Open Access Article
Open Access ArticleOcular protein optineurin shows reversibility from unfolded states and exhibits chaperone-like activity†
Anjali Dixit a,
Ankan Chakraborty
a,
Ankan Chakraborty a,
Jyoti Rani Nath
a,
Jyoti Rani Nath b,
Pramit K. Chowdhury
b,
Pramit K. Chowdhury b and
Bishwajit Kundu
b and
Bishwajit Kundu *a
*a
aKusuma School of Biological Sciences, Indian Institute of Technology, Delhi, India. E-mail: bkundu@bioschool.iitd.ac.in
bDepartment of Chemistry, Indian Institute of Technology, Delhi, India
First published on 28th February 2023
Abstract
Optineurin (OPTN) is a multifunctional, ubiquitously expressed cytoplasmic protein, mutants of which are associated with primary open-angle glaucoma (POAG) and amyotrophic lateral sclerosis (ALS). The most abundant heat shock protein crystallin, known for its remarkable thermodynamic stability and chaperoning activity, allows ocular tissues to withstand stress. The presence of OPTN in ocular tissues is intriguing. Interestingly, OPTN also harbors heat shock elements in its promoter region. Sequence analysis of OPTN exhibits intrinsically disordered regions and nucleic acid binding domains. These properties hinted that OPTN might be endowed with sufficient thermodynamic stability and chaperoning activity. However, these attributes of OPTN have not yet been explored. Here, we studied these properties through thermal and chemical denaturation experiments and monitored the processes using CD, fluorimetry, differential scanning calorimetry, and dynamic light scattering. We found that upon heating, OPTN reversibly forms higher-order multimers. OPTN also displayed a chaperone-like function by reducing the thermal aggregation of bovine carbonic anhydrase. It regains its native secondary structure, RNA-binding property, and melting temperature (Tm) after refolding from a thermally as well as chemically denatured state. From our data, we conclude that OPTN, with its unique ability to revert from the stress-mediated unfolded state and its unique chaperoning function, is a valuable protein of the ocular tissues.
1. Introduction
Optineurin is a 67 kDa cytoplasmic protein expressed ubiquitously in various tissues.1,2 The name ‘Optineurin’ has been derived from OPTIc NEURopathy INducing protein. The mutants of OPTN are linked with primary open-angle glaucoma (POAG), a pathological condition that causes irreversible blindness.3,4 OPTN has been suggested as a neuroprotective protein.5 Two mutants of OPTN, the E478G, and Q398X, are found associated with a neurodegenerative disease called amyotrophic lateral sclerosis (ALS).6Besides its disease association, OPTN is a versatile protein that participates in numerous biological functions. It serves as a prominent autophagy adaptor protein by binding to the cargoes, promoting their fusion with autophagosomes, before their degradation.7 It also plays a role in golgi maintenance and vesicular trafficking.8,9 The POAG-associated E50K mutant causes failure in the transport of cargos, golgi fragmentation, and apoptotic cell death.10 OPTN also negatively regulates TNF-α mediated NF-kB activation, inhibits necrosis, and provides cellular protection.11,12 Structurally it is composed of a leucine zipper motif, a zinc finger motif, a ubiquitin-binding domain (UBD), multiple coiled–coiled motifs, and an LC3-interacting region (LIR).1 Each of these domains enables OPTN to bind to several other proteins such as Rab8, TANK binding kinase-1 (TBK1), Myosin VI, Huntington, serine/threonine kinase receptor-interacting protein 1 (RIP1), etc.11,13–15 OPTN has been proposed to provide cytoprotection against toxic aggregates of mutant huntingtin and mutant ataxin-3.16 It was also reported that the ALS-associated mutants of OPTN promotes the aggregation of ubiquitinated TDP-43.17 Besides, OPTN also undergoes post-translation modification, which helps downstream signalling.18 Reports suggest that OPTN is expressed in ocular tissues and eye lenses.2 While extensive research has been done to date to elucidate the function of OPTN in glaucoma and/or ALS, no group has yet focused on the possible role of OPTN in terms of its thermal stability and chaperoning activity in other tissues.
Crystallins are highly stable lens proteins formed and maintained over a lifetime.19 They exhibit exceptional stability and maintain solubility even under extreme environmental conditions such as UV radiation, dryness, and elevated temperature.18,19 Owing to the presence of heat shock elements in their genes' promoter region, they are also categorized as small heat shock proteins (HSPs).19,20 Another class is the 70 kDa heat shock family of molecular chaperones. These are ubiquitously expressed in various tissues, including the ocular tissues, and are highly conserved.21,22 HSPs are upregulated in response to stress and play critical roles in multiple cellular events like protein folding, protein translocation, and protein degradation.23
Interestingly, heat shock elements have also been identified in the promoter sequence of OPTN.1 These heat shock elements might upregulate OPTN transcription in response to environmental stress. We hypothesize that OPTN, with its multi-domain, multi-interactive property, might function as a stress-related protein that works as a chaperone to protect the ocular and other tissues from UV, temperature, and other stress-related damages.
To test this hypothesis, we studied the thermodynamic properties of OPTN, which could shed some light on its importance in different tissues. Here we show that OPTN is reversible from thermally or chemically denatured states allowing it to act as a sink to withstand various cellular stress. We also demonstrated a chaperone-like activity of OPTN, making it a crucial protein for eye lens and other tissues. Together our study highlights OPTN as a stress-tolerant, chaperone-like protein, ubiquitously present in different tissues which equip the cells to survive environmental stresses.
2. Results
2.1 Purification and identification of optineurin
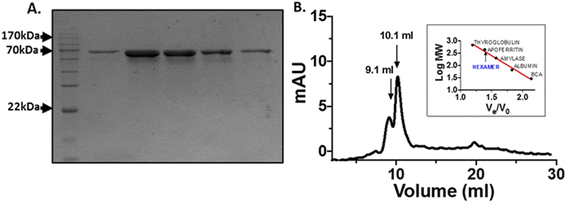
The His-tagged human OPTN was expressed and purified from BL21-gold (DE3) E. coli cells using the Ni-NTA affinity chromatography following standard protocol. The fractions were eluted by running a linear gradient from 50–500 mM imidazole concentration. A single band close to 70 kDa, equivalent to OPTN molecular weight of 67 kDa was observed on SDS-PAGE (Fig. 1A). The identity of this protein was confirmed by mass spectrometry and peptide mass fingerprinting analysis (Fig. S1†). The protein was analyzed by passing it through a Superdex 200 gel filtration column. Two peaks observed at 9.1 ml and 10.1 ml, respectively, corresponded to approximately 450 kDa and 700 kDa, indicating OPTN's natively hexameric or decameric nature (Fig. 1B).2.2 Structural changes in OPTN upon thermal denaturation
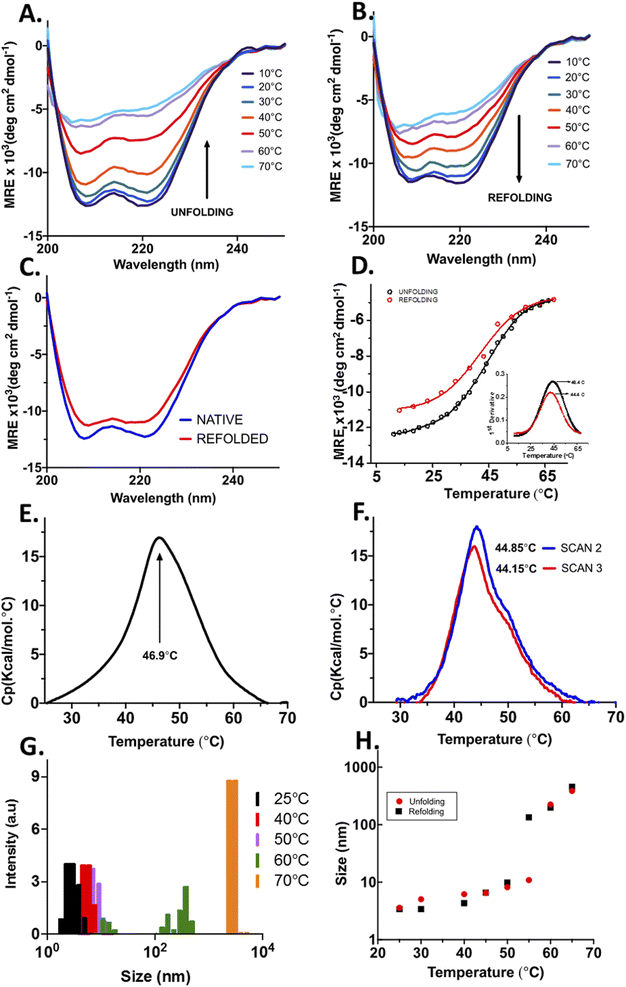
The structural content and thermal stability of OPTN were studied using circular dichroism spectroscopy. The CD spectrum of OPTN under native conditions showed two minima at 208 nm and 222 nm, representing a predominantly α-helical secondary structure. The secondary structural content estimated using the K2D3 software yielded ∼68% helix and 9% β-sheet.24 Upon gradually heating the protein at an increment of 2 °C, unfolding commences around 30 °C. Further increase in temperature resulted in a gradual decrease in the CD signal representing progressive unfolding. Nearly overlapping curves were obtained over the temperature range of 66–70 °C, signifying the completion of the unfolding transition, thereby giving rise to the denatured ensemble (Fig. 2A). We also observed an isodichroic point at 202 nm, suggestive of a two-state transition.25 The apparent thermodynamic parameters were obtained by fitting the CD-based thermal unfolding data to eqn (3) with the thermodynamic parameters provided in Table 1. Tm, the midpoint of thermal denaturation, was found to be ∼46.4 °C (Fig. 2D).| Tm (°C) | 46.4 ± 0.2 |
| ΔH (kcal mol−1) | 63.1 ± 0.8 |
| ΔCp (kcal K−1 mol−1) | 2.7 ± 0.1 |
| ΔS (kcal K−1 mol−1) | 0.20 ± 0.01 |
Temperature-mediated denaturation of OPTN using differential scanning calorimetry (DSC) was also performed. The Tm calculated from the curve was 46.9 ± 0.07 °C (Fig. 2E), close to that obtained from the equilibrium unfolding experiments.
To study whether the thermally denatured OPTN forms any oligomers, we measured its hydrodynamic radius using DLS after heating at various temperatures. The native OPTN at 25 °C displayed a size of around 6.5 nm. Above this temperature, a gradual increase in the size was observed. At 60 °C, the protein appeared to distribute into two distinct populations, one below 100 nm and another above 500 nm. At 70 °C a remarkable increase in the size of OPTN was observed, suggestive of the formation of very large oligomers (Fig. 2G).
2.3 Structural reversibility of OPTN from the thermally denatured state
When OPTN was cooled gradually from the 70 °C state, the CD spectrum started showing some secondary structural signatures below 60 °C. When the temperature was lowered further to 40 °C, a reversal of the CD signal was observed. Further reduction of temperature to 10 °C resulted in a near complete renaturation with the spectrum resembling that of the native protein (Fig. 2B). An isodichroic point was also obtained upon refolding. Structural content of the refolded protein was estimated to be 68% α-helix and 9% β-sheet respectively, close to that of the native protein before thermal denaturation (Fig. 2C).The ability of OPTN to undergo complete refolding was also confirmed by performing DSC on the protein that was cooled to 25 °C after heat treatment. Two subsequent scans were taken from which Tm values of 44.85 ± 0.04 °C and 44.15 ± 0.05 °C were obtained (Fig. 2F). These values are close to that of the native protein, further confirming significant reversibility.
We also measured the change in the size of OPTN with the reduction in temperature using DLS. Upon cooling from 70 °C the oligomers started dispersing into smaller-sized ensembles. When cooled down to 25 °C, the hydrodynamic radius of the protein resembled the size of the native protein, confirming complete reversibility (Fig. 2H).
2.4 Equilibrium unfolding and refolding of OPTN by chemical denaturation monitored by CD spectroscopy
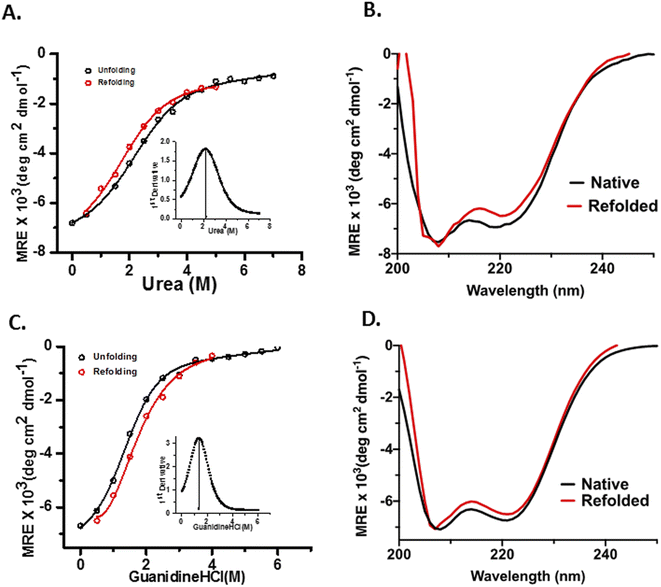
OPTN was subjected to different concentrations of urea (0–7 M) and GdHCl (0–5 M) and its unfolding transition was plotted using the CD signal at 222 nm as a function of urea or GdHCl and fitted using eqn (7) (see materials and methods). Similarly, a refolding curve was obtained by a stepwise dilution of the denaturant (Fig. 3). Complete unfolding of OPTN was observed in the presence of 5 M urea or 4 M GdHCl. They showed reversible sigmoidal transitions that could be fitted to a two-state model (Fig. 3A and C). Size exclusion chromatography performed at different denaturant concentrations resulted in gradual shifting of the primary hexameric peak to more expanded species (Fig. S3†). This is an interesting observation as it suggests that the hexameric OPTN is resistant to denaturant-mediated disassembly into monomeric subunits. The inability to dissociate into monomeric subunits (which should have appeared at a much higher elution volume) indicates that all denaturant-mediated transitions induce a conversion from the native hexameric (n = 6) to unfolded hexameric (n = 6) state. Based on this, we have analysed our data considering no change in the assembly of the protein and using a two-state model. The cm values for urea-induced unfolding and refolding were found to be 2.22 ± 0.11 M and 1.72 ± 0.11 M, respectively, as calculated from the first derivative of the unfolding transition curves (Fig. 3A, inset). Similarly, the GdHCl-induced unfolding and refolding yielded the cm values of 1.33 ± 0.06 M and 1.39 ± 0.07 M, respectively (Fig. 3C, inset). The free energy of the unfolding reaction [ΔG(D)] as a function of the denaturant was calculated using eqn (8) (see materials and methods). Extrapolation of the curve to 0 M denaturant yielded the ΔG(H2O) values of ∼1.7 kcal mol−1 and ∼1.6 kcal mol−1 for urea and GdHCl, respectively (Table 2).| Denaturant | ΔG(H2O) (kcal mol−1) | m (kcal mol−1 M−1) | cm (M) |
|---|---|---|---|
| Urea | 1.69 ± 0.08 | 0.76 ± 0.04 | 2.22 ± 0.11 |
| GdHCl | 1.62 ± 0.08 | 1.20 ± 0.06 | 1.33 ± 0.06 |
m-values have been shown to have a strong correlation with the change in accessible surface area (ΔASA)of proteins in general, with larger ΔASA resulting in larger m-values.26 For the 450 kDa hexameric OPTN, the m-value obtained for both urea and GdHCl are quite less as compared to that of a 450 kDa monomeric protein, implying that the oligomeric nature of the assembly prevents a significant change in the surface area as one progresses to the denatured ensemble. This is in further agreement with our hypothesis that the process of chemical denaturation does not perturb the hexameric state of OPTN. Moreover, the higher m-value of GdHCl is in line with the fact that GdHCl is a more potent denaturant than urea, in complete agreement with previous literature studies.
2.5 Reversibility of RNA-binding by OPTN after thermal denaturation
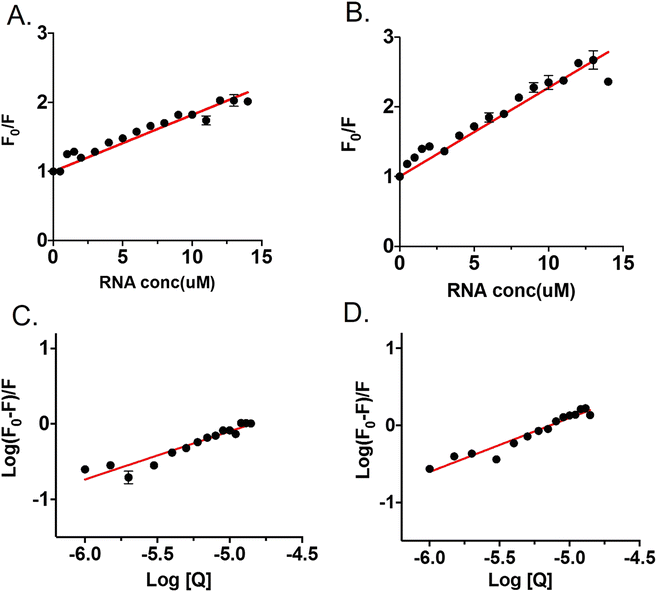
Fluorometric titration was performed with RNA against 7.5 μM OPTN. We observed a quenching in the intrinsic fluorescence of OPTN with increasing concentrations of RNA. From the Stern–Volmer plot, eqn (9) (see material and methods), the quenching constant was obtained (Fig. 4A). Eqn (10) was used to calculate the number of binding sites and the binding constant (Fig. 4C). The number of binding sites in folded OPTN was found to be 0.94 ± 0.04. When the experiment was repeated at 60 °C with partially denatured OPTN, we observed an inconsistent profile that could not be fitted to any binding model (Fig. S4†). However, when cooled back to 25 °C, the refolded OPTN showed consistent fluorescence quenching with an increase in RNA concentration, similar to the native OPTN (Fig. 4B and D). The calculated parameters (quenching constant, binding site and the binding constant) for heated and cooled OPTN were very similar to that observed for the native state, suggestive of refolding back into the native state (Table 3).| Native | Refolded | |
|---|---|---|
| KSV (μM) | 0.08 ± 0.001 | 0.13 ± 0.01 |
| K (10−6 mol L−1) | 4.61 ± 0.22 | 4.49 ± 0.24 |
| n | 0.94 ± 0.04 | 0.86 ± 0.04 |
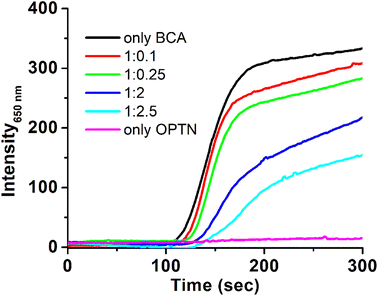
2.6 Prevention of thermal aggregation of BCA II by OPTN
To study if OPTN has any chaperoning activity, its effect on the thermal aggregation of bovine carbonic anhydrase II (BCA II) was monitored. BCA II when heated alone showed increased scattering due to the formation of larger-sized aggregates. However, aggregation decreased substantially in the presence of different concentrations of OPTN. The lag phase of initiation of Rayleigh scattering increased from 110 seconds for the BCA II alone, to 140 seconds for 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (BCAII
2 (BCAII![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OPTN) ratio. Intermittent ratios yielded intermittent results (Fig. 5). A significant decrease in the final scattering value was also observed in the presence of OPTN, suggesting a substantial reduction in aggregation.
OPTN) ratio. Intermittent ratios yielded intermittent results (Fig. 5). A significant decrease in the final scattering value was also observed in the presence of OPTN, suggesting a substantial reduction in aggregation.
3. Discussion
Transgenic mice expressing OPTN in the lens fiber remain localized to the lens cytoplasm.2 During early fetal development, with the onset of fiber differentiation, cells of the mammalian eye lens lose their mitochondria, ribosomes, and nuclei.19 While this is required to maintain transparency, the cells are crippled in their ability to synthesize or degrade proteins. In this scenario, the presence of OPTN in the ocular tissues raised our interest in its specific role. OPTN is also expressed in non-lenticular tissues, and its expression increases in some neurological diseases.1,4 Very little is known about the thermodynamic properties and function of optineurin (OPTN) in the context of its expression in the ocular or other tissues.Many of the designated cellular functions of OPTN are comparable to those carried out by HSPs. Sequence alignment of OPTN with other HSPs indicated that it does not contain the typical α-crystallin domain (Fig. S5†). However, with its nucleic acid (NA) binding domains, OPTN is more like the HSP 70. This prompted us to analyze the thermodynamic parameters of OPTN and evaluate whether it has any chaperoning activity comparable with other HSPs.
Under physiological conditions, OPTN exhibited hexameric assembly.27 Spectroscopic examination using CD depicted OPTN as primarily an α-helical protein consistent with previous reports (Fig. 2A).28 In contrast, our in silico analysis using online disorder prediction software like the IUPred2, PONDR, and the PONDR-FIT suggested the presence of ∼30% intrinsically disordered regions (IDRs) in OPTN (Fig. S2†).29–31 This difference could be because the in silico prediction considers OPTN's monomeric form. In contrast, CD provides averaged solution structural information where OPTN exists as an oligomer (hexamer or decamer, also observed through GFC and DLS). Having a well-established multifunctional role, OPTN is likely to dynamically shuffle between helical or disordered conformations, depending on whether it is involved in associative interaction with other partners. This finding indirectly supports that there might be a functional similarity between OPTN and HSPs, as IDRs have been reported in the structural core of HSP 27, 60, 70, and 90.32 They have also been found to form large oligomeric assemblies parallel to what we found in OPTN.33
To probe the unfolding thermodynamics and nature of the unfolding transition, we carried out equilibrium unfolding experiments using chemical denaturants wherein oligomers were not observed. With reversibility being observed in these equilibrium unfolding experiments, OPTN's N → U transition fitted to a two-state model giving rise to a ΔG(H2O) value close to 1.60 kcal mol−1. Moreover, the fact that the ΔG(H2O) values are quite close for both urea and GdHCl imply that electrostatic interactions do not play a major role in the unfolding of OPTN, with urea being neutral in nature, thereby providing insights into the forces holding the hexameric assembly together.
The hydrodynamic radius of native OPTN measured through DLS was 6.5 nm, confirming its hexameric state. The size increases with an increase in temperature. The size ranged from 250–890 nm at 65 °C, and at 70 °C it further increased to form poly dispersed oligomers ranging between 750–3000 nm. Remarkably, none of these oligomers crashed out of solution. Such heat-induced oligomeric transition has been reported for HSP 90 above 50 °C, with oligomers remaining in solution without precipitation.34 Interestingly, the oligomers formed by OPTN through weak non-covalent interactions were transient and spontaneously reverted to their native hexameric form upon cooling. This confirmed that OPTN undergoes a reversible thermal unfolding transition. CD and DSC experiments confirmed the reversibility of OPTN from the heat-denatured state. The Tm calculated from the unfolding and refolding curves by all these techniques was found to be around 46–47 °C. Since heat-denaturation of OPTN was always associated with the oligomeric association, as observed from the DLS data, it was not easy to make an accurate thermodynamic assessment of the thermally induced unfolding transitions, in particular, the unfolded baseline. Consequently, based on the fact that the chemical denaturation experiments pointed towards a native hexameric to unfolded hexameric transition (Fig. S3†), we assumed a similar two-state transition during thermal unfolding. However, it should be noted that the signal beyond 50 °C will have appreciable contributions from the higher-order reversible transient oligomers, thereby affecting the extracted thermodynamic parameters. It should also be noted that thermal denaturation of OPTN is associated with oligomeric assembly, and therefore is a concentration-dependent phenomenon. The thermodynamic parameters calculated here is at one protein concentration. A thorough investigation in future may therefore be undertaken at multiple protein concentrations to validate these findings.
The localization of OPTN in stress granules and the presence of a leucine-zipper and zinc-finger domains in its structure hinted toward its possible nucleic acid-binding function. A long non-coding RNA MalL1 has been reported to bind and stabilize OPTN.35 OPTN showed a good binding affinity with RNA with a Kd of 4.61 ± 0.22 μM. Interestingly, HSP70 has also been reported to bind U-rich RNA and stabilize mRNA, bringing functional parallelism between the two proteins.36 The refolded OPTN could bind RNA with almost the same affinity as the native protein, suggesting the regaining of the conserved nucleotide-binding domain (Fig. 4).
Interestingly, the OPTN promoter showed the presence of heat shock elements (HSEs) i.e., conserved regions which are binding sites for heat shock factors.1 This suggests that OPTN might be a heat shock protein (HSP) whose expression could be elevated during heat stress. In a way, it is similar to the multiple HSEs reported in the promoter region of HSP70.37 Based on these similarities between OPTN and HSPs in their cellular location, renaturation potential, multimeric nature, presence of HSE in promoters, and other thermodynamic parameters we were motivated to look for OPTN's chaperoning activity. The significant reduction in the thermal aggregation of BCA II with increasing concentration of OPTN confirmed its chaperoning activity. The function of OPTN is similar to the one displayed by the N-terminal domain of Pyrococcus furiosus L-aspargainase (NPfA), where reduction in aggregation was observed in a dose dependant manner.38 In these experiments, although there was an increase in the size of OPTN with temperature (analogous to the one observed in DLS), the relatively very low concentrations made the overall effect insignificant (Fig. S6†).
4. Conclusion
Our data reveal that the hexameric assembly of OPTN remains intact during the large thermal and chemical denaturation ranges employed here, signifying that over the physiologically admissible temperature variation (which is much smaller), the shift of OPTN from its functional form is negligible, the same having important functional consequences for its chaperone-like action. Therefore, we conclude that OPTN is a protein with significant structural reversibility after denaturation. OPTN is a versatile protein that could mould into a conformational multimer. We found that it interacts with RNA in its native state and also it regains this functionality after refolding. Reversible thermal transition to multimeric form is of special interest as it displays chaperone-like activity, presumably to perform a myriad of cellular functions. The thermodynamic properties of OPTN presented here lays the foundation for further explorations and validation with additional experiments. We also show a strong and logical correlation of its existence in the ocular tissues, the lens, and various other tissues where the OPTN's chaperoning and stress-absorbing role is of special significance.5. Materials and methods
5.1 Purification of optineurin
The optineurin clone pDEST17-hOPTN was a gift from Jon Ashwell (Addgene plasmid #23053; http://n2t.net/addgene: 23053; RRID: Addgene_23053). The clone was transformed into the BL21-Gold (DE3) E. coli expression host. Cells were grown in LB media (HiMedia, India) containing 100 mM ampicillin (Biobasic Inc.) in an incubator shaker (New Brunswick™ Innova® 44/44R) at 37 °C till an OD600 of 0.6, following induction with 1 mM IPTG (Biobasic Inc.) cells were allowed to grow further for 16 hours, before harvesting. The protein expression was checked by boiling small culture aliquots in an SDS-loading buffer and running on 12% SDS-PAGE. The cells were harvested by centrifugation. The pellet was sonicated in lysis buffer (50 mM sodium phosphate, 300 mM NaCl, and 10 mM imidazole, pH 8.0). The 6× His-tagged OPTN was purified under native conditions using Ni-NTA affinity chromatography. Washing and elution were done with imidazole concentrations ranging from 50–500 mM. Eluted fractions showing OPTN band on SDS-PAGE were pooled and dialyzed against 50 mM sodium phosphate and 50 mM NaCl, pH 8.0. Dialysed sample was concentrated using a 30 kDa cut-off centricon and stored at −80 °C freezer until further use.5.2 Thermal unfolding and refolding of OPTN
The thermal midpoint (Tm), enthalpy (ΔH), and entropy (ΔS) of unfolding and the change in heat capacity (ΔCp) were obtained by fitting the following equations to the data:39
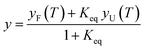 | (1) |
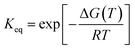 | (2) |
ΔG(T) = ΔH + ΔCp(T − Tm) − T(ΔS + ΔCp![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln(T/Tm)) ln(T/Tm))
| (3) |
| yF(T) = aF + bF(T) | (4) |
| yU(T) = aU + bU(T) | (5) |
The Stokes–Einstein equation was used to calculate the hydrodynamic radii (Rh)
| Rh = kBT/6πηDT | (6) |
The data obtained was fitted using a two-state non-linear regression equation, considering the pre and post-transition baselines using the following equation;40
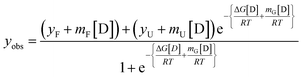 | (7) |
| ΔG[D] = ΔG(H2O) − m[D] | (8) |
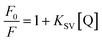 | (9) |
To analyse the binding interaction between OPTN and RNA, the binding constant was determined from the static quenching interaction. The following equation42 was used to determine the equilibrium between the free and bound species;
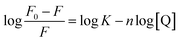 | (10) |
To assess RNA binding with denatured OPTN, the fluorescence titration experiments were simultaneously performed at 60 °C.
To assess the functional reversibility of OPTN from a thermally denatured state, the heated and cooled protein was again titrated with an increasing concentration of RNA, and the binding constants in each case were determined as mentioned above.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1, 1
0.1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.25, and 1
0.25, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) were prepared in 50 mM sodium phosphate, 100 mM NaCl, pH 7.4. The samples were then taken in a cuvette and placed in a pre-heated sample chamber at 65 °C in a PerkinElmer LS55 spectrofluorometer. The Rayleigh scattering was measured by keeping both the excitation and emission wavelengths at 650 nm and the excitation and emission slits fixed at 5 nm and 10 nm, respectively. Data was plotted as a function of time.
2) were prepared in 50 mM sodium phosphate, 100 mM NaCl, pH 7.4. The samples were then taken in a cuvette and placed in a pre-heated sample chamber at 65 °C in a PerkinElmer LS55 spectrofluorometer. The Rayleigh scattering was measured by keeping both the excitation and emission wavelengths at 650 nm and the excitation and emission slits fixed at 5 nm and 10 nm, respectively. Data was plotted as a function of time.Author contributions
Anjali Dixit: conceptualization, data curation, investigation, formal analysis validation, writing – original draft, writing – review and editing. Ankan Chakraborty: investigation, visualization; writing – review and editing. Jyoti Rani Nath; investigation, analysis, Pramit K Chowdhury: writing – review and editing, formal analysis. Bishwajit Kundu: conceptualization, funding acquisition, project administration, supervision, visualization; writing – review and editing.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The authors thank the Indian Institute of Technology Delhi for infrastructural support. AD acknowledges the fellowship support from the University Grant Commission, Govt. of India. AD also acknowledges Shubham Vashishtha, Dr Dushyant Garg, and Dr Kritika Singh for technical discussions. The authors acknowledge Dr Bhumika Ray and Nilanjana Bose for providing BCA II.References
- H. Ying and B. Y. J. T. Yue, Cellular and molecular biology of optineurin, Int Rev Cell Mol Biol., 2012, 294, 223–258, DOI:10.1016/B978-0-12-394305-7.00005-7.
- M. Kroeber, A. Ohlmann, P. Russell and E. R. Tamm, Transgenic studies on the role of optineurin in the mouse eye, Exp. Eye Res., 2006, 82(6), 1075–1085, DOI:10.1016/J.EXER.2005.11.004.
- T. Rezaie, A. Child and R. Hitchings, et al., Adult-onset primary open-angle glaucoma caused by mutations in optineurin, Science, 2002, 295, 1077–1079, DOI:10.1126/science.1066901.
- S. Zhang, Z. Shao and X. Liu, et al., The E50K optineurin mutation impacts autophagy-mediated degradation of TDP-43 and leads to RGC apoptosis in vivo and in vitro, Cell Death Discovery, 2021, 7(1), 1–13, DOI:10.1038/s41420-021-00432-0.
- A. Markovinovic, R. Cimbro, T. Ljutic, J. Kriz, B. Rogelj and I. Munitic, Optineurin in amyotrophic lateral sclerosis: Multifunctional adaptor protein at the crossroads of different neuroprotective mechanisms, Prog. Neurobiol., 2017, 154, 1–20, DOI:10.1016/J.PNEUROBIO.2017.04.005.
- H. Maruyama, H. Morino and H. Ito, et al., Mutations of optineurin in amyotrophic lateral sclerosis, Nature, 2010, 465(7295), 223–226, DOI:10.1038/nature08971.
- P. Wild, H. Farhan and D. G. mcewan, et al., Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth, Science, 2011, 333(6039), 228–233, DOI:10.1126/science.1205405.
- D. A. Sahlender, R. C. Roberts and S. D. Arden, et al., Optineurin links myosin VI to the Golgi complex and is involved in Golgi organization and exocytosis, J. Cell Biol., 2005, 169(2), 285–295, DOI:10.1083/JCB.200501162.
- D. d. Toro, J. Alberch and F. Lazaro-Dieguez, et al., Mutant huntingtin impairs post-Golgi trafficking to lysosomes by delocalizing optineurin/Rab8 complex from the Golgi apparatus, Mol. Biol. Cell, 2009, 20(5), 1478–1492, DOI:10.1091/mbc.E08-07-0726.
- G. Swarup and Z. Sayyad, Altered functions and interactions of glaucoma-associated mutants of optineurin, Front. Immunol., 2018, 9, 1287, DOI:10.3389/FIMMU.2018.01287/BIBTEX.
- G. Zhu, C. J. Wu, Y. Zhao and J. D. Ashwell, Optineurin negatively regulates tnfα- induced NF-kappab activation by competing with NEMO for ubiquitinated RIP, Curr. Biol., 2007, 17(16), 1438–1443, DOI:10.1016/J.CUB.2007.07.041.
- A. Nagabhushana, M. Bansal and G. Swarup, Optineurin is required for CYLD-dependent inhibition of tnfα-induced NF-κb activation, PLoS One, 2011, 6(3), e17477, DOI:10.1371/JOURNAL.PONE.0017477.
- J. S. Y. Au, C. Puri, G. Ihrke, J. Kendrick-Jones and F. Buss, Myosin VI is required for sorting of AP-1B–dependent cargo to the basolateral domain in polarized MDCK cells, J. Cell Biol., 2007, 177(1), 103–114, DOI:10.1083/JCB.200608126.
- S. Morton, L. Hesson, M. Peggie and P. Cohen, Enhanced binding of TBK1 by an optineurin mutant that causes a familial form of primary open angle glaucoma, FEBS Lett., 2008, 582(6), 997–1002, DOI:10.1016/J.FEBSLET.2008.02.047.
- T. A. Ryan and D. A. Tumbarello, Optineurin: a coordinator of membrane-associated cargo trafficking and autophagy, Front. Immunol., 2018, 9, 1024, DOI:10.3389/FIMMU.2018.01024.
- S. C. Moharir, A. K. Raghawan, R. Ramaswamy and G. Swarup, Autophagy-independent cytoprotection by optineurin from toxicity of aggregates formed by mutant huntingtin and mutant ataxin-3, J. Biochem., 2022, 171(5), 555–565, DOI:10.1093/jb/mvac011.
- T. Kakihana, M. Takahashi, Y. Katsuragi, S. I. Yamashita, J. Sango, T. Kanki, O. Onodera and M. Fujii, The optineurin/TIA1 pathway inhibits aberrant stress granule formation and reduces ubiquitinated TDP-43, iScience, 2021, 24(7), 102733, DOI:10.1016/j.isci.2021.102733.
- H. Ying, X. Shen, B. Park and B. Y. J. T. Yue, Posttranslational modifications, localization, and protein interactions of optineurin, the product of a glaucoma gene, PLoS One, 2010, 5(2), e9168, DOI:10.1371/JOURNAL.PONE.0009168.
- H. Bloemendal, W. de Jong, R. Jaenicke, N. H. Lubsen, C. Slingsby and A. Tardieu, Ageing and vision: structure, stability and function of lens crystallins, Prog. Biophys. Mol. Biol., 2004, 86(3), 407–485, DOI:10.1016/J.PBIOMOLBIO.2003.11.012.
- K. W. Roskamp, C. N. Paulson, W. D. Brubaker and R. W. Martin, Function and aggregation in structural eye lens crystallins, Acc. Chem. Res., 2020, 53(4), 863–874, DOI:10.1021/ACS.ACCOUNTS.0C00014/ASSET/IMAGES/ACS.ACCOUNTS.0C00014.SOCIAL.JPEG_V03.
- R. Klemenz, E. Frohli, R. H. Steiger, R. Schafer and A. Aoyama, αB-crystallin is a small heat shock protein, Proc. Natl. Acad. Sci. U. S. A., 1991, 88(9), 3652–3656, DOI:10.1073/PNAS.88.9.3652.
- M. Bagchi, M. Katar and H. Maisel, Heat shock proteins of adult and embryonic human ocular lenses, J. Cell. Biochem., 2002, 84(2), 278–284, DOI:10.1002/JCB.10023.
- J. G. Kiang and G. C. Tsokos, Heat shock protein 70 kDa: molecular biology, biochemistry, and physiology, Pharmacol. Ther., 1998, 80(2), 183–201, DOI:10.1016/S0163-7258(98)00028-X.
- C. Louis-Jeune, M. A. Andrade-Navarro and C. Perez-Iratxeta, Prediction of protein secondary structure from circular dichroism using theoretically derived spectra, Proteins, 2012, 80(2), 374–381, DOI:10.1002/PROT.23188.
- J. W. Nelson and N. R. Kallenbach, Stabilization of the ribonuclease S-peptide α-helix by trifluoroethanol, Proteins, 1986, 1(3), 211–217, DOI:10.1002/PROT.340010303.
- J. K. Myers, C. Nick Pace and J. Martin Scholtz, Denaturant m values and heat capacity changes: relation to changes in accessible surface areas of protein unfolding, Protein Sci., 1995, 4(10), 2138–2148, DOI:10.1002/PRO.5560041020.
- X. Shen, H. Ying and Y. Qiu, et al., Processing of optineurin in neuronal cells, J. Biol. Chem., 2011, 286(5), 3618, DOI:10.1074/JBC.M110.175810.
- H. Ying and J. T. Yue BY, Mammalian expression and biophysical examination of human wild type optineurin protein, Invest. Ophthalmol. Visual Sci., 2013, 54, 1610–1616 Search PubMed.
- B. Mészáros, G. Erdös and Z. Dosztányi, IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding, Nucleic Acids Res., 2018, 46(W1), W329–W337, DOI:10.1093/NAR/GKY384.
- P. Romero, Z. Obradovic, C. Kissinger, J. E. Villafranca and A. K. Dunker, Identifying disordered regions in proteins from amino acid sequence, Conf. Neural Networks, 1997, 1, 90–95, DOI:10.1109/ICNN.1997.611643.
- B. Xue, R. L. Dunbrack, R. W. Williams, A. K. Dunker and V. N. Uversky, PONDR-FIT: A meta-predictor of intrinsically disordered amino acids, Biochim. Biophys. Acta, Proteins Proteomics, 2010, 1804(4), 996–1010, DOI:10.1016/J.BBAPAP.2010.01.011.
- M. Sudnitsyna, E. Mymrikov, A. Seit-Nebi and B. Gusev N, The role of intrinsically disordered regions in the structure and functioning of small heat shock proteins, Curr. Protein Pept. Sci., 2012, 13(1), 76–85, DOI:10.2174/138920312799277875.
- A. P. Arrigo, Human small heat shock proteins: Protein interactomes of homo- and hetero-oligomeric complexes: An update, FEBS Lett., 2013, 587(13), 1959–1969, DOI:10.1016/J.FEBSLET.2013.05.011.
- M. Haslbeck, J. Peschek, J. Buchner and S. Weinkauf, Structure and function of α-crystallins: Traversing from in vitro to in vivo, Biochim. Biophys. Acta, 2016, 1860(1), 149–166, DOI:10.1016/J.BBAGEN.2015.06.008.
- M. Aznaourova, H. Janga and S. Sefried, et al., Noncoding RN.A MaIL1 is an integral component of the TLR4–TRIF pathway, Proc. Natl. Acad. Sci. U. S. A., 2020, 117(16), 9042–9053, DOI:10.1073/PNAS.1920393117.
- A. Kishor, E. J. F. White, A. E. Matsangos, Z. Yan, B. Tandukar and G. M. Wilson, Hsp70's RNA-binding and mRNA-stabilizing activities are independent of its protein chaperone functions, J. Biol. Chem., 2017, 292(34), 14122–14133, DOI:10.1074/JBC.M117.785394.
- D. D. Mosser, N. G. Theodorakis and R. I. Morimoto, Coordinate changes in heat shock element-binding activity and HSP70 gene transcription rates in human cells, Mol. Cell. Biol., 1988, 8(11), 4736–4744, DOI:10.1128/MCB.8.11.4736-4744.1988.
- R. Tomar, D. K. Garg, R. Mishra, A. K. Thakur and B. Kundu, N-terminal domain of Pyrococcus furiosus l-asparaginase functions as a non-specific, stable, molecular chaperone, FEBS J., 2013, 280(11), 2688–2699, DOI:10.1111/febs.12271.
- A. Malik, J. Kundu, S. K. Mukherjee and P. K. Chowdhury, Myoglobin unfolding in crowding and confinement, J. Phys. Chem. B, 2012, 116(43), 12895–12904, DOI:10.1021/JP306873V/SUPPL_FILE/JP306873V_SI_001.PDF.
- M. M. Santoro and D. W. Bolen, Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl alpha-chymotrypsin using different denaturants, Biochemistry, 1988, 27(21), 8063–8068, DOI:10.1021/BI00421A014/ASSET/BI00421A014.FP.PNG_V03.
- E. Bentmann, M. Neumann, S. Tahirovic, R. Rodde, D. Dormann and C. Haass, Requirements for stress granule recruitment of fused in sarcoma (FUS) and TAR DNA-binding Protein of 43 kDa (TDP-43), J. Biol. Chem., 2012, 287(27), 23079, DOI:10.1074/JBC.M111.32875.
- V. D. Suryawanshi, L. S. Walekar, A. H. Gore, P. v. Anbhule and G. B. Kolekar, Spectroscopic analysis on the binding interaction of biologically active pyrimidine derivative with bovine serum albumin, J. Pharm. Anal., 2016, 6(1), 56–63, DOI:10.1016/J.JPHA.2015.0.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ra07931c |
| This journal is © The Royal Society of Chemistry 2023 |