Graphene-based biosensors for detecting coronavirus: a brief review
Filimon Hadish
Abrha
*ab,
Tadele Hunde
Wondimu
bc,
Mebrahtu Hagos
Kahsay
 de,
Fetene
Fufa Bakare
de,
Fetene
Fufa Bakare
 bc,
Dinsefa Mensur
Andoshe
b and
Jung Yong
Kim
bc,
Dinsefa Mensur
Andoshe
b and
Jung Yong
Kim
 *bc
*bc
aDepartment of Chemistry, College of Natural and Computational Sciences, Aksum University, Aksum 1010, Ethiopia
bDepartment of Materials Science and Engineering, Adama Science and Technology University, Adama 1888, Ethiopia. E-mail: filimon.hadish@astu.edu.et; jungyong.kim@astu.edu.et
cCenter of Advanced Materials Science and Engineering, Adama Science and Technology University, Adama 1888, Ethiopia
dDepartment of Applied Chemistry, College of Natural and Computational Sciences, Mekelle University, Mekelle 231, Ethiopia
eDepartment of Applied Chemistry, Adama Science and Technology University, Adama 1888, Ethiopia
First published on 25th October 2023
Abstract
The coronavirus (SARS-CoV-2) disease has affected the globe with 770![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 437
437![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 327 confirmed cases, including about 6
327 confirmed cases, including about 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 956
956![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 deaths, according to the World Health Organization (WHO) as of September 2023. Hence, it is imperative to develop diagnostic technologies, such as a rapid cost-effective SARS-CoV-2 detection method. A typical biosensor enables biomolecule detection with an appropriate transducer by generating a measurable signal from the sample. Graphene can be employed as a component for ultrasensitive and selective biosensors based on its physical, optical, and electrochemical properties. Herein, we briefly review graphene-based electrochemical, field-effect transistor (FET), and surface plasmon biosensors for detecting the SARS-CoV-2 target. In addition, details on the surface modification, immobilization, sensitivity and limit of detection (LOD) of all three sensors with regard to SARS-CoV-2 were reported. Finally, the point-of-care (POC) detection of SARS-CoV-2 using a portable smartphone and a wearable watch is a current topic of interest.
900 deaths, according to the World Health Organization (WHO) as of September 2023. Hence, it is imperative to develop diagnostic technologies, such as a rapid cost-effective SARS-CoV-2 detection method. A typical biosensor enables biomolecule detection with an appropriate transducer by generating a measurable signal from the sample. Graphene can be employed as a component for ultrasensitive and selective biosensors based on its physical, optical, and electrochemical properties. Herein, we briefly review graphene-based electrochemical, field-effect transistor (FET), and surface plasmon biosensors for detecting the SARS-CoV-2 target. In addition, details on the surface modification, immobilization, sensitivity and limit of detection (LOD) of all three sensors with regard to SARS-CoV-2 were reported. Finally, the point-of-care (POC) detection of SARS-CoV-2 using a portable smartphone and a wearable watch is a current topic of interest.
Introduction
The coronavirus (COVID-19) pandemic has infected several billion people, causing over a million deaths worldwide, and the economic fallout has resulted in a global panic.1 The severe acute respiratory syndrome (SARS)-associated coronavirus pandemic has not only led to an unexpected public health crisis but also severely hampered the global economy, with analysts measuring its detrimental impact compared to the Second World War.2 Moreover, it affects birds and mammals3,4 and usually causes mild respiratory diseases in humans. However, strains have emerged, such as 2002–2004 SARS and Middle East respiratory syndrome (MERS), causing outbreaks of this lethal respiratory disease.3 The causative agent SARS-CoV-2 indicates coronavirus disease 2019 (COVID-19). In addition to SARS and MERS, swine acute diarrhea syndrome (SADS) harmed thousands of human lives in 2017. SADS agents are known to originate from bats. Its viruses are highly pathogenic to humans and livestock.5 Therefore, as these rapidly spreading viruses continue to increase, quick diagnostic techniques are critical to reduce and prevent the spread and transmission of these viruses.Thus far, diagnostic techniques, such as clustered regularly interspaced short palindromic repeats (CRISPR),6 reverse transcription polymerase chain reaction (RT-PCR), lateral flow assays (LFA), lateral flow immunoassay, and reverse transcription loop-mediated isothermal amplification (RT-LAMP), have been developed as detection methods for COVID-19. Although these methods exhibit superior analytical sensitivity, they have disadvantages because they require skilled personnel to operate complex analytical instruments and they are highly susceptible to other interference materials, resulting in either false positives or false negatives. Moreover, the aforementioned methods are incompatible with point-of-care testing, which poses a challenge to their portability.7
To improve these diagnostic methods, real-time, ultrafast, and low-cost sensors have been studied and developed.8,9 For example, sensors that incorporate nanomaterials, such as carbon nanotubes, multiwall carbon nanotubes, carbon dots, graphene, and transition metal dichalcogenides (TDMs: WS molybdenum disulfide, MoS2 molybdenum diselenide, WS2 tungsten disulfide and WSe2 tungsten diselenide), have been investigated.10 In particular, the latter nanomaterials have been widely studied owing to their applications in energy storage, catalyst, nanomedicine, optics and electronics.11 However, most of these materials contain elements that are toxic to the environment and human beings. Among the elements commonly used in the synthesis of MXenes, prolonged exposure to chromium, tungsten, and molybdenum could result in physical, muscular, and neurological degenerative diseases.12 Although these elements are essential for biochemical and physiological purposes,13 it would be very difficult to apply them to implantable sensors. This is because excessive ingestion of molybdenum can lead to serious bone and joint deformity, liver and kidney failure, and sterility.14 Beyond safety precaution synthesis of high quality, continuous, and spatially uniform single-layer TDMs, MXene is a challenging material that poses a large initial resistance15 unlike graphene, which has been in the limelight of research for over two decades. Therefore, considering the safety, ease of synthesis, bio-functionalization16–18 and biocompatibility,19,20 graphene is a well-studied material for detecting coronavirus, among the other nanomaterials.21
The biosensor is mainly composed of three parts: the biological detection component, transducer, and processing unit.22,23 Through this mini-review, we mainly discuss graphene-based biosensors, such as electrochemical, field-effect transistor (FET) and surface plasmon resonance (SPR) sensors (Scheme 1). Notably, although FET could be considered an electrochemical sensor if there is electrochemical doping,24,25 instead of an electrostatic sensor, we reviewed it separately based on the particular device structure and physics. Beginning with a general overview of the different variants (families of coronavirus), these biosensors are briefly covered, focusing on recent progress along with fabrication processes and the performance of the devices. Finally, we summarize the results accompanying our perspectives.
SARS-CoV-2
Thus far, alpha, beta, gamma, delta, and omicron (α, β, δ, γ, and o) variants of coronaviruses that mutate over time were identified.26 Although a family of the virus belonging to α-/β-variants infects mammals, including humans, the γ-/δ-variants mainly transmit infections to birds. Here, α- and β-variants correspond to severe acute respiratory syndrome coronavirus (SARS-CoV-2) and middle-east respiratory syndrome coronavirus (MERS-CoV), respectively. Specifically, SARS-CoV-2 belongs to the nidovirales family of coronaviridae, resulting in the main cause of mortality.27 As shown in Fig. 1a, the structure of SARS-CoV-2 is composed of nucleocapsid (N), membrane (M), envelope (E), ssRNA (+), and spikes (S1 & S2), among which the protein spikes serve as a binding site and fuse to the host cell receptor.28 Here, the SARS-CoV-2 name itself is known to originate from the typical spike (S) proteins that are ∼20 nm long and scarcely embedded in the lipid bilayer membrane. The epitope or antigenic determinant is a group of amino acids or other chemical groups29,30 known as the receptor-binding domain (RBD) and receptor-binding motif (RBM) of SARS-CoV-2. They stay on top of the S proteins with 25 nm space apart and enable the virus to interact with the angiotensin converting enzyme 2 (ACE2) receptor of the host cell.31 Specifically, RBD serves as the primary target antigen for neutralizing antibodies.32 | ||
| Fig. 1 (a) Schematic of the SARS-CoV-2 virus structure.28 (b) Immobilization of SARS-CoV-2 at the functionalized GO surface, subsequently followed by reduction of S and N proteins of SARS-CoV-2 virus (reprinted with permission;35 Copyright © 2021, American Chemical Society). | ||
Mutation of SARS-CoV-2 to a different variant is induced in its spike proteins, occurring mostly in the RBD part. The mutation allows the virus to remain immune to different vaccination trials for a significant time.33,34 However, the epitope sitting on the spike S protein is a large group of amino acids. Whether these amino acids belong to primary, secondary or tertiary structures, their terminal chemical compounds are rich in amide, carboxyl, amine, sulfide, and many other functional groups. These moieties are known to be favorable for sharing electrons through covalent bonds with the functionalized graphene surface. This indicates that SARS-CoV-2 can be easily detected in graphene-based biosensors. However, the bio-functionalization process (specific antibody attachment) on top of the 2D-graphene surface should be helpful for high-accuracy SARS-CoV-2 viral target biosensor applications through antibody–antigen complex formation, resulting in high sensitivity and selectivity.
Graphene and graphene oxide
Graphene is a 2D honeycomb crystal lattice with sp2pz hybridized carbon, which contains two atoms per unit cell. Here, the electrons in sp2 orbital form three covalent σ-bonds although the π-electrons in pz-orbitals are non-bonded and delocalized in a single flat layer of carbon atoms. Hence, the 2D delocalized π-orbitals show a conical shape in the electronic structure where the conduction and valence bands touch at the Diract point, resulting in a zero energy gap.36,37 Over the past two decades, graphene has played a crucial role in the field of smart materials owing to its unique electrical, optical, thermal, and mechanical properties.38–43 However, limitations, such as zero bandgap44 and non-emissive surface,45 hinder further applications of graphene in the pristine state. Therefore, the surface of graphene has been usually modified through chemical functionalization,46 substitutional doping,47–49 microwave irradiation,50,51 downstream oxygen plasma,52 hydrothermal method, and liquid exfoliation.53 In this circumstance, it should be very important to understand the chemical/physical attachment of various atoms and molecules to the surface of the π-conjugated graphene.54 In the case of non-covalent bonding, such as π-orbital interactions (e.g., π–π, H–π, gas–π, and cation/anion–π), the binding energies could be electrostatic, dispersion, and induction.18 However, these intermolecular interactions depend on the physical state (e.g., stabilization or destabilization) of the graphene surface, indicating that it is not sustainable but transient. However, the covalent functionalization of graphene surfaces with oxygen is an important chemical approach to surface engineering.55 For example, graphene oxide (GO) has a versatile surface with functional groups, such as hydroxyl, carboxyl, epoxide, and carbonyl,56,57 which are useful for the covalent immobilization of bio-receptors.58,59 Consequently, graphene-based biosensors have been used for detecting glucose (diabetes),52,60,61 nucleic acid (DNA/RNA),62–64 and various pathogenic bacteria/viruses.65–69Intermolecular interactions between graphene derivative and SARS-CoV-2
A well-designed graphene surface can have a binding function similar to that of a target molecule. Fukuda et al.35 investigated the antiviral effect of GO nano-sheets on SARS-CoV-2. Here, the plus-charged spikes of the virus were attracted to the minus-charged GO surface. As shown in Fig. 1b, SARS-CoV-2 proteins were decomposed through adsorption and concomitant inactivation on top of the GO surface. Importantly, graphene has a significant capacity to capture viral antibodies to its 2D surface in addition to the aforementioned antibacterial and antiviral properties. Moreover, functionalized graphene can effectively detect a viral-structured protein as a target, enabling a large-scale diagnosis for public health. Accordingly, graphene-based biosensors are suitable for SARS-CoV-2 detection, as emphasized by Yasri and Wiwanitkit.70 The biofunctionalization of graphene-based devices affords the enhancement of biocompatibility and bio-recognition ability with high sensitivity and selectivity. Therefore, the direct immobilization of the virus on top of the GO or graphene surface clearly highlights the potential of graphene-based SARS-CoV-2 biosensors. Table 1 displays the versatile graphene-based SARS-CoV-2 biosensors using the detection method, sample type, and limit of detection (LOD).| Graphene modification | Method of detection | Target/analyte | Sample type | LOD | Ref. |
|---|---|---|---|---|---|
| PBSAE = 1-pyrenebutyric acid N-hydroxysuccinimide ester, G = graphene; MXene = 2D transition-metal carbides; Ti2C = Di-titanium carbide; SWCNTs = single wall carbon nanotube; PI = polyimide; LSG = laser-scribed graphene; AuNS = gold nanostructures; Cys = cysteamine; EDC = 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide; NHS = N-hydroxy succinimide; ZnS = Zinc sulfide; GCE = glassy carbon electrode; PMB = poly methylene blue; PILs = poly ionic liquids, rGO = reduced graphene oxide; AuNPs = gold nanoparticles; RIU = refractive index unit, WS2 = tungsten disulfide; KNbO3 = potassium niobate; BP = black phosphorus; Blue P = blue phosphorus; UTM = universal transport medium. | |||||
| PBASE/G/SiO2/Si | FET | SARS-CoV-2 antigen | Nasopharyngeal swab specimens in UTM | 1 and 100 fg mL−1 | 71 |
| MXene (Ti2C)/G/SiO2/Si | FET | SARS-CoV-2 antigen | Artificial saliva and PBS | 1 fg mL−1 | 72 |
| AgGO/SiO2/Si | FET | SARS-CoV-2 antigen | PBS | 2.1 × 10−18 M | 73 |
| GO/Pt/PdNp/SiO2/Si | FET biosensor | SARS-CoV-2 antigen | PBS | 1 fg mL−1 | 74 |
| PBASE/G/SiO2/Si | Surface plasmon sensor | SARS-CoV-2 antigen | Artificial saliva, buffer | 1 and 3.75 fg mL−1 | 75 |
PI/LSG/AuNS/Cys/EDC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NHS NHS |
Electrochemical biosensor | SARS-CoV-2 antigen | Blood serum in PBS | 2.9 ng mL−1 | 76 |
| Au/Ti/EG/6H-SiC | Electrical transduction | SARS-CoV-2 antigen | Mid-turbinate swabs, saliva and exhaled breath aerosol samples in PBS | 1 ag mL−1 | 77 |
| ZnS/G/GCE | Electrochemical biosensor | SARS-CoV-2 nucleic acids | Hybridized DNA in 1.0 M KCl and 0.2 M K 3[Fe(CN)6] solutions | 2.1 × 10−20 M | 78 |
| G/Au/MgF2 | Surface plasmon sensor | Ethyl butanoate in SARS-CoV-2 | Exhaled breath | 0.0224 RIU | 79 |
| PMB/PILs/rGO/AuNPs | Miniature electrochemical biosensor (acupuncher) | SARS-CoV-2 | PBS | 38 pg mL−1 | 80 |
| G/paper | Electrochemical biosensor | Anti-SARS-CoV-2 monoclonal antibody CR3022 | RBD in PBS | 2 fg mL−1 | 81 |
Electrochemical biosensors
Electrochemical biosensors are suitable for ultrasensitive point-of-care (POC) SARS-CoV-2 detection at the time and place of patient care. They are cost-effective and simple for rapid and high-sensitivity detection.82 Basically, electrochemical sensors rely on the measurement of an electrical signal recorded by an electrochemical transducer. They can be categorized as amperometric, potentiometric, conductometric, and impedimetric depending on the type of signal.83 These biosensors have been widely used to detect biomaterials, such as nucleic acids, proteins, small molecular antibodies, and viruses.84For instance, Pan and coworkers developed rapid (less than 5 min), ultrasensitive, and quantitative detection of SARS-CoV–2 using an antisense oligonucleotide-directed electrochemical biosensor chip, affording a sensitivity of 231 (copies per μL)−1 and LOD of 6.9 copies per μL (Fig. 2). Fig. 2a shows the schematic configuration of the biosensor, targeting viral nucleocapsid phosphoprotein (N-gene) using highly specific antisense oligonucleotides (short single-stranded deoxyribonucleic acid; ssDNA) on top of the gold nanoparticle (AuNP)-modified graphene/paper platform. Fig. 2b shows the operation principle of this electrochemical biosensor. First, the infected samples are collected from the nasal swab or saliva of the patients with COVID-19. Second, the viral SARS-CoV-2 ribonucleic acid (RNA) is extracted from the samples. Third, viral RNA is added on top of the ssDNA-AuNP/graphene/paper platform. Fourth, wait 5 min for the incubation time. Finally, the electrical signals (voltage and/or current) are recorded. Fig. 2c shows two kinds of configurations without and with AuNPs on graphene/paper substrate. As shown in Fig. 2d, when AuNPs were employed, the sensing signal (here, the voltage difference) was much stronger, indicating that AuNPs are very useful components of electrochemical biosensors. Fig. 2e shows the positive/negative decision method using the complexation of ssDNA on AuNPs with the viral RNA. Importantly, the sensing signal is also dependent on the particle size of AuNPs, as shown in Fig. 2f, indicating that the large surface area of AuNPs capped with ssDNA (the immobilized recognizing element) is a key factor for the effective sensing of viral RNA.
 | ||
| Fig. 2 (a) Structure of the electrochemical biosensor. (b) Operation principle of the electrochemical biosensor. (c) Schematic representation of the proposed concept behind the enhancement in electrochemical response using gold nanoparticles when capped with ssDNA probes. (d) Relative change in the biosensor output voltage for the two sensor configurations. (e) Decision of Covid-19 negative and positive. (f) Sensor output response with varying sizes of functionalized spherical gold nanoparticles. Reprinted with permission from ACS Nano.85 | ||
A resistive model was employed to explain the above sensing platform, in which the change in voltage (ΔV) across the electrochemical sensor chip is given by
 | (1) |
Recently, the laser-scribed graphene (LSG)-based biosensor has gained significant attention as a miniaturized electrochemical POC immunoassay.86 Underpinning the evolution, Beduk et al.76 established an LSG biosensor fabricated through the electrodeposited 3-dimensional gold nanostructures (AuNSs) on top of the LSG for detecting SARS-CoV-2. As shown in Fig. 3, this electrochemical immunosensor can be integrated into a smartphone (Fig. 3a) or wearable watch (Fig. 3b), which can provide a user friendly POC diagnostic platform. Specifically, the second-generation custom with the brand name of ‘KAUSTat’ (Fig. 3a) has a poly-potentiostat device composed of built-in memory, battery, bluetooth, mini-USB connector, SD-card slot, and connectable add-on device, which affords multiple amperometric and voltammetric measurements.76
 | ||
Fig. 3 (a) Portable handmade POC potentiostat connected to a smartphone via a USB-C connection to record the signal. (b) Representation of KAUSTat used as a standalone watch-like device showing (i) LED on and (ii) LED off for indication. DPVs of the LSG/AuNS immunosensor presenting ΔIox (the change in oxidation current) after each experimental step and the measurement of 100 ng mL−1 SARS-CoV-2 spike protein by (c) PalmSens potentiostat and (d) portable handmade POC potentiostat. (e) Response of the LSG/AuNS immunosensor in the presence of SARS-CoV-2, IgM/IgG (7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), IgM, IgG/IgM (3 3), IgM, IgG/IgM (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7), IgG/IgM, and IgG (p value > 0.05). SARS-CoV-2 protein (150 ng mL−1) and interfering proteins (immunoglobins, IgG and IgM) were used for the selectivity test. (f) Proposed LSG/AuNS immunosensor in response to different concentrations of the SARS-CoV-2 spike protein vs. the LSG reference electrode. (g) Representation of SARS-CoV-2 detection in human blood serum. (Reprinted from76 by the grant of the PMC Open Access Subset from Analytical Chemistry.) 7), IgG/IgM, and IgG (p value > 0.05). SARS-CoV-2 protein (150 ng mL−1) and interfering proteins (immunoglobins, IgG and IgM) were used for the selectivity test. (f) Proposed LSG/AuNS immunosensor in response to different concentrations of the SARS-CoV-2 spike protein vs. the LSG reference electrode. (g) Representation of SARS-CoV-2 detection in human blood serum. (Reprinted from76 by the grant of the PMC Open Access Subset from Analytical Chemistry.) | ||
The differential pulse voltammetry (DPV) (Fig. 3c and d) response for various stages of the fabricated electrode showed a significant increase in the oxidation current density by electrodepositing AuNPs and cysteamine onto the LSG electrode. This increase was attributed to the high conductivity (owing to the surface area and catalytic activity of the AuNPs) and the polarizable ammonium groups on the electrode. Although the presence of graphene in the fabricated electrode increases electron transfer, the bulk structures of antibody-analyte (SARS-CoV-2) binding may partially hinder the electron transfer process at the surface of the electrode, leading to a decrease in the oxidation current (Fig. 3c). However, it is notable that the performance of the portable handmade POC potentiostat (Fig. 3d) showed a comparable function compared to the commercially available PalmSens electrochemical workstation (Fig. 3c).
Fig. 3e shows the response of this electrochemical immunosensor in the presence of SARS-CoV-2, IgM/IgG (7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), IgM, IgG/IgM (3
3), IgM, IgG/IgM (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7), IgG/IgM, and IgG (p value > 0.05), for which SARS-CoV-2 protein (150 ng mL−1) and interfering proteins (immunoglobins, IgG and IgM) were used for the selectivity test. The electrochemical tests carried out using the DPV method (Fig. 3f) for a wide range of SARS-CoV-2 S1 spike proteins confirmed that the device can detect low concentrations and achieve an LOD of 2.9 ng mL−1, as depicted in the inset in Fig. 5f. Moreover, the clinical RT-PCR positive and negative serum samples at 10 and 25% dilution provided an LOD of 3.8 and 8 ng mL−1 as well as 16 and 10 ng mL−1, respectively. Then, the authors suggested that this kind of electrochemical biosensor (see the schematic explanation in Fig. 3g) offers a portable, easy-to-use, and reliable POC diagnostic alternative platform for future applications.76
7), IgG/IgM, and IgG (p value > 0.05), for which SARS-CoV-2 protein (150 ng mL−1) and interfering proteins (immunoglobins, IgG and IgM) were used for the selectivity test. The electrochemical tests carried out using the DPV method (Fig. 3f) for a wide range of SARS-CoV-2 S1 spike proteins confirmed that the device can detect low concentrations and achieve an LOD of 2.9 ng mL−1, as depicted in the inset in Fig. 5f. Moreover, the clinical RT-PCR positive and negative serum samples at 10 and 25% dilution provided an LOD of 3.8 and 8 ng mL−1 as well as 16 and 10 ng mL−1, respectively. Then, the authors suggested that this kind of electrochemical biosensor (see the schematic explanation in Fig. 3g) offers a portable, easy-to-use, and reliable POC diagnostic alternative platform for future applications.76
Kim and coworkers77 reported a real-time ultra-sensitive detection of SARS-CoV-2 using a quasi-free-standing epitaxial graphene-based biosensor. Despite simple and low-cost fabrication techniques, this biosensor can rapidly detect both the mid-turbinate swabs and the exhaled breath aerosol samples of a SARS-CoV-2 infected patient with an LOD of 1 ag mL−1 (a spike protein antigen concentration). Interestingly, the potential application of a portable device was demonstrated by a fast response time of 0.6 sec to a human saliva sample in real-time detection. Another competent candidate among the graphene-based SARS-CoV-2 electrochemical biosensors is a miniaturized stainless steel acupuncture needle reported by Yang et al.80 Compared to other electrochemical sensors, the small size and pinpoint nature of the acupuncture needle electrode was produced by successive deposition of AuNPs, rGO, poly(methylene blue) (PMB), and poly(ionic liquid)s. The specificity and stability of the sensor were tested using versatile biomolecules, such as bovine serum albumin (BSA), immunoglobulin G (IgG), immunoglobulin M (IgM), glutathione (GSH), and L-tryptophan (L-Trp), along with SARS-CoV-2-S protein for about 15 days. The same value after 7 days and 76% retention after 15 days indicate that the developed biosensor has high specificity and stability. Finally, a 38 pg mL−1 LOD is the lowest sensitivity compared to the molecularly imprinted biosensors prior to January 2023. Jaewjaroenwattana et al.81 reported a graphene ink printed on a disposable paper substrate as an electrochemical platform for the detection of SARS-CoV-2. Surprisingly, the three-electrode system sensor incorporated a cheap Whatman filter paper and a cost-effective plant-based anti-SARS-CoV-2 monoclonal antibody. The antibody was directly immobilized on the graphene-inked working electrode surface through covalent bonding. The electrochemical detection of the spike protein of SARS-CoV-2 was performed using a DPV. Consequently, the screen-printed sensor produced 2.0 fg mL−1 LOD with high specificity. Similarly, a metal graphene hybridized (e.g. ZnS/G) ultrasensitive electrochemical biosensor for the rapid detection of SARS-COV-2 was developed by Sarwar et al.78 The nanocomposite sensor was fabricated by employing a step microwave-based non-equilibrium heating route on the glassy carbon electrode. Based on the previous electrochemical measurements in the testbed of biosensors, the LOD was 4.453 × 10–20 M for the synthetic DNA and 4.453 × 10–20 M for the SARS-CoV-2 standard, respectively.
Field-effect transistor (FET) biosensors
Field effect transistor (FET) biosensors are very widely used, highly sensitive,70 ease to handle, and robust87,88 compared to other techniques in the detection of the SARS-CoV-2 pandemic/epidemic. Versatile FETs, such as metal oxide semiconductor FET,89 metal nanowire FET,90 and disposable modular transistors,91,92 have been reported as effective biosensors for the detection of SARS-CoV-2. Among these FETs, the graphene-based FET biosensors exhibited excellent electro-catalytic activity for high-sensitivity detection attributed to the unique physical, chemical, and electrical properties of graphene materials.82,93 Particularly, the graphene-functionalized surface provides a suitable antibody–antigen immobilization environment and builds an amplified capacitance between bio-receptors and transducers.72,94,95 Convincingly, the immobilized single-stranded DNA (ssDNA) on the graphene surface serves as a macromolecular antibody for detecting the SARS-COV-2 antigen. These interactions between the ssDNA-functionalized graphene and the SARS-COV-2 RNA may shift in Dirac voltage (i.e., the minimum current point in the plot of IDSvs. VG, where IDS is the drain current and VG is the gate voltage) during the operation of graphene-based FET.96 In addition to the experimental advancement, Toral-Lopez et al.97 proposed a multi-scale simulation approach on graphene-based biological (Bio) FET. The author was motivated to carry out the study from the perspective that the Bio-FET takes advantage of other devices because the gate metal can detect minute amounts of ions or biomolecules. During the simulation, a single-layered graphene channel was mounted on top of the Si/SiO2 layers, and then the complex formation between the human Angiotensin Converting Enzyme 2 (ACE2) and the virus spike protein S1 receptor binder domain (S1RBD) was investigated as a function of the Bio-FET. Employing a self-consistent simulation, the quantitative and qualitative sensitivity variation across different charge distribution zones (Dirac voltage, p-branch and n-branch) was observed by detecting ACE2-S1RBD using the graphene-based Bio-FET. The maximum current output from an n-branch belongs to the positive bias voltage in the Bio-FET.97 However, both the utilization of unfunctionalized pristine graphene and the inconsiderate incubation condition of the virus are the partial limitations of this theoretical study.Fig. 4a displays the schematic DNA aptamer-conjugated graphene-based FET sensor platform for the ultrasensitive detection of SARS-CoV-2 and its variants using patient samples.99 As shown in Fig. 4b, when the SARS-CoV-2 virus was detected by the antibody of FET, the Dirac voltage (p- to n-channel transition point) is shifted to the right direction in the IDSvs. VG transfer curve, indicating that the electron concentrations are reduced at the graphene-sample fluid interface through the binding between antibody (DNA aptamer) and antigen (SARS-CoV-2). Fig. 3c shows the output characteristics (IDSvs. VDS, where VDS is the drain voltage) of the graphene-based FET with an increasing negative voltage of VG (i.e., hole induction at the p-channel). When VG = 0, there is a linear increase in IDS as VDS increases, indicating that there are free carriers at the graphene-sample interface (due to the zero bandgap effect). The shift of Dirac voltage (ΔVG) was tested for the different variants of SARS-CoV-2. As a result, Fig. 4e demonstrates that ΔVG is dependent on the species of the virus variant. Finally, Fig. 4f summarizes the sensing mechanism of graphene-based FET biosensors.99 However, it is notable that graphene (zero bandgap semiconductor) exhibits a special behavior when employed as an FET channel layer. It shows a transition from the p-channel to the n-channel, i.e., ambipolar behavior,100,101 as shown in Fig. 4b. In addition, the output characteristic curve does not show a saturation but rather a linear increase even at VG = 0 (Fig. 4c), indicating that the functionalized graphene is in a charge-doped state. Comparatively, it is notable that the organic semiconductor (e.g., π-conjugated small molecules and polymers) shows a threshold voltage (Vth) in the transfer curve after filling the trap states at the semiconductor-insulator interface.102,103 In addition, the charge mobility (μ) could be easily estimated using a FET testbed according to the following equations:
 | (2) |
 | (3) |
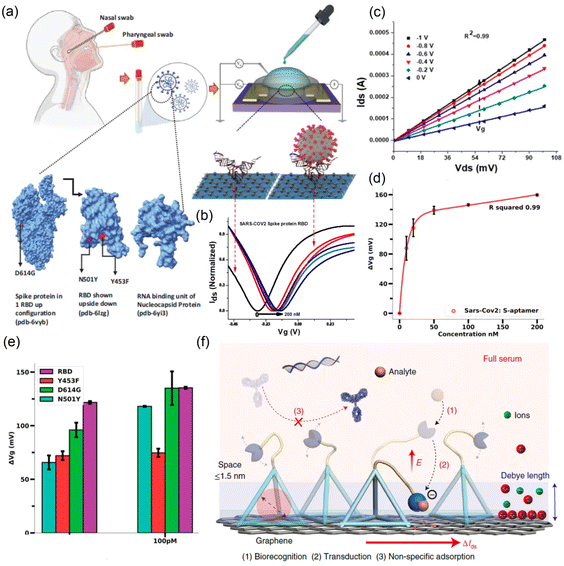 | ||
| Fig. 4 (a) Schematic aptamer graphene-based FET sensor diagnostic system for ultrasensitive detection of SARS-CoV-2 using patient samples. (b) SARS-CoV-2 receptor-binding domain-mediated transfer curve analysis of Aptamer-S derivatized graphene-based FET. (c) Output characteristics (IDSvs. VDS) of graphene FET derivatized using 1-pyrenebutyric acid N-hydroxysuccinimide ester (PBASE, 5 mM) were analyzed by sweeping VGS from 0 to 1 V at 0.2 V steps and variable drain source voltage (VDS) of 0 to 100 mV with incremental steps of 10 mV. (d) Receptor-binding domain concentration (0 to 200 nM)-dependent sensor response on the Aptamer-S-derivatized FET. (e) Aptamer-S-derivatized graphene-based FET sensors were able to detect the B.1.1.7 variant (N501Y), mink-related mutation (Y453F), and mutation at the S2 domain (D614G) of SARS-Cov-2. (Reprinted98 from PNAS). (f) Sensing mechanism of graphene-based FET biosensors. Owing to their high coverage of the surface, rigid bases avoid non-specific adsorption. The probes on the flexible cantilevers specifically recognize the targets. During electrostatic actuation, recognition events are detected in the g-FET channel, leading to efficient biorecognition and signal transduction. (Reprinted99 from Nature Biomedical Engineering.) | ||
Interestingly, Seo et al..71 fabricated a FET-based biosensor in which the SARS-CoV-2 spike ‘antibody’ is functionalized to a graphene sheet. The FET device could rapidly detect the cultured and nasopharyngeal swab specimens from SARS-CoV-2 patients at low concentrations of 1 fg mL−1 in PBS. Further, the FET biosensor detected SARS-CoV-2 in both culture medium and clinical samples with LOD: 1.6 × 101 plaque-forming unit (pfu) mL−1 and 2.42 × 102 copies per mL, respectively.71 Another real-time SARS-CoV-2 detection via GO-based FET biosensors decorated with bimetallic platinum and palladium (Pt/Pd) nanoparticles was reported by Wasfi et al.74 The authors underlined that the ‘magnetic’ spike antibody is useful to increase the sensitivity of SARS-CoV-2 antigen detection. Here, the FET device with the GO channel creates a suitable surface for the direct immobilization of specific antibodies against the SARS-CoV-2 spike protein. The high sensitivity with an LOD of up to 1 fg mL−1 towards the SARS-CoV-2 antigen in phosphate buffered saline (PBS) indicates the high performance of this FET biosensor for SARS-CoV-2 diagnosis.
Surface plasmon resonance biosensors
The surface plasmon resonance (SPR) sensor is an optical sensor,104,105 affording rapid detection, accuracy, high sensitivity, and portability.106,107 The light source108 and geometric configuration109 are the governing criteria for SPR sensors. Commonly, the performances of SPR sensors are evaluated using the following equations: | (4) |
 | (5) |
 | (6) |
For example, to increase light absorption for the detection of ultra-low SARS-COV-2, Hossain and Talukder113 reported the analytical and numerical calculations of graphene-based SPR sensors by integrating them with tungsten disulfide (WS2), potassium niobate (KNbO3), black phosphorus (BP), and blue phosphorus (BlueP). In the Kretschmann configuration,104 the positions of the incident light source, reflection and transmission detection planes for sensitivity characterization were simulated using a finite difference time domain (FDTD) technique. The sensor loaded with ACE-2 antibody on top of the plasmon sensor was in contact with a 1.70 nm thick graphene for both maximizing FWHM and reducing the R-profile. The designed sensor having stable binding kinetics towards loaded SARS-CoV-2 reached a LOD of 1 fM concentration (approximately 6 × 105 molecules) with a sensitivity of 201 degrees per RIU and 140 RIU-1 FOM. According to Hossain and Talukder,113 the sensors could be useful for detecting SARS-CoV-2 and other biological samples. Similarly, Payandehpeyman et al.114 developed graphene-based nanomechanical resonator sensors (i.e., detecting a change in the mechanical response of the system when a foreign SARS-CoV-2 object is added) based on antibody–antigen interactions. For this simulation, the variables were the single-layer graphene sheet (SLGS) size, aspect ratio and boundary conditions, antibody concentration, and virus number variation. Accordingly, they measured the frequency shift and relative frequency. The result confirms that the simulated biosensor can detect SARS-CoV-2 with LOD 10 virus/test with high quality.114 Captivatingly, the plasmonic-graphene biosensor with a four-layer configuration (Al, Au, SiO2, and graphene) can detect SARS-CoV-2 with a sensitivity of 664.8 nm RIU−1 according to Negahdar et al.'s work.115 In the case of Taya et al.'s contribution,116 their SPR sensor is composed of three layers: Ag, BiFeO3, and graphene. Here, the insertion of BiFeO3 into the layered structure affords not only a high refractive index and low loss but also an extremely high sensitivity of 293 deg RIU−1.
Du et al.117 developed a tunable plasmonic biosensor using a 2D-twisted bilayer graphene superlattice on top of the plasmonic Au layer (Fig. 5a) that can produce an ultralow reflectivity of 2.2038 × 10–9 and large Goos–Hänchen (GH) shift of 4.4785 × 104 μm. For a fixed RI variation of 0.0012 RIU, the device with 44 nm AU and 55.3° twisted bilayer graphene could produce a high GH sensitivity of 3.9570 × 107 μm RIU−1, affording the detection of 2.0 nM SARS-CoV-2 via a linear variation of 3 × 10–7 RIU.
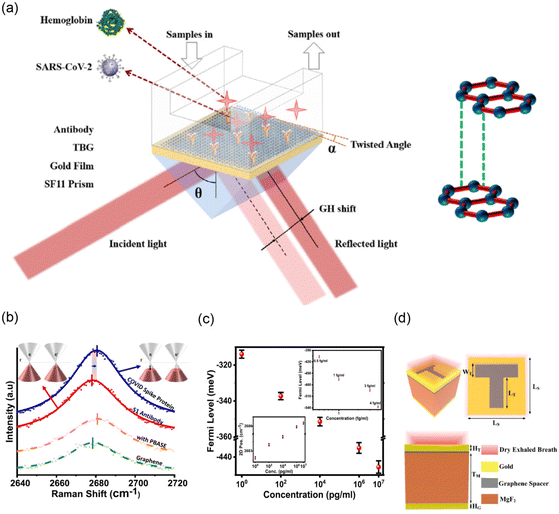 | ||
| Fig. 5 (a) Schematic diagram of twisted bilayer graphene (TBG)-enhanced SPR biosensor. (Reprinted from publication111 copyright @ 2023, with permission from DE GRUYTER.) (b) Depicted COVID spike protein detection via graphene phononics in PBS and 2D Raman peak of graphene (green), PBASE-modified graphene (orange), graphene-PBASE-antibody (red), and graphene-PBASE-antibody-spike protein (blue) structures and the attachment of PBASE p-dopes graphene, and the subsequent attachment of antibody n-dopes. (c) The 2D peak position (inset (bottom, left)) and the Fermi level of graphene change with various concentration ranges of the COVID spike protein in PBS. Similarly, the inset (top, right) shows the change in the Fermi level of graphene at low concentrations of spike protein in an artificial saliva medium. (Reprinted from75 by the grant of the PMC Open Access Subset from ACS Nano.) (d) Refractive index sensor designed using a metasurface-based slotted T-shape perfect absorber (reprinted from publication79 copyright @ 2022, with permission from Elsevier). | ||
Nguyen et al.75 reported a SARS-CoV-2 spike protein-induced phononic modification in antibody-coupled graphene for viral detection applications. In their work (Fig. 5b), graphene, 1-pyrenebutanoic acid succinimidyl ester (PBASE)-modified graphene, graphene-PBASE-antibody, and graphene PBASE-antibody-spike protein phononic were studied using Raman spectroscopy as a detection platform. In all cases, the sensitivity is determined based on the change in the graphene 2D-phononic vibrational mode. As shown in Fig. 5b, after coupling the spike protein on the graphene PBASE, a blue shift on the 2D peak as a result of the p-doping of graphene was observed. This phenomenon can be directly correlated with the change in the Fermi level of graphene by p-doping. Consequently, the sensor's sensitivity (Fig. 5c) was measured by the shift in the Fermi level as a function of SARS-CoV-2 spike protein concentrations in phosphate-buffered saline (PBS). The chemio-phononic system can be detected at LODs of ∼1 fg mL−1 in PBS and ∼3.75 fg mL−1 in artificial saliva (see Table 1). The authors suggested that their detection method could be further modified to diagnose other COVID viral variants and diseases.
Furthermore, Patel et al.79 proposed a novel refractive index sensor (RIS) for the detection of ethyl butanoate (CH3CH2CH2COOCH2CH3) arising from the exhaled breath of SARS-CoV-2-infected persons, which is based on graphene's characteristics, such as its high surface area and easy functionalization. Fig. 5d shows the metasurface-based slotted T-shape perfect absorber RIS with a configuration of Au/MgF2/graphene/Au. Finally, they obtained the highest sensitivity of 2500 nm RIU−1 with an LOD of 0.0224 RIU when the concentration of exhaled breath of SARS-CoV-2 was changed.
Conclusions and perspectives
Herein, we briefly reviewed the recent progress on graphene-based biosensors (e.g., electrochemical, FET, and SPR) with the function of recognition and transduction for detecting the SARS-CoV-2 target. Compared to the current polymerase chain reaction (PCR) test for COVID-19, the aforementioned methods have a huge potential to provide a rapid, accurate and simple detection at low cost. Furthermore, these new diagnostic methods can secure a point-of-care (POC) platform, also called near-patient testing, to control the spread of the variant SARS-CoV-2 virus. For this purpose, if biosensors were integrated into portable devices, such as smartphones and wearable watches, it would result in real-time SARS-CoV-2 detection as an ideal scenario. Apparently, graphene with a 2D-hexagonal honeycomb lattice is believed to play an important role in this sensor technology. However, its special characteristics, such as 2D (a limited surface area) and zero bandgap (the presence of free charge carriers, as in doped semiconductors), could simultaneously act as some disadvantages. Hence, for further improving the sensitivity of graphene-based biosensors, the 2D/3D nanostructural control on top of the 2D-graphene layer should be recommended. To this end, a gold (Au) nanoparticular structure with a high surface area should be continuously useful for sufficiently functionalizing antibodies (e.g., ssDNA aptamer), detecting SARS-CoV-2 targets through specific affinity by forming an antibody–antigen complex. Finally, the R&D activities, both experimental and theoretical, for eventual mass-production of biosensors with low cost, stability and high performance, should be continued in this post-pandemic era to tackle the coronavirus-like infectious disease in the future.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
The project (EDCF L/A No. ETH-6) entitled “Establishment of Centers of Excellence for Adama Science and Technology University” through international collaboration between Ethiopia and South Korea is appreciated.References
- J. P. A. Ioannidis, Eur. J. Clin. Invest., 2020, 50, e13423 CrossRef CAS PubMed.
- E. Morales-Narváez and C. Dincer, Biosens. Bioelectron., 2020, 163, 112274 CrossRef PubMed.
- T. S. Fung and D. X. Liu, Annu. Rev. Microbiol., 2019, 73, 529–557 CrossRef CAS PubMed.
- H. A. Rothan and S. N. Byrareddy, J. Autoimmun., 2020, 109, 102433 CrossRef CAS PubMed.
- Y. Fan, K. Zhao, Z.-L. Shi and P. Zhou, Viruses, 2019, 11, 210 CrossRef CAS PubMed.
- Z. Huang, D. Tian, Y. Liu, Z. Lin, C. J. Lyon, W. Lai, D. Fusco, A. Drouin, X. Yin, T. Hu and B. Ning, Biosens. Bioelectron., 2020, 164, 112316 CrossRef CAS PubMed.
- G. A. Naikoo, F. Arshad, I. U. Hassan, T. Awan, H. Salim, M. Z. Pedram, W. Ahmed, V. Patel, A. S. Karakoti and A. Vinu, Bioeng. Transl. Med., 2022, 7, e10305 CrossRef CAS PubMed.
- E. Borberg, E. Granot and F. Patolsky, Nat. Commun., 2022, 13, 6375 CrossRef CAS PubMed.
- X. Chen, Y. Liu, X. Zhan, Y. Gao, Z. Sun, W. Wen and W. Zheng, Bioengineered, 2022, 9, 548 CrossRef CAS PubMed.
- D. M. Nguyen and T. T. T. Toan, ECS Sens. Plus, 2002, 1, 031603 CrossRef.
- Y. Gogotsi and B. Anasori, ACS Nano, 2019, 13, 8491–8494 CrossRef CAS PubMed.
- M. Jaishankar, T. Tseten, N. Anbalagan, B. B. Mathew and K. N. Beeregowda, Interdiscip. Toxicol., 2014, 7, 60–72 CrossRef PubMed.
- P. B. Tchounwou, C. G. Yedjou, A. K. Patlolla and D. J. Sutton, Exper. Suppl., 2012, 101, 133–164 Search PubMed.
- C.-W. Wang, C. Liang and H.-J. Yeh, Chemosphere, 2016, 147, 82–87 CrossRef CAS PubMed.
- X. Wu, H. Zhang, J. Zhang and X. W. Lou, Adv. Mater., 2021, 33, 2008376 CrossRef CAS PubMed.
- Y. Wang, Z. Li, J. Wang, J. Li and Y. Lin, Trends Biotechnol., 2011, 29, 205–212 CrossRef CAS PubMed.
- S. Di, Y. Qian, L. Wang and Z. Li, J. Mater. Sci., 2022, 57, 3085–3113 CrossRef CAS.
- V. Georgakilas, M. Otyepka, A. B. Bourlinos, V. Chandra, N. Kim, K. C. Kemp, P. Hobza, R. Zboril and K. S. Kim, Chem. Rev., 2012, 112, 6156–6214 CrossRef CAS PubMed.
- C. Liao, Y. Li and S. C. Tjong, Int. J. Mol. Sci., 2018, 19, 3564 CrossRef PubMed.
- S. Syama and P. V. Mohanan, Int. J. Biol. Macromol., 2016, 86, 546–555 CrossRef CAS PubMed.
- V. Palmieri and M. Papi, Nano Today, 2020, 33, 100883 CrossRef CAS PubMed.
- S. J. Updike and G. P. Hicks, Nature, 1967, 214, 986–988 CrossRef CAS PubMed.
- F. Maddalena, M. J. Kuiper, B. Poolman, F. Brouwer, J. C. Hummelen, D. M. d. Leeuw, B. D. Boer and P. W. M. Blom, J. Appl. Phys., 2010, 108, 124501 CrossRef.
- J. D. Yuen, A. S. Dhoot, E. B. Namdas, N. E. Coates, M. Heeney, I. McCulloch, D. Moses and A. J. Heeger, J. Am. Chem. Soc., 2007, 129, 14367–14371 CrossRef CAS PubMed.
- L. G. Kaake, Y. Zou, M. J. Panzer, C. D. Frisbie and X.-Y. Zhu, J. Am. Chem. Soc., 2007, 129, 7824–7830 CrossRef CAS PubMed.
- V. M. C. H. Dias, A. F. Oliveira, A. K. B. B. Marinho, C. E. D. Santos Ferreira, C. E. F. Domingues, C. M. C. B. Fortaleza, C. F. D. L. Vidal, C. M. D. M. Carrilho, D. O. B. P. Pinheiro, D. B. de Assis, E. A. Medeiros, K. M. L. Morejón, L. Weissmann, L. Michelin, M. Carneiro, M. D. S. P. Nogueira, P. R. D. de Oliveira, R. J. Buralli, R. S. B. Stucchi, R. S. Lins, S. F. Costa and A. Chebabo, Braz. J. Infect. Dis., 2022, 26, 102703 CrossRef PubMed.
- K. Alyafei, R. Ahmed, F. F. Abir, M. E. H. Chowdhury and K. K. Naji, Comput. Biol. Med., 2022, 149, 106070 CrossRef CAS PubMed.
- J. Sengupta and C. M. Hussain, Inorganics, 2023, 11, 197 CrossRef CAS.
- C. S. Feldkamp and J. L. Carey, in Immunoassay, ed. E. P. Diamandis and T. K. Christopoulos, Immunoassay, Academic Press, San Diego, CA, USA, 1996, pp. 5–24 Search PubMed.
- C. C. Smith, K. S. Olsen, K. M. Gentry, M. Sambade, W. Beck, J. Garness, S. Entwistle, C. Willis, S. Vensko, A. Woods, M. Fini, B. Carpenter, E. Routh, J. Kodysh, T. O'Donnell, C. Haber, K. Heiss, V. Stadler, E. Garrison, A. M. Sandor, J. P. Y. Ting, J. Weiss, K. Krajewski, O. C. Grant, R. J. Woods, M. Heise, B. G. Vincent and A. Rubinsteyn, Genome Med., 2021, 13, 101 CrossRef CAS PubMed.
- M. F. Bachmann, M. O. Mohsen, L. Zha, M. Vogel and D. E. Speiser, npj Vaccines, 2021, 6, 2 CrossRef CAS PubMed.
- C. Rosadas, M. Khan, E. Parker, F. Marchesin, K. Katsanovskaja, M. Sureda-Vives, N. Fernandez, P. Randell, R. Harvey, A. Lilley, B. H. L. Harris, M. Zuhair, M. Fertleman, S. Ijaz, S. Dicks, C.-E. Short, R. Quinlan, G. P. Taylor, K. Hu, P. McKay, A. Rosa, C. Roustan, M. Zuckerman, K. El Bouzidi, G. Cooke, B. Flower, M. Moshe, P. Elliott, A. J. Spencer, T. Lambe, S. C. Gilbert, H. Kingston, J. K. Baillie, P. J. M. Openshaw, M. G. Semple, P. Cherepanov, M. O. McClure and R. S. Tedder, J. Virol. Methods, 2022, 302, 114475 CrossRef CAS PubMed.
- L. B. Shrestha, N. Tedla and R. A. Bull, Front. Immunol., 2021, 12, 752003 CrossRef CAS PubMed.
- L. Min and Q. Sun, Front. Mol. Biosci., 2021, 8, 671633 CrossRef CAS PubMed.
- M. Fukuda, M. S. Islam, R. Shimizu, H. Nasser, N. N. Rabin, Y. Takahashi, Y. Sekine, L. F. Lindoy, T. Fukuda, T. Ikeda and S. Hayami, ACS Appl. Nano Mater., 2021, 4, 11881–11887 CrossRef CAS PubMed.
- M. I. Katsnelson, Mater. Today, 2007, 10, 20–27 CrossRef CAS.
- C. N. R. Rao, K. Biswas, K. S. Subrahmanyam and A. Govindaraj, J. Mater. Chem., 2009, 19, 2457–2469 RSC.
- G. Yang, L. Li, W. B. Lee and M. C. Ng, Sci. Technol. Adv. Mater., 2018, 19, 613–648 CrossRef CAS PubMed.
- C. Soldano, A. Mahmood and E. Dujardin, Carbon, 2010, 48, 2127–2150 CrossRef CAS.
- P. Avouris and C. Dimitrakopoulos, Mater. Today, 2012, 15, 86–97 CrossRef CAS.
- C. N. R. Rao, A. K. Sood, R. Voggu and K. S. Subrahmanyam, J. Phys. Chem. Lett., 2010, 1, 572–580 CrossRef CAS.
- M. B. Burkholder, F. B. A. Rahman, E. H. Chandler, J. R. Regalbuto, B. F. Gupton and J. M. M. Tengco, Carbon Trends, 2022, 9, 100196 CrossRef CAS.
- B. Aïssa, N. K. Memon, A. Ali and M. K. Khraisheh, Front. Mater., 2015, 2, 58 Search PubMed.
- Y. Duan, C. D. Stinespring and B. Chorpening, ChemistryOpen, 2015, 4, 642–650 CrossRef CAS PubMed.
- P. Tian, L. Tang, K. S. Teng and S. P. Lau, Mater. Today Chem., 2018, 10, 221–258 CrossRef CAS.
- R. Zan, A. Altuntepe and S. Erkan, Phys. E, 2021, 128, 114629 CrossRef CAS.
- H. Rasuli, R. Rasuli, M. Alizadeh and G. BoonTong, Results Phys., 2020, 18, 103200 CrossRef.
- P.-C. Lin, R. Villarreal, S. Achilli, H. Bana, M. N. Nair, A. Tejeda, K. Verguts, S. De Gendt, M. Auge, H. Hofsäss, S. De Feyter, G. Di Santo, L. Petaccia, S. Brems, G. Fratesi and L. M. C. Pereira, ACS Nano, 2021, 15, 5449–5458 CrossRef CAS PubMed.
- F. Hadish, M.-H. Chiang, Y.-F. Hsieh, S.-Y. Wu and S. Jou, Adv. Mater. Sci. Eng., 2022, 2022, 4574772 Search PubMed.
- D. R. Yanti, U. Hikmah, A. Prasetyo and E. Hastuti, IOP Conf. Ser.: Earth Environ. Sci., 2020, 456, 012008 Search PubMed.
- M. Lee and S.-M. Paek, Nanomaterials, 2022, 12, 1507 CrossRef CAS PubMed.
- F. Hadish, S. Jou, B.-R. Huang, H.-A. Kuo and C.-W. Tu, J. Electrochem. Soc., 2017, 164, B336 CrossRef CAS.
- J. H. Jeong, S. Kang, N. Kim, R. Joshi and G.-H. Lee, Phys. Chem. Chem. Phys., 2022, 24, 10684–10711 RSC.
- C. Sainz-Urruela, S. Vera-López, M. Paz San Andrés and A. M. Díez-Pascual, J. Mol. Liq., 2022, 357, 119104 CrossRef CAS.
- Z. Guo, S. Chakraborty, F. A. Monikh, D.-D. Varsou, A. J. Chetwynd, A. Afantitis, I. Lynch and P. Zhang, Adv. Biol., 2021, 5, 2100637 CrossRef CAS PubMed.
- D. Chen, H. Feng and J. Li, Chem. Rev., 2012, 112, 6027–6053 CrossRef CAS PubMed.
- D. A. Dikin, S. Stankovich, E. J. Zimney, R. D. Piner, G. H. B. Dommett, G. Evmenenko, S. T. Nguyen and R. S. Ruoff, Nature, 2007, 448, 457–460 CrossRef CAS PubMed.
- K. Chaudhary, K. Kumar, P. Venkatesu and D. T. Masram, Adv. Colloid Interface Sci., 2021, 289, 102367 CrossRef CAS PubMed.
- M. K. Rabchinskii, S. A. Ryzhkov, N. A. Besedina, M. Brzhezinskaya, M. N. Malkov, D. Y. Stolyarova, A. F. Arutyunyan, N. S. Struchkov, S. D. Saveliev, I. D. Diankin, D. A. Kirilenko, S. I. Pavlov, D. V. Potorochin, F. Roth, M. V. Gudkov, A. A. Gulin, P. Cai, Z. Liu, A. V. Golovin and P. N. Brunkov, Carbon, 2022, 196, 264–279 CrossRef CAS.
- F. Wang, L. Liu and W. J. Li, IEEE Trans. NanoBiosci., 2015, 14, 818–834 Search PubMed.
- C. Zhang, Z. Zhang, Q. Yang and W. Chen, Electroanalysis, 2018, 30, 2504–2524 CrossRef CAS.
- M. T. Hwang, M. Heiranian, Y. Kim, S. You, J. Leem, A. Taqieddin, V. Faramarzi, Y. Jing, I. Park, A. M. van der Zande, S. Nam, N. R. Aluru and R. Bashir, Nat. Commun., 2020, 11, 1543 CrossRef CAS PubMed.
- Y. Bai, T. Xu and X. Zhang, Micromachines, 2020, 11, 60 CrossRef PubMed.
- A. Balaji, S. Yang, J. Wang and J. Zhang, Biosensors, 2019, 9, 74 CrossRef CAS PubMed.
- J. Lee, K. Takemura and E. Y. Park, Sens. Actuators, B, 2018, 276, 254–261 CrossRef CAS.
- K. Navakul, C. Sangma, P.-t. Yenchitsomanus, S. Chunta and P. A. Lieberzeit, Anal. Bioanal. Chem., 2021, 413, 6191–6198 CrossRef CAS PubMed.
- R. Dutta, K. Rajendran, S. K. Jana, L. M. Saleena and S. Ghorai, Biosensors, 2023, 13, 349 CrossRef CAS PubMed.
- S. Afsahi, M. B. Lerner, J. M. Goldstein, J. Lee, X. Tang, D. A. Bagarozzi, D. Pan, L. Locascio, A. Walker, F. Barron and B. R. Goldsmith, Biosens. Bioelectron., 2018, 100, 85–88 CrossRef CAS PubMed.
- T. Ando, NPG Asia Mater., 2009, 1, 17–21 CrossRef.
- S. Yasri and V. Wiwanitkit, Sens. Int., 2022, 3, 100171 CrossRef PubMed.
- G. Seo, G. Lee, M. J. Kim, S.-H. Baek, M. Choi, K. B. Ku, C.-S. Lee, S. Jun, D. Park, H. G. Kim, S.-J. Kim, J.-O. Lee, B. T. Kim, E. C. Park and S. I. Kim, ACS Nano, 2020, 14, 5135–5142 CrossRef CAS PubMed.
- Y. Li, Z. Peng, N. J. Holl, M. R. Hassan, J. M. Pappas, C. Wei, O. H. Izadi, Y. Wang, X. Dong, C. Wang, Y.-W. Huang, D. Kim and C. Wu, ACS Omega, 2021, 6, 6643–6653 CrossRef CAS PubMed.
- B. V. Krsihna, S. Ahmadsaidulu, S. S. T. Teja, D. Jayanthi, A. Navaneetha, P. R. Reddy and M. D. Prakash, Silicon, 2022, 14, 5913–5921 CrossRef CAS.
- A. Wasfi, F. Awwad, N. Qamhieh, B. Al Murshidi, A. R. Palakkott and J. G. Gelovani, Sci. Rep., 2022, 12, 18155 CrossRef CAS PubMed.
- N. H. L. Nguyen, S. Kim, G. Lindemann and V. Berry, ACS Nano, 2021, 15, 11743–11752 CrossRef CAS PubMed.
- T. Beduk, D. Beduk, J. I. de Oliveira Filho, F. Zihnioglu, C. Cicek, R. Sertoz, B. Arda, T. Goksel, K. Turhan, K. N. Salama and S. Timur, Anal. Chem., 2021, 93, 8585–8594 CrossRef CAS PubMed.
- S. Kim, H. Ryu, S. Tai, M. Pedowitz, J. R. Rzasa, D. J. Pennachio, J. R. Hajzus, D. K. Milton, R. Myers-Ward and K. M. Daniels, Biosens. Bioelectron., 2022, 197, 113803 CrossRef CAS PubMed.
- S. Sarwar, M.-C. Lin, C. Amezaga, Z. Wei, E. Iyayi, H. Polk, R. Wang, H. Wang and X. Zhang, Adv. Compos. Hybrid Mater., 2023, 6, 49 CrossRef CAS PubMed.
- S. K. Patel, J. Surve, J. Parmar, K. Aliqab, M. Alsharari and A. Armghan, Diamond Relat. Mater., 2023, 132, 109644 CrossRef CAS PubMed.
- X. Yang, Z.-Z. Yin, G. Zheng, M. Zhou, H. Zhang, J. Li, W. Cai and Y. Kong, Bioelectrochemistry, 2023, 151, 108375 CrossRef CAS PubMed.
- J. Jaewjaroenwattana, W. Phoolcharoen, E. Pasomsub, P. Teengam and O. Chailapakul, Talanta, 2023, 251, 123783 CrossRef CAS PubMed.
- T. Ji, Z. Liu, G. Wang, X. Guo, S. Akbar khan, C. Lai, H. Chen, S. Huang, S. Xia, B. Chen, H. Jia, Y. Chen and Q. Zhou, Biosens. Bioelectron., 2020, 166, 112455 CrossRef CAS PubMed.
- F. Mollarasouli, S. Kurbanoglu and S. A. Ozkan, Biosensors, 2019, 9, 86 CrossRef CAS PubMed.
- Z. Zhao, C. Huang, Z. Huang, F. Lin, Q. He, D. Tao, N. Jaffrezic-Renault and Z. Guo, TrAC, Trends Anal. Chem., 2021, 139, 116253 CrossRef CAS PubMed.
- M. Alafeet, K. Dighe, P. Moitra and D. Pan, ACS Nano, 2020, 14, 17028–17045 CrossRef PubMed.
- D. Sadighbayan, A. Minhas-Khan and E. Ghafar-Zadeh, IEEE Trans. NanoBiosci., 2022, 21, 232–245 CAS.
- N. Alnaji, A. Wasfi and F. Awwad, Sci. Rep., 2023, 13, 4485 CrossRef CAS PubMed.
- N. H. Al-Hardan, M. Firdaus-Raih, M. A. A. Hamid and A. Jalar, Crit. Rev. Solid State Mater. Sci., 2023 DOI:10.1080/10408436.2023.2169657.
- M. T. Amen, T. T. Pham, E. Cheah, D. P. Tran and B. Thierry, Molecules, 2022, 27, 7952 CrossRef CAS PubMed.
- A. Wasfi, F. Awwad, J. G. Gelovani, N. Qamhieh and A. I. Ayesh, Nanomaterials, 2022, 12, 2638 CrossRef CAS PubMed.
- C.-W. Chiu, M. Xian, J. L. Stephany, X. Xia, C.-C. Chiang, F. Ren, C.-T. Tsai, S.-S. Shan, Y.-T. Liao, J. F. Esquivel-Upshaw, S. R. Rananaware, L. T. Nguyen, N. C. Macaluso, P. K. Jain, M. N. Cash, C. N. Mavian, M. Salemi, M. E. Leon, C.-W. Chang, J. Lin and S. J. Pearton, J. Vac. Sci. Technol., B: Nanotechnol. Microelectron.: Mater., Process., Meas., Phenom., 2022, 40, 023204 CAS.
- C.-C. Chiang, C.-W. Chiu, F. Ren, C.-T. Tsai, Y.-T. Liao, J. F. Esquivel-Upshaw and S. J. Pearton, J. Vac. Sci. Technol., B: Nanotechnol. Microelectron.: Mater., Process., Meas., Phenom., 2022, 41, 012204 Search PubMed.
- S. Sreejith, J. Ajayan, J. M. Radhika, B. Sivasankari, S. Tayal and M. Saravanan, Measurement, 2023, 206, 112202 CrossRef.
- P. K. Sharma, E.-S. Kim, S. Mishra, E. Ganbold, R.-S. Seong, A. K. Kaushik and N.-Y. Kim, ACS Sens., 2021, 6, 3468–3476 CrossRef CAS PubMed.
- N. Alnaji, A. Wasfi and F. Awwad, Sci. Rep., 2023, 13, 4485 CrossRef CAS PubMed.
- I. Park, J. Lim, S. You, M. T. Hwang, J. Kwon, K. Koprowski, S. Kim, J. Heredia, S. A. Stewart de Ramirez, E. Valera and R. Bashir, ACS Sens., 2021, 6, 4461–4470 CrossRef CAS PubMed.
- A. Toral-Lopez, D. B. Kokh, E. G. Marin, R. C. Wade and A. Godoy, Nanoscale Adv., 2022, 4, 3065–3072 RSC.
- D. K. Ban, T. Bodily, A. G. Karkisaval, Y. Dong, S. Natani, A. Ramanathan, A. Ramil, S. Srivastava, P. Bandaru, G. Glinsky and R. Lal, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2206521119 CrossRef CAS PubMed.
- L. Wang, X. Wang, Y. Wu, M. Guo, C. Gu, C. Dai, D. Kong, Y. Wang, C. Zhang, D. Gu, C. Fan, Y. Xie, Z. Zhu, Y. Liu and D Wei, Nat. Biomed. Eng., 2022, 6, 276–285 CrossRef CAS PubMed.
- J. Y. Kim and C. D. Frisbie, Correlation of phase behavior and charge transport in conjugated polymer/fullerene blends, J. Phys. Chem. C, 2008, 112, 17726–17736 CrossRef CAS.
- J. Y. Kim, H. Cho, S. Noh, Y. Lee, Y. M. Nam, C. Lee and W. H. Jo, Charge transport in amorphous low bandgap conjugated polymer/fullerene films, J. Appl. Phys., 2012, 111, 043710 CrossRef.
- Z. Bao and J. Locklin, Organic Field Effect Transistor, CRC press, Boca Raton, FL, 2007 Search PubMed.
- A. J. Heeger, N. S. Sariciftci and E. B. Namdas, Semiconducting and Metallic Polymers, Oxford University Press, Oxford, UK, 2010 Search PubMed.
- E. Kretschmann and H. Raether, Z. Naturforsch., 1968, 23a, 2135–2136 CrossRef.
- B. Liedberg, C. Nylander and I. Lunström, Sens. Actuators, 1983, 4, 299–304 CrossRef CAS.
- B. A. Prabowo, A. Purwidyantri and K. C. Liu, Biosensors, 2018, 8, 80 CrossRef CAS PubMed.
- W. Xiaoying, M. Mingqiang, W. Xueliang and W. Shoujuan, in Analytical Chemistry, ed. S. Abhay Nanda, IntechOpen, Rijeka, 2020, DOI:10.5772/intechopen.92549, p. Ch. 5.
- J. Homola, Chem. Rev., 2008, 108, 462–493 CrossRef CAS PubMed.
- Q. Duan, Y. Liu, S. Chang, H. Chen and J.-h. Chen, Sensors, 2021, 21, 5262 CrossRef PubMed.
- P. S. Pandey, S. K. Raghuwanshi, A. Shadab, M. T. I. Ansari, U. K. Tiwari and S. Kumar, IEEE Sens. J., 2022, 22, 13800–13810 CAS.
- T. B. Akib, S. F. Mou, M. M. Rahman, M. M. Rana, M. R. Islam, I. M. Mehedi, M. A. P. Mahmud and A. Z. Kouzani, Sensors, 2021, 21, 3491 CrossRef PubMed.
- S. Mostufa, T. B. A. Akib, M. M. Rana, I. M. Mehedi, U. M. Al-Saggaf, A. U. Alsaggaf, M. U. Alsaggaf and M. S. Alam, Opt. Continuum., 2022, 1, 494–515 CrossRef CAS.
- M. M. Hossain and M. A. Talukder, PLoS One, 2023, 18, e0284812 CrossRef CAS PubMed.
- J. Payandehpeyman, N. Parvini, K. Moradi and N. Hashemian, ACS Appl. Nano Mater., 2021, 4, 6189–6200 CrossRef CAS PubMed.
- R. Negahdari, E. Rafiee and Z. Kordrostami, Plasmonics, 2023, 18, 1325–1335 CrossRef CAS PubMed.
- S. A. Taya, M. G. Daher, A. H. M. Almawgani, A. T. Hindi, S. H. Zyoud and I. Colak, Plasmonics, 2023, 18, 1441–1448 CrossRef CAS PubMed.
- F. Du, K. Zheng, S Zeng and Y. Yuan, Nanophotonics, 2023, 12, 1271–1284 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2023 |







