Open Access Article
Open Access ArticleA rapid abiotic/biotic hybrid sandwich detection for trace pork adulteration in halal meat extract†
Chehasan
Cheubong
 ab,
Hirobumi
Sunayama
ab,
Hirobumi
Sunayama
 *a,
Eri
Takano
*a,
Eri
Takano
 a,
Yukiya
Kitayama
c,
Hideto
Minami
a,
Yukiya
Kitayama
c,
Hideto
Minami
 a and
Toshifumi
Takeuchi
a and
Toshifumi
Takeuchi
 *ade
*ade
aGraduate School of Engineering, Kobe University, 1-1, Rokkodai-cho, Nada-ku, Kobe 657-8501, Japan. E-mail: takeuchi@gold.kobe-u.ac.jp; sunayama@penguin.kobe-u.ac.jp
bDepartment of Chemistry, Faculty of Science and Technology, Rajamangala University of Technology, Thanyaburi, Pathumthani 12110, Thailand
cGraduate School of Engineering, Osaka Metropolitan University, 1-1, Gakuen-cho, Naka-ku, Sakai, Osaka 599-8531, Japan
dCenter for Advanced Medical Engineering Research & Development (CAMED), Kobe University, 1-5-1, Minatojimaminami-machi, Chuo-ku, Kobe 650-0047, Japan
eInnovation Commercialization Division, Kobe University, 1-1, Rokkodai-cho, Nada-ku, Kobe 657-8501, Japan
First published on 24th August 2023
Abstract
In this study, we prepared molecularly imprinted polymer nanogels with good affinity for the Fc domain of immunoglobulin G (IgG) using 4-(2-methacrylamidoethylaminomethyl) phenylboronic acid as a modifiable functional monomer for post-imprinting in-cavity modification of a fluorescent dye (F-Fc-MIP-NGs). A novel nanogel-based biotic/abiotic hybrid sandwich detection system for porcine serum albumin (PSA) was developed using F-Fc-MIP-NGs as an alternative to a secondary antibody for fluorescence detection and another molecularly imprinted polymer nanogel capable of recognizing PSA (PSA-MIP-NGs) as a capturing artificial antibody, along with a natural antibody toward PSA (Anti-PSA) that was used as a primary antibody. After incubation of PSA and Anti-PSA with F-Fc-MIP-NGs, the PSA/Anti-PSA/F-Fc-MIP-NGs complex was captured by immobilized PSA-MIP-NGs for fluorescence measurements. The analysis time was less than 30 min for detecting pork adulteration of 0.01 wt% in halal beef and lamb meats. The detection limit was comparable to that of frequently used immunoassays. Therefore, we believe that this method is a promising, sensitive, and rapid detection method for impurities in real samples and could be a simple, inexpensive, and rapid alternative to conventional methods that have cumbersome procedures of 4 hours or more.
Introduction
The fast-growing market for halal meat has necessitated the development of a halal biomarker for halal meat authentication.1 One of the most common contaminations of halal meat products is pork meat, which poses a severe concern for Muslims.2 A fast, simple, sensitive, and selective sensing technique for porcine serum albumin (PSA) from pork would help manufacturers ensure their products are free from pork and suitable for Muslims to consume. Currently, immunoassays, which are the popular analytical methods for detecting protein biomarkers for disease diagnosis3 and food analysis,4 are most commonly used to detect pork contamination in halal meat products owing to their high sensitivity and specificity.5 However, these methods are cost-intensive and time-consuming, and natural antibodies and conjugated enzymes usually used for these assays have low stabilities. Furthermore, two or more antibodies and labeled antibodies are necessary to achieve susceptible and selective detection of target marker proteins. These limitations make the process time-consuming and expensive.Molecularly imprinted polymers (MIPs) prepared by the copolymerization of functional monomers, co-monomers, and cross-linkers in the presence of target molecules have already been used as artificial antibody alternatives to natural antibodies for various applications owing to their ease of preparation, high stability, and low cost.6 This process aims to overcome the known drawbacks. The MIP-based sensors are considered promising alternatives to immunosensors for sensitive and selective detection of protein targets.7 Previously, we have prepared MIP-NGs by molecular imprinting using pyrrolidyl acrylate as a functional monomer8 and developed a PSA-MIP-NG-based quartz crystal microbalance (QCM) sensor capable of detecting PSA in meat extracts.9 The selectivity of the sensor for PSA was high; less than 10% affinity of PSA obtained from other animal serum albumins with >80% homology in amino acid sequences. Nevertheless, the sensitivity was lower than that of immunosensors due to the limitation of the QCM device. Later, we developed a highly sensitive MIP-based fluorescence sensor to rapidly detect pork adulteration in halal meat extracts.10 For high-sensitivity recognition of target proteins, fluorescent PSA-MIP-NGs (F-PSA-MIP-NGs) targeting PSA were developed via molecular imprinting and post-imprinting modification (PIM)11 using 4-[2-(N-methacrylamido)ethylaminomethyl] benzoic acid as a functional monomer,12 which can create modifiable sites for fluorescence dye introduction in molecularly imprinted nanocavities. Since PIM-based functionalization of imprinted cavities was targeted to functional groups on functional monomer residues within the cavity, the undesired introduction of functional groups, such as fluorescent dyes outside the cavity that induces non-specific signals and high background noise, was suppressed. The F-PSA-MIP-NG-based fluorescence sensor exhibited excellent sensitivity with a limit of detection (LOD) of 2.5 ng mL−1 and allowed fast detection of pork adulteration in meat extracts. However, the performance of the sensors was still not comparable to that of immunosensors.
An effective strategy to amplify the signal and increase the sensitivity of detecting target molecules in immunoassays is to use conjugated secondary antibodies that recognize the specific crystallizable region (Fc domain) of immunoglobulin G (IgG) and capture primary antibodies with their antigen-binding domains. Immunosensor formats with various secondary antibody labels, such as enzyme immunosensors, radioimmunosensors, chemiluminescent immunosensors, and fluorescent immunosensors, have been developed to improve the sensor signal responses.13 Developing antibodies labeled with fluorescent dyes, radioisotopes, biotin, avidin, and enzymes, without compromising the intrinsic molecular recognition capacity of the antibodies is challenging. In contrast, synthetic polymer-based MIPs can be easily labeled using PIMs. MIPs are widely used as alternatives to natural antibodies for protein recognition in MIP-based sensors. However, to the best of our knowledge, MIPs have not yet been used as secondary antibody mimics for sensor signal amplification. Previous reports on different imprinting techniques have indicated that MIPs for whole IgG or Fc domains were the most promising artificial antibodies for application in protein purification and biosensors due to their selective recognition and sensitive detection of target molecules.14 Moreover, imprinting the Fc domain exhibited strong binding affinity for IgG in both humans and goats, such as protein A.15
Recently, our group successfully synthesized MIP-NGs using 3-methacrylamidophenylboronic acid as a functional monomer (no PIM available), targeting the Fc domain by precipitation polymerization of stealth nanocarriers with specific adsorption of IgG.16 It had an excellent recognition capability for IgG, resulting in the regulation of the protein corona formed on the MIP-NGs. This observation inspired us to explore a novel sensitive MIP-based sensor similar to conventional immunoassays using Fc domain-imprinted polymer nanogels bearing a fluorescence function (F-Fc-MIP-NGs) as conjugated secondary antibody mimics to increase the signal response for detecting pork adulteration in halal food.
Herein, we present a novel abiotic/biotic hybrid sandwich detection system that combines the specificity of the natural antibody and a diverse functionalization flexibility of the synthetic polymer-based receptor. The proposed system for specific detection of pork adulteration consists of MIP-NGs for capturing target PSA (PSA-MIP-NGs), a polyclonal anti-PSA antibody (Anti-PSA) as a primary antibody, and fluorescent MIP-NGs for Fc domain of IgG recognition as a secondary detection antibody. First, F-Fc-MIP-NGs were prepared via molecular imprinting and PIM, where 4-(2-methacrylamidoethylaminomethyl) phenylboronic acid (MAPBA)17 was used as a functional monomer with a modifiable site for PIM, and ATTO 647N NHS was introduced as a fluorescent reporter dye into the Fc domain-imprinted cavity (Fig. S1†). Second, the fluorescent signaling ability of the prepared F-Fc-MIP-NGs was investigated using the Fc domain, the deglycosylated Fc domain, whole IgG, and albumin. Finally, the functionality of the abiotic/biotic hybrid sandwich detection system was demonstrated, using the previously reported PSA-imprinted polymer nanogels to capture target PSA;9 Anti-PSA was used as the primary antibody, and F-Fc-MIP-NGs were used as the secondary antibody mimic. The sensitivity, selectivity, accuracy, stability, recovery, and feasibility of the proposed sandwich system for detecting pork adulteration for halal meat sensing applications were evaluated (Scheme 1).
Results and discussion
F-Fc-MIP-NGs was developed as an artificial secondary antibody capable of detecting the Fc domain for the abiotic/biotic hybrid sandwich detection. MAPBA was used as the functional monomer, which can bind to the Fc domain glycan via covalent boronate esters and bind to template molecules via non-covalent interactions, including electrostatic and hydrophobic interactions. The Fc domain as a template molecule for synthesizing Fc-MIP-NGs was prepared by a usual method. Briefly, human IgG was digested using papain and cysteine (Fig. S2a†),18a and then purification was conducted (Fig. S3 and S4†). The deglycosylated Fc domain was also prepared by glycosidase digestion18b (Fig. S2b and S4†). Under the polymerization condition, the CD spectra of the template molecule maintained their original secondary structure, as the Fc domain template was not denatured during polymerization (Fig. S5†). NIP-NGs were also prepared without adding the template molecule.After polymerization, the obtained NGs were purified by size exclusion and ion-exchange chromatography, as reported previously9 (Fig. S6†). The removal rate of the Fc domain from the prepared NGs was >80%, which was comparable to what was observed in our two previously reported studies,9,10 confirming the potential ability to rebind with the target molecule (Fig. S7†). The particle sizes of Fc-MIP-NGs and Fc-NIP-NGs appeared to be 21.1 nm and 17.8 nm, respectively. After the purification, the zeta potential of Fc-MIP-NGs was shifted from 4.4 mV to 19.7 mV (Fig. S8†), indicating that the template molecule Fc domain was successfully removed from the NG matrix—morphology of the MIP-NGs was observed by transmission electron microscopy (Fig. S9†).
For PIM, Fc-MIP-NGs were incubated with ATTO 647N NHS as the fluorescent reporter molecule for conjugation by the secondary amino group within the imprinted nanocavity of Fc-MIP-NGs. NIP-NGs were also treated with the fluorescent reporter molecule (F-NIP-NGs). The fluorescence intensity of ATTO 647N in F-Fc-MIP-NGs, F-NIP-NGs, and untreated NGs was investigated at an excitation wavelength of 647 nm, and a maximum peak at 668 nm was observed. The fluorescence spectrum of ATTO 647N was found only in F-Fc-MIP-NGs and F-NIP-NGs (Fig. S10†), suggesting that conjugation of the fluorescent reporter into the secondary amino group of NGs via PIM was successfully demonstrated.
The binding affinity and selectivity of F-Fc-MIP-NGs toward the Fc domain were investigated by fluorescence changes on a gold-coated sensor chip with immobilized F-Fc-MIP-NGs, using a custom-made liquid-handling robot equipped with a fluorescence microscope as previously reported (Fig. S11†).10,17,19 The fluorescence intensity derived from the introduced ATTO 647N in the nanocavities of F-Fc-MIP-NGs increased after immobilization (Fig. S12†), indicating that F-Fc-MIP-NGs were successfully immobilized onto the gold-coated sensor chip. Incubation with different concentrations of the Fc domain showed that the fluorescence intensity of the sensor chip increased with an increase in the concentration of the Fc domain. In contrast, NIP-NGs for the Fc domain provided lower signal responses than that provided by F-Fc-MIP-NGs at all concentrations (Fig. 1). The apparent binding constant (Ka) of F-Fc-MIP-NGs was estimated to be 6.20 × 107 M−1 (Fig. S13†), while that of F-NIP-NGs was 6.15 × 106 M−1. The results confirm the imprinting effect. The generated nanocavity can bind the Fc domain and transduce the binding event into fluorescence changes by molecular imprinting and PIMs.
 | ||
| Fig. 1 Binding behavior of the Fc domain (0–1600 nM) toward F-Fc-MIP-MGs and F-NIP-NGs immobilized sensor chips. Error bars (standard deviations) were obtained from triplicate experiments. | ||
The relative fluorescence intensities of F-Fc-MIP-NGs and F-NIP-NGs toward the Fc domain and structurally related reference proteins, including the deglycosylated Fc domain, whole IgG, and PSA at 100 nM, were investigated to evaluate selectivity. The selectivity factors of F-Fc-MIP-NGs toward these reference proteins, i.e., the deglycosylated Fc domain, whole IgG, and PSA, were estimated to be 0.49, 0.67, and 0.25, respectively. The selectivity factors of the reference proteins were lower than those of the Fc domain, indicating that F-Fc-MIP-NGs provided high selectivity for the Fc domain (Fig. 2a). The signal responses of the deglycosylated Fc domain, where the glycan was depleted from the polypeptide chain, and whole IgG for F-Fc-MIP-NGs, were higher than those of PSA, suggesting that F-Fc-MIP-NGs can selectively bind to the Fc domain of IgG. The apparent Ka values of F-Fc-MIP-NGs toward the deglycosylated Fc domain and whole IgG were estimated to be 3.02 × 107 M−1 and 5.06 × 107 M−1, respectively (Fig. S14†). The Ka value toward the deglycosylated Fc domain was half that of the glycosylated Fc domain, indicating that the interaction between MAPBA residues and the sugar chain on the Fc domain is essential for specific recognition. Furthermore, these Ka values were higher than those of NIP-NGs, indicating that the functional monomer MAPBA can interact with the sugar chain moiety and the polypeptide moiety in the Fc domain. In contrast, Fc domain selectivity was not observed in F-NIP-NGs. The selectivity factors of the deglycosylated Fc domain, whole IgG, and PSA at 100 nM were estimated to be 0.76, 2.37, and 1.51, respectively (Fig. 2b). These results indicated that the Fc domain-specific recognition nanocavities of F-Fc-MIP-NGs were completed by the molecular imprinting process and allowed ATTO 647N to be introduced into the nanocavities via the PIMs.
To develop an abiotic/biotic hybrid sandwich detection system for pork adulteration, the detection conditions were optimized considering four parameters: effective blocking reagents, the concentration of Anti-PSA, the concentration of F-Fc-MIP-NGs, and the binding time. The effect of blocking reagents was examined by incubation with PBS, Protein free PBS-blocking buffer, 0.1% and 0.5% (w/v) bovine serum albumin (BSA), and 0.5% (w/v) skimmed milk for 60 min after immobilization of PSA-MIP-NGs. A blank measurement with premix solutions containing Anti-PSA and F-Fc-MIP-NGs without PSA was performed to evaluate the appropriate blocking solution. After binding under the blank experiments, 0.5% w/v BSA blocking reagent was finally selected, because it provided the lowest relative fluorescence intensity change with blank measurement (Fig. S15†). Using the appropriate blocking reagent, the concentration of Anti-PSA in the premix solution was optimized (Fig. S16a†). The highest response was at 0.1 μg mL−1 Anti-PSA. The lower response values were observed at high concentrations of Anti-PSA owing to the self-binding or overlapping of antibodies in the solution.
The concentration of F-Fc-MIP-NGs mixed with 0.1 μg mL−1 Anti-PSA was optimized (Fig. S16b†). A concentration of 100 μg mL−1 F-Fc-MIP-NGs was selected, because it yielded the most significant response with a smaller amount of F-Fc-MIP-NGs. The response was not significantly different upon increasing the concentration of F-Fc-MIP-NGs above 100 μg mL−1. Thus, an Anti-PSA concentration of 0.1 μg mL−1 and an F-Fc-MIP-NGs concentration of 100 μg mL−1 were ultimately selected as the optimum conditions for use in further experiments.
The incubation time for PSA binding under the optimized concentration of Anti-PSA and F-Fc-MIP-NGs with 10 nM PSA in the premix solution were optimized (Fig. S17†). The optimized binding time was determined to be 30 min, because it yielded the highest response with the minimum incubation time. The results indicated that the proposed abiotic/biotic hybrid sandwich detection system provided a more rapid PSA detection compared to the conventional ELISA-based sandwich assay (4 h analysis time).
To confirm the potential of this detection system in the decreased analysis time from conventional 4 h to 30 min, the binding isotherm and sensitivity of the step-by-step immobilization of PSA, Anti-PSA, and F-Fc-MIP-NGs with a multi-washing system on the sensor chip with immobilized PSA-MIP-NGs were investigated (Fig. S18†). The binding isotherm and sensitivity of the proposed system with step-by-step immobilization were similar to those of the premix solution immobilization. As shown in Fig. S19,† the relative fluorescence intensity of the blank solution of step-by-step immobilization was higher than that of the premix solution immobilization due to high non-specific binding with multi-step immobilization and washing. The results confirmed that the proposed sandwich detection for PSA with the premix solution exhibited higher affinity and sensitivity for PSA and provided a more rapid detection of PSA compared to the step-by-step immobilization frequently used for ELISA.
A novel abiotic/biotic hybrid sandwich detection system for PSA was constructed under the optimized conditions using PSA-MIP-NGs as a capture antibody mimic and F-Fc-MIP-NGs as a fluorescent secondary antibody mimic. To develop a detection system with a sensitivity comparable to that of ELISA, natural Anti-PSA was adopted to increase the selectivity of PSA detection. The gold-coated sensor chips immobilized with PSA-MIP-NGs were incubated with the premix solution of Anti-PSA and F-Fc-MIP-NGs at different concentrations of PSA (0–100 nM), and the fluorescence intensity derived from bound F-Fc-MIP-NGs on the sensor chip was measured. The binding isotherm of the PSA detection presented a linear calibration range of 0.01–10 nM with r2 = 0.969 (Fig. S20†). The LOD of PSA for the developed system was calculated according to 3 SD m−1, where m is the slope of the linear part of the binding isotherm and SD is the standard deviation for 0 nM PSA. The LOD was determined to be 10 pM (0.5 ng mL−1). The sensitivity of the PSA abiotic/biotic hybrid sandwich detection system was higher than that of our previous PSA-MIP-NG-based QCM9 and fluorescent10 sensors. This detection system also exhibited the greatest sensitivity of MIP-based sensors for PSA detection compared to earlier reports,20 confirming that combining the advantages of artificial and natural antibodies can improve the sensitivity of PSA detection.
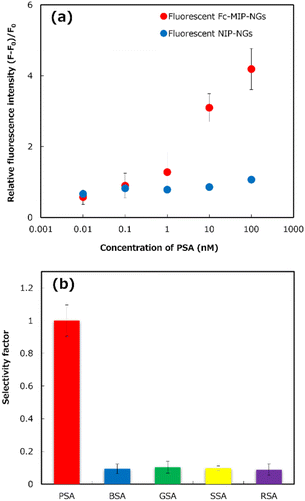
The binding behavior of PSA to the prepared abiotic/biotic hybrid sandwich detection using mixed F-Fc-MIP-NGs and F-NIP-NGs in premix solutions was investigated (Fig. 3a). The relative fluorescence intensity of PSA showed concentration-dependent binding to the proposed system with a premix solution containing Fc-MIP-NGs, such that the relative fluorescence intensities increased with increasing PSA concentration in solution. However, the response of F-NIP-NGs was lower at almost concentrations, suggesting that the premix solution containing F-NIP-NGs had low selectivity for the developed abiotic/biotic hybrid sandwich detection. These results indicate that the proposed sensor's high affinity for PSA was increased via the molecular imprinting process.
To illustrate the selectivity of the PSA abiotic/biotic hybrid sandwich detection, the immobilized PSA-MIP-NGs on the gold-coated sensor chip were incubated with a premix solution containing 10 nM PSA or four animal serum albumins (BSA, bovine serum albumin; GSA, goat serum albumin; SSA, sheep serum albumin; and RSA, rabbit serum albumin) that served as potential interference matrices, Anti-PSA, and F-Fc-MIP-NGs. As shown in Fig. 3b, the premixed solution containing other animal serum albumins exhibited a much lower selectivity factor than PSA, confirming that the proposed sensor had high selectivity for PSA. This proposed system provided the highest selectivity for PSA detection, compared with our MIP-based sensors reported previously.9,10 The results indicated that the developed system's selectivity was increased by combining the abiotic antibodies and the biotic antibody for capturing and detecting PSA in solution.
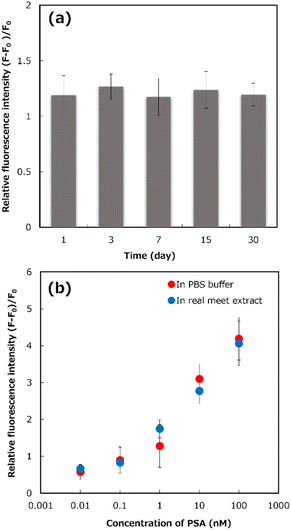
To evaluate the precision of the proposed system, the reproducibility of self-assembled monolayer preparation, capture PSA MIP-NG immobilization, and the blocking process was demonstrated using seven different premix solutions containing 1 nM PSA. The results, shown in Table S1,† show that the developed sensor's reproducibility was acceptable, with a relative standard deviation (%RSD) of 7.5%. The stability of the developed system was evaluated by detecting 1 nM PSA using immobilized PSA-MIP-NGs on the gold-coated sensor chip after storage for 30 days at 4 °C (Fig. 4a). After 30 days, the relative fluorescence intensity was not significantly different from that on the day of preparation. The results indicate that the sensor was stable and capable of PSA detection for at least one month.
An appropriate dilution of the meat extract was evaluated to detect PSA in real meat extract samples (Table S2†) to reduce the matrix interference effect for PSA detection. Different dilutions of real meat extract samples in PBS from 1- to 500-fold were prepared to detect 1 nM PSA using the detection system compared to the control solution (PBS). As shown in Fig. S21,† the relative fluorescence intensity in the system using undiluted and 10-fold diluted samples was much lower than that in the control solution, because of the interference matrix effects of the real meat extract samples, suggesting that PSA detection in the detection system was interrupted by the matrices in the meat extract samples. In contrast, the responses of 100- and 500-fold diluted meat extract samples were not significantly different from that of the control solution, confirming that the appropriate dilution of real meat extract sample for PSA detection using the developed system was a 100-fold dilution.
To demonstrate the accuracy of the detection system, a recovery test by spiking various concentrations of PSA (0–100 nM) into 100-fold diluted meat extract samples was performed (Fig. 4b). The fluorescent responses of the spiked real meat extracts for the developed system was similar to that of the PBS solution. Based on the binding profiles, the recovery rate results of the developed system, shown in Table S3,† were 90–136%. These results implied that PSA could be detected by the developed system, even in real meat extracts.
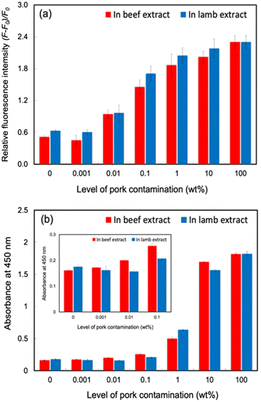
Under optimum conditions, the proposed system detected pork adulteration in halal meat extract samples (beef and lamb). Simulated pork-contaminated halal meat extract samples were prepared by mixing diluted pork extract and halal meat extracts solutions in the ranges of 0, 0.001, 0.01, 0.1, 1, 10, and 100 wt%. As shown in Fig. 5a, the fluorescent responses of the proposed sensor increased with increasing pork adulteration levels in beef and lamb extract samples. However, the response signals of 0.001% pork adulteration in halal meat extracts were close to those of the negative controls (100% of beef and lamb extracts) with overlapping error bars. Significant response signals were found for the 0.01 wt% contaminated samples, with non-overlapping error bars for the non-contaminated halal meat extract, indicating that the detection limit of pork adulteration in the two halal meat extracts for the proposed system was 0.01 wt%. With extremely low LODs of pork adulteration at 0.01 wt%, the potential of the proposed detection system for commercial use for identifying pork adulteration in halal raw meat samples is significant. Compared to our previous MIP-based methods for detecting pork adulteration in meat extracts,9,10 the abiotic/biotic hybrid sandwich detection system developed in this study provided increased sensitivity and selectivity. The sensitivity of the proposed system was also higher than those of previously developed immunoassay methods of pork adulteration detection based on indirect ELISA (0.5% w/w),5a sandwich ELISA (0.1% w/w),5b,e and lateral flow immunosensor (0.1% w/w).5d It was comparable to the highest sensitivity of pork adulteration detection using the enzyme immunoassay method with the same detection limit of 0.01 wt%.5f Moreover, the advantages of our developed detection system are the high sensitivity and selectivity and the reduced analysis time (from 4 h to 30 min) of detection of pork adulteration in halal meat extracts.
The effectiveness of the developed system in detecting pork adulteration in halal meat extract samples was compared with an immunosensor based on a sandwich ELISA. PSA solution at different dilutions in PBS buffer were used as control samples, and the detection limit was determined to be 0.01 wt% (Fig. S22a and b†). As shown in Fig. 5b, the ELISA method could not detect pork adulteration halal meat extract at 0.001 wt% simulated pork adulteration. At 0.01% pork adulteration, only pork contaminated with beef extract had a significantly different optical density (OD) value compared to the negative control, but pork adulteration in lamb extract was not detected. The observation may be due to the interference effect of the matrices in the raw lamb extracts. The results indicated that our detection system provided a sensitivity comparable to that of the ELISA standard method for detecting pork adulteration in halal meat extracts more rapidly.
Conclusion
This study used a combination of artificial and natural antibodies to develop a novel abiotic/biotic hybrid sandwich system for sensitively detecting pork adulteration in halal meat extracts. F-Fc-MIP-NGs were successfully developed using molecular imprinting and PIM with high affinity and selectivity for the Fc domain. They were used as a fluorescent secondary antibody mimic for the abiotic/biotic hybrid sandwich detection system. The system enhanced the sensitivity and selectivity of the MIP-based sensor. The developed system exhibited extremely sensitive detection of PSA with an exceptionally low LOD (0.5 ng mL−1) and excellent selectivity compared to other animal serum albumins (BSA, GSA, SSA, and RSA). Good analytical performance with 30 min analysis time, 7.5% RSD of repeatability, and >30 days of stability were achieved. The proposed detection system allowed the rapid detection of low levels of pork adulteration (0.01 wt%) in halal meat extract in 30 min, which was eight times faster than ELISA. Our detection system has a high potential for application in halal biomarker sensing owing to its rapid detection, sensitivity, specificity, easy preparation, and low cost. As various MIP-NGs can be easily prepared by changing the template molecules used and most of the corresponding polyclonal antibodies would be available commercially, the proposed abiotic/biotic hybrid sandwich detection system could be a powerful tool for establishing systems for detecting target proteins in broad areas, including food analysis, food control, diagnosis, and environmental analysis.Experimental section
The abiotic/biotic antibody hybrid sandwich detection was performed using the following procedure: (1) PSA-MIP-NGs were immobilized on a sensor chip, and the blocking process was performed using 0.5% w/v BSA; (2) the cocktail solution was prepared by mixing equal volumes of PBS containing 0.1 μg mL−1 of Anti-PSA with 100 μg mL−1 of F-Fc-MIP-NGs; (3) PSA was then added to the reaction mixture, followed by incubation for 30 min; (4) the premixed cocktail solution was dropped onto the PSA-MIP-NGs-immobilized sensor chip and incubated for 30 min; (5) after washing with pure water (3 × 500 μL) and PBS (3 × 500 μL), the sensor chip was inserted into a designed flat-type pipette tip, the fluorescence intensity was measured using a custom-made liquid-handling robot equipped with a fluorescence microscope. The relative fluorescence intensity was calculated using of the following equation: (F − F0)/F0, where F0 and F are the fluorescence intensities before and after incubation with PSA, respectively.Meat extract samples were prepared. Chopped raw meat (1 g) was mixed with 5 mL PBS, and then homogenized using a benchtop homogenizer (Polytron PT 1600 E, Kinematica AG, Luzern, Switzerland) for 2 min (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm), followed by centrifugation for 30 min at 4 °C (3 × 16
000 rpm), followed by centrifugation for 30 min at 4 °C (3 × 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g). The clear supernatant was collected and filtered thrice through a 0.2 μm polytetrafluoroethylene (PTFE) filter (DISMIC-13HP, Toyo Roshi Kaisha Ltd, Tokyo, Japan). The filtered meat extract samples were then used to measure total protein concentration using a NanoDrop One UV/Vis Spectrophotometer at 280 nm. The meat extract samples were then stored at −20 °C until use.
000g). The clear supernatant was collected and filtered thrice through a 0.2 μm polytetrafluoroethylene (PTFE) filter (DISMIC-13HP, Toyo Roshi Kaisha Ltd, Tokyo, Japan). The filtered meat extract samples were then used to measure total protein concentration using a NanoDrop One UV/Vis Spectrophotometer at 280 nm. The meat extract samples were then stored at −20 °C until use.
Author contributions
H. S. and T. T. conceived the project. C. C., H. S., E. T., and Y. K. performed the experiments. All authors contributed to the analysis and the interpretation of the results. The manuscript was written by C. C. and revised by H. S. and T. T. All authors have approved the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by JSPS KAKENHI (JP 18H05398 and JP 23H01775) and the Research Program of “Dynamic Alliance for Open Innovation Bridging Human, Environment and Materials” in “Network Joint Research Center for Materials and Devices”.References
- (a) S. Qureshi, M. Jamal, M. Qureshi, M. Rauf, B. Syed, M. Zulfiqar and N. Chand, J. Anim. Plant Sci., 2012, 2292, 79 Search PubMed; (b) E. Izberk-Bilgin and C. C. Nakata, Bus. Horiz., 2016, 59, 285–292 CrossRef.
- O. Al-Jowder, E. K. Kemsley and R. H. Wilson, Food Chem., 1997, 59, 195–201 CrossRef CAS.
- (a) B. V. Chikkaveeraiah, A. A. Bhirde, N. Y. Morgan, H. S. Eden and X. Chen, ACS Nano, 2012, 6, 6546–6561 CrossRef CAS PubMed; (b) S. Teepoo, N. Mhadbamrung, S. Moonmungmee and C. Cheubong, J. Bionanosci., 2016, 10, 63–68 CrossRef CAS.
- (a) B. Prieto-Simón, N. M. Bandaru, C. Saint and N. H. Voelcker, Biosens. Bioelectron., 2015, 67, 642–648 CrossRef PubMed; (b) J. H. Oh and M. K. Park, Food Control, 2016, 59, 780–786 CrossRef CAS.
- (a) F. C. Chen and Y. H. Hsieh, J. AOAC Int., 2000, 83, 79–85 CrossRef CAS PubMed; (b) L. Liu, F. C. Chen, J. L. Dorsey and Y.-H. P. Hsieh, J. Food Sci., 2006, 71, 1–6 CrossRef; (c) S. A. Lim and M. U. Ahmed, Food Chem., 2016, 206, 197–203 CrossRef PubMed; (d) B. Kuswandi, A. A. Gani and M. Ahmad, Food Biosci., 2017, 19, 1–6 CrossRef CAS; (e) C. P. Thienes, J. Masiri, L. A. Benoit, B. Barrios-Lopez, S. A. Samuel, D. P. Cox, A. P. Dobritsa, C. Nadala and M. Samadpour, J. AOAC Int., 2018, 101, 817–823 CrossRef CAS PubMed; (f) J. Mandli, I. El Fatimi, N. Seddaoui and A. Amine, Food Chem., 2018, 255, 380–389 CrossRef CAS PubMed.
- (a) K. Haupt, Molecular Imprinting, Springer-Verlag, Berlin, 2012 CrossRef; (b) R. Schirhagl, Anal. Chem., 2014, 86, 250–261 CrossRef CAS PubMed; (c) T. Takeuchi, T. Hayashi, S. Ichikawa, A. Kaji, M. Masui, H. Matsumoto and R. Sasao, Chromatography, 2016, 37, 43–64 CrossRef CAS; (d) M. Komiyama, T. Mori and K. Ariga, Bull. Chem. Soc. Jpn., 2018, 91, 1075–1111 CrossRef CAS; (e) K. Haupt, P. X. M. Rangel and B. Tse Sum Bui, Chem. Rev., 2020, 120(17), 9554–9582 CrossRef CAS PubMed; (f) Y. He and Z. Lin, J. Mater. Chem. B, 2022, 10, 6571–6589 RSC; (g) B. Tse Sum Bui, A. Mier and K. Haupt, Small, 2023, 19, 2206453 CrossRef CAS PubMed.
- (a) Y. Inoue, A. Kuwahara, K. Ohmori, H. Sunayama, T. Ooya and T. Takeuchi, Biosens. Bioelectron., 2013, 48, 113–119 CrossRef CAS PubMed; (b) Y. Suga, H. Sunayama, T. Ooya and T. Takeuchi, Chem. Commun., 2013, 49, 8450–8452 RSC; (c) T. Guo, Q. L. Deng, Z. G. Fang, C. C. Liu, X. Huang and S. Wang, Biosens. Bioelectron., 2015, 74, 498–503 CrossRef CAS PubMed; (d) L. Chen, X. Wang, W. Lu, X. Wu and J. Li, Chem. Soc. Rev., 2016, 45, 2137–2211 RSC; (e) G. Rijun, J. Hui, G. Huijun and W. Zonghua, Biosens. Bioelectron., 2018, 100, 56–70 CrossRef PubMed; (f) C. Lucia, P. Chiara, B. Paolo and A. M. Bossia, Talanta, 2018, 178, 772–779 CrossRef PubMed.
- (a) T. Takeuchi, Y. Kitayama, R. Sasao, T. Yamada, K. Toh, Y. Matsumoto and K. Kataoka, Angew. Chem., Int. Ed., 2017, 56, 7088–7092 CrossRef CAS PubMed; (b) T. Morishita, A. Yoshida, N. Hayakawa, K. Kiguchi, C. Cheubong, H. Sunayama, Y. Kitayama and T. Takeuchi, Langmuir, 2020, 36, 10674–10682 CrossRef CAS PubMed; (c) N. Hayakawa, T. Yamada, Y. Kitayama and T. Takeuchi, ACS Appl. Polym. Mater., 2020, 2(4), 1465–1473 CrossRef CAS.
- C. Cheubong, A. Yoshida, Y. Mizukawa, N. Hayakawa, M. Takai, T. Morishita, Y. Kitayama, H. Sunayama and T. Takeuchi, Anal. Chem., 2020, 92, 6401–6407 CrossRef CAS PubMed.
- C. Cheubong, E. Takano, Y. Kitayama, H. Sunayama, K. Minamoto, R. Takeuchi, S. Furutani and T. Takeuchi, Biosens. Bioelectron., 2021, 172, 112775 CrossRef CAS PubMed.
- (a) T. Takeuchi, T. Mori, A. Kuwahara, T. Ohta, A. Oshita, H. Sunayama, Y. Kitayama and T. Ooya, Angew. Chem., Int. Ed., 2014, 53, 12765–12770 CrossRef CAS PubMed; (b) T. Takeuchi and H. Sunayama, Chem. Commun., 2018, 54, 6243–6251 RSC; (c) H. Sunayama and T. Takeuchi, Chromatography, 2021, 42, 73–81 CrossRef CAS; (d) K. Mori, M. Hirase, T. Morishige, E. Takano, H. Sunayama, Y. Kitayama, S. Inubushi, R. Sasaki, M. Yashiro and T. Takeuchi, Angew. Chem., Int. Ed., 2019, 58, 1612–1615 CrossRef CAS PubMed; (e) T. Takeuchi, K. Mori, H. Sunayama, E. Takano, Y. Kitayama, T. Shimizu, Y. Hirose, S. Inubushi, R. Sasaki and H. Tanino, J. Am. Chem. Soc., 2020, 142, 6617–6624 CrossRef CAS PubMed.
- (a) H. Sunayama, T. Ooya and T. Takeuchi, Biosens. Bioelectron., 2010, 26, 458–462 CrossRef CAS PubMed; (b) H. Sunayama, T. Ohya and T. Takeuchi, Chem. Commun., 2014, 50, 1347 RSC; (c) H. Matsumoto, H. Sunayama, Y. Kitayama, E. Takano and T. Takeuchi, Sci. Technol. Adv. Mater., 2019, 20, 305–312 CrossRef CAS PubMed; (d) K. Tsutsumi, H. Sunayama, Y. Kitayama, E. Takano, Y. Nakamachi, R. Sasaki and T. Takeuchi, Adv. NanoBiomed Res., 2021, 1, 2000079 CrossRef CAS.
- Y. Li, Y. Sun, R. C. Beier, H. Lei, S. J. Gee, B. D. Hammock, H. Wang, Z. Wang, X. Sun, Y. Shen, J. Yang and Z. Xu, Trends Anal. Chem., 2017, 88, 25–40 CrossRef CAS.
- (a) N. Bereli, G. Ertürk, M. A. Tümer, R. Say and A. Denizli, Biomed. Chromatogr., 2013, 27, 599–607 CrossRef CAS PubMed; (b) A. Tretjakov, V. Syritski, J. Reut, R. Boroznjak, O. Volobujeva and A. Öpik, Microchim. Acta, 2013, 180, 1433–1442 CrossRef CAS; (c) A. Tretjakov, V. Syritski, J. Reut, R. Boroznjak and A. Öpik, Anal. Chim. Acta, 2016, 902, 182–188 CrossRef CAS PubMed; (d) R. Bai, Y. Sun, M. Zhao, Z. Han, J. Zhang, Y. Sun, W. Dong and S. Li, Talanta, 2021, 226, 122160 CrossRef CAS PubMed.
- E. Moczko, A. Guerreiro, C. Cáceres, E. Piletska, B. Sellergren and S. A. Piletsky, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2019, 1124, 1–6 CrossRef CAS PubMed.
- N. Hayakawa, Y. Kitayama, K. Igarashi, Y. Matsumoto, E. Takano, H. Sunayama and T. Takeuchi, ACS Appl. Mater. Interfaces, 2022, 14, 16074–16081 CrossRef CAS PubMed.
- T. Saeki, E. Takano, H. Sunayama, Y. Kitayama and T. Takeuchi, J. Mater. Chem. B, 2020, 8, 7987 RSC.
- (a) K. L. Bennett, S. V. Smith, R. J. Truscott and M. M. Sheil, Anal. Biochem., 1997, 245, 17–27 CrossRef CAS PubMed; (b) T. J. Kutzner, A. M. Higuero, M. Süßmair, J. Kopitz, M. Hingar, N. Díez-Revuelta, G. G. Caballero, H. Kaltner, I. Lindner, J. Abad-Rodríguez, D. Reusch and H. J. Gabius, Biochim. Biophys. Acta, Gen. Subj., 2020, 1864, 129449 CrossRef CAS PubMed.
- (a) E. Takano, N. Shimura, Y. Ujima, H. Sunayama, Y. Kitayama and T. Takeuchi, ACS Omega, 2019, 4, 1487–1493 CrossRef CAS; (b) E. Takano, N. Shimura, T. Akiba, Y. Kitayama, H. Sunayama, K. Abe, K. Ikebukuro and T. Takeuchi, Microchim. Acta, 2017, 184, 1595–1601 CrossRef CAS.
- (a) Q. R. Li, K. G. Yang, J. X. Liu, L. H. Zhang, Z. Liang, Y. K. Zhang and S. Li, Microchim. Acta, 2016, 183, 345–352 CrossRef CAS; (b) Y. P. Qin, H. Y. Wang, X. W. He, W. Y. Li and Y. K. Zhang, Talanta, 2018, 185, 620–627 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3nr02863a |
| This journal is © The Royal Society of Chemistry 2023 |