Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Self-assembled three-dimensional hydrogels based on graphene derivatives and cerium oxide nanoparticles: scaffolds for co-culture of oligodendrocytes and neurons derived from neural stem cells†
Yurena
Polo
 a,
Jon
Luzuriaga
a,
Jon
Luzuriaga
 b,
Sergio
Gonzalez de Langarica
b,
Sergio
Gonzalez de Langarica
 c,
Beatriz
Pardo-Rodríguez
c,
Beatriz
Pardo-Rodríguez
 b,
Daniel E.
Martínez-Tong
d,
Christos
Tapeinos
b,
Daniel E.
Martínez-Tong
d,
Christos
Tapeinos
 ef,
Irene
Manero-Roig
ef,
Irene
Manero-Roig
 bg,
Edurne
Marin
bg,
Edurne
Marin
 c,
Jone
Muñoz-Ugartemendia
c,
Jone
Muñoz-Ugartemendia
 c,
Gianni
Ciofani
c,
Gianni
Ciofani
 e,
Gaskon
Ibarretxe
e,
Gaskon
Ibarretxe
 b,
Fernando
Unda
b,
Fernando
Unda
 b,
Jose-Ramon
Sarasua
b,
Jose-Ramon
Sarasua
 c,
Jose Ramon
Pineda
c,
Jose Ramon
Pineda
 *bh and
Aitor
Larrañaga
*bh and
Aitor
Larrañaga
 *c
*c
aPolimerbio, Donostia-San Sebastian, Spain
bCell Signaling Lab, Department of Cell Biology and Histology, Faculty of Medicine and Nursing, University of the Basque Country (UPV/EHU), Leioa, Spain. E-mail: joseramon.pinedam@ehu.eus; Tel: +34 946 012 426
cGroup of Science and Engineering of Polymeric Biomaterials (ZIBIO Group), Department of Mining, Metallurgy Engineering and Materials Science, POLYMAT, University of the Basque Country (UPV/EHU), Bilbao, Spain. E-mail: aitor.larranagae@ehu.eus; Tel: +34 946 013 935
dPolymers and advanced materials: Physics, Chemistry and Technology, University of the Basque Country (UPV/EHU), Donostia-San Sebastian, Spain & Centro de Física de Materiales (UPV/EHU-CSIC), Donostia-San Sebastian, Spain
eSmart Bio-Interfaces, Istituto Italiano di Tecnologia, Pontedera, PI, Italy
fDivision of Pharmacy and Optometry, School of Health Sciences, Faculty of Biology, Medicine and Health, University of Manchester, Manchester, UK
gUniversité de Bordeaux IINS – UMR 5297, Bordeaux, France
hAchucarro Basque Center for Neuroscience Fundazioa, Leioa, Spain
First published on 8th February 2023
Abstract
Stem cell-based therapies have shown promising results for the regeneration of the nervous system. However, the survival and integration of the stem cells in the neural circuitry is suboptimal and might compromise the therapeutic outcomes of this approach. The development of functional scaffolds capable of actively interacting with stem cells may overcome the current limitations of stem cell-based therapies. In this study, three-dimensional hydrogels based on graphene derivatives and cerium oxide (CeO2) nanoparticles are presented as prospective supports allowing neural stem cell adhesion, migration and differentiation. The morphological, mechanical and electrical properties of the resulting hydrogels can be finely tuned by controlling several parameters of the self-assembly of graphene oxide sheets, namely the amount of incorporated reducing agent (ascorbic acid) and CeO2 nanoparticles. The intrinsic properties of the hydrogels, as well as the presence of CeO2 nanoparticles, clearly influence the cell fate. Thus, stiffer adhesion substrates promote differentiation to glial cell lineages, while softer substrates enhance mature neuronal differentiation. Remarkably, CeO2 nanoparticle-containing hydrogels support the differentiation of neural stem cells to neuronal, astroglial and oligodendroglial lineage cells, promoting the in vitro generation of nerve tissue grafts that might be employed in neuroregenerative cell therapies.
1. Introduction
Neurological disorders cause death or disability to more than 94 million people worldwide every year and this number is expected to rise to 103 million by 2030.1,2 Among them, stroke is the main cause of chronic impairment and the third leading cause of mortality worldwide.3 Stroke, like traumatic injuries, causes the death of the neural tissue in the affected area, but also provokes a dysfunction and secondary apoptotic death of the surrounding cells, inhibiting efficient restoration or functional recovery of the damaged tissue.3,4 Despite their huge socioeconomic impact, there is no available treatment for these conditions nowadays.5 Therefore, there is a need to explore new therapies that replace the injured neural network and promote the integration of new neurons and glial cells into the central nervous system (CNS).Among the different approaches to regenerate the CNS, cell-based therapies, particularly those based on neural stem cells (NSCs), offer the most straightforward alternative to reestablish a functional neural network by producing therapeutic factors, promoting the self-restoration of the damaged tissue and ultimately replacing the lost neural cells.4,6 However, the integration of the cells into the host CNS remains challenging.7 In this regard, tissue engineering offers the possibility to combine NSCs with scaffolds to enhance cell integration on the damaged area.6 Within the recently coined field of materiobiology, scaffolds are considered multifunctional devices with the capability to finely tune biological functions.8 In the particular case of nervous system regeneration, materials based on graphene derivatives have attracted considerable attention9 thanks to the possibility to create moldable platforms (e.g., with tailored chemical, mechanical and electrical features) to promote the adhesion and differentiation of NSCs towards functional glial and neuronal lineages.10,11 Graphene consists of a single layer of carbon atoms organized in a honeycomb lattice monolayer that can be arranged either in two- (2D) or three-dimensional (3D) scaffolds, thus resembling the complex architecture of the extracellular matrix.12,13 Although most of the studies with NSCs have been performed in 2D graphene derivative-based scaffolds, recently, 3D scaffolds have been reported to better promote the proliferative ability of NSCs, while maintaining similar adhesion features.14 Moreover, the physical properties of these 3D scaffolds including stiffness or pore geometry can modulate the adhesion, proliferation and differentiation capabilities of the NSCs.13 Understanding how these features interact in the formation of complex neural networks that support mature neuronal and glial interplay will be vital to ensure the successful integration of the NSCs into the graphene derivative-based 3D scaffolds.
Three-dimensional composite scaffolds offer several benefits concerning single material scaffolds since they allow a simple modulation of their physicochemical, mechanical and electrical properties by simply modifying their composition while exploiting potential synergistic effects among their constituents.15 Herein, the combination of 3D scaffolds made of graphene-derivatives and cerium oxide (CeO2) nanoparticles will be explored as a platform for the regeneration of the CNS. CeO2 nanoparticles comprise a cubic fluorite arrangement that acts as redox reaction sites thanks to the oxygen deficiencies at the nanoscale order.16 This oxygen deprivation endows antioxidant features to the CeO2 nanoparticles that resemble the activity of antioxidant natural enzymes (i.e., superoxide dismutase (SOD) and catalase (CAT)), which is beneficial for the promotion of the angiogenesis and the restoration of the neural architecture.17,18 Accordingly, CeO2 nanoparticles have been reported to provide neuroprotective effects, as demonstrated on an adult spinal cord neuron model19 and in vivo on a pharmacologically induced brain oxidative stress model.20
In this study, we combine the potential of graphene-based materials to promote the adhesion, proliferation and differentiation of NSCs, with the additional antioxidant and neuroprotective effects associated with CeO2 nanoparticles as a prospective approach for the restoration of the injured CNS. To achieve this aim, we engineered and characterized 3D hydrogels based on the combination of graphene derivatives and CeO2 nanoparticles with tunable stiffness, porous geometry and electrical conductivity. We further studied the adhesion, integration and differentiation capabilities of the NSCs towards neuronal, astroglial, and oligodendroglial lineages at different time points. This allowed us to establish heterocellular cultures for in vitro studies that mimicked the in vivo CNS tissue architecture.
2. Materials and methods
2.1. Fabrication of the hydrogels
Graphene oxide (GO) solution (4 mg ml−1 aqueous dispersion from Graphenea, Spain) was pre-diluted with distilled water to a concentration of 2 mg ml−1 and sonicated for 30 min. Cerium oxide (CeO2) nanoparticles (246 mg ml−1, cerium(IV) oxide, 20% in water from Thermo Fisher Scientific, USA) were also sonicated for 30 min to ensure a homogeneous dispersion. For those hydrogels containing CeO2 nanoparticles, the two dispersions were mixed in the corresponding GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CeO2 weight ratios (10
CeO2 weight ratios (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.25; 10
0.25; 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5; 10
0.5; 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.6; 10
0.6; 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.75 or 10
0.75 or 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and the mixture was then incubated under magnetic stirring for 3 h at 90 °C to facilitate the electrostatic interactions between the negatively charged GO sheets and the positively charged CeO2 nanoparticles. Samples containing only GO were also treated for 3 h at 90 °C to ensure equal thermal treatment in all the samples. Afterwards, L-ascorbic acid (AsA) (Sigma Aldrich, Spain) was added to reduce the GO sheets and enable hydrogel formation. The corresponding GO
1) and the mixture was then incubated under magnetic stirring for 3 h at 90 °C to facilitate the electrostatic interactions between the negatively charged GO sheets and the positively charged CeO2 nanoparticles. Samples containing only GO were also treated for 3 h at 90 °C to ensure equal thermal treatment in all the samples. Afterwards, L-ascorbic acid (AsA) (Sigma Aldrich, Spain) was added to reduce the GO sheets and enable hydrogel formation. The corresponding GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA weight ratio (1
AsA weight ratio (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; 1
1; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4; 1
4; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) was mixed for 30 min under stirring and the self-assembly of the GO was promoted by incubating the solution at 60 °C overnight. The medium of the resulting hydrogels was replaced with distilled water every 12 h for 7 days to ensure the complete elimination of the unreacted AsA. Hydrogels either were lyophilized for characterization or sterilized with 70% ethanol every 12 h for five times. Sterilized hydrogels were washed with sterile phosphate buffered saline (PBS) every 12 h for four times and finally replaced by Neurocult basal medium (STEMCELL Technologies, Canada) one day prior to cell culture experiments.
10) was mixed for 30 min under stirring and the self-assembly of the GO was promoted by incubating the solution at 60 °C overnight. The medium of the resulting hydrogels was replaced with distilled water every 12 h for 7 days to ensure the complete elimination of the unreacted AsA. Hydrogels either were lyophilized for characterization or sterilized with 70% ethanol every 12 h for five times. Sterilized hydrogels were washed with sterile phosphate buffered saline (PBS) every 12 h for four times and finally replaced by Neurocult basal medium (STEMCELL Technologies, Canada) one day prior to cell culture experiments.
2.2. Physico-chemical and functional characterization
2.3. Cell culture
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA (GO
AsA (GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and four times more AsA (GO
1) and four times more AsA (GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) were selected for cell experiments. The effect of the incorporation of CeO2 nanoparticles was studied with the GO
4) were selected for cell experiments. The effect of the incorporation of CeO2 nanoparticles was studied with the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 hydrogel containing a proportion of GO
4 hydrogel containing a proportion of GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CeO2 of 10
CeO2 of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.25 (GO
0.25 (GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25).
4 + CeO2 0.25).
As the hydrogels have different diameters, 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells were seeded on the GO
000 cells were seeded on the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 30
1 and 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells were seeded on the GO
000 cells were seeded on the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 and GO
4 and GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 hydrogels to keep a similar cell density. After 24 h, medium was changed to NeuroCult™ differentiation medium and cells were cultured as previously described.11
4 + CeO2 0.25 hydrogels to keep a similar cell density. After 24 h, medium was changed to NeuroCult™ differentiation medium and cells were cultured as previously described.11
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; GO
1; GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 and GO
4 and GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 hydrogels and subsequently fixed for 1 h at room temperature using 2% glutaraldehyde (cat# 50-262-19, Fisher Scientific, Pittsburgh, USA) diluted in 0.1 M cacodylate buffer (cat# C0250, Sigma-Aldrich, Spain). Cacodylate buffer and isosmolar sucrose (cat# S7903, Sigma-Aldrich, Spain) were then used to rinse the fixative solution. The samples were post-fixed for 1 h in the dark using 1% osmium tetraoxide (OsO4) in cacodylate (cat# O5500, Sigma-Aldrich, Spain). Then, OsO4 was eliminated by rinsing for 10 min each hydrogel with cacodylate buffer. Hydrogels were dehydrated with increasing series of EtOH (30°, 50°, 70°, 90°, 96°, 100°, and 2 × 100° absolute) for 20 min each and air dried for 5 h. Afterwards, conductive cement was used to mount the hydrogels on the scanning electron microscope supports. Finally, gold nanoparticles were flashed in an argon environment to create a metallic coating. Samples were observed using a Hitachi S-3400 N scanning electron microscope (Hitachi, Tokio, Japan).
4 + CeO2 0.25 hydrogels and subsequently fixed for 1 h at room temperature using 2% glutaraldehyde (cat# 50-262-19, Fisher Scientific, Pittsburgh, USA) diluted in 0.1 M cacodylate buffer (cat# C0250, Sigma-Aldrich, Spain). Cacodylate buffer and isosmolar sucrose (cat# S7903, Sigma-Aldrich, Spain) were then used to rinse the fixative solution. The samples were post-fixed for 1 h in the dark using 1% osmium tetraoxide (OsO4) in cacodylate (cat# O5500, Sigma-Aldrich, Spain). Then, OsO4 was eliminated by rinsing for 10 min each hydrogel with cacodylate buffer. Hydrogels were dehydrated with increasing series of EtOH (30°, 50°, 70°, 90°, 96°, 100°, and 2 × 100° absolute) for 20 min each and air dried for 5 h. Afterwards, conductive cement was used to mount the hydrogels on the scanning electron microscope supports. Finally, gold nanoparticles were flashed in an argon environment to create a metallic coating. Samples were observed using a Hitachi S-3400 N scanning electron microscope (Hitachi, Tokio, Japan).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, ab6142, Abcam, United Kingdom). Cell commitment towards neuronal lineage was determined by positive staining for DCX (1
200, ab6142, Abcam, United Kingdom). Cell commitment towards neuronal lineage was determined by positive staining for DCX (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300, sc8066, Santa Cruz, USA) and MAP2 (1
300, sc8066, Santa Cruz, USA) and MAP2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, ab5392, Abcam, United Kingdom). On the other hand, the commitments towards astroglial and oligodendroglial cell lineages were determined using GFAP (1
500, ab5392, Abcam, United Kingdom). On the other hand, the commitments towards astroglial and oligodendroglial cell lineages were determined using GFAP (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, G9269, Sigma-Aldrich, USA), S100β (1
500, G9269, Sigma-Aldrich, USA), S100β (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, Dako Cytomation, Denmark) and Olig2 (1
200, Dako Cytomation, Denmark) and Olig2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, MABN50, Millipore, USA). Alexa Fluor donkey anti-mouse, anti-rabbit, anti-goat or anti-chicken secondary antibodies coupled with 488 or 594 fluorophores (1
200, MABN50, Millipore, USA). Alexa Fluor donkey anti-mouse, anti-rabbit, anti-goat or anti-chicken secondary antibodies coupled with 488 or 594 fluorophores (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, Life Technologies, USA) and goat anti-chicken Texas Red (1
200, Life Technologies, USA) and goat anti-chicken Texas Red (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, ab6875, Abcam, United Kingdom) were used as secondary antibodies. To study the migration of cells into the hydrogels, serial cuts of 250 μm thickness slices of previously immunostained and 1% agarose embedded hydrogels were made and collected using a Microm HM650 V vibratome (Microm, International GmbH, Walldorf, Germany).
200, ab6875, Abcam, United Kingdom) were used as secondary antibodies. To study the migration of cells into the hydrogels, serial cuts of 250 μm thickness slices of previously immunostained and 1% agarose embedded hydrogels were made and collected using a Microm HM650 V vibratome (Microm, International GmbH, Walldorf, Germany).
| Primers | Sequence 5′–3′ | Lenght | Annealing | Amplicon (bp) |
|---|---|---|---|---|
| Gapdh (upstream) | GTATGACTCCACTCACGGCAA | 21 | 61.8 | 274 |
| Gapdh (downstream) | CTTCCCATTCTCGGCCTTG | 19 | 60.2 | 274 |
| Nestin (upstream) | CCCTGAAGTCGAGGAGCTG | 19 | 61.4 | 166 |
| Nestin (downstream) | CTGCTGCACCTCTAAGCGA | 19 | 61.7 | 166 |
| Gfap (upstream) | CTGGACTGCGTCATTTTCCC | 20 | 59.2 | 256 |
| Gfap (downstream) | CGATGGAGCCTCAGGGATGA | 20 | 61.1 | 256 |
| S100β (upstream) | TGGCTGCGGAAGTTGAGATT | 20 | 59.9 | 84 |
| S100β (upstream) | ATGGCTCCCAGCAGCTAAAG | 20 | 60.1 | 84 |
| Olig2 (upstream) | GTGGATGCTTATTACAGACC | 20 | 56.1 | 94 |
| Olig2 (downstream) | ACCTTCCGAATGTGAATTAG | 20 | 58.1 | 94 |
| Map2 (upstream) | GAAGAAACAGCTAATCTGCC | 20 | 58.1 | 423 |
| Map2 (downstream) | CTCTTGCTTATTCCATCAGTG | 21 | 59.0 | 423 |
| Dcx (upstream) | TCAGCATCTCCACCCAACCA | 20 | 61.1 | 94 |
| Dcx (downstream) | TTGTGCTTTCCCGGTTGACA | 20 | 60.3 | 94 |
For the quantitative PCR, 4.5 μL of SsoAdvanced Universal SYBR® Green Supermix (1725271; BioRad, Hercules, California, USA) were mixed with 0.5 μL of primers (0.3125 M), 0.3 μL of cDNA (1.5 ng μL−1) and the necessary nuclease free water to reach 10 μL final volume reaction per well. Each primer was evaluated for optimal efficacy (>90%) and single product amplification using the melting curve approach. The 2−ΔΔCt technique was employed to determine the relative expression of each gene, with Gapdh serving as internal control.11,21 qPCR was carried out in triplicate using an ABI PRISM® 7000 (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Data were examined using the CFX Manager™ program. For statistical analysis, 3 independent hydrogels were analyzed, and each of the samples measured in triplicate.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; GO
1; GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 and GO
4 and GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 hydrogels. After 14 days in vitro, NSCs were labeled with 50 μM of the 2′,7′-dichlorodihydrofluorescein diacetate cell-permeable probe (H2DCFDA, Invitrogen, USA) for 30 min at 37 °C and 5% CO2 to evaluate the accumulation of reactive oxygen species (ROS) within cells. Fluorescence intensity of 3 independent samples of each condition was determined on Fluoroskan Ascent plate fluorimeter (Thermo Labsystems, Santa Rosa, California, USA) to determine the fluorescence intensity (λex = 488 nm/λem = 520 nm) in the area.
4 + CeO2 0.25 hydrogels. After 14 days in vitro, NSCs were labeled with 50 μM of the 2′,7′-dichlorodihydrofluorescein diacetate cell-permeable probe (H2DCFDA, Invitrogen, USA) for 30 min at 37 °C and 5% CO2 to evaluate the accumulation of reactive oxygen species (ROS) within cells. Fluorescence intensity of 3 independent samples of each condition was determined on Fluoroskan Ascent plate fluorimeter (Thermo Labsystems, Santa Rosa, California, USA) to determine the fluorescence intensity (λex = 488 nm/λem = 520 nm) in the area.
3. Results and discussion
3.1. Effect of AsA on the morphology, mechanical and electrical conductivity of the self-assembled three-dimensional hydrogels
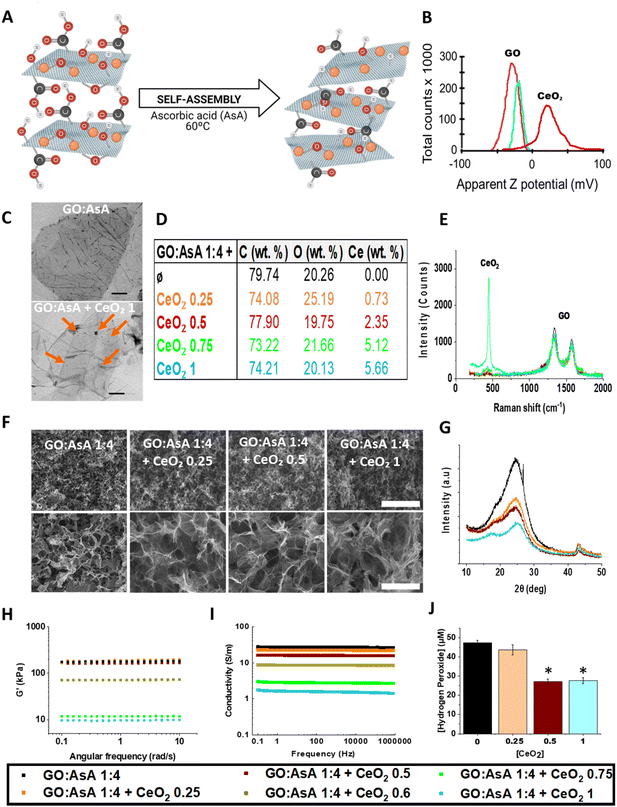
Graphene oxide (GO) is a carbon-based material bearing oxygenated functional groups, which improve the solubility and induce the expansion of the interlayer distance in aqueous solutions.22 The reduction of the GO provokes the deletion of oxygen and other atomic-scale lattice defects, thus enhancing the hydrophobic nature and promoting the accumulation of the sheets via non-covalent interactions (Fig. 1A).22,23 GO can be reduced in several ways, including chemical, thermal, electrochemical or light-driven approaches.22,24 Chemical reduction, in combination with thermal annealing, results more effective in repairing vacancy defects and removing out-of-plane carbonyl groups.22 Herein, we combined the chemical reduction with an overnight thermal treatment at 60 °C, using AsA as a reducing agent to overcome the limitations of more toxic alternatives.25 This represents an easy and quick manner to synthesize 3D hydrogels by self-assembly with high C/O ratios without compromising mechanical and electrical properties.25,26The absence or presence of oxygen functionalities will therefore have a direct impact on the final properties of the hydrogels.27 The different proportions of GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA allowed us to modulate both the reduction level and the arrangement of the GO sheets, which determined the pore size of the hydrogels (Fig. 1B). Scanning electron micrographs revealed the large porous structures formed by atomic wide walls of GO sheets. Increasing the amount of AsA diminished the repulsion forces between the GO sheets and enabled the formation of more compact structures with smaller pores. The GO
AsA allowed us to modulate both the reduction level and the arrangement of the GO sheets, which determined the pore size of the hydrogels (Fig. 1B). Scanning electron micrographs revealed the large porous structures formed by atomic wide walls of GO sheets. Increasing the amount of AsA diminished the repulsion forces between the GO sheets and enabled the formation of more compact structures with smaller pores. The GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 hydrogel showed the largest pores (7.3 ± 0.6 μm) with respect to the other two formulations (i.e., GO
1 hydrogel showed the largest pores (7.3 ± 0.6 μm) with respect to the other two formulations (i.e., GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 showing pores of 3.9 ± 0.2 μm & 1
4 showing pores of 3.9 ± 0.2 μm & 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 with pores of 4.3 ± 0.5 μm). In agreement with these results, X-ray diffraction demonstrated that the addition of an increasing amount of AsA triggered the reduction of GO sheets, resulting in the displacement of the diffraction peak to higher values (i.e., from 10° in the commercial GO to 24° in the GO
10 with pores of 4.3 ± 0.5 μm). In agreement with these results, X-ray diffraction demonstrated that the addition of an increasing amount of AsA triggered the reduction of GO sheets, resulting in the displacement of the diffraction peak to higher values (i.e., from 10° in the commercial GO to 24° in the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 & 1
4 & 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) (Fig. 1C). According to Bragg's law, the interlayer distance of the commercial GO was 0.85 nm and decreased till 0.77 nm in the GO
10) (Fig. 1C). According to Bragg's law, the interlayer distance of the commercial GO was 0.85 nm and decreased till 0.77 nm in the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 sample. The chemical reduction of GO stimulated the self-assembly of the reduced GO sheets thanks to the reduction of hydroxyl, epoxy, carboxyl and carbonyl groups,28 that enabled the formation of new π–π binding sites between the GO sheets. The deletion of oxygenated functional groups also raised the hydrophobicity of the graphene sheets. The combined effect of these two events provoked a random overlapping of flexible graphene sheets, thus favouring the self-assembly of the 3D hydrogels. Remarkably, the addition of more AsA decreased the interlayer distance in the GO
1 sample. The chemical reduction of GO stimulated the self-assembly of the reduced GO sheets thanks to the reduction of hydroxyl, epoxy, carboxyl and carbonyl groups,28 that enabled the formation of new π–π binding sites between the GO sheets. The deletion of oxygenated functional groups also raised the hydrophobicity of the graphene sheets. The combined effect of these two events provoked a random overlapping of flexible graphene sheets, thus favouring the self-assembly of the 3D hydrogels. Remarkably, the addition of more AsA decreased the interlayer distance in the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 and GO
4 and GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 hydrogels till 0.37 nm in both cases, but no differences were observed between these two formulations, suggesting that the reduction level was similar in both cases. These results were further corroborated by XPS (ESI Fig. 1†). The addition of AsA at a GO
10 hydrogels till 0.37 nm in both cases, but no differences were observed between these two formulations, suggesting that the reduction level was similar in both cases. These results were further corroborated by XPS (ESI Fig. 1†). The addition of AsA at a GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio clearly reduced the area associated to oxygen functionalities with respect to the original GO. Oxygen functionalities were further reduced in the GO
1 ratio clearly reduced the area associated to oxygen functionalities with respect to the original GO. Oxygen functionalities were further reduced in the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 ratio but no significant differences were observed when the GO
4 ratio but no significant differences were observed when the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA ratio was increased to 1
AsA ratio was increased to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, suggesting that the maximum level of reduction had been reached. As observed, the reduced graphene oxide hydrogels still contain oxygen functionalities in their structure, which are absent in the structure of pure graphite.
10, suggesting that the maximum level of reduction had been reached. As observed, the reduced graphene oxide hydrogels still contain oxygen functionalities in their structure, which are absent in the structure of pure graphite.
The mechanical properties of the hydrogels were measured by rheology. The GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 hydrogel showed the lowest shear modulus (G′) (22.8 ± 0.3 kPa), which in viscous materials is directly correlated with the elastic capabilities (Fig. 1D). The GO
1 hydrogel showed the lowest shear modulus (G′) (22.8 ± 0.3 kPa), which in viscous materials is directly correlated with the elastic capabilities (Fig. 1D). The GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 and GO
4 and GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 presented increased shear modulus (178.4 ± 2.8 kPa vs. 186.4 ± 2.6 kPa), demonstrating the modulation of the mechanical properties in almost one order of magnitude by controlling the reduction level through simply modifying the amount of AsA added. As shown in other studies and by us, once the maximum reduction level is achieved, the mechanical properties of the hydrogels remain stable.29 Although the human brain presents a low shear modulus of around 1–2.5 kPa,30 other central nervous system areas like the spinal cord recorded shear modulus of 250–300 kPa
10 presented increased shear modulus (178.4 ± 2.8 kPa vs. 186.4 ± 2.6 kPa), demonstrating the modulation of the mechanical properties in almost one order of magnitude by controlling the reduction level through simply modifying the amount of AsA added. As shown in other studies and by us, once the maximum reduction level is achieved, the mechanical properties of the hydrogels remain stable.29 Although the human brain presents a low shear modulus of around 1–2.5 kPa,30 other central nervous system areas like the spinal cord recorded shear modulus of 250–300 kPa![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 31 and the peripheral nervous system like ulnar and median nerves register a shear modulus around 10–20 kPa,32 which are similar to the mechanical properties presented by our hydrogels. Besides, it is reported the acquirement of well differentiated neural cultures of stem cells in vitro and in vivo with scaffolds that present even higher stiffness values,33 suggesting that all the hydrogels exhibited mechanical properties compatible with neural differentiation of stem cells.
31 and the peripheral nervous system like ulnar and median nerves register a shear modulus around 10–20 kPa,32 which are similar to the mechanical properties presented by our hydrogels. Besides, it is reported the acquirement of well differentiated neural cultures of stem cells in vitro and in vivo with scaffolds that present even higher stiffness values,33 suggesting that all the hydrogels exhibited mechanical properties compatible with neural differentiation of stem cells.
GO is a poor electrical conductor due to the lack of percolating conduits between sp2 carbon atoms that act as electron carriers in graphene. The reduction process with AsA induces the deletion of oxygen functionalities and raises the amount of sp2 or π–π binding sites which consequently increases the conductivity of the material.34 In accordance with the data of other studies,35 the electrical conductivity of the hydrogels increased with the addition of AsA. The GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 hydrogel exhibited the lowest electrical conductivity (0.6 S m−1), which increased in GO
1 hydrogel exhibited the lowest electrical conductivity (0.6 S m−1), which increased in GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (27 S m−1) and GO
4 (27 S m−1) and GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (35 S m−1) (Fig. 1E). Accordingly, we were able to enhance the electrical conductivity thanks to the deletion of atomic-scale lattice defects of the GO sheets via the thermochemical reduction process.22 According to these results, other groups have reported electrical conductivities between 0.045 and 600 S m−1 on graphene-based hydrogels.36–38 Remarkably, as it has been previously reported, the impedance or the electrical conductivity are constant in graphene oxide as usually observed for highly conducting systems.39,40 The human brain has an electrical conductivity of around 0.33 S m−1, where grey matter exhibits a conductivity of around 0.47 ± 0.24 S m−1 and white matter around 0.22 ± 0.17 S m−1.41,42 However, materials showing electrical conductivity values between 0.08–1.3 S m−1 or even higher values are able to electrically stimulate neurons.43,44 Therefore, all our hydrogels showed conductivity values compatible with neural stimulation.
10 (35 S m−1) (Fig. 1E). Accordingly, we were able to enhance the electrical conductivity thanks to the deletion of atomic-scale lattice defects of the GO sheets via the thermochemical reduction process.22 According to these results, other groups have reported electrical conductivities between 0.045 and 600 S m−1 on graphene-based hydrogels.36–38 Remarkably, as it has been previously reported, the impedance or the electrical conductivity are constant in graphene oxide as usually observed for highly conducting systems.39,40 The human brain has an electrical conductivity of around 0.33 S m−1, where grey matter exhibits a conductivity of around 0.47 ± 0.24 S m−1 and white matter around 0.22 ± 0.17 S m−1.41,42 However, materials showing electrical conductivity values between 0.08–1.3 S m−1 or even higher values are able to electrically stimulate neurons.43,44 Therefore, all our hydrogels showed conductivity values compatible with neural stimulation.
3.2. Effect of CeO2 nanoparticles on the morphology, mechanical, electrical and antioxidant properties of the self-assembled three-dimensional hydrogels
Cerium oxide (CeO2) nanoparticles are widely known for possessing antioxidant and neuroprotective properties.45 Here, CeO2 nanoparticles were incorporated during the self-assembly of the graphene-based hydrogels, a process that is favored by electrostatic interactions between the negatively charged GO sheets (−29.1 ± 0.36 mV) and the positively charged CeO2 nanoparticles (24.3 ± 0.57 mV), as demonstrated by dynamic light scattering (DLS) (Fig. 2A and B).To confirm the presence of CeO2 nanoparticles in our samples, we performed transmission electron microscopy (TEM). TEM micrographs showed the GO sheets successfully decorated with CeO2 nanoparticles prior to their reduction with AsA (Fig. 2C). Moreover, the amount of CeO2 nanoparticles on the surface of the GO sheets increased with the addition of more CeO2 to the samples (data not shown). These results were further corroborated with Raman spectroscopy and energy-dispersive X-ray spectrometry (EDX) (Fig. 2D, E and ESI Fig. 2†). Raman spectroscopy demonstrated that all the samples that contained CeO2 nanoparticles showed a band at 453 cm−1 that can be ascribed to the first order scattering of CeO2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 46 and increased in intensity as the amount of CeO2 nanoparticles raised. As expected, the GO
46 and increased in intensity as the amount of CeO2 nanoparticles raised. As expected, the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 sample only exhibited the characteristic D and G bands (1350 and 1580 cm1 respectively) which are related to the disarranged sp2-hybridized carbon structure and expansion of the C–C bond in graphitic materials.11,47 In accordance with these results, EDX demonstrated the presence of Ce in the samples containing CeO2 nanoparticles and presented similar proportions for carbon (C) (73.22–79.74%) and oxygen (O) (19.75–25.19%) in all the samples.
4 sample only exhibited the characteristic D and G bands (1350 and 1580 cm1 respectively) which are related to the disarranged sp2-hybridized carbon structure and expansion of the C–C bond in graphitic materials.11,47 In accordance with these results, EDX demonstrated the presence of Ce in the samples containing CeO2 nanoparticles and presented similar proportions for carbon (C) (73.22–79.74%) and oxygen (O) (19.75–25.19%) in all the samples.
SEM images demonstrated that, despite the addition of CeO2 nanoparticles, hydrogels preserved their highly porous structures formed by atomic wide walls of GO sheets (Fig. 2F). The estimated pore size was, however, slightly reduced after the incorporation of CeO2 nanoparticles. Accordingly, the estimated pore size were 3.9 ± 0.2 μm, 2.1 ± 0.1 μm, 2.6 ± 0.2 μm and 2.7 ± 0.2 μm for GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, GO
4, GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25, GO
4 + CeO2 0.25, GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.5 and GO
4 + CeO2 0.5 and GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 1, respectively. The XRD showed a peak at 24° in all the samples (Fig. 2G), demonstrating that the interlayer distance was not affected by the addition of CeO2 nanoparticles. Nevertheless, the intensity of the peak dropped down with the addition of CeO2 nanoparticles to the samples in a dose dependent manner, denoting an interlayer distortion of the GO sheets, mediated by the CeO2 nanoparticles. Remarkably, in all the samples there was a peak at 43° which corresponded to the plane of the graphene layer48 and indicated that the CeO2 nanoparticles were localized on the interlayer space without inducing any variation in the GO sheets.49
4 + CeO2 1, respectively. The XRD showed a peak at 24° in all the samples (Fig. 2G), demonstrating that the interlayer distance was not affected by the addition of CeO2 nanoparticles. Nevertheless, the intensity of the peak dropped down with the addition of CeO2 nanoparticles to the samples in a dose dependent manner, denoting an interlayer distortion of the GO sheets, mediated by the CeO2 nanoparticles. Remarkably, in all the samples there was a peak at 43° which corresponded to the plane of the graphene layer48 and indicated that the CeO2 nanoparticles were localized on the interlayer space without inducing any variation in the GO sheets.49
The mechanical behavior of the hydrogels decorated with CeO2 nanoparticles was again measured by rheology. The GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 and GO
4 + CeO2 0.25 and GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.5 (187.7 ± 7.5 kPa and 164.6 ± 1.6, respectively) showed a shear modulus similar to the GO
4 + CeO2 0.5 (187.7 ± 7.5 kPa and 164.6 ± 1.6, respectively) showed a shear modulus similar to the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 hydrogel (178.4 ± 2.8 kPa). A gradual decrease on the mechanical properties was however observed in the GO
4 hydrogel (178.4 ± 2.8 kPa). A gradual decrease on the mechanical properties was however observed in the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.6 (71.6 ± 0.7 kPa), GO
4 + CeO2 0.6 (71.6 ± 0.7 kPa), GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.75 (11.8 ± 0.1 kPa) and GO
4 + CeO2 0.75 (11.8 ± 0.1 kPa) and GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 1 (9.7 ± 0.1 kPa) hydrogels (Fig. 2H). The fact that the shear modulus decreased with the addition of more CeO2 nanoparticles to the samples indicated the possibility to tune the mechanical properties of the hydrogel by just modifying the CeO2 nanoparticle concentration. These results are in line with those obtained by other groups using dopant nanoparticle substances like platinum50 and can be explained due to the less organized structures associated to the incorporation of the CeO2 nanoparticles in the interlayer space of the GO sheets, as suggested by XRD.
4 + CeO2 1 (9.7 ± 0.1 kPa) hydrogels (Fig. 2H). The fact that the shear modulus decreased with the addition of more CeO2 nanoparticles to the samples indicated the possibility to tune the mechanical properties of the hydrogel by just modifying the CeO2 nanoparticle concentration. These results are in line with those obtained by other groups using dopant nanoparticle substances like platinum50 and can be explained due to the less organized structures associated to the incorporation of the CeO2 nanoparticles in the interlayer space of the GO sheets, as suggested by XRD.
We also measured the electrical properties of the hydrogels. GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 and GO
4 + CeO2 0.25 and GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.5 (22 S m−1 and 17 S m−1) maintained almost the same electrical conductivity of the GO
4 + CeO2 0.5 (22 S m−1 and 17 S m−1) maintained almost the same electrical conductivity of the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (27 S m−1) (Fig. 2I). However, the addition of an increasing amount of CeO2 nanoparticles resulted in a decline of the electrical conductivity of the GO
4 (27 S m−1) (Fig. 2I). However, the addition of an increasing amount of CeO2 nanoparticles resulted in a decline of the electrical conductivity of the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.6, GO
4 + CeO2 0.6, GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.75 and GO
4 + CeO2 0.75 and GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 1 hydrogels (8.5 S m−1; 2.8 S m−1 and 1.6 S m−1 respectively), which may be attributed to the more disordered arrangement of the GO sheets due to the incorporation of the CeO2 nanoparticles in the interlayer space. Nevertheless, the electrical values obtained proved that all our hydrogels could potentially trigger the electrical excitation of the seeded neural cells.43,44
4 + CeO2 1 hydrogels (8.5 S m−1; 2.8 S m−1 and 1.6 S m−1 respectively), which may be attributed to the more disordered arrangement of the GO sheets due to the incorporation of the CeO2 nanoparticles in the interlayer space. Nevertheless, the electrical values obtained proved that all our hydrogels could potentially trigger the electrical excitation of the seeded neural cells.43,44
It is well known that cerium oxide can exhibit +3 and +4 states that support the formation of CeO2 and CeO2−x and provides antioxidant properties.51 CeO2 nanoparticles resemble the antioxidant enzymes superoxide dismutase (SOD) and catalase (CAT) and, hence, scavenge reactive oxygen species (ROS) and free radicals51,52 in physiological conditions.45,53,54 Here, we applied a concentration of 50 μM hydrogen peroxide (H2O2) and measured the antioxidant capabilities of the hydrogels containing increasing amounts of CeO2 nanoparticles. It was reported in the literature that this H2O2 concentration is able to mimic the pro-oxidative environment found in vivo which may cause a detrimental effect on important cellular structures, thus leading to oxidative distress.55 As expected, all the hydrogels containing CeO2 nanoparticles were able to reduce the hydrogen peroxide concentration in a dose dependent manner. In contrast, the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 sample had no antioxidant properties, demonstrating that the CeO2 nanoparticles were responsible of the decline on the hydrogen peroxide concentration (Fig. 2J). These results are in agreement with other studies were CeO2 nanoparticles have demonstrated to possess antioxidant properties in vitro and in vivo, creating a more favorable microenvironment for angiogenesis and nerve reconstruction, resulting accordingly in a neuroprotective effect.18,56
4 sample had no antioxidant properties, demonstrating that the CeO2 nanoparticles were responsible of the decline on the hydrogen peroxide concentration (Fig. 2J). These results are in agreement with other studies were CeO2 nanoparticles have demonstrated to possess antioxidant properties in vitro and in vivo, creating a more favorable microenvironment for angiogenesis and nerve reconstruction, resulting accordingly in a neuroprotective effect.18,56
3.3. Adhesion and integration of NSCs on the hydrogels
Materials based on graphene derivatives, when arrested in hydrogel form, have been shown not only to minimize the direct toxicity on cells, but also to promote the growth and differentiation of neural cells.57 Herein, we explored the effect of different configurations of GO-based hydrogels with varying morphological, mechanical and electrical properties (which are dependent on one another) on the neural commitment of NSCs. Thus, the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 hydrogel, which exhibited the largest pores and lowest stiffness and electrical conductivity, was compared with the GO
1 hydrogel, which exhibited the largest pores and lowest stiffness and electrical conductivity, was compared with the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, which exhibited smaller pores, but higher stiffness and electrical conductivity. To compare the influence of the CeO2 nanoparticles on the neural differentiation, the GO
4, which exhibited smaller pores, but higher stiffness and electrical conductivity. To compare the influence of the CeO2 nanoparticles on the neural differentiation, the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 hydrogel was selected, which exhibited morphological, electrical and mechanical properties comparable to the GO
4 + CeO2 0.25 hydrogel was selected, which exhibited morphological, electrical and mechanical properties comparable to the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 one.
4 one.
NSCs were seeded directly on the hydrogels and, as shown by SEM micrographs, were able to attach without a need of Fetal Bovine Serum (FBS) supplementation or extracellular matrix (ECM)-derived compound coating in 24 h (ESI Fig. 3†). They could even migrate inside the hydrogels after 7 days in culture (ESI Fig. 4†), demonstrating a good infiltration of the cells in the material. Indeed, NSC survival, adhesion and infiltration are crucial to facilitate the bench to clinic implementation of these hydrogels. Besides, the use of ECM-derived coatings like laminin have been correlated with cell proliferation of brain cancers such ependymoma or glioblastoma.58 Thus, by eluding its use, we also avoid the possibility of having degradation products that could represent a lethal risk in clinical approaches.59 In the same way, fetal serum supplementation in cell culture has been ascribed to be strongly immunogenic in both rodents and humans. Hence, by eluding its use in here, we also maximize the bench to clinic translation potential of our systems.
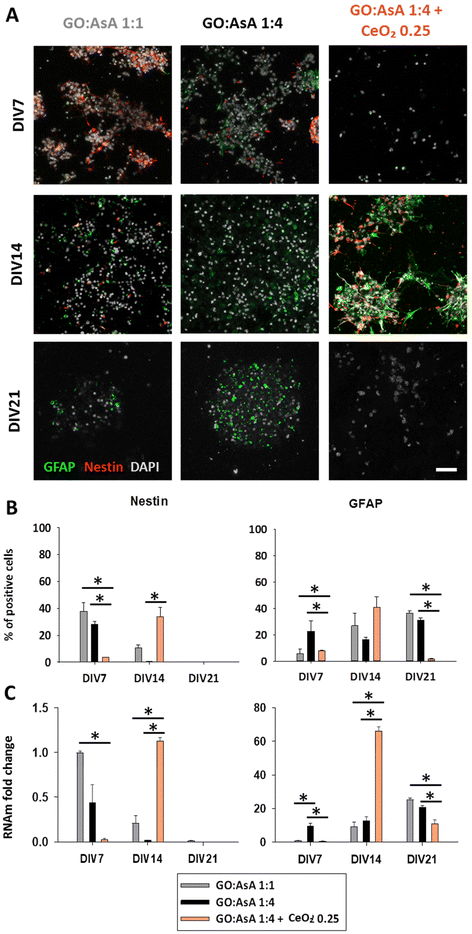
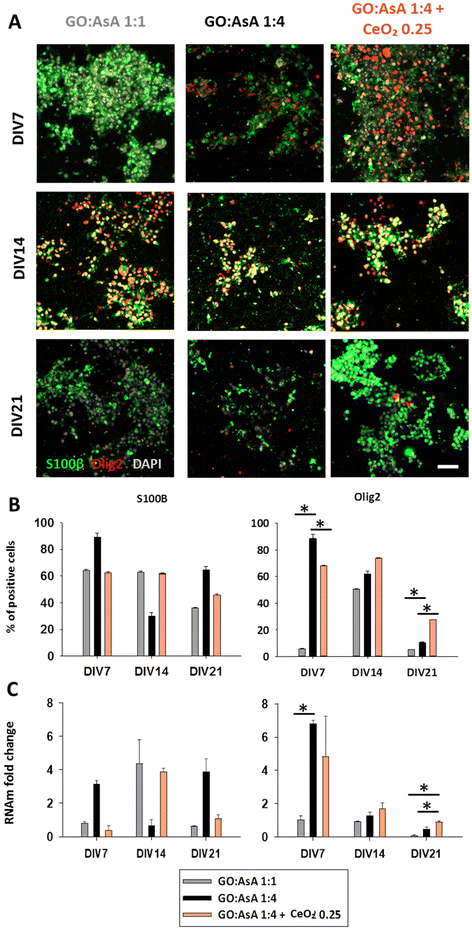
To assess the impact of the hydrogels on the establishment of a heterogeneous culture for future neural regeneration, after 7, 14 and 21 days in vitro (DIV), cells were fixed and immunostained for neural stem (Nestin), astroglial (GFAP and S100β), oligodendroglial (Olig2) or neuronal lineage markers (DCX and MAP2) to study the effect of each hydrogel on the differentiation fate of the NSCs. Additionally, RNA was extracted and quantitative retro transcriptase polymerase chain reaction (qPCR) was performed against Nestin, GFAP, S100β and MAP2 to better characterize the gene expression profile at messenger RNA (mRNA) level and corroborate the immunolabeling results.
3.4. Stemness and glial cell differentiation of NSCs on the hydrogels
The maintenance of the NSC phenotype of the seeded cells was assessed by the presence of the intermediate filament Nestin. Nestin is a marker of neural precursor cells in embryonic and adult CNS tissue60 and it is also considered to be a multipotent stem cell marker with crucial regulatory roles on the proliferation of immature cells61 including neural progenitor cells.62,63 Thus, non-differentiated NSCs are characterized by the expression of both Nestin and GFAP markers, but during the differentiation process, Nestin is progressively downregulated and replaced by other markers of more differentiated cells.64 We found that GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 hydrogel showed a more rapid decrease in the proportion of the Nestin positive stem cell population, suggesting a faster differentiation of the seeded NSCs to mature neuronal and glial cell lineages (3.7 ± 0.2%, p < 0.05), followed by the GO
4 + CeO2 0.25 hydrogel showed a more rapid decrease in the proportion of the Nestin positive stem cell population, suggesting a faster differentiation of the seeded NSCs to mature neuronal and glial cell lineages (3.7 ± 0.2%, p < 0.05), followed by the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (28.4 ± 2.2%, p < 0.05) and the GO
4 (28.4 ± 2.2%, p < 0.05) and the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (37.8 ± 6.4%, p < 0.05) at DIV7 (Fig. 3B). To further corroborate our findings, the messenger mRNA was analysed by qPCR. GO
1 (37.8 ± 6.4%, p < 0.05) at DIV7 (Fig. 3B). To further corroborate our findings, the messenger mRNA was analysed by qPCR. GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 exhibited very little Nestin expression at DIV7 (0.03 ± 0.01, fold change, p < 0.05) with respect to the control GO
4 + CeO2 0.25 exhibited very little Nestin expression at DIV7 (0.03 ± 0.01, fold change, p < 0.05) with respect to the control GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 hydrogel (Fig. 3C). By DIV21, Nestin expression was progressively lost in all the conditions, showing the exhaustion and eventual replacement of the stem cell population with mature differentiated neuronal and glial cells, in the presence of differentiation inducing media. Interestingly, in GO
1 hydrogel (Fig. 3C). By DIV21, Nestin expression was progressively lost in all the conditions, showing the exhaustion and eventual replacement of the stem cell population with mature differentiated neuronal and glial cells, in the presence of differentiation inducing media. Interestingly, in GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 hydrogels the proportion of Nestin positive cells still showed a significant peak at DIV14 (34.0 ± 6.7%, p < 0.05) compared to the other two conditions (GO
4 + CeO2 0.25 hydrogels the proportion of Nestin positive cells still showed a significant peak at DIV14 (34.0 ± 6.7%, p < 0.05) compared to the other two conditions (GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 10.6 ± 2.2% and GO
1 10.6 ± 2.2% and GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 0.6 ± 0.1%, p < 0.05) which had almost completely depleted the non-differentiated NSC population by that time (Fig. 3B). This suggested that between DIV7 and DIV14 the NSCs seeded on the GO
4 0.6 ± 0.1%, p < 0.05) which had almost completely depleted the non-differentiated NSC population by that time (Fig. 3B). This suggested that between DIV7 and DIV14 the NSCs seeded on the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 hydrogel still underwent several rounds of cell division, contrary to the other two hydrogels. This result seems to be related to a higher antioxidant capacity, which is associated with a better survival and maintenance of self-renewal in many types of stem cells, including NSCs.65 At mRNA level, Nestin expression was also clearly increased at DIV14 in the GO
4 + CeO2 0.25 hydrogel still underwent several rounds of cell division, contrary to the other two hydrogels. This result seems to be related to a higher antioxidant capacity, which is associated with a better survival and maintenance of self-renewal in many types of stem cells, including NSCs.65 At mRNA level, Nestin expression was also clearly increased at DIV14 in the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 hydrogels, in comparison to GO
4 + CeO2 0.25 hydrogels, in comparison to GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and GO
1 and GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (Fig. 3C). Thus, the NSC population was preserved for longer times up to DIV14 in the GO
4 (Fig. 3C). Thus, the NSC population was preserved for longer times up to DIV14 in the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 hydrogel, a fact which could be attributed to its antioxidant properties.65 It is well known that inflammation and oxidative stress, which are intimately linked to each other,66 are detrimental to NSC activation and neurogenesis.67 Therefore, the improved antioxidant capacity of the GO
4 + CeO2 0.25 hydrogel, a fact which could be attributed to its antioxidant properties.65 It is well known that inflammation and oxidative stress, which are intimately linked to each other,66 are detrimental to NSC activation and neurogenesis.67 Therefore, the improved antioxidant capacity of the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 hydrogel would also contribute to a maintenance of non-differentiated NSCs on the medium term (DIV14), despite their enhanced response to the initial wave of cell differentiation triggered by the soluble factors of the culture medium at DIV7. Non-differentiated NSCs can secrete neurotrophic factors that protect the developing neural tissue and help on its regeneration process.68 Therefore, the better preservation of stem cells on the GO
4 + CeO2 0.25 hydrogel would also contribute to a maintenance of non-differentiated NSCs on the medium term (DIV14), despite their enhanced response to the initial wave of cell differentiation triggered by the soluble factors of the culture medium at DIV7. Non-differentiated NSCs can secrete neurotrophic factors that protect the developing neural tissue and help on its regeneration process.68 Therefore, the better preservation of stem cells on the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 hydrogel might be helpful for the generation of a functional neural tissue in vitro.
4 + CeO2 0.25 hydrogel might be helpful for the generation of a functional neural tissue in vitro.
NSC are remnant cells of the CNS development restricted in neurogenic niches and maintain the ability to differentiate towards neuronal and glial lineages.69 A balanced neuronal and glial cell differentiation is important for the long-term survival of the NSC-derived cell cultures due to the supporting function of the glial cells over the new-born neurons on the regulation of their oxidative and metabolic balance, and the neurophysiological processes of ion and neurotransmitter uptake and release, among others.11,70
We also characterized the NSC differentiation process towards astroglial lineages, by the loss of Nestin, the increase of the expression of glial fibrillary acidic protein (GFAP) and the appearance of the S100 calcium-binding protein β (S100β) marker during the astroglial differentiation (Fig. 3 and 4). The percentage of GFAP positive cells at DIV7 indicated that GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 substrate promoted more astroglial differentiation (22.6 ± 8.2%, p < 0.05) than GO
4 substrate promoted more astroglial differentiation (22.6 ± 8.2%, p < 0.05) than GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (5.6 ± 3.4%, p < 0.05) or GO
1 (5.6 ± 3.4%, p < 0.05) or GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 (3.7 ± 0.2%, p < 0.05) hydrogels (Fig. 3B). At mRNA level, GO
4 + CeO2 0.25 (3.7 ± 0.2%, p < 0.05) hydrogels (Fig. 3B). At mRNA level, GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 also exhibited a higher expression of GFAP (9.7 ± 1.7, fold change, p < 0.05), in comparison with GO
4 also exhibited a higher expression of GFAP (9.7 ± 1.7, fold change, p < 0.05), in comparison with GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (1.0 ± 0.2, fold change, p < 0.05) and GO
1 (1.0 ± 0.2, fold change, p < 0.05) and GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 (0.7 ± 0.1, fold change, p < 0.05) (Fig. 3C). These results suggested that, as previously described, stiffer substrates like GO
4 + CeO2 0.25 (0.7 ± 0.1, fold change, p < 0.05) (Fig. 3C). These results suggested that, as previously described, stiffer substrates like GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 promoted the differentiation towards glial lineages compared to softer ones.71,72
4 promoted the differentiation towards glial lineages compared to softer ones.71,72
However, the presence of CeO2 nanoparticles could modulate the final fate of this astroglial phenotype into not only astrocytes, but also other types of glial cells or even the enhanced maintenance of non-differentiated stem cell phenotypes. In this sense, GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 exhibited an augmented GFAP expression on mRNA level at DIV14 (66.3 ± 2.4, fold change, p < 0.05), in comparison with GO
4 + CeO2 0.25 exhibited an augmented GFAP expression on mRNA level at DIV14 (66.3 ± 2.4, fold change, p < 0.05), in comparison with GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (9.3 ± 2.8, fold change, p < 0.05) and GO
1 (9.3 ± 2.8, fold change, p < 0.05) and GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (12.7 ± 2.6, fold change, p < 0.05) (Fig. 3C). However, this expression was correlated with a similar increase on Nestin mRNA level, suggesting a possible expansion of the NSC population, rather than an astroglial cell differentiation. Indeed, the co-expression of GFAP and Nestin has been ascribed to the stem phenotype of neural cells and their proliferation.73,74
4 (12.7 ± 2.6, fold change, p < 0.05) (Fig. 3C). However, this expression was correlated with a similar increase on Nestin mRNA level, suggesting a possible expansion of the NSC population, rather than an astroglial cell differentiation. Indeed, the co-expression of GFAP and Nestin has been ascribed to the stem phenotype of neural cells and their proliferation.73,74
Interestingly, at DIV21 we found an abundant GFAP positive astroglial cell subpopulation with no Nestin expression in GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (36.5 ± 1.8%, p < 0.05) and GO
1 (36.5 ± 1.8%, p < 0.05) and GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (31.1 ± 1.7%, p < 0.05) hydrogels, compared to GO
4 (31.1 ± 1.7%, p < 0.05) hydrogels, compared to GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 hydrogels where those GFAP + cells were relatively much less frequent (1.7 ± 0.1%, p < 0.05) (Fig. 3B). At mRNA level, GO
4 + CeO2 0.25 hydrogels where those GFAP + cells were relatively much less frequent (1.7 ± 0.1%, p < 0.05) (Fig. 3B). At mRNA level, GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 also exhibited a higher GFAP expression with respect to DIV7 (20.8 ± 0.9, fold change, p < 0.05), almost equaling the levels of the GO
1 also exhibited a higher GFAP expression with respect to DIV7 (20.8 ± 0.9, fold change, p < 0.05), almost equaling the levels of the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (25.5 ± 0.8, fold change, p < 0.05). Remarkably, GO
4 (25.5 ± 0.8, fold change, p < 0.05). Remarkably, GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 exhibited a lower GFAP mRNA expression (11.2 ± 2.2, fold change, p < 0.05) further corroborating the finding of the immunofluorescence assays (Fig. 3C).
4 + CeO2 0.25 exhibited a lower GFAP mRNA expression (11.2 ± 2.2, fold change, p < 0.05) further corroborating the finding of the immunofluorescence assays (Fig. 3C).
From the perspective of tissue engineering therapies, the presence of GFAP positive astrocytes may pose both advantages and disadvantages. On the one hand, mature astrocytes are an important supporting glial cell of the CNS, sustaining neuronal metabolism and function. On the other hand, astroglial cells have also been described to be involved in glial scar formation after CNS injury.75 Scar tissue has a dual component: first, the glial scar formed by glial precursors, reactive astrocytes and microglia found at the periphery of the lesion, and second, the fibrotic scar composed by phagocytic cells and fibroblasts at the lesion core.76 This biological process has been reported detrimental for an effective reinnervation of the damaged CNS.77 Taking into account that reactive astroglial phenotypes can be induced when these cells are exposed to a damaged CNS environment, the presence of high amounts of astrocytes (GFAP positive astroglial subpopulation) in the tissue engineered grafts may constitute a limitation for therapeutic purposes.76
During the maturation of the astrocytes, together with the expression of GFAP, cells acquire the expression of S100β.78,79 All the tested hydrogels presented a mature astroglial-like lineage population over time since the proportion of S100β positive cells were always higher than that of GFAP positive cells in all the conditions at protein level (Fig. 4A and B). qPCR results further corroborated the presence of S100β protein in all the conditions and demonstrated a higher expression of S100β mRNA on GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (3.9 ± 0.8 fold change, p < 0.05) at DIV21, compared to GO
4 (3.9 ± 0.8 fold change, p < 0.05) at DIV21, compared to GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (0.6 ± 0.0 fold change, p < 0.05) and GO
1 (0.6 ± 0.0 fold change, p < 0.05) and GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 (1.1 ± 0.2 fold change, p < 0.05) (Fig. 4C), suggesting a possible establishment of a mature astroglial differentiation on the stiffer hydrogels with smaller pores and greater mechanical and electrical properties. It is noteworthy that S100β, apart from mature astroglial cells, is also a marker of oligodendroglial lineage cells.80 Oligodendrocyte precursors are known to express both S100β and Olig2 markers.80 Therefore, in view of this result, we also studied the expression of oligodendrocyte transcription factor Olig2 on the cell-seeded hydrogels. The expression of Olig2 in oligodendrocyte progenitors has been reported to be increased during the remyelination process of injured axons in multiple sclerosis (MS),81 and is characteristic of an in situ expanding oligodendrocyte population. At DIV7, the proportion of the oligodendroglial lineage population on the GO
4 + CeO2 0.25 (1.1 ± 0.2 fold change, p < 0.05) (Fig. 4C), suggesting a possible establishment of a mature astroglial differentiation on the stiffer hydrogels with smaller pores and greater mechanical and electrical properties. It is noteworthy that S100β, apart from mature astroglial cells, is also a marker of oligodendroglial lineage cells.80 Oligodendrocyte precursors are known to express both S100β and Olig2 markers.80 Therefore, in view of this result, we also studied the expression of oligodendrocyte transcription factor Olig2 on the cell-seeded hydrogels. The expression of Olig2 in oligodendrocyte progenitors has been reported to be increased during the remyelination process of injured axons in multiple sclerosis (MS),81 and is characteristic of an in situ expanding oligodendrocyte population. At DIV7, the proportion of the oligodendroglial lineage population on the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (5.9 ± 0.4%, p < 0.05) was also lower compared to GO
1 (5.9 ± 0.4%, p < 0.05) was also lower compared to GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (88.5 ± 3.0%, p < 0.05) and GO
4 (88.5 ± 3.0%, p < 0.05) and GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 (68.2 ± 0.2%, p < 0.05) (Fig. 4B). These results were further corroborated at mRNA level, where GO
4 + CeO2 0.25 (68.2 ± 0.2%, p < 0.05) (Fig. 4B). These results were further corroborated at mRNA level, where GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 at DIV7 exhibited a significantly higher Olig2 expression (6.8 ± 0.2, fold change, p < 0.05) compared to GO
4 at DIV7 exhibited a significantly higher Olig2 expression (6.8 ± 0.2, fold change, p < 0.05) compared to GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, demonstrating again that stiffer substrates promoted a faster differentiation towards glial cell lineages (Fig. 4C).
1, demonstrating again that stiffer substrates promoted a faster differentiation towards glial cell lineages (Fig. 4C).
Interestingly, GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 was able to support much better the oligodendroglial cell population for long culture periods until DIV21 (27.6 ± 0.2%, p < 0.05) compared to GO
4 + CeO2 was able to support much better the oligodendroglial cell population for long culture periods until DIV21 (27.6 ± 0.2%, p < 0.05) compared to GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (5.4 ± 0.1%, p < 0.05) and GO
1 (5.4 ± 0.1%, p < 0.05) and GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (10.3 ± 0.5%, p < 0.05) (Fig. 4B), although at mRNA level all the hydrogels presented a decay on Olig2 expression (Fig. 4C). These results suggested that the presence of CeO2 nanoparticles might have helped on the establishment of a mature and healthy oligodendroglial lineage subpopulation expressing Olig2 protein within the hydrogel. Oligodendrocytes are known to be a particularly sensitive cell type to oxidative stress.82,83 Although the antioxidant effect of CeO2 0.25 using a concentrated source of exogenous H2O2 was limited, it might modulate the physiological levels of intracellular free radicals, which may explain the beneficial effect of the CeO2 nanoparticles addition on oligodendroglial survival in the scaffolds. Indeed, CeO2 nanoparticles have been reported to possess antioxidant and neuroprotective capabilities and even attenuate the inflammation and help on the recovery of demyelinating pathologies like MS,84 further suggesting their implication on oligodendrocyte function and survival. Our results also show that, even if no statistical difference was observed for the GO
4 (10.3 ± 0.5%, p < 0.05) (Fig. 4B), although at mRNA level all the hydrogels presented a decay on Olig2 expression (Fig. 4C). These results suggested that the presence of CeO2 nanoparticles might have helped on the establishment of a mature and healthy oligodendroglial lineage subpopulation expressing Olig2 protein within the hydrogel. Oligodendrocytes are known to be a particularly sensitive cell type to oxidative stress.82,83 Although the antioxidant effect of CeO2 0.25 using a concentrated source of exogenous H2O2 was limited, it might modulate the physiological levels of intracellular free radicals, which may explain the beneficial effect of the CeO2 nanoparticles addition on oligodendroglial survival in the scaffolds. Indeed, CeO2 nanoparticles have been reported to possess antioxidant and neuroprotective capabilities and even attenuate the inflammation and help on the recovery of demyelinating pathologies like MS,84 further suggesting their implication on oligodendrocyte function and survival. Our results also show that, even if no statistical difference was observed for the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + 0.25 CeO2 formulation on H2O2 reduction, the added CeO2 nanoparticles managed to decrease the accumulation of intracellular reactive oxygen species in NSCs (GO
4 + 0.25 CeO2 formulation on H2O2 reduction, the added CeO2 nanoparticles managed to decrease the accumulation of intracellular reactive oxygen species in NSCs (GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 100.0 ± 5.8, GO
1 100.0 ± 5.8, GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 88.7 ± 6.0 and GO
4 88.7 ± 6.0 and GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 48.7 ± 2.2, p < 0.05) (ESI Fig. 5†), enhancing the survival and modulating the differentiation pattern of these cells. This may be ascribed to the reduction of other reactive oxygen species or via other mechanisms beyond the scope of this study.
4 + CeO2 0.25 48.7 ± 2.2, p < 0.05) (ESI Fig. 5†), enhancing the survival and modulating the differentiation pattern of these cells. This may be ascribed to the reduction of other reactive oxygen species or via other mechanisms beyond the scope of this study.
Overall, our results indicated that stiffer substrates like GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 promoted astroglial and oligodendrogial differentiation. Nevertheless, both GO
4 promoted astroglial and oligodendrogial differentiation. Nevertheless, both GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and GO
1 and GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 hydrogels were able to support a GFAP positive astrocyte-like subpopulation. Remarkably, the addition of CeO2 nanoparticles on the GO
4 hydrogels were able to support a GFAP positive astrocyte-like subpopulation. Remarkably, the addition of CeO2 nanoparticles on the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 hydrogel induced a much better maintenance of both Nestin + stem cell populations and also Olig2 + oligodendroglial lineage cell populations for longer culture periods until DIV21.
4 + CeO2 hydrogel induced a much better maintenance of both Nestin + stem cell populations and also Olig2 + oligodendroglial lineage cell populations for longer culture periods until DIV21.
3.5. Neuronal differentiation pattern and establishment of neuron-oligodendrocyte co-cultures on graphene-derivatives and CeO2 nanoparticles-based hydrogels
Glial cells play a key role on regeneration, but mature and immature neurons are also needed to promote and coordinate innervation and, hence, the regeneration process.85 The presence of neuronal progenitor cells also needs to be assessed to predict an effective neuroregeneration outcome. Hence, we performed immunofluorescence labeling to detect the immature neuronal marker doublecortin (DCX).86 DCX expression is restricted to neuronal lineage immature cells and multipotent-precursors in both the developing and adult brain87 and also in regions of adult neurons undergoing a plastic reorganization of their dendrites.88 We found that the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 hydrogel was unable to support DCX positive cells at any of the periods tested and GO
4 hydrogel was unable to support DCX positive cells at any of the periods tested and GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 only supported them until DIV7 (ESI Fig. 6†). By contrast, GO
1 only supported them until DIV7 (ESI Fig. 6†). By contrast, GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 was able to maintain a substantial population of DCX positive cells until DIV21 (28.4 ± 0.3%, p < 0.05) (ESI Fig. 6B†). The qPCR further corroborated these findings, where GO
4 + CeO2 0.25 was able to maintain a substantial population of DCX positive cells until DIV21 (28.4 ± 0.3%, p < 0.05) (ESI Fig. 6B†). The qPCR further corroborated these findings, where GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 (4.4 ± 0.9, p < 0.05) exhibited 4 times more DCX mRNA at DIV14 than GO
4 + CeO2 0.25 (4.4 ± 0.9, p < 0.05) exhibited 4 times more DCX mRNA at DIV14 than GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (0.7 ± 0.1) and GO
1 (0.7 ± 0.1) and GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (0.2 ± 0.0) and 38 times more at DIV21 (GO
4 (0.2 ± 0.0) and 38 times more at DIV21 (GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 (38.3 ± 10.3, p < 0.05) (ESI Fig. 6C†). These results may indicate the presence of DCX + multipotent-precursors and are in accordance with the increase in Nestin positive cells that occurred in GO
4 + CeO2 0.25 (38.3 ± 10.3, p < 0.05) (ESI Fig. 6C†). These results may indicate the presence of DCX + multipotent-precursors and are in accordance with the increase in Nestin positive cells that occurred in GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 at DIV14. The persistence of DCX positive population in the GO
4 + CeO2 0.25 at DIV14. The persistence of DCX positive population in the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 hydrogels at DIV21 may be attributed to a better oxidative balance and long-term survival of neuronal lineage cells in culture, which are another very sensitive cell type to oxidative stress.82
4 + CeO2 0.25 hydrogels at DIV21 may be attributed to a better oxidative balance and long-term survival of neuronal lineage cells in culture, which are another very sensitive cell type to oxidative stress.82
The functionality and successful engraftment of bioengineered nerve tissues implies a balanced generation of both mature and immature glial and neuronal cells. Hence, we also studied the expression of other mature neuronal markers like microtubule-associated protein 2 (MAP2), which is expressed in the dendrites of fully mature neurons.89 The percentage of MAP2 positive neurons in our hydrogels was much higher in GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 (35.4 ± 0.2%, p < 0.05), than in GO
4 + CeO2 0.25 (35.4 ± 0.2%, p < 0.05), than in GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (0.04 ± 0.03% p < 0.05) and GO
1 (0.04 ± 0.03% p < 0.05) and GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (0.03 ± 0.02%, p < 0.05), which exhibited almost no mature neuronal generation at DIV7 (Fig. 5A and B). These results were further corroborated by qPCR, where GO
4 (0.03 ± 0.02%, p < 0.05), which exhibited almost no mature neuronal generation at DIV7 (Fig. 5A and B). These results were further corroborated by qPCR, where GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 exhibited a higher expression for MAP2 (26.1 ± 7.5 fold change, p < 0.05) compared to GO
4 + CeO2 0.25 exhibited a higher expression for MAP2 (26.1 ± 7.5 fold change, p < 0.05) compared to GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and GO
1 and GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 at DIV7 (Fig. 5C). On the contrary, no MAP2 positive cells were ever found at any time point on GO
4 at DIV7 (Fig. 5C). On the contrary, no MAP2 positive cells were ever found at any time point on GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 hydrogel, but qPCR results showed an increase on MAP2 expression at DIV21 (2.3 ± 0.7 fold change p < 0.05 compared to DIV7), suggesting a possible delay of the neuronal differentiation on stiffer substrates with smaller pores and greater electrical conductivity due to the favored differentiation towards astroglial lineages.71,72 These results also corroborated the positive effect of the addition of CeO2 nanoparticles for a quicker neuronal differentiation over GO based 3D materials.
4 hydrogel, but qPCR results showed an increase on MAP2 expression at DIV21 (2.3 ± 0.7 fold change p < 0.05 compared to DIV7), suggesting a possible delay of the neuronal differentiation on stiffer substrates with smaller pores and greater electrical conductivity due to the favored differentiation towards astroglial lineages.71,72 These results also corroborated the positive effect of the addition of CeO2 nanoparticles for a quicker neuronal differentiation over GO based 3D materials.
Interestingly, GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 showed an increase on MAP2 positive cells (22.9 ± 0.4%, p < 0.05) at DIV14, and at DIV21 on mRNA content (9.4 ± 0.8 fold change, p < 0.05 compared to DIV7) (Fig. 5B and C). These results are in agreement with DCX results in GO
1 showed an increase on MAP2 positive cells (22.9 ± 0.4%, p < 0.05) at DIV14, and at DIV21 on mRNA content (9.4 ± 0.8 fold change, p < 0.05 compared to DIV7) (Fig. 5B and C). These results are in agreement with DCX results in GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 at DIV7, which exhibited an increase in DCX positive cells. It might be assumed that some of these progenitor cells would proceed through their differentiation process to eventually give rise to mature neurons at DIV14 and the higher expression of MAP2 at DIV21 and corroborates the findings of other studies in the literature, where softer substrates were reported to promote the differentiation towards neuronal lineages.71,72
1 at DIV7, which exhibited an increase in DCX positive cells. It might be assumed that some of these progenitor cells would proceed through their differentiation process to eventually give rise to mature neurons at DIV14 and the higher expression of MAP2 at DIV21 and corroborates the findings of other studies in the literature, where softer substrates were reported to promote the differentiation towards neuronal lineages.71,72
However, the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 hydrogel was unable to support the survival of the MAP2 positive cells for longer periods (Fig. 5B). On the contrary, the GO
1 hydrogel was unable to support the survival of the MAP2 positive cells for longer periods (Fig. 5B). On the contrary, the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 hydrogel managed to support a subpopulation of MAP2 positive mature neurons until DIV21. Our findings suggested that the addition of CeO2 nanoparticles to the hydrogels allowed them to support the terminal differentiation of NSCs towards fully mature neurons. It is noteworthy that here CeO2 nanoparticles were physically attached to the hydrogels, thus possibly preventing their cellular internalization and possible detrimental effects on the neuronal lineage differentiation of the seeded NSCs, as it has been suggested in other studies.90 Moreover, in accordance with our study, CeO2 nanoparticles have also been shown to protect cells against oxidative stress, improving neuronal function and delaying neuronal death after a traumatic brain injury both in vitro and in vivo.91
4 + CeO2 0.25 hydrogel managed to support a subpopulation of MAP2 positive mature neurons until DIV21. Our findings suggested that the addition of CeO2 nanoparticles to the hydrogels allowed them to support the terminal differentiation of NSCs towards fully mature neurons. It is noteworthy that here CeO2 nanoparticles were physically attached to the hydrogels, thus possibly preventing their cellular internalization and possible detrimental effects on the neuronal lineage differentiation of the seeded NSCs, as it has been suggested in other studies.90 Moreover, in accordance with our study, CeO2 nanoparticles have also been shown to protect cells against oxidative stress, improving neuronal function and delaying neuronal death after a traumatic brain injury both in vitro and in vivo.91
As previously stated, a glial and neuronal equilibrium is primordial for cell survival and functionality of nerve tissues.11 Herein, in the absence of CeO2 nanoparticles (GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 hydrogels), the larger pores and lower stiffness and electrical conductivity of the GO
4 hydrogels), the larger pores and lower stiffness and electrical conductivity of the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 hydrogel enhanced the generation of neuronal lineage cells with respect to the smaller pores and greater mechanical and electrical properties of the GO
1 hydrogel enhanced the generation of neuronal lineage cells with respect to the smaller pores and greater mechanical and electrical properties of the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, where there is a preferred differentiation towards glial lineages at shorter periods (Fig. 5D). But, remarkably, at DIV21, in both hydrogels, GO
4, where there is a preferred differentiation towards glial lineages at shorter periods (Fig. 5D). But, remarkably, at DIV21, in both hydrogels, GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and GO
1 and GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, astrocytes were the major persisting cell type, despite the detection of some populations of neuronal lineage cells at shorter time points, DIV7 and DIV14. This result comes in agreement with the fact that astrocytes are the metabolically most resistant (least demanding) cell type of the CNS, whose high endogenous antioxidant and glycolytic capacity endows them with a higher ability to survive in adverse conditions.83,92,93 It is very likely that most of the DCX+ and MAP2+ neuronal cells that were being generated in the GO
4, astrocytes were the major persisting cell type, despite the detection of some populations of neuronal lineage cells at shorter time points, DIV7 and DIV14. This result comes in agreement with the fact that astrocytes are the metabolically most resistant (least demanding) cell type of the CNS, whose high endogenous antioxidant and glycolytic capacity endows them with a higher ability to survive in adverse conditions.83,92,93 It is very likely that most of the DCX+ and MAP2+ neuronal cells that were being generated in the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 hydrogel at DIV7 and DIV14 eventually perished at DIV21, because of an insufficient antioxidant capacity to support the increased mitochondrial oxidative phosphorylation that comes along with mature neuronal differentiation.94 Interestingly, in the presence of CeO2 nanoparticles, the GO
1 hydrogel at DIV7 and DIV14 eventually perished at DIV21, because of an insufficient antioxidant capacity to support the increased mitochondrial oxidative phosphorylation that comes along with mature neuronal differentiation.94 Interestingly, in the presence of CeO2 nanoparticles, the GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25 hydrogel supported the co-generation of both MAP2 positive mature neuronal lineage cells together with Olig2 positive oligodendroglial lineage cells until DIV21. The results of this work clearly encourage the incorporation of neuroprotective and antioxidant systems like CeO2 nanoparticles trapped in tissue engineering scaffolds to boost survival of these two extremely necessary and highly vulnerable cell types of the CNS. The balanced generation of neurons, astrocytes and oligodendrocytes within the bioengineered construct is fundamental for an eventual success of CNS regeneration therapies. It should be emphasized that once a balanced oligodendrocytes, neurons and astrocytes population has been established within the graft, the close contact of the three different cell types may protect each other and improve xenocell survival prior to the integration into the host tissue.95,96 Indeed, several studies highlighted the necessary integrin mediated connexion between neuronal axons and oligodendrocytes for the survival of both neuronal and oligodendroglial cells in vitro.97,98 Moreover, oligodendrocytes have also been shown to be a glial cell subpopulation that play a key role on axonal regeneration,98,99 hence the importance of preserving both neurons and oligodendrocytes together in the same bioengineered construct. Here we present a 3D hydrogel based on graphene derivatives and cerium oxide nanoparticles as a promising therapeutic tool for neurodegenerative and demyelinating pathologies involving neuronal and/or oligodendroglial cell death.
4 + CeO2 0.25 hydrogel supported the co-generation of both MAP2 positive mature neuronal lineage cells together with Olig2 positive oligodendroglial lineage cells until DIV21. The results of this work clearly encourage the incorporation of neuroprotective and antioxidant systems like CeO2 nanoparticles trapped in tissue engineering scaffolds to boost survival of these two extremely necessary and highly vulnerable cell types of the CNS. The balanced generation of neurons, astrocytes and oligodendrocytes within the bioengineered construct is fundamental for an eventual success of CNS regeneration therapies. It should be emphasized that once a balanced oligodendrocytes, neurons and astrocytes population has been established within the graft, the close contact of the three different cell types may protect each other and improve xenocell survival prior to the integration into the host tissue.95,96 Indeed, several studies highlighted the necessary integrin mediated connexion between neuronal axons and oligodendrocytes for the survival of both neuronal and oligodendroglial cells in vitro.97,98 Moreover, oligodendrocytes have also been shown to be a glial cell subpopulation that play a key role on axonal regeneration,98,99 hence the importance of preserving both neurons and oligodendrocytes together in the same bioengineered construct. Here we present a 3D hydrogel based on graphene derivatives and cerium oxide nanoparticles as a promising therapeutic tool for neurodegenerative and demyelinating pathologies involving neuronal and/or oligodendroglial cell death.
Overall, our results showed that hydrogels based on graphene-derivatives supported both glial and neuronal lineage differentiation of NSCs in short-term cultures. Softer substrates like GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with larger pores and lower electrical conductivities promoted cell differentiation towards neuronal lineages, while stiffer substrates like GO
1 with larger pores and lower electrical conductivities promoted cell differentiation towards neuronal lineages, while stiffer substrates like GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 with smaller pores and greater electrical properties enhanced glial cell differentiation. However, for long-term cultures, graphene derivatives-based hydrogels alone were unable to sustain a balanced long-term survival of neurons and oligodendrocytes. In contrast, thanks to the antioxidant and neuroprotective capabilities of CeO2 nanoparticles embedded on GO
4 with smaller pores and greater electrical properties enhanced glial cell differentiation. However, for long-term cultures, graphene derivatives-based hydrogels alone were unable to sustain a balanced long-term survival of neurons and oligodendrocytes. In contrast, thanks to the antioxidant and neuroprotective capabilities of CeO2 nanoparticles embedded on GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25, this hydrogel was able to support the generation of astroglial, oligodendroglial and neuronal cells until DIV21, providing a promising approach for CNS regeneration therapies.
4 + CeO2 0.25, this hydrogel was able to support the generation of astroglial, oligodendroglial and neuronal cells until DIV21, providing a promising approach for CNS regeneration therapies.
4. Conclusions
Herein, we present a simple and scalable method to fabricate 3D hydrogels based on graphene derivatives and CeO2 nanoparticles promoted by a self-assembly process. The resulting hydrogels showed highly porous structures with tunable electrical, morphological and mechanical properties that can be finely regulated by the addition of AsA and CeO2 nanoparticles. The final properties of the scaffolds, together with the advanced functionalities provided by the CeO2 nanoparticles, clearly determined the fate of NSCs, which were seeded on the hydrogels without any ECM-like compound coating or FBS supplementation. Accordingly, we found that softer hydrogels with larger pores and lower electrical conductivity (i.e., GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) induced an increase of neuronal lineage differentiation, whereas the stiffer ones with smaller pores and greater electrical properties (i.e., GO
1) induced an increase of neuronal lineage differentiation, whereas the stiffer ones with smaller pores and greater electrical properties (i.e., GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) promoted glial cell lineage differentiation. Remarkably, the hydrogel containing CeO2 nanoparticles (i.e., GO
4) promoted glial cell lineage differentiation. Remarkably, the hydrogel containing CeO2 nanoparticles (i.e., GO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AsA 1
AsA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 + CeO2 0.25) was the only one allowing the long-term establishment of a mature co-culture containing abundant populations of both neuronal and oligodendroglial lineage cells, which are the two most delicate and difficult to sustain cell types of the CNS. Our findings provide valuable insight on the creation and optimization of differentiated glial and neuronal 3D co-culture systems that enable the integration of both neurons and oligodendrocytes in a same hydrogel scaffold, which could be very useful for future CNS regeneration therapies.
4 + CeO2 0.25) was the only one allowing the long-term establishment of a mature co-culture containing abundant populations of both neuronal and oligodendroglial lineage cells, which are the two most delicate and difficult to sustain cell types of the CNS. Our findings provide valuable insight on the creation and optimization of differentiated glial and neuronal 3D co-culture systems that enable the integration of both neurons and oligodendrocytes in a same hydrogel scaffold, which could be very useful for future CNS regeneration therapies.
Author contributions
Y.P.: conceptualization, investigation, writing – original draft; J.L.: investigation, writing – review & editing; S.G.L.: investigation; B.P.R.: investigation; D.E.M.T.: investigation; C.T.: investigation; I.M.R.: investigation; E.M.: investigation; J.M.U.: investigation; G.C.: investigation, supervision; G.I.: conceptualization, writing – review & editing, funding acquisition; F.U.: funding acquisition; J.R.S.: conceptualization, funding acquisition, supervision; J.R.P.: conceptualization, investigation, writing – review & editing, supervision, funding acquisition; A.L.: conceptualization, investigation, writing – review & editing, supervision, funding acquisition.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
We would like to thank Jorge Fernandez (Polimerbio SL) for the support in this project. SGIker technical services (UPV/EHU) are gratefully acknowledged for XPS, XRD, SEM and TEM support. We would like to thank Ricardo Andrade and Alejandro Díez, responsible of the High-Resolution Analytical Microscopy Service in Biomedicine (SGIker UPV/EHU), for their invaluable help for the electron microscopy and cell videorecording assistance. We would like to thank Laura Escobar for confocal microscopy in the Achucarro Basque Center for Neuroscience Fundazioa and Fabrice Cordelières (Bordeaux Imaging Center) for the macro developed for ImageJ.This work has been funded by the Basque Government (GV/EJ) Department of Education (GIC21/131 IT1766-22, IT1751-22), Health Department (RIS3, 2021333012), Grants PID2019-106236GB-I00 and PID2019-104766RB-C21 funded by MCIN/AEI/10.13039/501100011033. Grant RYC-2013-13450 funded by MCIN/AEI/10.13039/501100011033 and by “ESF investing in your future” by the “European Union” and Achucarro Seed-Fund 003 (JRP), the University of the Basque Country (UPV/EHU) by GIU19/040, GIU 20/050, PPGA 20/22, COLAB19/03 and IKERTU-2020.0155. GV/EJ, Hazitek ZE-2019/00012-IMABI and ELKARTEK KK-2019/00093. Polimerbio and Y. P. have a Bikaintek PhD grant (20-AFW2-2018-00001). Part of this research was performed within the framework of the European Union's Horizon 2020 research and innovation program under the Marie Skłodowska-Curie Grant Agreement No. 793644 (BIONICS).
References
- V. L. Feigin, A. A. Abajobir, K. H. Abate, F. Abd-Allah, A. M. Abdulle, S. F. Abera, G. Y. Abyu, M. B. Ahmed, A. N. Aichour, I. Aichour, M. T. E. Aichour, R. O. Akinyemi, S. Alabed, R. Al-Raddadi, N. Alvis-Guzman, A. T. Amare, H. Ansari, P. Anwari, J. Ärnlöv, H. Asayesh, S. W. Asgedom, T. M. Atey, L. Avila-Burgos, E. Frinel, G. A. Avokpaho, M. R. Azarpazhooh, A. Barac, M. Barboza, S. L. Barker-Collo, T. Bärnighausen, N. Bedi, E. Beghi, D. A. Bennett, I. M. Bensenor, A. Berhane, B. D. Betsu, S. Bhaumik, S. M. Birlik, S. Biryukov, D. J. Boneya, L. N. B. Bulto, H. Carabin, D. Casey, C. A. Castañeda-Orjuela, F. Catalá-López, H. Chen, A. A. Chitheer, R. Chowdhury, H. Christensen, L. Dandona, R. Dandona, G. A. de Veber, S. D. Dharmaratne, H. P. Do, K. Dokova, E. R. Dorsey, R. G. Ellenbogen, S. Eskandarieh, M. S. Farvid, S.-M. Fereshtehnejad, F. Fischer, K. J. Foreman, J. M. Geleijnse, R. F. Gillum, G. Giussani, E. M. Goldberg, P. N. Gona, A. C. Goulart, H. C. Gugnani, R. Gupta, V. Hachinski, R. Gupta, R. R. Hamadeh, M. Hambisa, G. J. Hankey, H. A. Hareri, R. Havmoeller, S. I. Hay, P. Heydarpour, P. J. Hotez, M. B. Jakovljevic, M. Javanbakht, P. Jeemon, J. B. Jonas, Y. Kalkonde, A. Kandel, A. Karch, A. Kasaeian, A. Kastor, P. N. Keiyoro, Y. S. Khader, I. A. Khalil, E. A. Khan, Y.-H. Khang, A. Tawfih, A. Khoja, J. Khubchandani, C. Kulkarni, D. Kim, Y. J. Kim, M. Kivimaki, Y. Kokubo, S. Kosen, M. Kravchenko, R. V. Krishnamurthi, B. K. Defo, G. A. Kumar, R. Kumar, H. H. Kyu, A. Larsson, P. M. Lavados, Y. Li, X. Liang, M. L. Liben, W. D. Lo, G. Logroscino, P. A. Lotufo, C. T. Loy, M. T. Mackay, H. M. A. E. Razek, M. M. A. E. Razek, A. Majeed, R. Malekzadeh, T. Manhertz, L. G. Mantovani, J. Massano, M. Mazidi, C. McAlinden, S. Mehata, M. M. Mehndiratta, Z. A. Memish, W. Mendoza, M. A. Mengistie, G. A. Mensah, A. Meretoja, H. B. Mezgebe, T. R. Miller, S. R. Mishra, N. M. Ibrahim, A. Mohammadi, K. E. Mohammed, S. Mohammed, A. H. Mokdad, M. Moradi-Lakeh, I. M. Velasquez, K. I. Musa, M. Naghavi, J. W. Ngunjiri, C. T. Nguyen, G. Nguyen, Q. L. Nguyen, T. H. Nguyen, E. Nichols, D. N. A. Ningrum, V. M. Nong, B. Norrving, J. J. N. Noubiap, F. A. Ogbo, M. O. Owolabi, J. D. Pandian, P. G. Parmar, D. M. Pereira, M. Petzold, M. R. Phillips, M. A. Piradov, R. G. Poulton, F. Pourmalek, M. Qorbani, A. Rafay, M. Rahman, M. H. Rahman, R. K. Rai, S. Rajsic, A. Ranta, S. Rawaf, A. M. N. Renzaho, M. S. Rezai, G. A. Roth, G. Roshandel, E. Rubagotti, P. Sachdev, S. Safiri, R. Sahathevan, M. A. Sahraian, A. M. Samy, P. Santalucia, I. S. Santos, B. Sartorius, M. Satpathy, M. Sawhney, M. I. Saylan, S. G. Sepanlou, M. A. Shaikh, R. Shakir, M. Shamsizadeh, K. N. Sheth, M. Shigematsu, H. Shoman, D. A. S. Silva, M. Smith, E. Sobngwi, L. A. Sposato, J. D. Stanaway, D. J. Stein, T. J. Steiner, L. J. Stovner, R. S. Abdulkader, C. E. Szoeke, R. Tabarés-Seisdedos, D. Tanne, A. M. Theadom, A. G. Thrift, D. L. Tirschwell, R. Topor-Madry, B. X. Tran, T. Truelsen, K. B. Tuem, K. N. Ukwaja, O. A. Uthman, Y. Y. Varakin, T. Vasankari, N. Venketasubramanian, V. V. Vlassov, F. Wadilo, T. Wakayo, M. T. Wallin, E. Weiderpass, R. Westerman, T. Wijeratne, C. S. Wiysonge, M. A. Woldu, C. D. A. Wolfe, D. Xavier, G. Xu, Y. Yano, H. H. Yimam, N. Yonemoto, C. Yu, Z. Zaidi, M. E. S. Zaki, J. R. Zunt, C. J. L. Murray and T. Vos, Lancet Neurol., 2017, 16, 877–897 CrossRef PubMed.
- World Health Organization - Neurological disorders public health challenges.pdf, (n.d.). https://www.who.int/mental_health/neurology/neurological_disorders_report_web.pdf (accessed July 2, 2021).
- P. M. Rossini, C. Calautti, F. Pauri and J.-C. Baron, Lancet Neurol., 2003, 2, 493–502 CrossRef PubMed.
- I. Fischer, J. N. Dulin and M. A. Lane, Nat. Rev. Neurosci., 2020, 21, 366–383 CrossRef CAS PubMed.
- X. Tian, T. Fan, W. Zhao, G. Abbas, B. Han, K. Zhang, N. Li, N. Liu, W. Liang, H. Huang, W. Chen, B. Wang and Z. Xie, Bioact. Mater., 2021, 6, 2854–2869 CrossRef CAS PubMed.
- S. Jarrin, S. Cabré and E. Dowd, Neurochem. Int., 2021, 144, 104971 CrossRef CAS PubMed.
- S. Grade and M. Götz, npj Regener. Med., 2017, 2, 1–11 CrossRef PubMed.
- Y. Li, Y. Xiao and C. Liu, Chem. Rev., 2017, 117, 4376–4421 CrossRef CAS PubMed.
- R. Kumar, R. Rauti, D. Scaini, M. Antman-Passig, O. Meshulam, D. Naveh, L. Ballerini and O. Shefi, Adv. Funct. Mater., 2021, 31, 2104887 CrossRef CAS.
- A. Capasso, J. Rodrigues, M. Moschetta, F. Buonocore, G. Faggio, G. Messina, M. J. Kim, J. Kwon, E. Placidi, F. Benfenati, M. Bramini, G.-H. Lee and N. Lisi, Adv. Funct. Mater., 2021, 31, 2005300 CrossRef CAS.
- Y. Polo, J. Luzuriaga, J. Iturri, I. Irastorza, J. L. Toca-Herrera, G. Ibarretxe, F. Unda, J.-R. Sarasua, J. R. Pineda and A. Larrañaga, Nanomedicine, 2021, 31, 102314 CrossRef CAS PubMed.
- C. Tapeinos, Adv. NanoBiomed Res., 2021, 1, 2000059 CrossRef.
- Q. Ma, L. Yang, Z. Jiang, Q. Song, M. Xiao, D. Zhang, X. Ma, T. Wen and G. Cheng, ACS Appl. Mater. Interfaces, 2016, 8, 34227–34233 CrossRef CAS PubMed.
- Q. Fang, Y. Zhang, X. Chen, H. Li, L. Cheng, W. Zhu, Z. Zhang, M. Tang, W. Liu, H. Wang, T. Wang, T. Shen and R. Chai, Front. Bioeng. Biotechnol., 2020, 7, 436 CrossRef PubMed.
- N. Sultana, M. I. Hassan and M. M. Lim, Composite Synthetic Scaffolds for Tissue Engineering and Regenerative Medicine, Springer International Publishing, Cham, 2015 Search PubMed.
- K. R. Singh, V. Nayak, T. Sarkar and R. P. Singh, RSC Adv., 2020, 10, 27194–27214 RSC.
- C. Xu and X. Qu, NPG Asia Mater., 2014, 6, e90–e90 CrossRef CAS.
- Y. Qian, Q. Han, X. Zhao, H. Li, W.-E. Yuan and C. Fan, iScience, 2019, 12, 216–231 CrossRef CAS PubMed.
- M. Das, S. Patil, N. Bhargava, J.-F. Kang, L. M. Riedel, S. Seal and J. J. Hickman, Biomaterials, 2007, 28, 1918–1925 CrossRef CAS PubMed.
- A. Ranjbar, S. Soleimani Asl, F. Firozian, H. Heidary Dartoti, S. Seyedabadi, M. Taheri Azandariani and M. Ganji, J. Mol. Neurosci., 2018, 66, 420–427 CrossRef CAS PubMed.
- K. J. Livak and T. D. Schmittgen, Methods, 2001, 25, 402–408 CrossRef CAS PubMed.
- S. Pei and H.-M. Cheng, Carbon, 2012, 50, 3210–3228 CrossRef CAS.
- V. Agarwal and P. B. Zetterlund, Chem. Eng. J., 2021, 405, 127018 CrossRef CAS.
- Y. Shang, D. Zhang, Y. Liu and C. Guo, Bull. Mater. Sci., 2015, 38, 7–12 CrossRef CAS.
- C. K. Chua and M. Pumera, Chem. Soc. Rev., 2013, 43, 291–312 RSC.
- N. Ormategui, A. Veloso, G. P. Leal, S. Rodriguez-Couto and R. Tomovska, ACS Appl. Mater. Interfaces, 2015, 7, 14104–14112 CrossRef CAS PubMed.
- K. K. H. De Silva, H.-H. Huang and M. Yoshimura, Appl. Surf. Sci., 2018, 447, 338–346 CrossRef CAS.
- O. C. Compton and S. T. Nguyen, Small, 2010, 6, 711–723 CrossRef CAS PubMed.
- Y. Xu, K. Sheng, C. Li and G. Shi, ACS Nano, 2010, 4, 4324–4330 CrossRef CAS PubMed.
- E. Nicolas, S. Callé, S. Nicolle, D. Mitton and J.-P. Remenieras, Ultrasonics, 2018, 84, 119–125 CrossRef PubMed.
- S. Cheng, E. C. Clarke and L. E. Bilston, Med. Eng. Phys., 2008, 30, 1318–1337 CrossRef PubMed.
- Z. Ma, S. Hu, J. S. Tan, C. Myer, N. M. Njus and Z. Xia, J. Biomed. Mater. Res., Part A, 2013, 101, 2718–2725 CrossRef PubMed.
- A. F. Girão, J. Sousa, A. Domínguez-Bajo, A. González-Mayorga, I. Bdikin, E. Pujades-Otero, N. Casañ-Pastor, M. J. Hortigüela, G. Otero-Irurueta, A. Completo, M. C. Serrano and P. A. A. P. Marques, ACS Appl. Mater. Interfaces, 2020, 12, 38962–38975 CrossRef PubMed.
- S. Chae, T.-H. Le, C. S. Park, Y. Choi, S. Kim, U. Lee, E. Heo, H. Lee, Y. A. Kim, O. S. Kwon and H. Yoon, Nanoscale, 2020, 12, 13351–13359 RSC.
- S. Rao, J. Upadhyay, K. Polychronopoulou, R. Umer and R. Das, J. Compos. Sci., 2018, 2, 25 CrossRef.
- Z. Tan, S. Ohara, H. Abe and M. Naito, RSC Adv., 2014, 4, 8874–8878 RSC.
- X. Jing, H.-Y. Mi, B. N. Napiwocki, X.-F. Peng and L.-S. Turng, Carbon, 2017, 125, 557–570 CrossRef CAS.
- J. Park, J. H. Choi, S. Kim, I. Jang, S. Jeong and J. Y. Lee, Acta Biomater., 2019, 97, 141–153 CrossRef CAS PubMed.
- J. E. Hirsch, Phys. C, 1992, 199, 305–310 CrossRef CAS.
- Y. Nioua, S. El Bouazzaoui, B. M. G. Melo, P. R. S. Prezas, M. P. F. Graça, M. E. Achour, L. C. Costa and C. Brosseau, J. Mater. Sci., 2017, 52, 13790–13798 CAS.
- L. Koessler, S. Colnat-Coulbois, T. Cecchin, J. Hofmanis, J. P. Dmochowski, A. M. Norcia and L. G. Maillard, Hum. Brain Mapp., 2017, 38, 974–986 CrossRef PubMed.
- H. McCann, G. Pisano and L. Beltrachini, Brain Topogr., 2019, 32, 825–858 CrossRef PubMed.
- P. Zarrintaj, S. Manouchehri, Z. Ahmadi, M. R. Saeb, A. M. Urbanska, D. L. Kaplan and M. Mozafari, Carbohydr. Polym., 2018, 187, 66–84 CrossRef CAS PubMed.
- A. Saberi, F. Jabbari, P. Zarrintaj, M. R. Saeb and M. Mozafari, Biomolecules, 2019, 9, 448 CrossRef CAS PubMed.
- A. Y. Estevez and J. S. Erlichman, Nanomedicine, 2014, 9, 1437–1440 CrossRef CAS PubMed.
- J. Cui and G. A. Hope, J. Spectrosc., 2015, 2015, e940172 Search PubMed.
- J.-B. Wu, M.-L. Lin, X. Cong, H.-N. Liu and P.-H. Tan, Chem. Soc. Rev., 2018, 47, 1822–1873 RSC.
- B. Gupta, N. Kumar, K. Panda, V. Kanan, S. Joshi and I. Visoly-Fisher, Sci. Rep., 2017, 7, 45030 CrossRef CAS PubMed.
- Y. Yang, C. Tian, L. Sun, R. Lü, W. Zhou, K. Shi, K. Kan, J. Wang and H. Fu, J. Mater. Chem. A, 2013, 1, 12742 RSC.
- Y. Si and E. T. Samulski, Chem. Mater., 2008, 20, 6792–6797 CrossRef CAS.
- S. M. Hirst, A. S. Karakoti, R. D. Tyler, N. Sriranganathan, S. Seal and C. M. Reilly, Small, 2009, 5, 2848–2856 CrossRef CAS PubMed.
- G. Song, N. Cheng, J. Zhang, H. Huang, Y. Yuan, X. He, Y. Luo and K. Huang, Catalysts, 2021, 11, 1123 CrossRef CAS.
- C. Korsvik, S. Patil, S. Seal and W. T. Self, Chem. Commun., 2007, 1056–1058 RSC.
- S. M. Hirst, A. Karakoti, S. Singh, W. Self, R. Tyler, S. Seal and C. M. Reilly, Environ. Toxicol., 2013, 28, 107–118 CrossRef CAS PubMed.
- H. Sies, Redox Biol., 2017, 11, 613–619 CrossRef CAS PubMed.
- G. Ciofani, G. G. Genchi, I. Liakos, V. Cappello, M. Gemmi, A. Athanassiou, B. Mazzolai and V. Mattoli, Pharm. Res., 2013, 30, 2133–2145 CrossRef CAS PubMed.
- J. Yi, G. Choe, J. Park and J. Y. Lee, Polym. J., 2020, 52, 823–837 CrossRef CAS.
- J. D. Lathia, M. Li, P. E. Hall, J. Gallagher, J. S. Hale, Q. Wu, M. Venere, E. Levy, M. R. S. Rani, P. Huang, E. Bae, J. Selfridge, L. Cheng, H. Guvenc, R. E. McLendon, I. Nakano, A. E. Sloan, H. S. Phillips, A. Lai, C. L. Gladson, M. Bredel, S. Bao, A. B. Hjelmeland and J. N. Rich, Ann. Neurol., 2012, 72, 766–778 CrossRef CAS PubMed.
- C. A. Gregory, E. Reyes, M. J. Whitney and J. L. Spees, Stem Cells, 2006, 24, 2232–2243 CrossRef CAS PubMed.
- U. Lendahl, L. B. Zimmerman and R. D. McKay, Cell, 1990, 60, 585–595 CrossRef CAS PubMed.
- S. Suzuki, J. Namiki, S. Shibata, Y. Mastuzaki and H. Okano, J. Histochem. Cytochem., 2010, 58, 721–730 CrossRef CAS PubMed.
- A. Bernal and L. Arranz, Cell. Mol. Life Sci., 2018, 75, 2177–2195 CrossRef CAS PubMed.
- J. Dahlstrand, M. Lardelli and U. Lendahl, Brain Res. Dev. Brain Res., 1995, 84, 109–129 CrossRef CAS PubMed.
- F. Doetsch, J. M. García-Verdugo and A. Alvarez-Buylla, J. Neurosci., 1997, 17, 5046–5061 CrossRef CAS PubMed.
- S. Shaban, M. W. A. El-Husseny, A. I. Abushouk, A. M. A. Salem, M. Mamdouh and M. M. Abdel-Daim, Oxid. Med. Cell. Longevity, 2017, 2017, 5032102 Search PubMed.
- J. Lugrin, N. Rosenblatt-Velin, R. Parapanov and L. Liaudet, Biol. Chem., 2014, 395, 203–230 CrossRef CAS PubMed.
- C. T. Ekdahl, J.-H. Claasen, S. Bonde, Z. Kokaia and O. Lindvall, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 13632–13637 CrossRef CAS PubMed.
- L. Ottoboni, B. von Wunster and G. Martino, Front. Neurol., 2020, 11, 148 CrossRef PubMed.
- F. H. Gage and S. Temple, Neuron, 2013, 80, 588–601 CrossRef CAS PubMed.
- K. H. Lee, M. Cha and B. H. Lee, Int. J. Mol. Sci., 2021, 22, 13315 CrossRef CAS PubMed.
- Q. Ma, L. Yang, Z. Jiang, Q. Song, M. Xiao, D. Zhang, X. Ma, T. Wen and G. Cheng, ACS Appl. Mater. Interfaces, 2016, 8, 34227–34233 CrossRef CAS PubMed.
- K. Saha, A. J. Keung, E. F. Irwin, Y. Li, L. Little, D. V. Schaffer and K. E. Healy, Biophys. J., 2008, 95, 4426–4438 CrossRef CAS PubMed.
- S. Sergent-Tanguy, D. C. Michel, I. Neveu and P. Naveilhan, J. Neurosci. Res., 2006, 83, 1515–1524 CrossRef CAS PubMed.
- D. C. Lagace, M. C. Whitman, M. A. Noonan, J. L. Ables, N. A. DeCarolis, A. A. Arguello, M. H. Donovan, S. J. Fischer, L. A. Farnbauch, R. D. Beech, R. J. DiLeone, C. A. Greer, C. D. Mandyam and A. J. Eisch, J. Neurosci., 2007, 27, 12623–12629 CrossRef CAS PubMed.
- M. V. Sofroniew and H. V. Vinters, Acta Neuropathol., 2010, 119, 7–35 CrossRef PubMed.
- H. Kawano, J. Kimura-Kuroda, Y. Komuta, N. Yoshioka, H. P. Li, K. Kawamura, Y. Li and G. Raisman, Cell Tissue Res., 2012, 349, 169–180 CrossRef PubMed.
- A. McKeon and E. E. Benarroch, Neurology, 2018, 90, 925–930 CrossRef PubMed.
- E. Raponi, F. Agenes, C. Delphin, N. Assard, J. Baudier, C. Legraverend and J.-C. Deloulme, Glia, 2007, 55, 165–177 CrossRef PubMed.
- J. R. Pineda, M. Daynac, A. Chicheportiche, A. Cebrian-Silla, K. Sii Felice, J. M. Garcia-Verdugo, F. D. Boussin and M.-A. Mouthon, EMBO Mol. Med., 2013, 5, 548–562 CrossRef CAS PubMed.
- G. Santos, A. Barateiro, D. Brites and A. Fernandes, Front. Cell. Neurosci., 2020, 14, 279 CrossRef CAS PubMed.
- A. Wegener, C. Deboux, C. Bachelin, M. Frah, C. Kerninon, D. Seilhean, M. Weider, M. Wegner and B. Nait-Oumesmar, Brain J. Neurol., 2015, 138, 120–135 CrossRef PubMed.
- G. Ibarretxe, M. V. Sánchez-Gómez, M. R. Campos-Esparza, E. Alberdi and C. Matute, Glia, 2006, 53, 201–211 CrossRef PubMed.
- S. K. Thorburne and B. H. J. Juurlink, J. Neurochem., 1996, 67, 1014–1022 CrossRef CAS PubMed.
- E. Eitan, E. R. Hutchison, N. H. Greig, D. Tweedie, H. Celik, S. Ghosh, K. W. Fishbein, R. G. Spencer, C. Y. Sasaki, P. Ghosh, S. Das, S. Chigurapati, J. Raymick, S. Sarkar, S. Chigurupati, S. Seal and M. P. Mattson, Exp. Neurol., 2015, 273, 151–160 CrossRef CAS PubMed.
- N. Pirotte, N. Leynen, T. Artois and K. Smeets, Dev. Biol., 2016, 409, 4–15 CrossRef CAS PubMed.
- M. E. van Strien, S. A. van den Berge and E. M. Hol, Cell Res., 2011, 21, 1523–1525 CrossRef CAS PubMed.
- T. L. Walker, T. Yasuda, D. J. Adams and P. F. Bartlett, J. Neurosci., 2007, 27, 3734–3742 CrossRef CAS PubMed.
- T. Kremer, R. Jagasia, A. Herrmann, H. Matile, E. Borroni, F. Francis, H. G. Kuhn and C. Czech, PLoS One, 2013, 8, e59269 CrossRef CAS PubMed.
- J. G. Izant and J. R. McIntosh, Proc. Natl. Acad. Sci. U. S. A., 1980, 77, 4741–4745 CrossRef CAS PubMed.
- A. R. Gliga, K. Edoff, F. Caputo, T. Källman, H. Blom, H. L. Karlsson, L. Ghibelli, E. Traversa, S. Ceccatelli and B. Fadeel, Sci. Rep., 2017, 7, 9284 CrossRef PubMed.
- Z. S. Bailey, E. Nilson, J. A. Bates, A. Oyalowo, K. S. Hockey, V. S. S. S. Sajja, C. Thorpe, H. Rogers, B. Dunn, A. S. Frey, M. J. Billings, C. A. Sholar, A. Hermundstad, C. Kumar, P. J. VandeVord and B. A. Rzigalinski, J. Neurotrauma, 2020, 37, 1452–1462 CrossRef PubMed.
- B. H. J. Juurlink, S. K. Thorburne and L. Hertz, Glia, 1998, 22, 371–378 CrossRef CAS PubMed.
- A. Almeida, J. Almeida, J. P. Bolaños and S. Moncada, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 15294–15299 CrossRef CAS PubMed.
- C. Maffezzini, J. Calvo-Garrido, A. Wredenberg and C. Freyer, Cell. Mol. Life Sci., 2020, 77, 2483–2496 CrossRef CAS PubMed.
- M. J. Rigby, T. M. Gomez and L. Puglielli, Front. Neurosci., 2020, 14, 203 CrossRef PubMed.
- A. Amaral, T. Meisingset, M. Kotter and U. Sonnewald, Front. Endocrinol., 2013, 4, 54 CAS.
- E. E. Frost, P. C. Buttery, R. Milner and C. ffrench-Constant, Curr. Biol., 1999, 9, 1251–12S1 CrossRef CAS PubMed.
- L. Gang, Y. Yao, Y. Liu, Y. Li, K. Yang, L. Lu, Y. Cheng, X. Chen and Y. Tu, Neural Regener. Res., 2015, 10, 1612–1616 CrossRef PubMed.
- K. A. Chamberlain, S. E. Nanescu, K. Psachoulia and J. K. Huang, Neuropharmacology, 2016, 110, 633–643 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2nr06545b |
| This journal is © The Royal Society of Chemistry 2023 |