Open Access Article
Open Access ArticleSolvent-driven biotoxin into nano-units as a versatile and sensitive SERS strategy†
Yizhuang Chengabc,
Miao Qina,
Pan Li a and
Liangbao Yang
a and
Liangbao Yang *ab
*ab
aInstitute of Health and Medical Technology, Hefei Institutes of Physical Science, Chinese Academy of Sciences, Hefei 230031, China. E-mail: lbyang@iim.ac.cn
bUniversity of Science & Technology of China, Hefei 230026, Anhui, China
cHefei Cancer Hospital, Chinese Academy of Sciences, Hefei 230031, Anhui, China
First published on 3rd February 2023
Abstract
In recent years, marine biotoxins have posed a great threat to fishermen, human security and military prevention and control due to their diverse, complex, toxic and widespread nature, and the development of rapid and sensitive methods is essential. Surface-enhanced Raman spectroscopy (SERS) is a promising technique for the rapid and sensitive in situ detection of marine biotoxins due to its advantages of rapid, high sensitivity, and fingerprinting information. However, the complex structure of toxin molecules, small Raman scattering cross-section and low affinity to conventional substrates make it difficult to achieve direct and sensitive SERS detection. Here, we generate a large number of active hotspot structures by constructing monolayer nanoparticle films with high density hotspots, which have good target molecules that can actively access the hotspot structures using nanocapillaries. In addition, the efficient and stable signal can be achieved during dynamic detection, increasing the practicality and operability of the method. This versatile SERS method achieves highly sensitive detection of marine biotoxins GTX and NOD, providing good prospects for convenient, rapid and sensitive SERS detection of marine biotoxins.
Contamination of the natural environment with biotoxins is an increasing concern worldwide because of their high toxicicity and quick acting impact on environmental safety and public health. Marine biotoxins are highly active and specific metabolic components found in marine organisms and are generally highly toxic, and mainly produced by algae or phytoplankton. In most eutrophic waters, where algae are abundant and dominant, algal toxins can enter the human body through the digestive tract, causing diarrhoea, neuroparalysis, liver damage and in severe cases, poisoning and even death. Among the algal toxins, cyanobacterial toxins are the most common, mainly including hepatotoxins, neurotoxins and endotoxins.1–3 Among them, gonyautoxin (GTX) mainly acts on the nervous system, selectively inhibiting Na+ channels on the surface of nerve cells, etc. Accidental ingestion of shellfish containing this toxin can lead to neurotoxicity and, in severe cases, death due to respiratory muscle paralysis.4,5 Hepatotoxins include microcysin (MC), nodularin (NOD) and others. MC cause liver and kidney damage by inhibiting the activity of protein phosphatases 1 and 2A.6 The NOD toxin has been shown to cause hepatotoxicity through inhibition of phosphoproteins 1A and 2A (PP1 and PP2A), leading to a significant use of important cellular proteins, and is therefore considered a potential tumour promoter and carcinogen.1,7,8 Thus, to ensure the marine environment and food safety, it is important to develop highly sensitive and reliable method of quick detection of biotoxins. High performance liquid chromatography (HPLC)9 and liquid chromatography/mass spectrometry (LC-MS)10 provide accurate and sensitive analytical methods for the identification and quantification of toxins, but their instruments are expensive and require specialist personnel to operate them, and the memory effect of the mass spectrometry ion source reduces the accuracy and sensitivity of subsequent determinations of toxins. These have somewhat limited its greater breakthrough in bioassay.
Owing to its high sensitivity and specificity, surface-enhanced Raman spectroscopy (SERS) has been acted as the powerful analytical technology to apply into various fields. It can provide the fingerprint information of molecules and high sensitivity with level of single molecules.11,12 Therefore, it is of great significance and application value to establish a new SERS strategy to achieve efficient detection of toxins. However, SERS-based detection of marine biotoxins is still in its infancy, mainly due to the complex structure of toxin molecules, their large molecular weight and small Raman scattering cross section and low affinity to conventional substrates, thus making it difficult to achieve direct and highly sensitive SERS detection.13 Using cysteine-modified gold nanoparticles (Cys-AuNPs) as a SERS probe to capture STX molecules through electrostatic interactions generated by multiple hydrogen bonds between Cys and stenotoxin molecules.13 Zhu et al. determined the feasibility of using a SERS immunoassay to detect microcystin LR. Gold nanoparticles were assembled into nanorod chains to obtain an ultra-sensitive assay for microcystin LR with a detection limit as low as 5 pg mL−1 and a detection time of 15 min.14 Brezestean reported a complete theoretical DFT-vibrational Raman characterization. The development of a direct, rapid and convenient SERS assay for marine biotoxins is therefore essential.
Over the past few decades, efforts have been made to construct an orderly arrangement of large-area hot spot structures to improve the sensitivity of SERS methods. Progress has been made mainly in the design of various nanostructured SERS substrates with different sizes and uniform gaps.15–19 Liquid–liquid interfacial self-assembly has attracted attention in the preparation of controlled, flexible arrays of periodic nanostructures.20,21 However, for SERS measurements, the solid SERS substrates obtained typically exhibit poorer reproducibility and stability than colloidal SERS substrates.18,22 Recent studies have shown that in the presence of solution, nanocapillary interaction can effectively pump target molecules into the nanogap, enabling trace detection of a wide range of molecules, independent of their binding affinity to the substrate.23,24 The small scattering cross section of marine biotoxin molecules has a weak affinity for conventional noble metal substrates, and it remains a challenge to get the toxin molecules into the hotspot structure efficiently for highly sensitive SERS detection. Li25 constructed single hot-spot platforms from nanoparticles and nanowires, which provide “nano-channels” for capturing molecules due to capillary self-priming. High quality SERS spectra of a wide range of molecules with different properties can be generated on a single hotspot structure with reliable reproducibility and excellent sensitivity. Ge26 prepared large-area monolayer silver nanoparticle films by liquid–liquid interface assembly and covered them on the solution of the target to be measured. During the transition from the wet state to the dry state, the monolayer nanoparticle films formed a nano-capillary pump with capillary action, and the solution carried the target molecules continuously through the nanoparticle gap to achieve highly sensitive SERS detection of multiple molecules. Nanocapillary action plays an active role in the active transport of target molecules to hotspot structures, potentially solving the problem of small Raman cross-sections of biotoxins with poor affinity to conventional noble metal substrates.
In this study, inspired by the concept of molecular motion, a nanocapillary pump model was designed and constructed to achieve active capture of biotoxin molecules by hotspot structures. Large-area nanoparticle films were prepared using a liquid–liquid interface assembly method. During the evaporation of the solution, the gap between the nanoparticles gradually decreases and the target molecules continuously enter the gap along with the solution due to the nanocapillary effect. This work opens up a new and versatile method for the active capture of marine biotoxin molecules by SERS hotspot structures, as shown in Scheme 1.
 | ||
| Scheme 1 Schematic diagram of liquid–liquid assembly and SERS strategy of biotoxin with capillary force. | ||
The SERS platform of liquid/liquid interfacial self-assembly of nanoparticles in cyclohexane/water was inspired by a report in the literature.27 The preparation process is shown in Fig. S1.† Specifically, an appropriate volume of H2O was added to the cell until the bottom of the vessel was filled. Then, cyclohexane was dropped over the layer of water and an incompatible cyclohexane–water liquid interface appeared. Subsequently, nanoparticles were centrifuged to collect the precipitants and then diluted with ethanol, which was added slowly to the interface. Ultimately, the nanoparticles spontaneously self-assembled into a highly close-packed film over a large area at the cyclohexane/water liquid interface with the evaporation of cyclohexane (Fig. S2†).
One of the structures required for SERS substrates is precious metal nanoparticles with nanoscale gaps, and two-dimensional (2D) hexagonal arrays of closely spaced nanoparticles (NPs) are among the promising SERS substrates.21,28,29 The organization of nanoparticles into 2D densely packed films at the liquid–liquid interface is a well-established technique.30,31 The principle of liquid–liquid interface assembly is mainly due to the fact that the addition of ethanol to the sol gradually decreases the surface charge, most likely due to the competitive adsorption of ethanol molecules, displacing the surface capping agent from the gold surface.32,33 After further addition of ethanol, about half of the original charge density is retained and the gold sol remains stable.34,35 However, when a layer of cyclohexane is introduced above this solution to form a liquid–liquid interface, the gold nanocrystals rapidly adsorb to the water/cyclohexane interface.36 During this process, the number of hydrophilic nanoparticles escaping from the aqueous phase to the two-phase interface depends on the volume of ethanol added.34,37 This interfacial retention also depends on the contact angle of the particle with the water/oil interface. When the particle is hydrophilic or hydrophobic, it has a contact angle of less than or greater than 90° in the water/oil system and is located entirely within the water or oil phase.34,37 When its contact angle is about 90°, the particle prefers to reside at the interface.30,38 Other organic solvents (e.g., methanol, acetone and isopropanol) were also effective in driving silver or gold nanoparticles to the water–oil interface. In addition, it was obvious that method of liquid–liquid interfacial self-assembly is an ideal strategy to achieve SERS substrates and the nanoparticle films can be easily transferred on to wafer.
We have performed liquid–liquid interface self-assembly of Au cube, Au octahedron, Au tetradecahedron, Ag NPs, Au NRs, and Au NPs using a general self-assembly method, the morphology is shown in Fig. 1 and Fig. S3.† Based on the self-assembly results, all the nanoparticles can be assembled into 2D nanoparticle films, and the nanoparticles can also have high density of small gaps, which is expected to be used for ultrasensitive SERS detection.
 | ||
| Fig. 1 SEM images of Au cube (a), Au octahedron (b), Au tetradecahedron (c), Ag NPs (d), Au NRs (e) and Au NPs (f). | ||
Reproducibility and stability were also important indicators for evaluating the performance of the SERS substrate. 5 μL of CV aqueous solution (10−11 M) was added dropwise to the Au NRs substrate, and 15 points were randomly selected for testing, as displayed in Fig. 2a. The SERS intensity of each characteristic peak remained unchanged, and the relative standard deviation (RSD) of the intensity (1619 cm−1) was 11.997% (Fig. 2b). This shows that the prepared Au NRs substrate has good reproducibility.
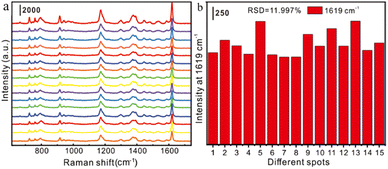 | ||
| Fig. 2 (a)Reproducibility characterization of gold nanorod substrates for detection of 10−11 M CV; (b). relative standard deviation (RSD) at the 1619 cm−1 characteristic peak of the CV molecule. | ||
To analyse that the target molecules can actively enter the small gap structure of gold nanorods, we analysed the force of the target solution in the nanogap. As the solution gradually evaporates, the liquid bridge caused by the surface tension of nanoparticles leads to the gradual reduction of the gap and the formation of hot spot structure, as shown in Fig. 3. The interaction force between two nanorods can be expressed in terms of the van der Waals potential E(r) = −A/(12πd2);26 and the relationship between the action potential energy and the action force is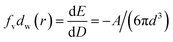 .26 In the formula, A is the Hamaker constant of substance, which is defined as
.26 In the formula, A is the Hamaker constant of substance, which is defined as 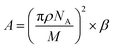 ; where ρ stands for the density of substance, in kg m−3, NA stands for Avogadro's constant, which is 6.02 × 1023. M represents the relative molecular mass of the substance in kg mol; d is the distance between the two nanoparticles in m. When the solution is in the nanogap, the Hamaker constant for the interaction between the liquid and the nanoparticles is
; where ρ stands for the density of substance, in kg m−3, NA stands for Avogadro's constant, which is 6.02 × 1023. M represents the relative molecular mass of the substance in kg mol; d is the distance between the two nanoparticles in m. When the solution is in the nanogap, the Hamaker constant for the interaction between the liquid and the nanoparticles is  .For the system with water as the solvent, As = 6.3 × 10−20 J, Al = 4.0 × 10−20 J. The calculation shows that the Hamerck constant is 5.0 × 10–20 J for the interaction between aqueous solution and nanoparticle gap. From the expression of interaction potential energy between nanoparticles and aqueous solution, the smaller the gap between the nanoparticles (i.e. the smaller the value of d), the greater the van der Waals force on the aqueous solution. By analysing the distribution of the gap between nanoparticles and van der Waals force, it is concluded that the van der Waals force can be as high as −1010 N m−2 near nanoparticles. Researchers used molecular dynamics methods to study the action range of van der Waals force at about 10 nm26. At this gap scale, the aqueous solution is bound in the gap of nanoparticles by a large van der Waals force, and exists in the form of thin film water.39
.For the system with water as the solvent, As = 6.3 × 10−20 J, Al = 4.0 × 10−20 J. The calculation shows that the Hamerck constant is 5.0 × 10–20 J for the interaction between aqueous solution and nanoparticle gap. From the expression of interaction potential energy between nanoparticles and aqueous solution, the smaller the gap between the nanoparticles (i.e. the smaller the value of d), the greater the van der Waals force on the aqueous solution. By analysing the distribution of the gap between nanoparticles and van der Waals force, it is concluded that the van der Waals force can be as high as −1010 N m−2 near nanoparticles. Researchers used molecular dynamics methods to study the action range of van der Waals force at about 10 nm26. At this gap scale, the aqueous solution is bound in the gap of nanoparticles by a large van der Waals force, and exists in the form of thin film water.39
The target molecule follows the solution largely in the gap of the nanoparticles, providing the prerequisite for sensitive detection by SERS.
In this experimental system, a monolayer of nanoparticles assembled from gold nanorods can be seen as a nanocapillary pump structure formed by a tight stack of multiple nanorods. The nanoparticles are placed close to each other to form small gaps of different structures, and the force analysis of the different gap shapes is shown in Fig. 3b and c. In the case of two nanorods forming the gap structure shown in Fig. 3b, the motion law of the solution in the nanoscale gap can be based on the Young–Laplace equation.23,40,41
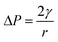 | (1) |
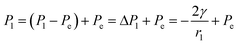 | (2) |
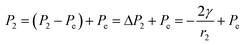 | (3) |
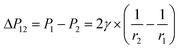 , from Fig. 3b, it was seen that r2> r1, ΔP12 < 0, that is, the pressure of the aqueous solution at position 2 is greater than that at position 1, and the solution will flow from position 2 to position 1. When the stacked gold nanorod structure forms the structure shown in Fig. 3c, the pressure in the region of different positions (position 1 and position 3 in the figure) can be calculated according to eqn (4) and (5):
, from Fig. 3b, it was seen that r2> r1, ΔP12 < 0, that is, the pressure of the aqueous solution at position 2 is greater than that at position 1, and the solution will flow from position 2 to position 1. When the stacked gold nanorod structure forms the structure shown in Fig. 3c, the pressure in the region of different positions (position 1 and position 3 in the figure) can be calculated according to eqn (4) and (5):
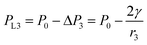 | (4) |
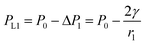 | (5) |
Because the radius of curvature r3 > r1, the internal fluid pressure at position 3 is greater than the internal fluid pressure at position 1 (PL3>PL3). The target solution will flow from 3 to the small gap 1. In summary, the target solution will continuously flow from the high-pressure region to the low-pressure region (small gap region), and the solvent will carry the analyte into the optimal hot spot region in the process. Thus, the solution tends to flow from the large gap to the small gap in the nano-gap until evaporation is complete. During this process, the target molecule is in the same path as the solution and continuously enters the hot spot with the solvent, which provides the prerequisite for performing SERS high sensitivity detection.
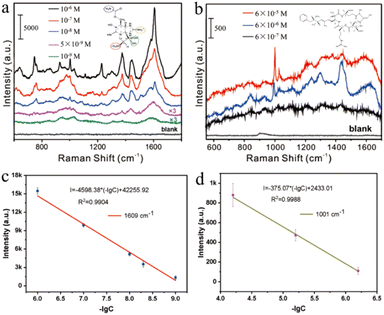
In previous studies, researchers often used functionalized SERS substrates or methods to achieve sensitive detection of target molecules. Although the molecular weight of marine biotoxins is large, their affinity to nanoparticles is weak, and it is difficult to detect them using conventional SERS methods. Here, we detect marine biotoxins by actively capturing target molecules with small interstitial structures formed by monolayer gold nanorods. A nano capillary pump model with hot spots was constructed, and target molecules were driven into hot spots during the solvent–evaporation process, realizing efficient detection. GTX and NOD are two common marine biological toxins, which cause great harm to human health. Fig. 4a indicates the sensitive detection of GTX with 1 nM level. 746 cm−1 represents the deformation vibration of C–H–O group, 926 cm−1 represents the symmetric stretching vibration of C–C–N group, 1375 cm−1 and 1446 cm−1 represent the ring stretching vibration, and 1609 cm−1 represents the stretching vibration of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, as shown in Fig. S4 and Table S1,† which is consistent with the literature report.43 As Fig. 4b shows, NOD (6 × 10−7 M) has been successfully analysed. Characteristic peaks 1001 cm−1, 1030 cm−1, 1215 cm−1, and 1600 cm−1 correspond to the phenyl ring breathing vibration, the C–H deformation of the phenyl ring, the CH2 wagging vibration, and the phenyl ring C–C stretch vibration respectively, as shown in Table S2†.44 The characteristic peak can be clearly discerned even when the GTX and NOD concentrations were reduced to 10−9 M and 10−7 M. Furthermore, we performed a quantitative analysis of the characteristic peak at 1609 cm−1 and 1001 cm−1, and the fitted linear regression equation were I1609 = −4598.38 (−lg[CGTX]) + 42
N, as shown in Fig. S4 and Table S1,† which is consistent with the literature report.43 As Fig. 4b shows, NOD (6 × 10−7 M) has been successfully analysed. Characteristic peaks 1001 cm−1, 1030 cm−1, 1215 cm−1, and 1600 cm−1 correspond to the phenyl ring breathing vibration, the C–H deformation of the phenyl ring, the CH2 wagging vibration, and the phenyl ring C–C stretch vibration respectively, as shown in Table S2†.44 The characteristic peak can be clearly discerned even when the GTX and NOD concentrations were reduced to 10−9 M and 10−7 M. Furthermore, we performed a quantitative analysis of the characteristic peak at 1609 cm−1 and 1001 cm−1, and the fitted linear regression equation were I1609 = −4598.38 (−lg[CGTX]) + 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 255.92 and I1001 = −375.07 (−lg[CNOD]) + 2433.01. It can be concluded from the above equation that the linear correlation coefficient of GTX is 0.9904 and the LOD is 6.47 × 10−10 M. Similarly, NOD's linear correlation coefficient of the equation was 0.9988 and the LOD was 3.26 × 10−7 M. On the other hand, the GTX with 1 × 10−9 M and NOD with 1 × 10−7 M were also calculated. As shown in Fig. S5,† the SERS features of GTX and NOD is difficult to distinct in lower concentration, thus the detection limits of GTX and NOD were 5 × 10−9 M and 6 × 10−7 M in the manuscript. The above results show that the method exhibited high sensitivity for marine biotoxin detection.
255.92 and I1001 = −375.07 (−lg[CNOD]) + 2433.01. It can be concluded from the above equation that the linear correlation coefficient of GTX is 0.9904 and the LOD is 6.47 × 10−10 M. Similarly, NOD's linear correlation coefficient of the equation was 0.9988 and the LOD was 3.26 × 10−7 M. On the other hand, the GTX with 1 × 10−9 M and NOD with 1 × 10−7 M were also calculated. As shown in Fig. S5,† the SERS features of GTX and NOD is difficult to distinct in lower concentration, thus the detection limits of GTX and NOD were 5 × 10−9 M and 6 × 10−7 M in the manuscript. The above results show that the method exhibited high sensitivity for marine biotoxin detection.
In addition, we have performed relevant assays that simulate real-world conditions, as shown in Fig. S6.† Microcystins (MC-LR) are a class of cyclic heptapeptide hepatotoxins and the World Health Organization (WHO) recommends a maximum permissible concentration of MC-LR in drinking water of 1 μg L−1.45 Therefore, trace detection of MC-LR has an important role in the field of environmental safety and human health. Using the method described above, no significant characteristic peaks were detected in the substrate, while characteristic peaks at 1017, 1190, 1223 and 1625 cm−1 were detected when the microcystin standard solution was added to the system and were also detected at as low as 2.5 × 10−7 M, as shown in Fig. S6a.† Among them, the 1017 cm−1 characteristic peak was attributed to the C–C symmetric stretching vibration on the benzene ring; 1190 cm−1 attributed to the -NH3+ distortion and wobble vibration on L-leucine leucine; 1223 cm−1 attributed to the twisted wobble vibration of CH2 on the glutamic acid residue of L-glutamic acid; 1625 cm−1 attributed to the –COO– stretching vibration.45,46 Further direct detection of MC-LR by simulated addition to seawater allowed the detection of MC-LR down to 5 × 10−6 M. The actual detection results show that this SERS method for automatic capture of target molecules with small gaps is promising for practical applications and promises highly sensitive, fast and convenient direct detection in real systems.
In this paper, a SERS method for the active capture of marine biotoxin molecules in small gaps based on a nano-capillary model was developed. A monolayer nanoparticle film with a high density of hot spots was constructed at the interface using a liquid–liquid interface self-assembly method. During the process of the nanoparticle film from wet to dry, the gap of the nanoparticles continuously decreases and a large number of hotspots are formed. Due to the pressure difference at the gaps, the target molecules inevitably move towards the smaller gaps, effectively activating the hot spots. The detection limits for the weaker affinity marine biotoxin molecules GTX and NOD were 5 × 10−9 M and 6 × 10−7 M, respectively. More importantly, a practical system of 2.5 × 10−6 M MC-LC in seawater was achieved. It provides a theoretical and technical basis for the rapid and convenient detection of biotoxins. In the future, the selectivity of analytical method should be considered.
Author contributions
L. B. Y. conceived and designed the experiments. Y. Z. C. performed material synthesis, structural characterization and Raman measurements and wrote the paper. M. Q. and P. L. performed material synthesis. All authors discuss the results and commented on the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the Anhui Provincial Natural Science Foundation Project (2208085MB31) and Anhui Provincial Key R&D Program (No. 202104d07020002).References
- D. I. Melnikova and T. Y. MagarlamovJo, Toxins, 2022, 14(8), 576 CrossRef CAS PubMed.
- J. L. Banach, E. F. Hoek-van den Hil and H. J. van der Fels-KlerxJo, Compr. Rev. Food Sci. F, 2020, 19, 332–364 CrossRef CAS PubMed.
- F. Farabegoli, L. Blanco, L. P. Rodriguez, J. M. Vieites and A. G. CabadoJo, Mar. Drugs, 2018, 16(6), 188 CrossRef PubMed.
- N. Estrada, E. J. Nunez-Vazquez, A. Palacios, F. Ascencio, L. Guzman-Villanueva and R. G. ContrerasJo, Front. Immunol., 2021, 12, 634497 CrossRef CAS PubMed.
- Z. C. Wei, W. Ding, M. L. Li, J. X. Shi, H. Z. Wang, Y. R. Wang, Y. B. Li, Y. Q. Xu, J. J. Hu, Z. M. Bao and X. L. HuJo, Toxins, 2022, 14(2), 108 CrossRef CAS PubMed.
- B. Zegura, A. Straser and M. FilipicJo, Mutat. Res., Rev. Mutat., 2011, 727, 16–41 CrossRef CAS PubMed.
- V. G. Christensen and E. KhanJo, Sci. Total Environ., 2020, 736, 139515 CrossRef CAS PubMed.
- I. A. Brezestean, A. M. R. Gherman, A. Colnita, N. E. Dina, C. Muller Molnar, D. Marconi, V. Chis, I. L. David and S. Cinta-PinzaruJo, Int. J. Mol. Sci., 2022, 23, 15741 CrossRef CAS PubMed.
- L. E. LlewellynJo, Nat. Prod. Rep., 2006, 23, 200–222 RSC.
- C. Dell'Aversano, L. Tattaglione, G. Polito, K. Dean, M. Giacobbe, S. Casabianca, S. Capellacci, A. Penna and A. D. Turner Jo, Chemosphere, 2019, 215, 881–892 CrossRef PubMed.
- I. Alessandri and J. R. LombardiJo, Chem. Rev., 2016, 116, 14921–14981 CrossRef CAS PubMed.
- C. Zhang, Z. X. Li, S. Qiu, W. X. Lu, M. R. Shao, C. Ji, G. C. Wang, X. F. Zhao, J. Yu and Z. LiJo, Nanophotonics, 2022, 11, 33–44 CrossRef CAS.
- C. Cao, P. Li, H. Liao, J. Wang, X. Tang and L. Yang, Anal. Bioanal. Chem., 2020, 412, 4609–4617 CrossRef CAS PubMed.
- Y. Y. Zhu, H. Kuang, L. G. Xu, W. Ma, C. F. Peng, Y. F. Hua, L. B. Wang and C. L. XuJo, J. Mater. Chem., 2012, 22, 2387–2391 RSC.
- X. Dai, W. H. Fu, H. Y. Chi, V. S. Mesias, H. N. Zhu, C. W. Leung, W. Liu and J. Q. HuangJo, Nat. Commun., 2021, 12, 1292 CrossRef CAS PubMed.
- N. H. Kim, W. Hwang, K. Baek, M. R. Rohman, J. Kim, H. W. Kim, J. Mun, S. Y. Lee, G. Yun, J. Murray, J. W. Ha, J. Rho, M. Moskovits and K. KimJo, J. Am. Chem. Soc., 2018, 140, 4705–4711 CrossRef CAS PubMed.
- X. Z. Qiao, B. S. Su, C. Liu, Q. Song, D. Luo, G. Mo and T. WangJo, Adv. Mater., 2018, 30, 1702275 CrossRef PubMed.
- H. Dang, S. G. Park, Y. Wu, N. Choi, J. Y. Yang, S. Lee, S. W. Joo, L. X. Chen and J. ChooJo, Adv. Funct. Mater., 2021, 31, 2105703 CrossRef CAS.
- M. Arabi, A. Ostovan, Y. Q. Wang, R. C. Mei, L. W. Fu, J. H. Li, X. Y. Wang and L. X. ChenJo, Nat. Commun., 2022, 13, 5757 CrossRef CAS PubMed.
- M. D. Scanlon, E. Smirnov, T. J. Stockmann and P. PeljoJo, Chem. Rev., 2018, 118, 3722–3751 CrossRef CAS PubMed.
- H. Q. Liu, J. Y. Zeng, L. P. Song, L. L. Zhang, Z. H. Chen, J. H. Li, Z. D. Xiao, F. M. Su and Y. J. HuangJo, Nanoscale Horiz., 2022, 7, 554–561 RSC.
- L. B. Yang, P. Li, H. L. Liu, X. H. Tang and J. H. LiuJo, Chem. Soc. Rev., 2015, 44, 2837–2848 RSC.
- M. Qin, M. H. Ge, P. Li, S. Y. Chen, G. Y. Huang, X. H. Tong, W. Han, D. L. Ren, Y. He, D. Y. Lin, L. B. Yang and Z. Q. TianJo, Adv. Opt. Mater., 2022, 10, 2200551 CrossRef CAS.
- W. L. Wang, S. H. Pu, W. Y. Hu, J. L. Gu, B. Ren, Z. Q. Tian and G. K. LiuJo, Chem. Commun., 2022, 58, 3953–3956 RSC.
- P. Li, X. N. Yan, F. Zhou, X. G. Tang, L. B. Yang and J. H. LiuJo, J. Mater. Chem. C, 2017, 5, 3229–3237 RSC.
- M. H. Ge, P. Li, G. L. Zhou, S. Y. Chen, W. Han, F. Qin, Y. M. Nie, Y. X. Wang, M. Qin, G. Y. Huang, S. F. Li, Y. T. Wang, L. B. Yang and Z. Q. TianJo, J. Am. Chem. Soc., 2021, 143, 7769–7776 CrossRef CAS PubMed.
- S. Y. Liu, X. D. Tian, Y. Zhang and J. F. LiJo, Anal. Chem., 2018, 90, 7275–7282 CrossRef CAS PubMed.
- D. Dashen, Y. Lim Wei, D. M. Smilgies, S. Kae Jye, S. Qianqian and C. WenlongJo, Nanoscale, 2018, 10, 5065–5071 RSC.
- Z. Sun, A. Umar, J. Zeng, X. Luo, L. Song, Z. Zhang, Z. Chen, J. Li, F. Su and Y. HuangJo, ACS Appl. Nano Mater., 2022, 5, 1220–1231 CrossRef CAS.
- S. Gwo, H.-Y. Chen, M.-H. Lin, L. Sun and X. LiJo, Chem. Soc. Rev., 2016, 45, 5672–5716 RSC.
- Q. Guo, M. Xu, Y. Yuan, R. Gu and J. YaoJo, Langmuir, 2016, 32, 4530–4537 CrossRef CAS PubMed.
- S. J. Guo and S. J. DongJo, J. Mater. Chem., 2011, 21, 16704–16716 RSC.
- F. Reincke, S. G. Hickey, W. K. Kegel and D. VanmaekelberghJo, Angew. Chem., Int. Ed., 2004, 43, 458–462 CrossRef CAS PubMed.
- H. W. Duan, D. Y. Wang, D. G. Kurth and H. MohwaldJo, Angew. Chem., Int. Ed., 2004, 43, 5639–5642 CrossRef CAS PubMed.
- H.-B. Yao, L.-B. Mao, Y.-X. Yan, H.-P. Cong, X. Lei and S.-H. YuJo, ACS Nano, 2012, 6, 8250–8260 CrossRef CAS PubMed.
- C. C. Li, Y. K. Xu, X. Y. Li, Z. W. Ye, C. Y. Yao, Q. L. Chen, Y. F. Zhang and S. E. J. BellJo, Adv. Mater. Interfaces, 2020, 7(14), 2000391 CrossRef CAS.
- R. Aveyard, B. P. Binks, J. H. Clint, P. D. I. Fletcher, T. S. Horozov, B. Neumann, V. N. Paunov, J. Annesley, S. W. Botchway, D. Nees, A. W. Parker, A. D. Ward and A. N. BurgessJo, Phys. Rev. Lett., 2002, 88(24), 246102 CrossRef CAS PubMed.
- Y. J. Li, W. I. J. Huang and S. G. SunJo, Angew, 2006, 45, 2537–2539 CrossRef CAS PubMed.
- N. Mitarai and F. NoriJo, Adv. Phys., 2007, 55, 1–45 CrossRef.
- E. Dujardin, T. W. Ebbesen, H. Hiura and K. TanigakiJo, Science, 1994, 265, 1850–1852 CrossRef CAS PubMed.
- N. R. Tas, P. Mela, T. Kramer, J. W. Berenschot and A. van den BergJo, Nano Lett., 2003, 3, 1537–1540 CrossRef CAS.
- L. F. Scatena, M. G. Brown and G. L. RichmondJo, Science, 2001, 292, 908–912 CrossRef CAS PubMed.
- C. Muller, B. Glamuzina, I. Pozniak, K. Weber, D. Cialla, J. Popp and S. C. PinzaruJo, Talanta, 2014, 130, 108–115 CrossRef CAS PubMed.
- R. A. Halvorson, W. N. Leng and P. J. VikeslandJo, Anal. Chem., 2011, 83, 9273–9280 CrossRef CAS PubMed.
- S. He, W. Xie, S. Fang, D. Zhou, K. Djebbi, Z. Zhang, J. Du, C. Du and D. WangJo, Talanta, 2019, 195, 401–406 CrossRef CAS PubMed.
- R. A. Halvorson, W. Leng and P. J. VikeslandJo, Anal. Chem., 2011, 83, 9273–9280 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: [DETAILS]. See DOI: https://doi.org/10.1039/d2ra07216e |
| This journal is © The Royal Society of Chemistry 2023 |