Open Access Article
Open Access ArticleNatural sesquiterpene quinone/quinols: chemistry, biological activity, and synthesis†
Xin-Hui
Tian
 ab,
Li-Li
Hong
ab,
Li-Li
Hong
 a,
Wei-Hua
Jiao
a,
Wei-Hua
Jiao
 a and
Hou-Wen
Lin
a and
Hou-Wen
Lin
 *a
*a
aMarine Drugs Research Center, Department of Pharmacy, Ren Ji Hospital, School of Medicine, State Key Laboratory of Microbial Metabolism, Shanghai Jiao Tong University, Shanghai 200127, P. R. China. E-mail: franklin67@126.com
bInstitute of Interdisciplinary Integrative Medicine Research, Shanghai University of Traditional Chinese Medicine, Shanghai 201203, P. R. China. E-mail: tianxinhui@126.com
First published on 13th January 2023
Abstract
Covering: 2010 to 2021
Sesquiterpene quinone/quinols (SQs) are characterized by a C15-sesquiterpenoid unit incorporating a C6-benzoquinone/quinol moiety. Numerous unprecedented carbon skeletons have been constructed with various connection patterns between the two parts. The potent anti-cancer, anti-inflammatory, anti-microbial, anti-viral, and fibrinolytic activities of SQs are associated with their diverse structures. The representative avarol has even entered the stage of clinical phase II research as an anti-HIV agent, and was developed as paramedic medicine against psoriasis. This review provides an overall summary of 558 new natural SQs discovered between 2010 and 2021, including seven groups and sixteen structure-type subgroups, which comprehensively recapitulates their chemical structures, spectral characteristics, source organisms, biological activities, synthesis, and biosynthesis, aiming to expand the application scope of this unique natural product resource.
1. Introduction
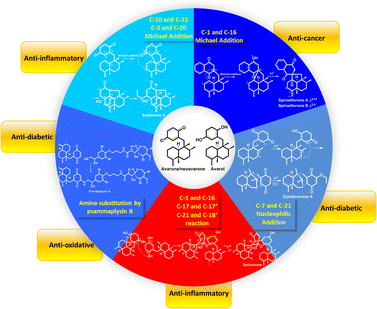
Terpenoid quinone/quinols, which are generally known as meroterpenoids, are hybrids of terpenoids and quinone/quinols. Sesquiterpene quinone/quinols (SQs) are the most common meroterpenoids found in nature. They have unique frameworks due to the structural variation of the sesquiterpene and quinone/quinol moieties, and varied connection patterns between the two units. Most of these compounds may be derived from polyketide synthase- and terpenoid synthase-mediated biosynthetic pathways, and others may be derived from shikimate-terpenoid adducts, which are the most common type of non-polyketide-derived meroterpenoids.1,2 SQs exhibit potent biological activities related to the redox characteristics and electron transfer function of the quinone/quinol group, and the hydrophobic–hydrophilic balance of the sesquiterpenoid unit. For example, avarol from Dysidea avara exhibits anti-HIV, anti-leukemic, and anti-parasitic activities, and has been used as paramedic medicine against psoriasis by the German company KliniPharm.3–10 Ilimaquinone from Hippospongia metachromia induces Golgi vesiculation and fragmentation, which may be closely related to its diverse biological activities, including anti-HIV, cytotoxic, anti-inflammatory, and anti-microbial properties.11–16 These natural SQs have proven to be significant resources for drug discovery. Marcos et al. and Capon reviewed the structure, bioactivities, source organisms, and stereochemical investigations of marine-derived SQs in 1995 and 2010, respectively.17,18 During the past decade, an increasing number of new SQs have emerged due to rapid advances in discovery methods, such as genome mining, biotransformation, microbial co-culture, chemistry first and MS/MS-based molecular networking, and fluorescent image-based high-content screening techniques, fast-growing improvement in analytic techniques such as NMR, MS, and single crystal X-ray diffraction, and great progress in diving equipment and collection tools used for marine organisms and microorganisms.19–29 However, there have been no systematic reviews on these newly discovered SQs, and there are only some reviews on meroterpenoids that cover part of the SQ family.30–32In this article, we provide 558 new SQs discovered during 2010–2021 and comprehensively describe their source organisms, chemical structure, bioactivities, and synthesis. SQs are extensively distributed in marine sponges, algae, ascidians, coral, fungi, and plants (Fig. 1). The metabolites from marine and marine-derived fungi account for 60.2% of the total SQs, and sponges are the predominant sources, producing 38.0% of these SQ metabolites. The fungi of Stachybotrys sp. and Ganoderma sp. produced 130 and 82 new compounds, respectively, and the sponge of Dysidea sp. produced 84 new compounds, ∼53% of the total number of SQs, which is the hottest research target.
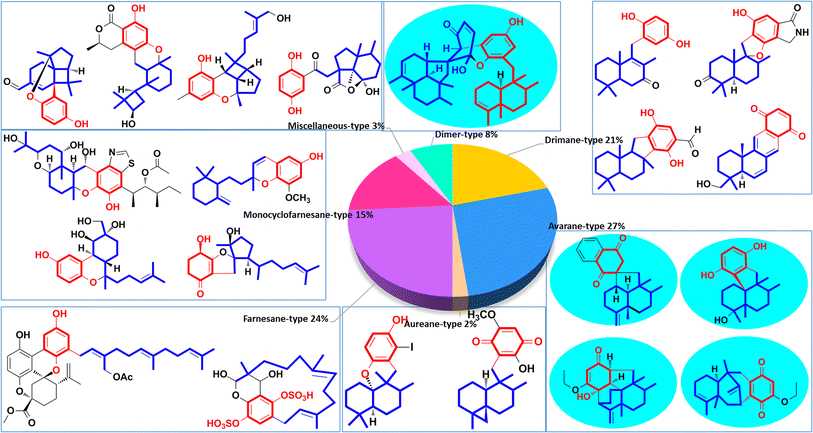
These natural SQs are divided into seven groups and sixteen subgroups based on the skeletons of the sesquiterpenoid subunits (Fig. 2), including five groups for drimane-type (tauranin-type, phenylspirodrimane-type, chrodrimanin-type, dasyscyphin-type, and hymenopsin-type), five groups for avarane-type (typical avarane-type, dactyloquinone-type, dysideanone-type, spiroetherane-type, and dysidavarane-type), four groups for monocyclofarnesane type (cochlioquinone-type, metachromiane-type, bisabolane-type, and tricycloalternarene-type), and two groups for dimer-type (true and pseudo SQ-dimers). The drimane-type (118 compounds, 21%), avarane-type (152, 27%), and farnesane-type (134, 24%) comprise an enormous proportion of SQs, followed by monocyclofarnesane-type (83, 15%) and dimer-type (47, 8%). Avarane-type SQs only exist in marine sponges, and more than half have been reported from Dysidea sp. The dysideanone-, septosone-, dysidavarane, dysiarenone-, and spiroetherane-type SQs in the blue shadow of Fig. 2 were all discovered from Dysidea sponge by H. W. Lin's group.33–37 The SQ second metabolites of fungi include drimane-, aureane-, farnesane-, monocyclofarnesane-, bisabolane-, and dimer-type. Most phenylspirodrimanedrimane-type analogs are isolated from the genus Stachybotrys, about half of the farnesane-type and all bisabolane-type analogues are reported from the genus Ganoderma.
Pharmaceutical studies have indicated that 48% of SQs were biologically active with ∼34.3% of the active compounds (268 compounds) displaying anti-cancer activity, followed by anti-inflammatory (13.1%), anti-microbial (12.6%), anti-diabetic (7.2%), anti-oxidant (6.7%), anti-viral (6.0%), reno-protective (5.2%), fibrinolysis (4.5%), and other (10.4%) activities (Fig. 3). The anti-cancer SQs are more active toward lung cancer cells (17%), breast cancer cells (11%), intestinal cancer cells (9%), liver cancer cells (8.5%), leukemia cells (7.7%), cervical cancer cells (6.1%), ovarian cancer cells (5.7%), and gastric cancer cells (5.0%) (Fig. 3). The anticancer and fibrolytic activities seem to be more attractive and visible. Thus, many in vivo and in vitro activity and mechanistic studies have been carried out. However, further biological studies on many promising new drug candidates have been limited due to the scarcity of samples obtained from natural resources. The chemical synthetic and biosynthetic routes outlined in this review will provide inspiration for synthetic design and enhance the understanding of the biomedical potential and biosynthetic logic of SQs.
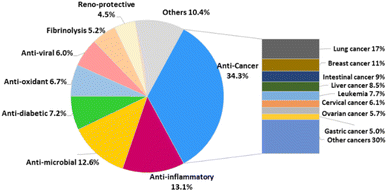 | ||
| Fig. 3 Proportion of each activity compared to the whole occurrence of activities observed for sesquiterpene quinone/quinols isolated from nature. | ||
2. Structures, classifications, and distributions of natural SQs
We have classified SQs into seven groups, five of which have a unified sesquiterpenoid skeleton, one type consists of miscellaneous sesquiterpenoid skeletons, and the last type consists of SQ dimers. Some groups of SQs contain subtypes because new carbon skeletons are built when the sesquiterpenoid and quinone units are connected in different ways. We have summarized advanced discovering techniques, common structural characteristics, and typical NMR, MS spectral signals for each type of SQs, which will help researchers efficiently recognize the unknown constituents. The determination of the absolute stereochemistry of SQs has also been included because it is crucial for their bioactivity and further synthetic design.2.1. Drimane-type SQs
Drimane-type SQs are the most common structures found in nature, and the sesquiterpene unit features a drimane skeleton containing trans- or cis-fused ring junctions. They contain five subtypes including tauranin-type, phenylspirodrimane-type, chrodrimanin-type, dasyscyphin-type, and hymenopsin-type based on the connection modes between drimane and the C-6 benzene moiety.2.2. Avarane-type SQs
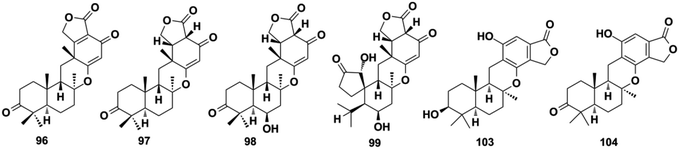
The sesquiterpenoid part of avarane-type SQs possess a rearranged drimane scaffold, which are also known as the 4,9-friedodrimane-type, in which one methyl of drimane migrates from the C-4 to C-5 position and the other methyl migrates from the C-10 to the C-9 position. The earliest and most well-known examples of this skeleton type are avarol and avarone, which were isolated from Dysidea avara.3 Avarane-type SQs exist as metabolites, which are only derived from sponges, such as Dysidea sp., Dactylospongia sp., Spongia sp., Smenospongia sp., and Hyrtios sp. The H. W. Lin's group have intensively studied the chemical constitution of the above sponges and reported 124 new SQs since the year of 2012, accounting for ∼80% of all avarane-type SQs reported to date.33–37,79,84,86–99 Among these compounds, eight were new skeletons, namely, “Dysiarenone,” “Septosone,” “Frondoplysin,” “Dysideanone,” “Spiroetherane,” and “Dysidavarone,” and rated as “hot off the press” by Natural Product Reports.33–37,97,100–104| Comp. | δ H-10 (J in Hz) | δ C-12 | Comp. | δ H-10 (J in Hz) | δ C-12 |
|---|---|---|---|---|---|
| a Cis-fused A/B ring. b Trans-fused A/B ring. | |||||
144![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a a |
1.09 (br d 6.3) | 32.5 | 202b | 0.78 (dd 11.6, 1.8) | 20.5 |
145![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a a |
1.10 (br d 6.3) | 32.5 | 203b | 1.04 (m) | 19.9 |
146![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a a |
1.11 (br d 6.3) | 32.5 | 204b | 1.03 (m) | 19.8 |
147![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a a |
1.11 (br d 5.9) | 32.5 | 208b | 0.93 (dd 12.8, 2.3) | 20.6 |
148![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a a |
1.11 (br d 6.2) | 32.5 | 209b | 1.25 (m) | 21.6 |
149![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a a |
1.32 (br d 5.7) | 32.4 | 210b | 1.21 (m) | 21.3 |
200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a a |
1.19 (br d 5.8) | 33.5 | 213b | 0.78 (dd 12.0, 2.0) | 20.8 |
229![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a a |
1.40 (d 6.6) | 33.2 | 217b | 0.92 (d 13.0) | 20.6 |
231![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a a |
1.17 (d 6.0) | 33.3 | 218b | 0.92 (dd 9.5, 1.5) | 20.6 |
232![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a a |
1.19 (m) | 33.4 | 220b | 0.78 (dd 11.5, 2.0) | 20.6 |
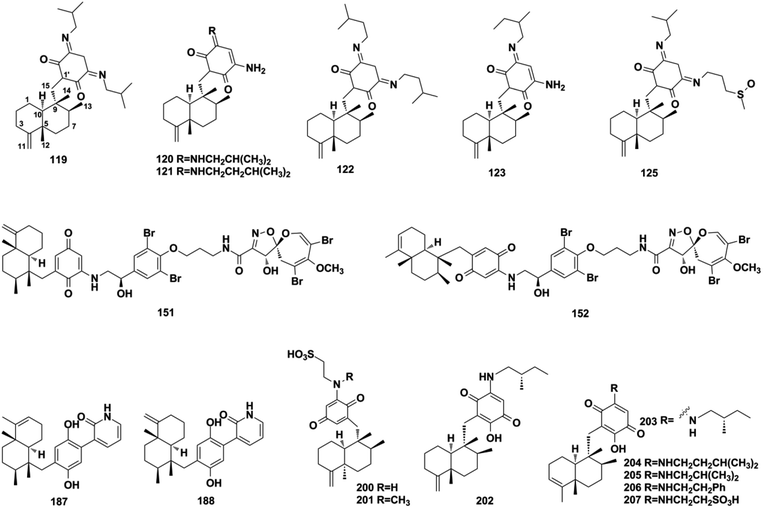
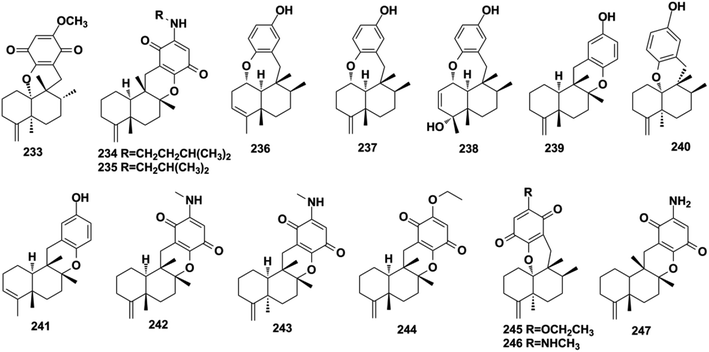
Using a hemisynthetic phishing probe coupled with MS/MS molecular networking, dactylocyanines A–H (119–126), which possesses an unexpected zwitterionic diamino-meta-quinonoid blue scaffold, have been isolated from Dactylospongia metachromia.24 Fucci2 is a newly-developed fluorescent cell cycle probe; a fluorescent image-based high-content screening technique using HeLa/Fucci2 cells was developed to explore cell cycle inhibitors from Dactylospongia metachromia, and three new compounds, namely, neoisosmenospongine (127), nakijiquinone I (128), and (−)-dictyoceratin-C (129), were discovered.21 More compounds were obtained from Dactylospongia sp., which were described as dactylospongins A–D (130–133), ent-melemeleone B (134), melemeleones C–D (135–136), dysidaminone N (137), nakijinol B (138), smenospongines B–C (139–140), 19-methoxy-dictyoceratin A (141), 5,8-di-epi-ilimaquinone (142), nakijiquinone V (143), 5-epi-nakijiquinones S, Q, T, U, and N (144–148), and 5-epi-nakijinols C–D (149–150).16,84,98,105–107 In addition to Dactylospongia sp., Dysidea sp. has been found to be an important source of avarane-type SQ metabolites. LC-MS–guided fractionation of Dysidea frondosa produced a pair of bioconjugates, frondoplysin A and B (151–152), which are composed of a sesquiterpene quinone and an unusual psammaplysin alkaloid.97 Seventeen structural analogs, dysidaminones A–M (153–165) and 19-methyl and 18-, 19-, and 18-phenethyl-substituted aminoavarones (166–169) were isolated from the sponge Dysidea fragilis.93 The same research group has also reported cinerols A–K (170–180) from Dysidea cinerea, 8-ethoxyneoavarone (181), 19-ethoxyneoavarone (182), 18-ethoxyavarone (183), 19-ethoxyavarone (184), and (−)-N-methylmelemeleone A (185) from Dysidea avara, dysicigyhone A (186) from Dysidea septosa, and the first examples of terpene-polyketide-pyridine hybrid metabolites, dysivillosins A–D (187–190) from Dysidea villosa.87,88,94,95,108Dysidea sp. sponges have also yielded (+)-19-methylaminoavarone (191), 20-methoxyneoavarone (192), 17-O-acetylavarol (193), 17-O-acetylneoavarol (194), 18-aminoarenarone (195), 19-aminoarenarone (196), 18-methylaminoarenarone (197), 19-methylaminoarenarone (198), smenospongimine (199), and melemeleones C and D (200–201).55,109–112 We noticed that compounds 200 and 201 were both reported as new compounds; however, they are isomers of the reported compounds in terms of the stereogenic centers at C-5, C-8, C-9, and C-10, and the double bond positions in the sesquiterpenoid part.84,109 Nakijiquinones L, K, N, O, Q, and R (202–207) were discovered from collections of Okinawan marine sponges of the family Spongiidae, and they possess unique side chains derived from amino acids.113 The chromatographic fractionation of the extract from Spongia sp. furnished langconols A–C (208–210), langcoquinones A–F (211–216), 18-deoxy-18-formamidodictyoceratin B (217), 18-deoxy-18-(2-hydroxyacetyl)aminodictyoceratin B (218), dictyoceratin D (219), N-methyl-ent-smenospongine (220), and N-methyl-5-epi-smenospongine (221).79,114–116 The chemical investigation of sponges Hyrtios sp., Smenospongia cerebriformis, and Aka coralliphaga, afforded nakijinols F and G (222–223), smenohaimiens C–E (224–226), and akadisulfate B (227), respectively.56,96,117 Biotransformation has been employed to identify drug leads from the oxidative incubation mixture of Smenospongia aurea, S. cerebriformis, and Verongula rigida, followed by the identification of the structurally new nakijinol E (228), 5-epi-nakijinol E (229), nakijinone A (230), 5-epi-nakijinone A (231), and 5-epi-20-O-ethylsmenoquinone (232).19
2.3. Aureane-type SQs
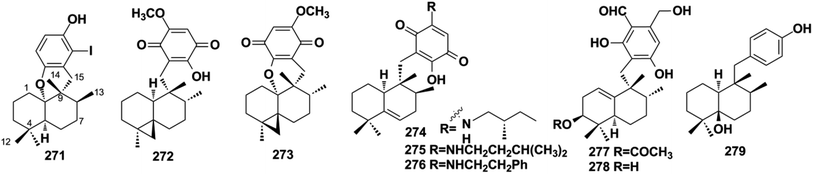
Aureane-type SQs are the methyl (5 → 4)-abeo products of avarane-type SQs, which feature a gem-dimethyl at the C-4 position. The rare iodo-substituted 6′-iodoaureol (271) was isolated from the sponge Smenospongia sp.123 Bioassay-guided isolation of the active extract of Dactylospongia elegans resulted in 4,5-di-epi-dactylospongiaquinone (272) and 10,17-O-cyclo-4,5-di-epi-dactylospongiaquinone (273), both with a cyclopropyl ring system characterized by two doublet signals observed at δH < 1 ppm, J = 4.0 Hz for CH2-12.16 Nakijiquinone J (274) with (S)-2-methylbutylamine and nakijiquinones M and P (275, 276) isolated from the sponges of the Spongiidae family exist as amine forms.113 Myrothecisins C and D (277–278) are isolated from the medical plant-derived fungus Myrothecium sp. OUCMDZ-2784.70 Yahazunol B (279) is the chemical constituent of Spongia pertusa Esper.792.4. Farnesane-type SQs
Farnesane-type SQs are characterized by a farnesane-type sesquiterpenoid long-chain attached to the quinol unit. A total of 35 papers have described 122 new farnesane-type SQs isolated from the marine sponge Aka coralliphagum, the medicinal plant Rhododendron anthopogonoides, and five genera of fungi, including Ganoderma, Stachybotrys, Penicillium, Albatrellus, and Alternaria. Almost all of the metabolites from the Stachybotrys species share a common isoindolinone motif. There is a fascinating stereochemical anomaly in which most of the SQs isolated from Ganoderma species are found to be enantiomers or racemates, which may be due to the flexibility of the aliphatic chain, and γ-lactones or cyclic ethers are usually formed in these species.
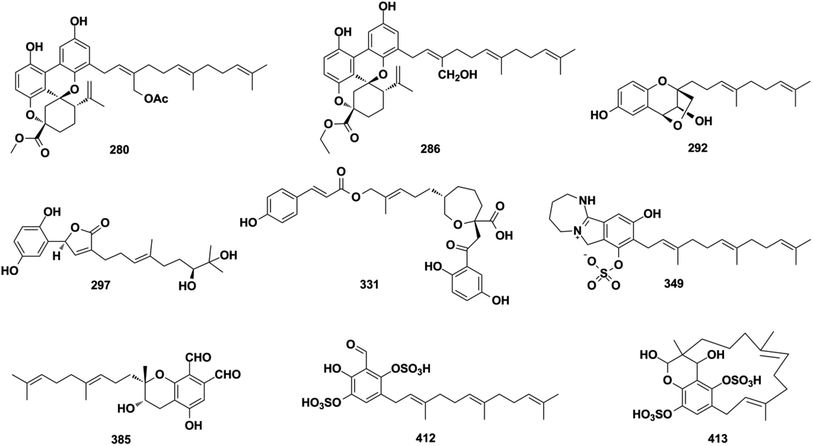
Ganoderma fungi contain more than 80 species, many of which have been used as folk medicine in Asian countries since ancient times. An in-depth investigation has been carried out on Ganoderma species by Y. X. Cheng's group and they reported cochlearoids A, B, and E (280–282), cochlearoids H–J (283–285), (±)-cochlearoids N–Q (286–289), cochlearols C, D, R, Q (290–293), gancochlearols D, E, and I (294–296), and ganomycin K and F (297–298) from Ganoderma cochlear; lucidumones B–H (299–305), dayaolingzhiols C–E (306–308), ganodermaone B (309), and chizhine F (310) from Ganoderma lucidum; and (±)-zizhines A–F (311–316) from Ganoderma sinensis.124–135 Compounds 280–282 and 286–289 possess a unique methanobenzo[c]oxocino[2,3,4-ij]-isochromene scaffold.125,126,128 The stereogenic centers of 292 were assigned as 1′S, 2′S, and 3′S using a combination of MAD, Δδmax, and R2 analysis of its 1H NMR chemical shifts in CHCl3, DMSO, and MeOH, and the NMR calculation method was proven to be an effective tool for determining the absolute configuration of compounds that lack efficient ROESY data.130,131,136 The first stereochemical establishment of compound 297 was assigned as 1R,10S using the Rh2(OCOCF3)4-induced ECD method according to the bulkiness rule.127 There are also many other studies focusing on the constituents of Ganoderma sp., such as ganomycin C (317), cochlearins B–E and I (318–322) from G. cochlear, ganofuran B (323) from G. lucidum, ganoduriporols C–L (324–333) from Ganoderma ahmadii, and ganoresinains A, B, and E (334–336) from G. resinaceum, among which the unique oxepane was formed in the structures of compounds 331–333.137–142 Serial fractionation and purification of the marine fungi Stachybotrys chartarum and Stachybotrys longispora has led to the identification of isoindolinone chartarutines A–H (337–344), stachybotrysams A–D (345–348), stachybotrin G (349), and FGFC2 4–7 (350–354), and orsellinic acid-based stachybonoids A–C (355–357).25,26,61,65,143,144 It is worth mentioning that stachybotrin G reported as compounds 349 and 84 have totally different structures.71,143 A total of 27 Stachybotrys Microspora Triprenyl Phenols (SMTP) congeners (358–384) with characteristic tricyclic γ-lactam, geranylmethyl, and N-linked side chain, along with their precursor pre-SMTP (385), have been reported from amine-fed cultures of Stachybotrys microspore.145–148 Verruculide B (386) was obtained from ascidian-derived Penicillium verruculosum.76 Diethyl sulfate (DES) mutagenesis is an effective method used to activate the silent pathways and exploit new metabolites of fungi, and chrysomutanin (387) was separated from the DES-mutant Penicillium chrysogenum.28 Amyloid-β aggregation inhibition activity-guided fractionation of Albatrellus yasudae has afforded scutigeric acid (388), albatrelactone methyl ester (389), albatrelactone (390), 10′,11′-dihydroxygrifolic acid (391), 2-hydroxy-1-methoxy neogrifolin (392), and 9′-keto-grifolic acid (393).149,150 Bicycloalternarenes A–F (394–399), monocycloalternarenes A–D (400–403), and tricycloalternarene J (404) are produced by the sponge-derived fungus Alternaria sp.151,152Rhododendron anthopogonoides Maxim has been used as a herbal medicine for rheumatoid arthritis and chronic bronchitis, and four enantiomers (±)-anthoponoids E–H (405–408) and (±)-daurichromene D (409) were isolated from its twigs and leaves by chiral-phase HPLC using n-hexane/isopropanol as the eluent.153 Four sulfated SQs, siphonodictyals E1–E4 (410–413), have been isolated from the sponge Aka coralliphagum and the proposed biosynthetic pathway showed that the macrocycle in 413 may be derived from 412via an aldol addition reaction.81
2.5. Monocyclofarnesane-type SQs
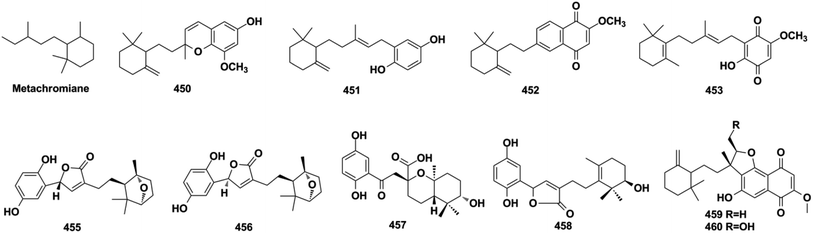
The sesquiterpenoid moiety of monocyclo-farnesane-type SQs may be derived from cyclization and methyl migration of their farnesyl precursor. Monocyclofarnesane-type SQs are mainly classified into cochlioquinone-type, metachromiane-type, bisabolane-type, and tricycloalternarene-type based on the skeleton of the sesquiterpenoid unit. These compounds are widely distributed in fungi and marine sponges.2.6. Miscellaneous SQs
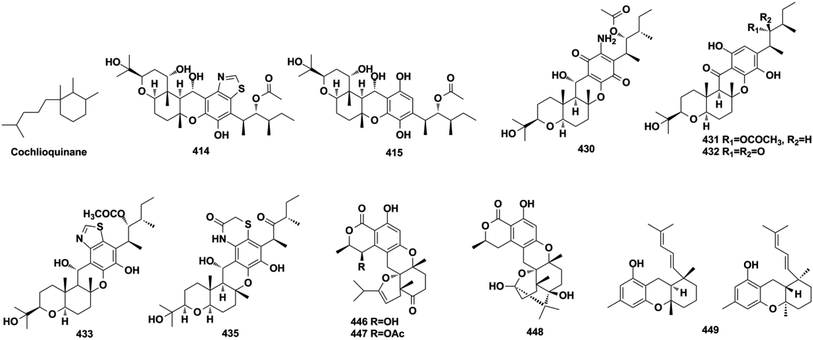
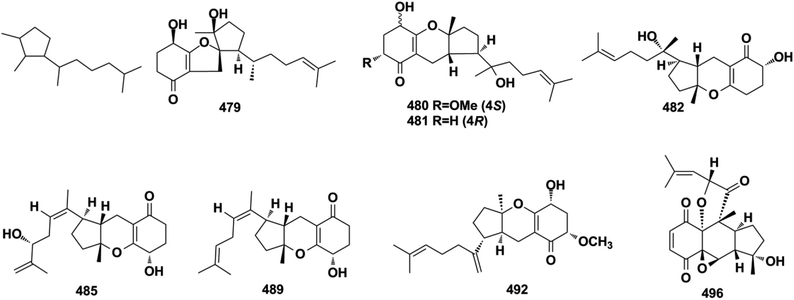
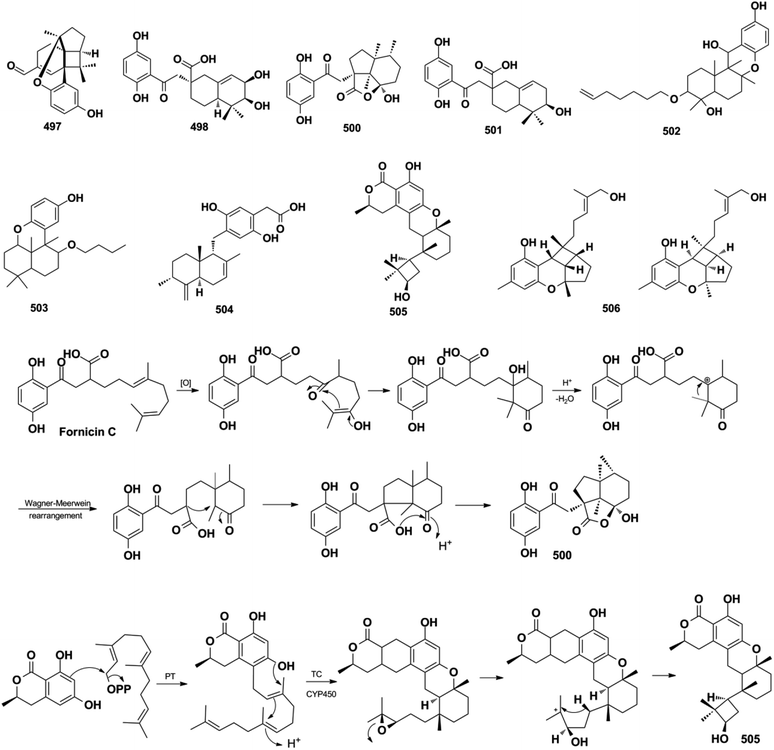
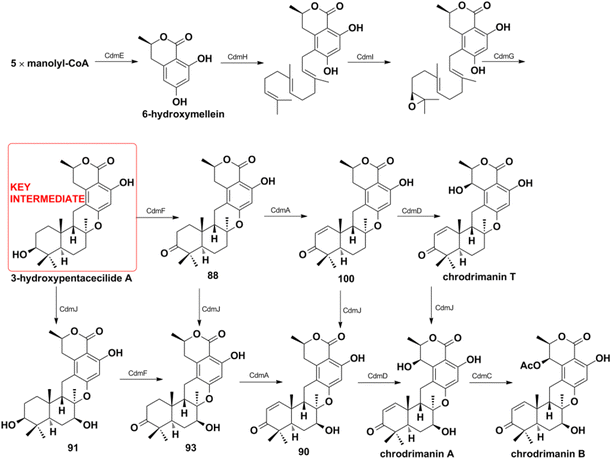
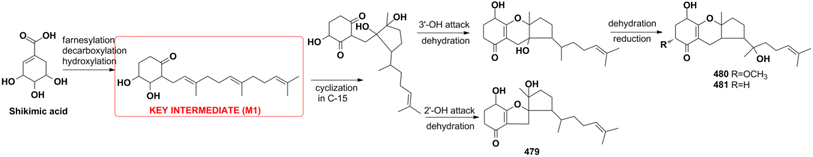
Miscellaneous SQs usually have rearranged sesquiterpenoid skeletons. Cochlearols B, N, and O (497–499), ganodermaone A (500), and ganoresinain D (501) have been isolated from G. cochlea and G. resinaceum.130,133,142,169 Compound 500 has a rearranged carbon backbone that may be derived from fornicin C, which was also purified from Ganoderma sp. via nucleophilic addition, Wagner–Meerwein rearrangement, cyclization, and esterification (Scheme 3).133 The sesquiterpenoid moiety of 3-(hept-36-enyloxy)-decahydro-4,6a,12a,12b-tetramethyl-1H-benzo[a]xanthene-4,10,12-triol (502), 1-butoxy-4,4,11b,11c-tetramethyl-decahydrobenzo[kl]xanthen-10-ol (503) from the macroalga Gracilaria salicornia, and peyssonoic acid B (504) from the macroalga Peyssonnelia sp. feature a rearranged drimane skeleton, and the rearrangement may be catalyzed by S-adenosyl methionine-mediated methyl transfer reactions.51,52 Talaromyolide D (505) from Talaromyces sp. CX11 has been proposed to originate from 6-hydroxymellein via cytochrome P450-mediated epoxidation, epoxide ring opening, and cation intermediate deprotonation (Scheme 3).159 Optically pure (±)-anthoponoids B–D (506–508), (±)-fastinoids A and B (509–510), and (±)-rubiginosin A (511) have been isolated from Rhododendron anthopogonoides using chiral HPLC.153,1702.7. SQ dimers
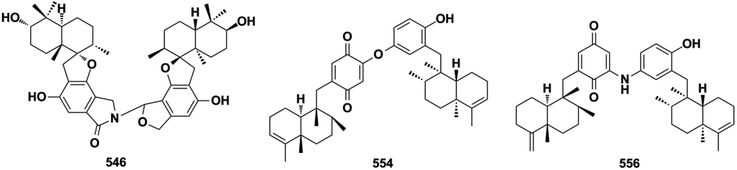
SQ dimers are classified into true and pseudo-types according to the dimerization patterns of the two monomers.171–173 Their molecular weights are in the range of 630–900, which is twice that of their corresponding monomers. In true SQ-dimers, two units of SQs are directly connected via one or two C–C bonds. In the pseudo-type, the monomers are linked by ether, amine, or alkyl amine bridges. These SQ dimers are mainly distributed in the fungi of Stachybotrys sp. and are widely found in the sponges of the genera Dysidea, Dactylospongia, and Smenospongia. Most of these monomers are phenylspirodrimane-type SQs.3. Biological activities and mechanisms of action
The natural products provide numerous medicines for disease treatment since ancient times and the natural sesquiterpenod quinone/quinols (SQs) are proved to be important in the new drug discovery process. A fascinating example is avarol that was developed as a topical ointment for psoriasis. It acts by downregulating the inflammatory cytokines of TNF-α, NF-κB, as well as eicosanoid and superoxide in psoriatic skin.183,184 It also inhibits the growth of erythroleukemia cells by inducing single-strand breaks of DNA.6 In addition, avarol exerts potent anti-HIV activities by inhibiting reverse transcriptase and augmenting immune responses proteins selectively.4,5 However, the anti-HIV studies of avarol by the University Medicine of the Johannes Gutenberg-University Mainz was terminated in clinical phase II. Avarone is the oxidative derivative of avarol, which exhibits 70% curative ratio against leukemia mice at 10 mg kg−1 and increases their life span by 146% when the treatment began at day 1, being a promising anti-cancer drug candidate.185 We may obtain SQs with potential druggability by structural optimization in the far future. The structural diversity of SQs contributes to their broad range of bioactivities. Approximately 48% of all SQ metabolites display potential bioactivity, such as anti-cancer compounds, mainly from Stachybotrys sp. and Dysidea sp., anti-inflammatory compounds mostly distributed in Dysidea sp., anti-microbial components mainly found in fungi, anti-oxidant compounds mainly isolated from Ganoderma cochlea, anti-viral metabolites purified from Stachybotrys sp. and Talaromyces sp., reno-protective metabolites reported from Ganoderma sp., and fribrinolytic products discovered from Stachybotrys sp.3.1. Anti-cancer activity
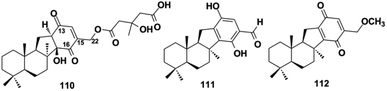
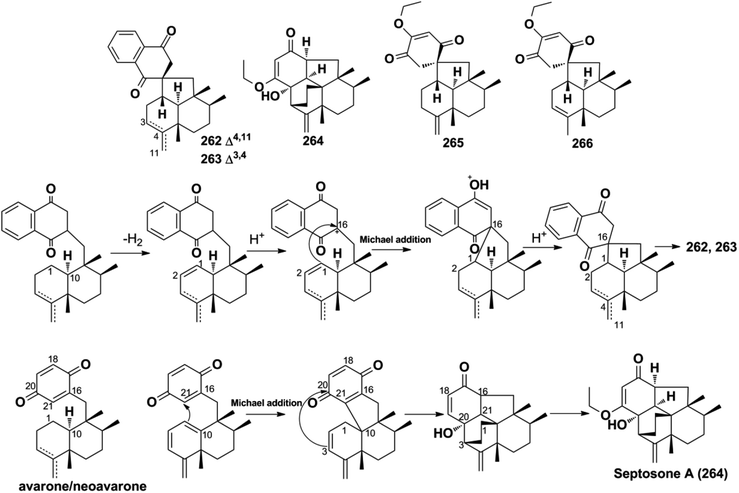
More than one-third of active SQs exhibit anti-cancer activity, providing a rich source of anti-cancer agents (Table 2). They exert anti-tumor activity via cytotoxicity, inducing cancer cell apoptosis, cell cycle arrest, preventing migration, inhibiting tumor-associated protein kinases, and blocking angiogenesis. Smenospongine from the sponge Spongia pertusa has been identified as a promising candidate agent for breast cancer treatment because significant anti-tumor effects have been observed in mouse xenograft tumor models. It downregulates stem cell markers Nanog, Sox2, and Bmil, induces DNA damage, initiates caspase-dependent apoptosis, and arrests the G0/G1 phase of breast cancer cells MCF7-Nanog via the p38/AMPKα pathway.91 The SQ dimers bistachybotrysins A–B, D–E, M (514–515, 517–518, 525) from S. chartarum are cytotoxic on HCT116, NCI-H460, BGC823, Daoy, and HepG2 cells with IC50 of 1.8–8.9 μM.175,177,178 Epoxyphomalin D (22) displays highly potent cytotoxicity by inhibiting the proliferation of 34 types of cancer cells, including liver cancer, melanoma, ovarian cancer, and renal cancer cells with IC50 values in the range of 0.72–13.52 μM.49 The overgrowth of angiogenesis from an existing vascular system is usually found in solid tumors, and preventing angiogenesis is an important anti-cancer approach. Spiroetherones A and B (262–263) from Dysidea etheria inhibit the angiogenesis of zebrafish with IC50 values of 2.4 ± 0.3 and 8.7 ± 0.6 μM, respectively; compound 262 is also cytotoxic toward NCI-H929, A549, HepG2, and SK-OV-3 cancer cells with IC50 values in the range of 7.4–12.2 μM, exhibiting a combined anti-cancer mechanism.34 Chartarolides A and B (34–35) and 5-epi-nakijiquinone N (148) inhibit the proliferation of HCT-116, HepG2, BGC-823, NCI-H1625, A2780, MCF-7, and L5178Y cancer cell lines with IC50 values in the range of 1.3–5.5 μM, which may be mechanistically related to the inhibition of a group of cancer-related kinases, including FGFR3, IGF1R, PDGFRb, TrKB, ALK, FAK, IGF1-R, SRC, and VEGF-R2 (IC50 = 0.97–11.3 μM).62,105 While 5-epi-nakijinol C (149), cinerol F (175), and 20-demethoxy-20-methylaminodactyloquinone D (242) only inhibit tumor-related kinases of ALK, FAK, IGF1-R, VEGF-R2, SHP-1, and CDK-2 with IC50 values at 2.78–7.78 μM.79,94,105 Metastasis occurs when cancer spreads from the primary location to secondary sites and is the major driving force of cancer patient mortality. (−)-Lucidumone B (299), (−)-lucidumone D (301), (+)-lucidumone F (303), and (+)-lucidumone H (305) selectively inhibit Kyse30 cell migration at a nontoxic concentration of 20 μM and their anti-metastatic effects are related to the downregulation of N-cadherin.135 Gancochlearol I (296) suppresses the epithelial–mesenchymal transition of triple-negative breast cancer cells (MDA-MB-231) by downregulating the key factors of TWIST1 and SNAI1 at 20 μM, providing valuable drug candidates for the treatment of cancer metastasis.127 Nuclear factor κB (NF-κB) plays a key role in the proliferation, migration, and apoptosis of cancer cells, and is an important target for anti-cancer drug discovery. Dysidaminones C, E, H, and J (155, 157, 160, and 162), and 18-aminoavarone (167) inhibit NF-κB in HEK293 cells with IC50 values in the range of 0.05–0.27 μM, and they are also cytotoxic toward NCI-H929, HepG2, B16F10, and SK-OV-3 cancer cells with IC50 values in the range of 0.45–9.65 μM. Structure–activity relationship analysis has shown that the 18-amino group in the quinone unit is essential for cytotoxicity and NF-κB inhibitory activity.93 The WNT pathway is commonly activated in many cancers and its activation is dependent on β-catenin-related gene transcription. (+)-5-epi-20-O-ethylsmenoquinone (232) inhibits the proliferation of CRT-positive SW480 and HCT116 colon cancer cells via the inhibition of Wnt/β-catenin and accelerating cytosolic β-catenin degradation at 1.5–6.0 μM.19 Stachybochartin C (537) time- and dose-dependently inhibits colony formation (0.5–2.0 μM) and proliferation (1.5–50 μM) in U-2OS cells, and the remarkably increased caspase-8, caspase-9, and PARP indicate that this effect was associated with caspase-dependent apoptosis.67 Many SQs exhibit multiple bioactivities. 19-O-methylpelorol (113), dysiquinol D (239), dysiherbol A (248), and gancochlearol I (296) display anti-tumor (IC50 = 0.58–9.2 μM) and anti-inflammatory activities (IC50 = 0.49–9.2 μM).84,87,92,127 Dasyscyphin C (110), langcoquinones C and D (213–214), aureol B (240), and (+)-cochlearoid P (288) exhibit both cytotoxic (IC50 = 4.1–9.6 μM) and anti-microbial activities (MIC = 2–8 μg mL−1, 6.25–12.5 μM).83,109,115,116,126 Phomoarcherin B (104) exerted inhibition on five human epidermoid carcinoma and adenocarcinoma cells (IC50 = 0.1–9.4 μg mL−1) and antimalarial activity against P. falciparum with IC50 = 0.79 μg mL−1.77 Bistachybotrysin M (525) prevents HCT116, NCI-H460, BGC823, Daoy, and HepG2 cancer cell proliferation with IC50 values in the range of 1.8–3.5 μM, and it also exerts neuroprotective effects toward glutamate-induced toxicity (17.4% at 10 μM) that is comparable to reservatrol (16.1% at 10 μM).175 Although smenospongine B (139), nakijiquinone L (202), langconol C (210), langcoquinones A–D (211–214), and metachromin V (451) are cytotoxic toward several cancer cells with IC50 or GI50 values in the range of 4.8–10.0 μM, the lack of selectivity in normal cell lines (GI50 on CHO-K1 = 2.1–3.0 μM, IC50 on WI-38 = 5.6–8.8 μM) may make them difficult to develop into new anti-cancer drugs in the future.106,115,116,162| Comp./pos. | IC50 | Cells | Ref. |
|---|---|---|---|
| a Positive control | |||
| Epoxyphomalin D (22) | IC50 = 0.72–13.52 μM | T24, BXF 1218L, CNXF 498NL, SF268, HCT116, HT29, GXF 251L, HNXF 536L, LXFL 1121L, LXFA 526L, LXFL 529L, LXFA 629L, H460, MAXF 401NL, MCF-7, MEXF 276L, MEXF 394NL, MEXF 462NL, MEXF 514L, MEXF 520L, OVXF 1619L, OVXF 899L, OVCAR3, PAXF 1657L, PANC1, 22RV1, DU145, PC3M, PXF 1752L, RXF 1781L, RXF 393NL, RXF 486L, RXF 944L, UXF 11138L | 49 |
| Chartarolide A (34) | IC50 = 1.3–5.5 μM | HCT-116, HepG2, BGC-823, NCI-H1650, A2780, MCF-7 | 62 |
| IC50 = 2.6–9.1 μM | FGFR3, IGF1R, PDGFRb, TrKB | ||
| Chartarolide B (35) | IC50 = 1.6–4.8 μM | HCT-116, HepG2, BGC-823, NCI-H1650, A2780, MCF-7 | |
| IC50 = 4.9–11.3 μM | FGFR3, IGF1R, TrKB | ||
| Chartarolide C (36) | IC50 = 5.4–12.5 μM | HCT-116, HepG2, BGC-823, NCI-H1650, A2780, MCF-7 | |
| Taxola | IC50 = 0.001–0.09 μM | ||
| Satratoxin Ha | IC50 < 0.5 μM | FGFR3, IGF1R, PDGFRb, TrKB | |
| Phomoarcherin B (104) | IC50 = 0.1–9.4 μg mL−1 | KKU-100, KKU-M139, KKU-M156, KKU-M214, KB | 77 |
| Ellipticinea | IC50 = 0.19–7.11 μg mL−1 | ||
| Dasyscyphin F (111) | IC50 = 4.1–8.2 μM | MDA-MB-231, MDA-MB-435 | 83 |
| Taxola | IC50 = 0.002–0.007 μM | ||
| 19-O-methylpelorol (113) | IC50 = 9.2 μM | PC-9 | 84 |
| Smenospongine B (139) | GI50 = 6.0–10.0 μM | SF-268, MCF-7, H460, HT-29 | 106 |
| GI50 = 3.0 μM | CHO-K1 (normal cells) | ||
| 5-Epi-nakijiquinone N (148) | IC50 = 0.97–3.03 μM | ALK, FAK, IGF1-R, SRC, VEGF-R2 | 105 |
| IC50 = 1.8 μM | L5178Y | ||
| 5-Epi-nakijinol C (149) | IC50 = 3.31–7.78 μM | ALK, FAK, IGF1-R, VEGF-R2 | |
| Kahalalide Fa | IC50 = 4.3 μM | L5178Y | |
| Dysidaminone C (155) | IC50 = 0.57–8.71 μM | NCI-H929, HepG2, B16F10, SK-OV-3//TNF-α induced HEK293/NF-κB cells | 93 |
| IC50 = 0.22 μM | |||
| Dysidaminone E (157) | IC50 = 0.94–9.52 μM | ||
| IC50 = 0.27 μM | |||
| Dysidaminone H (160) | IC50 = 0.62–9.65 μM | ||
| IC50 = 0.23 μM | |||
| Dysidaminone J (162) | IC50 = 0.45–2.89 μM | ||
| IC50 = 0.11 μM | |||
| 18-Aminoavarone (167) | IC50 = 0.68–8.15 μM | ||
| IC50 = 0.05 μM | |||
| 5-Fluorouracila | IC50 = 0.39–7.64 μM | ||
| Rocaglamidea | IC50 = 0.12 μM | ||
| Cinerol F (175) | IC50 = 2.78 ± 0.06 μM | SHP-1 | 94 |
| (−)-20-Methoxyneoavarone (192) | IC50 = 0.9–4.6 μM | A549, HeLa, HCT-116, Jurkat, K562, BEL-7402 | 110 |
| Adriamycina | IC50 < 1 μM | ||
| Nakijiquinone L (202) | IC50 = 4.8–8.9 μM | A549, MCF-7, HeLa/WI-38 | 115 |
| IC50 = 6.9 μM | |||
| Langconol C (210) | IC50 = 5.0–7.8 μM | ||
| IC50 = 8.7 μM | |||
| Langcoquinone A (211) | IC50 = 7.9–9.9 μM | ||
| IC50 = 8.4 μM | |||
| Langcoquinone B (212) | IC50 = 6.2–8.7 μM | ||
| IC50 = 8.8 μM | |||
| Langcoquinone C (213) | IC50 = 6.6–9.6 μM | ||
| IC50 = 8.2 μM | |||
| 5-FUa | IC50 = 4.7–5.6 μM | ||
| IC50 = 6.8 μM | |||
| Langcoquinone D (214) | IC50 = 5.9–8.9 μM | A549, MCF-7, HeLa/WI-38 | 116 |
| IC50 = 5.6 μM | |||
| 5-Fua | IC50 = 5.5–6.5 μM | ||
| IC50 = 5.6 μM | |||
| (+)-5-Epi-20-O-ethylsmenoquinone (232) | IC50 = 3.24 μM | SW480 | 19 |
| IC50 = 2.95 μM | HCT116 | ||
| Dysiquinol D (239) | IC50 = 2.81 ± 0.01 μM | NCI-H929 | 87 |
| Rocaglamidea | IC50 = 3.94 ± 0.02 μM | ||
| Aureol B (240) | IC50 = 4.8–5.9 μM | K562, A549 | 109 |
| DOXa | IC50 = 0.8–1.6 μM | ||
| 20-Demethoxy-20-methylaminodactyloquinone D (242) | K d = 4.8 μM | CDK-2 | 79 |
| Dysiherbol A (248) | IC50 = 0.58 ± 0.02 μM | NCI-H929 | 92 |
| Avarola | IC50 = 3.32 ± 0.1 μM | ||
| Rocaglamidea | IC50 = 3.9 ± 0.1 μM | ||
| Spiroetherone A (262) | IC50 = 2.4 ± 0.3 μM | Zebrafish model | 34 |
| IC50 = 7.4–12.2 μM | NCI-H929, A549, HepG2, SK-OV-3 | ||
| Spiroetherone B (263) | IC50 = 8.7 ± 0.6 μM | Zebrafish model | 34 |
| (−)-Cochlearoid N (286) | IC50 = 7.68 μM | K562 | 126 |
| (+)-Cochlearoid P (288) | IC50 = 6.63 μM | ||
| Taxola | IC50 = 0.0023 μM | ||
| Metachromin V (451) | GI50 = 3.2–10.0 μM | SF-268, MCF-7, H460, HT-29/CHO-K1 (normal cells) | 162 |
| GI50 = 2.1 μM | |||
| Paclitaxela | GI50 = 0.01–0.02 μM | ||
| GI50 = 5.9 μM | |||
| Staurosporinea | GI50 = 0.04–11.0 μM | ||
| GI50 = 0.13 μM | |||
| Bistachybotrysin A (514) | IC50 = 2.8–7.5 μM | HCT116, NCI-H460, Daoy | 177 |
| Bistachybotrysin B (515) | IC50 = 4.2–6.6 μM | NCI-H460, BGC823, Daoy | |
| Bistachybotrysin D (517) | IC50 = 6.8–7.5 μM | HCT116, HepG2 | 178 |
| Bistachybotrysin E (518) | IC50 = 6.7–8.9 μM | HCT116, BGC823 | |
| Bistachybotrysin M (525) | IC50 = 1.8–3.5 μM | HCT116, NCI-H460, BGC823, Daoy, HepG2 | 175 |
| Taxola | IC50 = 0.0002–0.01 μM | HCT116, NCI-H460, BGC823, Daoy, HepG2 | 175, 177 and 178 |
| Paclitaxela | IC50 = 0.002–0.038 μM | ||
| Stachybochartin C (537) | IC50 = 11.6 ± 1.6 μM | MDA-MB-231/U2-OS | 67 |
| IC50 = 14.5 ± 3.1 μM | |||
| Stachybochartin D (538) | IC50 = 10.4 ± 0.9 μM | ||
| IC50 = 9.2 ± 0.1 μM | |||
| Cisplatina | IC50 = 11.3 ± 0.6 μM | ||
| IC50 = 5.9 ± 1.3 μM | |||
| Doxorubicina | IC50 = 1.0 ± 0.1 μM | ||
| IC50 = 1.2 ± 0.9 μM | |||
3.2. Anti-inflammatory activity
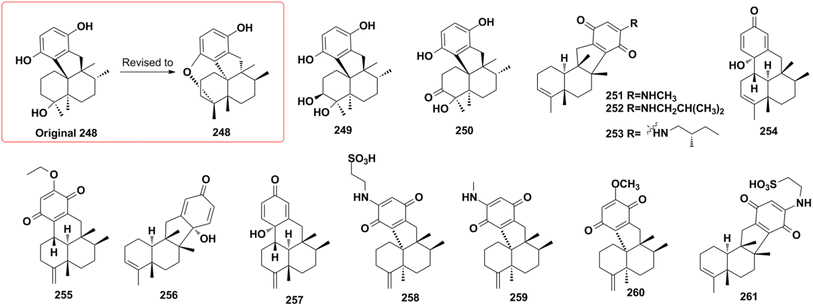
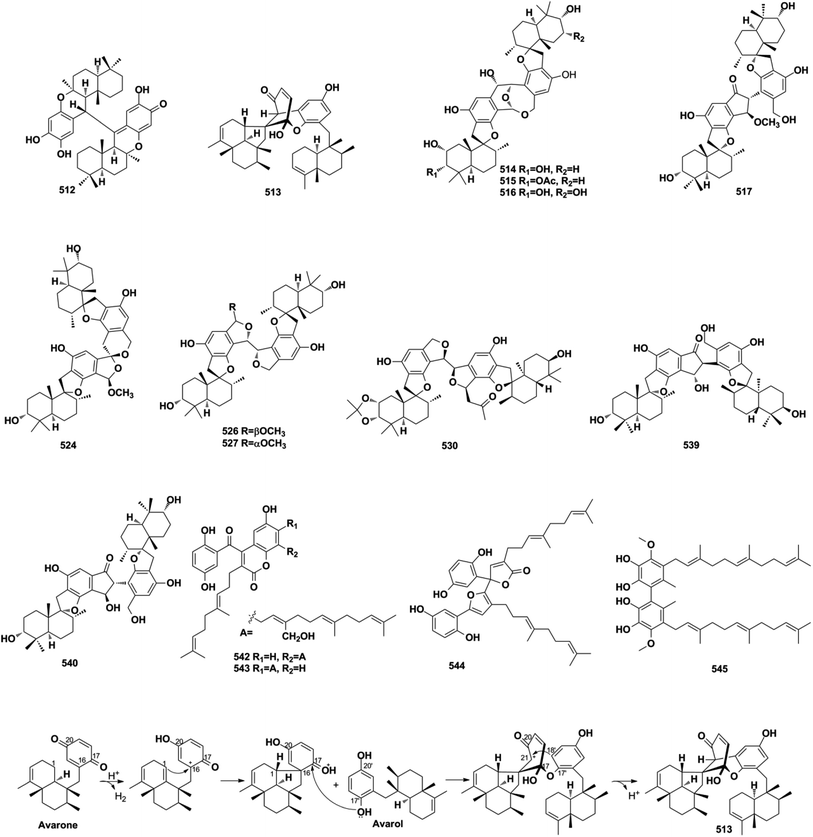
About half of the anti-inflammatory SQs possess avarane-type structure. 19-O-methylpelorol (113), dactylospongins A–B, D (130–131, 133), ent-melemeleone B (134), and dysidaminone N (137) inhibit the production of inflammatory cytokines IL-6, IL-8, IL-1β, and PEG2 in LPS-induced THP-1 cells with IC50 of 5.1–9.2 μM.84 Dysiherbol A (248) inhibits NF-κB production in TNF-α-induced RAW264.7 cells with an IC50 value of 0.49 μM, which is 10- and 20-fold more potent than that of dysiherbols B and C (249–250), respectively, indicating that the increased degree of oxidation at C-3 dramatically decreases the anti-inflammatory activity.92 Dysifragilone A (251) has comparable anti-inflammatory activity with that of hydrocortisone succinate, which inhibits the enzymatic activity and protein expression of iNOS and COX-2, the release of downstream NO, PGE2, and IL-6 by selectively blocking the p38/MAPK pathway in LPS-stimulated RAW264.7 cells.90,99 At concentrations of 2.5–10 μM, septosone A (264) exhibits anti-inflammatory activity comparable to that of indomethacin in alleviating migration and decreasing the number of macrophages surrounding zebrafish neuromasts.36 The gancochlearol E (295), (+)-gancochlearol I (296), (−)-gancochlearol I (296), gancochlearol G (455), and gancochlearol H (456) with γ-valerolactone unit inhibit the COX-2 enzyme with IC50 of 1.03–2.02 μM (IC50 of pos.celecoxib = 0.03 μM), and the inhibitory effect of both enantiomers of 296 shows little difference.127 The inhibitory effects of (+)-anthoponoid E (405), (−)-anthoponoid G (407), and anthoponoid H (408) on IL-1β and NF-κB support the traditional use of Rhododendron anthopogonoides for rheumatoid arthritis.153 The simultaneous inhibitory effect of COX-2/5-LOX is important for lipoxin synthesis to resolve inflammation. The 3-(hept-36-enyloxy)-decahydro-4,6a,12a,12b-tetramethyl-1H-benzo[a]xanthene-4,10,12-triol (502) significantly inhibits COX-2, 5-LOX, and COX-1 with IC50 values in the range of 1.33–1.57 mM, and the calculated binding energy of −12.30 kcal mol−1 with 5-LOX further substantiates its anti-inflammatory potential.52 The anti-inflammatory effect of the SQ dimer dysiarenone (513) increases 10 times when compared with that of avarol, indicating the importance of the unique 2-oxaspiro[bicyclo[3.3.1]-nonane-9,10-cyclopentane] scaffold. Further mechanism study indicated that 513 dose-dependently inhibited COX-2 enzyme expression and the corresponding PGE2 production in LPS-induced RAW264.7 cells without affecting COX-1 expression at 0–8 μM.373.3. Anti-microbial activity
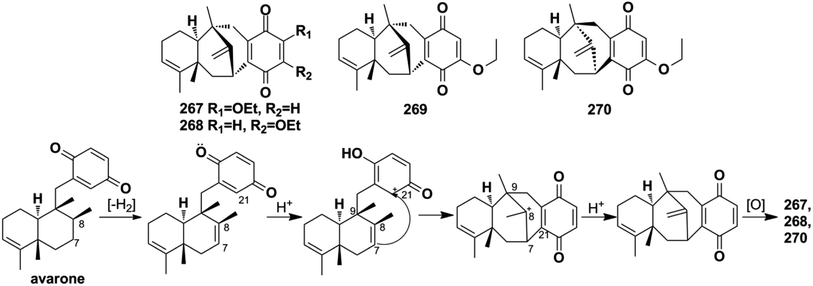
Three-quarters of anti-microbial SQs are sensitive to the Staphylococcus and Bacillus strains. Aureol B (240, MIC = 1–8 μg mL−1), with the key quinol functional group being considerably more active than melemeleones C–D (135–136), and cycloaurenones A–C (258–260) bearing the quinone moiety for inhibiting the growth of bacterial S. aureus, B. subtilis, S. enterica, and P. hauseri, and the fungus T. rubrum, may be attributed to the high amount of reactive oxygen radicals produced by the quinol unit.109,186 The SQ dimers stachyin B (546) and chartarlactams Q, S (547, 549) inhibit S. aureus, methicillin-resistant S. aureus, B. subtilis, and S. epidermidis with IC50 of 1.02–1.75 μM and 4–8 μg mL−1, which are comparable to the positive control chloramphenicol (IC50 = 1.45–2.46 μM, 1.0 μg mL−1); this may be due to the superior ability of the dimers in binding double sites in the target.59,181 Both (+)-cochlearoid P (288) and (−)-cochlearoid (288) inhibit the growth of S. aureus with IC50 < 10 μM, while the activity of (+)-cochlearoid O (287) (IC50 = 17.99 μM) decreased when compared with its enantiomer (−)-cochlearoid O (287) (IC50 = 6.28 μM), indicating the significant influence of the farnesyl position (IC50 of pos.ciprofloxacin = 0.21 μM).126 The typical avarane-type SQs of smenospongimine (199), nakijiquinone L (202), and langcoquinones A–D (211–214) are sensitive to the strains of both B. subtilis and S. aureus with MIC values of 3.13 μg mL−1 or 6.25–25.0 μM (MIC of pos.chloromycetin/ampicillin = 3.13 μg mL−1, 6.25–25.0 μM).55,113–1163.4. Anti-diabetic activity
PTP1B is considered a potential target in type II diabetes because of its negative regulatory effect on insulin receptors. Seventeen SQs, including verruculide A (100), frondoplysins A and B (151–152), cinerols A–C, and F (170–172, and 175), 17-O-acetylavarol (193), 17-O-acetylneoavarol (194), nakijinol G (223), avapyran (241), dysidavarone A (267), and ganoduriporols C, D, F, G, and H (324, 325, and 327–329), exhibit potent anti-diabetic activity by inhibiting PTP1B with IC50 values in the range of 0.39–20.0 μM.33,76,94,96,97,111,137,141 Among these compounds, frondoplysin A–B (151–152) display more potent activity than the positive control (IC50/oleanolicacid = 3.7 ± 0.03 μM) in inhibiting PTP1B with IC50 of 0.39 ± 0.04 μM and 0.65 ± 0.03 μM, and 151 is proved to be a mixed PTP1B inhibitor by detailed enzymatic kinetic study.97 Molecular docking showed that ganoduriporol F (327) forms H-bonds with ALA-217, GLN-266, and ARG-24, which are located in the active site of PTP1B.137 In addition, craterellin A (12) significantly inhibits the 11β-HSD2 enzyme that regulates the glucose-raising hormone with an IC50 value of 1.5 μg mL−1, and myrothecisin C (277) inhibits α-glucosidase with an IC50 value of 5.8 μM.44,703.5. Anti-oxidant activity
A transgenic fluorescent zebrafish experiment has shown that frondoplysin A (151) exhibits anti-oxidative activity over five times stronger than that of vitamin C at nontoxic concentrations in the range of 10–40 μM.97 Dysidaminone H (160) protects against oxidative injury in HaCaT cells via the activation of Nrf2/ARE/HO-1, which is mediated by the phosphorylation of AMPK/ERK at concentrations of 2.5–10 μM.89 Puupehenol (107) has been shown to be a potent anti-oxidant with an FRAP value of 2500 at 1.0 mg mL−1 (pos.vitaminC 870 at 1.0 mg mL−1).80 The anti-oxidant activities of 13-[[2-(hexyloxy)-2,5,5,8a-tetramethyldecahydro-1-naphthalenyl](methoxy)methyl] benzenol (29), akadisulfate B (227), ganomycin C (317), cochlearins B–E, and I (318–322), ganocochlearins A, C, and D (468–470), cochlearin A (471), 3-(hept-36-enyloxy)-decahydro-4,6a,12a,12b-tetramethyl-1H-benzo[a]xanthene-4,10,12-triol (502), 1-butoxy-4,4,11b,11c-tetramethyl-decahydrobenzo[kl]xanthen-10-ol (503), and diplopuupehenone (512) have been tested using DPPH+, ABTS+, and OH radical scavenging assay, and they show comparable anti-oxidant activities with the positive controls trolox, vitamin C, α-tocopherol, and butylated hydroxytoluene.52,56,138,1393.6. Anti-viral activity
Stachybotrin D (51) has been found to be a novel non-nucleoside reverse transcriptase inhibitor of both wild-type HIV-1 and five NNRTI-resistant HIV-1 strains with EC50 values in the range of 6.2–23.8 μM (EC50 of pos.nevirapine = 0.23–51.9 μM).64 Stachybotrysins A, B, and G (66, 67, and 72) and stachybotrysams A–C (345–347) show inhibitory effects on HIV-1 virus with IC50 values in the range of 1.0–19.6 μM (IC50 of pos.efavirenz = 2.0–4.0 nM).60,61 Although chartarutines B, G, and H (338, 343, and 344; IC50 = 5.57–5.58 μM) exhibit weaker anti-HIV activity than efavirenz (IC50 = 0.65 μM, CC50 = 40), their lower cytotoxicity (CC50 > 100) suggests that they are worthy of further investigation as anti-HIV candidates.25 Chartarlactam T (550) blocks ZIKU virus replication by targeting the NS5 and E proteins at a concentration of 10 μM.181 Stachybotrysins A and F (66 and 71), and stachybotrysin (77) effectively inhibit influenza A virus with IC50 values in the range of 12.4–18.7 μM (IC50 of pos.ribavirion = 2.0–4.0 nM).60,69 Talaromyolide K (448) dose-dependently inhibits pseudorabies virus at concentration in the range of 3.13–50 μg mL−1 and prevents pseudorabies virus-causing cell injury with low toxicity to the host cells.1603.7. Fibrinolytic activity
SMTPs metabolites activate the fibrinolytic system by inducing conformational changes in plasminogen, in which the ionizable group in the N-linked side chain is essential for their fibrinolytic activity.146 As the most representative congener, SMTP-7 has been proven to be a promising thrombolytic candidate for the treatment of ischemic stroke and cerebral infarction in rodents and primates.187,188 The SMTP dimer FGFC1 (553) acts as a promising thrombolytic agent by dose-dependently hydrolyzing fibrin at concentrations in the range of 5–25 μmol L−1, effectively dissolving pulmonary thrombus in the rat lung without risk of hemorrhagic activity at 5–25 mg kg−1.182,189 FGFC6 and FGFC7 (353 and 354) show competitive fibrinolytic activities with relative activities of 6.90 and 3.86 at 0.025 g L−1 (pos. 6.46), whereas FGFC4 and FGFC5 were inactive, which may be due to the loss of the carboxyl acid group in the N-linked chain.26 SMTPs-19, 22, 25, and 43 (366, 369, 372, and 380) bearing phenylamine-based side chains are as potent as SMTP-7 (Emax/EC10 = 1.57-fold μM−1) in enhancing urokinase-catalyzed plasminogen activation (Emax/EC10 = 1.16–1.57-fold μM−1).145,1473.8. Reno-protective activity
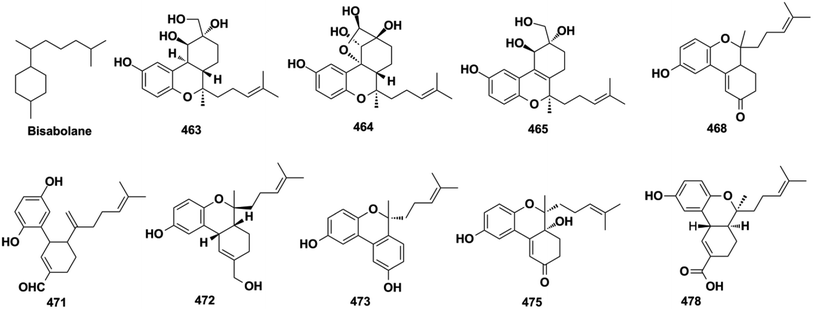
The excessive activation of TGF-β reduces the degradation of extracellular matrix components, which are the hallmarks of renal fibrosis. The overexpression of fibronectin is a common feature of renal disease. Cochlearoids H and I (283–284), (+)-cochlearol S (472), (−)-cochlearol U (474), (+)-cochlearol U (474), (−)-cochlearol Y (476), and cochlearoids F and G (542 and 543) exhibit significant reno-protective activity by inhibiting the viability of TGF-β1-induced NRK-49F cells and fibronectin production in TGF-β1-induced HKC-8 cells at nontoxic concentrations in the range of 2.5–20 μM.130,131 Cochlearol B (497) inhibits collagen I, fibronectin, and αSMA in TGF-β1-induced NRK-52E cells at concentrations in the range of 5–20 μM, and this action is dependent on the disruption of Smad2 and Smad3 activation.169 (−)-Gancochlearols J and K (463 and 464) and ganodermaone B (309) dose-dependently inhibit the viability of TGF-β1-induced NRK-52e cells by attenuating extracellular matrix collagen I and fibronectin at nontoxic concentrations in the range of 10–40 μM.133,136 MCP-1 is considered to be a new diagnostic marker and therapeutic target for progressive renal injury, chizhine F (310) dose-dependently reduces fibronectin and MCP-1 expression in high-glucose-stimulated rat mesangial cells at concentrations in the range of 2.5–10 μM.134 These reno-protective components from Ganoderma fungi provide evidence for their use in kidney disorders.3.9. Other activities
19-Methoxy-9,15-ene-puupehenol (109) exhibits anti-atherosclerosis activity by effectively upregulating SR-B1 (130%) in HepG2 cells at a concentration of 1.78 μM (pos.trichostatinA = 5.25 μM, 100%), and it satisfies both Veber's and Lipinski's rule for drug-like molecules with oral bioavailability.78 Acetylcholinesterase (AChE) inhibitors are the drugs of choice for the treatment of Alzheimer's disease. Dayaolingzhiols D and E (307–308) inhibit AChE with IC50 values of 8.52 ± 1.90 and 7.37 ± 0.52 μM (pos.tacrine = 7.37 ± 0.52 μM), respectively.132 Scutigeric acid (388) and bis-2-hydroxy-1-methoxy neogrifolin (545) exhibit anti-Alzheimer's disease activity by inhibiting Aβ aggregation with IC50 values of 6.6 and 12.3 μM (pos.myricetin = 7.4 and 9.9 μM), respectively, and compound 388 also inhibits β-secretase with an IC50 of 1.6 μM (pos.myricetin = 2.8 μM).149,150 Cochlearoid A (280) has potent activity toward Cav3.1 TTCC through channel gating modulation (IC50 = 11.4 μM, Hill coefficient = 1.6; IC50 of pos.mibefradil = 10.4 μM, Hill coefficient = 1.6), indicating its possible application in neurological disorders.125 Arthripenoid C (416) possesses immune-suppressive activity by inhibiting ConA-induced T cells and the corresponding production of TNF-α and IFN-γ with an IC50 value of 4.2–8.8 μM.20 Dysivillosins A–D (187–190) exhibits anti-allergic activity by inhibiting the release of β-hexosaminidase and downregulating the production of LTB4 and IL-4 in antigen-stimulated RBL-2H3 mast cells at concentrations in the range of 6.0–19.9 μM, and the action of compound 187 was triggered by suppressing the Syk/PLCγ1 pathway.95 Chartarlactams D–F, K, L, N, and O (39–41, 46, 541, 48, and 49) exhibit potent lipid-lowering effects at 10 μM, as assessed by Oil Red O staining, and compounds 40 and 41 inhibit both intracellular triglyceride and total cholesterol levels in HepG2 cells.63 Stachybotrysin (77) attenuated RANKL-induced osteoclast differentiation by suppressing c-Fos, ERK, JNK, and p38 while activating NFATc1 at 5.0 μg mL−1.69 Gancochlearol C (544) inhibits the tuberculosis-related enzyme NAT2 with an IC50 value of 5.29 ± 0.1 μM.128 Chrodrimanins D–F (89–91) display insecticidal activity against silkworms with LD50 values of 20, 10, and 50 μg g−1, respectively.734. Total synthesis and biosynthesis
The stereo-controlled synthesis of SQs with attractive structures and exciting bioactivities usually takes many years. Therefore only a few compounds discovered in the recent ten years have been successfully synthesized. For example, avarol and avarone were first discovered in 1974, then they were synthesized by the groups of EA Theodorakis, SM Hecht, AS Sarma, and T Katoh during the years 1982–2008.190–194 EA theodorakis developed an enantioselective synthesis of avarol and avarone by constructing the entire framework through C11–C1′ bond and using Barton's radical decarboxylation method. In addition, the biosynthetic strategy can also produce scarce natural products, which acts more efficiently by providing a concise cascade reaction. In the process of exploring new biosynthetic methods, new pathways, gene clusters, and the encoded enzyme functions are discovered.20,27,195,1964.1. Total synthesis
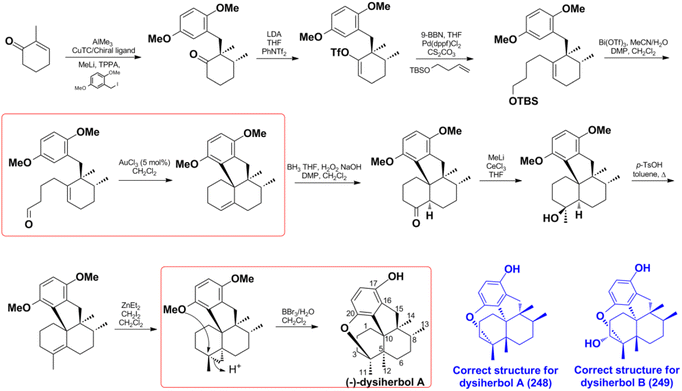
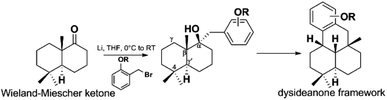
Z. Y. Lu's group disclosed a different synthetic strategy for dysiherbol A (248); the compound was obtained in the racemic form, and the revised structure of 248 is the same as that of Schmalz's group (Scheme 6).121,122 Two of the most important steps are to connect the terpene and quinol moieties stereoselectively using Wieland–Miescher ketone and bulky benzyl bromide, and to construct the 6/6/5/6-fused tetracyclic skeleton using intramolecular Heck reaction. The 6/6/5/6-tetracyclic intermediate-1 can be obtained in the presence of Pd2(dba)3, SPhos, and Et3N, irrespective of the presence of axial-OBn or ketone group at the C8 position. However, the glycol acetal on C4 must be removed because the methoxyl on C5′ and the acetal would be very close when the cyclization reaction occurs. Dysiherbol (248) comes from intermediate-2 through three different pathways including (1) directly from intermediate-2 using BBr3 at 23 °C, (2) olefin hydration mediated by Mn(dpm)3, PhSiH3, O2, and PPh3, intramolecular etherification, and then demethylation, (3) etherification and then demethylation. Z. Y. Lu and C. K. Chong highlighted the above synthetic method.198 Echavarren et al. constructed the 6/6/5/6 carbocyclic core for dysiherbols A–C (248–250) via gold(I)-promoted (3 + 2) cycloaddition between terminal allenes and dimethoxyphenyl carbenes, and then AIBN-triggered radical cyclization.199,200
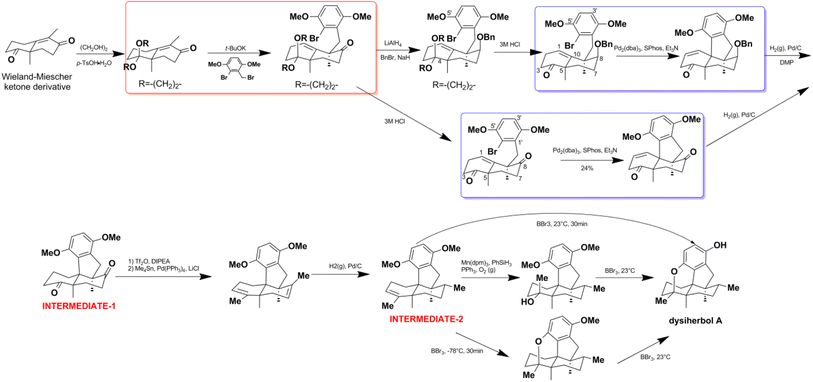
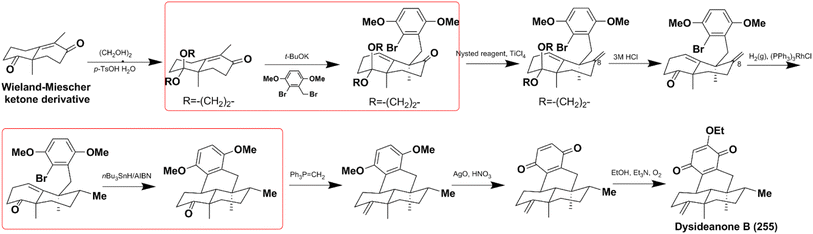
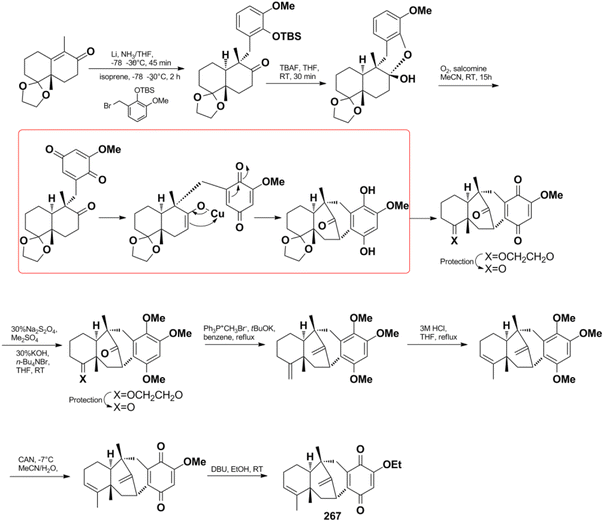
Jana et al. successfully constructed dysideanone's fused 6/6/6/6 framework by applying the intramolecular γ-arylation of cycloalkanols starting from Wieland–Miescher ketone derivative.201–203 Computational investigation proved that the regioselective γ-arylation of cycloalkanols are more preferred by 3.4 kcal mol−1 when there are two methyl substituents on C-4 (Scheme 8).204
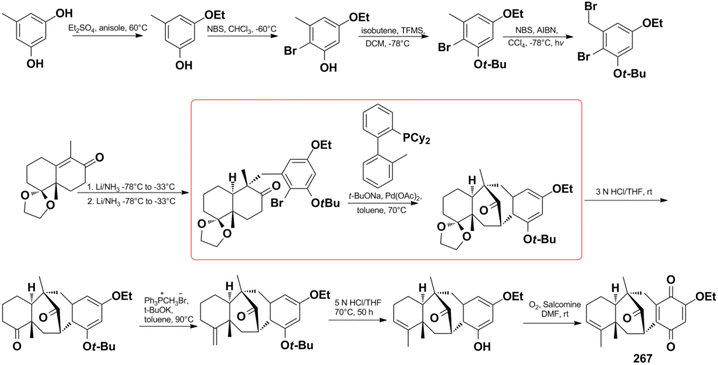
Menche et al. also successfully synthesized dysidavarone A (267) in 10 steps with a total yield of 11% (Scheme 10). They built the core tetracyclic structure via the stereoselective reaction of a Wieland–Miescher-type ketone and benzyl bromide, followed by the pivotal Pd(OAc)2 ligand-catalyzed intramolecular ketone α-arylation reaction.206 The synthetic method confirmed the unique 3D structure of dysidavarone A and provided another concise route for constructing complex fused ring systems of this type of congeners.
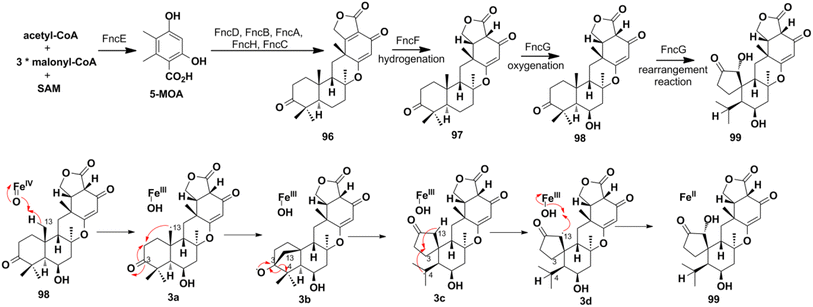
4.2. Biosynthesis
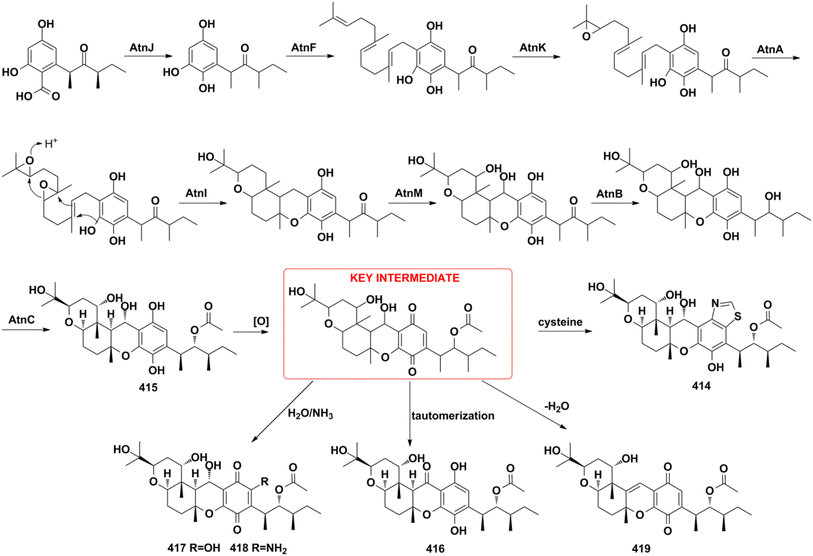
5. Conclusions and perspective
Natural SQs are hybrids of sesquiterpenoid and quinone/quinol, and they produce numerous drug candidates that are yet to be exploited. They have unique advantages in developing drug candidates for the above reasons. First is that the intrinsic structure feature makes SQs easily meet the requirement of drug-like rules. The anti-atherogenic 19-methoxy-9,15-ene-puupehenol (109) was reported to satisfy two more requirements for drug-like molecules with oral bioavailability according to both Veber's rule and Lipinski's rule.78 The second is the significant bioactivity of SQs. Nearly half of all natural SQs have been proven to be active in vitro or in vivo. The third point is safety. SQs exhibit significant anti-cancer, anti-inflammatory, and anti-viral activities without affecting normal cells or tissues. The novel sPLA2 inhibitor bolinaquinone acts as a potent anti-inflammatory agent on acute and chronic rat models without affecting the protective levels in other non-infected tissues and exhibits important protection against body and spleen weight loss, unlike the clinical drug dexamethasone.207 The last feature is the multitarget features of SQs. Easily formed and highly reactive semiquinone radicals help SQs to participate in multiple pathological processes. SMTP-7 is effective in treating thrombotic stroke in rodents and primates by modulating plasminogen conformation, which is a neuroprotective mechanism neutralizing cytokine-mediated pro-inflammation factors, reducing ROS formation and enhancing clot clearance.145,187,188,208–212 These multitarget, multilink regulatory characteristics make SMTP-7 a possible candidate drug for thrombolytic therapy.The only two reviews present a total of 285 marine environment SQ metabolites mostly containing the bicyclic sesquiterpenoid unit before the year 2010.17,18 Including the 558 SQs described in this review, a total of 843 natural SQs have been discovered up to the year 2021. The increasing interest on SQs makes them increasingly important in the research area of natural products chemistry. The novel discovery strategies including multi-omics, gene cluster reconstruction, genic mutation, heterologous expression, and characteristic stereochemical, NMR, and MS features of each type of SQs provided will help accelerate SQs discovery and elucidate their structures more effectively. In the recent ten years, 60.2% new SQs were isolated from marine-derived fungi and marine organisms, implying the predominance of marine resources in new drug development. The SQs mainly distribute in relatively limited species, such as the fungi of Stachybotrys sp. and Ganoderma sp., and the sponge of Dysidea sp., displaying a significant chemotaxonomic role. The natural SQs are first systematically classified into seven types and sixteen subtypes in this review. Among them, the drimane-type, avarane-type, farnesane-type, and monocyclofarnesane-type occupy the largest proportion of the total SQs, accounting for 87%. All of the avarane-type SQs are produced by marine sponges, whereas most phenylspirodrimanedrimane-type and farnesane-type SQ analogs are isolated from the fungi of Stachybotrys and Ganoderma, respectively. There are many unprecedented skeletons in avarane-type SQs, and these unique metabolites indicate the amazing variety of metabolic pathways that exist in marine sponges. The phenylspirodrimanedrimane-type SQ monomers tend to dimerize in Stachybotrys fungi under the catalysis of specific enzymes, and these metabolic enzymes and the related functional gene clusters are worthy of further study. Many SQs possess intriguing biological and pharmacological activities, as well as fascinating structural scaffolds that are crucial for new drug development. Spiroetherone A (262) with unique spiro[4,5]decane skeleton exhibits combined anti-cancer effect by inhibiting angiogenesis and cancer cell proliferation.34 Dysifragilone A (251) and septosone A (264) that featur a characteristic 6/6/6/6-fused tetracyclic ring system and pentacyclo[6.6.2.13,1401,1002,7]heptadecane ring system have comparable anti-inflammatory activity with that of hydrocortisone succinate and indomethacin, respectively.36,90 SMTP-7 and dimeric FGFC1 (553) have been proved to be potent anti-stroke leads by dissolving thrombus in vivo.182,187–189 Furthermore, the insecticidal chrodrimanins D–F (89–91), antimalarial phomoarcherin B (104), and anti-microbial SQs have significant ecological function in protecting the host from alien attack.73,77 However, further biological studies on these compounds are severely restricted by their scarcity. The proposed biosynthetic pathways, chemical synthetic, and enzyme catalyzed biosynthetic routes outlined in this review will help solve the problem of sample scarcity. In addition, biotransformation and microbial co-cultures have provided a new approach to enrich trace constituents. More samples are needed to reveal novel bioactivity and help enhance the understanding of the pharmacokinetic, pharmacodynamics, and safety characteristics of SQs. In most studies, other important bioactivities of the SQs could not be evaluated comprehensively due to the limitation of activity testing platform and the prevalence of anti-cancer activity evaluation systems. It is also necessary to screen SQs with novel targets and strengthen the interdisciplinary cooperation between natural product chemistry and synthesis, analysis, pharmacology, and genomics.
6. Author contributions
Xin-Hui Tian: writing review and editing. Li-Li Hong & Wei-Hua Jiao: data curation. Hou-Wen Lin: project administration and supervision.7. Conflicts of interest
There are no conflicts to declare.8. Acknowledgements
This study was funded by National Natural Science Foundation of China (No. 82204233, 22137006, 82022068, 41906075). National Key Research and Development Program of China (No. 2022YFC2804100). Innovative Research Team of High-level Local Universities in Shanghai (No. SHSMU-ZDCX20212702).9. References
- R. Geris and T. J. Simpson, Nat. Prod. Rep., 2009, 26, 1063–1094 RSC.
- M. H. Jiang, Z. E. Wu, L. Liu and S. H. Chen, Org. Biomol. Chem., 2021, 19, 1644–1704 RSC.
- L. Minale, R. Riccio and G. Sodano, Tetrahedron Lett., 1974, 38, 3401–3404 CrossRef.
- W. E. G. Muller, C. Sobel, B. Diehl-Seifert, A. Maidhof and H. C. Schroder, Biochem. Pharmacol., 1987, 36, 1489–1494 CrossRef CAS PubMed.
- P. S. Sarin, D. Sun, A. Thornton and W. E. Muller, J. Natl. Cancer Inst., 1987, 78, 663–666 CAS.
- W. E. G. Muller, D. Sladic, R. K. Zahn, K. H. Bassler, N. Dogovic, H. Gerner, M. J. Gasic and H. C. Schroder, Cancer Res., 1987, 47, 6565–6571 CAS.
- H. C. Schroder, R. Wenger, H. Gerner, P. Reuter, Y. Kuchino, D. Sladic and W. E. G. Muller, Cancer Res., 1989, 49, 2069–2076 CAS.
- C. Imperatore, R. Gimmelli, M. Persico, M. Casertano, A. Guidi, F. Saccoccia, G. Ruberti, P. Luciano, A. Aiello, S. Parapini, S. Avunduk, N. Basilico, C. Fattorusso and M. Menna, Mar. Drugs, 2020, 18, 112 CrossRef CAS PubMed.
- D. Sipkema, R. Osinga, W. Schatton, D. Mendola, J. Tramper and R. H. Wijffels, Biotechnol. Bioeng., 2005, 90, 201–225 CrossRef CAS PubMed.
- W. Schatton, M. Schatton and R. Pietschmann, EU Pat., EP1391197B1, 2006 Search PubMed.
- R. T. Luibrand, T. R. Erdman, J. J. Vollmer and P. J. Scheuer, Tetrahedron, 1979, 35, 609–612 CrossRef CAS.
- R. J. Capon and J. K. Macleod, J. Org. Chem., 1987, 52, 5059–5060 CrossRef CAS.
- P. A. Takizawa, J. K. Yucel, B. Veit, D. J. Faulkner, T. Deerinck, G. Soto, M. Ellisman and V. Malhotra, Cell, 1993, 73, 1079–1090 CrossRef CAS PubMed.
- B. Veit, J. K. Yucel and V. Malhotra, J. Cell Biol., 1993, 122, 1197–1206 CrossRef CAS PubMed.
- S. Park, E. Yun, I. H. Hwang, S. Yoon, D. E. Kim, J. S. Kim, M. Na, G. Y. Song and S. Oh, Mar. Drugs, 2014, 12, 3231–3244 CrossRef CAS PubMed.
- L. Du, Y. D. Zhou and D. G. Nagle, J. Nat. Prod., 2013, 76, 1175–1181 CrossRef CAS PubMed.
- R. J. Capon, Stud. Nat. Prod. Chem., 1995, 289–326 CAS.
- I. S. Marcos, A. Conde, R. F. Moro, P. Basabe, D. Diez and J. G. Urones, Mini-Rev. Org. Chem., 2010, 7, 230–254 CrossRef CAS.
- I. H. Hwang, J. Oh, W. Zhou, S. Park, J. H. Kim, A. G. Chittiboyina, D. Ferreira, G. Y. Song, S. Oh, M. K. Na and M. T. Hamann, J. Nat. Prod., 2015, 78, 453–461 CrossRef CAS PubMed.
- X. Zhang, T. T. Wang, Q. L. Xu, Y. Xiong, L. Zhang, H. Han, K. Xu, W. J. Guo, Q. Xu, R. X. Tan and H. M. Ge, Angew. Chem., Int. Ed., 2018, 57, 8184–8188 CrossRef CAS PubMed.
- Y. Hitora, A. Sejiyama, K. Honda, Y. Ise, F. Losung, R. E. P. Mangindaan and S. Tsukamoto, Bioorg. Med. Chem., 2021, 31, 115968 CrossRef CAS PubMed.
- J. Y. Han, J. Y. Zhang, Z. J. Song, G. L. Zhu, M. M. Liu, H. Q. Dai, T. Hsiang, X. T. Liu, L. X. Zhang, R. J. Quinn and Y. J. Feng, Appl. Microbiol. Biotechnol., 2020, 104, 3835–3846 CrossRef CAS PubMed.
- F. Zhang, M. Zhao, D. R. Braun, S. S. Ericksen, J. S. Piotrowski, J. Nelson, J. Peng, G. E. Ananiev, S. Chanana, K. Barns, J. Fossen, H. Sanchez, M. G. Chevrette, I. A. Guzei, C. G. Zhao, L. Guo, W. P. Tang, C. R. Currie, S. R. Rajski, A. Audhya, D. R. Andes and T. S. Bugni, Science, 2020, 370, 974–978 CrossRef CAS PubMed.
- N. Bonneau, G. Chen, D. Lachkar, A. Boufridi, J. F. Gallard, P. Retailleau, S. Petek, C. Debitus, L. Evanno, M. A. Beniddir and E. Poupon, Chem.–Eur. J., 2017, 23, 14454–14461 CrossRef CAS PubMed.
- Y. Li, D. Liu, S. Cen, P. Proksch and W. H. Lin, Tetrahedron, 2014, 70, 7010–7015 CrossRef CAS.
- Y. Yin, Q. Fu, W. H. Wu, M. H. Cai, X. S. Zhou and Y. X. Zhang, Mar. Drugs, 2017, 15, 214 CrossRef PubMed.
- D. X. Yan and Y. D. Matsuda, Org. Lett., 2021, 23, 3211–3215 CrossRef CAS PubMed.
- H. Qiao, S. H. Zhang, Y. Dong, Y. Yang, R. Xu, B. Chen, Y. Wang, T. J. Zhu, C. B. Cui, G. G. Zhang and C. W. Li, Nat. Prod. Res., 2020, 1–8 Search PubMed.
- D. Arora, P. Gupta, S. Jaglan, C. Roullier, O. Grovel and S. Bertrand, Biotechnol. Adv., 2020, 40, 107521 CrossRef PubMed.
- M. Menna, C. Imperatore, F. D'Aniello and A. Aiello, Mar. Drugs, 2013, 11, 1602–1643 CrossRef CAS PubMed.
- P. A. García, Á. P. Hernández, A. S. Feliciano and M. Á. Castro, Mar. Drugs, 2018, 16, 292 CrossRef PubMed.
- M. Nazir, M. Saleem, M. I. Tousif, M. A. Anwar, F. Surup, I. Ali, D. Wang, N. Z. Mamadalieva, E. Alshammari, M. L. Ashour, A. M. Ashour, I. Ahmed, Elizbit, I. R. Green and H. Hussain, Biomol, 2021, 11, 957 CAS.
- W. H. Jiao, X. J. Huang, J. S. Yang, F. Yang, S. J. Piao, H. Gao, J. Li, W. C. Ye, X. S. Yao, W. S. Chen and H. W. Lin, Org. Lett., 2012, 14, 202–205 CrossRef CAS PubMed.
- W. H. Jiao, Q. H. Xu, J. Cui, R. Y. Shang, Y. Zhang, J. B. Sun, Q. Yang, K. C. Liu and H. W. Lin, Org. Chem. Front., 2020, 7, 368–373 RSC.
- W. H. Jiao, T. T. Xu, H. B. Yu, G. D. Chen, X. J. Huang, F. Yang, Y. S. Li, B. N. Han, X. Y. Liu and H. W. Lin, J. Nat. Prod., 2014, 77, 346–350 CrossRef CAS PubMed.
- Y. H. Gui, W. H. Jiao, M. Zhou, Y. Zhang, D. Q. Zeng, H. R. Zhu, K. C. Liu, F. Sun, H. F. Chen and H. W. Lin, Org. Lett., 2019, 21, 767–770 CrossRef CAS PubMed.
- W. H. Jiao, B. H. Cheng, G. D. Chen, G. H. Shi, J. Li, T. Y. Hu and H. W. Lin, Org. Lett., 2018, 20, 3092–3095 CrossRef CAS PubMed.
- K. Kawashima, K. Nakanishi and H. Nishikawa, Chem. Pharm. Bull., 1964, 12, 796–803 CrossRef CAS PubMed.
- S. M. Fang, C. B. Cui, C. W. Li, C. J. Wu, Z. J. Zhang, L. Li, X. J. Huang and W. C. Ye, Mar. Drugs, 2012, 10, 1266–1287 CrossRef CAS PubMed.
- X. P. Lin, Q. Y. Wu, Y. Y. Yu, Z. Liang, Y. H. Liu, L. L. Zhou, L. Tang and X. F. Zhou, Sci. Rep., 2017, 7, 10757 CrossRef PubMed.
- X. P. Lin, X. F. Zhou, F. Z. Wang, K. S. Liu, B. Yang, X. W. Yang, Y. Peng, J. Liu, Z. Ren and Y. H. Liu, Mar. Drugs, 2012, 10, 106–115 CrossRef CAS PubMed.
- Y. Fu, P. Wu, J. H. Xue and X. Y. Wei, J. Nat. Prod., 2014, 77, 1791–1799 CrossRef CAS PubMed.
- Y. Fu, P. Wu, J. H. Xue, H. X. Li and X. Y. Wei, Mar. Drugs, 2015, 13, 3360–3367 CrossRef CAS PubMed.
- L. Zhang, Y. Shen, F. Wang, Y. Leng and J. K. Liu, Phytochemistry, 2010, 71, 100–103 CrossRef CAS PubMed.
- C. J. Zheng, C. L. Shao, M. Chen, Z. G. Niu, D. L. Zhao and C. Y. Wang, Chem. Biodiversity, 2015, 12, 1407–1414 CrossRef CAS PubMed.
- C. Li, D. H. Li, S. X. Cai, F. P. Wang, X. Xiao and Q. G. Gu, Acta Pharmacol. Sin., 2010, 45, 1275–1278 Search PubMed.
- L. E. Schmidt, S. T. Deyrup, J. Baltrusaitis, D. C. Swenson, D. T. Wicklow and J. B. Gloer, J. Nat. Prod., 2010, 73, 404–408 CrossRef CAS PubMed.
- A. Hirose, H. Maeda, A. Tonouchi, T. Nehira and M. Hashimoto, Tetrahedron, 2014, 70, 1458–1463 CrossRef CAS.
- I. E. Mohamed, S. Kehraus, A. Krick, G. M. Konig, G. Kelter, A. Maier, H. H. Fiebig, M. Kalesse, N. P. Malek and H. Gross, J. Nat. Prod., 2010, 73, 2053–2056 CrossRef CAS PubMed.
- C. J. Chen, Y. Q. Zhou, X. X. Liu, W. J. Zhang, S. S. Hu, L. P. Lin, G. M. Huo, R. H. Jiao, R. X. Tan and H. M. Ge, Tetrahedron Lett., 2015, 56, 6183–6189 CrossRef CAS.
- A. L. Lane, L. Mular, E. J. Drenkard, T. L. Shearer, S. Engel, S. Fredericq, C. R. Fairchild, J. Prudhomme, K. L. Roch, M. E. Hay, W. Aalbersberg and J. Kubanek, Tetrahedron, 2010, 66, 455–461 CrossRef CAS PubMed.
- K. Chakraborty, T. Antony and M. Joy, Algal Res., 2019, 40, 101472 CrossRef.
- F. Ishibashi, S. Sato, K. Sakai, S. Hirao and K. Kuwano, Biosci., Biotechnol., Biochem., 2013, 77, 1120–1122 CrossRef CAS PubMed.
- M. Kumagai, K. Nishikawa, H. Matsuura, T. Umezawa, F. Matsuda and T. Okino, Molecules, 2018, 23, 1214 CrossRef PubMed.
- X. Zhang, H. Y. Xu, A. M. Huang, L. Wang, Q. Wang, P. Y. Cao and P. M. Yang, Chem. Pharm. Bull., 2016, 64, 1036–1042 CrossRef CAS PubMed.
- L. K. Shubina, A. I. Kalinovsky, T. N. Makarieva, S. N. Fedorov, S. A. Dyshlovoy, P. S. Dmitrenok, I. I. Kapustina, E. Mollo, N. K. Utkina, V. B. Krasokhin, V. A. Denisenko and V. A. Stonik, Nat. Prod. Commun., 2012, 7, 487–490 CrossRef CAS PubMed.
- H. Liu, X. M. Li, Y. Liu, P. Zhang, J. N. Wang and B. G. Wang, J. Nat. Prod., 2016, 79, 806–811 CrossRef CAS PubMed.
- A. R. Wang, Y. B. Xu, Y. X. Gao, Q. Huang, X. Luo, H. M. An and J. Y. Dong, Phytochem. Rev., 2015, 14, 623–655 CrossRef CAS.
- B. Wu, V. Oesker, J. Wiese, S. Malien, R. Schmaljohann and J. F. Imhoff, Mar. Drugs, 2014, 12, 1924–1938 CrossRef PubMed.
- J. L. Zhao, J. M. Feng, Z. Tan, J. M. Liu, J. Y. Zhao, R. D. Chen, K. B. Xie, D. W. Zhang, Y. Li, L. Y. Yu, X. G. Chen and J. G. Dai, J. Nat. Prod., 2017, 80, 1819–1826 CrossRef CAS PubMed.
- J. L. Zhao, J. M. Liu, Y. Shen, Z. Tan, M. Zhang, R. D. Chen, J. Y. Zhao, D. W. Zhang, L. Y. Yu and J. G. Dai, Phytochem. Lett., 2017, 20, 289–294 CrossRef CAS.
- D. Liu, Y. Li, X. D. Li, Z. B. Cheng, J. Huang, P. Proksch and W. H. Lin, Tetrahedron Lett., 2017, 58, 1826–1829 CrossRef CAS.
- Y. Li, C. M. Wu, D. Liu, P. Proksch, P. Guo and W. H. Lin, J. Nat. Prod., 2014, 77, 138–147 CrossRef CAS PubMed.
- X. H. Ma, L. T. Li, T. J. Zhu, M. Y. Ba, G. Q. Li, Q. Q. Gu, Y. Guo and D. H. Li, J. Nat. Prod., 2013, 76, 2298–2306 CrossRef CAS PubMed.
- P. P. Zhang, Y. F. Li, C. X. Jia, J. J. Lang, S. I. Niaz, J. Li, J. Yuan, J. C. Yu, S. H. Chen and L. Liu, RSC Adv., 2017, 7, 49910 RSC.
- W. X. Chunyu, Z. G. Ding, M. G. Li, J. Y. Zhao, S. J. Gu, Y. Gao, F. Wang, J. H. Ding and M. L. Wen, Helv. Chim. Acta, 2016, 99, 583–587 CrossRef CAS.
- H. Zhang, M. H. Yang, F. F. Zhuo, N. Gao, X. B. Cheng, X. B. Wang, Y. H. Pei and L. Y. Kong, RSC Adv., 2019, 9, 3520 RSC.
- M. X. Hua, W. M. Zheng, K. H. Sun, X. F. Gu, X. M. Zeng, H. T. Zhang, T. H. Zhong, Z. Z. Shao and Y. H. Zhang, Nat. Prod. Res., 2019, 33, 386–392 CrossRef PubMed.
- J. W. Kim, S. K. Ko, H. M. Kim, G. H. Kim, S. Son, G. S. Kim, G. J. Hwang, E. S. Jeon, K. S. Shin, I. J. Ryoo, Y. S. Hong, H. Oh, K. H. Lee, N. K. Soung, D. Hashizume, T. Nogawa, S. Takahashi, B. Y. Kim, H. Osada, J. H. Jang and J. S. Ahn, J. Nat. Prod., 2016, 79, 2703–2708 CrossRef CAS PubMed.
- Y. C. Xu, C. Wang, H. S. Liu, G. L. Zhu, P. Fu, L. P. Wang and W. M. Zhu, Mar. Drugs, 2018, 16, 363 CrossRef CAS PubMed.
- D. Zhou, L. J. Li, H. Qi, J. J. Pan, H. Zhang, J. D. Wang and W. S. Xiang, J. Antibiot., 2015, 68, 339–341 CrossRef CAS PubMed.
- H. Hayashi, Y. Oka, K. Kai and K. Akiyama, Biosci., Biotechnol., Biochem., 2012, 76, 745–748 CrossRef CAS PubMed.
- H. Hayashi, Y. Oka, K. Kai and K. Akiyama, Biosci., Biotechnol., Biochem., 2012, 76, 1765–1768 CrossRef CAS PubMed.
- T. El-Elimat, M. Figueroa, H. A. Raja, S. Alnabulsi and N. H. Oberlies, Tetrahedron Lett., 2021, 72, 153067 CrossRef CAS PubMed.
- Y. M. Fu, C. H. Li, J. Zhu, L. T. Zhang, Y. Wang, Q. L. Chen, L. Xu, S. Y. Zhang, Y. Fang and T. Liu, Biochem. Syst. Ecol., 2020, 93, 104186 CrossRef CAS.
- H. Yamazaki, W. Nakayama, O. Takahashi, R. Kirikoshi, Y. Izumikawa, K. Iwasaki, K. Toraiwa, K. Ukai, H. Rotinsulu, D. S. Wewengkang, D. A. Sumilat, R. E. P. Mangindaan and M. Namikoshi, Bioorg. Med. Chem. Lett., 2015, 25, 3087–3090 CrossRef CAS PubMed.
- C. Hemtasin, S. Kanokmedhakul, K. Kanokmedhakul, C. Hahnvajanawong, K. Soytong, S. Prabpai and P. Kongsaeree, J. Nat. Prod., 2011, 74, 609–613 CrossRef CAS PubMed.
- H. A. Wahab, N. B. Pham, T. S. T. Muhammad, J. N. A. Hooper and R. J. Quinn, Mar. Drugs, 2017, 15, 1–10 Search PubMed.
- J. Li, B. B. Gu, F. Sun, J. R. Xu, W. H. Jiao, H. B. Yu, B. N. Han, F. Yang, X. C. Zhang and H. W. Lin, J. Nat. Prod., 2017, 80, 1436–1445 CrossRef CAS PubMed.
- K. Hagiwara, J. E. G. Hernandez, M. K. Harper, A. Carroll, C. A. Motti, J. Awaya, H. Y. Nguyen and A. D. Wright, J. Nat. Prod., 2015, 78, 325–329 CrossRef CAS PubMed.
- Q. Göthel and M. Köck, Beilstein J. Org. Chem., 2014, 10, 613–621 CrossRef PubMed.
- V. R. D. L. Parra, V. Mierau, T. Anke and O. Sterner, Tetrahedron, 2006, 62, 1828–1832 CrossRef.
- L. Flores-Bocanegra, M. Augustinovic, H. A. Raja, S. J. Kurina, A. C. Maldonado, J. E. Burdette, J. O. Falkinham, C. J. Pearce and N. H. Oberlies, Tetrahedron Lett., 2021, 68, 152896 CrossRef CAS PubMed.
- J. Li, F. Yang, Z. Wang, W. Wu, L. Liu, S. P. Wang, B. X. Zhao, W. H. Jiao, S. H. Xu and H. W. Lin, Org. Biomol. Chem., 2018, 16, 6773–6782 RSC.
- P. L. Winder, H. L. Baker, P. Linley, E. A. Guzmán, S. A. Pomponi, M. C. Diaz, J. K. Reed and A. E. Wright, Bioorg. Med. Chem., 2011, 19, 6599–6603 CrossRef CAS PubMed.
- J. Li, W. Wu, F. Yang, L. Liu, S. P. Wang, W. H. Jiao, S. H. Xu and H. W. Lin, Chem. Biodiversity, 2018, 15, e1800078 CrossRef PubMed.
- W. H. Jiao, T. T. Xu, B. B. Gu, G. H. Shi, Y. Zhu, F. Yang, B. N. Han, S. P. Wang, Y. S. Li, W. Zhang, J. Li and H. W. Lin, RSC Adv., 2015, 5, 87730–87738 RSC.
- Y. H. Gui, L. Liu, W. Wu, Y. Zhang, Z. L. Jia, Y. P. Shi, H. T. Kong, K. C. Liu, W. H. Jiao and H. W. Lin, Bioorg. Chem., 2020, 94, 103435 CrossRef CAS PubMed.
- L. Liu, W. Wu, J. Li, W. H. Jiao, L. Y. Liu, J. Tang, L. Liu, F. Sun, B. N. Han and H. W. Lin, Biomed. Pharmacother., 2018, 100, 417–425 CrossRef CAS PubMed.
- W. H. Jiao, T. T. Xu, F. Zhao, H. Gao, G. H. Shi, J. Wang, L. L. Hong, H. B. Yu, Y. S. Li, F. Yang and H. W. Lin, Eur. J. Org. Chem., 2015, 2015, 960–966 CrossRef CAS.
- J. Tang, W. Wu, F. Yang, L. Y. Liu, Z. Yang, L. Liu, W. Z. Tang, F. Sun and H. W. Lin, Cancer Med., 2018, 7, 3965–3976 CrossRef CAS PubMed.
- W. H. Jiao, G. H. Shi, T. T. Xu, G. D. Chen, B. B. Gu, Z. Wang, S. Peng, S. P. Wang, J. Li, B. N. Han, W. Zhang and H. W. Lin, J. Nat. Prod., 2016, 79, 406–411 CrossRef CAS PubMed.
- W. H. Jiao, T. T. Xu, H. B. Yu, F. R. Mu, J. Li, Y. S. Li, F. Yang, B. N. Han and H. W. Lin, RSC Adv., 2014, 4, 9236 RSC.
- W. H. Jiao, J. Li, D. Wang, M. M. Zhang, L. Y. Liu, F. Sun, J. Y. Li, R. J. Capon and H. W. Lin, J. Nat. Prod., 2019, 82, 2586–2593 CrossRef CAS PubMed.
- W. H. Jiao, B. H. Cheng, G. H. Shi, G. D. Chen, B. B. Gu, Y. J. Zhou, L. L. Hong, F. Yang, Z. Q. Liu, S. Q. Qiu, Z. G. Liu, P. C. Yang and H. W. Lin, Sci. Rep., 2017, 7, 8947 CrossRef PubMed.
- J. Wang, F. R. Mu, W. H. Jiao, J. Huang, L. L. Hong, F. Yang, Y. Xu, S. P. Wang, F. Sun and H. W. Lin, J. Nat. Prod., 2017, 80, 2509–2514 CrossRef CAS PubMed.
- W. H. Jiao, J. Li, M. M. Zhang, J. Cui, Y. H. Gui, Y. Zhang, J. Y. Li, K. C. Liu and H. W. Lin, Org. Lett., 2019, 21, 6190–6193 CrossRef CAS PubMed.
- H. B. Yu, Z. F. Yin, B. B. Gu, J. P. Zhang, S. P. Wang, F. Yang and H. W. Lin, Nat. Prod. Res., 2021, 35, 1620–1626 CrossRef CAS PubMed.
- H. X. Li, Q. Zhang, X. Jin, X. W. Zou, Y. X. Wang, D. X. Hao, F. H. Fu, W. H. Jiao, C. X. Zhang, H. W. Lin, K. Matsuzaki and F. Zhao, Mol. Med. Rep., 2018, 17, 674–682 CAS.
- R. A. Hill and A. Sutherland, Nat. Prod. Rep., 2018, 35, 702–706 RSC.
- R. A. Hill and A. Sutherland, Nat. Prod. Rep., 2019, 36, 556–560 RSC.
- R. A. Hill and A. Sutherland, Nat. Prod. Rep., 2019, 36, 1378–1382 RSC.
- R. A. Hill and A. Sutherland, Nat. Prod. Rep., 2012, 29, 435–439 RSC.
- R. A. Hill and A. Sutherland, Nat. Prod. Rep., 2014, 31, 706–710 RSC.
- G. Daletos, N. J. D. Voogd, W. E. G. Müller, V. Wray, W. H. Lin, D. Feger, M. Kubbutat, A. H. Aly and P. Proksch, J. Nat. Prod., 2014, 77, 218–226 CrossRef CAS PubMed.
- S. P. B. Ovenden, J. L. Nielson, C. H. Liptrot, R. H. Willis, D. M. Tapiolas, A. D. Wright and C. A. Motti, J. Nat. Prod., 2011, 74, 65–68 CrossRef CAS PubMed.
- W. Balansa, U. Mettal, Z. G. Wuisan, A. Plubrukarn, F. G. Ijong, Y. Liu and T. F. Schäberle, Mar. Drugs, 2019, 17, 158 CrossRef CAS PubMed.
- A. N. E. S. Hamed, W. Wätjen, R. Schmitz, Y. Chovolou, R. A. Edrada-Ebel, D. T. A. Youssef, M. S. Kamel and P. Proksch, Nat. Prod. Commun., 2013, 8, 289–292 CrossRef.
- C. K. Kim, J. K. Woo, S. H. Kim, E. Cho, Y. J. Lee, H. S. Lee, C. J. Sim, D. C. Oh, K. B. Oh and J. Shin, J. Nat. Prod., 2015, 78, 2814–2821 CrossRef CAS PubMed.
- X. C. Luo, P. L. Li, K. Y. Wang, N. J. D. Voogd, X. L. Tang and G. Q. Li, Nat. Prod. Res., 2021, 35, 2866–2871 CrossRef CAS PubMed.
- D. B. Abdjul, H. Yamazaki, O. Takahashi, R. Kirikoshi, K. Ukai and M. Namikoshi, J. Nat. Prod., 2016, 79, 1842–1847 CrossRef CAS PubMed.
- N. K. Utkina, V. A. Denisenko and V. B. Krasokhin, J. Nat. Prod., 2010, 73, 788–791 CrossRef PubMed.
- Y. Takahashi, M. Ushio, T. Kubota, S. Yamamoto, J. Fromont and J. Kobayashi, J. Nat. Prod., 2010, 73, 467–471 CrossRef CAS PubMed.
- H. M. Nguyen, T. Ito, N. N. Win, T. Kodama, V. Q. Hung, H. T. Nguyen and H. Morita, Phytochem. Lett., 2016, 17, 288–292 CrossRef CAS.
- H. M. Nguyen, T. Ito, S. Kurimoto, M. Ogawa, N. N. Win, V. Q. Hung, H. T. Nguyen, T. Kubota, J. Kobayashi and H. Morita, Bioorg. Med. Chem. Lett., 2017, 27, 3043–3047 CrossRef CAS PubMed.
- T. Ito, H. M. Nguyen, N. N. Win, H. Q. Vo, H. T. Nguyen and H. Morita, J. Nat. Med., 2018, 72, 298–303 CrossRef PubMed.
- P. V. Kiem, L. T. Huyen, D. T. Hang, N. X. Nhiem, B. H. Tai, H. L. T. Anh, P. V. Cuong, T. H. Quang, C. V. Minh, N. V. Dau, Y. A. Kim, L. Subedi, S. Y. Kim and S. H. Kim, Bioorg. Med. Chem. Lett., 2017, 27, 1525–1529 CrossRef CAS PubMed.
- H. Mitome, T. Nagasawa, H. Miyaoka, Y. Yamada and R. W. M. V. Soest, J. Nat. Prod., 2001, 64, 1506–1508 CrossRef CAS PubMed.
- H. Mitome, T. Nagasawa, H. Miyaoka, Y. Yamada and R. W. M. V. Soest, Tetrahedron, 2002, 58, 1693–1696 CrossRef CAS.
- L. T. Huyen, D. T. Hang, N. X. Nhiem, B. H. Tai, H. L. T. Anh, T. H. Quang, P. H. Yen, C. V. Minh, N. V. Dau and P. V. Kiem, Nat. Prod. Commun., 2017, 12, 477–478 CrossRef PubMed.
- J. Baars, I. Grimm, D. Blunk, J. M. Neudörfl and H. G. Schmalz, Angew. Chem., Int. Ed., 2021, 60, 14915–14920 CrossRef CAS PubMed.
- C. K. Chong, Q. L. Zhang, J. Ke, H. M. Zhang, X. D. Yang, B. J. Wang, W. Ding and Z. Y. Lu, Angew. Chem., Int. Ed., 2021, 60, 13807–13813 CrossRef CAS PubMed.
- H. Prawat, C. Mahidol, W. Kaweetripob, S. Wittayalai and S. Ruchirawat, Tetrahedron, 2012, 68, 6881–6886 CrossRef CAS.
- W. W. Cao, Q. Luo, Y. X. Cheng and S. M. Wang, Fitoterapia, 2016, 110, 110–115 CrossRef CAS PubMed.
- F. J. Zhou, Y. Nian, Y. M. Yan, Y. Gong, Q. Luo, Y. Zhang, B. Hou, Z. L. Zuo, S. M. Wang, H. H. Jiang, J. Yang and Y. X. Cheng, Org. Lett., 2015, 17, 3082–3085 CrossRef CAS PubMed.
- F. Y. Qin, Y. M. Yan, Z. C. Tu and Y. X. Cheng, J. Asian Nat. Prod. Res., 2019, 21, 542–550 CrossRef CAS PubMed.
- Y. P. Li, X. T. Jiang, F. Y. Qin, H. X. Zhang and Y. X. Cheng, Bioorg. Chem., 2021, 109, 104706 CrossRef CAS PubMed.
- L. Z. Cheng, F. Y. Qin, X. C. Ma, S. M. Wang, Y. M. Yan and Y. X. Cheng, Molecules, 2018, 23, 1797 CrossRef PubMed.
- M. Dou, R. T. Li and Y. X. Cheng, Chin. Herb. Med., 2016, 8, 85–88 CrossRef CAS.
- X. L. Wang, Z. H. Wu, L. Di, F. J. Zhou, Y. M. Yan and Y. X. Cheng, Phytochemistry, 2019, 162, 199–206 CrossRef CAS PubMed.
- X. L. Wang, F. J. Zhou, M. Dou, Y. M. Yan, S. M. Wang, L. Di and Y. X. Cheng, Bioorg. Med. Chem. Lett., 2016, 26, 5507–5512 CrossRef CAS PubMed.
- Q. Luo, Z. L. Yang and Y. X. Cheng, Tetrahedron, 2019, 75, 2910–2915 CrossRef CAS.
- J. J. Zhang, F. Y. Qin, X. H. Meng, Y. M. Yan and Y. X. Cheng, Bioorg. Chem., 2020, 100, 103930 CrossRef CAS PubMed.
- Q. Luo, X. L. Wang, L. Di, Y. M. Yan, Q. Lu, X. H. Yang, D. B. Hu and Y. X. Cheng, Tetrahedron, 2015, 71, 840–845 CrossRef CAS.
- D. Cai, J. J. Zhang, Z. H. Wu, F. Y. Qin, Y. M. Yan, M. Zhang and Y. X. Cheng, Bioorg. Chem., 2021, 110, 104774 CrossRef CAS PubMed.
- X. H. Meng, F. Y. Qin, X. T. Jiang, Y. Li and Y. X. Cheng, Bioorg. Chem., 2021, 112, 104950 CrossRef CAS PubMed.
- J. C. Guo, F. D. Kong, Q. Y. Ma, Q. Y. Xie, R. S. Zhang, H. F. Dai, Y. G. Wu and Y. X. Zhao, Front. Chem., 2020, 8, 279 CrossRef CAS PubMed.
- X. R. Peng, J. Q. Liu, C. F. Wang, Z. H. Han, Y. Shu, X. Y. Li, L. Zhou and M. H. Qiu, Food Chem., 2015, 171, 251–257 CrossRef CAS PubMed.
- X. R. Peng, X. Wang, L. Chen, H. Yang, L. Li, S. Y. Lu, L. Zhou and M. H. Qiu, Fitoterapia, 2018, 127, 286–292 CrossRef CAS PubMed.
- M. Adams, M. Christen, I. Plitzko, S. Zimmermann, R. Brun, M. Kaiser and M. Hamburger, J. Nat. Prod., 2010, 73, 897–900 CrossRef CAS PubMed.
- J. C. Guo, Q. Y. Ma, F. D. Kong, Q. Y. Xie, L. M. Zhou, Q. Ding, Y. G. Wu and Y. X. Zhao, Chin. J. Org. Chem., 2019, 39, 3264–3268 CrossRef CAS.
- X. Q. Chen, L. X. Chen, S. P. Li and J. Zhao, Phytochem. Lett., 2017, 22, 214–218 CrossRef CAS.
- X. H. Ma, H. T. Wang, F. Li, T. J. Zhu, Q. Q. Gu and D. H. Li, Tetrahedron, 2015, 56, 7053–7055 CrossRef CAS.
- R. H. Guo, Y. T. Zhang, D. Duan, Q. Fu, X. Y. Zhang, X. W. Yu, S. J. Wang, B. Bao and W. H. Wu, Chin. J. Chem., 2016, 34, 1194–1198 CrossRef CAS.
- H. Koide, K. Hasegawa, N. Nishimura, R. Narasaki and K. Hasumi, J. Antibiot., 2012, 65, 361–367 CrossRef CAS PubMed.
- K. Hasegawa, H. Koide, W. M. Hu, N. Nishimura, R. Narasaki, Y. Kitano and K. Hasumi, J. Antibiot., 2010, 63, 589–593 CrossRef CAS PubMed.
- H. Koide, R. Narasaki, K. Hasegawa, N. Nishimura and K. Hasumi, J. Antibiot., 2012, 65, 91–93 CrossRef CAS PubMed.
- Y. Nishimura, E. Suzuki, K. Hasegawa, N. Nishimura, Y. Kitano and K. Hasumi, J. Antibiot., 2012, 65, 483–485 CrossRef CAS PubMed.
- M. Akiba, K. Kinoshita, Y. Kino, J. Sato and K. Koyama, Bioorg. Med. Chem. Lett., 2020, 30, 126808 CrossRef CAS PubMed.
- Y. Masuda, K. Fujihara, S. Hayashi, H. Sasaki, Y. Kino, H. Kamauchi, M. Noji, J. Satoh, T. Takanami, K. Kinoshita and K. Koyama, J. Nat. Prod., 2021, 84, 1748–1754 CrossRef CAS PubMed.
- G. J. Zhang, G. W. Wu, T. J. Zhu, T. Kurtán, A. Mándi, J. Y. Jiao, J. Li, X. Qi, Q. Q. Gu and D. H. Li, J. Nat. Prod., 2013, 76, 1946–1957 CrossRef CAS PubMed.
- X. Shi, W. Wei, W. J. Zhang, C. P. Hua, C. J. Chen, H. M. Ge, R. X. Tan and R. H. Jiao, J. Asian Nat. Prod. Res., 2015, 17, 143–148 CrossRef CAS PubMed.
- Q. Shi, T. T. Li, Y. M. Wu, X. Y. Sun, C. Lei, J. Y. Li and A. J. Hou, Phytochemistry, 2020, 180, 112524 CrossRef CAS PubMed.
- Y. Long, T. Tang, L. Y. Wang, B. He and K. Gao, J. Nat. Prod., 2019, 82, 2229–2237 CrossRef CAS PubMed.
- Y. Long, T. Tang, L. Y. Wang, B. He and K. Gao, J. Nat. Prod., 2019, 82, 3205 CrossRef CAS PubMed.
- D. S. Manamgoda, L. Cai, A. H. Bahkali, E. Chukeatirote and K. D. Hyde, Fungal Diversity, 2011, 51, 3–42 CrossRef.
- M. Wang, Z. H. Sun, Y. C. Chen, H. X. Liu, H. H. Li, G. H. Tan, S. N. Li, X. L. Guo and W. M. Zhang, Fitoterapia, 2016, 110, 77–82 CrossRef CAS PubMed.
- Q. Y. Qi, L. Huang, L. W. He, J. J. Han, Q. Chen, L. Cai and H. W. Liu, Chem. Biodiversity, 2014, 11, 1892–1899 CrossRef CAS PubMed.
- X. Cao, Y. T. Shi, X. D. Wu, K. W. Wang, S. H. Huang, H. X. Sun, J. S. Dickschat and B. Wu, Org. Lett., 2019, 21, 6539–6542 CrossRef CAS PubMed.
- X. Cao, Y. T. Shi, S. H. Wu, X. D. Wu, K. W. Wang, H. X. Sun, S. He, J. S. Dickschat and B. Wu, Tetrahedron, 2020, 76, 131349 CrossRef CAS.
- Y. Hitora, K. Takada, Y. Ise, S. P. Woo, S. Inoue, N. Mori, H. Takikawa, S. Nakamukai, S. Okada and S. Matsunaga, Bioorg. Med. Chem., 2020, 28, 115233 CrossRef CAS PubMed.
- S. P. B. Ovenden, J. L. Nielson, C. H. Liptrot, R. H. Willis, D. M. Tapiolas, A. D. Wright and C. A. Motti, J. Nat. Prod., 2011, 74, 1335–1338 CrossRef CAS PubMed.
- Y. X. Song, H. B. Huang, Y. C. Chen, J. Ding, Y. Zhang, A. J. Sun, W. M. Zhang and J. H. Ju, J. Nat. Prod., 2013, 76, 2263–2268 CrossRef CAS PubMed.
- F. L. Li, Y. Tang, W. G. Sun, J. K. Guan, Y. Y. Lu, S. T. Zhang, S. Lin, J. P. Wang, Z. X. Hu and Y. H. Zhang, Bioorg. Chem., 2019, 92, 103279 CrossRef CAS PubMed.
- L. Wang, J. Y. Jiao, D. Liu, X. M. Zhang, J. Li, Q. Che, T. J. Zhu, G. J. Zhang and D. H. Li, Chem. Biodiversity, 2020, 17, e2000226 CAS.
- U. Sommart, V. Rukachaisirikul, K. Trisuwan, K. Tadpetch, S. Phongpaichit, S. Preedanon and J. Sakayaroj, Phytochem. Lett., 2012, 5, 139–143 CrossRef CAS.
- M. Carbone, L. Nunez-Pons, M. Paone, F. Castelluccio, C. Avila and M. Gavagnin, Tetrahedron, 2012, 68, 3541–3544 CrossRef CAS.
- M. Carbone, L. Nunez-Pons, M. Paone, F. Castelluccio, C. Avila and M. Gavagnin, Tetrahedron, 2012, 68, 8515 CrossRef CAS.
- M. Dou, L. Di, L. L. Zhou, Y. M. Yan, X. L. Wang, F. J. Zhou, Z. L. Yang, R. T. Li, F. F. Hou and Y. X. Cheng, Org. Lett., 2014, 16, 6064–6067 CrossRef CAS PubMed.
- G. H. Huang, C. Lei, K. X. Zhu, J. Y. Li, J. Li and A. J. Hou, Chin. J. Nat. Med., 2019, 17, 0963–0969 CAS.
- Z. J. Zhan, Y. M. Ying, L. F. Ma and W. G. Shan, Nat. Prod. Rep., 2011, 28, 594–629 RSC.
- L. F. Ma, Y. L. Chen, W. G. Shan and Z. J. Zhan, Nat. Prod. Rep., 2020, 37, 999–1030 RSC.
- Y. H. Ma, X. X. Dou and X. H. Tian, Phytochem. Rev., 2020, 19, 983–1043 CrossRef CAS.
- N. K. Utkina, V. A. Denisenko and V. B. Krasokhin, Tetrahedron Lett., 2011, 52, 3765–3768 CrossRef CAS.
- J. M. Liu, X. N. Jia, J. L. Zhao, J. M. Feng, M. H. Chen, R. D. Chen, K. B. Xie, D. W. Chen, Y. Li, D. Zhang, Y. Peng, S. Y. Si and J. G. Dai, Org. Chem. Front., 2020, 7, 531–542 RSC.
- J. M. Feng, M. Zhang, X. N. Jia, J. L. Zhao, R. D. Chen, K. B. Xie, D. W. Chen, Y. Li, J. M. Liu and J. G. Dai, Fitoterapia, 2019, 136, 104158 CrossRef CAS PubMed.
- J. L. Zhao, J. M. Feng, Z. Tan, J. M. Liu, M. Zhang, R. D. Chen, K. B. Xie, D. W. Chen, Y. Li, X. G. Chen and J. G. Dai, Bioorg. Med. Chem. Lett., 2018, 28, 355–359 CrossRef CAS PubMed.
- M. Zhang, J. M. Feng, X. N. Jia, J. L. Zhao, J. M. Liu, R. D. Chen, K. B. Xie, D. W. Chen, Y. Li, D. Zhang and J. G. Dai, Chin. Chem. Lett., 2019, 30, 435–438 CrossRef CAS.
- Z. G. Ding, J. H. Ding, J. Y. Zhao, W. X. Chunyu, M. G. Li, S. J. Gu, F. Wang and M. L. Wen, Fitoterapia, 2018, 125, 94–97 CrossRef CAS PubMed.
- Z. G. Ding, J. Y. Zhao, J. H. Ding, W. X. Chunyu, M. G. Li, S. J. Gu, F. Wang and M. L. Wen, Nat. Prod. Res., 2018, 32, 2370–2374 CrossRef CAS PubMed.
- D. Liu, Y. Li, X. C. Guo, W. Ji and W. H. Lin, Chem. Biodiversity, 2020, 17, e2000170 CAS.
- G. Wang, W. H. Wu, Q. G. Zhu, S. Q. Fu, X. Y. Wang, S. T. Hong, R. H. Guo and B. Bao, Chin. J. Chem., 2015, 33, 1089–1095 CrossRef CAS.
- M. Amigó, M. Payá, A. Braza-Boïls, S. D. Rosa and M. C. Terencio, Life Sci., 2008, 82, 256–264 CrossRef PubMed.
- M. L. Ferrandiz, M. J. Sanz, G. Bustos, M. Payfi, M. J. Alcaraz and S. D. Rosa, Eur. J. Pharmacol., 1994, 253, 75–82 CrossRef CAS PubMed.
- W. E. G. Muller, A. Maidhof, R. K. Zahn, H. C. Schroder, M. J. Gasic, D. Heidemann, A. Bernd, B. Kurelec, E. Eich and G. Seibert, Cancer Res., 1985, 45, 4822–4826 CAS.
- J. M. Miguel del Corral, M. Gordaliza, M. A. Castro, M. M. Mahiques, P. Chamorro, A. Molinari, M. D. García-Grávalos, H. B. Broughton and A. S. Feliciano, J. Med. Chem., 2001, 44, 1257–1267 CrossRef CAS PubMed.
- T. Hashimoto, K. Shibata, K. Nobe, K. Hasumi and K. Honda, J. Pharmacol. Sci., 2010, 114, 41–49 CrossRef CAS PubMed.
- T. Miyazaki, Y. Kimura, H. Ohata, T. Hashimoto, K. Shibata, K. Hasumi and K. Honda, Stroke, 2011, 42, 1097–1104 CrossRef CAS PubMed.
- T. Yan, W. H. Wu, T. W. Su, J. J. Chen, Q. G. Zhu, C. Y. Zhang, X. Y. Wang and B. Bao, Arch. Pharmacal Res., 2015, 38, 1530–1540 CrossRef CAS PubMed.
- T. T. Ling, A. X. Xiang and E. A. Theodorakis, Angew. Chem., Int. Ed., 1999, 20, 3089–3091 CrossRef.
- T. T. Ling, E. Poupon, E. J. Rueden, S. H. Kim and E. A. Theodorakis, J. Am. Chem. Soc., 2002, 124, 12261–12267 CrossRef CAS PubMed.
- E. P. Locke and S. M. Hecht, Chem. Commun., 1996, 24, 2717–2718 RSC.
- A. S. Sarma and P. Chattopadhyay, J. Org. Chem., 1982, 47, 1727–1731 CrossRef CAS.
- J. Sakurai, T. Oguchi, K. Watanabe, H. Abe, S. I. Kanno, M. Ishikawa and T. Katoh, Chem.–Eur. J., 2008, 14, 829–837 CrossRef CAS PubMed.
- J. H. George, Acc. Chem. Res., 2021, 54, 1843–1855 CrossRef CAS PubMed.
- T. X. Bai, Z. Y. Quan, R. Zhai, T. Awakawa, Y. Matsuda and I. Abe, Org. Lett., 2018, 20, 7504–7508 CrossRef CAS PubMed.
- S. M. Shen, G. Appendino and Y. W. Guo, Nat. Prod. Rep., 2022, 39, 1803–1832 RSC.
- C. K. Chong and Z. Y. Lu, Synlett, 2021, 32, 1777–1783 CrossRef CAS.
- X. Yin, M. Mato and A. M. Echavarren, Angew. Chem., Int. Ed., 2017, 56, 14591–14595 CrossRef CAS PubMed.
- X. Yin, M. Mato and A. M. Echavarren, Angew. Chem., 2017, 129, 14783–14787 CrossRef.
- M. A. Haque and C. K. Jana, Chem.–Eur. J., 2017, 23, 13300–13304 CrossRef CAS PubMed.
- M. A. Haque and C. K. Jana, Chem.–Eur. J., 2019, 25, 12849 CrossRef CAS PubMed.
- M. A. Haque, B. L. Sailo, G. Padmavathi, A. B. Kunnumakkara and C. K. Jana, Eur. J. Med. Chem., 2018, 160, 256–265 CrossRef CAS PubMed.
- S. De, E. Mahal, M. A. Haque, C. K. Jana and D. Koley, J. Org. Chem., 2021, 86, 1133–1140 CrossRef CAS PubMed.
- Y. Fukui, K. Narita and T. Katoh, Chem.–Eur. J., 2014, 20, 2436–2439 CrossRef CAS PubMed.
- B. Schmalzbauer, J. Herrmann, R. Muller and D. Menche, Org. Lett., 2013, 15, 964–967 CrossRef CAS PubMed.
- R. Lucas, C. Giannini, M. V. D'Auria and M. Paya, J. Pharmacol. Exp. Ther., 2003, 304, 1172–1180 CrossRef CAS PubMed.
- K. Shibata, T. Hashimoto, K. Nobe, K. Hasumi and K. Honda, Naunyn-Schmiedeberg's Arch. Pharmacol., 2011, 384, 103–108 CrossRef CAS PubMed.
- Y. Akamatsu, A. Saito, M. Fujimura, H. Shimizu, M. Mekawy, K. Hasumi and T. Tominaga, Neurosci. Lett., 2011, 503, 110–114 CrossRef CAS PubMed.
- K. Shibata, T. Hashimoto, K. Nobe, K. Hasumi and K. Honda, Naunyn-Schmiedeberg's Arch. Pharmacol., 2010, 382, 245–253 CrossRef CAS PubMed.
- W. M. Hu, R. Narasaki, N. Nishimura and K. Hasumi, Thromb. J., 2012, 10, 1–9 CrossRef PubMed.
- H. Sawada, N. Nishimura, E. Suzuki, J. Zhuang, K. Hasegawa, H. Takamatsu, K. Honda and K. Hasumi, J. Cereb. Blood Flow Metab., 2014, 34, 235–241 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2np00045h |
| This journal is © The Royal Society of Chemistry 2023 |