.81 (m, 2H), 4.63–4.51 (m, 4H), 3.85–3.75 (m, 4H), 3.34–3.32 (m, 4H), 2.92 (s, 2H), 2.45–2.41 (m, 4H), 2.39 (s, 6H), 2.11–2.09 (m, 4H), 1.57–1.52 (m, 4H); 13C NMR (CDCl3, 100 MHz) δ 143.8, 141.9, 139.5, 137.3, 130.2, 130.1, 129.9, 128.7, 128.6, 128.3, 128.2, 128.1, 128.0, 126.7, 125.8, 94.25, 71.2, 69.9, 69.1, 64.6, 54.73, 44.3, 30.5, 29.8, 21.6 and 14.2. ESI-MS m/z: [M + Na]+ calcd For C46H56N2O9S2H, 845.3505; found: 845.3532.
G1-Glycoconjugate (5)
A mixture of PETIM G1-(NH2)4 4 (8 mg, 0.014 mmol) and sugar vinyl sulfoxide 1 (38.5 mg, 0.084 mmol) in MeOH/H2O (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (1.5 mL) was added with NaHCO3 (2 mg) and stirred at 45 °C for 15 h. The crude reaction mixture was evaporated, resuspended in H2O (2 mL) and followed by washing with ethyl acetate (10 × 2 mL). The aqueous layer was concentrated in vacuo to afford compound 5, as a yellowish white gum (22 mg, 81%). 1H NMR (500 MHz, CD3OD): δ 7.69–7.60 (m, 13H), 7.33–7.20 (m, 23H), 6.69–6.54 (m, 4H), 5.13 (b, 4H), 4.61–4.44 (m, 8H), 3.71 (b, 7H), 3.63 (m, 7H), 3.45 (br. s, 21H), 3.00–2.67 (m, 9H), 2.38–2.34 (m, 24H), 1.77–1.47 (m, 19H). 13C NMR (CD3OD, 125 MHz) δ 138.9, 131.1, 129.8, 129.6, 129.1, 128.2, 127.9, 127.0, 95.3, 72.4, 71.2, 70.8, 69.6, 64.8, 61.0, 51.9, 37.7, 31.0, 28.4, 26.8, 23.9, 23.7, 21.5, 21.3, 18.7. MALDI-MS m/z: [M + Na]+ calcd for C110H148N6O21S4H2, 2018.9737; found: 2018.330.
1) (1.5 mL) was added with NaHCO3 (2 mg) and stirred at 45 °C for 15 h. The crude reaction mixture was evaporated, resuspended in H2O (2 mL) and followed by washing with ethyl acetate (10 × 2 mL). The aqueous layer was concentrated in vacuo to afford compound 5, as a yellowish white gum (22 mg, 81%). 1H NMR (500 MHz, CD3OD): δ 7.69–7.60 (m, 13H), 7.33–7.20 (m, 23H), 6.69–6.54 (m, 4H), 5.13 (b, 4H), 4.61–4.44 (m, 8H), 3.71 (b, 7H), 3.63 (m, 7H), 3.45 (br. s, 21H), 3.00–2.67 (m, 9H), 2.38–2.34 (m, 24H), 1.77–1.47 (m, 19H). 13C NMR (CD3OD, 125 MHz) δ 138.9, 131.1, 129.8, 129.6, 129.1, 128.2, 127.9, 127.0, 95.3, 72.4, 71.2, 70.8, 69.6, 64.8, 61.0, 51.9, 37.7, 31.0, 28.4, 26.8, 23.9, 23.7, 21.5, 21.3, 18.7. MALDI-MS m/z: [M + Na]+ calcd for C110H148N6O21S4H2, 2018.9737; found: 2018.330.
G2-Glycoconjugate (7)
A mixture of PETIM G2-(NH2)8 6 (10 mg, 0.007 mmol) and sugar vinyl sulfoxide 1 (38.5 mg, 0.084 mmol) in MeOH/H2O (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (1.5 mL) was added with NaHCO3 (2 mg) and stirred at 45 °C for 24 h. The crude reaction mixture was evaporated, resuspended in H2O (2 mL) and followed by washing with ethyl acetate (10 × 2 mL). The aqueous layer was concentrated in vacuo to afford compound 7, as a yellowish white gum (18 mg, 78%). 1H NMR (500 MHz, CD3OD): δ 7.73–7.68 (m, 18H), 7.48–7.38 (m, 51H), 6.81 (app. s., 2H), 6.59 (app. d, J = 8 Hz, 6H), 5.36–5.26 (m, 7H), 4.18–4.17 (m, 2H), 4.93–4.85 (m, 5H), 3.74 (s, 11H), 3.63 (s, 97H), 3.39 (s, 3H), 3.20–3.13 (m, 12H), 2.85–2.76 (m, 6H), 2.61–2.55 (m, 2H), 2.45–2.42 (m, 54H), 2.26–2.21 (m, 1H), 2.20 (s, 9H), 1.80 (s, 14H), 1.58–1.27 (br. m, 14H), 1.11–0.94 (m, 14H). 13C NMR (CD3OD, 125 MHz) δ 169.1, 147.5, 142.1, 141.3, 140.4, 130.1, 128.9, 128.5, 128.1, 127.9, 127.5, 127.4, 126.0, 125.6, 93.9, 72.9, 72.7, 70.0, 68.7, 68.3, 61.4, 60.2, 59.1, 38.2, 31.8, 29.8, 29.5, 26.6, 22.9, 20.7, 20.2, 20.0.
1) (1.5 mL) was added with NaHCO3 (2 mg) and stirred at 45 °C for 24 h. The crude reaction mixture was evaporated, resuspended in H2O (2 mL) and followed by washing with ethyl acetate (10 × 2 mL). The aqueous layer was concentrated in vacuo to afford compound 7, as a yellowish white gum (18 mg, 78%). 1H NMR (500 MHz, CD3OD): δ 7.73–7.68 (m, 18H), 7.48–7.38 (m, 51H), 6.81 (app. s., 2H), 6.59 (app. d, J = 8 Hz, 6H), 5.36–5.26 (m, 7H), 4.18–4.17 (m, 2H), 4.93–4.85 (m, 5H), 3.74 (s, 11H), 3.63 (s, 97H), 3.39 (s, 3H), 3.20–3.13 (m, 12H), 2.85–2.76 (m, 6H), 2.61–2.55 (m, 2H), 2.45–2.42 (m, 54H), 2.26–2.21 (m, 1H), 2.20 (s, 9H), 1.80 (s, 14H), 1.58–1.27 (br. m, 14H), 1.11–0.94 (m, 14H). 13C NMR (CD3OD, 125 MHz) δ 169.1, 147.5, 142.1, 141.3, 140.4, 130.1, 128.9, 128.5, 128.1, 127.9, 127.5, 127.4, 126.0, 125.6, 93.9, 72.9, 72.7, 70.0, 68.7, 68.3, 61.4, 60.2, 59.1, 38.2, 31.8, 29.8, 29.5, 26.6, 22.9, 20.7, 20.2, 20.0.
G3-Glycoconjugate (9)
A mixture of PETIM G3-(NH2)16 8 (10 mg, 0.003 mmol) and sugar vinyl sulfoxide 1 (33 mg, 0.072 mmol) in aq. MeOH/H2O (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (1.5 mL) was added with NaHCO3 (2 mg) and stirred at 45 °C for 24 h. The crude reaction mixture was evaporated, resuspended in H2O (2 mL) and followed by washing with ethyl acetate (10 × 2 mL). The aqueous layer was concentrated in vacuo to afford compound 9, as a yellowish white gum (20 mg, 74%). 1H NMR (400 MHz, CD3OD): δ 7.74–7.66 (m, 57H), 7.42–7.25 (m, 114H), 6.75–6.60 (m, 15H), 5.19 (b, 16H), 4.67–4.58 (m, 15H), 4.50 (br. s, 15H), 3.77–3.68 (m, 25H), 3.54 (br. s, 121H), 3.36–3.33 (m, 9H), 3.09–3.06 (m, 138H), 2.64–2.61 (m, 8H), 2.46–2.40 (m, 150H), 2.25–2.19 (m, 17H), 1.90–1.87 (m, 62H). 13C NMR (CD3OD, 100 MHz) δ 138.9, 131.1, 129.8, 129.6, 129.2, 129.1, 128.2, 126.9, 95.4, 72.4, 70.8, 69.3, 67.9, 64.8, 60.3, 59.3, 55.9, 52.8, 52.0, 39.0, 30.9, 28.7, 26.5, 23.9, 21.5, 21.3.
1) (1.5 mL) was added with NaHCO3 (2 mg) and stirred at 45 °C for 24 h. The crude reaction mixture was evaporated, resuspended in H2O (2 mL) and followed by washing with ethyl acetate (10 × 2 mL). The aqueous layer was concentrated in vacuo to afford compound 9, as a yellowish white gum (20 mg, 74%). 1H NMR (400 MHz, CD3OD): δ 7.74–7.66 (m, 57H), 7.42–7.25 (m, 114H), 6.75–6.60 (m, 15H), 5.19 (b, 16H), 4.67–4.58 (m, 15H), 4.50 (br. s, 15H), 3.77–3.68 (m, 25H), 3.54 (br. s, 121H), 3.36–3.33 (m, 9H), 3.09–3.06 (m, 138H), 2.64–2.61 (m, 8H), 2.46–2.40 (m, 150H), 2.25–2.19 (m, 17H), 1.90–1.87 (m, 62H). 13C NMR (CD3OD, 100 MHz) δ 138.9, 131.1, 129.8, 129.6, 129.2, 129.1, 128.2, 126.9, 95.4, 72.4, 70.8, 69.3, 67.9, 64.8, 60.3, 59.3, 55.9, 52.8, 52.0, 39.0, 30.9, 28.7, 26.5, 23.9, 21.5, 21.3.
Biological studies
The antibacterial efficacies of amine-terminated dendrimers and the corresponding glycoconjugates were evaluated on the growth of E. coli DH5α (Gram-negative), P. aeruginosa (Schroeter) Migula 27853 (Gram-negative), S. aureus Rosenbach 25923 (Gram-positive) and M. smegmatis mc2 155 (acid fast bacilli). All the bacterial strains were maintained in specific growth medium. P. aeruginosa and S. aureus were cultivated in Mueller-Hinton (MH) and Tryptic Soy Broth (TSB), respectively. E. coli, P. aeruginosa and S. aureus strains were grown in LB (Luria-Bertani) broth, whereas M. smegmatis and M. tuberculosis H37Rv strains was grown in MB7H9 medium (Difco) and supplemented with glucose (2%), Tween 80 (0.05%) and 10% OADC only for M. tuberculosis at 37 °C. Kanamycin (25 μg mL−1) was used as positive control in the bacterial growth experiments. For growth assay, each compound (25 μg mL−1) was added at 0 h and the growth of cells was maintained under shaking condition at 37 °C. The bacterial growth was measured by optical density (OD) changes of the cultures at 600 nm. OD values for a specific time point were plotted and analyzed by Graph Pad Prism software. The growth inhibition assays for all bacterial species were performed three independent triplicates. Growth medium without any dendrimer compounds was taken as the positive control in all the experiments.Minimum inhibitory concentration (MIC) estimation of the dendrimers
Determination of the MIC of M. smegmatis and M. tuberculosis H37Rv was confirmed by cell viability study using 96-well Microplate Alamar Blue Assay (MABA). The water solubility, permeability inside cell, low toxicity towards bacterial cell and stability in cell culture makes it perfect as visual indicator for the analysis of viability of the bacterial cell culture. Resazurin is a blue non-fluorescent dye, turns fluorescent pink dye resorufin when reduced. So, the reduction of resazurin (blue, non-fluorescent) to resorufin (pink, fluorescent) indicates metabolic impairment of the cells. Thus, the visual change of the colour is used for the assay to determine the minimum inhibitory concentration (MIC). The determination of MIC for PETIM and sugar vinyl sulfoxide modified PETIM was conducted by 96-well Microplate Alamar Blue Assay. The protocol of the assay is well established and commonly as resazurin microtiter assay (REMA) method.64–66 Briefly, M. smegmatis and M. tuberculosis H37Rv cells (at 0.05 OD) was taken in a 96-well flat-bottom microtiter plate, where PETIM and modified PETIM dendrimers were added to maintain a final volume of 200 μL. The starting concentrations of the dendrimers were 200 μg mL−1 and 400 μg mL−1, for M. smegmatis and M. tuberculosis H37Rv, respectively. The compounds were diluted serially up to concentrations 0.39 μg mL−1. The cultures were allowed to grow by incubating for 48 h and 6 days for M. smegmatis and M. tuberculosis H37Rv, respectively, at 37 °C. After that, resazurin solution (0.04%) was added to the culture and incubated further for another 4 h for M. smegmatis and 24 h for M. tuberculosis H37Rv cells. The colour remained blue if the growth of the bacteria cell was inhibited. A negative control with no dendrimers added was used to compare with the dendrimer treated bacteria culture. The experiments were conducted in triplicate.Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We are grateful to Department of Biotechnology, New Delhi, for a research support (Grant Number BT/PR33123/MED/29/1497/2020). B. S. and K. D. are grateful to IISc, Bangalore, CSIR, New Delhi, for research fellowships. AM acknowledge fellowship support from the DBT-Research Associateship (RA) Programme in Life Sciences. We thank Prof. Jayanta Chatterji, IISc, Bangalore, for providing S. aureus and P. aeruginosa strains.References
- K. Ohtsubo and J. D. Marth, Cell, 2006, 126, 855 CrossRef CAS PubMed.
- C. Reily, T. J. Stewart, M. B. Renfrow and J. Novak, Nat. Rev. Nephrol., 2019, 15, 346 CrossRef PubMed.
- K. Handa and S.-i. Hakomori, Glycoconjugate J., 2017, 34, 693 CrossRef CAS PubMed.
- J. M. Baskina and C. R. Bertozzi, QSAR Comb. Sci., 2007, 26, 1211 CrossRef.
- C. Unverzagt and Y. Kajihara, Chem. Soc. Rev., 2013, 42, 4408 RSC.
- J. W. Wehner, M. Hartmann and T. K. Lindhorst, Carbohydr. Res., 2013, 371, 22 CrossRef CAS PubMed.
- R. D. Astronomo and D. R. Burton, Nat. Rev. Drug Discovery, 2010, 9, 308 CrossRef CAS PubMed.
- S. Feng, C. Xiong, S. Wang, Z. Guo and G. Gu, ACS Infect. Dis., 2019, 5, 1423 CrossRef CAS PubMed.
- B. Liet, E. Laigre, D. Goyard, B. Todaro, C. Tiertant, D. Boturyn, N. Berthet and O. Renaudet, Chem.–Eur. J., 2019, 25, 15508 CrossRef CAS PubMed.
- N. T. Supekar, V. Lakshminarayanan, C. J. Capicciotti, A. Sirohiwal, C. S. Madsen, M. A. Wolfert, P. A. Cohen, S. J. Gendler and G.-J. Boons, ChemBioChem, 2018, 19, 121 CrossRef CAS PubMed.
- K. Streichert, C. Seitz, E. Hoffmann, I. Boos, W. Jelkmann, T. Brunner, C. Unverzagt and M. Rubini, ChemBioChem, 2019, 20, 1914 CrossRef CAS PubMed.
- B. Sarkar and N. Jayaraman, Front. Chem., 2020, 8(1–29), 570185 CrossRef CAS PubMed.
- G. C. Daskhan, H.-T. T. Tran, P. J. Meloncelli, T. L. Lowary, L. J. West and C. W. Cairo, Bioconjugate Chem., 2018, 29, 343 CrossRef CAS PubMed.
- M. G. García, J. M. Benito, A. P. Butera, C. O. Mellet, J. M. G. Fernández and J. L. J. Blanco, J. Org. Chem., 2012, 77, 1273 CrossRef PubMed.
- M. Wolfenden, J. Cousin, P. N. Makker, A. Raz and M. J. Cloninger, Molecules, 2015, 20, 7059 CrossRef CAS PubMed.
- C. Fessele, S. Wachtler, V. Chandrasekaran, C. Stiller, T. K. Lindhorst and A. Krueger, Eur. J. Org. Chem., 2015, 5519 CrossRef CAS.
- P. Wu, M. Malkoch, J. N. Hunt, R. Vestberg, E. Kaltgrad, M. G. Finn, V. V. Fokin, K. B. Sharpless and C. J. Hawker, Chem. Commun., 2005, 5775 RSC.
- M. G. Cuesta, D. Goyard, E. Nanba, K. Higaki, J. M. G. Fernández, O. Renaudet and C. O. Mellet, Chem. Commun., 2019, 55, 12845 RSC.
- L. G. Weaver, Y. Singh, G. Vamvounis, M. F. Wyatt, P. L. Burn and J. T. Blanchfield, Polym. Chem., 2014, 5, 1173 RSC.
- X. Li, T. Fang and G.-J. Boons, Angew. Chem., Int. Ed., 2014, 53, 7179 CrossRef CAS.
- S. Munneke, E. M. Dangerfield, B. L. Stocker and M. S. M. Timmer, Glycoconjugate J., 2017, 34, 633 CrossRef CAS PubMed.
- B. Sarkar, A. Mahapa, D. Chatterji and N. Jayaraman, Biochemistry, 2019, 58, 3561 CrossRef CAS PubMed.
- D. Astruc, L. Liang, A. Rapakousiou and J. Ruiz, Acc. Chem. Res., 2012, 45, 630 CrossRef CAS PubMed.
- D. A. Tomalia, J. B. Christensen and U. Boas, in Dendrimers, Dendrons and Dendritic Polymers, Cambridge University Press, Cambridge, UK, 2012 Search PubMed.
- R. K. O'Reilly, C. J. Hawker and K. L. Wooley, Chem. Soc. Rev., 2006, 35, 1068 RSC.
- S. Svenson and D. A. Tomalia, Adv. Drug Delivery Rev., 2012, 64, 102 CrossRef.
- D. Astruc, E. Boisselier and C. Ornelas, Chem. Rev., 2010, 110, 1857 CrossRef CAS PubMed.
- L. Röglin, E. H. M. Lempens and E. W. Meijer, Angew. Chem., Int. Ed., 2011, 50, 102 CrossRef PubMed.
- A.-M. Caminade, A. Ouali, R. Laurent, C.-O. Turrin and J.-P. Majoral, Chem. Soc. Rev., 2015, 44, 3890 RSC.
- N. Jayaraman, Chem. Soc. Rev., 2009, 38, 3463 RSC.
- S. Mignani, J. Rodrigues, R. Roy, X. Shi, V. Ceña, S. E. Kazzouli and J.-P. Majoral, Drug Discovery Today, 2019, 24, 1184 CrossRef CAS PubMed.
- L. I. F. Moura, A. Malfanti, C. Peres, A. I. Matos, E. Guegain, V. Sainz, M. Zloh, M. J. Vicent and H. F. Florindo, Mater. Horiz., 2019, 6, 1956 RSC.
- M. L. Wolfenden and M. J. Cloninger, Bioconjugate Chem., 2006, 17, 958 CrossRef CAS PubMed.
- C. Sehad, T. C. Shiao, L. M. Sallam, A. Azzouz and R. Roy, Molecules, 2018, 23, 1890 CrossRef PubMed.
- S. Mignani, V. D. Tripathi, D. Soam, R. P. Tripathi, S. Das, S. Singh, R. Gandikota, R. Laurent, A. Karpus, A.-M. Caminade, A. Steinmetz, A. Dasgupta, K. K. Srivastava and J.-P. Majoral, Biomacromolecules, 2021, 22, 2659 CrossRef CAS PubMed.
- A. Castonguay, E. Ladd, T. G. M. van de Ven and A. Kakkar, New J. Chem., 2012, 36, 199 RSC.
- M. A. Mintzer, E. L. Dane, G. A. O'Toole and M. W. Grinstaff, Mol. Pharm., 2012, 9, 342 CrossRef CAS.
- I. H. Bermejo, J. M. H. Ros, L. S. García, M. Maly, C. V. Expósitoa, J. Soliveri, F. J. de la Mata, J. L. C. Patiño, J.-P. Serrano, J. S. Nieves and R. Gómez, Eur. Polym. J., 2018, 101, 159 CrossRef.
- A. Felczak, N. Wrońska, A. Janaszewska, B. Klajnert, M. Bryszewska, D. Appelhans, B. Voit, S. Rózalska and K. Lisowska, New J. Chem., 2012, 36, 2215 RSC.
- K. Ciepluch, B. Maciejewska, K. Gałczyńska, D. K. Ciepluch, M. Bryszewska, D. Appelhans, Z. D. Kawa and M. Arabski, Bioorg. Chem., 2019, 91(1–6), 103121 CrossRef CAS PubMed.
- S. Alfei and A. M. Schito, Nanomaterials, 2020, 10(1–52), 2022 CrossRef CAS PubMed.
- M. Á. Ortega, A. G. Merino, O. F. Martínez, J. R. Ruiz, L. Pekarek, L. G. Guijarro, N. G. Honduvilla, M. A. Mon, J. Buján and S. G. Gallego, Pharmaceutics, 2020, 12(1–27), 874 CrossRef CAS PubMed.
- D. Alkekhia and A. Shukla, J. Biomed. Mater. Res., Part A, 2019, 107A, 1324 CrossRef PubMed.
- D. Pranantyo, L. Q. Xu, Z. Hou, E.-T. Kang and M. B.-C. Park, Polym. Chem., 2017, 8(1–10), 3364 RSC.
- B. Wang, R. S. Navath, A. R. Menjoge, B. Balakrishnan, R. Bellair, H. Dai, R. Romero, S. Kannan and R. M. Kannan, Int. J. Pharm., 2010, 395, 298 CrossRef CAS PubMed.
- T. R. Krishna, S. Jain, U. S. Tatu and N. Jayaraman, Tetrahedron, 2005, 61, 4281 CrossRef CAS.
- U. P. Thankappan, S. N. Madhusudana, A. Desai, G. Jayamurugan, Y. B. R. D. Rajesh and N. Jayaraman, Bioconjugate Chem., 2011, 22, 115 CrossRef CAS PubMed.
- A. Lakshminarayanan, V. K. Ravi, R. Tatineni, Y. B. R. D. Rajesh, V. Maingi, K. S. Vasu, N. Madhusudhan, P. K. Maiti, A. K. Sood, S. Das and N. Jayaraman, Bioconjugate Chem., 2013, 24, 1612 CrossRef CAS PubMed.
- A. Lakshminarayanan, B. U. Reddy, N. Raghav, V. K. Ravi, A. Kumar, P. K. Maiti, A. K. Sood, N. Jayaraman and S. Das, Nanoscale, 2015, 7, 16921 RSC.
- C. Z. Chen and S. L. Cooper, Adv. Mater., 2000, 12, 843 CrossRef CAS.
- H. C. Hansen, S. Haataja, H. J. Finne and G. Magnusson, J. Am. Chem. Soc., 1997, 119, 6974 CrossRef CAS.
- A. M. Holmes, J. R. Heylings, K. W. Wan and G. P. Moss, Int. J. Antimicrob. Agents, 2019, 53, 500 CrossRef CAS PubMed.
- T. R. Krishna and N. Jayaraman, J. Org. Chem., 2003, 68, 9694 CrossRef CAS PubMed.
- A. Mukherjee and N. Jayaraman, Tetrahedron, 2012, 68, 8746 CrossRef CAS.
- A. Mukherjee and N. Jayaraman, Carbohydr. Res., 2013, 380, 51 CrossRef CAS PubMed.
- M. Touaibia and R. Roy, Mini-Rev. Med. Chem., 2007, 7, 1270 CrossRef CAS.
- J. Rojo and R. Delgado, Anti-Infect. Agents Med. Chem., 2007, 6, 151 CrossRef CAS.
- A. I. Lopez, R. Y. Reins, A. M. McDermott, B. W. Trautner and C. Cai, Mol. BioSyst., 2009, 5, 1148 RSC.
- C. Z. Chen, S. L. Cooper and N. C. B. Tan, Chem. Commun., 1999, 1585 RSC.
- P. Ortega, J. L. C. Patino, M. A. M. Fernandez, J. Soliveri, R. Gomez and F. J. de la Mata, Org. Biomol. Chem., 2008, 6, 3264 RSC.
- L. J. Alderwick, J. Harrison, G. S. Lloyd and H. L. Birch, Cold Spring Harbor Perspect. Med., 2015, 5(1–15), a021113 CrossRef PubMed.
- T. J. Silhavy, D. Kahne and S. Walker, Cold Spring Harbor Perspect. Biol., 2010, 2(1–16), a000414 Search PubMed.
- A. K. Mishra, N. N. Driessen, B. J. Appelmelk and G. S. Besra, FEMS Microbiol. Rev., 2011, 35, 1126 CrossRef CAS PubMed.
- S. N. Rampersad, Sensors, 2012, 12, 12347 CrossRef CAS PubMed.
- K. R. Gupta, S. Kasetty and D. Chatterji, Appl. Environ. Microbiol., 2015, 81, 2571 CrossRef CAS PubMed.
- M. Elshikh, S. Ahmed, S. Funston, P. Dunlop, M. McGaw, R. Marchant and I. M. Banat, Biotechnol. Lett., 2016, 38, 1015 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ra08196b |
| This journal is © The Royal Society of Chemistry 2023 |