Open Access Article
Open Access ArticleCopper(II) and silver(I) complexes with dimethyl 6-(pyrazine-2-yl)pyridine-3,4-dicarboxylate (py-2pz): the influence of the metal ion on the antimicrobial potential of the complex†
Tina P. Andrejević a,
Ivana Aleksic
a,
Ivana Aleksic b,
Jakob Kljun
b,
Jakob Kljun c,
Marta Počkaj
c,
Marta Počkaj c,
Matija Zlatar
c,
Matija Zlatar d,
Sandra Vojnovic
d,
Sandra Vojnovic b,
Jasmina Nikodinovic-Runic
b,
Jasmina Nikodinovic-Runic b,
Iztok Turel
b,
Iztok Turel *c,
Miloš I. Djuran
*c,
Miloš I. Djuran *e and
Biljana Đ. Glišić
*e and
Biljana Đ. Glišić *a
*a
aDepartment of Chemistry, Faculty of Science, University of Kragujevac, R. Domanovića 12, 34000 Kragujevac, Serbia. E-mail: biljana.glisic@pmf.kg.ac.rs
bInstitute of Molecular Genetics and Genetic Engineering, University of Belgrade, Vojvode Stepe 444a, 11042 Belgrade, Serbia
cFaculty of Chemistry and Chemical Technology, University of Ljubljana, Večna Pot 113, Ljubljana, SI-1000, Slovenia. E-mail: Iztok.Turel@fkkt.uni-lj.si
dDepartment of Chemistry, University of Belgrade-Institute of Chemistry, Technology and Metallurgy, Njegoševa 12, 11000 Belgrade, Serbia
eSerbian Academy of Sciences and Arts, Knez Mihailova 35, 11000 Belgrade, Serbia. E-mail: milos.djuran@pmf.kg.ac.rs
First published on 1st February 2023
Abstract
Dimethyl 6-(pyrazine-2-yl)pyridine-3,4-dicarboxylate (py-2pz) was used as a ligand for the synthesis of new copper(II) and silver(I) complexes, [CuCl2(py-2pz)]2 (1), [Cu(CF3SO3)(H2O)(py-2pz)2]CF3SO3·2H2O (2), [Ag(py-2pz)2]PF6 (3) and {[Ag(NO3)(py-2pz)]·0.5H2O}n (4). The complexes were characterized by spectroscopic and electrochemical methods, while their structures were determined by single crystal X-ray diffraction analysis. The X-ray analysis revealed the bidentate coordination mode of py-2pz to the corresponding metal ion via its pyridine and pyrazine nitrogen atoms in all complexes, while in polynuclear complex 4, the heterocyclic pyrazine ring of one py-2pz additionally behaves as a bridging ligand between two Ag(I) ions. DFT calculations were performed to elucidate the structures of the investigated complexes in solution. The antimicrobial potential of the complexes 1–4 was evaluated against two bacterial (Pseudomonas aeruginosa and Staphylococcus aureus) and two Candida (C. albicans and C. parapsilosis) species. Silver(I) complexes 3 and 4 have shown good antibacterial and antifungal properties with minimal inhibitory concentration (MIC) values ranging from 4.9 to 39.0 μM (3.9–31.2 μg mL−1). All complexes inhibited the filamentation of C. albicans and hyphae formation, while silver(I) complexes 3 and 4 had also the ability to inhibit the biofilm formation process of this fungus. The binding affinity of the complexes 1–4 with calf thymus DNA (ct-DNA) and bovine serum albumin (BSA) was studied by fluorescence emission spectroscopy to clarify the mode of their antimicrobial activity. Catechol oxidase biomimetic catalytic activity of copper(II) complexes 1 and 2 was additionally investigated by using 3,5-di-tert-butylcatechol (3,5-DTBC) and o-aminophenol (OAP) as substrates.
Introduction
Antimicrobial resistance to the currently used antimicrobials has become a global problem, which significantly influences the ability of humans to prevent and treat an enormously increasing number of bacterial and fungal infections and affects the success of cancer chemotherapy.1 Infections that are caused by multidrug resistant pathogens are associated with longer hospital stays and worse clinical outcomes, such as morbidity and mortality in the infected patients and increased costs on the healthcare system.2 In the Review on Antimicrobial Resistance, commissioned by the UK Government, it is estimated that antimicrobial resistance will lead to the death of 10 million people per year by 2050.3 Although this report has been criticized regarding the estimated quantification of morbidity and mortality,4 there is no doubt that the spread of antimicrobial resistance is an urgent concern, which requires a global and coordinated action plan, including the development of new antimicrobials with different and more effective mechanisms of action.5,6A major problem in the field of development of new antimicrobial agents is the fact that approximately 25% of the compounds currently in clinical trials are entirely new structural classes, while the remaining 75% are derivatives of the already used drugs and represent a short-term solution of the problem.7 Moreover, all these compounds in both preclinical and clinical development are purely organic and have simple one- or two-dimensional shapes.8 On the contrary, metal complexes can adopt different geometries and generally possess three-dimensional shape, which can contribute to their improved clinical success rates, because molecular shape is one of the most important factors in molecular recognition by a biomolecule.9–14 In addition, metal complexes exert different modes of action, including ligand exchange or release, formation of reactive oxygen species (ROS), redox activation and catalytic generation of toxic species, which are difficult or even impossible to be achieved with organic compounds alone.15,16 Recently, a comprehensive overview of the developments in the design of metal complexes with antifungal activity have been provided by Gasser et al.,17 highlighting the combination of different metal ions with antifungally active organic molecules, leading to the increased bioavailability, uptake and efficacy of an antifungal agent. This concept can be also successfully used for the development of novel agents for the treatment of cancer and bacterial and parasitic infections.18 Moreover, Frei et al. have assessed the antibacterial and antifungal activity of 906 metal-containing compounds and found an impressive success rate in comparison to purely organic compounds, confirming that the combination of an extended arsenal of potential modes of action with a three-dimensional shape makes them potential antimicrobial drug candidates with some crucial advantages.8 It is important to mention that a plethora of metal-based drugs have been studied also as anticancer and antiparasitic agents, antivirals, as adjuvants to elicit an immune response and for many other indications; nevertheless, only a few of them have been successfully introduced into the clinic.19,20 Most of the investigated metal complexes act as prodrugs, which can be transformed into the active species either by ligand exchange or redox reactions, and are likely to be multi-targeting in its action.19
In the last few years, the antimicrobial activity of different copper(II) complexes has been extensively investigated with some promising results.1,21–24 For instance, a few copper(II) complexes with 2-acetylpyridine-N-substituted thiosemicarbazonates have shown significant antimicrobial potential against methicillin resistant Staphylococcus aureus (MRSA), Klebsiella pneumoniae and Candida albicans, which is comparable to or even better than that of the gentamicin and amphotericin B, used as reference compounds.21 A good antibacterial activity against a range of Gram-negative (Salmonella typhimurium, Pseudomonas aeruginosa, Escherichia coli and K. pneumoniae) and Gram-positive (MRSA and Staphylococcus aureus) bacteria was also shown by copper(II) complexes with N,N′-diarylformamidine dithiocarbamate ligands, surpassing in many cases the activity of antibiotic ciprofloxacin.22 Moreover, two copper(II) complexes, [Cu(gluc)(hpb)(H2O)]gluc and [Cu(gluc)(hpbc)(H2O)]gluc (hpb is 2-(2′-pyridyl)benzimidazole, hpbc is 5-chloro-2-(2′-pyridyl)benzimidazole, gluc is D-gluconic acid), have shown good inhibitory activity against three Gram-positive bacteria (S. aureus, Bacillus subtilis, Listeria monocytogenes) and one Gram-negative bacterium (E. coli), as well as good cytotoxic activity against the tested cancer cell lines.23 On the other hand, no antibacterial effect was observed for five copper(II) complexes with pyridine-4,5-dicarboxylate esters, while two of them, namely [Cu(NO3)2(py-2metz)(H2O)] and [CuCl2(py-2metz)]n (py-2metz is dimethyl 2-(4-methylthiazol-2-yl)pyridine-4,5-dicarboxylate), have shown a moderate antifungal activity with a minimal inhibitory concentration (MIC) of 31.25 μg mL−1 against C. albicans.24 Similarly, none of the five copper(II) complexes with aromatic nitrogen-containing heterocycles (N-heterocycles), e.g. pyrimidine, pyrazine, quinazoline and phthalazine, manifested significant antimicrobial activity against the investigated bacterial and Candida strains.25 Nevertheless, these complexes were able to inhibit bacterial quorum sensing (QS) and, reducing virulence without a pronounced effect on the bacterial growth, they can offer a lower risk for resistance development.25
Following the use of silver(I) sulfadiazine as a broad-spectrum topical antibiotic for some burn wounds,26 different classes of silver(I) complexes have attracted attention for their antimicrobial properties, such as those with N-heterocyclic carbenes (NHCs) and N-heterocycles as ligands.27–30 Even though silver(I) complexes have shown significant growth inhibitory effects against Gram-positive and Gram-negative bacteria and fungi, their exact mode of action remained not well understood. Nevertheless, the most reported one includes a slow release of the active Ag(I) ions, which further react with the thiol or other key functional groups of proteins and enzymes,31–33 leading to the denaturation of proteins and the impairing of the membrane function.34,35 Besides that, Ag(I) ions can produce ROS, which target lipids, nucleic acids and proteins, causing their malfunction.36 Moreover, treatment of bacteria with silver(I) ions was found to induce the condensation of DNA molecules in the cytoplasm, resulting in the loss of DNA replication ability and, consequently, in the death of bacteria.33,34 Although the mechanism of action of silver(I) complexes is not well understood, it has been proposed that the antimicrobial activity of these complexes is mainly connected to their solubility, stability, lipophilicity, redox ability, and rate of Ag(I) ions release.33 All these factors are directed by the choice of suitable ligands, and they are fundamental for maintaining the bioavailability of complex during an extended period, thus preventing reinfection or resistance development.
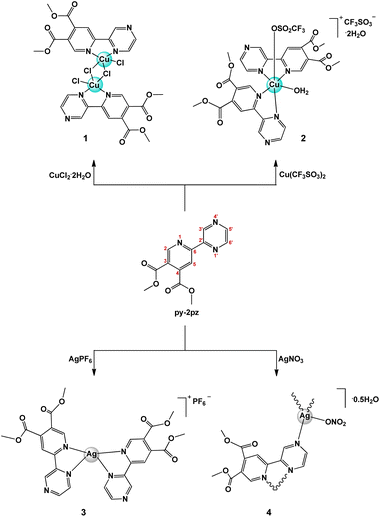
Based on the all above reasons, in the present study, new copper(II) and silver(I) complexes, namely [CuCl2(py-2pz)]2 (1), [Cu(CF3SO3)(H2O)(py-2pz)2]CF3SO3·2H2O (2), [Ag(py-2pz)2]PF6 (3) and {[Ag(NO3)(py-2pz)]·0.5H2O}n (4), were synthesized with dimethyl 6-(pyrazine-2-yl)pyridine-3,4-dicarboxylate (py-2pz) as ligand (Scheme 1). Two different metal ions were chosen to check how they can influence on the antimicrobial activity of the resulted metal complex. Moreover, py-2pz was chosen as a versatile ligand which can behave both as bridging and chelating bidentate ligand (Scheme 1). Structurally similar ligands such as derivatives of different pyridine-4,5-dicarboxylate esters, dimethyl 2-(thiazol-2-yl)pyridine-4,5-dicarboxylate (py-2tz), dimethyl 2-(4-methylthiazol-2-yl)pyridine-4,5-dicarboxylate (py-2metz) and dimethyl 2,2′-bipyridine-4,5-dicarboxylate (py-2py), were previously used for the synthesis of silver(I) complexes, which have shown the significant activity against cow mastitis associated pathogens,37 of ruthenium(II) complexes as inhibitors of aldo-keto reductases38 and 15-lipoxygenase-1,39 and of the abovementioned copper(II) complexes,24 as well as antifungal zinc(II) complex.40 The antimicrobial activity of the synthesized complexes 1–4 were evaluated against two bacterial (P. aeruginosa and S. aureus) and two Candida (C. albicans and C. parapsilosis) species, as well as their toxicity on the human normal fibroblast cell line (MRC-5). With the aim to obtain a more detailed information on the antifungal potential of the complexes, their inhibitory activity on C. albicans filamentation and biofilm formation was investigated. The DNA/BSA binding properties of the complexes were investigated by fluorescence emission spectroscopic method, while oxidase catalytic activity of copper(II) complexes 1 and 2 was studied by 3,5-di-tert-butylcatechol (3,5-DTBC) and o-aminophenol (OAP) as substrates.
Results and discussion
Synthesis and structural characterization of complexes 1–4
Dimethyl 6-(pyrazine-2-yl)pyridine-3,4-dicarboxylate (py-2pz) (Scheme 1), which contains two aromatic nitrogen-containing heterocyclic rings, pyridine and pyrazine, is potentially good chelating ligand for different metal ions. In order to investigate the coordination mode and binding ability of py-2pz ligand to the metal ions, in the present study this compound has been prepared by following the previously reported procedure41 and its reactions with different copper(II) and silver(I) salts have been investigated.Copper(II) and silver(I) complexes, [CuCl2(py-2pz)]2 (1), [Cu(CF3SO3)(H2O)(py-2pz)2]CF3SO3·2H2O (2), [Ag(py-2pz)2]PF6 (3) and {[Ag(NO3)(py-2pz)]·0.5H2O}n (4) were obtained in a good yield in ethanol via the aerobic reaction of py-2pz ligand with an equimolar amount of the corresponding CuX2 (X = Cl− (1) and CF3SO3− (2)) or AgX (X = PF6− (3) and NO3− (4)) salt at room temperature (Scheme 1). They were characterized by elemental analysis, molar conductivity measurement, spectroscopic (IR, UV-Vis, 1H NMR for 3 and 4) and electrochemical (cyclic voltammetry) methods, while their crystal structures were determined by single crystal X-ray crystallography. DFT calculations were performed to elucidate the structures of the synthesized complexes 1–4 in solution.
Solid state studies
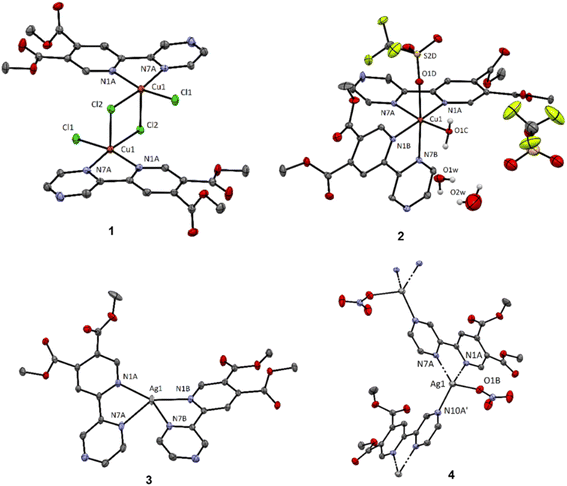
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 and with one coordinated triflate ligand and one co-crystallized as counter-anion (Fig. 1). The octahedral coordination sphere is completed with a coordinated water molecule which is part of a hydrogen bond network consisting of two additional solvate water molecules. Electronic effects result in slight elongation of the axial Cu–N bond by approximately 0.25 Å and a significant elongation of the Cu–Otriflate bond (2.39 Å; selected bond lengths and angles are summarized in the caption of Fig. 1). The triflate anion is highly disordered and we were not able to appropriately model the fragment to completely fit the residual electron density. Thus, a second structural model was proposed, using the Solvent mask function within Olex2-1.3 (ref. 43) to remove the residual electron density and calculate the void. The integrated electron density corresponds to 78 electrons within a 170 Å3. The electron count corresponds to the triflate ion; however, the void is indeed too spacious allowing for a highly disordered structure (21.25 Å3 per non-hydrogen atom). For more details see ESI.†
2 and with one coordinated triflate ligand and one co-crystallized as counter-anion (Fig. 1). The octahedral coordination sphere is completed with a coordinated water molecule which is part of a hydrogen bond network consisting of two additional solvate water molecules. Electronic effects result in slight elongation of the axial Cu–N bond by approximately 0.25 Å and a significant elongation of the Cu–Otriflate bond (2.39 Å; selected bond lengths and angles are summarized in the caption of Fig. 1). The triflate anion is highly disordered and we were not able to appropriately model the fragment to completely fit the residual electron density. Thus, a second structural model was proposed, using the Solvent mask function within Olex2-1.3 (ref. 43) to remove the residual electron density and calculate the void. The integrated electron density corresponds to 78 electrons within a 170 Å3. The electron count corresponds to the triflate ion; however, the void is indeed too spacious allowing for a highly disordered structure (21.25 Å3 per non-hydrogen atom). For more details see ESI.†
In the case of silver(I) complexes, the use of silver(I) hexafluorophosphate results in a four-coordinated complex 3 with metal-to-ligand molar ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, having an intermediate geometry between square planar and tetrahedral with
2, having an intermediate geometry between square planar and tetrahedral with 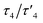 values44 of 0.51/0.47. τ4 value is 0 and 1 for ideal square planar and tetrahedral geometry, respectively.44 Reaction of py-2pz with silver(I) nitrate leads to the formation of a four-coordinated polymeric species 4, however in contrast to the previously reported structures of pyridine-4,5-dicarboxylate ester ligands bearing a second pyridyl/thiazolyl/2-methylthiazolyl ring,37 the nitrate in the complex 4 acts as terminal monodentate ligand and the polymeric species is bridged by the binding of a second silver(I) ion to the N10 nitrogen atom of the pyrazine ring. The geometry in this case is a slightly distorted tetrahedron with
values44 of 0.51/0.47. τ4 value is 0 and 1 for ideal square planar and tetrahedral geometry, respectively.44 Reaction of py-2pz with silver(I) nitrate leads to the formation of a four-coordinated polymeric species 4, however in contrast to the previously reported structures of pyridine-4,5-dicarboxylate ester ligands bearing a second pyridyl/thiazolyl/2-methylthiazolyl ring,37 the nitrate in the complex 4 acts as terminal monodentate ligand and the polymeric species is bridged by the binding of a second silver(I) ion to the N10 nitrogen atom of the pyrazine ring. The geometry in this case is a slightly distorted tetrahedron with  values44 of 0.76/0.73.
values44 of 0.76/0.73.
Solution studies
1H NMR spectra for silver(I) complexes 3 and 4 were recorded in DMSO-d6 with the aim to confirm the bidentate coordination of the py-2pz to the Ag(I) ion in these complexes (1H NMR spectra are given in ESI†). For this purpose, the 1H NMR spectra of the silver(I) complexes were compared with that of the uncoordinated py-2pz ligand (ESI†). In the spectra of complexes, the number of signals is the same as that of py-2pz, confirming its bidentate coordination to the Ag(I) ion. However, the proton resonances in the spectra of complexes are almost at the same chemical shifts compared to those of the ligand, which is an expected spectroscopic behavior of the silver(I) complexes in DMSO-d6 and can be a consequence of the fast ligand exchange on the NMR timescale.49The UV-Vis spectra of complexes 1–4 were recorded in DMSO at room temperature. In the visible region of the spectra of copper(II) complexes 1 and 2, a low intensity band (ε = 102.0 and 43.8 M−1 cm−1) assigned to d–d transition appears at 875.0 and 805.0 nm, is observed (Fig. S1†).50 On the other hand, silver(I) complexes 3 and 4 exhibit the absorbance peak at 296.0 nm, which is caused by the intraligand charge transfer transitions.51 The absorbance peaks for both complexes show a blue shift compared to that for the free py-2pz ligand (λ = 326.0 nm) (Fig. S2†).
Further, the UV-Vis spectra of the complexes were recorded in the presence of DMSO (a solvent used for the preparation of stock solutions for the biological experiments) and in DMSO/PBS solution (PBS is phosphate buffered saline; Fig. S3†), immediately after their dissolution, as well as after 48 h. Since no significant changes (shift of the absorbance maximum or the appearance of new bands) were observed in the UV-Vis spectra during time of measurements, it can be proposed that the py-2pz ligand remains coordinated to the corresponding metal ion in solution.
The molar conductivity measurements undoubtedly confirm the constitution of copper(II) complexes 1 and 2, and are in accordance with the fact that 1 behaves as nonelectrolyte, while 2 is 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 type of electrolyte.52 On the other hand, the values of molar conductivity for 3 and 4 are not conclusive and only suggest the presence of ionic species in solution, which is expected for 3.
1 type of electrolyte.52 On the other hand, the values of molar conductivity for 3 and 4 are not conclusive and only suggest the presence of ionic species in solution, which is expected for 3.
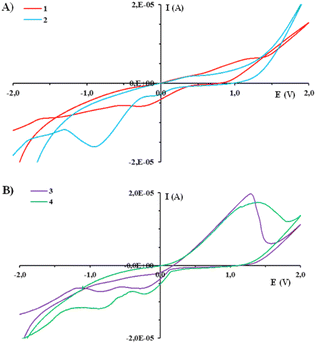
The electrochemical behavior of metal complexes is of great importance for a better understanding of their redox stability, as well as biological activity and mode of interaction with biologically important biomolecules.53,54 The cyclic voltammograms of complexes 1–4 and py-2pz were recorded in DMSO at glassy carbon (GC) electrode in DMSO and 0.1 M tetrabutylammoniumhexafluorophosphate (TBAHP) as a supporting electrolyte, under following conditions, Ebegin = −2.0 V and Eend = 2.0 V (Fig. 2 and S4†). It is important to mention that py-2pz ligand is redox inactive in the applied voltage range (Fig. S4†). As can be seen from Fig. 2A, in the cyclic voltammogram of complex 1, only one reduction peak at −0.27 V is observed, which can be attributed to the Cu(II) → Cu(I) process, while the complex 2 showed two reduction peaks, at −0.05 and −0.89 V for Cu(II) → Cu(I) and Cu(I) → Cu(0) reduction processes, respectively, which were assigned in accordance with the literature data for the previously reported copper(II) complexes, including those with the structurally similar pyridine-4,5-dicarboxylate esters.24,55,56 The presence of only one reduction peak in the voltammogram of complex 1 can be the consequence of its higher redox stability compared to that of 2.
On the other hand, silver(I) complexes 3 and 4 in anodic direction show one oxidation peak Ia corresponding to the Ag(I) → Ag(II) process (Fig. 2B).37,57–59 When the cyclic voltammogram was recorded in the cathodic direction, two reduction peaks Ic,1 and Ic,2 were observed, which can be attributed to the Ag(II) → Ag(I) and Ag(I) → Ag(0) processes, respectively (Fig. 2B), in accordance with the other silver(I) complexes which electrochemical behavior was previously reported.37,57–59
Computational results
DFT calculations were performed to clarify the structures of complexes 1–4 in DMSO solution. For this purpose, we relied on the thermodynamics of possible reactions starting from the X-ray determined structures.60 Free energy changes, ΔrG (298 K), for the formation of probable coordination structures were investigated (Table 1). ΔrG (298 K) was calculated based on the difference in Gibbs free energies of products and reactants at ZORA-M06-2X/TZP-COSMO(DMSO)//ZORA-BP86-D4/TZP-COSMO(DMSO) level of theory.| Reaction | ΔrG (298 K) |
|---|---|
| [CuCl2(py-2pz)]2 (1) ⇌ 2 [CuCl2(py-2pz)] | −3.75 |
| [Cu(CF3SO3)(H2O)(py-2pz)2]+ (2) ⇌ [Cu(H2O)(py-2pz)2]2+ + CF3SO3− | −2.01 |
| [Cu(CF3SO3)(H2O)(py-2pz)2]+ (2) ⇌ [Cu(CF3SO3)(py-2pz)2]+ + H2O | −3.58 |
| [Cu(CF3SO3)(H2O)(py-2pz)2]+ (2) ⇌ [Cu(py-2pz)2]2+ + CF3SO3− + H2O | −4.86 |
| [Ag(py-2pz-N,N′)2]+ (3) ⇌ [Ag(py-2pz-N,N′)(py-2pz-N)]+ | +11.26 |
| [Ag(NO3)(py-2pz-N,N′)(py-2pz-N)] (4) ⇌ [Ag(py-2pz-N,N′)(py-2pz-N)]+ + NO3− | +4.04 |
For complex 1, the formation of tetra-coordinate, mononuclear [CuCl2(py-2pz)] species (Fig. 3) is thermodynamically favoured. Two Cu(II) centers in 1 are very weakly ferromagnetically coupled, with the exchange constant J = 0.3 cm−1 calculated in broken-symmetry formalism at ZORA-M06-2X/TZP//X-ray level of theory. Unpaired electrons are in the local dx2–y2 orbital, and the spin density is mainly located in the equatorial plane of the subunits that are parallelly displaced. Delocalization of the spin density is towards directly coordinated equatorial donor atoms (Fig. S5†).
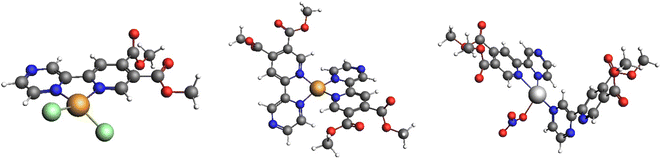 | ||
| Fig. 3 The structures of [CuCl2(py-2pz)] (left), [Cu(py-2pz)2]2+ (middle) and [Ag(NO3)(py-2pz-N,N′)(py-2pz-N)] (right) optimized at the ZORA-BP86-D4/TZP-COSMO(DMSO) level of theory. | ||
In the case of complex 2, dissociation of weakly coordinated CF3SO3− and of monodentate aqua ligand were considered. Thermodynamically the formation of tetra-coordinate [Cu(py-2pz)2]2+ is preferred (Fig. 3). For polymeric structure 4, a model complex [Ag(NO3)(py-2pz-N,N′)(py-2pz-N)] was considered, built from the corresponding X-ray structure. In this structure, one py-2pz ligand is bidentate (py-2pz-N,N′), while the other is monodentate (py-2pz-N) (Fig. 3). Dissociation of monodentate NO3− is disadvantageous according to positive ΔrG. In complex 3, both py-2pz ligands are bidentately coordinated to the Ag(I) center. Structure 3 is the most stable, with two py-2pz ligands coordinated to Ag(I) center, ignoring possible polymerization.
Biological studies
| Microorganism/cell | Compound/MIC and IC50, μM (μg mL−1) | |||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | py-2pz | AgSD | |
| a Results are given as mean of three independent measurements with standard error being between 1–3%. | ||||||
| S. aureus ATCC 25923 | >613.2 (>500) | >519.6 (>500) | 39.0 (31.2) | 34.5 (15.6) | >1830 (>500) | 75 (26.8) |
| P. aeruginosa PAO1 | 153.3 (125) | 259.8 (250) | 19.5 (15.6) | 8.6 (3.9) | >1830 (>500) | 25 (8.9) |
| C. albicans ATCC 10231 | 613.2 (500) | >519.6 (>500) | 4.9 (3.9) | 8.6 (3.9) | >1830 (>500) | 10 (3.6) |
| C. parapsilosis ATCC 22019 | 613.2 (500) | 519.6 (500) | 4.9 (3.9) | 8.6 (3.9) | >1830 (>500) | 2.5 (0.89) |
| MRC-5 cells | 85.8 (70) | 129.9 (125) | 31.3 (25) | 16.6 (7.5) | 439.2 (120) | 10 (3.6) |
As can be seen from Table 2, silver(I) complexes 3 and 4 showed a good antifungal activity on both tested Candida strains (C. albicans and C. parapsilosis) with MIC values of 4.9 and 8.6 μM (3.9 μg mL−1), respectively, while no significant anti-Candida activity was observed for copper(II) complexes 1 and 2. Both silver(I) complexes 3 and 4 are more active against C. albicans than the clinically used AgSD complex (Table 2). Regarding antibacterial activity, polynuclear silver(I) complex 4 with coordinated py-2pz and nitrate was the most active, with MIC value of 8.6 μM (3.9 μg mL−1) against P. aeruginosa PAO1, while slightly reduced activity was noted for 3 against the same strain with MIC being 19.5 μM (15.6 μg mL−1; this complex is mononuclear and contains two bidentately coordinated py-2pz ligands). Moreover, both investigated copper(II) complexes 1 and 2 show only a moderate activity against P. aeruginosa PAO1. The activity of the complexes against this microbial species is of particular interest, since it is a human pathogen, responsible for severe infections, including those associated with cystic fibrosis, burn wounds and the use of catheters, and is a major cause of nosocomial infections.63–65 The corresponding py-2pz ligand did not affect the microbial growth at 1830 μM (500 μg mL−1), indicating that the observed activity of the silver(I) complexes is due to the presence of Ag(I) ions. As it was mentioned in the Introduction, the antimicrobial activity of silver(I) complexes is connected to their ability to release free Ag(I) ions, which show the activity through different pathways.33 In general, the antimicrobial activity of silver(I) complexes 3 and 4 is similar to that previously reported for those with pyridine-4,5-dicarboxylate esters,37 which are structural analogues of py-2pz. Moreover, silver(I) complexes with pyridinecarboxylates (picolinate, nicotinate and dipicolinate) have shown a significant activity against C. parapsilosis (MIC = 5 μM), with fungistatic effect, while the antibacterial activity against S. aureus was higher (MIC = 10 μM)66 in comparison to 3 and 4. A similar antimicrobial activity against C. parapsilosis and S. aureus was also shown by polymeric silver(I) complexes with pyridine-3,5-dicarboxylate67 and pyridine-2-sulfonate.68
Regarding previously reported copper(II) complexes, those with pyridine-4,5-dicarboxylate esters did not affect bacterial growth under the tested conditions, and only moderate antifungal activity against C. albicans ATCC 10231 was noted.24 On the other hand, copper(II) complexes with 2-hydroxypyridine, 2-aminopyridine and pyridine-2-carboxylic acid inhibited the growth of B. subtilis and partially inhibited the growth of C. albicans ATCC 90028,69 while [Cu(2,2′-bipy)Cl2(thiouracil)] (2,2′-bipy is 2,2′-bipyridine) has shown fungicidal and fungistatic activities against 21 clinical isolates of Candida spp.70
Antiproliferative activity of both copper(II) and silver(I) complexes have been previously investigated on different human cells.71,72 In the present study, with the aim to check the therapeutic potential of complexes 1–4, their antiproliferative activity on the human normal fibroblast cell line (MRC-5) was tested. The obtained IC50 values (μM and μg mL−1; IC50 is defined as the concentration inhibiting 50% of cell growth after 48 h treatment with the tested compounds) are given in Table 2. From the presented data, it can be concluded that the silver(I) complexes 3 and 4 have the positive values of selectivity indices (SI; the ratio between IC50 and MIC values), with the best therapeutic profile (SI = 6.4) in respect to MRC-5 observed for 3 against both Candida spp. (Table 2). Moreover, the complexes 3 and 4 are 3.1 and 1.7-fold less toxic against MRC-5, respectively, than AgSD.
The investigated copper(II) complexes 1 and 2 also show effects on the viability of the MRC-5 cells, whereby complex 1 appeared to be more cytotoxic (Table 2). However, the cytotoxic activity of py-2pz ligand against MRC-5 cells was 120 μM, suggesting that some of the effects observed for the complexes are due to this ligand.45 Similarly, IC50 values in the range 24.0–289.0 μM were previously reported for copper(II) complexes with aromatic N-heterocycles on the same cell line.25 Contrary to this, copper(II) complexes of the Casiopeínas® family, [Cu(L-N,N′)(X-N,O)]NO3 and [Cu(L-N,N′)(Y-O,O′)]NO3, L is 2,2′-bipyridine, 1,10-phenanthroline or their substituted derivatives, X is an essential amino acid or peptide and Y is acetylacetonate or salicylaldehydate,73–77 and copper(II) complexes with 2,2′:6′,2′′-terpyridine (terpy) or its 4′-substituted derivatives78–82 have shown remarkable antiproliferative activity in several cell lines.
Yeast-to-hyphae transition is also the early step in Candida biofilm formation.84 Biofilms represent the most common mode of fungal growth and have importance in nosocomial infections, due to their high antimicrobial resistance.88 Having established that silver(I) complexes 3 and 4 have shown significant anti-Candida activity and that they completely inhibit the formation of hyphae, their ability to effect C. albicans biofilm formation process was analyzed (Table S1†). When MIC of the complexes was applied, the percentage of the formed C. albicans biofilms was 51% and 62% for complexes 3 and 4, respectively. No activity of complex 4 was observed at lower concentrations, while complex 3 had an impact on biofilm formation even at 0.99 μM (0.44 μg mL−1).
Interaction with biomolecules
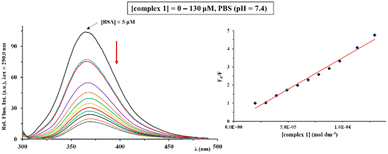
The emission spectra of BSA were recorded in the absence and presence of an increasing amount of the complexes 1–4, in the wavelength range from 295.0 to 500.0 nm, after excitation of the protein at λex = 290.0 nm. Fig. 5 illustrates the behavior of BSA solution after the addition of an increasing concentration (0–130 μM) of copper(II) complex 1. As can be seen, BSA shows a very intensive fluorescence at 365.0 nm in the PBS solution (pH = 7.4), which mainly originates from tryptophan, tyrosine, and phenylalanine residues in this biomolecule.97 After the addition of complexes 1–4 to BSA solution of a constant concentration (5 μM), a decrease of fluorescence intensity at 365.0 nm was observed, because of their binding to BSA (Fig. 5).
The data calculated by Stern–Volmer and Scatchard equations (Stern–Volmer constants (Ksv), quenching rate constants (Kq), binding constants (KA) and the number of binding sites per BSA (n)) are presented in Table 3. The Ksv values follow the order 1 > 3 > 2 > 4, indicating that silver(I) complex 4 has a lowest affinity for BSA compared to the remaining three complexes. The values of Kq constants of the complexes are significantly higher than 2 × 1010 M−1 s−1, being in accordance with a static quenching mechanism.98 The KA values of the complexes are in the range 2.15 × 104 to 6.03 × 106 M−1 and they show the affinity of the complexes to bind to SA for their transport towards the biological targets. However, these constants are much lower than those for the strongest noncovalent interaction (∼1015 M−1),99 suggesting the reversible binding of the complexes to BSA and their release upon arrival at the target sites. The number of binding sites per BSA molecule suggests the binding of complexes to one site per protein molecule.
| Complex | Ksv (M−1) | Hypochromism (%) | Kq (M−1 s−1) | KA (M−1) | n |
|---|---|---|---|---|---|
| 1 | (8.74 ± 0.13) × 105 | 84.4 | 8.74 × 1013 | 6.03 × 106 | 1.55 |
| 2 | (2.28 ± 0.06) × 105 | 84.4 | 2.28 × 1013 | 5.81 × 105 | 1.17 |
| 3 | (5.41 ± 0.11) × 105 | 79.4 | 5.41 × 1013 | 3.08 × 106 | 1.48 |
| 4 | (2.10 ± 0.02) × 104 | 60.5 | 2.10 × 1012 | 2.15 × 104 | 1.02 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) between the hydrophobic octanol phase and the hydrophilic water phase shows the ability of a compound to be transported through the cell membrane.102 The values of log
P) between the hydrophobic octanol phase and the hydrophilic water phase shows the ability of a compound to be transported through the cell membrane.102 The values of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P for clinically used drugs, including metal-based compounds in the Comprehensive Medicinal Chemistry database are in range from −0.4 to 5.6.103 The presently synthesized copper(II) (1 and 2) and silver(I) (3 and 4) complexes have the values in this ideal lipophilicity range, from 0.86 to 2.28. Complex 3 has a greater log
P for clinically used drugs, including metal-based compounds in the Comprehensive Medicinal Chemistry database are in range from −0.4 to 5.6.103 The presently synthesized copper(II) (1 and 2) and silver(I) (3 and 4) complexes have the values in this ideal lipophilicity range, from 0.86 to 2.28. Complex 3 has a greater log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value than the remaining complexes, suggesting its distribution mainly in the octanol phase.
P value than the remaining complexes, suggesting its distribution mainly in the octanol phase.Except log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value of a potentially new therapeutic agent, its molecular weight should be taken into consideration according to the Lipinski's rule of five.104 According to this rule, it is highly desirable that the molecular weight of orally bioavailable drugs does not exceed 500 due to their easier uptake. In the case of the presently synthesized complexes, the molecular weight of 1–3 is higher than 500, and approximately 500 for 4. However, this does not limit their potential application in therapeutic purposes since there are many examples of significantly larger molecules that can be efficiently transported into cells.105 Moreover, it was found that metal-based drugs often do not follow the Lipinski's rule of five and that in the case of these drugs, molecular volume should be considered as an alternative of molecular weight.19
P value of a potentially new therapeutic agent, its molecular weight should be taken into consideration according to the Lipinski's rule of five.104 According to this rule, it is highly desirable that the molecular weight of orally bioavailable drugs does not exceed 500 due to their easier uptake. In the case of the presently synthesized complexes, the molecular weight of 1–3 is higher than 500, and approximately 500 for 4. However, this does not limit their potential application in therapeutic purposes since there are many examples of significantly larger molecules that can be efficiently transported into cells.105 Moreover, it was found that metal-based drugs often do not follow the Lipinski's rule of five and that in the case of these drugs, molecular volume should be considered as an alternative of molecular weight.19
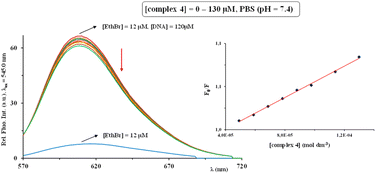
The interaction of the complexes 1–4 with ct-DNA was investigated by the addition of increasing amounts of investigated complexes 1–4 (0–190 μM) into the EthBr-ct-DNA adduct solution ([ct-DNA]/[EthBr] = 10). The behavior of the latter solution in the presence of complexes was monitored through the changes of the fluorescence intensity at λex = 545.0 nm. It is important to note that none of the tested complexes show fluorescence at room temperature in solution or in the presence of DNA at λex = 545.0 nm. The fluorescence emission spectra for EthBr–ct-DNA system in the absence and presence of increasing concentrations of silver(I) complex 4 are shown in Fig. 6.
In all cases, after addition of the investigated complexes, a decrease in fluorescence intensity was observed, indicating their interaction with EthBr–ct-DNA. This decrease can be the consequence of EthBr displacement by the complex, or the binding of a complex to EthBr–ct-DNA system, leading to the formation of a new non-fluorescent adduct.98 Since KA values of the investigated complexes (Table 4) are much lower than that for EthBr (KA = 2 × 106 M−1)98 and that the percentage of hypochromism is less than 30% (for DNA intercalator lucigenin is 50%),108 the possible mode of their binding to ct-DNA is non-intercalative (electrostatic). Moreover, the interaction between ct-DNA and 1–4 is a dominantly static quenching process, what can be concluded from the values of Kq constants (Table 4), which are higher than the limiting diffusion rate constant of this biomolecule (2 × 1010 M−1 s−1).98
| Complex | Ksv (M−1) | Hypochromism (%) | Kq (M−1 s−1) | KA (M−1) | n |
|---|---|---|---|---|---|
| 1 | (1.67 ± 0.01) × 103 | 28.2 | 1.67 × 1011 | 3.67 × 102 | 0.80 |
| 2 | (2.63 ± 0.01) × 102 | 5.4 | 2.63 × 1010 | 4.22 × 101 | 0.50 |
| 3 | (1.50 ± 0.01) × 103 | 17.9 | 1.50 × 1011 | 1.37 × 103 | 0.99 |
| 4 | (1.14 ± 0.01) × 103 | 15.4 | 1.14 × 1011 | 2.00 × 103 | 1.06 |
Catalytic activity of copper(II) complexes 1 and 2
Mimicking the oxidase catalytic activity of the presently synthesized copper(II) complexes 1 and 2 represents also the aim of this study. Hence, two copper(II)-containing enzymes, catechol oxidase (CAO) and phenoxazinone synthase (PHS) were chosen for this purpose. These two enzymes catalyze the aerobic oxidation of a wide range of biologically significant phenolic derivatives.109 The schematic presentation of the reactions catalyzed by these two oxidases is shown in Scheme S1.† Catechol oxidase enzyme belongs to the type-3 copper proteins characterized by a dinuclear copper active site which is EPR silent in the native met CuII–CuII form, due to a strong antiferromagnetic coupling between the μ-OH-bridged Cu(II) ions.110,111 This enzyme catalyzes the oxidation of catechols to the corresponding quinones, that undergo auto polymerization process, leading to the formation of the brown pigment melanin, which is responsible for protection of damaged tissues against pathogens.112 Furthermore, one of the major multicopper oxidase enzyme, which belongs to the type-2 copper proteins is phenoxazinone synthase, which is found naturally in the bacterium Streptomyces antibioticus.113,114 This enzyme catalyzes the oxidative coupling of o-aminophenols, resulting in the production of the phenoxazinone chromophore in the final stage of the biosynthesis of actinomycin D, which is a naturally occurring antitumor agent.115 The synthesis and investigation of functional model transition metal complexes for metalloenzymes that mimic the oxidase activity is therefore of profound importance for the development of new and efficient bioinspired catalysts for different oxidation reactions. This study helps not only to understand the biochemical phenomena in the natural systems but also clues to the development of small molecule catalysts for specific oxidation reaction under mild conditions.116 In the light of the abovementioned facts, various dinuclear as well as mononuclear copper(II) complexes have been recently shown as successful models for both catechol oxidase and phenoxazinone synthase metalloenzymes.116–118In order to investigate the catalytic activity of the copper(II) complexes 1 and 2 for CAO activity, 3,5-di-tert-butylcatechol (3,5-DTBC) was used as a substrate. This substrate has the bulky groups, which prevent ring opening and shows an increase in the absorbance at 400.0 nm, because of the formation of 3,5-di-tert-butylquinone (3,5-DTBQ).119 The catalytic activity of the synthesized complexes was investigated by following the oxidation of 3,5-DTBC to 3,5-DTBQ by mixing the methanolic solution of the substrate and the solution of the corresponding complex in DMSO/methanol (v/v 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
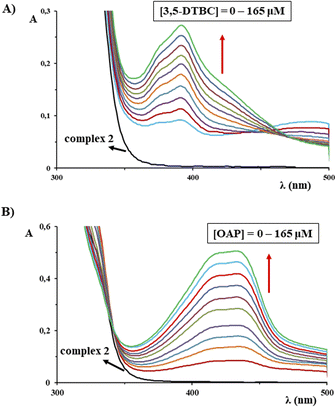
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9) under aerobic conditions at room temperature by UV-Vis spectrophotometry. As can be seen from Fig. 7A for complex 2, a gradual increase in absorbance at 400.0 nm is in accordance with significant oxidase activity for the oxidation of 3,5-DTBC to 3,5-DTBQ.
9) under aerobic conditions at room temperature by UV-Vis spectrophotometry. As can be seen from Fig. 7A for complex 2, a gradual increase in absorbance at 400.0 nm is in accordance with significant oxidase activity for the oxidation of 3,5-DTBC to 3,5-DTBQ.
A similar experiment was performed to examine the catalytic activity of the complexes 1 and 2 for PHS activity, i.e. in the process of the catalytic oxidation of o-aminophenol (OAP) to 2-aminophenoxazine-3-one (APX). An increase in the absorbance at 435.0 nm, was observed in the UV-Vis spectra for both complexes, confirming the conversion of OAP to APX and their catalytic activity (Fig. 7B).119 It is also important to note that the reference experiment in the absence of copper(II) catalyst was performed under the investigated reaction conditions, and the amount of the oxidized products 3,5-DTBQ and APX can be neglected in comparison to that formed in the presence of complexes 1 and 2.
Experimental
Materials and instrumentation
Copper(II) salts (CuCl2·2H2O and Cu(CF3SO3)2), silver(I) salts (AgPF6 and AgNO3), silver(I) sulfadiazine (AgSD), ethanol, methanol, acetone, dimethyl sulfoxide (DMSO), deuterated DMSO, phosphate buffered saline (PBS), bovine serum albumin (BSA), calf thymus DNA (ct-DNA), ethidium bromide (EthBr), n-octanol, 3,5-di-tert-butylcatechol (3,5-DTBC) and o-aminophenol (OAP) were obtained from Sigma-Aldrich (St. Louis, Missouri, USA). All chemicals (reagents and solvents) were used as supplied without purification.Elemental microanalysis for carbon, hydrogen and nitrogen of the synthesized copper(II) and silver(I) complexes was performed by the Microanalytical Laboratory, Faculty of Chemistry, University of Belgrade. The IR spectra in the 4000–450 cm−1 region were recorded using PerkinElmer Spectrum 100 spectrometer by KBr pellet technique (abbreviations used: vs for very strong, s for strong, m for medium, w for weak). 1H NMR spectra for py-2pz and silver(I) complexes 3 and 4 were recorded on a Bruker Avance III spectrometer at 500 MHz at room temperature using tetramethylsilane as the internal standard. 5.0 mg of each compound was dissolved in 0.6 mL of DMSO-d6 and transferred into a 5 mm NMR tube. Chemical shifts (δ) are expressed in ppm and coupling constants (J) are given in Hz. The UV-Vis spectra were recorded on a Shimadzu double-beam spectrophotometer after dissolving the copper(II) and silver(I) complexes in DMSO, as well as after 48 h standing at ambient temperature, over the wavelength range of 1100–200 nm. The concentrations of the solutions used for these measurements were 1 × 10−2 M (1 and 2), 2.63 × 10−5 M (3) and 5.21 × 10−5 M (4). Moreover, the solution behavior of complexes 1–4 was studied by UV-Vis measurements in DMSO/PBS (v/v is 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for 1 and 2, 1
1 for 1 and 2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 42.9 for 3 and 1
42.9 for 3 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 23.1 for 4) over 48 h at room temperature. Molar conductivity measurements were measured at room temperature on a digital conductivity-meter Crison Multimeter MM 41. The concentration of the solutions of complexes in DMSO used for these measurements was 1 × 10−3 M. The cyclic voltammetry (CV) measurements were performed using a potentiostat/galvanostat AutoLab PGSTAT204. The cell (5.0 mL) consisted of three electrode system, glassy carbon (GC) electrode as a working electrode, Ag/AgCl (saturated KCl) as a reference electrode and platinum wire as a counter electrode. All reported potentials are referred versus the Ag/AgCl (saturated KCl) reference electrode. The fluorescence emission spectra for DNA/BSA interactions of the complexes were recorded using Jasco FP-6600 spectrophotometer.
23.1 for 4) over 48 h at room temperature. Molar conductivity measurements were measured at room temperature on a digital conductivity-meter Crison Multimeter MM 41. The concentration of the solutions of complexes in DMSO used for these measurements was 1 × 10−3 M. The cyclic voltammetry (CV) measurements were performed using a potentiostat/galvanostat AutoLab PGSTAT204. The cell (5.0 mL) consisted of three electrode system, glassy carbon (GC) electrode as a working electrode, Ag/AgCl (saturated KCl) as a reference electrode and platinum wire as a counter electrode. All reported potentials are referred versus the Ag/AgCl (saturated KCl) reference electrode. The fluorescence emission spectra for DNA/BSA interactions of the complexes were recorded using Jasco FP-6600 spectrophotometer.
Synthesis of dimethyl 6-(pyrazine-2-yl)pyridine-3,4-dicarboxylate
Dimethyl 6-(pyrazine-2-yl)pyridine-3,4-dicarboxylate (py-2pz) used for the synthesis of copper(II) and silver(I) complexes were prepared by the method previously described in the literature.41 The purity of the synthesized compound was confirmed by elemental microanalysis and 1H NMR spectroscopy.Synthesis of complexes 1–4
Copper(II) complexes, [CuCl2(py-2pz)]2 (1) and [Cu(CF3SO3)(H2O)(py-2pz)2]CF3SO3·2H2O (2), and silver(I) complexes [Ag(py-2pz)2]PF6 (3) and {[Ag(NO3)(py-2pz)]·0.5H2O}n (4) were synthesized according to the modified procedures for the preparation of copper(II) and silver(I) complexes with pyridine-4,5-dicarboxylate esters as ligands.24,37The solution of 1.0 mmol of py-2pz (273.2 mg) in 10.0 mL of ethanol was added slowly under stirring to the solution containing an equimolar amount of the corresponding metal salt (170.5 mg of CuCl2·2H2O for 1, 361.7 mg of Cu(CF3SO3)2 for 2, 252.8 mg of AgPF6 for 3 and 169.8 mg of AgNO3 for 4) in 5.0 mL of ethanol. The reaction mixture was stirred for 3 h on a magnetic stirrer at room temperature (in dark for 3 and 4). Complexes 1 and 3 were obtained after recrystallization of obtained solid products in a mixture of methanol and water (v/v 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and acetone, respectively, while complexes 2 and 4 crystallized from the mother ethanol solution after evaporation at room temperature for 3–5 days. The crystals suitable for single crystal X-ray diffraction analysis were filtered off and dried on room temperature. Yield (calculated on the basis of py-2pz): 256.8 mg (63%) for 1, 283.9 mg (59%) for 2, 283.8 mg (71%) for 3 and 252.6 mg (57%) for 4.
1) and acetone, respectively, while complexes 2 and 4 crystallized from the mother ethanol solution after evaporation at room temperature for 3–5 days. The crystals suitable for single crystal X-ray diffraction analysis were filtered off and dried on room temperature. Yield (calculated on the basis of py-2pz): 256.8 mg (63%) for 1, 283.9 mg (59%) for 2, 283.8 mg (71%) for 3 and 252.6 mg (57%) for 4.
Anal. calcd for 1 = C26H22Cl4Cu2N6O8 (MW = 815.37): C, 38.30; H, 2.72; N, 10.31. Found: C, 38.17; H, 2.77; N, 10.46%. IR (KBr, ν, cm−1): 3085w, 3049w (ν(Car–H)), 2956w (ν(C–H)), 1749vs, 1726vs (ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)), 1615w, 1556m, 1438m, 1429m, 1410m, 1385m (νas(Car
O)), 1615w, 1556m, 1438m, 1429m, 1410m, 1385m (νas(Car![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Car) and νas(Car
Car) and νas(Car![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)), 1306vs, 1281vs, 1270vs (ν(C–O)), 795m (γ(Car–H)). UV-Vis (DMSO, λmax, nm): 875.0 (ε = 1.02 × 102 M−1 cm−1). ΛM (DMSO): 27.4 Ω−1 cm2 mol−1.
N)), 1306vs, 1281vs, 1270vs (ν(C–O)), 795m (γ(Car–H)). UV-Vis (DMSO, λmax, nm): 875.0 (ε = 1.02 × 102 M−1 cm−1). ΛM (DMSO): 27.4 Ω−1 cm2 mol−1.
Anal. calcd for 2 = C28H28CuF6N6O17S2 (MW = 962.22): C, 34.95; H, 2.93; N, 8.73. Found: C, 34.78; H, 3.01; N, 8.82%. IR (KBr, ν, cm−1): 3475br (ν(O–H)), 3115w, 3093w, 3059w (ν(Car–H)), 2959w, 2932w (ν(C–H)), 1730vs (ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)), 1647w, 1618m, 1592w, 1560m, 1536w, 1497w, 1489w, 1440m, 1416m, 1390m (νas(Car
O)), 1647w, 1618m, 1592w, 1560m, 1536w, 1497w, 1489w, 1440m, 1416m, 1390m (νas(Car![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Car) and νas(Car
Car) and νas(Car![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)), 1315s (ν(C–O)), 1285vs, 1255vs (νas(SO3)), 1226s (νs(CF3)), 1163s, 1107m (νas(CF3)), 1084w, 1052m, 1030s (νs(SO3)), 797m (γ(Car–H)). UV-Vis (DMSO, λmax, nm): 805.0 (ε = 43.8 M−1 cm−1). ΛM (DMSO): 84.1 Ω−1 cm2 mol−1.
N)), 1315s (ν(C–O)), 1285vs, 1255vs (νas(SO3)), 1226s (νs(CF3)), 1163s, 1107m (νas(CF3)), 1084w, 1052m, 1030s (νs(SO3)), 797m (γ(Car–H)). UV-Vis (DMSO, λmax, nm): 805.0 (ε = 43.8 M−1 cm−1). ΛM (DMSO): 84.1 Ω−1 cm2 mol−1.
Anal. calcd for 3 = C26H22AgN6F6O8P (MW = 799.33): C, 39.07; H, 2.77; N, 10.51. Found: C, 39.25; H, 2.72; N, 10.58%. IR (KBr, ν, cm−1): 3109w (ν(Car–H)), 2963w, 2932w (ν(C–H)), 1733vs (ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)), 1607m, 1562w, 1552w, 1489w, 1441m, 1412w, 1375w (νas(Car
O)), 1607m, 1562w, 1552w, 1489w, 1441m, 1412w, 1375w (νas(Car![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Car) and νas(Car
Car) and νas(Car![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)), 1314s, 1301s, 1279s, 1250m (ν(C–O)), 839vs (ν(P–F)), 796m (γ(Car–H)). 1H NMR (500 MHz, DMSO-d6): δ = 3.92 (d, J = 6.1 Hz, 6H, Ar–COOCH3), 8.58 (d, J = 0.6 Hz, 1H, H5), 8.84 (m, 2H, H5′ and H6′), 9.17 (d, J = 0.7 Hz, 1H, H2), 9.60 (d, J = 0.9 Hz, 1H, H3′) ppm. UV-Vis (DMSO, λmax, nm): 296.0 (ε = 3.3 × 104 M−1 cm−1). ΛM (DMSO): 30.9 Ω−1 cm2 mol−1.
N)), 1314s, 1301s, 1279s, 1250m (ν(C–O)), 839vs (ν(P–F)), 796m (γ(Car–H)). 1H NMR (500 MHz, DMSO-d6): δ = 3.92 (d, J = 6.1 Hz, 6H, Ar–COOCH3), 8.58 (d, J = 0.6 Hz, 1H, H5), 8.84 (m, 2H, H5′ and H6′), 9.17 (d, J = 0.7 Hz, 1H, H2), 9.60 (d, J = 0.9 Hz, 1H, H3′) ppm. UV-Vis (DMSO, λmax, nm): 296.0 (ε = 3.3 × 104 M−1 cm−1). ΛM (DMSO): 30.9 Ω−1 cm2 mol−1.
Anal. calcd for 4 = C13H12AgN4O7.5 (MW = 452.14): C, 34.53; H, 2.68; N, 12.39. Found: C, 34.72; H, 2.58; N, 12.46%. IR (KBr, ν, cm−1): 3072w, 3034w, 3009w (ν(Car–H)), 2957m, 2929m (ν(C–H)), 1733vs, 1720vs (ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)), 1660w, 1636w, 1623w, 1598m, 1556m, 1439m (νas(Car
O)), 1660w, 1636w, 1623w, 1598m, 1556m, 1439m (νas(Car![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Car) and νas(Car
Car) and νas(Car![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)), 1384s, 1368s (νas(NO3)), 1332s, 1315m, 1291vs, 1280vs, 1263s (ν(C–O)), 798m (γ(Car–H)). 1H NMR (500 MHz, DMSO-d6): δ = 3.92 (d, J = 6.1 Hz, 6H, Ar–COOCH3), 8.58 (d, J = 0.6 Hz, 1H, H5), 8.84 (s, 2H, H5′ and H6′), 9.18 (d, J = 0.6 Hz, 1H, H2), 9.60 (d, J = 2.6 Hz, 1H, H3′) ppm. UV-Vis (DMSO, λmax, nm): 296.0 (ε = 1.6 × 104 M−1 cm−1). ΛM (DMSO): 37.6 Ω−1 cm2 mol−1.
N)), 1384s, 1368s (νas(NO3)), 1332s, 1315m, 1291vs, 1280vs, 1263s (ν(C–O)), 798m (γ(Car–H)). 1H NMR (500 MHz, DMSO-d6): δ = 3.92 (d, J = 6.1 Hz, 6H, Ar–COOCH3), 8.58 (d, J = 0.6 Hz, 1H, H5), 8.84 (s, 2H, H5′ and H6′), 9.18 (d, J = 0.6 Hz, 1H, H2), 9.60 (d, J = 2.6 Hz, 1H, H3′) ppm. UV-Vis (DMSO, λmax, nm): 296.0 (ε = 1.6 × 104 M−1 cm−1). ΛM (DMSO): 37.6 Ω−1 cm2 mol−1.
Crystallographic data collection and refinement of the structures
X-ray diffraction data was collected on an Oxford Diffraction SuperNova diffractometer with Mo/Cu microfocus X-ray source with mirror optics and an Atlas detector at 150(2) K. The structures were solved in Olex2 graphical user interface43 by direct methods implemented in SHELXT and refined by a full-matrix least-squares procedure based on F2 using SHELXL.120 All non-hydrogen atoms were refined anisotropically. The hydrogen atoms were placed at calculated positions and treated using appropriate riding models. Additional details on structural properties and refinement details are given in the ESI (Tables S2 and S3†).Computational details
All DFT calculations have been performed with the ADF121,122 engine in Amsterdam Modeling Suite (version 2022.102).123 The all-electron triple-zeta Slater-type orbitals plus one polarization function (TZP) basis set was used for all atoms. Relativistic effects were accounted for by the scalar-relativistic Zeroth-Order Regular Approximation (ZORA).124–126 Open-shell copper(II) complexes 1 and 2 are treated with unrestricted DFT formalism. Geometry optimizations with the Quasi-Newton method and delocalized coordinates127 were performed using general gradient approximation consisting of Becke's exchange128 and Perdew's correlation129 with Grimme's fourth generation dispersion energy corrections,130 i.e. BP86-D4 density functional. The COSMO solvation model,131,132 as implemented in ADF,133 with DMSO as solvent was used. Analytical harmonic frequencies134–136 were calculated at the same level of theory. Vibrational analysis with free rotor interpolation corrections as proposed by Grimme137 for low-frequency contributions (frequency cut-off 100 cm−1) has been used to evaluate zero-point and entropic effects to the Gibbs free energy at 298 K. Because vibrational analysis is done at a standard state of 1 atm, conversion to 1 mol dm−3 solution standard state is done (correction of 1.89 kcal mol−1 to the free energies at 298 K). Electronic energies used to calculate the Gibbs free energy were evaluated with M06-2X138,139 meta-hybrid functional at ZORA-BP86-D4/TZP-COSMO (DMSO) geometries. The exchange coupling constant J in complex 1 was calculated with broken-symmetry DFT formalism140–144 at ZORA-M06-2X/TZP level of theory at X-ray geometry according to the Yamaguchi approach.145 Broken symmetry solutions are obtained from the high-spin states with the spin-flip method. Before single-point calculations on the dinuclear Cu(II) complex from the X-ray structure, the positions of hydrogen atoms were optimized at the ZORA-BP86-D4/DZP level of theory. The Fast Inertial Relaxation Engine146 based optimizer in Cartesian coordinates was used for hydrogen optimization. The LibXC library147 was used for all calculations employing M06-2X functional. The Cartesian coordinates of all the optimized structures are available in the ESI.†Evaluation of antimicrobial potential of complexes 1–4
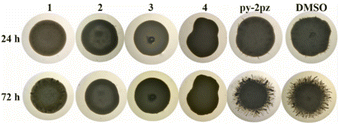
For determination of minimal inhibitory concentration (MIC) values of complexes 1–4, standardized broth microdilution methodology was used, recommended by the National Committee for Clinical Laboratory Standards (M07-A8) for bacteria and Standards of European Committee on Antimicrobial Susceptibility Testing (v 7.3.1: method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for yeasts) for Candida spp.148 In this study, two bacterial strains (Pseudomonas aeruginosa NCTC 10332 and Staphylococcus aureus ATCC 25923) and two fungal strains (Candida albicans ATCC 10231 and Candida parapsilosis ATCC 22019) were used (NCTC is National Collection of Type Cultures, ATCC is American Type Culture Collection). All tested compounds were firstly dissolved in DMSO at a concentration of 50 mg mL−1, to reach the highest tested concentration of 500 μg mL−1 (making sure that the final concentration of the DMSO in the assay was lower than 1%, v/v). The inoculums were 5 × 105 colony forming units, cfu mL−1, for bacteria and 1 × 105 cfu mL−1 for fungal species. The positive control was silver(I) sulfadiazine (AgSD) as the most widely used antimicrobial agent based on the silver(I). The MIC value was recorded as the lowest concentration that inhibited the growth after 24 h at 37 °C, using the Tecan Infinite 200 Pro multiplate reader (Tecan Group Ltd, Männedorf, Switzerland).In vitro toxicity assessment
Antiproliferative activity of complexes 1–4 was determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay149 on human lung fibroblasts cells (MRC-5), obtained from the American Type Culture Collection (ATCC). The inoculums of 1 × 104 cells per well were cultured in the complete RPMI 1640 medium (RPMI is Roswell Park Memorial Institute; Gibco™ by Thermo Fischer Scientific, CE) as a monolayer and further incubated with the tested compounds at a concentration from a maximum of 120 μM, in a humidified atmosphere of 95% air and 5% CO2 at 37 °C for 48 h. The extent of MTT reduction was measured spectrophotometrically at 540.0 nm using Tecan Infinite 200 Pro multiplate reader (Tecan Group Ltd, Männedorf, Switzerland). Cytotoxicity was expressed as the concentration of the tested compound, which inhibits the cell growth by 50% (IC50) in comparison with the negative control (DMSO-treated cells).Serratia marcescens ATCC 27117 was used for the assessment of influence of complexes 1–4 on prodigiosin production. The methodology was the same as for the C. violaceum CV026 assay, but no autoinducer was needed.153
Inhibition of violacein and prodigiosin production was defined as the presence of blurry white hallows around discs containing compound, measured in mm and it was distinguished from the zones of clearance that indicated growth inhibition of the tested strains.
DNA binding study
| F0/F = 1 + Kqτ0[complex] = 1 + Ksv[complex] |
log(F0 − F)/F = log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KA + n KA + n![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log[complex] log[complex] |
Protein binding study
The protein binding study was performed by tryptophan fluorescence quenching experiments using BSA (5 μM) in PBS (pH 7.4). The quenching of the emission intensity of tryptophan residues of BSA at 365.0 nm was monitored using the increasing concentration of the complexes 1–4 (up to 130 μM). Fluorescence spectra were recorded in the range 295.0–500.0 nm with an excitation wavelength of 290.0 nm. The corresponding binding constants of the complexes (KA) and apparent binding sites (n) were calculated as it was explained for DNA binding study.97,155Lipophilicity assay
The lipophilicity of the complexes was determined by the flask-shaking method.100 The complexes 1–4 were dissolved in DMSO and added to water/n-octanol system. This mixture was vortexed for 1 h at 25 °C to allow partitioning. After that time, the solutions were left to stand for 24 h until the separation of two phases. The concentration of complexes in both phases was determined by measuring absorbance values using previously determined calibration curves. Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values were calculated according to the following equation:
P values were calculated according to the following equation:log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = log(co/cw) P = log(co/cw) |
Catalytic activity of copper(II) complexes 1 and 2
3,5-Di-tert-butylcatechol (3,5-DTBC) and o-aminophenol (OAP) were used as substrates for investigation of the oxidase biomimetic catalytic activity of copper(II) complexes 1 and 2 (Scheme S1†). The solutions of 3,5-DTBC and OAP were prepared in DMSO, while complexes were dissolved in DMSO/water (v/v 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9). The progress of the reaction was followed by UV-Vis spectrophotometry by monitoring the increase of the absorbance of the quinone band at 400.0 nm (ε = 1.9 × 103 M−1 cm−1)156 and amino phenoxazinone at 435.0 nm (ε = 9.095 × 103 M−1 cm−1)157 as a function of time.
9). The progress of the reaction was followed by UV-Vis spectrophotometry by monitoring the increase of the absorbance of the quinone band at 400.0 nm (ε = 1.9 × 103 M−1 cm−1)156 and amino phenoxazinone at 435.0 nm (ε = 9.095 × 103 M−1 cm−1)157 as a function of time.
Conclusions
In the present study, synthesis, structural characterization and biological evaluation of four new copper(II) and silver(I) complexes with dimethyl 6-(pyrazine-2-yl)pyridine-3,4-dicarboxylate (py-2pz), [CuCl2(py-2pz)]2 (1), [Cu(CF3SO3)(H2O)(py-2pz)2]CF3SO3·2H2O (2), [Ag(py-2pz)2]PF6 (3) and {[Ag(NO3)(py-2pz)]·0.5H2O}n (4) have been reported. The obtained spectroscopic and crystallographic results have shown that py-2pz ligand is bidentately coordinated to the Cu(II) and Ag(I) ions through its pyridine and pyrazine nitrogen atoms in all complexes, while in polynuclear complex 4, heterocyclic pyrazine ring of py-2pz additionally acts as a bridging ligand between two Ag(I) ions. According to the DFT results, studied copper(II) and silver(I) complexes in DMSO solution are most stable as tetra-coordinated species. Furthermore, DFT calculations revealed a weak ferromagnetic coupling between Cu(II) centers in the X-ray structure of dinuclear complex 1. Silver(I) complexes 3 and 4 have manifested a remarkable antifungal activity on both tested Candida strains with MIC values of 4.9 and 8.6 μM (3.9 μg mL−1), respectively, while no significant anti-Candida activity was observed for copper(II) complexes 1 and 2 (MIC values are higher than 500 μM). Moreover, the complexes 3 and 4 have shown a good antibacterial activity against P. aeruginosa PAO1 with MIC being 19.5 and 8.6 μM (15.6 and 3.9 μg mL−1, respectively), while the activity of 1 and 2 against this strain is moderate (MIC = 153.3 and 259.8 μM; 125 and 250 μg mL−1, respectively). The significantly higher antimicrobial potential of the synthesized silver(I) complexes can be attributed to the presence of Ag(I) ion, which can show its activity through different pathways, including the interactions with proteins, enzymes and DNA, as well as the ROS production. All synthesized complexes had the ability to inhibit the hyphae formation of C. albicans, while silver(I) complexes 3 and 4 also have shown the activity towards biofilms of this fungus. The complexes 1–4 can bind to the serum albumin tightly and reversibly (KA are in the range 2.15 × 104 to 6.03 × 106 M−1), while the interaction with DNA is rather electrostatic since their KA values are much lower than that for EthBr and that the percentage of hypochromism is less than 30%. Finally, two copper(II) complexes 1 and 2 have been shown as successful models for both copper(II)-containing metalloenzymes, catechol oxidase and phenoxazinone synthase.Overall, the obtained results confirm that the nature of metal ion coordinated to the planar organic molecule has a significant influence on the antimicrobial activity of the resulted metal complex. Thus, the coordination of the organic molecule (py-2pz) to the Ag(I) ion leads to the formation of significantly more active complexes against the microbial strains in respect to its coordination to Cu(II), what is in accordance with current use of silver and its compounds in medicine for treatment of various microbial infections. The latter finding along with those previously reported for the structurally similar ligands, such as derivatives of pyridine-4,5-dicarboxylate esters, should be taken into consideration in design of novel metal-based compounds for potential use in medicine.
Author contributions
Conceptualization, T. P. A., I. T., M. I. Dj. and B. Đ. G.; methodology, T. P. A., I. A., J. K., M. P. and S. V.; software, J. K., M. P. and M. Z.; validation, J. N. R., I. T., M. I. Dj. and B. Đ. G.; investigation, T. P. A., I. A., J. K., M. P., M. Z. and S. V.; resources, I. T. and M. I. Dj.; writing-original draft preparation, T. P. A., I. A., J. K., M. P., M. Z. and S. V.; writing-review and editing, J. N. R., I. T., M. I. Dj. and B. Đ. G.; visualization, T. P. A., I. A., J. K., M. P., M. Z. and S. V.; supervision, I. T., M. I. Dj. and B. Đ. G. All authors read and approved the final manuscript.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
This research has been financially supported by the Ministry of Education, Science and Technological Development of the Republic of Serbia (Agreements No. 451-03-68/2022-14/200026, 451-03-68/2022-14/200042 and 451-03-68/2022-14/200122) and by the Slovenian Research Agency (grant P1-0175). The EN→FIST Centre of Excellence, Trg OF 13, SI-1000 Ljubljana, Slovenia, is acknowledged for the use of the SuperNova diffractometer. This research has also received funding from the Serbian Academy of Sciences and Arts under project No. F128. MZ acknowledges the support of the Science Fund of the Republic of Serbia for the computational part of this work (project #7750288, Tailoring Molecular Magnets and Catalysts Based on Transition Metal Complexes – TMMagCat).Notes and references
- S. Nasiri Sovari and F. Zobi, Chemistry, 2020, 2, 418–452 CrossRef.
- M. Gajdács, E. Urbán, A. Stájer and Z. Baráth, Eur. J. Investig. Health Psychol. Educ., 2021, 11, 71–82 CrossRef PubMed.
- J. O'Neill, Review on Antimicrobial Resistance Antimicrobial Resistance: tackling a crisis for the health and wealth of nations, London: Review on Antimicrobial Resistance, 2014 Search PubMed.
- M. E. A. de Kraker, A. J. Stewardson and S. Harbarth, PLoS Med., 2016, 13, e1002184 CrossRef PubMed.
- F. Prestinaci, P. Pezzotti and A. Pantosti, Pathog. Global Health, 2015, 109, 309–318 CrossRef PubMed.
- C. J. L. Murray, K. S. Ikuta, F. Sharara, L. Swetschinski, G. R. Aguilar, A. Gray, C. Han, C. Bisignano, P. Rao, E. Wool, S. C. Johnson, A. J. Browne, M. G. Chipeta, F. Fell, S. Hackett, G. Haines-Woodhouse, B. H. K. Hamadani, E. A. P. Kumaran, B. McManigal, S. Achalapong, R. Agarwal, S. Akech, S. Albertson, J. Amuasi, J. Andrews, A. Aravkin, E. Ashley, F.-X. Babin, F. Bailey, S. Baker, B. Basnyat, A. Bekker, R. Bender, J. A. Berkley, A. Bethou, J. Bielicki, S. Boonkasidecha, J. Bukosia, C. Carvalheiro, C. Castañeda-Orjuela, V. Chansamouth, S. Chaurasia, S. Chiurchiù, F. Chowdhury, R. C. Donatien, A. J. Cook, B. Cooper, T. R. Cressey, E. Criollo-Mora, M. Cunningham, S. Darboe, N. P. J. Day, M. De Luca, K. Dokova, A. Dramowski, S. J. Dunachie, T. D. Bich, T. Eckmanns, D. Eibach, A. Emami, N. Feasey, N. Fisher-Pearson, K. Forrest, C. Garcia, D. Garrett, P. Gastmeier, A. Z. Giref, R. C. Greer, V. Gupta, S. Haller, A. Haselbeck, S. I. Hay, M. Holm, S. Hopkins, Y. Hsia, K. C. Iregbu, J. Jacobs, D. Jarovsky, F. Javanmardi, M. Khorana, N. Kissoon, E. Kobeissi, T. Kostyanev, K. Phommasone, S. Khuduwan, F. Krapp, R. Krumkamp, A. Kumar, H. H. Kyu, C. Lim, K. Lim, D. Limmathurotsakul, M. J. Loftus, M. Lunn, J. Ma, A. Manoharan, F. Marks, J. May, M. Mayxay, N. Mturi, T. Munera-Huertas, P. Musicha, L. A. Musila, M. M. Mussi-Pinhata, R. N. Naidu, T. Nakamura, R. Nanavati, S. Nangia, P. Newton, C. Ngoun, A. Novotney, D. Nwakanma, C. W. Obiero, T. J. Ochoa, A. Olivas-Martinez, P. Olliaro, E. Ooko, E. Ortiz-Brizuela, P. Ounchanum, G. D. Pak, J. L. Paredes, A. Y. Peleg, C. Perrone, T. Phe, N. Plakkal, A. Ponce-de-Leon, M. Raad, T. Ramdin, S. Rattanavong, A. Riddell, T. Roberts, J. V. Robotham, A. Roca, V. D. Rosenthal, K. E. Rudd, N. Russell, H. S. Sader, W. Saengchan, J. Schnall, J. A. G. Scott, S. Seekaew, M. Sharland, M. Shivamallappa, J. Sifuentes-Osornio, A. J. Simpson, N. Steenkeste, A. J. Stewardson, T. Stoeva, N. Tasak, A. Thaiprakong, G. Thwaites, C. Tigoi, C. Turner, P. Turner, H. R. van Doorn, S. Velaphi, A. Vongpradith, M. Vongsouvath, H. Vu, T. Walsh, J. L. Walson, S. Waner, T. Wangrangsimakul, P. Wannapinij, T. Wozniak, T. E. M. W. Young-Sharma, K. C. Yu, P. Zheng, B. Sartorius, A. D. Lopez, A. Stergachis, C. Moore, C. Dolecek and M. Naghavi, Lancet, 2022, 399, 629–655 CrossRef CAS PubMed.
- A. Frei, Antibiotics, 2020, 9, 90 CrossRef CAS PubMed.
- A. Frei, J. Zuegg, A. G. Elliott, M. Baker, S. Braese, C. Brown, F. Chen, C. G. Dowson, G. Dujardin, N. Jung, A. P. King, A. M. Mansour, M. Massi, J. Moat, H. A. Mohamed, A. K. Renfrew, P. J. Rutledge, P. J. Sadler, M. H. Todd, C. E. Willans, J. J. Wilson, M. A. Cooper and M. A. T. Blaskovich, Chem. Sci., 2020, 11, 2627–2639 RSC.
- F. Lovering, J. Bikker and C. Humblet, J. Med. Chem., 2009, 52, 6752–6756 CrossRef CAS PubMed.
- F. Lovering, MedChemComm, 2013, 4, 515–519 RSC.
- A. W. Hung, A. Ramek, Y. Wang, T. Kaya, J. A. Wilson, P. A. Clemons and D. W. Young, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 6799–6804 CrossRef CAS PubMed.
- W. H. B. Sauer and M. K. Schwarz, J. Chem. Inf. Comput. Sci., 2003, 43, 987–1003 CrossRef CAS PubMed.
- W. R. J. D. Galloway, A. Isidro-Llobet and D. R. Spring, Nat. Commun., 2010, 1, 80 CrossRef PubMed.
- C. N. Morrison, K. E. Prosser, R. W. Stokes, A. Cordes, N. Metzler-Nolte and S. M. Cohen, Chem. Sci., 2020, 11, 1216–1225 RSC.
- G. Gasser and N. Metzler-Nolte, Curr. Opin. Chem. Biol., 2012, 16, 84–91 CrossRef CAS PubMed.
- T. Gianferrara, I. Bratsos and E. Alessio, Dalton Trans., 2009, 7588–7598 RSC.
- Y. Lin, H. Betts, S. Keller, K. Cariou and G. Gasser, Chem. Soc. Rev., 2021, 50, 10346–10402 RSC.
- P. Chellan and P. J. Sadler, Chem.–Eur. J., 2020, 26, 8676–8688 CrossRef CAS PubMed.
- E. J. Anthony, E. M. Bolitho, H. E. Bridgewater, O. W. L. Carter, J. M. Donnelly, C. Imberti, E. C. Lant, F. Lermyte, R. J. Needham, M. Palau, P. J. Sadler, H. Shi, F.-X. Wang, W.-Y. Zhang and Z. Zhang, Chem. Sci., 2020, 11, 12888–12917 RSC.
- C. Imberti and P. J. Sadler, in Advances in Inorganic Chemistry, ed. P. J. Sadler and R. van Eldik, Academic Press, 2020, vol. 75, pp. 3–56 Search PubMed.
- M. Kaushal, T. S. Lobana, L. Nim, R. Bala, D. S. Arora, I. Garcia-Santos, C. E. Duff and J. P. Jasinski, New J. Chem., 2019, 43, 11727–11742 RSC.
- S. D. Oladipo, B. Omondi and C. Mocktar, Polyhedron, 2019, 170, 712–722 CrossRef CAS.
- D.-H. Cai, C.-L. Zhang, Q.-Y. Liu, L. He, Y.-J. Liu, Y.-H. Xiong and X.-Y. Le, Eur. J. Med. Chem., 2021, 213, 113182 CrossRef CAS PubMed.
- T. P. Andrejević, I. Aleksic, M. Počkaj, J. Kljun, D. Milivojevic, N. Lj. Stevanović, J. Nikodinovic-Runic, I. Turel, M. I. Djuran and B. Đ. Glišić, Dalton Trans., 2021, 50, 2627–2638 RSC.
- B. Đ. Glišić, I. Aleksic, P. Comba, H. Wadepohl, T. Ilic-Tomic, J. Nikodinovic-Runic and M. I. Djuran, RSC Adv., 2016, 6, 86695–86709 RSC.
- Z. Aziz, S. F. Abu and N. J. Chong, Burns, 2012, 38, 307–318 CrossRef CAS PubMed.
- A. Kascatan-Nebioglu, M. J. Panzner, C. A. Tessier, C. L. Cannon and W. J. Youngs, Coord. Chem. Rev., 2007, 251, 884–895 CrossRef CAS.
- N. A. Johnson, M. R. Southerland and W. J. Youngs, Molecules, 2017, 22, 1263 CrossRef PubMed.
- S. Medici, M. Peana, G. Crisponi, V. M. Nurchi, J. I. Lachowicz, M. Remelli and M. A. Zoroddu, Coord. Chem. Rev., 2016, 327–328, 349–359 CrossRef CAS.
- X. Liang, S. Luan, Z. Yin, M. He, C. He, L. Yin, Y. Zou, Z. Yuan, L. Li, X. Song, C. Lv and W. Zhang, Eur. J. Med. Chem., 2018, 157, 62–80 CrossRef CAS PubMed.
- S. Y. Liau, D. C. Read, W. J. Pugh, J. R. Furr and A. D. Russell, Lett. Appl. Microbiol., 1997, 25, 279–283 CrossRef CAS PubMed.
- C. Chiericatti, J. C. Basílico, M. L. Z. Basílico and J. M. Zamaro, Microporous Mesoporous Mater., 2014, 188, 118–125 CrossRef CAS.
- M. Claudel, J. V. Schwarte and K. M. Fromm, Chemistry, 2020, 2, 849–899 CrossRef CAS.
- Q. L. Feng, J. Wu, G. Q. Chen, F. Z. Cui, T. N. Kim and J. O. Kim, J. Biomed. Mater. Res., 2000, 52, 662–668 CrossRef CAS PubMed.
- W. K. Jung, H. C. Koo, K. W. Kim, S. Shin, S. H. Kim and Y. H. Park, Appl. Environ. Microbiol., 2008, 74, 2171–2178 CrossRef CAS PubMed.
- H.-J. Park, J. Y. Kim, J. Kim, J.-H. Lee, J.-S. Hahn, M. B. Gu and J. Yoon, Water Res., 2009, 43, 1027–1032 CrossRef CAS PubMed.
- T. P. Andrejević, D. Milivojevic, B. Đ. Glišić, J. Kljun, N. Lj. Stevanović, S. Vojnovic, S. Medic, J. Nikodinovic-Runic, I. Turel and M. I. Djuran, Dalton Trans., 2020, 49, 6084–6096 RSC.
- K. Traven, M. Sinreih, J. Stojan, S. Seršen, J. Kljun, J. Bezenšek, B. Stanovnik, I. Turel and T. L. Rižner, Chem.-Biol. Interact., 2015, 234, 349–359 CrossRef CAS PubMed.
- K. Traven, N. Eleftheriadis, S. Seršen, J. Kljun, J. Bezenšek, B. Stanovnik, I. Turel and F. J. Dekker, Polyhedron, 2015, 101, 306–313 CrossRef CAS.
- T. P. Andrejević, I. Aleksić, J. Kljun, B. V. Pantović, D. Milivojevic, S. Vojnovic, I. Turel, M. I. Djuran and B. Đ. Glišić, Inorganics, 2022, 10, 71 CrossRef.
- J. Bezenšek, B. Prek, U. Grošelj, M. Kasunič, J. Svete and B. Stanovnik, Tetrahedron, 2012, 68, 4719–4731 CrossRef.
- A. W. Addison, T. N. Rao, J. Reedijk, J. van Rijn and G. C. Verschoor, Dalton Trans., 1984, 1349–1356 RSC.
- O. V. Dolomanov, L. J. Bourhis, R. J. Gildea, J. A. K. Howard and H. Puschmann, J. Appl. Crystallogr., 2009, 42, 339–341 CrossRef CAS.
- L. Yang, D. R. Powell and R. P. Houser, Dalton Trans., 2007, 955–964 RSC.
- N. Lj. Stevanović, T. P. Andrejević, A. Crochet, T. Ilic-Tomic, N. S. Drašković, J. Nikodinovic-Runic, K. M. Fromm, M. I. Djuran and B. Đ. Glišić, Polyhedron, 2019, 173, 114112 CrossRef.
- H. El Hamdani, M. El Amane and C. Duhayon, J. Mol. Struct., 2018, 1155, 789–796 CrossRef CAS.
- Y. Nakajima, Y. Shiraishi, T. Tsuchimoto and F. Ozawa, Chem. Commun., 2011, 47, 6332–6334 RSC.
- A. S. Potapov, E. A. Nudnova, A. I. Khlebnikov, V. D. Ogorodnikov and T. V. Petrenko, Inorg. Chem. Commun., 2015, 53, 72–75 CrossRef CAS.
- U. Kalinowska-Lis, A. Felczak, L. Chęcińska, K. Zawadzka, E. Patyna, K. Lisowska and J. Ochocki, Dalton Trans., 2015, 44, 8178–8189 RSC.
- B. J. Hathaway, in Comprehensive Coordination Chemistry, ed. G. Wilkinson, R. D. Gillard and J. A. McCleverty, Pergamon, Oxford, 1987, vol. 5 Search PubMed.
- Y. Jiang, C.-F. Zhu, Z. Zheng, J.-B. He and Y. Wang, Inorg. Chim. Acta, 2016, 451, 143–147 CrossRef CAS.
- I. Ali, W. A. Wani and K. Saleem, Synth. React. Inorg. Met., 2013, 43, 1162–1170 CrossRef CAS.
- J. M. S. Cardoso, I. Correia, A. M. Galvão, F. Marques and M. F. N. N. Carvalho, J. Inorg. Biochem., 2017, 166, 55–63 CrossRef CAS PubMed.
- M. Sirajuddin, S. Ali and A. Badshah, J. Photochem. Photobiol., B, 2013, 124, 1–19 CrossRef CAS PubMed.
- A. P. Sandoval-Rojas, L. Ibarraa, M. T. Cortés, M. A. Macías, L. Suescun and J. Hurtado, J. Electroanal. Chem., 2017, 805, 60–67 CrossRef CAS.
- E. Franco, E. López-Torres, A. Mendiola and T. Sevilla, Polyhedron, 2000, 19, 441–451 CrossRef CAS.
- J.-Y. Wu, Y.-L. Pan, X.-J. Zhang, T. Sun, Y.-P. Tian, J.-X. Yang and Z.-N. Chen, Inorg. Chim. Acta, 2007, 360, 2083–2091 CrossRef CAS.
- P. Połczyński, R. Jurczakowski and W. Grochala, J. Phys. Chem. C, 2013, 117, 20689–20696 CrossRef.
- D. P. Ašanin, S. Skaro Bogojevic, F. Perdih, T. P. Andrejević, D. Milivojevic, I. Aleksic, J. Nikodinovic-Runic, B. Đ. Glišić, I. Turel and M. I. Djuran, Molecules, 2021, 26, 1871 CrossRef PubMed.
- N. Stevanović, M. Zlatar, I. Novaković, A. Pevec, D. Radanović, I. Z. Matić, M. Đorđić Crnogorac, T. Stanojković, M. Vujčić, M. Gruden, D. Sladić, K. Anđelković, I. Turel and B. Čobeljić, Dalton Trans., 2021, 51, 185–196 RSC.
- B. Đ. Glišić, L. Senerovic, P. Comba, H. Wadepohl, A. Veselinovic, D. R. Milivojevic, M. I. Djuran and J. Nikodinovic-Runic, J. Inorg. Biochem., 2016, 155, 115–128 CrossRef PubMed.
- S. Medici, M. Peana, V. M. Nurchi and M. A. Zoroddu, J. Med. Chem., 2019, 62, 5923–5943 CrossRef CAS PubMed.
- T. Bjarnsholt and M. Givskov, Anal. Bioanal. Chem., 2007, 387, 409–414 CrossRef CAS PubMed.
- K. Olejnickova, V. Hola and F. Ruzicka, Pathog. Dis., 2014, 72, 87–94 CAS.
- D. Savoia, Future Microbiol., 2014, 9, 917–928 CrossRef CAS PubMed.
- Z. Vargová, M. Almáši, D. Hudecová, D. Titková, I. Rostášová, V. Zeleňák and K. Györyová, J. Coord. Chem., 2014, 67, 1002–1021 CrossRef.
- S. H. Alisir, S. Demir, B. Sariboga and O. Buyukgungor, J. Coord. Chem., 2015, 68, 155–168 CrossRef CAS.
- M. Rendošová, Z. Vargová, D. Sabolová, N. Imrichová, D. Hudecová, R. Gyepes, B. Lakatoš and K. Elefantová, J. Inorg. Biochem., 2018, 186, 206–216 CrossRef PubMed.
- T. Suksrichavalit, S. Prachayasittikul, C. Nantasenamat, C. Isarankura-Na-Ayudhya and V. Prachayasittikul, Eur. J. Med. Chem., 2009, 44, 3259–3265 CrossRef CAS PubMed.
- F. G. da Silva Dantas, A. A. de Almeida-Apolonio, R. P. de Araújo, L. R. V. Favarin, P. F. de Castilho, F. de Oliveira Galvão, T. I. E. Svidzinski, G. A. Casagrande and K. M. P. de Oliveira, Molecules, 2018, 23, 1856 CrossRef PubMed.
- C. Santini, M. Pellei, V. Gandin, M. Porchia, F. Tisato and C. Marzano, Chem. Rev., 2014, 114, 815–862 CrossRef CAS PubMed.
- C. N. Banti and S. K. Hadjikakou, Metallomics, 2013, 5, 569–596 CrossRef CAS PubMed.
- J. Serment-Guerrero, P. Cano-Sanchez, E. Reyes-Perez, F. Velazquez-Garcia, M. E. Bravo-Gomez and L. Ruiz-Azuara, Toxicol. In Vitro, 2011, 25, 1376–1384 CrossRef CAS PubMed.
- R. Galindo-Murillo, J. C. García-Ramos, L. Ruiz-Azuara, T. E. Cheatham III and F. Cortés-Guzmán, Nucleic Acids Res., 2015, 43, 5364–5376 CrossRef CAS PubMed.
- J. Serment-Guerrero, M. E. Bravo-Gomez, E. Lara-Rivera and L. Ruiz-Azuara, J. Inorg. Biochem., 2017, 166, 68–75 CrossRef CAS PubMed.
- J. C. García-Ramos, A. G. Gutiérrez, A. Vázquez-Aguirre, Y. Toledano-Magaña, A. L. Alonso-Sáenz, V. Gómez-Vidales, M. Flores-Alamo, C. Mejía and L. Ruiz-Azuara, BioMetals, 2017, 30, 43–58 CrossRef PubMed.
- A. G. Gutiérrez, A. Vázquez-Aguirre, J. C. García-Ramos, M. Flores-Alamo, E. Hernández-Lemus, L. Ruiz-Azuara and C. Mejía, J. Inorg. Biochem., 2013, 126, 17–25 CrossRef PubMed.
- S. Roy, S. Saha, R. Majumdar, R. R. Dighe and A. R. Chakravarty, Polyhedron, 2010, 29, 2787–2794 CrossRef CAS.
- V. M. Manikandamathavan, V. Rajapandian, A. J. Freddy, T. Weyhermüller, V. Subramanian and B. U. Nair, Eur. J. Med. Chem., 2012, 57, 449–458 CrossRef CAS PubMed.
- J. Grau, R. F. Brissos, J. Salinas-Uber, A. B. Caballero, A. Caubet, O. Roubeau, L. Korrodi-Gregório, R. Pérez-Tomásc and P. Gamez, Dalton Trans., 2015, 44, 16061–16072 RSC.
- V. M. Manikandamathavan, M. Thangaraj, T. Weyhermuller, R. P. Parameswari, V. Punitha, N. N. Murthy and B. U. Nair, Eur. J. Med. Chem., 2017, 135, 434–446 CrossRef CAS PubMed.
- B. Đ. Glišić, J. Nikodinovic-Runic, T. Ilic-Tomic, H. Wadepohl, A. Veselinović, I. M. Opsenica and M. I. Djuran, Polyhedron, 2018, 139, 313–322 CrossRef.
- J. A. Romo, C. G. Pierce, A. K. Chaturvedi, A. L. Lazzell, S. F. McHardy, S. P. Saville and J. L. Lopez-Ribot, MBio, 2017, 8, e01991 CrossRef CAS PubMed.
- N. Božinović, S. Šegan, S. Vojnovic, A. Pavic, B. A. Šolaja, J. Nikodinovic-Runic and I. M. Opsenica, Chem. Biol. Drug Des., 2016, 88, 795–806 CrossRef PubMed.
- N. D. Savić, S. Vojnovic, B. Đ. Glišić, A. Crochet, A. Pavic, G. V. Janjić, M. Pekmezović, I. M. Opsenica, K. M. Fromm, J. Nikodinovic-Runic and M. I. Djuran, Eur. J. Med. Chem., 2018, 156, 760–773 CrossRef PubMed.
- N. Lj. Stevanović, I. Aleksic, J. Kljun, S. Skaro Bogojevic, A. Veselinovic, J. Nikodinovic-Runic, I. Turel, M. I. Djuran and B. Đ. Glišić, Pharmaceuticals, 2021, 14, 24 CrossRef PubMed.
- B. S. R. Mohamed, M. Subramanian and K. P. Shunmugiah, Appl. Microbiol. Biotechnol., 2014, 98, 6775–6785 CrossRef PubMed.
- H. T. Taff, K. F. Mitchell, J. A. Edward and D. R. Andes, Future Microbiol., 2013, 8, 1325–1337 CrossRef CAS PubMed.
- F. Imperi, F. Massai, C. R. Pillai, F. Longo, E. Zennaro, G. Rampioni, P. Visca and L. Leoni, Antimicrob. Agents Chemother., 2013, 57, 996–1005 CrossRef CAS PubMed.
- I. M. Stanojević, I. Aleksić, N. S. Drašković, B. Đ. Glišić, S. Vojnović and J. Nikodinović-Runić, J. Serb. Chem. Soc., 2017, 82, 1357–1367 CrossRef.
- A. Mohanty, C. H. Tan and B. Cao, Environ. Sci.: Nano, 2016, 3, 351–356 RSC.
- B. R. Singh, B. N. Singh, A. Singh, W. Khan, A. H. Naqvi and H. B. Singh, Sci. Rep., 2015, 5, 13719 CrossRef PubMed.
- K. Naik and M. Kowshik, J. Appl. Microbiol., 2014, 117, 972–983 CrossRef CAS PubMed.
- A. M. Merlot, D. S. Kalinowski and D. R. Richardson, Front. Physiol., 2014, 5, 299 Search PubMed.
- C. Tan, J. Liu, H. Li, W. Zheng, S. Shi, L. Chen and L. Ji, J. Inorg. Biochem., 2008, 102, 347–358 CrossRef CAS PubMed.
- X. M. He and D. C. Carter, Nature, 1992, 358, 209–215 CrossRef CAS PubMed.
- P. Smolénski, C. Pettinari, F. Marchetti, M. F. C. Guedes da Silva, G. Lupidi, G. V. B. Patzmay, D. Petrelli, L. A. Vitali and A. J. L. Pomberio, Inorg. Chem., 2015, 54, 434–440 CrossRef PubMed.
- Y. Shi, C. Guo, Y. Sun, Z. Liu, F. Xu, Y. Zhang, Z. Wen and Z. Li, Biomacromolecules, 2011, 12, 797–803 CrossRef CAS PubMed.
- O. H. Laitinen, V. P. Hytönen, H. R. Nordlund and M. S. Kulomaa, Cell. Mol. Life Sci., 2006, 63, 2992–3017 CrossRef CAS PubMed.
- C. A. Puckett and J. K. Barton, J. Am. Chem. Soc., 2007, 129, 46–47 CrossRef CAS PubMed.
- A. Ghezzi, M. Aceto, C. Cassino, E. Gabano and D. Osella, J. Inorg. Biochem., 2004, 98, 73–78 CrossRef CAS PubMed.
- L. Fetzer, B. Boff, M. Ali, M. Xiangjun, J.-P. Collin, C. Sirlin, C. Gaiddon and M. Pfeffer, Dalton Trans., 2011, 40, 8869–8878 RSC.
- A. K. Ghose, V. N. Viswanadhan and J. J. Wendoloski, J. Comb. Chem., 1999, 1, 55–68 CrossRef CAS PubMed.
- C. A. Lipinski, F. Lombardo, B. W. Dominy and P. J. Feeney, Adv. Drug Delivery Rev., 2012, 64, 4–17 CrossRef.
- S. Neidle, Therapeutic Applications of Quadruplex Nucleic Acids, Academic Press, 2012 Search PubMed.
- I. Turel and J. Kljun, Curr. Top. Med. Chem., 2011, 11, 2661–2687 CrossRef CAS PubMed.
- J. R. Lakowicz. Principles of Fluorescence Spectroscopy, Plenum Press, New York, 3rd edn, 2006 Search PubMed.
- H.-L. Wu, W.-Y. Li, X.-W. He, K. Miao and H. Liang, Anal. Bioanal. Chem., 2002, 373, 163–168 CrossRef CAS PubMed.
- J. S. Valentine, C. S. Foote, A. Greenberg and J. F. Liebman, Active oxygen in biochemistry, Blackie Academic and Professional, An Imprint of Chapman and Hall, London, 1993 Search PubMed.
- S. Torelli, C. Belle, S. Hamman, J.-L. Pierre and E. Saint-Aman, Inorg. Chem., 2002, 41, 3983–3989 CrossRef CAS PubMed.
- C. Gerdemann, C. Eicken and B. Krebs, Acc. Chem. Res., 2002, 35, 183–191 CrossRef CAS PubMed.
- I. A. Koval, P. Gamez, C. Belle, K. Selmeczi and J. Reedijk, Chem. Soc. Rev., 2006, 35, 814–840 RSC.
- A. W. Smith, A. Camara-Artigas, M. Wang, J. P. Allen and W. A. Francisco, Biochemistry, 2006, 45, 4378–4387 CrossRef CAS PubMed.
- C. E. Barry, P. G. Nayar and T. P. Begley, Biochemistry, 1989, 28, 6323–6333 CrossRef CAS PubMed.
- X. Mu, L. Song, Q. Li, R. Yin, X. Zhao and D. Wang, Int. J. Gynecol. Obstet., 2018, 143, 225–231 CrossRef CAS PubMed.
- M. Shyamal, T. K. Mandal, A. Panja and A. Saha, RSC Adv., 2014, 4, 53520–53530 RSC.
- A. E.-M. M. Ramadan, S. Y. Shaban, M. M. Ibrahim, S. A. Sallam, F. I. El-Shami and S. Al-Juaid, J. Mater. Sci., 2020, 55, 6457–6481 CrossRef CAS.
- K. S. Banu, M. Mukherjee, A. Guha, S. Bhattacharya, E. Zangrando and D. Das, Polyhedron, 2012, 45, 245–254 CrossRef CAS.
- N. Ch. Jana, P. Brandão, A. Frontera and A. Panja, Dalton Trans., 2020, 49, 14216–14230 RSC.
- G. M. Sheldrick, Acta Crystallogr., Sect. A: Found. Adv., 2015, 71, 3–8 CrossRef PubMed.
- G. te Velde, F. M. Bickelhaupt, E. J. Baerends, C. Fonseca Guerra, S. J. A. van Gisbergen, J. G. Snijders and T. Ziegler, J. Comput. Chem., 2001, 22, 931–967 CrossRef CAS.
- E. J. Baerends, T. Ziegler, A. J. Atkins, J. Autschbach, D. Bashford, O. Baseggio, A. Bérces, F. M. Bickelhaupt, C. Bo, P. M. Boerritger, L. Cavallo, C. Daul, D. P. Chong, D. v Chulhai, L. Deng, R. M. Dickson, J. M. Dieterich, D. E. Ellis, M. van Faassen, A. Ghysels, A. Giammona, S. J. A. van Gisbergen, A. Goez, A. W. Götz, S. Gusarov, F. E. Harris, P. van den Hoek, Z. Hu, C. R. Jacob, H. Jacobsen, L. Jensen, L. Joubert, J. W. Kaminski, G. van Kessel, C. König, F. Kootstra, A. Kovalenko, M. Krykunov, E. van Lenthe, D. A. McCormack, A. Michalak, M. Mitoraj, S. M. Morton, J. Neugebauer, V. P. Nicu, L. Noodleman, V. P. Osinga, S. Patchkovskii, M. Pavanello, C. A. Peeples, P. H. T. Philipsen, D. Post, C. C. Pye, H. Ramanantoanina, P. Ramos, W. Ravenek, J. I. Rodríguez, P. Ros, R. Rüger, P. R. T. Schipper, D. Schlüns, H. van Schoot, G. Schreckenbach, J. S. Seldenthuis, M. Seth, J. G. Snijders, M. Solà, S. M. M. Swart, D. Swerhone, G. te Velde, V. Tognetti, P. Vernooijs, L. Versluis, L. Visscher, O. Visser, F. Wang, T. A. Wesolowski, E. M. van Wezenbeek, G. Wiesenekker, S. K. Wolff, T. K. Woo and A. L. Yakovlev, ADF 2022.1, SCM, Theoretical Chemistry, Vrije Universiteit, Amsterdam, The Netherlands, 2022, https://www.scm.com Search PubMed.
- R. Rüger, M. Franchini, T. Trnka, A. Yakovlev, E. van Lenthe, P. Philipsen, T. van Vuren, B. Klumpers and T. Soini, AMS 2022.1, SCM, Theoretical Chemistry, Vrije Universiteit, Amsterdam, The Netherlands, 2022, https://www.scm.com Search PubMed.
- E. van Lenthe, E. J. Baerends and J. G. Snijders, J. Chem. Phys., 1993, 99, 4597–4610 CrossRef CAS.
- E. van Lenthe, E. J. Baerends and J. G. Snijders, J. Chem. Phys., 1994, 101, 9783–9792 CrossRef CAS.
- C. van Wüllen, J. Chem. Phys., 1998, 109, 392–399 CrossRef.
- M. Swart and F. M. Bickelhaupt, Int. J. Quantum Chem., 2006, 106, 2536–2544 CrossRef CAS.
- A. D. Becke, Phys. Rev. A, 1988, 38, 3098–3100 CrossRef CAS PubMed.
- J. P. Perdew, Phys. Rev. B, 1986, 33, 8822–8824 CrossRef PubMed.
- E. Caldeweyher, S. Ehlert, A. Hansen, H. Neugebauer, S. Spicher, C. Bannwarth and S. Grimme, J. Chem. Phys., 2019, 150, 154122 CrossRef PubMed.
- A. Klamt and G. Schoermann, J. Chem. Soc., Perkin Trans. 2, 1993, 799–805 RSC.
- A. Klamt, J. Phys. Chem., 1995, 99, 2224–2235 CrossRef CAS.
- C. C. Pye and T. Ziegler, Theor. Chem. Acc., 1999, 101, 396–408 Search PubMed.
- A. Bérces, R. M. Dickson, L. Fan, H. Jacobsen, D. Swerhone and T. Ziegler, Comput. Phys. Commun., 1997, 100, 247–262 CrossRef.
- H. Jacobsen, A. Bérces, D. P. Swerhone and T. Ziegler, Comput. Phys. Commun., 1997, 100, 263–276 CrossRef CAS.
- S. K. Wolff, Int. J. Quantum Chem., 2005, 104, 645–659 CrossRef CAS.
- S. Grimme, Chem.–Eur. J., 2012, 18, 9955–9964 CrossRef CAS PubMed.
- Y. Zhao and D. G. Truhlar, J. Chem. Phys., 2006, 125, 194101 CrossRef PubMed.
- Y. Zhao and D. G. Truhlar, Theor. Chem. Acc., 2008, 120, 215–241 Search PubMed.
- G. Jonkers, C. A. de Lange, L. Noodleman and E. J. Baerends, Mol. Phys., 1982, 46, 609–620 CrossRef CAS.
- L. Noodleman, J. Chem. Phys., 1981, 74, 5737–5743 CrossRef CAS.
- L. Noodleman and E. R. Davidson, Chem. Phys., 1986, 109, 131–143 CrossRef.
- L. Noodleman, J. G. Norman, J. H. Osborne, A. Aizman and D. A. Case, J. Am. Chem. Soc., 1985, 107, 3418–3426 CrossRef CAS.
- F. Neese, Coord. Chem. Rev., 2009, 253, 526–563 CrossRef CAS.
- T. Soda, Y. Kitagawa, T. Onishi, Y. Takano, Y. Shigeta, H. Nagao, Y. Yoshioka and K. Yamaguchi, Chem. Phys. Lett., 2000, 319, 223–230 CrossRef CAS.
- E. Bitzek, P. Koskinen, F. Gähler, M. Moseler and P. Gumbsch, Phys. Rev. Lett., 2006, 97, 170201 CrossRef PubMed.
- M. A. L. Marques, M. J. T. Oliveira and T. Burnus, Comput. Phys. Commun., 2012, 183, 2272–2281 CrossRef CAS.
- M. C. Arendrup, M. Cuenca-Estrella, C. Lass-Flörl and W. Hope, Clin. Microbiol. Infect., 2012, 18, E246–E247 CrossRef CAS PubMed.
- F. Ferrer, R. Fanciullino, G. Milano and J. Ciccolini, Clin. Pharmacol. Ther., 2020, 108, 458–470 CrossRef PubMed.
- H. Liu, J. Köhler and G. R. Fink, Science, 1994, 266, 1723–1726 CrossRef CAS PubMed.
- C. G. Pierce, P. Uppuluri, A. R. Tristan, F. L. Wormley Jr, E. Mowat, G. Ramage and J. L. Lopez-Ribot, Nat. Protoc., 2008, 3, 1494–1500 CrossRef CAS PubMed.
- K. H. McClean, M. K. Winson, L. Fish, A. Taylor, S. R. Chhabra, M. Camara, M. Daykin, J. H. Lamb, S. Swift, B. W. Bycroft, G. S. A. B. Stewart and P. Williams, Microbiology, 1997, 143, 3703–3711 CrossRef CAS PubMed.
- I. Aleksic, S. Šegan, F. Andrić, M. Zlatović, I. Moric, D. M. Opsenica and L. Senerovic, ACS Chem. Biol., 2017, 12, 1425–1434 CrossRef CAS PubMed.
- R. Bera, B. K. Sahoo, K. S. Ghosh and S. Dasgupta, Int. J. Biol. Macromol., 2008, 42, 14–21 CrossRef CAS PubMed.
- A. Wolfe, G. H. Shimer Jr. and T. Meehan, Biochem, 1987, 26, 6392–6396 CrossRef CAS PubMed.
- J. Reim and B. Krebs, J. Chem. Soc., Dalton Trans., 1997, 3793–3804 RSC.
- A. C. Sousa, M. C. Oliveira, L. O. Martins and M. P. Robalo, Green Chem., 2014, 16, 4127–4136 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: 1H NMR spectra for py-2pz, 3 and 4, Fig. S1–S6, Scheme S1, Tables S1–S3 and Cartesian coordinates of all DFT optimized structures. CCDC 2220146–2220150. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d2ra07401j |
| This journal is © The Royal Society of Chemistry 2023 |