Open Access Article
Open Access ArticleRegulating the surface topography of CpG nanoadjuvants via coordination-driven self-assembly for enhanced tumor immunotherapy†
Li
Zhang‡
abc,
Lingpu
Zhang‡
d,
Yuqi
Wang
e,
Kai
Jiang
e,
Chao
Gao
e,
Pengfei
Zhang
b,
Yujie
Xie
 f,
Bin
Wang
d,
Yun
Zhao
a,
Haihua
Xiao
f,
Bin
Wang
d,
Yun
Zhao
a,
Haihua
Xiao
 *d and
Jie
Song
*d and
Jie
Song
 *be
*be
aSchool of Life Science, Hangzhou Institute for Advanced Study, University of Chinese Academy of Sciences, Hangzhou, Zhejiang 310024, China
bHangzhou Institute of Medicine, Chinese Academy of Sciences, Hangzhou, Zhejiang 310022, China
cSchool of Pharmacy, Changzhou University, Changzhou, Jiangsu 213164, China
dBeijing National Laboratory for Molecular Sciences, State Key Laboratory of Polymer Physics and Chemistry, Institute of Chemistry, Chinese Academy of Sciences, Beijing 100190, China
eInstitute of Nano Biomedicine and Engineering, Department of Instrument Science and Engineering, School of Electronic Information and Electrical Engineering, Shanghai Jiao Tong University, Shanghai 200240, China. E-mail: sjie@sjtu.edu.cn
fSchool of Chemistry, University of Birmingham, Edgbaston, Birmingham, B15 2TT, UK
First published on 26th July 2023
Abstract
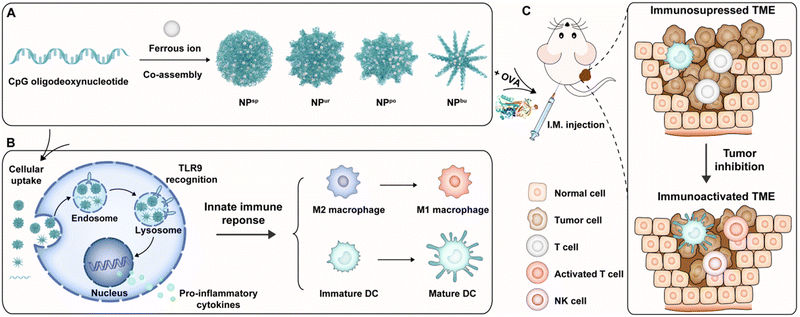
Immunoadjuvants play a key role in enhancing the efficacy of therapeutic tumor vaccines for treating malignant and recurrent cancers. However, due to the bottleneck in the rational design and mechanistic understanding of novel adjuvants, currently available immunoadjuvants in clinical practice are very limited. To boost adjuvant design and development, herein we propose a surface topography regulatory strategy for constructing novel adjuvants with improved adjuvant properties. One of the licensed adjuvants with a well-defined molecular mechanism of immune activation, cytosine–phosphate–guanine oligodeoxynucleotides (CpG ODNs), was used as the material framework. We constructed immunostimulatory CpG nanoparticles (CpG NPs) with different surface topographies by coordination-driven self-assembly between CpG ODNs and ferrous ions. These self-assembled CpG NPs combine the biological and physical activation abilities of innate immunity and can be used as adjuvants of tumor antigens for malignant tumor immunotherapy. The experimental results showed that these CpG NPs could rapidly enter innate immune cells and remold the tumor microenvironment (TME) to enhance anti-tumor immunotherapy via (i) inducing proinflammatory cytokine production; (ii) promoting the transformation of macrophages from immunosuppressed M2 types into immunoactivated M1 types; (iii) amplifying the antigen presentation of mature dendritic cells (DCs), and (iv) activating T cells in tumor sites. Among the prepared nanostructures, pompon-shaped nanoparticles (NPpo) showed the strongest adjuvant properties and anti-tumor immunotherapeutic effect as the adjuvant of ovalbumin in melanoma-bearing mice. Overall, this work provides an effective strategy for designing novel adjuvants for activating the immunosuppressed TME to enable better cancer immunotherapy.
1. Introduction
Tumor immunotherapy has revolutionized traditional cancer therapies and made remarkable clinical advances in recent years. As a new type of effective tumor immunotherapy, therapeutic tumor vaccines can activate or enhance the anti-tumor immune response of the body by expressing specific tumor antigens and then killing and eliminating tumor cells under the assistance of immunoadjuvants, showing great application prospects in the treatment of malignant tumors.1–3 Most tumor vaccines consist of one or more tumor antigenic components and immunoadjuvants that enhance the magnitude, breadth, and durability of the antigenic immune response.4–6 Therefore, immunoadjuvants are key components in improving vaccine effectiveness. However, due to safety concerns as well as the difficulties in the research of adjuvant design and immunologic mechanisms, the development of new adjuvants is very slow.7–9 Clinically approved adjuvants are far from meeting the needs of tumor therapy and vaccine development. Consequently, developing new, novel immunoadjuvants with high safety, excellent properties and a clear immunologic mechanism is of great significance for the development of therapeutic tumor vaccines and the immunotherapy of malignant tumors.Cytosine–phosphate–guanosine oligodeoxynucleotides (CpG ODNs) are one of the clinically licensed adjuvants with a well-defined molecular mechanism of immune activation, attracting extensive attention and continuous research in the field of immunotherapy.10–13 They can be recognized by toll-like receptor 9 (TLR-9) on endo/lysosomal membranes and induce the production of pro-inflammatory cytokines to successively activate both innate and adaptive immunity.14,15 However, owing to structural instability in the complex tumor microenvironment (TME), CpG adjuvants are currently only approved for hepatitis B and SARS-CoV-2 vaccines and not for tumor vaccines.16,17 Thus, in order to broaden their application, researchers have focused on improving the structural stability and prolonging the half-life of CpG ODNs through skeleton modification, protein/peptide binding or nanoparticle preparation.18–21 Nanoparticular adjuvants have the advantages of enhancing the immune response and reducing tolerance owing to their characteristics of small size and large specific surface area.22,23 With the in-depth study of nanoadjuvants, researchers found that the surface topography of nanoadjuvants can influence the adsorption of antigens and adhesion with immune cells and is another key factor in determining the efficacy of nanoadjuvants in addition to the composition and particle size.24–26 Thus, constructing CpG-based nanoparticles (CpG NPs) with desirable surface topography is an effective approach to improve the structural stability of CpG ODNs and enhance their adjuvant properties.
Emerging evidence indicates that nanoadjuvants with special surface topographies have unique performance advantages.27,28 Previous studies have reported that a gold nanostar adjuvant system with surface modification of CpG ODNs showed deep tumor penetration ability, induced a cascade immune response and significantly inhibited tumor growth.29–31 Compared to spheroidal nanostructures that accumulated mostly in the center of the endosome, star-shaped nanostructures accumulated primarily along the interior edges, which achieved a higher level of toll-like receptor 9 (TLR9) activation along with the enhanced secretion of pro-inflammatory cytokines and chemokines.32 Another study found that the nanospiky surface of TiO2 microparticles can activate and amplify the innate immune response by placing mechanical stress on the cells, stimulating potassium efflux and inflammasome activation during the phagocytosis of immune cells.27 These findings indicate that increased surface roughness of nanoadjuvants contributes to enhanced immune activities, thereby advancing our understanding of the mechanisms by which surface topography modulates immune responses. However, these nanoadjuvants were fabricated based on rigid nanoparticles and may have a significant impact on immune responses. Therefore, the structure–activity relationship between surface topography and immune activity of CpG-framed nanoadjuvants still needs to be explored.
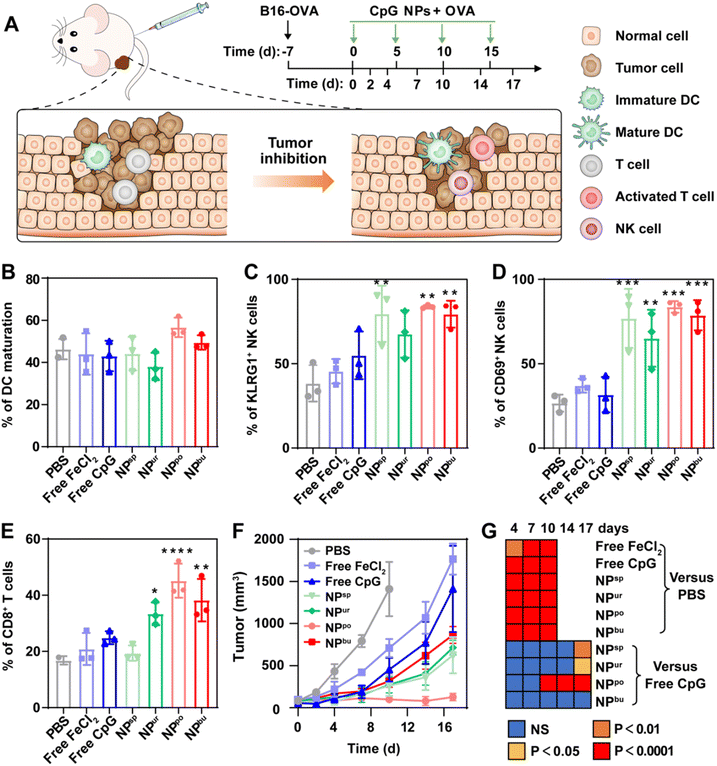
To understand the immunomodulatory capacity of surface topographies of nanoadjuvants on immune cells for cancer therapy, we designed a series of specific surface topographies on self-assembled CpG nanoparticles (CpG NPs) ranging from smooth to short plush to plush and finally to a spiky surface. They were named spherical, urchin-, pompon- and burr-shaped nanoparticles and denoted as NPsp, NPur, NPpo and NPbu for short, respectively (Scheme 1A). Due to the combination of physical and biological cues, the designed CpG NPs are believed to be able to modulate innate immune responses of the TME for enhanced anti-tumor immunotherapy. First, we tested the cytotoxicity, cellular uptake behavior and subcellular distribution of different nanostructures in RAW264.7 macrophages. Then, the polarization of macrophages and maturation of dendritic cells on different nanostructures were systematically examined by the evaluation of cytokine secretion and cell surface marker expression (Scheme 1B). Last, we assessed the anti-tumor efficacy and immunosuppressive TME activation of the obtained CpG NPs on a melanoma animal model by combining them with tumor model antigen ovalbumin (OVA) (Scheme 1C).
2. Experimental section
2.1. Preparation and characterization of CpG NPs
Inspired by the one-pot synthesis method reported by Li's team,33,34 we adopted a similar method to prepare CpG NPs with different surface topographies. Concretely, for the preparation of spherical CpG NPs, 10 μL CpG aqueous solution (100 μM) and 8 μL FeCl2 aqueous solution (50 mM) were first added into 82 μL pure water in a 200 μL PCR tube, immediately followed by a quick vortex to make sure all the ingredients mixed thoroughly. Next, the mixture was allowed to stand at 95 °C for 2 h. Finally, the product was obtained by cooling it to room temperature. The other three CpG NPs were prepared in double distilled water and Tris–HCl buffer solution (20 mM, pH = 7.4) supplemented with 10 mM MgCl2, respectively. The concentrations of CpG strands and FeCl2 in the reaction system are shown in Table S1.† The total volume of the reaction system was set at 100 μL.The product solution was deposited on a carbon-coated copper grid and subjected to TEM imaging (120 kV) and SEM imaging (2 kV) to characterize the surface topography. The hydrodynamic size and distribution of the prepared nanoparticles were measured by dynamic light scattering on a Zetasizer Nano ZS instrument (Malvern, UK). The characteristic absorption spectra of free CpG ODN solution and the above assembly solution were measured using a circular dichroism spectrometer in a heating process (25–95 °C) and a thermostatic process (95 °C) for 2 h, respectively.
2.2. Cell culture
RAW264.7 cells and Hacat used in the experiments were purchased from the Shanghai Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China). Cells were cultured in high-glucose DMEM supplemented with 10% FBS and 1% P/S in a 5% CO2, 37 °C incubator. BMMs were prepared by culturing mouse bone-marrow cells isolated from Balb/c mice with a 100 ng mL−1 macrophage colony-stimulating factor (Biolegend) in a cell culture medium for 7 d. BMDCs were prepared by culturing bone-marrow cells from Balb/c mice with 10 ng mL−1 of granulocyte-macrophage colony-stimulating factor (Biolegend) in a cell culture medium for 7 d. BMDCs were also sorted on a high-speed cell sorter based on DC marker CD11c on a BD FACSAria. All the procedures and experiments were performed in an endotoxin-free environment.2.3. MTT assays
Cells were seeded in 96 well plates at 1 × 104 cells per well for 12 h and then were cultured with free CpG, free FeCl2 and CpG NPs at a series of CpG concentration gradients (0–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nM) for 24 h. To the cells, 20 μL of MTT in PBS solution (2 mg mL−1) were added and incubated for 4 h at 37 °C. After incubation, the medium was removed and 100 μL dimethyl sulfoxide was added to completely dissolve formazan. Finally, the absorbance was measured using a microplate reader (Tecan, Switzerland) at a wavelength of 570 nm to calculate the cell survival rate. The values represented the mean ± SD of 5 replicates and were plotted relative to the untreated cells.
000 nM) for 24 h. To the cells, 20 μL of MTT in PBS solution (2 mg mL−1) were added and incubated for 4 h at 37 °C. After incubation, the medium was removed and 100 μL dimethyl sulfoxide was added to completely dissolve formazan. Finally, the absorbance was measured using a microplate reader (Tecan, Switzerland) at a wavelength of 570 nm to calculate the cell survival rate. The values represented the mean ± SD of 5 replicates and were plotted relative to the untreated cells.
2.4. Live/dead assays
To further determine cell viability, RAW264.7 cells were incubated with free CpG, free FeCl2 and CpG NPs at a dose of 1 nmol sample per well (CpG equivalent of 1000 nM) for 24 h. After the indicated times of treatment, cells were stained with calcein-AM to identify live cells (green fluorescence) and ethidium homodimer to exclude dead cells (red fluorescence). The results were analyzed by fluorescence microscopy (CSU-W1-SoRa, Nikon).2.5. Cellular uptake studies
RAW264.7 cells were cultured on a glass-bottom plate for 24 h, and then the FAM-labeled free CpG and CpG NPs were added and incubated with cells (CpG equivalent of 1000 nM) for 4 h in the experiments. The cell nucleus was stained with Hoechst 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 258 and imaged with a confocal fluorescence microscope (CSU-W1-SoRa, Nikon). To quantify the cellular uptake efficiency, flow cytometry (CytoFLEX LX, Beckman Coulter) was used to determine the green fluorescence intensity (corresponding to the amount of free CpG or CpG NPs) in each cell.
258 and imaged with a confocal fluorescence microscope (CSU-W1-SoRa, Nikon). To quantify the cellular uptake efficiency, flow cytometry (CytoFLEX LX, Beckman Coulter) was used to determine the green fluorescence intensity (corresponding to the amount of free CpG or CpG NPs) in each cell.
2.6. Intracellular distribution of CpG NPs
RAW264.7 cells were seeded in a 4-chamber glass plate at a density of 1 × 104 cells per well and subsequently incubated with FAM-labeled CpG NPs (CpG equivalent of 1000 nM) for 0.5, 1, 2, 4 and 6 h at 37 °C. After rinsing with PBS buffer 3 times, cells were fixed with paraformaldehyde for 15 min. The fixed cells were washed with PBS buffer 3 times and subsequently stained with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5 mixed solution of Hoechst 33
1.5 mixed solution of Hoechst 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 258 (20 μg mL−1) and LysoTracker Red DND-99 (100 nM) for 40 min at 37 °C. Finally, the above cells were observed using a fluorescence microscope fitted with the correct filter set. Multiple fluorescence images were collected from each sample group at each time point for co-location parameter analysis. Co-localization parameters were analyzed using the NIS-Elements analysis software. To comprehensively display the fluorescence distribution in cells, single cells in each test group were selected for multi-layer scanning to obtain Z-stack images (step size, 1 μm).
258 (20 μg mL−1) and LysoTracker Red DND-99 (100 nM) for 40 min at 37 °C. Finally, the above cells were observed using a fluorescence microscope fitted with the correct filter set. Multiple fluorescence images were collected from each sample group at each time point for co-location parameter analysis. Co-localization parameters were analyzed using the NIS-Elements analysis software. To comprehensively display the fluorescence distribution in cells, single cells in each test group were selected for multi-layer scanning to obtain Z-stack images (step size, 1 μm).
2.7. Gene profiling
RAW264.7 macrophages were first incubated in a 24-well plate with a density of 2 × 104. After cells adhered to the wall, culture media were replaced with IL-4 containing culture media with a concentration of 10 ng mL−1. After 20 h, free CpG, FeCl2 solution and CpG NPs were added to the cell culture media at a dose of 1 nmol CpG strands per well and a dose of 400 nmol FeCl2 solution. After another 20 h, mRNA in the cells was collected for reverse transcription and amplification with real-time qPCR analysis. Expressions of genes related to M1 and M2 macrophage subtypes were analyzed by quantitative real-time RT-PCR using the indicated primers in Table S3.† The data are presented as the fold change in gene expression normalized to an endogenous reference gene and relative to the untreated control.2.8. Cell-surface marker analysis
Cell surface markers were analyzed by flow cytometry (BD FACSAria). To this end, BMDCs were cultured with free CpG, FeCl2 solution and CpG NPs for 12 h. The cells were collected and stained with anti-MHC-II (25-9-17, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200), anti-CD40 (3/23, 1
200), anti-CD40 (3/23, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100), anti-CD80 (16-10A1, 1
100), anti-CD80 (16-10A1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100) and anti-CD86 (GL-1, 1
100) and anti-CD86 (GL-1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100) antibodies. All the antibodies were obtained from Biolegend. The stained cells were fixed, quantified by flow cytometry and analyzed with FlowJo software.
100) antibodies. All the antibodies were obtained from Biolegend. The stained cells were fixed, quantified by flow cytometry and analyzed with FlowJo software.
2.9. Animals
Balb/c mice were obtained from Beijing National Laboratory for Molecular Sciences and housed on a 12 h day/12 h night schedule (lights on from 19:00 to 07:00) at a constant temperature (22 ± 1 °C) and humidity (60%). Studies were approved by the Animal Care and Use Committee at the Institute of Chemistry, Chinese Academy of Sciences (No. 2022R0004). All the animals received humane care and every effort was made to minimize suffering in compliance with institutional and National Institutes of Health guidelines.2.10. Anti-tumor efficacy test
Balb/c mice (6 weeks, half male and half female) were subcutaneously inoculated with B16-OVA cells for tumor modeling first. After 7 days, the tumor-bearing mice were intramuscularly injected with PBS, free CpG, free FeCl2, and four types of CpG NPs (1.32 mM FeCl2 solution for free FeCl2, CpG equivalent of 3.3 μM for free CpG and CpG NPs) combined with OVA. The injection was given every 4 days for a total of 4 injections. Tumor volumes were monitored on days 0, 2, 4, 7, 10 and 14. After a 14-day test, ratios of the tumor-infiltrating mature DCs (CD80 and CD86), activated T cells (CD3 and CD8) and NK cells (KLRG1 and CD69) in melanoma in different treatment groups were detected by flow cytometry.2.11. In vivo biosafety evaluation
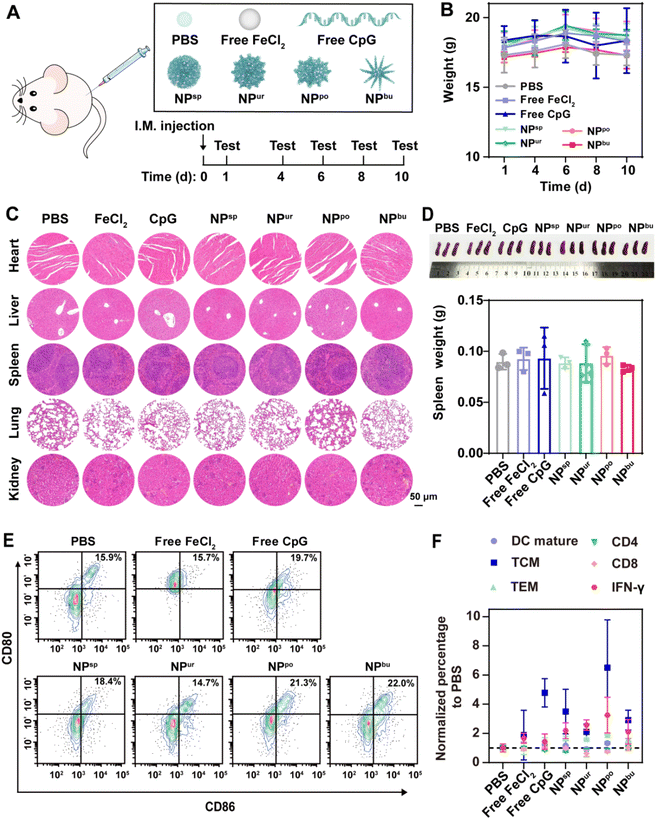
Balb/c mice (6 weeks, half male and half female) were intramuscularly injected with PBS, free CpG, free FeCl2, or self-assembled CpG NPs at an equivalent dose of CpG strands (10 μg per mouse). The injection amount is 1.32 mM FeCl2 solution for free FeCl2 and 3.3 μM equivalent CpG for free CpG and CpG NPs in 100 μL solution, respectively. The body weight of mice was monitored on days 1, 4, 6, 8 and 10. After 10 days, the mouse spleens were taken out, photographed and weighed. Then, the removed spleens were ground and utilized for parameter detections, including CD11b, CD80, and CD86 secreted from DCs and CD3, CD4, CD8, CD44, CD62L, and IFN-γ from T cells. The results were analyzed with flow cytometry and FlowJo software.2.12. Statistics and reproducibility
A two-tailed t-test was used to analyze the difference between sample and PBS groups, and a one-way analysis of variance (ANOVA) among multiple groups with a confidence interval of 95%. All the data are presented as the mean ± s.d. as indicated in the experiments. P values were calculated using PRISM software (GraphPad) and regarded as significant if less than 0.05. No animals were excluded from the analysis.3. Results and discussion
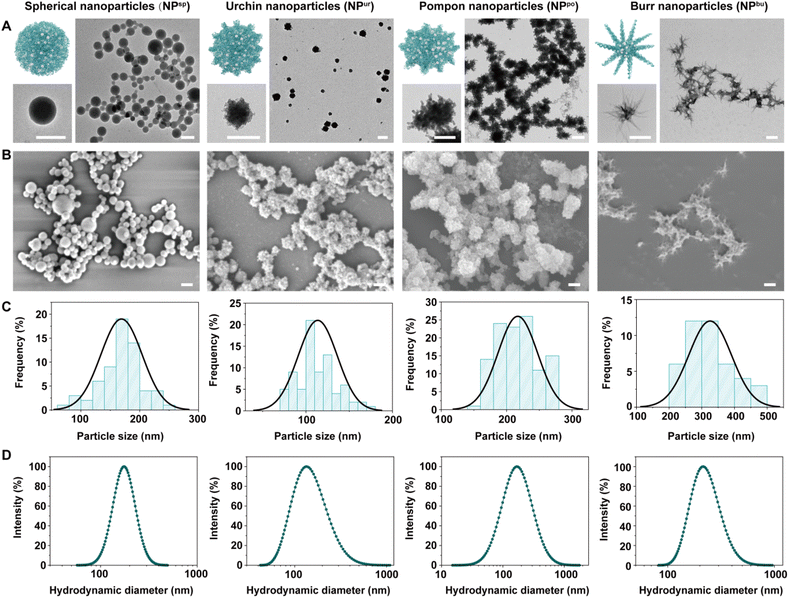
3.1. Preparation and characterization of CpG NPs
CpG NPs were fabricated via a one-pot synthesis method based on the co-assembly of CpG ODNs and ferrous ions. Briefly, the solution of CpG ODNs and FeCl2·4H2O was intensively mixed, heated to 95 °C and left for 2 h, and then cooled to room temperature to obtain the assembled CpG NPs. Various surface topographies were obtained via the regulation of the concentration of assembly materials and the ionic strength of the assembly solvent (Table S1†). We hypothesize that this is due to a corresponding adjustment in the competition and balance of intermolecular forces between the DNA–DNA base complementary pairing and the ferrous ion-DNA coordination.35,36Fig. 1A and B show the surface topography of different samples under transmission electron microscopy (TEM) and scanning electron microscopy (SEM). The diameter distribution of the obtained CpG NPs measured from the TEM results was shown in Fig. 1C, which was 169 ± 35 nm (NPsp), 114 ± 22 nm (NPur), 217 ± 30 nm (NPpo), and 325 ± 65 nm (NPbu), respectively. Fig. 1D shows the dynamic light scattering (DLS) results. Hydrodynamic diameters (Dh) and polydispersity indices (PDIs) of the CpG NPs in ultrapure water were measured as 173 nm (0.20), 132 nm (0.39), 168 nm (0.55), and 216 nm (0.26), respectively (Table S2†). Hence, the obtained CpG NPs have similar sizes (∼200 nm) and relatively narrow size distributions in an aqueous solution. Zeta potential measurement shows that the CpG ODNs were negatively charged while all the CpG NPs were positively charged (Fig. S1†). Furthermore, to test the structural stability of these nanoparticles in aqueous solutions, we tracked their size changes in ultrapure water and PBS buffer. As shown in Fig. S2,† CpG NPs retained their nanoscale diameters after 7 days of room temperature storage in both solutions.
Meanwhile, we monitored the conformational transformation of CpG ODNs during the co-assembly process using circular dichroism (CD) spectroscopy. Free CpG solution showed two negative CD signals at 210 and 250 nm and two positive signals at 220 and 275 nm (Fig. S3A†), which correspond to the characteristic absorption peaks of B-DNA conformation.37,38 With an increase in temperature (from 25 °C to 95 °C), the signal intensity of circular dichroism decreased gradually, but there was no deviation. The characteristic absorption peaks of CpG ODNs were retained in the mixed solution of CpG ODNs and ferrous ions, but the peak intensity dropped to negative values. With an increase in temperature of the mixed solution, the signal intensity decreased continuously, and splits and deviations occurred (Fig. S3B†). This was believed to be caused by the formation of CpG NPs and the reduction of free CpG ODNs. The corresponding ultraviolet-visible spectral results also showed the structural transformation of CpG ODNs and confirmed the successful co-assembly of CpG NPs.
3.2. Cytotoxicity, cellular uptake and subcellular distribution of CpG NPs
To explore the safe dose of CpG NPs for driving an anti-cancer immune response, we tested the cytotoxicity of CpG NPs toward RAW264.7 macrophages and human immortalized keratinocytes (Hacat). First, RAW264.7 macrophages were incubated with a gradient concentration (0–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nM) of free CpG, free FeCl2 and CpG NPs for 24 h. As shown in Fig. S4A,† macrophage viability decreased along with an increase in the sample concentration. Over 90% of RAW264.7 macrophages survived in all test groups at relatively low dosages (<500 nM) of equivalent CpG strands; over 75% of cells survived even at the higher dosage (2000 nM) of equivalent CpG strands. Subsequently, we tested the viability of Hacat incubated in the gradient concentration (0–2000 nM) of the above samples. Similarly, Hacat viability decreased slowly with an increase in the sample concentration, and the cell survival rate was more than 75% at a maximum concentration of 2000 nM (Fig. S4B†). According to these results, we adapted 1000 nM equivalent CpG strands, where cell activity was more than 80% in all sample groups, for the following cellular uptake and immune activation experiment. Fig. S4C† shows the live/dead cell-staining fluorescence images of RAW264.7 macrophages after incubation with CpG NPs at CpG equivalent 1000 nM for 24 h. The proportion of living cells in all sample groups was above 90% except for the pompon-shaped nanostructure (NPpo), further confirming the safety of the selected dose of CpG NPs.
000 nM) of free CpG, free FeCl2 and CpG NPs for 24 h. As shown in Fig. S4A,† macrophage viability decreased along with an increase in the sample concentration. Over 90% of RAW264.7 macrophages survived in all test groups at relatively low dosages (<500 nM) of equivalent CpG strands; over 75% of cells survived even at the higher dosage (2000 nM) of equivalent CpG strands. Subsequently, we tested the viability of Hacat incubated in the gradient concentration (0–2000 nM) of the above samples. Similarly, Hacat viability decreased slowly with an increase in the sample concentration, and the cell survival rate was more than 75% at a maximum concentration of 2000 nM (Fig. S4B†). According to these results, we adapted 1000 nM equivalent CpG strands, where cell activity was more than 80% in all sample groups, for the following cellular uptake and immune activation experiment. Fig. S4C† shows the live/dead cell-staining fluorescence images of RAW264.7 macrophages after incubation with CpG NPs at CpG equivalent 1000 nM for 24 h. The proportion of living cells in all sample groups was above 90% except for the pompon-shaped nanostructure (NPpo), further confirming the safety of the selected dose of CpG NPs.
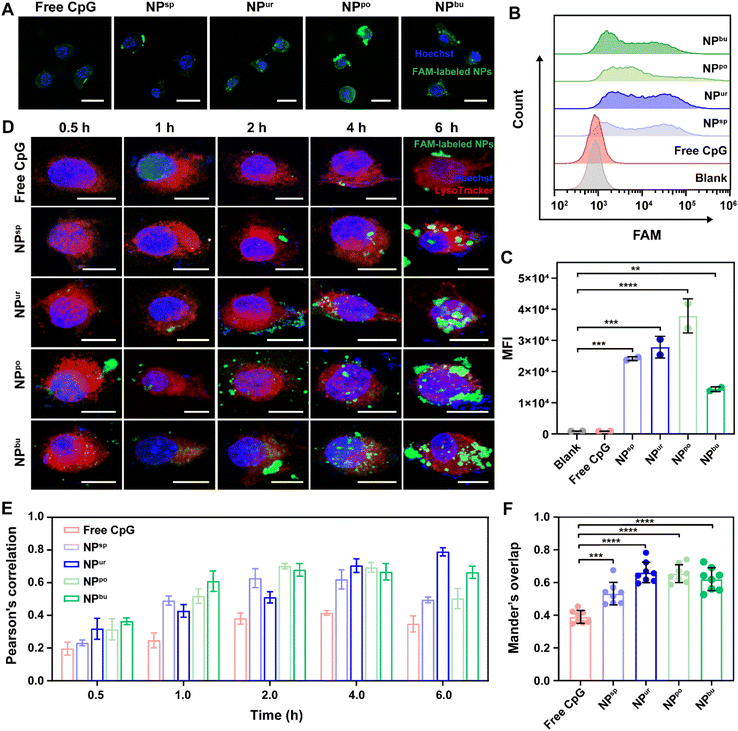
Cellular adhesion and internalization are the initial steps in immune cell activation by CpG NPs. To understand the effect of surface topography on the interaction between NPs and immune cells, the cellular uptake efficiency of CpG NPs in RAW264.7 macrophages was examined by confocal laser scanning microscopy (CLSM) and flow cytometry. First, 6-carboxy-fluorescein (FAM) labeled CpG ODNs were used to prepare fluorescent CpG NPs. Then, RAW264.7 macrophages were exposed to CpG ODNs/NPs in a serum culture medium at a safe dosage of 1000 nM for 4 h. As shown in Fig. 2A and S5,† FAM-labeled CpG ODNs/NPs (green fluorescence) were both observed to accumulate within cells after 4 h of incubation. To quantitatively estimate the amount of sample uptake by cells, we used flow cytometry to measure the fluorescence intensity of intracellular FAM labels. Fig. 2B and C show the mean fluorescence intensity of the internalized samples after 4 h of incubation, which are 914, 921, 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 132, 27
132, 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 848, 37
848, 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 865 and 14
865 and 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 366, respectively. Notably, NPpo exhibited the highest fluorescence intensity among the four CpG NPs, revealing that NPpo significantly increased the cellular uptake efficiency of CpG ODNs. We speculated that NPpo has the optimal specific surface area for cell adhesion and cell uptake. Furthermore, we used NanoLive to observe the real-time cellular uptake behaviors of CpG NPs by capturing a picture every 30 seconds within 2 h. To display the cell uptake process, we next integrated them into a video with 6 frames per second. As shown in Video S1 and S2,† we found that a non-spherical surface structure is more likely to cause the deformation and phagocytosis of macrophages. Together, the self-assembled CpG NPs with desirable surface topography can successfully improve the cellular uptake efficiency of CpG ODNs.
366, respectively. Notably, NPpo exhibited the highest fluorescence intensity among the four CpG NPs, revealing that NPpo significantly increased the cellular uptake efficiency of CpG ODNs. We speculated that NPpo has the optimal specific surface area for cell adhesion and cell uptake. Furthermore, we used NanoLive to observe the real-time cellular uptake behaviors of CpG NPs by capturing a picture every 30 seconds within 2 h. To display the cell uptake process, we next integrated them into a video with 6 frames per second. As shown in Video S1 and S2,† we found that a non-spherical surface structure is more likely to cause the deformation and phagocytosis of macrophages. Together, the self-assembled CpG NPs with desirable surface topography can successfully improve the cellular uptake efficiency of CpG ODNs.
Previous studies have demonstrated that CpG ODNs can be recognized by toll-like 9 receptors (TLR9) on endo/lysosomal membranes after internalization and induce a cascade immune response.39,40 To understand how the self-assembled CpG NPs affect TLR9 recognition to activate immunity, we next tracked their subcellular distributions by CLSM. As depicted in Fig. 2D and S7,† the green fluorescence intensity of FAM-labeled CpG strands increased with the extension of the incubation time of cells in samples. The overlap between FAM-labeled CpG NPs (green) and LysoTracker-labeled lysosomes (red) also changed with incubation time. To quantitatively analyze the association between CpG NPs and lysosomes, Pearson's correlation and Manders' overlap between them were analyzed using the NIS-Elements analysis software (Fig. S8†). As shown in Fig. 2E, Pearson's correlation values of samples increased with time during the first 4 h. After 4 h of incubation, the correlation values of all CpG NPs were higher than the correlation threshold of 0.5, indicating the successful co-localization of CpG NPs and lysosomes.41 To eliminate the influence of fluorescence intensity changes on co-localization parameters, we then used the Manders' overlap as an indicator to assess the co-localization between CpG NPs and lysosomes. It ranges from 0 to 1; zero correlates to no overlap and one correlates to complete overlap. As shown in Fig. 2F, after incubation for 4 h, the co-localization levels between CpG NPs and lysosomes were significantly higher than that between free CpG ODNs and lysosomes. Thus, we speculate that the self-assembled CpG NPs may facilitate immune activation via enhancing cell internalization and interaction with TLR9 on lysosome membranes.
3.3. Innate immune responses of CpG NPs
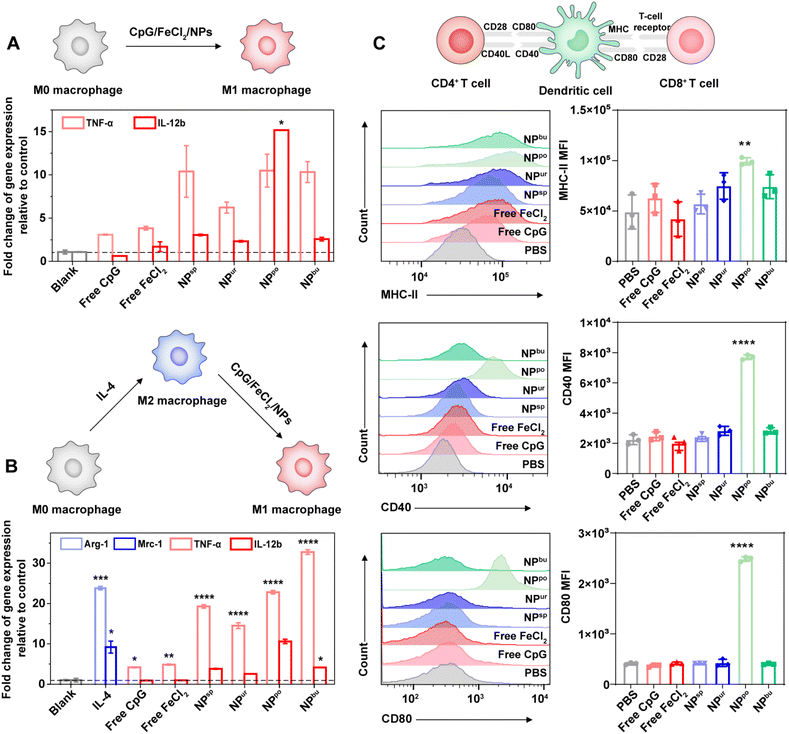
To evaluate the immune potency of the prepared CpG NPs, we detected and compared the innate immune response levels induced by CpG ODNs and CpG NPs. First, we incubated RAW264.7 macrophages with the safe dose (1000 nM) of CpG ODNs and CpG NPs for 20 h and then tested mRNA expression levels of their secreted proinflammatory cytokines. As shown in Fig. 3A, after treatment with CpG NPs, the expression levels of tumor necrosis factor (TNF)-α and interleukin (IL)-12b secreted by RAW264.7 macrophages were significantly higher than those of untreated cells. As the typical markers of inflammatory macrophages (M1 phenotype), expression levels of TNF-α and IL-12b represent levels of immune activation. In comparison to cells treated with free CpG and free FeCl2, CpG NP treated cells secreted more proinflammatory cytokines. This is probably caused by the stronger pro-inflammatory interaction due to the enhanced uptake amount of CpG NPs into cells compared with free CpG. Additionally, NPpo-treated cells secreted the most amount of TNF-α and IL-12b, which was beyond 10 and 15 times that of untreated cells, respectively. This is consistent with the above cellular uptake results. Considering the immunosuppressive microenvironment in tumors, we next analyzed macrophage polarization from the immunosuppressive M2 to the immunostimulatory M1 phenotype in the presence of free CpG, FeCl2 and CpG NPs. Thus, RAW264.7 macrophages were pre-induced to differentiate into the M2 phenotype by IL-4. As revealed in Fig. 3B, after cells were treated with 10 ng mL−1 IL-4 for 20 h, the gene expression levels of arginase (Arg)-1 and mannose receptor C type 1 (Mrc-1) were dramatically increased, indicating their successful differentiation into the M2 phenotype. As expected, a noticeable up-regulation of proinflammatory cytokine (TNF-α and IL-12b) expression was detected after stimulation with our CpG NPs for another 20 h. The expression levels of M2 markers (Arg-1 and Mrc-1) were also tested, showing their over 95% inhibition after incubation with CpG NPs (Table S4†). These results demonstrated that our designed CpG NPs can facilitate macrophage polarization from the immunosuppressive M2 to the immunostimulatory M1 phenotype, and their immune activation potency increased with the increased amount of cellular uptake.Dendritic cells (DCs) present the broadest range of foreign antigens to T cells. Once the NPs are internalized by DCs, it will induce the maturation of DCs and present them to naive helper CD4+ T cells and naive cytotoxic CD8+ T cells in immune organs and then activate them.42,43 Thus, the expression levels of the surface markers of mature DCs, major histocompatibility complex class II (MHC-II), CD40 and CD80, were tested via flow cytometry. After incubation with different samples at a concentration of 1000 nM, NPpo-treated DCs displayed extremely high mean fluorescence intensity (MFI) of MHC-II, CD40 and CD80 compared to those treated with other samples (Fig. 3C). This was probably caused by their enhanced cellular uptake amount. This data indicated that self-assembled CpG NPs with specific surface topography can significantly facilitate the maturation of DCs, thereby promoting the presentation of antigen by activated DCs to naive T cells. Together, our results suggested that the innate immune response was successfully amplified by regulating the surface topography of CpG NPs.
3.4. CpG NPs as a potent adjuvant for enhanced immunotherapy
To investigate the therapeutic efficacy of the CpG NPs as adjuvants of tumor antigens, melanoma-bearing mice were adopted to evaluate the ability of the prepared CpG NPs to inhibit tumor growth. Mice were first subcutaneously inoculated with B16-OVA cells for the construction of a melanoma cancer model. When the tumor volume reached ca. 100 mm3, free CpG, FeCl2, and CpG NPs were mixed with OVA model antigen and injected intramuscularly into the melanoma-bearing mice every 5 days, respectively. Tumor volumes were monitored every certain day as described in Fig. 4A. After a 17-day test, ratios of recruited mature DCs, activated T cells and natural killer (NK) cells in melanoma in different treatment groups were detected by flow cytometry. Gating strategies for them are shown in Fig. S9–S11.† Analysis of DC maturation showed no obvious difference among the experimental groups, except for a slight up-regulation induced by NPpo (Fig. 4B). NK cells are an essential component in the removal of tumors. As shown in Fig. 4C and D, in contrast with free CpG and FeCl2 treated groups, the numbers of activated KLRG1+ NK cells and CD69+ NK cells were greatly enhanced through the treatment of CpG NPs combined with OVA. Specifically, the percentage of KLRG1+ NK cells was separately raised from 38.3% in the PBS-treated group to 79.6%, 67.7%, 83.8% and 79.4% in the four CpG NPs-treated groups. Similarly, the percentage of CD69+ NK cells was separately raised from 26.7% in the PBS-treated group to 76.9%, 65.1%, 83.6% and 78.4% in the four CpG NP-treated groups. In addition, the NPpo-treated group had the highest fold increase with more than two- and three-fold increases in the number of the two NK cells, individually. Regarding activated T cells, CD3+CD8+ T cells were significantly increased in NPur, NPpo, and NPbu-treated groups, but there were no obvious changes in PBS, free CpG, free FeCl2 and NPsp-treated groups (Fig. 4E). Similarly, melanoma-bearing mice treated with NPpo and OVA revealed the highest percentage of activated T cells, about two-fold higher than that by the PBS, OVA plus free CpG, FeCl2 or NPsp treated groups and 1.5-fold higher than that by the OVA plus NPur or NPbu treated groups. Finally, as illustrated in Fig. 4F, the injection of OVA and NPpo almost completely blocked the tumor growth, whereas other treatment groups were much less effective. The immunotherapeutic benefit of OVA and NPpo was evident as early as 10 days after intramuscular injection and continued throughout the entire experimental period (Fig. 4G). In conclusion, NPpo synergistically worked with OVA to effectively promote DC maturation, activate NK cells and T cells in the tumor site, and inhibit tumor growth in melanoma-bearing mice.3.5. Biosafety evaluation
To evaluate the clinical relevance of the prepared CpG NPs, their in vivo biosafety was assessed in healthy mice with the administration of immunostimulatory doses (10 μg per mouse, 3.3 μM equivalent CpG strands). Beforehand, the reactive oxygen species (ROS) in human immortalized keratinocytes (Hacat) were measured because the presence of ferrous ions would generate hydroxyl radicals (˙OH) in a weakly acidic microenvironment.44–47 Hacat was incubated with free CpG, free FeCl2, and CpG NPs (CpG equivalent of 1000 nM) for 24 h. As shown in Fig. S12,† there was a relatively obvious increase in ROS levels in cells incubated with free FeCl2 and four CpG NPs, ranging from 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 to 15
500 to 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. Taking the average ROS level (11
000. Taking the average ROS level (11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 320) of PBS-treated cells as a control group, the ROS levels of free FeCl2-treated cells increased by 33.0% and those of four CpG NPs-treated cells increased by 19.5%, 27.3%, 30.6%, and 19.5%, respectively. The lower ROS levels of Hacat incubated with CpG NPs indicated controllable ROS and low ferroptosis probability in normal cells.
320) of PBS-treated cells as a control group, the ROS levels of free FeCl2-treated cells increased by 33.0% and those of four CpG NPs-treated cells increased by 19.5%, 27.3%, 30.6%, and 19.5%, respectively. The lower ROS levels of Hacat incubated with CpG NPs indicated controllable ROS and low ferroptosis probability in normal cells.
To further assess the in vivo biosafety of CpG NPs, healthy mice were intramuscularly injected with PBS, free CpG, free FeCl2, and self-assembled CpG NPs with an equivalent dose of CpG strands (Fig. 5A). The body weight of mice was monitored after injection. As shown in Fig. 5B, the weight of mice in the experimental groups and the control group exhibited the same trend. All the mice continued to gain weight during the first six days and then lost a little. Ten days after injection, major organs (heart, liver, spleen, lung and kidney) were sectioned and stained for histopathological analysis. As displayed in Fig. 5C, hematoxylin and eosin (H&E) stained histologic sections of mice treated with free CpG, free FeCl2, or CpG NPs showed no difference from those treated with PBS. Moreover, there was no significant difference between each group in the spleen weight (Fig. 5D). The spleen, as the largest immune organ, contains a large number of immune cells. It is the genesis center of cellular immunity and humoral immunity and plays an important role in anti-tumor efficacy. Therefore, mouse spleens were subsequently ground for immunological analysis to further evaluate the systematic immune safety of CpG NPs. Activation markers expressed on the cell membrane, such as CD11b, CD80, and CD86 secreted by DCs and CD3, CD4, CD8, and IFN-γ secreted by T cells were determined by flow cytometry based on the gating strategies shown in Fig. S13–S15†. As depicted in Fig. 5E, the average percentage of mature DCs increased from 15.9% in the PBS-treated group to 19.7%, 18.4%, 21.3% and 22.0% in the free CpG, NPsp, NPpo and NPbu-treated groups, respectively. The proportion of mature DCs was marginally reduced in the other two groups. These negligible changes indicated that CpG NPs have no adverse effect on the immune system of healthy mice. As for activated T cells, except for NPpo, no distinct changes can be observed in other groups. As shown in Fig. 5F, the percentages of central memory T cells (TCM), effector memory T cells (TEM) and interferon-gamma (IFN-γ) in the spleen of NPpo-treated mice increased slightly and remained below 20%, 40% and 6%, respectively; the percentages of CD4 and CD8 molecules on the surface of T cells decreased by 7.3 and 4.1%, respectively. Overall, CpG NPs have no adverse effects associated with autoimmune toxicity or systemic toxicities. These results declared the good biosafety of the prepared CpG NPs, providing their potential as therapeutic vaccine adjuvants for effective tumor immunotherapy.
4. Conclusions
In summary, we successfully constructed CpG NPs with tunable surface topographies by regulating the co-assembly conditions between CpG ODNs and ferrous ions. Compared to free CpG ODNs, the assembled CpG NPs exhibited enhanced cellular uptake behaviors and amplified innate immune responses (macrophage polarization and DC maturation). Significantly, CpG NPs with a pompon-shaped nanostructure (NPpo) showed higher cellular uptake efficiency and immunostimulatory capacity than other nanostructures. Furthermore, when combined with the tumor model antigen—OVA, NPpo showed the strongest anti-tumor efficacy on a B16-OVA melanoma-bearing mouse model, indicating the desired surface topography for an optimal adjuvant effect. Ultimately, in vitro and in vivo systematic toxicity assays indicated the favorable safety of the CpG NPs for biological applications. Together, this study demonstrated that CpG NPs with specific surface topography highly amplified the immune activation performance of the TME, which holds great promise as an effective immunological adjuvant in cancer therapy.Author contributions
Li Zhang: investigation, methodology, validation, formal analysis, data curation, visualization, writing – original draft. Lingpu Zhang: investigation, methodology, validation, formal analysis, data curation, visualization. Yuqi Wang: investigation, resources, writing – review & editing, visualization. Kai Jiang: investigation, resources. Chao Gao: investigation, resources. Pengfei Zhang: writing – review & editing, visualization. Yujie Xie: writing – review & editing, visualization. Bin Wang: writing – review & editing, visualization. Haihua Xiao: resources, writing – review & editing, visualization, supervision. Jie Song: conceptualization, methodology, resources, writing – review & editing, supervision, project administration, funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors are grateful for the financial support from the National Natural Science Foundation of China (No. 22161132008) and the National Key R&D Program of China (No. 2021YFF1200200 and 2022YFA1304500).Notes and references
- M. J. Lin, J. Svensson-Arvelund, G. S. Lubitz, A. Marabelle, I. Melero, B. D. Brown and J. D. Brody, Nat. Cancer, 2022, 3, 911–926 CrossRef CAS PubMed.
- A. York, Nat. Rev. Cancer, 2021, 21, 214–215 CrossRef CAS PubMed.
- J. D. Martin, H. Cabral, T. Stylianopoulos and R. K. Jain, Nat. Rev. Clinic. Oncol., 2020, 17, 251–266 CrossRef PubMed.
- J. Meng, P. Zhang, Q. Chen, Z. Wang, Y. Gu, J. Ma, W. Li, C. Yang, Y. Qiao and Y. Hou, Adv. Mater., 2022, 34, 2202168 CrossRef CAS PubMed.
- S. Liu, Q. Jiang, X. Zhao, R. Zhao, Y. Wang, Y. Wang, J. Liu, Y. Shang, S. Zhao, T. Wu, Y. Zhang, G. Nie and B. Ding, Nat. Mater., 2021, 20, 421–430 CrossRef CAS PubMed.
- B. Zhou, J. Liu, M. Lin, J. Zhu and W. R. Chen, Coordin. Chem. Rev., 2021, 442, 214009 CrossRef CAS.
- B. Pulendran, S. A. P and D. T. O'Hagan, Nat. Rev. Drug Discovery, 2021, 20, 454–475 CrossRef CAS PubMed.
- L.-X. Zhang, J. Hu, Y.-B. Jia, R.-T. Liu, T. Cai and Z. P. Xu, Nanoscale, 2021, 13, 7533–7549 RSC.
- Y. Li, C. Ayala-Orozco, P. R. Rauta and S. Krishnan, Nanoscale, 2019, 11, 17157–17178 RSC.
- J. Ming, J. Zhang, Y. Shi, W. Yang, J. Li, D. Sun, S. Xiang, X. Chen, L. Chen and N. Zheng, Nanoscale, 2020, 12, 3916–3930 RSC.
- R. Khalifehzadeh and H. Arami, Nanoscale, 2020, 12, 9603–9615 RSC.
- G. Del Giudice, R. Rappuoli and A. M. Didierlaurent, Semin. Immunol., 2018, 39, 14–21 CrossRef CAS PubMed.
- N. Hanagata, Int. J. Nanomed., 2017, 12, 515–531 CrossRef CAS PubMed.
- S. R. Paludan and A. G. Bowie, Immunity, 2013, 38, 870–880 CrossRef CAS PubMed.
- A. M. Krieg, Curr. Opin. Immunol., 2000, 12, 35–43 CrossRef CAS PubMed.
- R. N. Hyer and R. Janssen, Open Forum Infect. Dis., 2018, 5, S677–S678 CrossRef.
- S. Schillie, A. Harris, R. Link-Gelles, J. Romero, J. Ward and N. Nelson, Morb. Mortal. Wkly. Rep., 2018, 67(15), 455 CrossRef PubMed.
- M. H. Teplensky, M. Evangelopoulos, J. W. Dittmar, C. M. Forsyth, A. J. Sinegra, S. Wang and C. A. Mirkin, Nat. Biomed. Eng., 2023, 1–17 Search PubMed.
- H. Shan, W. Dou, Y. Zhang and M. Qi, Nanoscale, 2020, 12, 22268–22280 RSC.
- X. Dong, J. Liang, A. Yang, Z. Qian, D. Kong and F. Lv, Biomaterials, 2019, 209, 111–125 CrossRef CAS PubMed.
- J. Zhou, L. Sun, L. Wang, Y. Liu, J. Li, J. Li, J. Li and H. Yang, Angew. Chem., Int. Ed. Engl., 2019, 58, 5236–5240 CrossRef CAS PubMed.
- S. Zhang, Y. Feng, M. Meng, Z. Li, H. Li, L. Lin, C. Xu, J. Chen, K. Hao and Z. Tang, Biomaterials, 2022, 289, 121794 CrossRef CAS PubMed.
- A. Banstola, J.-H. Jeong and S. Yook, Acta biomater., 2020, 114, 16–30 CrossRef CAS PubMed.
- J. Tan, B. Ding, B. Teng, P. a. Ma and J. Lin, Adv. Funct. Mater., 2022, 32, 2111670 CrossRef CAS.
- J. Li, X. Jiang, H. Li, M. Gelinsky and Z. Gu, Adv. Mater., 2021, 33, 2004172 CrossRef CAS PubMed.
- Y. Zhu, H. Liang, X. Liu, J. Wu, C. Yang, T. M. Wong, K. Y. Kwan, K. M. Cheung, S. Wu and K. W. Yeung, Sci. Adv., 2021, 7, eabf6654 CrossRef CAS PubMed.
- J. Wang, H. J. Chen, T. Hang, Y. Yu, G. Liu, G. He, S. Xiao, B. R. Yang, C. Yang, F. Liu, J. Tao, M. X. Wu and X. Xie, Nat. Nanotechnol., 2018, 13, 1078–1086 CrossRef CAS PubMed.
- S. N. Christo, A. Bachhuka, K. R. Diener, A. Mierczynska, J. D. Hayball and K. Vasilev, Adv. Healthcare Mater., 2016, 5, 956–965 CrossRef CAS PubMed.
- A. Chen, L. Wu, Y. Luo, S. Lu, Y. Wang, Z. Zhou, D. Zhou, Z. Xie and J. Yue, Small, 2022, 18, 2200993 CrossRef CAS PubMed.
- L. Xu, X. Wang, W. Wang, M. Sun, W. J. Choi, J. Y. Kim, C. Hao, S. Li, A. Qu, M. Lu, X. Wu, F. M. Colombari, W. R. Gomes, A. L. Blanco, A. F. de Moura, X. Guo, H. Kuang, N. A. Kotov and C. Xu, Nature, 2022, 601, 366–373 CrossRef CAS PubMed.
- L. A. Dykman, S. A. Staroverov, A. S. Fomin, V. A. Khanadeev, B. N. Khlebtsov and V. A. Bogatyrev, Int. Immunopharmacol., 2018, 54, 163–168 CrossRef CAS PubMed.
- K. Lee, Z. N. Huang, C. A. Mirkin and T. W. Odom, Nano Lett., 2020, 20, 6170–6175 CrossRef CAS PubMed.
- M. Li, C. Wang, Z. Di, H. Li, J. Zhang, W. Xue, M. Zhao, K. Zhang, Y. Zhao and L. Li, Angew. Chem., Int. Ed. Engl., 2019, 58, 1350–1354 CrossRef CAS PubMed.
- J. Zhang, Z. Di, H. Yan, Y. Zhao and L. Li, Nano Lett., 2021, 21, 2793–2799 CrossRef CAS PubMed.
- H. Sigel, Biol. Trace Elem. Res., 1989, 21, 49–59 CrossRef CAS PubMed.
- H. Sigel and R. Griesser, Chem. Soc. Rev., 2005, 34, 875–900 RSC.
- F. R. Steven Daly and V. Gabelica, Science, 2020, 368, 1465–1468 CrossRef PubMed.
- B. Ranjbar and P. Gill, Chem. Biol. Drug Des., 2009, 74, 101–120 CrossRef CAS PubMed.
- U. Ohto, T. Shibata, H. Tanji, H. Ishida, E. Krayukhina, S. Uchiyama, K. Miyake and T. Shimizu, Nature, 2015, 520, 702–705 CrossRef CAS PubMed.
- D. M. Klinman, Nat. Rev. Immunol., 2004, 4, 249–258 CrossRef CAS PubMed.
- K. W. Dunn, M. M. Kamocka and J. H. McDonald, Am. J. Physiol. Cell Physiol., 2011, 300, C723–C742 CrossRef CAS PubMed.
- S. K. Wculek, F. J. Cueto, A. M. Mujal, I. Melero, M. F. Krummel and D. Sancho, Nat. Rev. Immunol., 2020, 20, 7–24 CrossRef CAS PubMed.
- R. L. Sabado, S. Balan and N. Bhardwaj, Cell Res., 2017, 27, 74–95 CrossRef CAS PubMed.
- H. Ranji-Burachaloo, P. A. Gurr, D. E. Dunstan and G. G. Qiao, ACS Nano, 2018, 12, 11819–11837 CrossRef CAS PubMed.
- Z. Tang, Y. Liu, M. He and W. Bu, Angew. Chem., Int. Ed. Engl., 2019, 58, 946–956 CrossRef CAS PubMed.
- Z. Tang, P. Zhao, H. Wang, Y. Liu and W. Bu, Chem. Rev., 2021, 121, 1981–2019 CrossRef CAS PubMed.
- F. Zhang, F. Li, G. H. Lu, W. Nie, L. Zhang, Y. Lv, W. Bao, X. Gao, W. Wei, K. Pu and H. Y. Xie, ACS Nano, 2019, 13, 5662–5673 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3na00322a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |