Open Access Article
Open Access ArticleSynthesis of 13C-methyl-labeled amino acids and their incorporation into proteins in mammalian cells†
Matteo
Borgini‡
 a,
Łukasz
Wieteska§
b,
Cynthia S.
Hinck
b,
Troy
Krzysiak
b,
Andrew P.
Hinck
a,
Łukasz
Wieteska§
b,
Cynthia S.
Hinck
b,
Troy
Krzysiak
b,
Andrew P.
Hinck
 *b and
Peter
Wipf
*b and
Peter
Wipf
 *a
*a
aDepartment of Chemistry, University of Pittsburgh, Pittsburgh, PA 15260, USA. E-mail: pwipf@pitt.edu; Tel: +1 (412) 624-8606
bDepartment of Structural Biology, University of Pittsburgh, Pittsburgh, PA 15260, USA. E-mail: ahinck@pitt.edu; Tel: +1 (412) 648-8533
First published on 7th November 2023
Abstract
Isotopic labeling of methyl-substituted proteinogenic amino acids with 13C has transformed applications of solution-based NMR spectroscopy and allowed the study of much larger and more complex proteins than previously possible with 15N labeling. Procedures are well-established for producing methyl-labeled proteins expressed in bacteria, with efficient incorporation of 13C-methyl labeled metabolic precursors to enable the isotopic labeling of Ile, Val, and Leu methyl groups. Recently, similar methodology has been applied to enable 13C-methyl labeling of Ile, Val, and Leu in yeast, extending the approach to proteins that do not readily fold when produced in bacteria. Mammalian or insect cells are nonetheless preferable for production of many human proteins, yet 13C-methyl labeling using similar metabolic precursors is not feasible as these cells lack the requisite biosynthetic machinery. Herein, we report versatile and high-yielding synthetic routes to 13C methyl-labeled amino acids based on palladium-catalyzed C(sp3)–H functionalization. We demonstrate the efficient incorporation of two of the synthesized amino acids, 13C-γ2-Ile and 13C-γ1,γ2-Val, into human receptor extracellular domains with multiple disulfides using suspension-cultured HEK293 cells. Production costs are reasonable, even at moderate expression levels of 2–3 mg purified protein per liter of medium, and the method can be extended to label other methyl groups, such as 13C-δ1-Ile and 13C-δ1,δ2-Leu. In summary, we demonstrate the cost-effective production of methyl-labeled proteins in mammalian cells by incorporation of 13C methyl-labeled amino acids generated de novo by a versatile synthetic route.
Introduction
Structural characterization of proteins by NMR has long relied on uniform labeling with 15N and 13C isotopes and assignment of signals based on transfer of spin coherence between connected backbone nuclei.1,2 Subsequent transfer of spin coherence to side chain nuclei can enable nearly complete assignment of the 1H, 13C, and 15N resonances and, in turn, full structure determination based on identification of inter-residue NOEs, residual dipolar couplings (RDCs), and other restraint types.3–6 In spite of the success of this approach, and its extension to larger proteins by deuteration7,8 and sensitivity enhancement by cross-correlation approaches,9,10 this strategy is still limited for larger proteins due to the unfavorable transverse spin relaxation properties of backbone and most sidechain nuclei. In addition, this approach is only applicable to proteins that can be produced with uniform 15N and 13C labeling and thus it is practically restricted to proteins that can be produced in bacteria or methylotrophic yeast, such as Pichia pastoris.11,12One method that has been impactful for overcoming challenges associated with unfavorable transverse spin relaxation of larger proteins is selective isotopic labeling of the sidechain methyl groups with 13C.13–16 Sidechain methyl groups have much more favorable transverse spin relaxation properties compared to backbone nuclei due to rapid (ps timescale) rotation around the three-fold methyl bond axis. Methyls also have other benefits for observation in larger proteins, including increased signal intensity due to the chemical equivalence of the three methyl protons and insensitivity to pH, enabling observation at higher pH values without accompanying loss of signal intensity due to solvent exchange. Robust procedures for selective isotopic labeling of the sidechain methyl groups of Ile, Leu, and Val with 13C are well-established for proteins expressed in bacteria. Typically, this involves inclusion of simpler 13C-methyl-labeled metabolic precursors, such as 3-13C-3,4,4′,4′′-2H-α-ketoisovalerate for Leu-δ1,δ2 and Val-γ1,γ2 or 4-13C-3,3′-2H-α-ketobutyrate for Ile-δ1 in 2H2O-based growth medium.14–18 Procedures have also been developed for the backbone-independent assignment of methyl signals, the most reliable of which is mutagenesis,19 though for larger proteins with many methyl groups this may be impractical, and other approaches that leverage existing structural information may be used to attain assignments.20–28
Methyl labeling, coupled with sensitivity enhancement by cross-correlation approaches, has had an enormous impact in terms of extending the applicability of NMR to study the structure and function of much larger and more complex systems than previously possible using uniform labeling with 15N, 13C, and 2H isotopes.14,16 Nonetheless, there are still many proteins, such as mammalian membrane receptors or secreted proteins that are heavily disulfide-bonded, that can be very difficult or impossible to express in bacteria, precluding methyl labeling using the aforementioned metabolic precursors. Recently, it was shown that yeast cultured on 2H2O-containing medium with 4-13C-α-ketobutyrate or 3-13C-α-ketoisovalerate could be used to generate Ile-δ1, Val-γ1,γ2, or Leu-δ1,δ2 labeled proteins, extending the strategy of labeling methyls with low-cost metabolic precursors to proteins that were previously inaccessible by bacterial expression.11,12
Methyl labeling in yeast, however, is not without limitations – for example, the labeling efficiency is lower than that possible by bacterial expression (ca. 45% for Ile-δ1, 74% for Val-γ1,γ2, and 10% for Leu-δ1,δ2), and for both Val-γ1,γ2 and Leu-δ1,δ2, achieving this level of efficiency requires cultivation under acidic conditions, which many membrane receptors or other complex human proteins might not tolerate.11,12 Moreover, many proteins are not correctly folded by the yeast protein production machinery; thus, mammalian cells, such as human embryonic kidney (HEK293) or Chinese hamster ovary (CHO) cells, remain the most suitable hosts for expression of mammalian proteins.29 Mammalian cells also maintain native post-translational modifications that may be critical to produce functionally active proteins. However, due to the inability of mammalian cells to synthesize most of the methyl-bearing amino acids, including Ile, Leu, and Val, from simple building blocks,30 it is not generally possible to generate methyl-labeled proteins in mammalian cells simply using the types of metabolic precursors described above.
One possible approach for producing 13C-methyl labeled amino acids in mammalian cells is to use alternative metabolic precursors, such as 13C uniformly labeled D-glucose, which can be used to label the methyl groups of alanine due to conversion of glucose to pyruvate and in turn alanine by aminotransferases.31 However, it is not possible to label other methyl-bearing amino acids using this approach, nor is it possible to achieve near complete labeling of alanine by metabolic labeling from 13C uniformly labeled D-glucose;31 thus, alternative approaches are needed. One such approach is to synthesize the requisite amino acids with 13C at the desired methyl positions and use these to construct medium that sustains the growth of CHO or HEK293 cells for protein expression. In the past, adoptions of this approach were limited due to low expression yields and the extended duration required to generate a stable transfected cell line that produced the target protein at high levels. However, advancements over the past two decades have significantly mitigated these challenges and newer expression systems based on modified versions of HEK293 or CHO have become an established method of choice for mammalian protein production.32 Furthermore, the inclusion of an N-terminal signal peptide together with either a hexahistidine or strep tag in the protein's sequence allows for a swift one-step purification process directly from the conditioned medium.
Labeling of the methione methyl group (13C-ε-Met) with high efficiency in mammalian cells is now relatively well established, which is not surprising, given the low cost of 13C-ε-Met and the generally lower amounts of this amino acid required in the medium compared to most other amino acids. In the past several years, there are reports of successfully labeling the methyl groups of other amino acids, such as Val and Leu,33,34 though this is not as widespread, presumably due to the high cost of these building blocks. Thus, although there has been progress towards producing 13C-methyl labeled amino acids in mammalian cells, one barrier that remains is the availability of low-cost synthetic routes to the requisite 13C methyl-labeled amino acids.
To address this challenge, we report here synthetic routes for 13C-γ2-Ile·HCl (1) and 13C-γ1,γ2-Val·HCl (2) using palladium-catalyzed C(sp3)–H functionalizations.35,36 The key step(s) were performed using an N-phthaloyl (NPhth) protected L-alanine (3) residue bearing an 8-aminoquinoline amide at the C-terminus as a directing auxiliary (DA) group. Iodomethane-13C was used for 13C-methyl-labeling. The introduction of the 13C methyl-labeled group demonstrated a high degree of regio- and stereoselectivity at the β-position due to the DA coordination to the transition metal, resulting in the anti-configuration in the case of 13C-γ2-Ile·HCl (1).37 The critical steps of the syntheses of 1 and 2 included the prevention of a potential erosion of diastereoselectivity at the epimerizable stereogenic carbons during the removal of DA and NPhth groups. Therefore, mild conditions were developed for the deprotection steps, suitable for the required synthetic scale. Starting with 1 to 2 g of iodomethane-13C, this synthetic route afforded 100 to 250 mg of the two target amino acids, 13C-γ2-Ile·HCl (1) and 13C-γ1,γ2-Val·HCl (2), and enabled the expression of 1 to 2 mg of two different 20 kDa disulfide-bonded receptors using suspension cultured HEK293 freestyle cells. The labeling efficiency of 13C-γ2-Ile and 13C-γ1,γ2-Val was found to be near 100% by comparison of the 13C-γ2-Ile and 13C-γ1,γ2-Val signal intensities relative to those in a natural abundance sample, and there was no apparent scrambling to other amino acids. Thus, we demonstrate the cost-effective and efficient production of 13C methyl-labeled proteins in mammalian cells, thereby expanding the benefits of methyl-labeling to larger and more challenging protein targets than previously feasible.
Results
Synthesis of 13C methyl-labeled γ2-Ile and γ1,γ2-Val
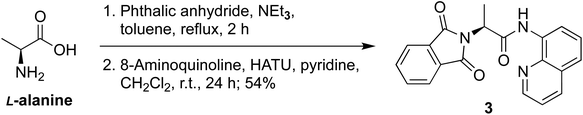
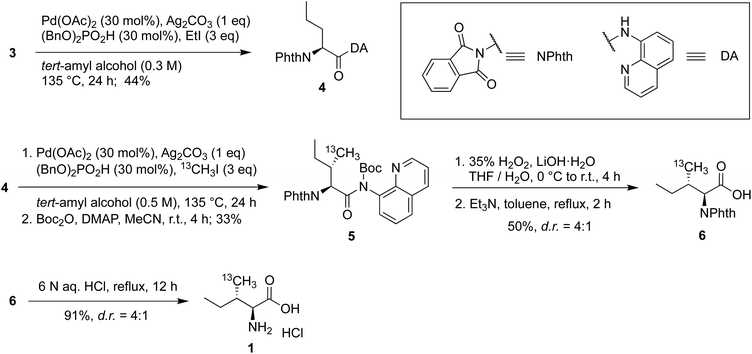
Both 13C methyl-labeled γ2-Ile·HCl (1) and 13C methyl-labeled γ1,γ2-Val·HCl (2) were obtained from the known intermediate 3, which was synthesized from L-alanine by protecting the amine as N-phthaloyl (NPhth) imide followed by coupling of the resulting carboxylic acid with 8-aminoquinoline.38–40 Intermediate 3 was obtained in 54% yield over two steps (Scheme 1).For the synthesis of 13C methyl-labelled γ2-Ile (1), we first performed a C(sp3)–H functionalization on intermediate 3 using similar reaction conditions to our previously reported protocol,41 which were obtained after an extensive optimization of literature conditions and scale-up,38,42 leading to the generation of the 2-aminovaleric backbone 4 in 44% yield (Scheme 2). The second functionalization allowed us to introduce the 13C methyl-group with the (S)-configuration in a good diastereomeric ratio and obtain the 13C methyl-labeled γ2-Ile scaffold. At this point, we faced the critical step to selectively remove the DA and NPhth groups without affecting the two stereocenters. Therefore, compound 5 was synthesized by Boc-protection of the previous intermediate in 33% yield over two steps, allowing the subsequent cleavage of the amide bond under mild conditions.43 Then, DA was removed using 35% H2O2 and LiOH, affording a mixture of the desired product and the carbamoylbenzoic acid by-product. The latter was converted into the desired phthalimide-protected analog 6 by treating the crude mixture with triethylamine in toluene at reflux in 50% yield, providing a 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of diastereomers. The deprotection of the phthalimide group of 6 was performed using well-established conditions to preserve the configuration at the α-stereocenter, affording 13C methyl-labeled γ2-Ile·HCl 1 in 91% yield.
1 ratio of diastereomers. The deprotection of the phthalimide group of 6 was performed using well-established conditions to preserve the configuration at the α-stereocenter, affording 13C methyl-labeled γ2-Ile·HCl 1 in 91% yield.
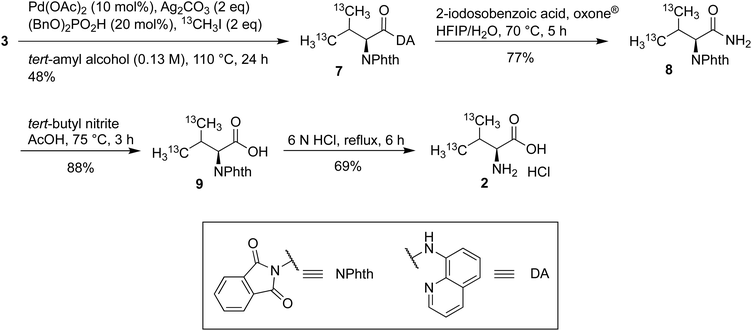
The synthesis of 13C methyl-labeled γ1,γ2-Val (2) commenced with the C(sp3)–H functionalization of 3 using iodomethane-13C to give the 13C methyl-labeled γ1,γ2-Val scaffold 7 in 48% yield (Scheme 3). The 8-aminoquinoline amide was converted into the primary amide 8 in 77% yield using 2-iodosobenzoic acid and oxone® and then into the corresponding carboxylic acid 9 using tert-butyl nitrite in acetic acid in 88% yield. These deprotection conditions were chosen to facilitate the purification of intermediates, and, simultaneously, avoid erosion of stereoselectivity.43,44 In the past several years, successful labeling of the methyl groups of other amino acids, such as Val, has been reported.31,45 The 13C methyl-labeled γ1,γ2-Val·HCl (2) was obtained by heating 9 at reflux in a 6 N HCl solution for 6 h.
Protein labelling
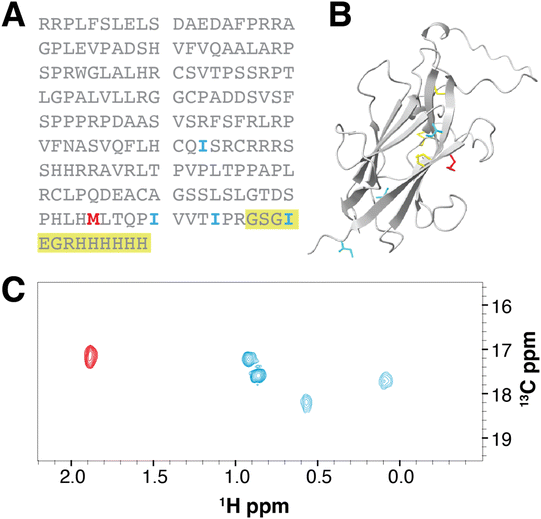
In this study, we choose as a model two related proteins: the C-terminal zona pellucida domain of the TGF-β co-receptor betaglycan, BGZPC,46,47 and a pituary-specific paralog known as TGFBRIII-like or R3like. The zona pellucida domain is a small (∼20 kDa) extracellular domain with 3 to 4 disulfide bonds that is present in proteins involved in various biological processes, including cell signaling and cell adhesion. The betaglycan ZPC domain is responsible for binding to the signaling proteins TGF-β and inhibinA and modulating signaling outcomes, whereas R3like binds and modulates signaling of inhibinB.48–50 Dysregulation of these interactions may contribute to the development of inflammatory disorders, cancer, or infertility. Despite significant efforts, structures of these receptor extracellular domains, which both contain three disulfide bonds, bound to their cognate signaling ligands are not known and thus our efforts are directed towards using NMR approaches to determine the structures of the corresponding ligand–receptor complexes. Although BGZPC can be produced in bacteria in form of inclusion bodies and refolded to a natively folded protein, the folding efficiency is poor and the production, refolding, and purification is complicated and lengthy, making it an expensive and difficult protein to produce using bacterial expression. R3like can also be produced in bacteria in the form of inclusion bodies, but we have been unable to obtain natively folded protein by refolding. BGZPC and R3like represent a common outcome where expression of disulfide-rich mammalian proteins can be difficult to obtain using the E. coli expression system. On the other hand, both proteins can be natively folded by HEK293 cells and secreted into the medium, from which they can be easily collected and purified using a tag-based affinity purification.BGZPC13C-γ2-Ile and 13C-γ1,γ2-Val labelling
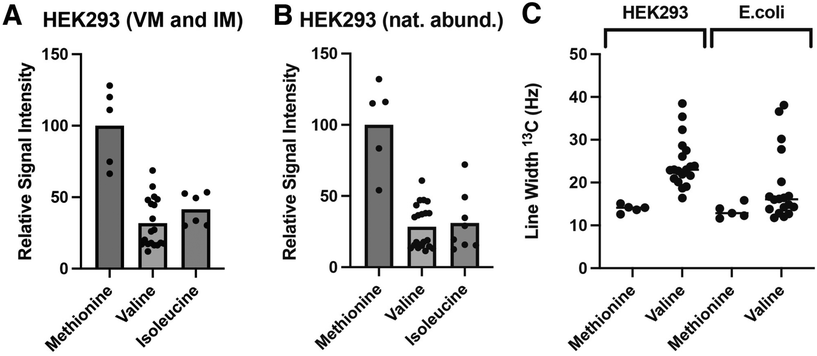
We have previously established a robust protocol for producing BGZPC in suspension cultured HEK293F cells,51 with a yield of approximately 4 mg of fully purified protein per liter of commercially available complete Hyclone TransFx-H liquid medium. We therefore adapted this procedure to produce methyl-labeled BGZPC since the production yield is reasonable and since this same growth medium is commercially available without amino acids. We showed through pilot studies that the production yield with medium constructed with amounts of unlabeled amino acids detailed in the Methods section afforded an overall yield of protein that was reduced by only a factor of 2 relative to commercially available complete medium. We therefore proceeded to produce BGZPC, with labeling of either synthesized 13C-γ2-Ile and commercially available 13C-ε-Met (BGZPC IM) or synthesized 13C-γ1,γ2-Val and commercially available 13C-ε-Met (BGZPC VM). In addition, since this protein can be also produced with some effort in E. coli, we decided to use this to our advantage to compare the 13C linewidths of the valine methyl 13C NMR signals from the incorporated amino acids to those obtained from the established bacterial ILVM method for 13C-methyl labeling with background deuteration. Notably, in both cases, we enriched the medium with a saturating concentration of 13C-ε-Met to serve as a reference for complete (100%) labeling.With the amounts of 13C-γ2-Ile and 13C-γ1,γ2-Val synthesized from 1–2 g scale quantities of the input low cost 13C reagent, iodomethane-13C (Table 1, upper portion), and with the production yields and medium composition noted above, we were able to readily produce sufficient quantities of fully purified BGZPC IM and BGZPC VM for several 300 μL NMR samples at a concentration of 50 μM (Table 1, lower portion). We further observed that the spectra of the labeled proteins yielded signals only in the chemical shift ranges expected for the type of label incorporated and that the number of signals observed was in accord with the number expected based on the amino acid sequence (13 Val, 6 Ile, and 5 Met), both for the control 13C-ε-Met and the synthesized 13C-γ2-Ile or 13C-γ1,γ2-Val (Fig. 1A–D). We also observe that the majority of the peak positions in the labeled samples match the signals in the much more concentrated natural abundance sample (Fig. 1E and F). Minor chemical shift differences visible on the spectra may originate from a small propensity to self-associate at the higher concentration, but not lower concentrations, variation in the glycosylation of the BGZPC domain at one of the two possible N-linked glycosylation sites that it possesses, or small difference in buffer conditions.
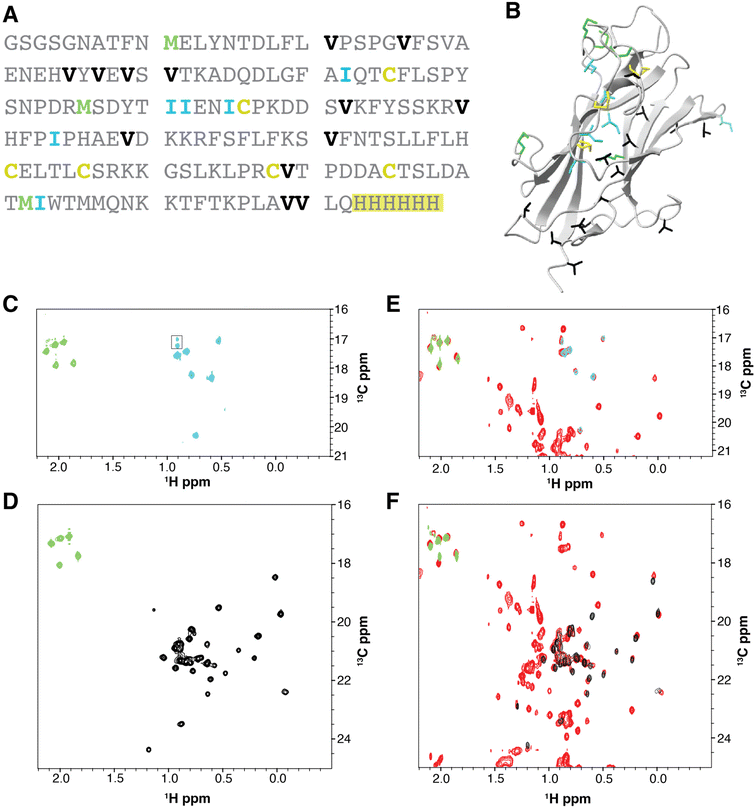 | ||
| Fig. 1 13C-Methyl-labeling of the Ile-γ2, Val-γ1,γ2, and Met-ε signals of BGZPC as expressed in suspension-cultured HEK293F cells. (A). Amino acid sequence of BGZPC. Ile, Val, and Met residues are highlighted in cyan, black, and green, respectively. The artificial C-terminal histidine tag used for purification is highlighted in yellow. (B). Structure of BGZPC (PDB 3QW9) highlighting the locations of its three disulfide bonds (yellow) and the Ile, Val, and Met residues (cyan, black, and green, respectively). (C) and (D). 1H–13C HSQC shift correlation spectrum of BGZPC labeled with 13C-γ2-Ile and 13C-ε-Met (C) or 13C-γ1,γ2-Val and 13C-ε-Met (D). Signals originating from 13C-γ2-Ile, 13C-γ1,γ2-Val, and 13C-ε-Met are shaded cyan, black, and green, respectively. (E) and (F). Reference 1H–13C HSQC spectra of BGZPC at natural abundance as produced using mammalian cell expression. HSQC spectra for 13C-γ2-Ile and 13C-ε-Met and 13C-γ1,γ2-Val and 13C-ε-Met labeled BGZPC (same spectra as shown in panels C and D, respectively) are overlaid on top of the reference 1H–13C HSQC spectrum (red) of BGZPC at natural abundance (E and F, respectively). | ||
| AA | Amount 13CH3I (g) | Amount AA synthesized (mg) |
|---|---|---|
| 13C-γ2-Ile | 2.29 | 236 |
| 13C-γ1,γ2-Val | 1.14 | 78 |
| Protein sample | Amount AA (mg) | Volume medium (mL) | Protein amount (mg) |
|---|---|---|---|
| BGZPC13C-γ2-Ile | 100 | 500 | 1.1 |
| BGZPC13C-γ1,γ2-Val | 78 | 420 | 0.8 |
| R3like 13C-γ2-Ile | 100 | 500 | 0.7 |
Production of secreted receptors in mammalian cells typically results in significant mass heterogeneity, due to heterogenous glycosylation. Production in glycosylation-deficient cell lines and aggressive treatment with deglycosidases, such as PNGaseF, can result in samples that approach the expected mass of the core protein and are more mass homogenous, yet it is nonetheless difficult to obtain a completely homogenous sample that can be used to accurately assess the incorporation efficiency of 13C- and 15N-labeled amino acids using mass spectrometry. Hence, as an alternative approach, we compared the relative signal intensities of 13C-ε-Met/13C-γ1,γ2-Val and 13C-ε-Met/13C-γ2-Ile in the BGZPC VM and BGZPC IM samples that we prepared relative to the same signals in a sample of BGZPC at natural abundance (Fig. 2A and B). Incorporation of 13C-ε-methionine into protein derived from mammalian cultures has also been shown to occur with an efficiency of 95% or greater.52 If so, this suggests that incorporation of both 13C-γ1,γ2-Val and 13C-γ2-Ile using our HEK293F expression system occurs with comparable efficiency based on the near equal Met/Val and Met/Ile signal intensity ratios relative those of the BGZPC natural abundance sample (Fig. 2A and B).
Selectively protonating 13C-methyl groups within a highly deuterated environment using the E. coli system has the advantage of reducing the dipolar relaxation contribution from neighboring protons. In accord with this, we observe that the 13C line widths for the 13C-γ1,γ2-Val signals in the E. coli produced deuterated ILVM sample (ESI, Fig. 1S†), which are generally more rigid and thus more susceptible to effects from dipolar broadening compared to that of the more flexible Met-ε signals, are on average only 1.2-fold greater than those of the corresponding 13C-ε-Met signals (Fig. 2C). In contrast, the 13C line widths for the 13C-γ1,γ2-Val signals in the mammalian produced protonated VM sample are roughly a factor of 1.6 greater than those of the corresponding 13C-ε-Met signals (Fig. 2C). Owing to the less crowded nature of 1H–13C methyl correlation spectra compared to 1H–15N amide correlation spectra, adverse consequences due to resonance overlap are reduced, enabling the clear and distinct discrimination of signals. Nonetheless, in the context of large proteins or protein complexes, it would be advisable to explore modifications in amino acid synthesis to facilitate the incorporation of deuterium in positions adjacent to the 13C-methyl site, both to reduce potential overlap and to increase sensitivity by sharpening the signals as recently shown for the incorporation of 13C-δ2-Leu in otherwise protonated background.45
R3like 13C-γ2-Ile labelling
Human R3like proved to be difficult to refold from reconstituted bacterially expressed inclusion bodies, despite significant structural similarity to the BGZPC domain as predicted by AlphaFold and a preserved number of internal disulfides. However, production and purification of R3like from HEK293 cell culture could be successfully accomplished,48 and using the same labeling strategy demonstrated for BGZPC, we succeeded in obtaining a sufficient quantity of 13C-ε-Met/13C-γ2-Ile R3like for several 50 μM NMR samples from 100 mg of 13C-γ2-Ile (Table 1, lower portion). Similar to that observed for BGZPC, we observe signals only in the chemical shift ranges expected for the incorporated labels and the number of observed signals matches expectations (4 Ile and 1 Met) (Fig. 3). Chemical shifts recorded for 13C-γ2-Ile suggest that the protein sample is natively folded.Discussion
Methyl-labeling has greatly extended the applicability of NMR to much larger and much more complex systems than what was possible using traditional approaches based upon observation and assignment of backbone 15N and 13C signals. However, methyl-labeling in mammalian cells is not possible using metabolic precursors, as it is widely done in bacteria and increasingly in yeast, due to the absence of the requisite biosynthetic machinery, though as recently shown uniformly 13C-labeled D-glucose can be used to label alanine methyl groups with 25% efficiency.31Herein, we report an efficient synthetic route to 13C-labeled amino acids based on the palladium-catalyzed C(sp3)–H functionalization of protected L-alanine and demonstrate its effectiveness through the synthesis of 13C-γ2-Ile and 13C-γ1,γ2-Val. We demonstrate that these amino acids can be incorporated into two different disulfide bonded receptor extracellular domains using suspension cultured HEK293 cells transiently transfected with the plasmid coding for the protein of interest. We observe by comparison of signal intensities to those in a natural abundance sample that the efficiency of labeling is near 100%. We also demonstrate there is little to no scrambling to other amino acids and that labeling is cost-efficient, with an input of 1 to 2 g of 13CH3I being adequate to generate sufficient 13C-methyl labeled amino acids to produce several 300 μL batches of 50 μM NMR samples. Importantly, the establishment of this low-cost and simple technology extends methyl-labeling approaches to many large and complex human proteins that cannot be easily produced in a native functional state by either bacterial or yeast expression.
For some applications, such as the determination of complex structures based on observation of inter-molecular inter-methyl NOEs, it may be important to achieve a higher density of signals than is possible with labeling of only 13C-γ2-Ile and 13C-γ1,γ2-Val. In light of this, it will be important to extend the synthetic methodology that we have developed to generate other types of 13C-methyl-labeled amino acids, such as 13C-δ1,δ2-Leu and 13C-γ1-Val. Owing to the versatile nature of the synthesis that we have developed, it may be possible to attain 13C-δ1,δ2-Leu by performing the C(sp3)–H functionalization of intermediate 3 using 2-iodopropane-1,3-13C2 instead of iodomethane-13C, or by using a γ-activating DA instead of the β-activating DA 8-aminoquinoline amide.5313C-γ1-Val can be obtained by a mono-functionalization of 3 using iodomethane-13C followed by a second C–H functionalization with unlabelled CH3I. Furthermore, it is feasible to attain other 13C-methyl-labeled amino acids, such as phenylalanine, tyrosine and tryptophan by using the corresponding 13C-labeled electrophiles iodobenzene, 4-iodophenol, and N-protected iodoindole, respectively.42
As it was demonstrated, in the absence of uniform deuteration, the linewidths for 13C-labeled methyl groups appeared broader compared to the highly deuterated background used in E. coli production. Alternative approaches will be required to attenuate transverse relaxation through dipolar coupling to other protons. One possible approach to attenuate relaxation, as recently shown for incorporation of 13C-δ2-Leu into an IgG,45 is to modify the synthesis to replace protons immediately adjacent to the 13C-methyl with deuterons. In the case of 13C-γ2-Ile and13C-γ1,γ2-Val this could possibly be done by palladium-catalyzed hydrogen/deuterium exchange on the NH Boc-deprotected intermediate of compound 5 and intermediate 7 in the presence of D2O, respectively.54
A further extension of the synthetic approach that we have developed is the synthesis of amino acids with fluorinated methyl groups, and the incorporation of these into proteins using bacterial, yeast, or mammalian cell expression systems. Due to the high sensitivity for detection of 19F, and the absence of background 19F signals in biological systems, this approach offers significant promise for studying protein structure and dynamics in living cells, as recently demonstrated by the successful detection of 19F-Trp- and 19F-Tyr-labeled proteins incorporated into mammalian cells.55
Experimental
General methods for chemical synthesis and analysis
Unless stated otherwise, all reactions were performed under an atmosphere of N2 that was passed through a column (10 × 2 cm) of Drierite®. Degassed solvents were prepared by freeze–pump–thaw techniques. All glassware and stir bars were flame-dried or stored in a drying oven prior to use. Reactions were monitored by TLC (thin layer chromatography) analysis using pre-coated silica gel 60 F254, and spots were visualized using UV light at 254 nm and 385 nm. Purifications by chromatography were performed on SiO2. 1H/13C NMR spectra were recorded on Bruker Avance 300/76 MHz, Bruker Avance 400/101 MHz, Bruker Avance 500/126 MHz or Bruker Avance 601/151 MHz instruments. High resolution mass spectra were obtained on a Micromass UK Limited, Q-TOF Ultima API or a Thermo Scientific Exactive Orbitrap LC-MS. IR spectra were obtained using neat samples on a PerkinElmer 100 IR ATR spectrometer. Melting points were obtained using a Mel-Temp instrument and are not corrected. Chemical shifts were reported in parts per million (ppm) with the residual solvent peak (CDCl3: 7.26 ppm for 1H, 77.16 ppm for 13C; DMSO-d6: 2.50 ppm for 1H, 39.52 ppm for 13C) used as the internal standard. Chemical shifts were tabulated as follows: chemical shift, multiplicity (s = singlet, d = doublet, t = triplet, q = quartet, h = sextet, dd = doublet of doublet, dt = doublet of triplet, ddd = doublet of doublet of doublet, tt = triplet of triplet, dtd = double of triplet of doublet, dddt = doublet of doublet of doublet of triplet, ddq = doublet of doublet of quartet, dtt = doublet of doublet of triplet, tqd = triplet of quartet of doublet, dpd = doubled of pentet of doubled, tdd = triplet of doublet of doublet, qdd = quartet of doublet of doublet, m = multiplet, br s = broad singlet), coupling constant(s), and integration.(S)-2-(1,3-Dioxoisoindolin-2-yl)-N-(quinolin-8-yl)propenamide (3)
A solution of phthalic anhydride (4.16 g, 28.1 mmol), L-alanine (2.50 g, 28.1 mmol, 1 eq.), and triethylamine (1.18 mL, 8.42 mmol, 0.3 eq.) in toluene (22 mL, 1.3 M) was heated at reflux for 2 h. The reaction mixture was cooled to room temperature and concentrated in vacuo. The crude residue was acidified by addition of 1 N aq. HCl (40 mL) and extracted with CH2Cl2 (2×). The combined organic layers were dried (MgSO4), filtered, and concentrated to give crude (S)-2-(1,3-dioxoisoindolin-2-yl)propanoic acid as a white solid that was used in the next step without further purification.A solution of (S)-2-(1,3-dioxoisoindolin-2-yl)propanoic acid (4.90 g, 22.4 mmol, 1 eq.) in CH2Cl2 (20 mL, 1.1 M) was treated sequentially with 8-aminoquinoline (2.53 g, 17.2 mmol, 0.77 eq.), pyridine (2.85 mL, 34.6 mmol, 1.55 eq.), and HATU (8.50 g, 22.4 mmol, 1 eq.). The reaction mixture was stirred at ambient temperature for 24 h. The deep brown solution was diluted with ethyl acetate, washed with sat. NaHCO3 and brine, dried (MgSO4), and purified by chromatography on SiO2 (2% acetone in CH2Cl2) to afford 3 (4.20 g, 12.2 mmol, 54%) as a white solid. Characterization data were in agreement with literature results.41 According to this experimental protocol, Wang et al. obtained 3 with an enantiomeric ratio (er) of 99.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.2.39
0.2.39
(S)-2-(1,3-Dioxoisoindolin-2-yl)-N-(quinolin-8-yl)pentanamide (4)
A mixture of amide 3 (0.50 g, 1.45 mmol, 1 eq.), Pd(OAc)2 (98 mg, 0.43 mmol, 30 mol%), Ag2CO3 (0.40 g, 1.45 mmol, 1 eq.), (BnO)2PO2H (123 mg, 0.434 mmol, 30 mol%), and ethyl iodide (356 μL, 4.34 mmol, 3 eq.) in tert-amyl alcohol (4.4 mL, 0.3 M) in a microwave glass vial (purged with argon and sealed with a PTFE cap) was heated at 135 °C for 24 h. The reaction mixture was filtered through a pad of Celite and concentrated in vacuo. The crude residue was purified by chromatography on SiO2 (25% acetone in hexanes) to afford 4 (0.24 g, 0.63 mmol, 44%) as a white solid. Characterization data were in agreement with literature results:381H NMR (500 MHz, Chloroform-d) δ 10.35 (s, 1 H; quinoline N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C
C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) –), 8.80–8.51 (m, 2 H; quinoline signals), 8.15 (dd, J = 8.3, 1.7 Hz, 1 H; quinoline signals), 7.90 (dt, J = 6.9, 3.5 Hz, 2 H; NPhth –CH
–), 8.80–8.51 (m, 2 H; quinoline signals), 8.15 (dd, J = 8.3, 1.7 Hz, 1 H; quinoline signals), 7.90 (dt, J = 6.9, 3.5 Hz, 2 H; NPhth –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C
C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) –C), 7.76 (dd, J = 5.5, 3.1 Hz, 2 H; NPhth –C
–C), 7.76 (dd, J = 5.5, 3.1 Hz, 2 H; NPhth –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–C), 7.51 (dd, J = 4.2, 3.1 Hz, 2 H; quinoline signals), 7.43 (dd, J = 8.3, 4.3 Hz, 1 H; quinoline signals), 5.15 (dd, J = 11.1, 5.2 Hz, 1 H; –C
CH–C), 7.51 (dd, J = 4.2, 3.1 Hz, 2 H; quinoline signals), 7.43 (dd, J = 8.3, 4.3 Hz, 1 H; quinoline signals), 5.15 (dd, J = 11.1, 5.2 Hz, 1 H; –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) CON–), 2.67–2.54 (m, 1 H; –C
CON–), 2.67–2.54 (m, 1 H; –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2CH–), 2.46–2.30 (m, 1 H; –C
2CH–), 2.46–2.30 (m, 1 H; –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2CH–), 1.47 (h, J = 7.4 Hz, 2 H; –C
2CH–), 1.47 (h, J = 7.4 Hz, 2 H; –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2CH3), 1.04 (t, J = 7.4 Hz, 3 H; –CH2C
2CH3), 1.04 (t, J = 7.4 Hz, 3 H; –CH2C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3).
3).
tert-Butyl ((2S,3S)-2-(1,3-dioxoisoindolin-2-yl)-3-(methyl-13C)pentanoyl)(quinolin-8-yl)carbamate (5)
A mixture of amide 4 (1.51 g, 4.05 mmol, 1 eq.), Pd(OAc)2 (273 mg, 1.21 mmol, 30 mol%), Ag2CO3 (1.12 g, 4.05 mmol, 1 eq.), (BnO)2PO2H (345 mg, 1.21 mmol, 30 mol%), and iodomethane-13C (753 μL, 12.1 mmol, 3 eq.) in tert-amyl alcohol (7.5 mL, 0.5 M) in a microwave glass vial (purged with argon and sealed with a PTFE cap) was heated at 135 °C for 24 h. The reaction mixture was filtered through a pad of Celite® (CH2Cl2 to rinse, 250 mL collection flask), and concentrated in vacuo. The crude residue was re-suspended in a mixture of 50% acetone/hexanes, filtered through a pad of SiO2 (50% acetone in hexanes to rinse, 500 mL collection flask) and concentrated in vacuo. The resulting crude mixture containing the desired intermediate, bis-alkylated byproduct, and remaining starting material was purified by reverse-phase chromatography (C18, 30 g cartridge, H2O/MeOH; 6 CV of 100% water, then 80 CV of gradient elution to 100% MeOH) to remove the remaining starting material. The resulting mixture of desired intermediate and bis-alkylated byproduct was used for the next step without further purification.A solution of this mixture (0.88 g, 2.26 mmol, 1 eq.) Boc2O (1.48 g, 6.79 mmol, 3 eq.), and DMAP (0.55 g, 4.53 mmol, 2 eq.) in anhydrous CH3CN (5 mL, 0.4 M) was stirred at room temperature for 4 h, concentrated in vacuo and purified by chromatography on SiO2 (10% acetone in hexanes) to afford 5 (1.90 g, 3.96 mmol, 33% yield over two steps) as a white foam: IR (ATR, CH2Cl2) 3061, 2933, 2971, 2876, 1740, 1716, 1500, 1469, 1369, 1293, 1265, 1254, 1153, 1125 cm−1; 1H NMR (500 MHz, Chloroform-d) δ 8.62 (br s, 1 H; quinoline signal), 8.10 (dd, J = 8.2, 1.8 Hz, 1 H; quinoline signal), 7.85 (dd, J = 5.5, 2.9 Hz, 2 H; NPhth –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C
C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) –C), 7.77 (dd, J = 8.3, 1.4 Hz, 1 H; quinoline signal), 7.76–7.67 (m, 3 H; quinoline signal overlapped with NPhth –C
–C), 7.77 (dd, J = 8.3, 1.4 Hz, 1 H; quinoline signal), 7.76–7.67 (m, 3 H; quinoline signal overlapped with NPhth –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–C), 7.54 (t, J = 7.8 Hz, 1 H; quinoline signal), 7.30 (td, J = 8.4, 4.1 Hz, 1 H; quinoline signal), 5.93–5.63 (m, 1 H; –C
CH–C), 7.54 (t, J = 7.8 Hz, 1 H; quinoline signal), 7.30 (td, J = 8.4, 4.1 Hz, 1 H; quinoline signal), 5.93–5.63 (m, 1 H; –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) CON–), 2.73–2.58 (m, 1 H; –C
CON–), 2.73–2.58 (m, 1 H; –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 13CH3), 1.57 (dpd, J = 14.5, 7.3, 3.3 Hz, 1 H; –C
13CH3), 1.57 (dpd, J = 14.5, 7.3, 3.3 Hz, 1 H; –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2CH3), 1.29 (d, J = 6.7 Hz, 1.5 H; –CH13C
2CH3), 1.29 (d, J = 6.7 Hz, 1.5 H; –CH13C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3), 1.17–0.93 (m, 11.5 H; –C(C
3), 1.17–0.93 (m, 11.5 H; –C(C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3)3 overlapped with –C
3)3 overlapped with –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2CH3 and –CH13C
2CH3 and –CH13C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3), 0.88 (t, J = 7.4 Hz, 3 H; –CH2C
3), 0.88 (t, J = 7.4 Hz, 3 H; –CH2C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3); 13C NMR (151 MHz, Chloroform-d) δ 171.9, 167.8, 152.7, 150.1, 134.1, 132.0, 129.2, 128.9, 128.1, 126.6, 123.5, 121.5, 83.0, 58.8, 27.4, 25.6, 23.1, 22.9, 17.0, 15.7, 11.8, 11.7, 11.5; HRMS (ESI+) m/z calcd for C2713CH30O5N3 [M + H]+, 489.2214; found, 489.2198.
3); 13C NMR (151 MHz, Chloroform-d) δ 171.9, 167.8, 152.7, 150.1, 134.1, 132.0, 129.2, 128.9, 128.1, 126.6, 123.5, 121.5, 83.0, 58.8, 27.4, 25.6, 23.1, 22.9, 17.0, 15.7, 11.8, 11.7, 11.5; HRMS (ESI+) m/z calcd for C2713CH30O5N3 [M + H]+, 489.2214; found, 489.2198.
(2S,3S)-2-(1,3-Dioxoisoindolin-2-yl)-3-(methyl-13C)pentanoic acid (6)
A solution of the Boc-protected intermediate 5 (1.80 g, 3.69 mmol, 1 eq.) in THF and water (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, THF/H2O, 7.2 mL, 0.5 M) was cooled to 0 °C and treated with 30% hydrogen peroxide (3.26 mL, 32.5 mmol, 8.8 eq.) and lithium hydroxide monohydrate (233 mg, 5.54 mmol, 1.5 eq.), stirred at 0 °C for 45 min, warmed to room temperature and stirred for an additional 3 h. The reaction mixture was quenched at 0 °C with 1.5 M aqueous sodium thiosulfate (2 mL) and concentrated under reduced pressure. The residue was washed with CH2Cl2 (2×), and the aqueous phase was then acidified to pH 1–2 with 10% aqueous HCl and extracted with EtOAc (2×). The combined organic extracts were dried (Na2SO4) and concentrated under reduced pressure. Both product and the ring-opened phthalimide byproduct were present and the mixture was directly used in the next step. A suspension of the crude mixture (0.27 g, 0.97 mmol, 1 eq.) and triethylamine (0.07 mL, 0.48 mmol, 0.5 eq.) in toluene (3 mL) was stirred for 2 h at reflux, cooled to room temperature and concentrated in vacuo. The crude residue was acidified by addition of 10% aqueous HCl and extracted with EtOAc (2×), dried (Na2SO4), filtered, and concentrated in vacuo. Purification by chromatography on SiO2 (2% MeOH in CH2Cl2) gave 6 as a white solid (0.42 g, 1.61 mmol, 50% yield over two steps in trans/cis = 4
1, THF/H2O, 7.2 mL, 0.5 M) was cooled to 0 °C and treated with 30% hydrogen peroxide (3.26 mL, 32.5 mmol, 8.8 eq.) and lithium hydroxide monohydrate (233 mg, 5.54 mmol, 1.5 eq.), stirred at 0 °C for 45 min, warmed to room temperature and stirred for an additional 3 h. The reaction mixture was quenched at 0 °C with 1.5 M aqueous sodium thiosulfate (2 mL) and concentrated under reduced pressure. The residue was washed with CH2Cl2 (2×), and the aqueous phase was then acidified to pH 1–2 with 10% aqueous HCl and extracted with EtOAc (2×). The combined organic extracts were dried (Na2SO4) and concentrated under reduced pressure. Both product and the ring-opened phthalimide byproduct were present and the mixture was directly used in the next step. A suspension of the crude mixture (0.27 g, 0.97 mmol, 1 eq.) and triethylamine (0.07 mL, 0.48 mmol, 0.5 eq.) in toluene (3 mL) was stirred for 2 h at reflux, cooled to room temperature and concentrated in vacuo. The crude residue was acidified by addition of 10% aqueous HCl and extracted with EtOAc (2×), dried (Na2SO4), filtered, and concentrated in vacuo. Purification by chromatography on SiO2 (2% MeOH in CH2Cl2) gave 6 as a white solid (0.42 g, 1.61 mmol, 50% yield over two steps in trans/cis = 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 diastereomeric ratio): 1H NMR (400 MHz, Chloroform-d, 60 °C) Major diastereomer: δ 7.88 (ddd, J = 5.4, 3.1, 1.1 Hz, 2 H; NPhth –CH
1 diastereomeric ratio): 1H NMR (400 MHz, Chloroform-d, 60 °C) Major diastereomer: δ 7.88 (ddd, J = 5.4, 3.1, 1.1 Hz, 2 H; NPhth –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C
C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) –C), 7.74 (ddd, J = 5.5, 3.1, 1.1 Hz, 2 H; NPhth –C
–C), 7.74 (ddd, J = 5.5, 3.1, 1.1 Hz, 2 H; NPhth –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–C), 4.73 (ddd, J = 8.6, 3.4, 1.1 Hz, 1 H; –C
CH–C), 4.73 (ddd, J = 8.6, 3.4, 1.1 Hz, 1 H; –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) CO2H–), 2.64–2.50 (m, 1 H; –C
CO2H–), 2.64–2.50 (m, 1 H; –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 13CH3), 1.53 (dtd, J = 13.6, 6.9, 4.1 Hz, 1 H; –C
13CH3), 1.53 (dtd, J = 13.6, 6.9, 4.1 Hz, 1 H; –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2CH3), 1.32 (d, J = 6.7 Hz, 1.5 H; –CH13C
2CH3), 1.32 (d, J = 6.7 Hz, 1.5 H; –CH13C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3), 1.20–1.07 (m, 1 H; –C
3), 1.20–1.07 (m, 1 H; –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2CH3), 1.04–0.97 (m, 1.5 H; –CH13C
2CH3), 1.04–0.97 (m, 1.5 H; –CH13C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3), 0.89 (td, J = 7.4, 1.1 Hz, 3 H; –CH2C
3), 0.89 (td, J = 7.4, 1.1 Hz, 3 H; –CH2C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3); 13C NMR (151 MHz, Chloroform-d) Major diastereomer: δ 173.6, 168.0, 134.5, 131.7, 123.8, 57.3, 34.5 (d, J = 35.3 Hz), 22.5 (d, J = 34.8 Hz), 16.9, 10.9 (d, J = 35.0 Hz).
3); 13C NMR (151 MHz, Chloroform-d) Major diastereomer: δ 173.6, 168.0, 134.5, 131.7, 123.8, 57.3, 34.5 (d, J = 35.3 Hz), 22.5 (d, J = 34.8 Hz), 16.9, 10.9 (d, J = 35.0 Hz).
γI-(13C)-L-Isoleucine·HCl (1)
A solution of 6 (0.40 g, 1.53 mmol, 1 eq.) in 6 N aq. HCl (20 mL) was heated at reflux for 12 h in a pressure tube. The precipitated phthalic acid was removed by extraction with AcOEt (3×). The water layer was freeze-dried to give 1 (236 mg, 1.40 mmol, 91%, in trans/cis = 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 diastereomeric ratio) as a white solid: αD25 +5.1 (c 0.1, H2O); 1H NMR (500 MHz, DMSO-d6) Major diastereomer δ 13.74 (s, 1 H; –CO2
1 diastereomeric ratio) as a white solid: αD25 +5.1 (c 0.1, H2O); 1H NMR (500 MHz, DMSO-d6) Major diastereomer δ 13.74 (s, 1 H; –CO2![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ), 8.37 (s, 3 H, –N
), 8.37 (s, 3 H, –N![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3+), 3.79 (s, 1 H, C
3+), 3.79 (s, 1 H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) CO2H), 1.92 (tdd, J = 9.1, 5.8, 3.1 Hz, 1 H; –C
CO2H), 1.92 (tdd, J = 9.1, 5.8, 3.1 Hz, 1 H; –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) (13CH3)), 1.48 (qdd, J = 12.9, 10.4, 6.5 Hz, 1 H; –C
(13CH3)), 1.48 (qdd, J = 12.9, 10.4, 6.5 Hz, 1 H; –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2CH3), 1.35–1.23 (m, 1 H; –C
2CH3), 1.35–1.23 (m, 1 H; –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2CH3), 1.05 (d, J = 6.9 Hz, 1.5 H; −13C
2CH3), 1.05 (d, J = 6.9 Hz, 1.5 H; −13C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3), 0.89 (td, J = 7.4, 4.4 Hz, 3 H; –C
3), 0.89 (td, J = 7.4, 4.4 Hz, 3 H; –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3), 0.80 (d, J = 6.9 Hz, 1.5 H; −13C
3), 0.80 (d, J = 6.9 Hz, 1.5 H; −13C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3); 13C NMR (126 MHz, DMSO-d6) Major diastereomer: δ 170.1 (d, J = 2.3 Hz), 56.0, 35.6 (d, J = 34.8 Hz), 21.6 (d, J = 34.7 Hz), 14.4, 11.5 (d, J = 2.2 Hz); HRMS (ESI+) m/z calcd for C513CH12O2N, 131.0896, found 131.0898.
3); 13C NMR (126 MHz, DMSO-d6) Major diastereomer: δ 170.1 (d, J = 2.3 Hz), 56.0, 35.6 (d, J = 34.8 Hz), 21.6 (d, J = 34.7 Hz), 14.4, 11.5 (d, J = 2.2 Hz); HRMS (ESI+) m/z calcd for C513CH12O2N, 131.0896, found 131.0898.
(S)-2-(1,3-Dioxoisoindolin-2-yl)-3-(methyl-13C)-N-(quinolin-8-yl)butanamide-4-13C (7)
A mixture of 3 (0.70 g, 2.03 mmol, 1 eq.), Pd(OAc)2 (50 mg, 0.20 mmol, 10 mol%), silver carbonate (1.13 g, 4.05 mmol, 2 eq.), (BnO)2PO2H (115 mg, 0.40 mmol, 20 mol%) was added to a flame-dried 75 mL pressure tube. Then, tert-amyl alcohol (15.6 mL, 0.13 M) was added followed by iodomethane-13C (0.25 mL, 4.05 mmol, 2 eq.). The reaction mixture was heated at 110 °C for 24 h, diluted with CH2Cl2, and then filtered through a pad of Celite® (CH2Cl2 to rinse), and concentrated in vacuo. The crude residue contained the desired product along with the mono-alkylated intermediate and the starting material and was re-subjected to the same reaction conditions to push the reaction to completion. After the work-up, the crude residue was purified by chromatography on SiO2 (10% acetone in hexanes) to give 7 (0.37 g, 0.98 mmol, 48%) as a pale-yellow solid: 1H NMR (400 MHz, Chloroform-d) δ 10.58 (s, 1 H; –CON![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) –), 8.85 (dd, J = 4.2, 1.7 Hz, 1 H; quinoline signal), 8.76 (dd, J = 5.0, 4.1 Hz, 1 H, quinoline signal), 8.14 (dd, J = 8.3, 1.7 Hz, 1 H, quinoline signal), 7.94–7.85 (m, 2 H; NPhth –CH
–), 8.85 (dd, J = 4.2, 1.7 Hz, 1 H; quinoline signal), 8.76 (dd, J = 5.0, 4.1 Hz, 1 H, quinoline signal), 8.14 (dd, J = 8.3, 1.7 Hz, 1 H, quinoline signal), 7.94–7.85 (m, 2 H; NPhth –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C
C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) –C), 7.79–7.69 (m, 2 H; NPhth –C
–C), 7.79–7.69 (m, 2 H; NPhth –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–C), 7.55–7.48 (m, 2 H; quinoline signal), 7.44 (dd, J = 8.3, 4.2 Hz, 1 H; quinoline signal), 4.70 (dt, J = 10.8, 2.8 Hz, 1 H; –C
CH–C), 7.55–7.48 (m, 2 H; quinoline signal), 7.44 (dd, J = 8.3, 4.2 Hz, 1 H; quinoline signal), 4.70 (dt, J = 10.8, 2.8 Hz, 1 H; –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) CONH–), 3.23 (dddt, J = 13.3, 10.2, 6.6, 3.3 Hz, 1 H; –C
CONH–), 3.23 (dddt, J = 13.3, 10.2, 6.6, 3.3 Hz, 1 H; –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) (13CH3)2), 1.39 (dd, J = 6.7, 5.3 Hz, 1.5 H; –CH13C
(13CH3)2), 1.39 (dd, J = 6.7, 5.3 Hz, 1.5 H; –CH13C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3), 1.15 (dd, J = 6.6, 5.2 Hz, 1.5 H; –CH13C
3), 1.15 (dd, J = 6.6, 5.2 Hz, 1.5 H; –CH13C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3), 1.07 (dd, J = 6.6, 5.3 Hz, 1.5 H; –CH13C
3), 1.07 (dd, J = 6.6, 5.3 Hz, 1.5 H; –CH13C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3), 0.84 (dd, J = 6.6, 5.2 Hz, 1.5 H; –CH13C
3), 0.84 (dd, J = 6.6, 5.2 Hz, 1.5 H; –CH13C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3); 13C NMR (101 MHz, Chloroform-d) δ 168.3, 167.0, 148.7, 138.9, 136.3, 134.4, 134.4, 131.8, 128.1, 127.4, 123.8, 122.1, 121.8, 117.1, 63.4, 27.0 (d, J = 35.0 Hz), 20.6, 19.8, 15.8, 11.3 (dd, J = 35.1, 1.6 Hz).
3); 13C NMR (101 MHz, Chloroform-d) δ 168.3, 167.0, 148.7, 138.9, 136.3, 134.4, 134.4, 131.8, 128.1, 127.4, 123.8, 122.1, 121.8, 117.1, 63.4, 27.0 (d, J = 35.0 Hz), 20.6, 19.8, 15.8, 11.3 (dd, J = 35.1, 1.6 Hz).
(S)-2-(1,3-Dioxoisoindolin-2-yl)-3-(methyl-13C)butanamide-4-13C (8)
A 50 mL round-bottom flask was charged with 7 (369 mg, 0.99 mmol, 1 eq.), 2-iodosobenzoic acid (78.3 mg, 0.30 mmol, 30 mol%), and oxone® (1.82 g, 2.96 mmol, 3 eq.), followed by a mixture of HFIP/H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 7.2 mL, 0.14 M). The reaction mixture was stirred at 70 °C for 5 h, quenched by the addition of sat. aq. NaHCO3 and extracted with CH2Cl2 (3×). The combined organic layers were concentrated in vacuo and the crude residue was purified by chromatography on SiO2 (30% hexanes in ethyl acetate) to give 8 (0.19 g, 0.76 mmol, 77%) as a white solid: 1H NMR (500 MHz, DMSO-d6) δ 7.92–7.84 (m, 4 H; NPhth signals), 7.47 (s, 1 H; –CON
1, 7.2 mL, 0.14 M). The reaction mixture was stirred at 70 °C for 5 h, quenched by the addition of sat. aq. NaHCO3 and extracted with CH2Cl2 (3×). The combined organic layers were concentrated in vacuo and the crude residue was purified by chromatography on SiO2 (30% hexanes in ethyl acetate) to give 8 (0.19 g, 0.76 mmol, 77%) as a white solid: 1H NMR (500 MHz, DMSO-d6) δ 7.92–7.84 (m, 4 H; NPhth signals), 7.47 (s, 1 H; –CON![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 7.09 (s, 1 H; –CON
2), 7.09 (s, 1 H; –CON![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 4.27 (dt, J = 8.6, 3.3 Hz, 1 H; –C
2), 4.27 (dt, J = 8.6, 3.3 Hz, 1 H; –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) CONH2), 2.73–2.61 (m, 1 H; –C
CONH2), 2.73–2.61 (m, 1 H; –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) (13CH3)2), 1.14 (dd, J = 6.7, 5.3 Hz, 1.5 H; –CH13C
(13CH3)2), 1.14 (dd, J = 6.7, 5.3 Hz, 1.5 H; –CH13C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3), 0.89 (ddd, J = 6.6, 5.1, 1.4 Hz, 3 H; –CH13C
3), 0.89 (ddd, J = 6.6, 5.1, 1.4 Hz, 3 H; –CH13C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3), 0.64 (dd, J = 6.8, 5.2 Hz, 1.5 H; –CH13C
3), 0.64 (dd, J = 6.8, 5.2 Hz, 1.5 H; –CH13C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3); 13C NMR (126 MHz, DMSO-d6) δ 169.4, 167.7, 134.5, 131.3, 123.1, 58.5, 26.9 (d, J = 34.8 Hz), 20.9, 19.3, 15.6 (d, J = 1.9 Hz), 11.2 (dd, J = 34.9, 2.0 Hz).
3); 13C NMR (126 MHz, DMSO-d6) δ 169.4, 167.7, 134.5, 131.3, 123.1, 58.5, 26.9 (d, J = 34.8 Hz), 20.9, 19.3, 15.6 (d, J = 1.9 Hz), 11.2 (dd, J = 34.9, 2.0 Hz).
(S)-2-(1,3-Dioxoisoindolin-2-yl)-3-(methyl-13C)butanoic-4-13C acid (9)
A stirred solution of the primary amide 8 (185 mg, 0.75 mmol, 1 eq.) in acetic acid (4 mL, 0.19 M) was dropwise treated with tert-butyl nitrite (149 μL, 1.13 mmol, 1.5 eq.) and stirred at 75 °C under air atmosphere for 3 h. The reaction progress was monitored by TLC. AcOH was removed by gently heating the flask to 40 °C while under high vacuum and cooling the receiver flask at −78 °C. The resulting residue was purified by chromatography on SiO2 (2% MeOH in CH2Cl2) to afford 9 (0.16 g, 0.66 mmol, 88%) as a white solid: 1H NMR (300 MHz, Chloroform-d) δ 7.88 (dd, J = 5.5, 3.1 Hz, 2 H; NPhth –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C
C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) –C), 7.75 (dd, J = 5.5, 3.0 Hz, 2 H; NPhth –C
–C), 7.75 (dd, J = 5.5, 3.0 Hz, 2 H; NPhth –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–C), 4.63 (dt, J = 8.5, 3.3 Hz, 1 H; –C
CH–C), 4.63 (dt, J = 8.5, 3.3 Hz, 1 H; –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) CO2H), 2.86–2.67 (m, 1 H; –C
CO2H), 2.86–2.67 (m, 1 H; –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) (13CH3)2), 1.38 (dd, J = 6.6, 5.3 Hz, 1.5 H; –CH(13C
(13CH3)2), 1.38 (dd, J = 6.6, 5.3 Hz, 1.5 H; –CH(13C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3)2), 1.13 (dd, J = 6.8, 5.2 Hz, 1.5 H; –CH(13C
3)2), 1.13 (dd, J = 6.8, 5.2 Hz, 1.5 H; –CH(13C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3)2), 0.96 (dd, J = 6.6, 5.3 Hz, 1.5 H; –CH(13C
3)2), 0.96 (dd, J = 6.6, 5.3 Hz, 1.5 H; –CH(13C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3)2), 0.71 (dd, J = 6.8, 5.2 Hz, 1.5 H; –CH(13C
3)2), 0.71 (dd, J = 6.8, 5.2 Hz, 1.5 H; –CH(13C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3)2); 13C NMR (76 MHz, Chloroform-d) δ 173.0, 167.9, 134.5, 131.8, 123.8, 57.9, 27.3 (d, J = 34.9 Hz), 21.0, 19.6, 16.0 (d, J = 1.9 Hz), 11.4 (dd, J = 34.9, 1.9 Hz).
3)2); 13C NMR (76 MHz, Chloroform-d) δ 173.0, 167.9, 134.5, 131.8, 123.8, 57.9, 27.3 (d, J = 34.9 Hz), 21.0, 19.6, 16.0 (d, J = 1.9 Hz), 11.4 (dd, J = 34.9, 1.9 Hz).
(2S)-2-Amino-3-(13C)methyl(13C)butanoic acid·HCl (2)
A solution of 9 (0.18 g, 0.73 mmol, 1 eq.) in 6 N aq. HCl (2.1 mL, 0.35 M) was heated at reflux for 6 h in a pressure tube. The precipitated phthalic acid was removed by extraction with AcOEt (3×). The water layer was freeze-dried to give 2 (78 mg, 0.50 mmol, 69%): αD25 +47.8 (c 0.12, H2O); 1H NMR (500 MHz, DMSO-d6) δ 8.36 (br s, 3 H; –N![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3+), 7.58–7.10 (m, 1 H; –CO2
3+), 7.58–7.10 (m, 1 H; –CO2![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ), 3.69 (q, J = 4.2 Hz, 1 H; –C
), 3.69 (q, J = 4.2 Hz, 1 H; –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) CO2H), 2.19 (ddq, J = 11.9, 9.1, 4.7, 3.7 Hz, 1 H; –C
CO2H), 2.19 (ddq, J = 11.9, 9.1, 4.7, 3.7 Hz, 1 H; –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) (13CH3)2), 1.10 (td, J = 7.0, 4.9 Hz, 3 H; −13CH3), 0.85 (td, J = 6.9, 4.9 Hz, 3 H; −13CH3); 13C NMR (126 MHz, DMSO-d6) δ 170.1, 57.4, 24.5 (d, J = 34.9 Hz), 18.1, 17.7, 14.3 (d, J = 2.2 Hz), 11.4 (dd, J = 34.9, 2.1 Hz); HRMS (ESI+) m/z calcd for C313C2H10O2N, 118.0773, found 118.0776.
(13CH3)2), 1.10 (td, J = 7.0, 4.9 Hz, 3 H; −13CH3), 0.85 (td, J = 6.9, 4.9 Hz, 3 H; −13CH3); 13C NMR (126 MHz, DMSO-d6) δ 170.1, 57.4, 24.5 (d, J = 34.9 Hz), 18.1, 17.7, 14.3 (d, J = 2.2 Hz), 11.4 (dd, J = 34.9, 2.1 Hz); HRMS (ESI+) m/z calcd for C313C2H10O2N, 118.0773, found 118.0776.
Mammalian plasmid expression constructs
Rat betaglycan ZPC domain (BGZPC)46,47 was expressed in freestyle suspension cultured HEK293 (HEK293F) cells (Invitrogen R79007) downstream of the rat serum albumin signal peptide and rat serum albumin and a thrombin cleavage site in pcDNA3.1+ (Invitrogen V79020). Mouse TGFβR3-like (R3like)48 was expressed similarly, but downstream of the rat serum albumin signal peptide alone. Hexahistidine tags were appended to the C-terminal of both BGZPC and R3like to enable facile purification. Coding sequences of BGZPC and R3like were obtained by gene synthesis (Twist Biosciences) and the full coding cassettes were inserted between the NheI and XhoI sites in pcDNA3.1+. Constructs were verified by Sanger DNA sequencing over the entire length of the coding sequence.Bacterial expression constructs
Rat betaglycan ZPC domain (BGZPC) was expressed in E. coli BL21(DE3) cells (EMD-Millipore 69450) downstream of thioredoxin, a hexahistidine tag, and a thrombin cleavage site in plasmid pET32a (EMD-Millipore 69015). The coding sequence for BGZPC was comprised of residues 590–757 of NCBI NP_058952 and a strepII tag, WSHPQFEK, was fused to its C-terminus. The coding sequence was obtained by gene synthesis (Twist Biosciences) and inserted between a KpnI site, which was introduced immediately following the last codon of the Trx-thrombin coding casette intrinsic to pET32a, and the XhoI site. Constructs were verified by Sanger DNA sequencing over the entire length of the coding sequence.Protein expression and isotope labeling in mammalian cells
HEK293 freestyle cells (293F) were maintained in a custom formulation of HyClone HyCell TransFx-H medium (Cytiva). The custom formulated medium was prepared by mixing (per liter): 9.2 g TransFx-H media purchased without glucose or amino acids, 6 g glucose (Millipore-Sigma G7021), 3.2 g sodium bicarbonate (Millipore-Sigma S5761), 1 g Pluronic F-68 (AppliChem Panreac A12880500), 586 mg L-glutamine (Millipore-Sigma G8540), 50 mg L-alanine (Millipore-Sigma A7469), 500 mg L-arginine (Millipore-Sigma A8094), 900 mg L-asparagine (Millipore-Sigma A4159), 200 mg L-aspartic acid (Millipore-Sigma A7219), 100 mg L-cysteine-HCl (Millipore-Sigma C7477), 300 mg L-glutamic acid (Millipore-Sigma G8415), 30 mg glycine (Millipore-Sigma G5417), 200 mg L-histidine-HCl (Millipore-Sigma H6034), 400 mg L-isoleucine (Alfa Aesar J63045), 500 mg L-leucine (Millipore-Sigma L6914), 500 mg L-lysine-HCl (Millipore-Sigma L8662), 115 mg L-methionine (Millipore-Sigma M5308), 200 mg L-phenylalanine (Millipore-Sigma P8740), 500 mg L-proline (Millipore-Sigma P5607), 500 mg L-serine (Millipore-Sigma S4311), 380 mg L-threonine (Millipore-Sigma T8441), 200 mg L-tryptophan (Millipore-Sigma T8941), 200 mg L-tyrosine disodium dihydrate (J.T. Baker 2094-05), and 370 mg L-valine (Millipore-Sigma V0513). A suspension culture was grown on an orbital shaker (125 rpm) in a 37 °C incubator with 8% CO2. Cells were passaged every 3–4 days by pelleting (8 min, 90g, 25 °C) and resuspending in fresh medium with a final density of 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mL.
000 cells per mL.
In preparation for transfection to produce 13C-γ2-Ile, 13C-ε-Met (IM) or 13C-γ1,γ2-Val, 13C-ε-Met (VM) labeled Alb-BGZPC or R3like, cells were pelleted and resuspended in fresh media at 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mL−1 in 50% of the desired volume for protein expression, termed the transfection volume, and allowed to continue to grow overnight. Three hours prior to transfection, the cells were pelleted and resuspended in fresh media, lacking the desired amino acid(s) to be labeled (dropout media), at 80% of the transfection volume using a density that would equate to 2
000 mL−1 in 50% of the desired volume for protein expression, termed the transfection volume, and allowed to continue to grow overnight. Three hours prior to transfection, the cells were pelleted and resuspended in fresh media, lacking the desired amino acid(s) to be labeled (dropout media), at 80% of the transfection volume using a density that would equate to 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500
500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mL for the transfection volume. Thirty minutes prior to transfection, the plasmid DNA (1.5 mg L−1 of transfection volume) and PEI (4.5 mg L−1 transfection volume, Polysciences 24765) were diluted in separate tubes with dropout media to 1
000 cells per mL for the transfection volume. Thirty minutes prior to transfection, the plasmid DNA (1.5 mg L−1 of transfection volume) and PEI (4.5 mg L−1 transfection volume, Polysciences 24765) were diluted in separate tubes with dropout media to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20. The PEI was added to the plasmid DNA and incubated at room temperature for 20 min. The DNA/PEI complex was then added along with media containing the labeled amino acids and an additional 4 mM L-glutamine to the cell suspension. Cells were allowed to grow overnight (14 h) before being diluted 2-fold with media containing the labeled amino acids, an additional 4 mM L-glutamine and valproic acid (1 mg mL−1, Alfa Aesar A12962-18). The transfected cells were allowed to grow for 4–5 days before the conditioned media was harvested. Synthesized methyl-labeled amino acids, 13C-δ1-Ile and 13C-γ1,γ2-Val, were used at half the concentration of corresponding unlabeled amino acids. 13C-ε-Met was used at the full concentration. Unlabeled Alb-BGZPC was produced using a similar procedure, but with expi293 suspension cultured HEK293 cells (Invitrogen A14527) and expi293 medium (Invitrogen A1435101), as previously described.56
20. The PEI was added to the plasmid DNA and incubated at room temperature for 20 min. The DNA/PEI complex was then added along with media containing the labeled amino acids and an additional 4 mM L-glutamine to the cell suspension. Cells were allowed to grow overnight (14 h) before being diluted 2-fold with media containing the labeled amino acids, an additional 4 mM L-glutamine and valproic acid (1 mg mL−1, Alfa Aesar A12962-18). The transfected cells were allowed to grow for 4–5 days before the conditioned media was harvested. Synthesized methyl-labeled amino acids, 13C-δ1-Ile and 13C-γ1,γ2-Val, were used at half the concentration of corresponding unlabeled amino acids. 13C-ε-Met was used at the full concentration. Unlabeled Alb-BGZPC was produced using a similar procedure, but with expi293 suspension cultured HEK293 cells (Invitrogen A14527) and expi293 medium (Invitrogen A1435101), as previously described.56
Protein expression and isotope labeling in E. coli
Trx-BGZPC fusion protein isotopically labeled with 13C-δ1-Ile, 13C-γ1,γ2-Val, 13C-δ1,δ2-Leu, and 13C-ε-Met (ILVM) was produced by inoculating colonies from freshly transformed BL21(DE3) cells into water-based M9 medium and incubated at 37 °C with shaking until the cell density reached OD600 0.2–0.3. The cells were pelleted and resuspended in an equal volume of M9 medium that contained 99.9% 2H2O (Cambridge Isotope Laboratories DLM-4) and incubated at 37 °C with shaking until the cell density reached OD600 0.6–0.65. Afterward, per liter of medium, 75 mg 4-13C-3,3′-2H-α-ketobutyric acid (Cambridge Isotope Laboratories CDLM-7318), 200 mg 3-13C-3,4,4′,4′′-2H-α-ketovaleric acid (Cambridge Isotope Laboratories CDLM-7317), and 50 mg 13C-ε-methionine (Cambridge Isotope Laboratories CLM-206) were added. Protein expression was induced 1 h later by adding 200 mg solid IPTG (Gold Bio, I2481) per 1 L of culture. Cells were harvested 6 h post induction by centrifugation.Purification of BGZPC and R3like from mammalian cells
Conditioned media containing secreted protein was diluted 1.5× with buffer A (25 mM Na2HPO4, 150 mM NaCl, 6 mM imidazole, 1 mM NiSO4 pH 8.0) and loaded onto a previously equilibrated HisPur NiNTA column (Thermo A50584). Protein was eluted with imidazole gradient 6 mM–500 mM over 10 column volumes (CV). Fractions containing albumin-BGZPC or R3like were pooled together and dialyzed against 20 mM HEPES, 150 mM NaCl, and concentrated. To separate BGZPC from albumin, 3 units of thrombin per mg of protein was added and the mixture was incubated with stirring overnight at 4 °C. The released BGZPC, and R3like, were further purified on a Superdex 75 26/60 size exclusion chromotography column (Cytiva 28-9893-34) equilibrated with dialysis buffer.Purification of BGZPC from E. coli
Bacterial pellet from 6 liters culture of overexpressed Trx-BGZPC was resuspended in 300 mL of disruption buffer (100 mM Tris-HCl, 1 mM EDTA, pH 8.3) and sonicated in 50 mL batches (2.5 min, 500 W, 50% power, 50% duty cycle) on ice and centrifuged. The pellet, which contained the Trx-BGZPC as inclusion bodies, was washed sequentially, first by resuspending them in disruption buffer containing 0.5 M NaCl and then disruption buffer containing 1% Trition X-100. Inclusion bodies were then solubilized overnight in 200 mL 8 M urea, 25 mM Tris, 1 mM EDTA, reduced by addition of 30 mM DTT for 30 min, and acidified with glacial acetic acid to pH 4.5. Mixture was concentrated ten-fold, dialyzed against 8 M urea, 25 mM sodium acetate pH 4.5 and added dropwise to folding buffer (0.1 M Tris, 0.25 M guanidine HCl, 0.5 M L-arginine, 2 mM reduced glutathione (Millipore-Sigma G4251), 0.5 mM oxidized glutathione (Millipore-Sigma G4501), pH 8.0) at a protein concentration of 0.1 mg mL−1. After stirring for 36 h at 4 °C, the folding mixture was concentrated, dialyzed against 0.1 M Tris, 0.15 M L-arginine pH 8.0, and BGZPC was cleaved from thioredoxin with 3U of thrombin per mg of fusion protein overnight at 4 °C with stirring. The digestion mixture was dialyzed against 25 mM sodium acetate pH 4.2 and diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with 8 M urea. This mixture was loaded onto a 1 cm × 8 cm Source S cation exchange column (Cytiva 17094410) equilibrated in 25 mM sodium acetate, 4 M urea, pH 4.2 and eluted with a linear salt gradient (0–1 M NaCl over ten column volumes). Fractions containing natively folded BGZPC, as assessed by NMR 1H–15N HSQC spectra, were further purified from mixture using streptactin-XT column (IBA Lifesciences 2-5030-002).
1 with 8 M urea. This mixture was loaded onto a 1 cm × 8 cm Source S cation exchange column (Cytiva 17094410) equilibrated in 25 mM sodium acetate, 4 M urea, pH 4.2 and eluted with a linear salt gradient (0–1 M NaCl over ten column volumes). Fractions containing natively folded BGZPC, as assessed by NMR 1H–15N HSQC spectra, were further purified from mixture using streptactin-XT column (IBA Lifesciences 2-5030-002).
Protein NMR spectra
Samples of 13C-methyl labeled BGZPC or R3like were dialyzed into 25 mM phosphate, 25 mM NaCl, pH 7.0, concentrated to a final protein concentration between 20–50 μM, and after adding 2H2O to 5%, samples were transferred to 5 mm susceptibility-matched NMR microtubes (Millipore-Sigma Z529451). A sample of unlabeled BGZPC was prepared similarly, but at a concentration of 700 μM. All NMR spectra were recorded using a Bruker AVII 800 MHz instrument equipped with a cryogenically cooled 1H{13C,15N} 5 mm Z-gradient TCI cryoprobe, except for the BGZPC ILVM spectrum which was recorded with a similarly configured Bruker AVII 900 MHz instrument. All spectra were recorded with a 1H–13C HSQC pulse sequence with a WATERGATE solvent suppression scheme with 100 complex points and sweep width of 3623 Hz in the 13C-dimension (36.2 Hz per point), except for the BGZPC ILVM spectrum which was recorded with 113 complex points and sweep width of 4075 Hz in the in the 13C-dimension (36.2 Hz per point). All datasets were processed using NMRPipe57 and analyzed with the CcpNmr analysis software package.58Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge financial support from the National Institutes of Health (GM058670 and CA233622). This project has also received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie Grant Agreement No. 893196 to Ł. Wieteska.References
- M. Ikura, L. E. Kay and A. Bax, Biochemistry, 1990, 29, 4659–4667 CrossRef CAS PubMed.
- L. E. Kay, M. Ikura, R. Tschudin and A. Bax, J. Magn. Reson., 1990, 89, 496–514 CAS.
- G. M. Clore and A. M. Gronenborn, Curr. Opin. Chem. Biol., 1998, 2, 564–570 CrossRef CAS PubMed.
- G. M. Clore and A. M. Gronenborn, Trends Biotechnol., 1998, 16, 22–34 CrossRef CAS PubMed.
- R. S. Lipsitz and N. Tjandra, Annu. Rev. Biophys. Biomol. Struct., 2004, 33, 387–413 CrossRef CAS PubMed.
- N. Tjandra, Structure, 1999, 7, R205–R211 CrossRef CAS PubMed.
- K. H. Gardner and L. E. Kay, Annu. Rev. Biophys. Biomol. Struct., 1998, 27, 357–406 CrossRef CAS PubMed.
- T. Yamazaki, W. Lee, C. H. Arrowsmith, D. R. Muhandiram and L. E. Kay, J. Am. Chem. Soc., 1994, 116, 11655–11666 CrossRef CAS.
- K. Pervushin, R. Riek, G. Wider and K. Wuthrich, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 12366–12371 CrossRef CAS PubMed.
- V. Dotsch and G. Wagner, Curr. Opin. Struct. Biol., 1998, 8, 619–623 CrossRef CAS PubMed.
- A. R. Pickford and J. M. O'Leary, Isotopic labeling of recombinant proteins from the methylotrophic yeast pichia pastoris, in Protein NMR techniques, ed. A. K. Downing, Humana Press, Totowa, NJ, 2004, pp. 17–33 Search PubMed.
- T. Sugiki, O. Ichikawa, M. Miyazawa-Onami, I. Shimada and H. Takahashi, Isotopic labeling of heterologous proteins in the yeast pichia pastoris and kluyveromyces lactis, in Protein NMR techniques, ed. A. Shekhtman and D. S. Burz, 2012, Humana Press, Totowa, NJ, pp. 19–36 Search PubMed.
- N. K. Goto and L. E. Kay, Curr. Opin. Struct. Biol., 2000, 10, 585–592 CrossRef CAS PubMed.
- V. Tugarinov and L. E. Kay, ChemBioChem, 2005, 6, 1567–1577 CrossRef CAS PubMed.
- A. M. Ruschak and L. E. Kay, J. Biomol. NMR, 2010, 46, 75–87 CrossRef CAS PubMed.
- R. Kerfah, M. J. Plevin, R. Sounier, P. Gans and J. Boisbouvier, Curr. Opin. Struct. Biol., 2015, 32, 113–122 CrossRef CAS PubMed.
- P. Gans, O. Hamelin, R. Sounier, I. Ayala, M. A. Dura, C. D. Amero, M. Noirclerc-Savoye, B. Franzetti, M. J. Plevin and J. Boisbouvier, Angew. Chem., Int. Ed., 2010, 49, 1958–1962 CrossRef CAS PubMed.
- N. K. Goto, K. H. Gardner, G. A. Mueller, R. C. Willis and L. E. Kay, J. Biomol. NMR, 1999, 13, 369–374 CrossRef CAS PubMed.
- C. Amero, M. Asuncion Durá, M. Noirclerc-Savoye, A. Perollier, B. Gallet, M. J. Plevin, T. Vernet, B. Franzetti and J. Boisbouvier, J. Biomol. NMR, 2011, 50, 229–236 CrossRef CAS PubMed.
- F. A. Chao, J. Kim, Y. Xia, M. Milligan, N. Rowe and G. Veglia, J. Magn. Reson., 2014, 245, 17–23 CrossRef CAS PubMed.
- F. A. Chao, L. Shi, L. R. Masterson and G. Veglia, J. Magn. Reson., 2012, 214, 103–110 CrossRef CAS PubMed.
- M. C. Clay, T. Saleh, S. Kamatham, P. Rossi and C. G. Kalodimos, Structure, 2022, 30, 69–79 CrossRef CAS PubMed.
- Y. R. Monneau, P. Rossi, A. Bhaumik, C. Huang, Y. Jiang, T. Saleh, T. Xie, Q. Xing and C. G. Kalodimos, J. Biomol. NMR, 2017, 69, 215–227 CrossRef CAS PubMed.
- S. Nerli, V. S. De Paula, A. C. McShan and N. G. Sgourakis, Nat. Commun., 2021, 12, 691 CrossRef CAS PubMed.
- I. Pritisanac, M. T. Degiacomi, T. R. Alderson, M. G. Carneiro, E. Ab, G. Siegal and A. J. Baldwin, J. Am. Chem. Soc., 2017, 139, 9523–9533 CrossRef CAS PubMed.
- I. Pritisanac, J. M. Wurz, T. R. Alderson and P. Guntert, Nat. Commun., 2019, 10, 4922 CrossRef PubMed.
- Y. Xu, M. Liu, P. J. Simpson, R. Isaacson, E. Cota, J. Marchant, D. Yang, X. Zhang, P. Freemont and S. Matthews, J. Am. Chem. Soc., 2009, 131, 9480–9481 CrossRef CAS PubMed.
- Y. Xu and S. Matthews, J. Biomol. NMR, 2013, 55, 179–187 CrossRef CAS PubMed.
- G. Walsh, Nat. Biotechnol., 2018, 36, 1136–1145 CrossRef CAS PubMed.
- J. Trolle, R. M. McBee, A. Kaufman, S. Pinglay, H. Berger, S. German, L. Liu, M. J. Shen, X. Guo, J. A. Martin, M. E. Pacold, D. R. Jones, J. D. Boeke and H. H. Wang, eLife, 2022, 11, e72847 CrossRef PubMed.
- M. J. Rogals, J.-Y. Yang, R. V. Williams, K. W. Moremen, I. J. Amster and J. H. Prestegard, Glycobiology, 2021, 31, 425–435 CrossRef CAS PubMed.
- E. Tan, C. S. H. Chin, Z. F. S. Lim and S. K. Ng, Front. Bioeng. Biotechnol., 2021, 9, 796991 CrossRef PubMed.
- F. Bumbak, A. C. Keen, N. J. Gunn, P. R. Gooley, R. A. D. Bathgate and D. J. Scott, Biochim. Biophys. Acta, Biomembr., 2018, 1860, 1372–1383 CrossRef CAS PubMed.
- J. Goncalves, M. Eilers, K. South, C. A. Opefi, P. Laissue, P. J. Reeves and S. O. Smith, Chapter seventeen – magic angle spinning nuclear magnetic resonance spectroscopy of g protein-coupled receptors, in Methods in enzymology, ed. P. M. Conn, Academic Press, 2013, pp. 365–389 Search PubMed.
- J. H. Docherty, T. M. Lister, G. McArthur, M. T. Findlay, P. Domingo-Legarda, J. Kenyon, S. Choudhary and I. Larrosa, Chem. Rev., 2023, 123, 7692–7760 CrossRef CAS PubMed.
- B. Liu, A. M. Romine, C. Z. Rubel, K. M. Engle and B.-F. Shi, Chem. Rev., 2021, 121, 14957–15074 CrossRef CAS PubMed.
- M. Zhang, S. Zhong, Y. Peng, J. Jiang, Y. Zhao, C. Wan, Z. Zhang, R. Zhang and A. Q. Zhang, Org. Chem. Front., 2021, 8, 133–168 RSC.
- S.-Y. Zhang, Q. Li, G. He, W. A. Nack and G. Chen, J. Am. Chem. Soc., 2013, 135, 12135–12141 CrossRef CAS PubMed.
- H. Wang, Y. Park, Z. Bai, S. Chang, G. He and G. Chen, J. Am. Chem. Soc., 2019, 141, 7194–7201 CrossRef CAS PubMed.
- G. Liao, X.-S. Yin, K. Chen, Q. Zhang, S.-Q. Zhang and B.-F. Shi, Nat. Commun., 2016, 7, 12901 CrossRef PubMed.
- M. Borgini and P. Wipf, Tetrahedron, 2022, 120, 132876 CrossRef CAS.
- B. Wang, W. A. Nack, G. He, S.-Y. Zhang and G. Chen, Chem. Sci., 2014, 5, 3952–3957 RSC.
- L. S. Fitzgerald and M. L. O'Duill, Chem. – Eur. J., 2021, 27, 8411–8436 CrossRef CAS PubMed.
- Z. Zhang, X. Li, M. Song, Y. Wan, D. Zheng, G. Zhang and G. Chen, J. Org. Chem., 2019, 84, 12792–12799 CrossRef CAS PubMed.
- S. Yanaka, H. Yagi, R. Yogo, M. Yagi-Utsumi and K. Kato, J. Biomol. NMR, 2018, 71, 193–202 CrossRef CAS PubMed.
- S. K. Kim, M. J. Whitley, T. C. Krzysiak, C. S. Hinck, A. B. Taylor, C. Zwieb, C.-H. Byeon, X. Zhou, V. Mendoza, F. López-Casillas, W. Furey and A. P. Hinck, Structure, 2019, 27, 1427–1442 CrossRef CAS PubMed.
- S. J. Lin, Y. Hu, J. Zhu, T. K. Woodruff and T. S. Jardetzky, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 5232–5236 CrossRef CAS PubMed.
- E. Brule, Y. Wang, Y. Li, Y. F. Lin, X. Zhou, L. Ongaro, C. A. I. Alonso, E. R. S. Buddle, A. L. Schneyer, C. H. Byeon, C. S. Hinck, N. Mendelev, J. P. Russell, M. Cowan, U. Boehm, F. Ruf-Zamojski, M. Zamojski, C. L. Andoniadou, S. C. Sealfon, C. A. Harrison, K. L. Walton, A. P. Hinck and D. J. Bernard, Sci. Adv., 2021, 7, eabl4391 CrossRef CAS PubMed.
- S. K. Kim, M. A. Henen and A. P. Hinck, Exp. Biol. Med., 2019, 244, 1547–1558 CrossRef CAS PubMed.
- Y. Li, J. Fortin, L. Ongaro, X. Zhou, U. Boehm, A. Schneyer, D. J. Bernard and H. Y. Lin, Endocrinology, 2018, 159, 4077–4091 CrossRef CAS PubMed.
- M. A. Henen, P. Mahlawat, C. Zwieb, R. B. Kodali, C. S. Hinck, R. D. Hanna, T. C. Krzysiak, U. Ilangovan, K. E. Cano, G. Hinck, M. Vonberg, M. McCabe and A. P. Hinck, J. Biol. Chem., 2019, 294, 3065–3080 CrossRef CAS PubMed.
- S. J. Archer, A. Bax, A. B. Roberts, M. B. Sporn, Y. Ogawa, K. A. Piez, J. A. Weatherbee, M. L. Tsang, R. Lucas, B. L. Zheng, J. Wenker and D. A. Torchia, Biochemistry, 1993, 32, 1152–1163 CrossRef CAS PubMed.
- S. Sen, J. Das and D. Maiti, Tetrahedron Chem., 2022, 1, 100005 CrossRef.
- F.-F. Sheng, J.-G. Gu, K.-H. Liu and H.-H. Zhang, J. Org. Chem., 2022, 87, 16084–16089 CrossRef CAS PubMed.
- W. Zhu, A. J. Guseman, F. Bhinderwala, M. Lu, X. C. Su and A. M. Gronenborn, Angew. Chem., Int. Ed., 2022, 61, e202201097 CrossRef CAS PubMed.
- E. J. Goebel, R. A. Corpina, C. S. Hinck, M. Czepnik, R. Castonguay, R. Grenha, A. Boisvert, G. Miklossy, P. T. Fullerton, M. M. Matzuk, V. J. Idone, A. N. Economides, R. Kumar, A. P. Hinck and T. B. Thompson, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 15505–15513 CrossRef CAS PubMed.
- F. Delaglio, S. Grzesiek, G. W. Vuister, G. Zhu, J. Pfeifer and A. Bax, J. Biomol. NMR, 1995, 6, 277–293 CrossRef CAS PubMed.
- W. F. Vranken, W. Boucher, T. J. Stevens, R. H. Fogh, A. Pajon, M. Llinas, E. L. Ulrich, J. L. Markley, J. Ionides and E. D. Laue, Proteins, 2005, 59, 687–696 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ob01320k |
| ‡ Current address: Department of Chemistry and Biochemistry, Augusta University, Augusta, GA 30912, USA. |
| § Current address: Francis Crick Institute, London, NW1 1AT, UK. |
| This journal is © The Royal Society of Chemistry 2023 |