Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
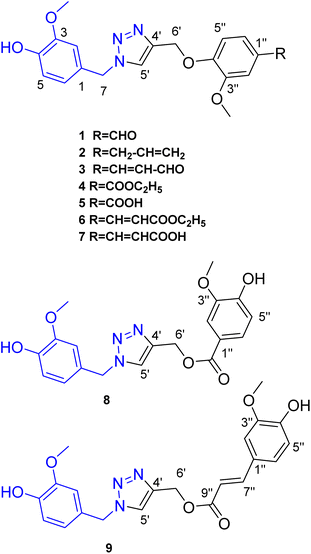
Click-designed vanilloid-triazole conjugates as dual inhibitors of AChE and Aβ aggregation†
Marwa Elsbaey *a,
Yasuhiro Igarashib,
Mahmoud A. A. Ibrahim
*a,
Yasuhiro Igarashib,
Mahmoud A. A. Ibrahim cd and
Eman Elattara
cd and
Eman Elattara
aPharmacognosy Department, Faculty of Pharmacy, Mansoura University, Mansoura 35516, Egypt. E-mail: marwaelsebay1611@mans.edu.eg
bBiotechnology Research Center and Department of Biotechnology, Toyama Prefectural University, 5180 Kurokawa, Imizu, Toyama, 939-0398, Japan
cComputational Chemistry Laboratory, Chemistry Department, Faculty of Science, Minia University, 61519, Egypt
dSchool of Health Sciences, University of KwaZulu-Natal, Westville, Durban 4000, South Africa
First published on 19th January 2023
Abstract
Based on their reported neuroprotective properties, vanilloids provide a good starting point for the synthesis of anti-Alzheimer's disease (AD) agents. In this context, nine new 1,2,3-triazole conjugates of vanilloids were synthesized via click chemistry. The compounds were tested for their effect on acetylcholine esterase (AChE) and amyloid-beta peptide (Aβ) aggregation. The triazole esters (E)-(1-(4-hydroxy-3-methoxybenzyl)-1H-1,2,3-triazol-4-yl)methyl 3-(4-hydroxy-3 methoxyphenyl)acrylate 9 and (1-(4-hydroxy-3-methoxybenzyl)-1H-1,2,3-triazol-4-yl)methyl-4-hydroxy-3-methoxybenzoate 8 displayed dual inhibitory activity for AChE and Aβ aggregation with IC50 values of 0.47/0.31 μM and 1.2/0.95 μM, respectively, as compared to donepezil (0.27 μM) and tacrine (0.41 μM), respectively. The results showed that the triazole ester moiety is more favorable for the activity than the triazole ether moiety. This could be attributed to the longer length of the spacer between the two vanillyl moieties in the triazole esters. Furthermore, the binding affinities and modes of the triazole esters 9 and 8 were examined against AChE and Aβ utilizing a combination of docking predictions and molecular dynamics (MD) simulations. Docking computations revealed promising binding affinity of triazole esters 9 and 8 as potential AChE, Aβ40, and Aβ42 inhibitors with docking scores of −10.4 and −9.4 kcal mol−1, −5.8 and −4.7 kcal mol−1, and −3.3 and −2.9 kcal mol−1, respectively. The stability and binding energies of triazole esters 9 and 8 complexed with AChE, Aβ40, and Aβ42 were measured and compared to donepezil and tacrine over 100 ns MD simulations. According to the estimated binding energies, compounds 9 and 8 displayed good binding affinities with AChE, Aβ42, and Aβ40 with average ΔGbinding values of −32.9 and −31.8 kcal mol−1, −12.0 and −10.5 kcal mol−1, and −20.4 and −16.6 kcal mol−1, respectively. Post-MD analyses demonstrated high steadiness for compounds 9 and 8 with AChE and Aβ during the 100 ns MD course. This work suggests the triazole conjugate of vanilloids as a promising skeleton for developing multi-target potential AD therapeutics.
1. Introduction
Alzheimer's disease (AD) is a progressive neurodegenerative disorder and is the most prevalent cause of dementia.1 It affects more than 40 million people worldwide.2 Even though the pathophysiology of AD has not been completely elucidated, cholinergic deficiency and amyloid-β peptide (Aβ) deposition are widely accepted as important features of the pathophysiology of AD,3 the pathological aggregates of Aβ deposits being known as senile plaques.4 Acetylcholine esterase (AChE) has been reported to consistently co-localize with Aβ deposits and induce their assembly by forming a complex with the growing fibrils.4The enzyme AChE is involved in the hydrolysis of the neurotransmitter acetylcholine (ACh). The profound loss of forebrain cholinergic neurons during the progression of AD, results in a progressive decline in acetylcholine. Current therapies are mostly based on AChE inhibitors (AChEI) to reverse the cholinergic deficit.5 Dual targeted inhibitors of AChE and Aβ aggregation are the main focus of AD paradigm.6 These drugs can be synthesized or harvested from nature, the advantage of the latter being the potential for chemical diversity, biological selectivity and favorable properties. Natural products and their derivatives represent more than 50% of the market pharmaceutics.7
Vanilloids are a group of natural products that are characterized by the presence of a vanillyl group. They include vanillin, vanillic acid, eugenol, capsaicin etc. Vanilloids attracted the attention of the authors because of their reported neuroprotective properties. Vanillin is reported to inhibit both Aβ aggregation and AChE and its profound antioxidant activity in neuroblastoma cells.8 Eugenol,9 vanillic acid,10 ferulic acid,11 curcumin, and capsaicin12 are reported to decrease or inhibit AChE. Vanillin,13 eugenol,14 ferulic acid, its derivatives,15 curcumin16 and capsaicin17 are also reported to inhibit Aβ aggregation. Vanillin derivatives have been reported as multi-target drugs for AD treatment.8,18 Several ferulic acid and its analogs based scaffolds were developed for management of AD.19,20 Consequently, the authors have selected the vanilloid pharmacophore to design potential anti-AD drugs. The triazole ring is reported as a good linker to combine pharmacophores into innovative bioactive functional molecules.21 Furthermore, several triazole-based compounds were reported as promising inhibitors against AD.22–25 In this context, we are encouraged to design new 1,2,3-triazole-conjugates of vanilloids and investigate their neuroprotective activity against AD. The molecular docking technique was utilized to anticipate the docking scores and poses of the most potent triazole esters 9 and 8 with AChE and Aβ40/42. The docked structures of compounds 9 and 8 complexed with AChE and Aβ40/42 were then subjected to MD over the simulation time of 100 ns. Structural and energetical analyses were utilized to examine the constancy of compounds 9 and 8 complexed with the investigated targets over 100 ns MD.
2. Results and discussion
2.1. Chemistry
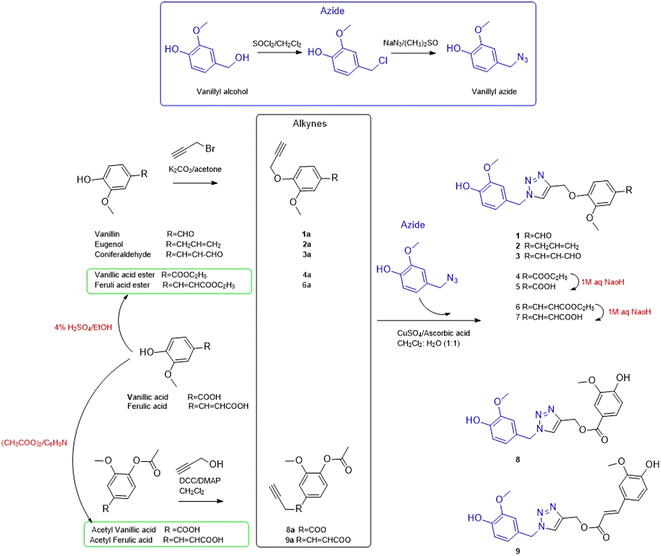
Based on the reported anti-Alzheimer properties of both of vanilloids and triazole-based compounds, the authors set a rationale for synthesis of triazole-conjugates of vanilloids via click chemistry. Nine new 1,2,3-triazole-conjugates of vanilloids (Fig. 1) were prepared via click chemistry (Scheme 1).For azide, vanillyl alcohol were purchased from Sigma-Aldrich. For alkynes, ferulic acid, and vanillin were purchased from Sigma-Aldrich. Coniferaldehyde and vanillic acid were previously isolated from Cocos nucifera L.26 Eugenol was extracted and purified from clove oil using 30% aqueous KOH, according to the literature.27 Vanillyl alcohol was converted to vanillyl azide and reacted with different alkynes. The monoalkynes were prepared from natural vanilloids, as will be described in the experimental part.
Compounds 1–3 were obtained via click reaction of vanillyl azide with the respective mono-alkyne ether. Compounds 4 and 6 were obtained via click reaction of vanillyl azide with the respective mono-alkyne ether after masking the carboxylic moiety with an ester. Compounds 5 and 7 were obtained from the ester hydrolysis of compounds 4 and 6, respectively. Compounds 8 and 9 were obtained via click reaction of vanillyl azide with the mono-alkyne ester after masking the hydroxyl group.
Investigation of the 1H-NMR Table 1 and APT Table 2 spectral data of the compounds confirmed the formation of a triazole moiety in each compound. The triazole moiety was characterized in 1H-NMR spectrum by the presence of the methine proton signal at δH ranging from 7.60 to 8.27, in addition to the methylene protons (–CH2O–) and (–CH2N–) at δH ranging from 5.19 to 5.40 and 5.35 to 5.48, respectively.
| H | 1 | 2a | 3a | 4 | 5 | 6 | 7 | 8a | 9 |
|---|---|---|---|---|---|---|---|---|---|
| a Is recorded in CDCl3. The rest of compounds are recorded in DMSO-d6.b Overlapped. | |||||||||
| Azide moiety | |||||||||
| 2 | 6.99, brs | 6.74, brs | 6.69, brs | 6.99, brs | 6.98, brs | 7.00, brs | 6.98, brs | 6.76 (1.8) | 6.98, brs |
| 5 | 6.73–6.79 | 6.88 (8) | 6.83 (8) | 6.74–6.77 | 6.77 | 6.76–6.79 | 6.76 | 6.88 (8) | 6.76–6.77 |
| 6 | 6.73–6.79 | 6.78 (8, 1.8) | 6.75 (2.0, 8) | 6.74–6.77 | 6.77 | 6.76–6.79 | 6.76 | 6.80 (8, 1.8) | 6.76–6.77 |
| 7 | 5.46, 2H, s | 5.38, 2H, s | 5.35, 2H, s | 5.46, 2H, s | 5.48, 2H, s | 5.45, 2H, s | 5.46, 2H, s | 5.40, 2H, s | 5.45, 2H, s |
| OCH3 | 3.74, s | 3.78, 3H, s | 3.75, 3H, s | 3.74, 3H, s | 3.74, 3H, s | 3.74, 3H, s | 3.73, 3H, s | 3.78, 3H, s | 3.74, 3H, s |
| OH | 9.14 | — | — | 9.15 | 9.14 | — | — | 9.14 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||||
| Triazole moiety | |||||||||
| 5′ | 8.27, s | 7.53, s | 7.52, s | 8.25, s | 8.25, s | 8.19, s | 8.23, s | 7.60, s | 8.19, s |
| 6′ | 5.23, 2H, s | 5.20, 2H, s | 5.23, 2H, s | 5.19, 2H, s | 5.20, 2H, s | 5.21, 2H, s | 5.14, 2H, s | 5.40, 2H, s | 5.21, 2H, s |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||||
| Alkyne moiety | |||||||||
| 2′′ | 7.37–7.39 | 6.69b | 6.99, brs | 7.44 (1.8) | 7.44, brs | 7.32, brs | 7.31, brs | 7.5 (1.8) | 7.32, brs |
| 5′′ | 7.37–7.39 | 6.92 (8.2) | 7.10 (8.3) | 7.57 (8.0, 1.8) | 7.55 (8.2) | 7.2b | 7.16–7.31 | 7.57 (8.0, 1.8) | 7.14b |
| 6′′ | 7.54 (8.2, 1.5) | 6.66–6.69b | 7.06b | 7.26 (8) | 7.24 (8.2) | 6.76–6.79b | 7.16–7.31 | 6.89 (8) | 7.12b |
| 7′′ | 9.84, s | 3.3 (6.7) | 7.32 (15.8) | — | — | 7.55 (16) | 7.52 (16) | — | 7.56 (15.9) |
| 8′′ | — | 5.92, m | 6.53 (7.7, 15.8) | — | — | 6.47 (16) | 6.45 (16) | — | 6.48 (15.9) |
| 9′′ | — | 5.06, m | 9.57 (7.7) | — | — | — | — | — | — |
| OCH3 | 3.79, 3H, s | 3.79, 3H, s | 3.80, 3H, s | 3.77, 3H, s | 3.76, 3H, s | 3.79, 3H, s | 3.76, 3H, s | 3.79, 3H, s | 3.79, 3H, s |
| CH2 | — | — | — | 4.27, 2H (7.1) | — | 4.17, 2H (7.1) | — | ||
| CH3 | — | — | — | 1.3, 3H (7.1) | — | 1.2, 3H, (7.1) | — | ||
| COOH | — | 12.7 | — | — | — | ||||
| 1 | 2a | 3a | 4 | 5 | 6 | 7 | 8a | 9 | |
|---|---|---|---|---|---|---|---|---|---|
| a Recorded in CDCl3. The rest of compounds are recorded in DMSO-d6. | |||||||||
| Azide moiety | |||||||||
| 1 | 126.5, qC | 126.1, qC | 126.1, qC | 126.5, qC | 126.5, qC | 126.6, qC | 126.5, qC | 126.0, qC | 126.6, qC |
| 2 | 112.6, CH | 112.3, CH | 110.9, CH | 112.6, CH | 112.6, CH | 112.7, CH | 112.6, CH | 110.9, CH | 112.7, CH |
| 3 | 147.6, qC | 147.2, qC | 147.2, qC | 147.7, qC | 147.6, qC | 147.7, qC | 147.6, qC | 147.2, qC | 147.7, qC |
| 4 | 146.7, qC | 146.4, qC | 146.4, qC | 146.7, qC | 146.7, qC | 146.7, qC | 146.7, qC | 146.4, qC | 146.7, qC |
| 5 | 115.5, CH | 114.8, CH | 114.8, CH | 115.5, CH | 115.5, CH | 115.4, CH | 115.5, CH | 114.9, CH | 115.5, CH |
| 6 | 121.1, CH | 121.6, CH | 121.8, CH | 121.1, CH | 121.1, CH | 121.1, CH | 121.1, CH | 121.7, CH | 121.2, CH |
| 7 | 53.0, CH2 | 54.2, CH2 | 54.5, CH2 | 52.9, CH2 | 52.9, CH2 | 53.0, CH2 | 52.9, CH2 | 54.3, CH2 | 52.9, CH2 |
| OCH3 | 55.6, CH3 | 55.9, CH3 | 56.09, CH3 | 55.6, CH3 | 55.6, CH3 | 55.62, CH3 | 55.6, CH3 | 56.09, CH3 | 55.7, CH3 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||||
| Triazole moiety | |||||||||
| 4′ | 142.1, qC | 144.8, qC | 144.2, qC | 142.3, qC | 142.4, qC | 142.6, qC | 142.6, qC | 143.4, qC | 142.3, qC |
| 5′ | 124.8, CH | 122.7, CH | 123.2, CH | 124.7, CH | 124.7, CH | 124.6, CH | 124.6, CH | 124.5, CH | 124.6, CH |
| 6′ | 61.7, CH2 | 63.4, CH2 | 63.0, CH2 | 61.6, CH2 | 61.6, CH2 | 61.6, CH2 | 61.6, CH2 | 57.7, CH2 | 57.0, CH2 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||||
| Alkyne moiety | |||||||||
| 1′′ | 129.9, qC | 133.9, qC | 127.8, qC | 122.4, qC | 123.4, qC | 127.4, qC | 127.6, qC | 121.5, qC | 125.5, qC |
| 2′′ | 109.6, CH | 110.9, CH | 110.3, CH | 111.8, CH | 112.1, CH | 110.6, CH | 110.5, CH | 112.0, CH | 111.2, CH |
| 3′′ | 149.3, qC | 149.5, qC | 149.8, qC | 148.6 qC | 148.5 qC | 149.2, qC | 149.2, qC | 148.0, qC | 148.0, qC |
| 4′′ | 152.8, qC | 145.9, qC | 150.6, qC | 151.5, qC | 151.3, qC | 149.6, qC | 149.4, qC | 150.5, qC | 149.5, qC |
| 5′′ | 112.5, CH | 114.5, CH | 113.7, CH | 112.5, CH | 112.4, CH | 115.7, CH | 115.5, CH | 114.3, CH | 115.6, CH |
| 6′′ | 125.9, CH | 120.5, CH | 123.4, CH | 122.9, CH | 123.0, CH | 122.7, CH | 122.4, CH | 125.7, CH | 123.3, CH |
| 7′′ | 191.5, CH | 39.9, CH2 | 152.8, CH | 165.5, qC | 167.1, qC | 144.5, CH | 143.9, CH | 166.3, qC | 145.6, CH |
| 8′′ | — | 137.5, CH | 127.1, CH | — | — | 113.4, CH | 113.1, CH | — | 114.0, CH |
| 9′′ | — | 115.8, CH2 | 193.7, CH | — | — | 166.6, qC | 168.0, qC | — | 166.4, qC |
| OCH3 | 55.5, CH3 | 55.8, CH3 | 56.05, CH3 | 55.5, CH3 | 55.4, CH3 | 55.58, CH3 | 55.5, CH3 | 55.8, CH3 | 55.6, CH3 |
| CH2 | — | — | — | 60.5, CH2 | — | 59.9, CH2 | — | — | — |
| CH3 | — | — | — | 14.3, CH3 | — | 14.0, CH3 | — | — | — |
Compound 1 was obtained via click reaction of vanillyl azide with mono-alkyne vanillin as colorless needles, yielding 16.2%. Its molecular formula was determined to be C19H19N3O5 from [M − H]−, [M + H]+ and [M + Na]+ at m/z 368.1248, 370.1365, and 392.1185, respectively, (calc. 368.1246, 370.1403 and 392.1222) in the HRMS (Fig. S3 and S4†). The 1H-NMR spectrum (Fig. S1†) revealed the presence of an aldehydic proton; a downfield proton; six aromatic protons, two methylene signals and two methoxy groups. The three aromatic protons at δH 6.99 (H-2) and 6.73–6.79 (H-5/6); the methylene at δH 5.46 (H-7) and the methoxy groups at δH 3.74 (3H, s) were assigned to the vanillyl moiety of the azide. This was confirmed by the APT signals at δC 147.6, 146.7, 126.5, 121.1, 115.5, 55.6, and 53.0. Meanwhile, the aldehyde proton at δH 9.84 (H-7′′), the remaining three aromatic protons at δH 3.37–7.39 (H-2′′/5′′) and 7.54 (H-6′′), and the methoxy group at δH 3.79 (3H, s) were assigned to the vanillin moiety from the alkyne. This was confirmed from the APT signals at δC 191.5, 152.8, 149.3, 129.9, 125.9, 112.5, 109.6 and 55.5. These data were compatible with those published for the vanillyl alcohol28 and vanillin29 moieties. The methylene at δH 5.23 (H-6′), the methine at δH 8.27 (H-5′), and the APT signals at δC 142.1, 124.8, and 61.7 were typical for triazole moiety.30 Hence it was concluded to be the new compound, 4-((1-(3-methoxy-4-hydroxybenzyl)-1H-1,2,3-triazol-4-yl)methoxy)-3-methoxybenzaldehyde.
Compound 2 was obtained via click reaction of vanillyl azide with mono-alkyne eugenol as brown amorphous residue, yield 19.0%. Its molecular formula was determined to be C21H23N3O4 from [M + H]+ and [M + Na]+ at m/z 382.1774 and 404.1589, respectively, (calc. 382.1767 and 404.1589) in the HRMS (Fig. S8†). The NMR data were similar for compound 1 except for replacing vanillin signals with eugenol signals. The eugenol moiety was characterized by the aromatic signals at δH 6.69, 6.92, and 6.66–6.69 and the allyl moiety at δH 5.92, 5.06, 3.3. Hence it was concluded to be the new compound, 4-((4-((4-allyl-2-methoxyphenoxy)methyl)-1H-1,2,3-triazol-1-yl)methyl)-2-methoxyphenol.
Compound 3 was obtained via click reaction of vanillyl azide with mono-alkyne coniferaldehyde as yellow amorphous residue, yielding 33.2%. Its molecular formula was determined to be C21H21N3O5 from [M + H]+ and [M + Na]+ at m/z 396.1501 and 418.1323, respectively, (calc. 396.1559 and 418.1379) in the HRMS (Fig. S12†); [M − H]− at m/z 394.1410 (calc. 394.1403) (Fig. S13†). The coniferaldehyde moiety was characterized by the aromatic signals at δH 7.10, 7.06, and 6.99; the olefinic protons at δH 7.32 and 6.53; and the aldehydic proton at δH 9.57. Hence it was concluded to be the new compound, 4-((1-(3-methoxy-4-hydroxybenzyl)-1H-1,2,3-triazol-4-yl)methoxy)-3-methoxyphenyl acrylaldehyde.
Compounds 4 and 8 were obtained via click reaction of vanillyl azide with the mono-alkyne ether of vanillic acid ester and the mono-alkyne ester of acetyl vanillic acid, respectively, as a white powder, yielding 63.8%; and white residue, yielding 21.5%.
Compound 4 was assigned to the molecular formula C21H23N3O6 from [M − H]− at m/z 412.1511 (calc. 412.1509) in the HRMS. The vanillic acid ester moiety was characterized by the aromatic signals at δH 7.57, 7.44, and 7.26 and the ethyl moiety at δH 4.27 (2H, q) and 1.30 (2H, t). It was identified as 4-((1-(3-methoxy-4-hydroxybenzyl)-1H-1,2,3-triazol-4-yl)methoxy)-3-methoxy-ethyl benzoate.
Compound 5 was obtained from the ester hydrolysis of compound 4, as white amorphous powder, yielding 95.2%. Compounds 5 and its corresponding ethyl ester 4 showed the same NMR data except for the presence of the characteristic signals of the ethyl moiety at δH 4.27 (2H, q) and 1.3 (2H, t). For the APT spectrum, the free carboxylic group in compound 5 resonated at a higher field at δC 167.1 compared to the corresponding ethyl ester 4, where it resonated at δC 165.5. The same pattern was observed for compound 7 and its corresponding ethyl ester 6. Compound 5 was identified as 4-((1-(3-methoxy-4-hydroxybenzyl)-1H-1,2,3-triazol-4-yl)methoxy)-3-methoxybenzoic acid.
The isomeric compounds 5 and 8 were assigned to the molecular formula C19H19N3O6 based on the molecular ion peak at 408.1170 and 408.1178, respectively. The triazole ester 8 showed a distinct upfield shift in C-6′ (δC 57.7) and C-7′′ (δC 166.3) as compared to the triazole ether 5 (δC 61.6) and (δC 167.1). Also, the chemical shift of the vanillic acid moiety was slightly different between the two compounds. The same pattern was observed for the isomeric compounds 7 and 9, where C-6′ and C-7′′resonated at δC 61.6/57.0 and 168.0/166.4, respectively. It was named (1-(4-hydroxy-3-methoxybenzyl)-1H-1,2,3-triazol-4-yl)methyl 4-hydroxy-3-methoxy benzoate.
Similarly, compounds 6 and 9 were obtained via click reaction of vanillyl azide with the mono-alkyne ether of ferulic acid ester and the mono-alkyne ester of acetyl ferulic acid, respectively, as a white powder, yielding 56.8%; and white powder yielded 68.0%.
Compound 6 was assigned to the molecular formula C23H25N3O6 from [M − H]− at m/z 438.1664, (calc. 438.1665) in the HRMS. The ferulic acid ester moiety was characterized by the aromatic protons at δH 7.32, 7.2, 6.76–6.79; the olefinic protons at δH 7.55 and 6.47; and the methoxy group at δH 3.79. It was named as ethyl 4-((1-(3-methoxy-4-hydroxybenzyl)-1H-1,2,3-triazol-4-yl)methoxy)-3-methoxyphenyl acrylate.
Compound 7 was obtained from the ester hydrolysis of compound 6, as white amorphous powder, yielding 43.4%. Both compounds displayed the same NMR data except for the presence of the characteristic signals of the ethyl moiety at δH 4.17 (2H, q), 1.2 (2H, t), and δC 59.9, 14.0. For the APT spectrum, the free carboxylic group in compound 7 resonated at a higher field at δC 168.0 as compared to the corresponding ethyl ester 6, where it resonated at δC 166.6. It was named as 4-((1-(3-methoxy-4-hydroxybenzyl)-1H-1,2,3-triazol-4-yl)methoxy)-3-methoxyphenyl acrylic acid.
The isomeric compounds 7 and 9 were assigned to the molecular formula C21H21N3O6 based on the molecular ion peak at 410.1438 and 410.1361 (calc. 410.1352), respectively. The triazole ester 9 showed a distinct upfield shift in C-6′ (δC 57.0) and C-9′ (δC 166.4) as compared to the triazole ether 7 (δC 61.6) and C-9′′(δC 168.0). It was named as (E)-(1-(4-hydroxy-3-methoxybenzyl)-1H-1,2,3-triazol-4-yl)methyl 3-(4-hydroxy-3 methoxyphenyl)acrylate.
2.2. Biological evaluation
The semi-synthetic compounds (1–9) and curcumin were evaluated for their inhibitory effect on acetylcholine esterase and β-amyloid aggregation Table 3. Curcumin was chosen because it is a natural vanilloid analogue with reported AChE and Aβ aggregation inhibition activity.| Compound | IC50 ± SD (μM) | |
|---|---|---|
| AChE | Aβ42 inhibition | |
| a The data are expressed as mean ± standard deviation. | ||
| 1 | 14.24 ± 0.72 | 4.85 ± 0.24 |
| 2 | 26.83 ± 1.35 | 22.74 ± 1.14 |
| 3 | 5.53 ± 0.28 | 2.86 ± 0.14 |
| 4 | 2.107 ± 0.12 | 1.59 ± 0.08 |
| 5 | 2.109 ± 0.11 | 1.48 ± 0.07 |
| 6 | 2.52 ± 0.13 | 3.06 ± 0.15 |
| 7 | 10.55 ± 0.53 | 4.48 ± 0.22 |
| 8 | 1.2 ± 0.06 | 0.95 ± 0.05 |
| 9 | 0.47 ± 0.02 | 0.31 ± 0.02 |
| Curcumin | 8.72 ± 0.44 | 13.39 ± 0.67 |
| Donepezil | 0.28 ± 0.01 | — |
| Tacrine | — | 0.41 ± 0.02 |
Regarding the acetylcholine esterase assay results, compound 9 was the most active, IC50 value of 0.47 ± 0.02 μM; compounds 8, 4, 5, 6, and 3 showed IC50 values ranging from 1.2 ± 0.06 to 5.53 ± 0.28 μM.
Compound 9 was the most active, showing an IC50 value of 0.47 ± 0.02 μM, which is about two times that of the standard donepezil, the IC50 value of 0.27 ± 0.01 μM. It is worth noting that its isomeric compound 7 was much less active, the IC50 value of 10.55 ± 0.53 μM. This may suggest that the triazole ester moiety is more favorable to the activity than the triazole ether moiety. This may be also confirmed by observing the IC50 values of the isomeric compounds 5 and 8; the triazole ester 8 displayed about half of the IC50 value of its isomeric triazole ether 5, IC50 value 1.2 ± 0.06 and 2.11 ± 0.11 μM, respectively.
Compound 4 showed nearly similar activity to compound 5, suggesting that a free or conjugated carboxylic group may have not impact on the activity. However, this pattern was not observed for 7 and its ethyl ester 6 compound 4 showed nearly similar activity to compound 5, suggesting that a free or conjugated carboxylic group may have no impact on the activity. However, this pattern was not observed for 7 and its ethyl ester 6; the ethyl ester derivative 6 displayed about the fifth of IC50 of the free form 7, IC50 value 2.52 ± 0.13 and 10.55 ± 0.53 μM, respectively. It is worth noting that compounds 9, 8, 4, 5, 6 and 3 were more active than curcumin, the IC50 value of 8.72 ± 0.1 μM. Compounds 7, 1, and 2 were much less active than curcumin showing the IC50 value of 10.55 ± 0.53, 14.24 ± 0.72 and 26.83 ± 1.35 μM. The hybrid containing the vanillin and the eugenol moiety was the least active.
The ethyl ester derivative 6 displayed about fifth of IC50 of the free form 7, the IC50 value of 2.52 ± 0.13 and 10.55 ± 0.53 μM, respectively. It is worth noting that compounds 9, 8, 4, 5, 6, and 3 were more active than curcumin, the IC50 value of 8.72 ± 0.1 μM. Compounds 7, 1, and 2 were much less active than curcumin showing the IC50 value of 10.55 ± 0.53, 14.24 ± 0.72, and 26.83 ± 1.35 μM. The hybrid containing the vanillin and the eugenol moiety was the least active.
For the amyloid-β aggregation assay, compound 9 was more active than tacrine; their IC50 values were 0.31 ± 0.02 and 0.41 ± 0.02 μM, respectively. Compound 8 was next in activity with the IC50 values of 0.95 ± 0.05 μM. It is worth noting that the triazole esters 9 and 8 were much more active than their isomeric triazole ethers 7 and 5, respectively. This may suggest that the triazole ester moiety is more favorable to the activity. Compound 5 and its ethyl ester 4 also showed comparable IC50 values of 1.48 ± 0.07 and 1.59 ± 0.08 μM, respectively. For compound 7 and its ethyl ester 6, they showed nearly similar IC50 values of 4.48 ± 0.22 and 3.06 ± 0.15 μM, respectively. Next in activity to compounds 5 and 4 was compound 3, showing the IC50 value of 2.86 ± 0.14 μM.
All compounds except for compound 2 were more active than curcumin, the IC50 value of 22.74 ± 1.14 and 13.39 ± 0.67 μM, respectively. The hybrid containing the eugenol moiety was the least active.
From the above results, it can be concluded that compounds 9 and 8 could act as dual inhibitors for AChE and Aβ aggregation with IC50 values 0.47/1.2 and 0.31/0.95 μM, respectively. Their promising activity over compounds 1–7, could be attributed to the longer length of the spacer between the two vanillyl moieties. Therefore, they hold a particular interest in developing new anti-Alzheimer drugs. The skeleton of 9 and 8 may offer some structural features for the development of dual inhibitors. Hence, they should be subjected to further investigation for designing novel anti-Alzheimer drugs. The results provides a preliminary idea about the anti-AD potential of vanilloid-triazole conjugates. However, a further extensive study is required to investigate their activity in vitro and in vivo, including the morphology of the Aβ-oligomers.
2.3. Molecular docking
The docking scores and poses of triazole esters 9 and 8 with AChE and Aβ40/42 were predicted using AutoDock4.2.6 software. The protended binding features and docking scores are shown in Fig. 2. As depicted in Fig. 2, triazole esters 9 and 8 unveiled good docking scores towards AChE and Aβ40 with values of −10.4 and −5.8 kcal mol−1 and −9.4 and −4.7 kcal mol−1, respectively. Triazole esters 9 and 8 were also investigated against the Aβ42 protein. For Aβ42, the docking scores were not promising compared to those against Aβ40, with values of −3.3 and −2.9 kcal mol−1 of compounds 9 and 8, respectively (Fig. 2). The good potentiality of compounds 9 and 8 may be imputed to their ability to form a variation of H-bonds, π-based, and other interactions with the key residues within the binding sites of AChE and Aβ40. More precisely, compound 9 demonstrated two hydrogen bonds with THR83 (2.27 Å) and ARG296 (1.86 Å) inside the binding site of AChE (Fig. 2). For Aβ40, compound 9 formed three hydrogen bonds with ASP1 (2.07, 2.80 Å) and LYS16 (1.76 Å). Compound 8 exhibited five hydrogen bonds with ASP1 (2.13 Å), GLU3 (3.06 Å), ASP7 (1.94, 2.14 Å), and GLY9 (2.84 Å) (Fig. 2). Although compound 8 could not form any hydrogen bond within the binding site of AChE, other noncovalent interactions were noticed, involving π–π stacking interactions with PHE297, TYR337, TYR341, and TRP286 (Fig. 2).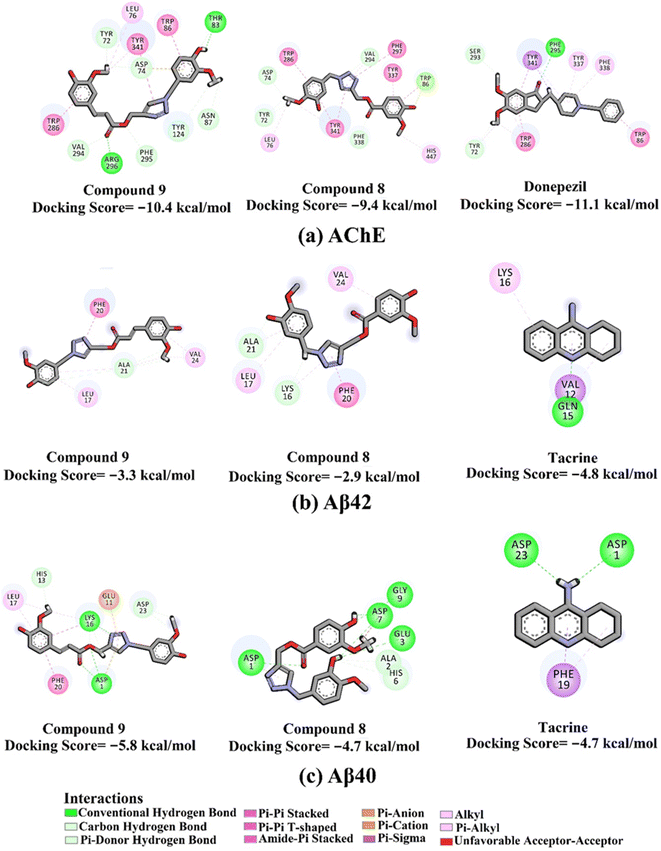 | ||
| Fig. 2 2D molecular interactions of triazole esters 9 and 8 and controls with (a) AChE (PDB ID: 4EY7), (b) Aβ42 (PDB ID: 1IYT), and (c) Aβ40 (PDB ID: 1BA4). | ||
Compared with compounds 9 and 8, donepezil displayed a similar docking score towards AChE with a value of −11.1 kcal mol−1, forming only one hydrogen bond with PHE295 and π–π stacking interactions with TRP86 and TRP286 (Fig. 2). On the other hand, tacrine exhibited one and two hydrogen bonds with GLN15 (1.85 Å) and ASP1 (2.08 Å) and GLY29 (1.92 Å) with the Aβ42 and Aβ40, respectively (Fig. 3).
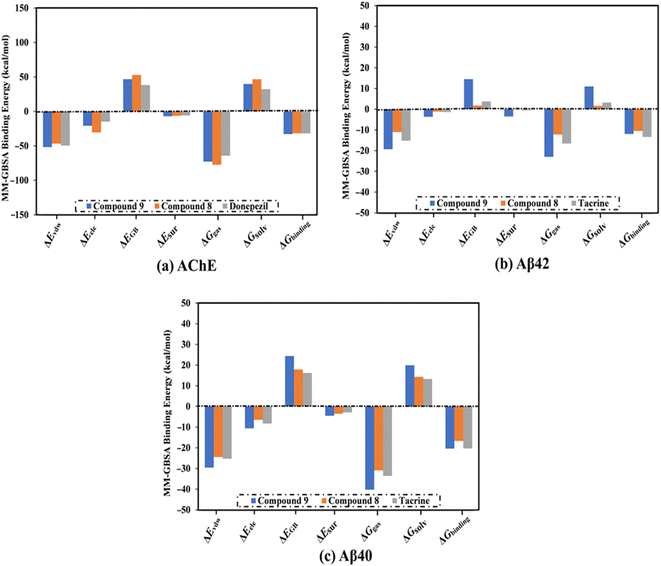 | ||
| Fig. 3 Decomposition of binding affinities for the studied ligands complexed with (a) AChE, (b) Aβ42, and (c) Aβ40 during a period of 100 ns MD. | ||
2.4. Molecular dynamics simulations
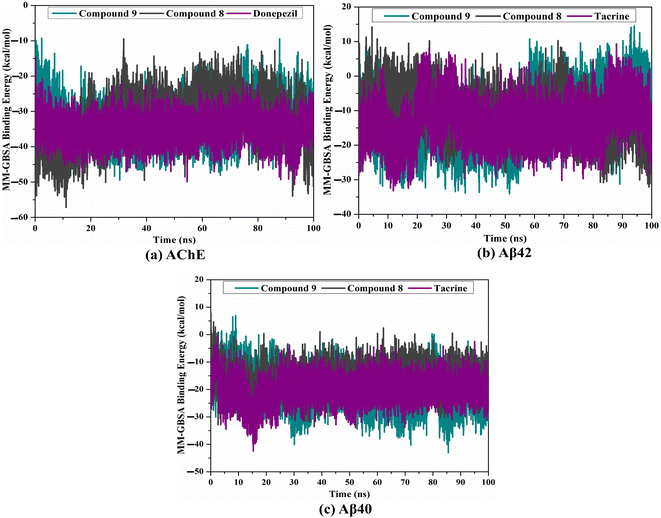
MD simulations were utilized to puzzle out the stabilization of the ligand–target complex, structural specifics, conformational elasticities, and the trustworthiness of ligand–target binding energy.31,32 Consequently, the inspected triazole esters 9 and 8 in complex with AChE and Aβ40/42 were submitted to MD simulations over 100 ns, followed by binding affinity evaluations. The estimated MM-GBSA binding affinities over 100 ns MD simulations are depicted in Fig. 3. As illustrated in Fig. 3, compounds 9 and 8 complexed with AChE exposed competitive binding affinities with an average ΔGbinding values of −32.9 and −31.8 kcal mol−1, respectively, compared to donepezil in complex with AChE (calc. −35.2 kcal mol−1). However, compounds 9 and 8 complexed with Aβ42 manifested appropriate binding energies with values of −12.0 and −10.5 kcal mol−1, respectively, compared to tacrine (calc. −13.4 kcal mol−1) (Fig. 2). A comparison of ΔGbinding values of compounds 9 and 8 complexed with Aβ42 and those with Aβ40 revealed the higher potency of compounds 9 and 8 with Aβ40 over Aβ42 with ΔGbinding values of −20.4 and −16.6 kcal mol−1, respectively (Fig. 3). The calculated MM-GBSA binding energies were in line with the IC50 values.To determine the most crucial interactions between ligand and target, binding affinities of the studied ligands in complex with AChE and Aβ40/42 were decomposed and illustrated in Fig. 3. As shown in Fig. 3, the binding energies of compounds 9 and 8 and donepezil complexed with AChE were dominated by Evdw interactions with values of −51.8, −46.7, and −49.5 kcal mol−1, respectively. Eele interactions were appropriate with values of −20.9, −30.6, and −14.8 kcal mol−1 for compounds 9 and 8 and donepezil complexed with AChE, respectively (Fig. 3).
For compounds 9 and 8 and tacrine complexed with Aβ42 and Aβ40, Evdw interactions were a significant contributor with values of −19.3, −11.0, and −15.2 kcal mol−1, and −29.6, −24.5, and −25.3 kcal mol−1, respectively (Fig. 3). Eele interactions were favorable with values of −3.7, −1.2, and −1.4 and −10.6, −6.5, and −8.3 kcal mol−1 for compounds 9 and 8 and tacrine complexed with Aβ42 and Aβ40, respectively (Fig. 3). These binding energies computations provided quantitative evidence of compounds 9 and 8 as anti-Alzheimer (AD) agents.
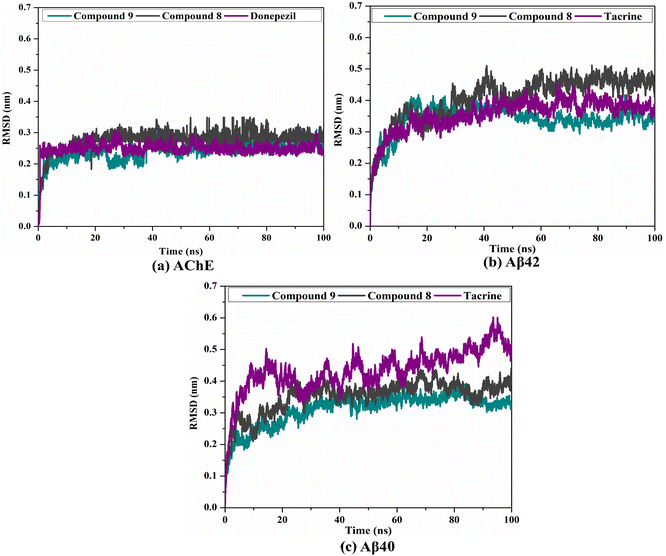
2.5. Post-MD analyses
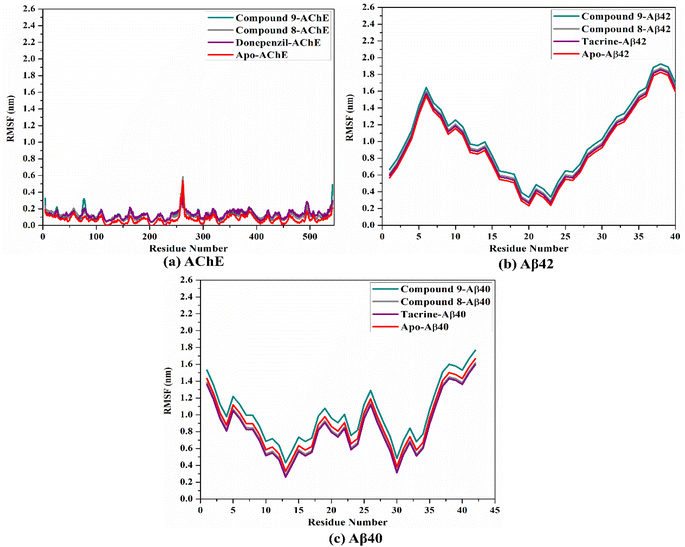
To confirm the steadiness of compounds 9 and 8 in complex with AChE and Aβ40/42, the complexes were investigated structurally and energetically during a period of 100 ns MD, and the results were compared to those of controls (i.e., donepezil and tacrine).3. Experimental
3.1. General experimental procedures
The progress of reactions and the purity of final products were monitored by thin layer chromatography (TLC), carried out using Merck precoated silica gel F254 plates (E-Merck, Germany) and using vanillin–sulfuric acid spray reagent. Column chromatography was carried out using silica gel G 60-230 (Merck, Germany). The solvents used included n-hexane, methylene chloride (CH2Cl2), and ethyl acetate (EtOAc) used were of reagent grade (El-Nasr Co., Abu Zaabal – Kalyoubia, Cairo, Egypt). 1H and APT spectra were measured in CDCl3 and DMSO-d6 using Bruker Avance III HD-400 spectrometer at 400 MHz for 1H and 100 MHz for APT in NMR unit, Faculty of Pharmacy, Mansoura, Egypt. Chemical shifts (δ) are expressed in ppm with reference to the residual solvent signal. Coupling constants (J values) are given in Hz. Melting points were determined on Stuart® melting point apparatus model SMP10 and are uncorrected. High-resolution mass (HR-ESI-MS) was measured using a Bruker microTOF mass spectrometer (Shimadzu, Tokyo, Japan). IR spectra were obtained using a Thermo Scientific Nicolet™ iS™ 10 FT-IR spectrometer instrument.3.2. Chemicals
Propargyl bromide, propargyl alcohol, vanillyl alcohol, ferulic acid, clove oil, sodium azide, N,N′-dicyclohexylcarbodiimide solution (DCC) and 4-(dimethylamino)pyridine (DMAP) and were purchased from Sigma-Aldrich St. Louis, USA. Vanillin (100% purity) was purchased from Eternal Pearl (The Zhonghua Chemical Factory, Zhejiang, China). Coniferaldehyde and vanillic acid were previously isolated from Cocos nucifera L., as reported.26 Eugenol was extracted and purified from clove oil using 30% aqueous KOH, according to the literature.27Vanillyl chloride (24.15 mmol) was dissolved in 5 ml DMSO, then sodium azide (48.3 mmol) was added. The reaction was stirred at room temperature and monitored by TLC. After completion, the reaction was quenched with water. The crude was extracted with ethyl acetate, dried over anhydrous sodium sulphate and concentrated under a vacuum.35 The azide was purified by silica gel column chromatography (2 cm × 28 cm, 35 g) using a gradient elution of ethyl acetate in hexane. The effluents, 50 ml each were collected, concentrated, and screened by TLC. Fractions with the same chromatographic pattern were pooled together. Fractions (7–15), eluted with 5–7% ethyl acetate in hexane, afforded vanillyl azide as a pure compound.
3.2.2.1. General procedure for preparation of alkynes by propargylic etherification. The start vanilloid was stirred with K2CO3 (3.5 equivalent) in dry acetone at room temperature for 2 hours. Propargyl bromide (1.3 equivalents) was added to the mixture was stirred under reflux at 80 °C for 10 h.36 After the completion of the reaction confirmed by TLC, the reaction was stopped by the addition of water. The solid salts were separated by filtration, and the product was extracted with ethyl acetate (3 × 50 ml). The ethyl acetate was concentrated under reduced pressure to give corresponding alkynes.
For vanillic or ferulic acid, before preparation of the corresponding propargyl ethers, the carboxylic group was masked by esterification.37 Vanillic (500 mg, 1.7 mmol) or ferulic acid (500 mg, 2.05 mmol) were dissolved in 25 ml absolute EtOH containing 1 ml conc. H2SO4 and refluxed.37
3.2.2.2. General procedure for preparation of alkynes by propargylic esterification. Prior to the preparation of the corresponding propargyl ester, the hydroxyl group was masked by acetylation. Vanillic (300 mg, 1.7 mmol) or ferulic acid (400 mg, 2.05 mmol), and pyridine (2 mmol), were dissolved in acetic anhydride (2 ml, 18.1 mol).38 The reaction was stirred for 12 h and monitored by TLC for completion.
Acetyl vanillic acid (360 mg, 1.7 mmol) or acetyl ferulic acid (415 mg, 1.75 mmol) and propargyl alcohol (1.7 mmol) were dissolved in dry CH2Cl2. To the stirred mixture, DCC (3.4 mmol) and DMAP (0.17 mmol) were added dropwise. The reaction mixture was monitored by TLC for completion. After 24 h stirring at room temperature, the reaction mixture was filtered over silica and the solution was washed with CH2Cl2 and concentrated under reduced pressure.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (1
H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). To this mixture, CuSO4·5H2O (0.15 eq.) and ascorbic acid (0.45 eq.) were added and stirred at room temperature. The reaction was monitored by TLC till completion. The reaction was quenched with distilled water and extracted with ethyl acetate, dried over anhydrous Na2SO4 and concentrated under a vacuum.39 The product was purified by silica gel column chromatography using gradient elution of ethyl acetate in hexane as to give 1,2,3-triazole derivatives.
1). To this mixture, CuSO4·5H2O (0.15 eq.) and ascorbic acid (0.45 eq.) were added and stirred at room temperature. The reaction was monitored by TLC till completion. The reaction was quenched with distilled water and extracted with ethyl acetate, dried over anhydrous Na2SO4 and concentrated under a vacuum.39 The product was purified by silica gel column chromatography using gradient elution of ethyl acetate in hexane as to give 1,2,3-triazole derivatives.3.3. Biological evaluation
3.4. In silico computations
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 energy evaluations and 250 independent runs were employed for docking computations. The remaining docking parameters were kept at the default settings. The AutoGrid program was employed to construct the grid maps. A grid box with dimensions 50 Å × 50 Å × 50 Å (x, y, z directions) was set around the binding pocket of AChE. The grid spacing value of 0.375 Å was utilized. The grid of AChE was positioned at the coordinates X = 11.367, Y = −56.25, and Z = −22.605. For Aβ40/42 targets, the AutoDock-based blind docking strategy was utilized in the current study.
000 energy evaluations and 250 independent runs were employed for docking computations. The remaining docking parameters were kept at the default settings. The AutoGrid program was employed to construct the grid maps. A grid box with dimensions 50 Å × 50 Å × 50 Å (x, y, z directions) was set around the binding pocket of AChE. The grid spacing value of 0.375 Å was utilized. The grid of AChE was positioned at the coordinates X = 11.367, Y = −56.25, and Z = −22.605. For Aβ40/42 targets, the AutoDock-based blind docking strategy was utilized in the current study.| ΔGbinding = Gcomplex − (Gligand + Gtarget) |
4. Conclusion
Based on their reported neuroprotective properties, the authors have selected the vanilloid pharmacophore to design potential anti-AD drugs. Nine new vanilloids hybrids (1–9) were semi-synthetized via click reactions of vanillyl azide and several vanilloid monoalkynes. Compounds 9 and 8 showed remarkable ACE and Aβ aggregation inhibition. The results suggested that the triazole ester moiety may be favorable for the activity over the triazole ether moiety. The results showed that compounds 9 and 8 are promising dual AChE/Aβ aggregation inhibitors. They may serve as potential leads for designing novel anti-Alzheimer agents.Author contributions
Marwa Elsbaey and Eman Elattar: conceptualization, investigation, methodology, writing – review & editing; Mahmoud Ibrahim: formal analysis, software, writing – review & editing; Yasuhiro Igarashi: supervision.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The computational work of this study was accomplished using the resources supported by the Science and Technology Development Fund, STDF, Egypt, Grants No. 5480 & 7972 (Granted to Mahmoud Ibrahim).References
- H. Balleza-Tapia, S. Crux, Y. Andrade-Talavera, P. Dolz-Gaiton, D. Papadia, G. Chen, J. Johansson and A. Fisahn, Elife, 2018, 7, e37703 CrossRef PubMed.
- Y. Yan, H. Yang, Y. Xie, Y. Ding, D. Kong and H. Yu, Neurochem. Res., 2020, 45, 989–1006 CrossRef CAS PubMed.
- X. Jin, M. Wang, J. Shentu, C. Huang, Y. Bai, H. Pan, D. Zhang, Z. Yuan, H. Zhang and X. Xiao, Oncol. Lett., 2020, 19, 1593–1601 CAS.
- A. Alvarez, C. Opazo, R. Alarcón, J. Garrido and N. C. Inestrosa, J. Mol. Biol., 1997, 272, 348–361 CrossRef CAS PubMed.
- M.-S. García-Ayllón, D. H. Small, J. Avila and J. Sáez-Valero, Front. Mol. Neurosci., 2011, 4, 22 Search PubMed.
- Y. Wang, H. Wang and H.-z. Chen, Curr. Neuropharmacol., 2016, 14, 364–375 CrossRef CAS PubMed.
- M. Campora, V. Francesconi, S. Schenone, B. Tasso and M. Tonelli, Pharmaceuticals, 2021, 14, 33 CrossRef CAS.
- L. Blaikie, G. Kay and P. K. T. Lin, Bioorg. Med. Chem. Lett., 2020, 30, 127505 CrossRef CAS PubMed.
- S. A. Adefegha, B. M. Okeke and G. Oboh, J. Food Biochem., 2021, 45, e13276 CrossRef CAS PubMed.
- J. C. H. Singh, R. M. Kakalij, R. P. Kshirsagar, B. H. Kumar, S. S. B. Komakula and P. V. Diwan, Pharm. Biol., 2015, 53, 630–636 CrossRef CAS PubMed.
- D. Szwajgier and K. Borowiec, J. Inst. Brew., 2012, 118, 40–48 CrossRef CAS.
- I. Orhan, Q. Naz, M. Kartal, F. Tosun and B. Şener, Z. Naturforsch., C: J. Biosci., 2007, 62, 684–688 CrossRef CAS PubMed.
- S. Abuhamdah, D. Thalji, N. M. Abuirmeileh, I. Alsalahat, A. N. Abuirmeileh and A. Bahnassi, Int. J. Pharmacol., 2017, 13(6) DOI:10.3923/ijp.2017.573.582.
- P. Taheri, P. Yaghmaei, H. S. Tehrani and A. Ebrahim-Habibi, Neurophysiology, 2019, 51, 114–119 CrossRef CAS.
- M. Kikugawa, H. Tsutsuki, T. Ida, H. Nakajima, H. Ihara and T. Sakamoto, Biosci., Biotechnol., Biochem., 2016, 80, 547–553 CrossRef CAS PubMed.
- P. H. Reddy, M. Manczak, X. Yin, M. C. Grady, A. Mitchell, S. Tonk, C. S. Kuruva, J. S. Bhatti, R. Kandimalla and M. Vijayan, J. Alzheimer's Dis., 2018, 61, 843–866 Search PubMed.
- J. Wang, B.-L. Sun, Y. Xiang, D.-Y. Tian, C. Zhu, W.-W. Li, Y.-H. Liu, X.-L. Bu, L.-L. Shen and W.-S. Jin, Transl. Psychiatry, 2020, 10, 1–12 CrossRef PubMed.
- M. Scipioni, G. Kay, I. L. Megson and P. K. T. Lin, MedChemComm, 2019, 10, 764–777 RSC.
- E.-J. Wang, M.-Y. Wu and J.-H. Lu, Cells, 2021, 10, 2653 CrossRef CAS PubMed.
- Y. P. Singh, H. Rai, G. Singh, G. K. Singh, S. Mishra, S. Kumar, S. Srikrishna and G. Modi, Eur. J. Med. Chem., 2021, 215, 113278 CrossRef CAS.
- K. Liu, W. Shi and P. Cheng, Dalton Trans., 2011, 40, 8475–8490 RSC.
- M. Xu, Y. Peng, L. Zhu, S. Wang, J. Ji and K. Rakesh, Eur. J. Med. Chem., 2019, 180, 656–672 CrossRef CAS.
- A. Kaur, S. Mann, A. Kaur, N. Priyadarshi, B. Goyal, N. K. Singhal and D. Goyal, Bioorg. Chem., 2019, 87, 572–584 CrossRef CAS PubMed.
- H. Karimi Askarani, A. Iraji, A. Rastegari, S. N. Abbas Bukhari, O. Firuzi, T. Akbarzadeh and M. Saeedi, BMC Chem., 2020, 14, 1–13 CrossRef PubMed.
- M. de Freitas Silva, E. Tardelli Lima, L. Pruccoli, N. G. Castro, M. J. R. Guimarães, F. M. da Silva, N. Fonseca Nadur, L. L. de Azevedo, A. E. Kümmerle and I. A. Guedes, Molecules, 2020, 25, 3165 CrossRef CAS PubMed.
- M. Elsbaey and B. Abdel, International Journal of Pharmacognosy and Phytochemical Research, 2017, 9, 1288–1292 Search PubMed.
- A. A. Khalil, U. ur Rahman, M. R. Khan, A. Sahar, T. Mehmood and M. Khan, RSC Adv., 2017, 7, 32669–32681 RSC.
- Y. Zhang, V. K. Thakur, Y. Li, T. F. Garrison, Z. Gao, J. Gu and M. R. Kessler, Macromol. Mater. Eng., 2018, 303, 1700278 CrossRef.
- P. Dwivedi, K. B. Mishra, B. B. Mishra and V. K. Tiwari, Glycoconjugate J., 2017, 34, 61–70 CrossRef CAS PubMed.
- M. Irfan, B. Aneja, U. Yadava, S. I. Khan, N. Manzoor, C. G. Daniliuc and M. Abid, Eur. J. Med. Chem., 2015, 93, 246–254 CrossRef CAS PubMed.
- M. De Vivo, M. Masetti, G. Bottegoni and A. Cavalli, J. Med. Chem., 2016, 59, 4035–4061 CrossRef CAS PubMed.
- J. E. Kerrigan, in In Silico Models for Drug Discovery, ed. S. Kortagere, Humana Press, Totowa, NJ, 2013, pp. 95–113, DOI:10.1007/978-1-62703-342-8_7.
- J. Li, L. Chen, X. Yan, Y. Li, D. Wei and D. Wang, J. Chem. Res., 2015, 39, 524–526 CrossRef CAS.
- J. A. Doiron, L. M. Leblanc, M. J. Hébert, N. A. Levesque, A. F. Paré, J. Jean-François, M. Cormier, M. E. Surette and M. Touaibia, Chem. Biol. Drug Des., 2017, 89, 514–528 CrossRef CAS.
- S. Howson and S. Peter, in Formation of benzyl azide from benzyl bromide; benzyl azide, ChemSpider-Synthetic, 2010, pp. 408 Search PubMed.
- T. Batool, N. Rasool, Y. Gull, M. Noreen, F.-u.-H. Nasim, A. Yaqoob, M. Zubair, U. A. Rana, S. U.-D. Khan and M. Zia-Ul-Haq, PLoS One, 2014, 9, e115457 CrossRef PubMed.
- Y. Belay, L.-C. Coetzee, D. B. G. Williams and A. Muller, Tetrahedron Lett., 2019, 60, 501–503 CrossRef CAS.
- M. Sova, A. Perdih, M. Kotnik, K. Kristan, T. L. Rižner, T. Solmajer and S. Gobec, Bioorg. Med. Chem., 2006, 14, 7404–7418 CrossRef CAS.
- E. M. Elattar, A. A. Galala, H. E. A. Saad and F. A. Badria, ChemistrySelect, 2022, 7, e202202194 CrossRef CAS.
- J. Cheung, M. J. Rudolph, F. Burshteyn, M. S. Cassidy, E. N. Gary, J. Love, M. C. Franklin and J. J. Height, J. Med. Chem., 2012, 55, 10282–10286 CrossRef CAS.
- O. Crescenzi, S. Tomaselli, R. Guerrini, S. Salvadori, A. M. D'Ursi, P. A. Temussi and D. Picone, Eur. J. Biochem., 2002, 269, 5642–5648 CrossRef CAS PubMed.
- M. Coles, W. Bicknell, A. A. Watson, D. P. Fairlie and D. J. Craik, Biochemistry, 1998, 37, 11064–11077 CrossRef CAS PubMed.
- M. H. Olsson, C. R. Sondergaard, M. Rostkowski and J. H. Jensen, J. Chem. Theory Comput., 2011, 7, 525–537 CrossRef CAS PubMed.
- OMEGA 2.5.1.4, OpenEye Scientific Software, Santa Fe, NM, USA, 2013 Search PubMed.
- P. C. Hawkins, A. G. Skillman, G. L. Warren, B. A. Ellingson and M. T. Stahl, J. Chem. Inf. Model., 2010, 50, 572–584 CrossRef CAS PubMed.
- SZYBKI 1.9.0.3, OpenEye Scientific Software, Santa Fe, NM, USA, 2016 Search PubMed.
- T. A. Halgren, J. Comput. Chem., 1999, 20, 720–729 CrossRef CAS PubMed.
- J. Gasteiger and M. Marsili, Tetrahedron, 1980, 36, 3219–3228 CrossRef CAS.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, J. Comput. Chem., 2009, 30, 2785–2791 CrossRef CAS PubMed.
- D. A. Case, R. M. Betz, D. S. Cerutti, T. E. Cheatham III, T. A. Darden, R. E. Duke, T. J. Giese, H. Gohlke, A. W. Goetz, N. Homeyer, S. Izadi, P. Janowski, J. Kaus, A. Kovalenko, T. S. Lee, S. LeGrand, P. Li, C. Lin, T. Luchko, R. Luo, B. Madej, D. Mermelstein, K. M. Merz, G. Monard, H. Nguyen, H. T. Nguyen, I. Omelyan, A. Onufriev, D. R. Roe, A. Roitberg, C. Sagui, C. L. Simmerling, W. M. Botello-Smith, J. Swails, R. C. Walker, J. Wang, R. M. Wolf, X. Wu, L. Xiao and P. A. Kollman, AMBER 2016, University of California, San Francisco, 2016 Search PubMed.
- M. A. A. Ibrahim, K. A. A. Abdeljawaad, A. H. M. Abdelrahman, L. A. Jaragh-Alhadad, H. F. Oraby, E. B. Elkaeed, G. A. H. Mekhemer, G. A. Gabr, A. M. Shawky, P. A. Sidhom, M. E. S. Soliman, M. F. Moustafa, P. W. Pare and M. F. Hegazy, Molecules, 2022, 27, 3104 CrossRef CAS PubMed.
- M. A. A. Ibrahim, E. A. A. Badr, A. H. M. Abdelrahman, N. M. Almansour, A. M. Shawky, G. A. H. Mekhemer, F. Alrumaihi, M. F. Moustafa and M. A. M. Atia, Cell Biochem. Biophys., 2021, 79, 189–200 CrossRef CAS PubMed.
- M. A. A. Ibrahim, E. A. A. Badr, A. H. M. Abdelrahman, N. M. Almansour, G. A. H. Mekhemer, A. M. Shawky, M. F. Moustafa and M. A. M. Atia, Mol. Inf., 2022, 41, e2060039 CrossRef PubMed.
- J. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman and D. A. Case, J. Comput. Chem., 2004, 25, 1157–1174 CrossRef CAS PubMed.
- J. A. Maier, C. Martinez, K. Kasavajhala, L. Wickstrom, K. E. Hauser and C. Simmerling, J. Chem. Theory Comput., 2015, 11, 3696–3713 CrossRef CAS PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery, J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, Ö. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski and D. J. Fox, Gaussian 09, Revision E01, Gaussian Inc., Wallingford CT, USA, 2009 Search PubMed.
- C. I. Bayly, P. Cieplak, W. D. Cornell and P. A. Kollman, J. Phys. Chem., 1993, 97, 10269–10280 CrossRef CAS.
- G. A. Ross, A. S. Rustenburg, P. B. Grinaway, J. Fass and J. D. Chodera, J. Phys. Chem. B, 2018, 122, 5466–5486 CrossRef CAS PubMed.
- Dassault Systèmes BIOVIA, B.D.S.V., version 2019, Dassault Systèmes BIOVIA, San Diego, CA, USA, 2019 Search PubMed.
- I. Massova and P. A. Kollman, Perspect. Drug Discovery Des., 2000, 18, 113–135 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ra07539c |
| This journal is © The Royal Society of Chemistry 2023 |