Open Access Article
Open Access ArticleOptical, structural, and biological properties of silver nanoclusters formed within the loop of a C-12 hairpin sequence†
Akhilesh Kumar
Gupta
a,
Nolan
Marshall
a,
Liam
Yourston
a,
Lewis
Rolband
b,
Damian
Beasock
b,
Leyla
Danai
b,
Elizabeth
Skelly
b,
Kirill A.
Afonin
 b and
Alexey V.
Krasnoslobodtsev
b and
Alexey V.
Krasnoslobodtsev
 *a
*a
aDepartment of Physics, University of Nebraska at Omaha, Omaha, NE 68182, USA. E-mail: akrasnos@unomaha.edu; Tel: +1402-554-3723
bNanoscale Science Program, Department of Chemistry, University of North Carolina at Charlotte, Charlotte, NC 28223, USA
First published on 12th June 2023
Abstract
Silver nanoclusters (AgNCs) are the next-generation nanomaterials representing supra-atomic structures where silver atoms are organized in a particular geometry. DNA can effectively template and stabilize these novel fluorescent AgNCs. Only a few atoms in size – the properties of nanoclusters can be tuned using only single nucleobase replacement of C-rich templating DNA sequences. A high degree of control over the structure of AgNC could greatly contribute to the ability to fine-tune the properties of silver nanoclusters. In this study, we explore the properties of AgNCs formed on a short DNA sequence with a C12 hairpin loop structure (AgNC@hpC12). We identify three types of cytosines based on their involvement in the stabilization of AgNCs. Computational and experimental results suggest an elongated cluster shape with 10 silver atoms. We found that the properties of the AgNCs depend on the overall structure and relative position of the silver atoms. The emission pattern of the AgNCs depends strongly on the charge distribution, while all silver atoms and some DNA bases are involved in optical transitions based on molecular orbital (MO) visualization. We also characterize the antibacterial properties of silver nanoclusters and propose a possible mechanism of action based on the interactions of AgNCs with molecular oxygen.
1 Introduction
Silver nanoclusters (AgNCs) are ultra-small metal nanoclusters consisting of just a few atoms of silver. The AgNCs represent supra-atomic metal assemblies where ligands stabilizing these supra-atomic structures influence the shapes and sizes of the AgNCs. One class of AgNCs is the DNA stabilized hybrid nanostructures, which involve cytosine-rich single-stranded sequences encapsulating silver. The optical properties of DNA-stabilized AgNCs (AgNC@DNA) are dictated by their size, shape, and stoichiometry.1 AgNCs comprising only a few silver atoms exhibit molecule-like optical properties where the continuous density of electronic energy states breaks up into discrete levels.2–9 High fluorescence quantum yield, easy synthetic procedure, and biocompatibility afforded by DNA make AgNC@DNA an attractive alternative to conventional fluorophores in the development of sensing modalities for the detection of heavy metals,10 low concentration levels of miRNA,11,12 cellular imaging,13 and various DNA sequences.14–16 The DNA template is essential for modulating various properties of AgNC@DNAs. DNA sequences not only contribute to the stabilization of AgNCs but also perturb the electronic states of the AgNC@DNA, dictating the absorption and fluorescence patterns observed.17–20 Various DNA sequences can be utilized to tune the optical properties of AgNC by adjusting their emission color in the UV,19,20 visible,19,21 and NIR parts of the electromagnetic spectrum.1,8,22Among the possible DNA sequences capable of templating optically active AgNCs are single stranded DNA,1,23 hairpin loops,24–26 i-motif,27,28 and G-quadruplexes.29,30 Hairpin loop structures tend to form a cytosine-rich pocket suitable for the formation of stable AgNCs.25,26,31 Our understanding of AgNCs' properties can be improved by complementing experimental results with computational studies.32 Both experimental and computational studies have already contributed to our understanding of the shape and size of the AgNC@DNA within the templating DNA sequence.33,34 However, structural details of the AgNC@DNA are still missing despite a few X-ray crystallography studies identifying the exact structures of different AgNC@DNA.8,9 Computational studies can bridge the gap in experimental techniques which still cannot reach the level of materials comprised of only a few atoms. Computational studies are expected, therefore, to uncover precise contributions of nucleobases to the structure and optical properties of AgNC@DNA. The intimate connection between the silver of the nanocluster and DNA nucleobases is important but poorly understood.35 Therefore, studying the details of DNA–silver contacts in AgNC@DNA is critical to further our understanding, ability to control various properties of AgNCs, and advancing their practical applications. One such practical application is found in the antibacterial activity of AgNC@DNA. We have previously demonstrated that CN hairpin loops of different sizes exhibit variable antibacterial activity.26 While active towards inhibiting the growth of bacteria, these AgNCs remain non-toxic to mammalian cells, suggesting a selective antibacterial mode of action. The degree of antibacterial activity was found to be insensitive to the size of the loop but linked to the optical response of AgNC@hpCN.26
In this study, we combine experimental and computational studies to further understand the structural and photophysical properties of AgNCs formed on hairpin looped structure with 12 cytosines in the loop (AgNCs@hpC12). Our results suggest that 10 atoms are involved in the formation of optically active AgNCs. Different involvement of 12 cytosines in the loop was identified as critical, important, or non-essential for the properties of the templated AgNCs. The connection of optical properties and biological properties of AgNC@hpC12 is further explored.
2 Experimental section
2.1 AgNCs@DNA synthesis
Oligonucleotides were purchased from Integrated DNA Technologies after having been desalted and lyophilized. Upon receipt, oligonucleotides were resuspended to a 100 μM concentration and used without additional purification. A single stranded DNA template, ssC12, was used as reported previously.20 A hairpin loop DNA template, hpC12, with the following sequence was used: 5′-TATCCGTCCCCCCCCCCCCACGGATA. The sequence and the length were chosen due to its well-documented ability to form fluorescent AgNCs.25 Different stem sequences of hpC12 were observed to have minimal effect on the binding sites in the C12 loop (refer to Fig. S4†). To prepare the AgNCs@DNA, 100 μM DNA template, double-deionized water (ddiH2O, ∼18 MΩ), 1 mM AgNO3, and 100 mM NH4OAc (pH 6.9) were combined to achieve the following final concentrations: CDNA = 10 μM, CAgNO3 = 120 μM, CNH4Ac = 4 mM. We followed a previously published protocol where the number of Ag+ was adjusted to match the number of cytosines in the loop according to n·AgNO3:Cn.26 Solutions were then vortexed, centrifuged, and incubated at 95 °C for 2 minutes. These solutions were quickly transferred to an ice bath and incubated for 20 minutes. NaBH4 solution was prepared fresh by dissolving NaBH4 granules in chilled water to a concentration of 10 mM and stored on ice until ready for use. The AgNC@DNA samples were then treated with NaBH4 using an equimolar amount as the Ag+. Silver control solution was prepared identically as the AgNC@DNA samples, with ddiH2O replacing the DNA template. The concentration of AgNO3 and DNA for AgNC synthesis were optimized by varying concentrations independently while observing fluorescence intensity and pattern in the EEM, Fig. S1.† Following the synthesis, all solutions were stored at 4 °C away from light for approximately 16 hours. To remove any excess silver from the AgNC@DNA solutions, they were washed three times with a total of 1.2 mL of 4 mM NH4OAc using 3 kDa molecular weight cut-off filters by centrifuging three times at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rcf at 4 °C. The final spin was continued until ∼50 μL of AgNC@DNA solution remained after approximately 25 minutes.
000 rcf at 4 °C. The final spin was continued until ∼50 μL of AgNC@DNA solution remained after approximately 25 minutes.
2.2 Fluorescence measurements
The excitation and emission spectra were acquired on a Duetta – Fluorescence and Absorbance Spectrometer (Horiba, Inc., Chicago, IL, USA). In all the measurements, the concentration of the templating sequence was kept the same at ∼10 μM. Fluorescence measurements were carried out in a Sub-Micro Fluorometer Cell, model 16.40F-Q-10 (from StarnaCells, Inc., Atascadero, CA, USA), at room temperature of ∼22 °C. The excitation–emission matrix spectra (EEMS) were recorded with 0.5 nm resolution. Fluorescence spectra were recorded with the emission wavelength range from 400 nm to 850 nm, the initial excitation wavelength was set to 350 nm, and the final excitation wavelength was set to 700 nm with an increment of 0.5 nm. Matrix data were then used for 2D contour plots using MagicPlot Pro software.2.3 Bacterial growth inhibition assays
K12 E. coli were grown, from single colonies, in lysogeny broth (LB) at 37 °C with 200 rpm constant shaking overnight in a GeneMate Incubated Shaker. The following morning, bacterial cultures were diluted to an optical density at 600 nm (OD600) between 0.016–0.019 using LB. 50 μL of diluted bacteria were added to a sterile 96-well black-walled plate. AgNC@hpC12, following the removal of excess silver, were added for a final concentration of 3 μM. For the Ag-control solutions, 6 μL were added to each well, replicating the same amount of silver that would be present in the AgNC@hpC12 solutions if all the silver had bound to the DNA templates. Carbenicillin was added as a positive control for a final concentration of 50 μg mL−1. In the buffer control wells, 10 μL of 4 mM NH4OAc buffer, pH 6.9, was added. All wells were adjusted to a final volume of 100 μL using LB. The lid of the plate was hydrophobically treated by filling it with 10 mL of 20% ethanol, 0.05% Triton-X100 for 30 seconds before the excess was poured off. The lid was then allowed to dry upright, in a biosafety cabinet for at least 30 minutes. The plate and lid were sealed with parafilm prior to microplate optical density measurements to prevent excess evaporation. Optical density measurements were obtained using a Tecan Spark microwell plate reader, with measurements taking place every 15 minutes at 600 nm for 22 hours. The temperature inside the plate reader was set to 37 °C, and the plate was shaken for 30 seconds after every measurement. A total of five biological repeats, with four technical repeats each, were completed. The growth curves are shown as the average OD600 at each time point with the standard error of the mean (SEM) of each measurement shown as a dotted line above and below each solid line in the same color. The mean and SEM of each growth curve was calculated using GraphPad Prism 9.2.4 Mammalian cell viability assays
HEK293-FT, HeLa, and MDA-MB-231 cell lines were maintained in Dulbecco's Modified Eagle Medium (DMEM, Gibco) containing 100 U per mL penicillin, 100 μg per mL streptomycin, and 10% heat-inactivated fetal bovine serum, at 37 °C and 5% CO2. Cells were seeded in 96-well plates at a density of 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well 24 hours prior to the introduction of treatments. The media was removed from each well and replaced with fresh media containing a final concentration of 3 μM AgNC@hpC12, 6 μL of Ag-control, or 10 μL of 4 mM NH4OAc buffer, pH 6.9 where indicated. The plates were then incubated for 24 hours at 37 °C, 5% CO2. Following this incubation, 20 μL of CellTiter 96® Aqueous One Solution Cell Proliferation Assay (MTS) were added to each well and returned to incubate at the same conditions for 75 minutes. The absorbance of 490 nm light was read for each well using a Tecan Spark microplate reader, with 16 reads per well being averaged to make the final value. Results are reported as the average of four biological repeats with three technical repeats each ± the standard deviation (SD). The mean and SD of each assay was calculated using GraphPad Prism 9.
000 cells per well 24 hours prior to the introduction of treatments. The media was removed from each well and replaced with fresh media containing a final concentration of 3 μM AgNC@hpC12, 6 μL of Ag-control, or 10 μL of 4 mM NH4OAc buffer, pH 6.9 where indicated. The plates were then incubated for 24 hours at 37 °C, 5% CO2. Following this incubation, 20 μL of CellTiter 96® Aqueous One Solution Cell Proliferation Assay (MTS) were added to each well and returned to incubate at the same conditions for 75 minutes. The absorbance of 490 nm light was read for each well using a Tecan Spark microplate reader, with 16 reads per well being averaged to make the final value. Results are reported as the average of four biological repeats with three technical repeats each ± the standard deviation (SD). The mean and SD of each assay was calculated using GraphPad Prism 9.
2.5 Atomic force microscopy imaging
AFM imaging of AgNCs@DNA was performed on MultiMode AFM Nanoscope IV system (Bruker Instruments, Santa Barbara, CA, USA) in tapping mode. Briefly, 5 μL of the AgNC@DNA solution were deposited on amino-propyl-silatrane (APS) modified mica for a total of 2 min.19,20,36 Excess sample was washed with DI water and gently dried under a flow of high purity argon gas and under vacuum overnight. AFM images in air were then recorded with a 1.5 Hz scanning rate using TESPA-300 probes from Bruker Nano, Inc. The probes have ∼320 kHz resonance frequency and a spring constant of about 40 N m−1. Images were processed and analyzed using Fiji (ImageJ) and plotted with MagicPlot Pro software. AFM imaging could be challenging method for unexperienced researchers due to specific surface chemistry required for reliable deposition of AgNC@DNA.36 Other alternative methods to assess heterogeneity of the samples might be considered, such as reversed-phase high-performance liquid chromatography (HPLC),37 gel electrophoresis,38 or Transmission Electron Microscopy (TEM).392.6 Energy-dispersive X-ray spectroscopy (EDS) analysis
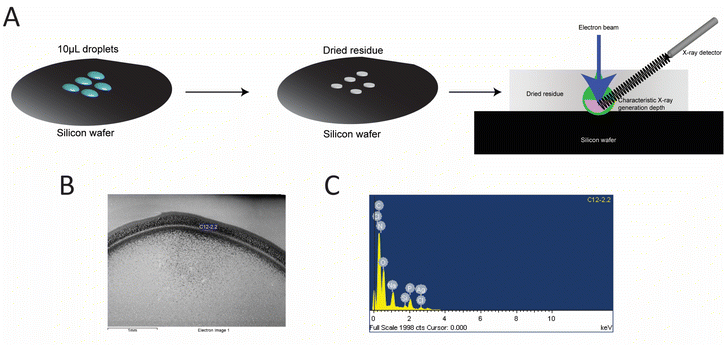
AgNC@hpC12 solutions prepared as described above were diluted to 50 μM after filtration. These solutions were deposited as two 10 μL droplets onto a polished Si wafer and allowed to dry overnight. Droplet residue was analyzed with a JEOL JSM 6480 SEM and an Oxford Instruments INCA EDS system. The probe energy was 5 keV. Atomic concentrations were calculated by the INCA software from P Kα and Ag Lα characteristic X-rays. Because of the variable drying pattern observed, sample data with a low substrate signal (<5% atomic percentage) was used for the statistical analysis. This limits data to droplet residue of sufficient thickness where nearly all the signal originates from the sample.2.7 Computational details of AgNCs construction and geometry optimization
The initial model of Ag10NC@hpC12 was constructed and edited in molecular editor Avogadro. All DFT calculations, including DFT geometry refinement and TD-DFT were performed in Gaussian 16. The hpC12 was constructed first and optimized with the Universal Force Field (UFF). Silver atoms were added to the loop connecting with N3 heteroatoms of cytosines in the loop at positions 1–5 and 7–11 (5′ to 3′) and optimized with UFF. The Ag–N3 bonding was maintained at positions 1–3, 9 and released at positions 4, 5, 7, 8, 10, 11 according to T-scanning (see Results section for details). Next, the geometry of the entire construct was optimized with the UFF in Avogadro with a 10 × 10−7 convergence factor. The geometry was further refined using the ONIOM method implemented in Gaussian 16. The “low-level” layer (DNA hairpin) remained geometrically “frozen”, while “high-level” layer (all silver atoms) were subjected to optimization using DFT with M062X density functional and LANL2DZ basis set. TD-DFT calculations were performed using the optimized structures to compute the nanoclusters' ground-to excited-state optical transitions simulating absorption spectrum of the AgNCs. TD-DFT calculations used M06-2X density functional and LANL2DZ basis set for calculations of both ground- and excited-states. Additionally, a solvent environment was simulated with the solvation model based on density (SMD) for water. Up to 96 excited states were calculated to cover the energy span of the visible and UV (up to 5 eV).3 Results and discussion
3.1 Cytosine-rich ssDNA templates
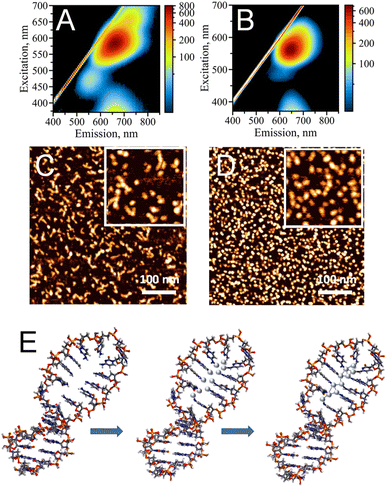
Single-stranded (ss), cytosine-rich DNA oligonucleotides have been demonstrated to be the most suitable capping agents for templating stable AgNC@DNA due to high affinity of cytosine's N3 heterocyclic atom to silver ions, Ag+.3,4 To date, various sequences have been reported to stabilize AgNCs with unique optical properties, including bright emission bands in the visible part of the spectrum.1 Distinct colors (e.g., blue, green, red) of AgNC@DNA have been observed based on prevalent emission wavelengths for a particular nanocluster20 making AgNC@DNA an exciting new emissive supra-atomic nanostructure suitable for bioimaging,40,41 biosensing,10,12 and even a bioactive material with antibacterial properties.26,42While single-stranded C-rich sequences have high affinity to silver ions, they can also form alternative non-canonical DNA structures. For example, a duplex stabilized by silver in a C–Ag–C pairing rather than canonical Watson–Crick pairing,43 or the i-motif where semi-protonated C-rich oligonucleotides are arranged in a tetra-stranded structure.44 Our previously reported data suggest that the formation of alternative DNA structures via strong silver mediated C–Ag–C base paring results in structural heterogeneity of AgNC@DNA populations.20 The uncontrollable template driven assembly of AgNCs leads to a heterogeneous population of the observed optical peaks (Fig. 1A) also suggesting the formation of various sizes and shapes of AgNCs as confirmed by AFM imaging (Fig. 1C). The challenge of AgNC@DNA's heterogeneity can be solved by rational design of the template involving, for example, hairpin-loop structures rather than sequences containing cytosine-rich ssDNAs (Fig. 1B). C-rich hairpin loop template is different from the simple single C-rich strand because it has less chances to form other secondary structures of DNA mediated by silver ions. While still providing the binding sites for Ag+, the loops also protect silver atoms while coordinating and trapping them. Atomic force microscopy imaging validated that the DNA hairpin-loop design removes structural ambiguity and yields a uniform functional population of the AgNC@DNA (Fig. 1D). AFM image analysis produced an almost ideal distribution of sizes for AgNC@hpC12 (Fig. S2A†). While AgNC@ssC12 template also produces similar to AgNC@hpC12 monomeric size, there is also a significant presence of larger structures as evident from the statistical distribution for AgNC@ssC12 complex shown in Fig. S2B.† In addition to structural homogeneity, AgNCs templated on hpC12 loops consistently produce an almost ideal single peak of “red” fluorescence with the maximum excitation wavelength at λEXC = 560 nm and maximum emission wavelength at λEM = 635 nm (Fig. 1B). This single peak, termed RI, also suggests the formation of a more homogeneous population of single AgNC type in terms of the emissive nanocluster's shape, size, and charge state. Although single, this RI emission peak for AgNC@hpC12 slightly extends to longer wavelengths as excitation increases. This observation implies a possibility of a small degree of AgNC heterogeneity or, alternatively, the shift might be associated with the red edge excitation shift phenomenon for this emissive RI state.19,20,45 Notably, both AgNC@ssC12 and AgNC@hpC12 exhibit emission observed for excitation wavelengths below 400 nm which can be attributed to the characteristic broad UV-excited emission commonly observed for DNA-templated AgNCs.18–20 Since the hpC12 looped templates resulted in more homogenous populations of AgNCs, we have focused on further understanding of AgNC fluorophores and their optical, structural, and biological properties using hpDNA structures.46,47Fig. 1E shows schematically the binding of silver atoms to N3 atom of C12 hairpin loop structure and subsequent formation of AgNC upon reduction with sodium borohydride.
3.2 Role of individual cytosines in the loop
The number of cytosines in the loop defines the fluorescence pattern of the AgNC@DNA (Fig. S3†).25,26 This number also dictates the size of the loop within which AgNC is hosted. hpC12 contains twelve cytosines and can potentially bind up to 12 silver ions considering a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
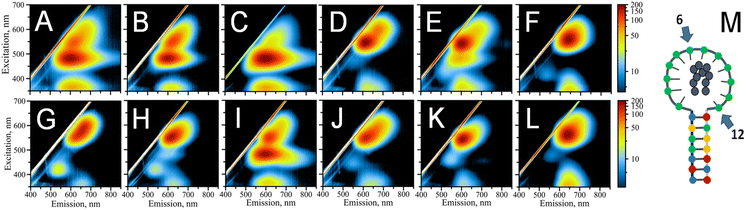
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry. The key factors in defining the photophysical properties of the clusters are the intimate interactions between silver atoms of the nanocluster and DNA ligand-bases.35 To probe the role of individual cytosines in the hpC12 loop for the stabilization of AgNC, we have replaced one cytosine at a time with a thymine base at all twelve positions. Thymine has the lowest binding affinity to silver48 and it does not coordinate the AgNC when the cluster is finally formed upon reduction.35 Furthermore, thymidine can be enzymatically removed from the DNA backbone without disturbing the environment of the cluster.49 Therefore, the C → T replacement was aimed at pinpointing the cytosines which are critical in stabilizing and coordinating AgNC. Unlike reducing the number of cytosines in the loop, which also reduces the size of the loop,25,26 C → T replacement preserves the same overall size of the loop, directly probing the effect of a particular cytosine base, especially since the size of thymine is similar to that of cytosine. The final composition of the resultant loop remained 12 bases in total “C11 + T1” with varying positions of the single thymine. The variants were termed T1–T12, with the number indicating thymine's position starting from the 5′ end of the loop next to the double-stranded stem. Fig. 2M shows a schematic of AgNC@hpC12, and Fig. 2A–L show the resultant excitation-emission maps for all twelve “C11 + T1” modified hpDNAs. Thymine single base replacement revealed the differences in the fluorescence pattern of AgNCs@hpDNA exhibited as obvious changes to the main RI peak, always very distinct for the AgNC@hpC12 loop. Two samples, T6 and T12, resemble single red peaks typical for the AgNC@hpC12 loop, suggesting that these two positions in the loop are less important in stabilizing the AgNCs as shown in Fig. 2F and L, respectively. The rest of the T-scans show various degrees of variation from the single RI peak. For example, the red peak is blue-shifted by ∼10 nm compared to T6, T12 variants, and the unmodified hpC12 loop. This peak has λEM = 620 nm and λEXC = 550 nm and is termed “red II” = RII to differentiate from “red I” = RI peak observed for the intact AgNC@hpC12. The most dramatic differences are observed for T1, T2, T3, and T9, where EEMs show dual peak featuring RII peak and dominating “orange” = O peak with emission at λMAX = 600 nm and excitation at λMAX = 485 nm, Fig. 2A–C, and I, respectively. This dramatic change in the fluorescence pattern suggests critical involvement of C1, C2, C3, and C9 bases in stabilization of AgNC. The rest of the variants, positions 4, 7, 8, 10, and 11, are all variations of the RII peak with only minor shifts of maxima for both excitation and emission. Perhaps, the only other position that features a substantial “orange” shoulder is T5. Positions 7 and 8 both have a faint green peak at λEXC/λEM = 420/535 nm. Interestingly, the excitation-emission pattern seems to repeat itself in an N + 6 manner. T3 is similar to T9, T4 to T10, T5 to T11, and T6 to T12. T6 and T12 samples form almost identical pattern as the intact AgNC@hpC12. Exceptions are T1–T7 and T2–T8 pairs for which the resemblance is minimal.
1 stoichiometry. The key factors in defining the photophysical properties of the clusters are the intimate interactions between silver atoms of the nanocluster and DNA ligand-bases.35 To probe the role of individual cytosines in the hpC12 loop for the stabilization of AgNC, we have replaced one cytosine at a time with a thymine base at all twelve positions. Thymine has the lowest binding affinity to silver48 and it does not coordinate the AgNC when the cluster is finally formed upon reduction.35 Furthermore, thymidine can be enzymatically removed from the DNA backbone without disturbing the environment of the cluster.49 Therefore, the C → T replacement was aimed at pinpointing the cytosines which are critical in stabilizing and coordinating AgNC. Unlike reducing the number of cytosines in the loop, which also reduces the size of the loop,25,26 C → T replacement preserves the same overall size of the loop, directly probing the effect of a particular cytosine base, especially since the size of thymine is similar to that of cytosine. The final composition of the resultant loop remained 12 bases in total “C11 + T1” with varying positions of the single thymine. The variants were termed T1–T12, with the number indicating thymine's position starting from the 5′ end of the loop next to the double-stranded stem. Fig. 2M shows a schematic of AgNC@hpC12, and Fig. 2A–L show the resultant excitation-emission maps for all twelve “C11 + T1” modified hpDNAs. Thymine single base replacement revealed the differences in the fluorescence pattern of AgNCs@hpDNA exhibited as obvious changes to the main RI peak, always very distinct for the AgNC@hpC12 loop. Two samples, T6 and T12, resemble single red peaks typical for the AgNC@hpC12 loop, suggesting that these two positions in the loop are less important in stabilizing the AgNCs as shown in Fig. 2F and L, respectively. The rest of the T-scans show various degrees of variation from the single RI peak. For example, the red peak is blue-shifted by ∼10 nm compared to T6, T12 variants, and the unmodified hpC12 loop. This peak has λEM = 620 nm and λEXC = 550 nm and is termed “red II” = RII to differentiate from “red I” = RI peak observed for the intact AgNC@hpC12. The most dramatic differences are observed for T1, T2, T3, and T9, where EEMs show dual peak featuring RII peak and dominating “orange” = O peak with emission at λMAX = 600 nm and excitation at λMAX = 485 nm, Fig. 2A–C, and I, respectively. This dramatic change in the fluorescence pattern suggests critical involvement of C1, C2, C3, and C9 bases in stabilization of AgNC. The rest of the variants, positions 4, 7, 8, 10, and 11, are all variations of the RII peak with only minor shifts of maxima for both excitation and emission. Perhaps, the only other position that features a substantial “orange” shoulder is T5. Positions 7 and 8 both have a faint green peak at λEXC/λEM = 420/535 nm. Interestingly, the excitation-emission pattern seems to repeat itself in an N + 6 manner. T3 is similar to T9, T4 to T10, T5 to T11, and T6 to T12. T6 and T12 samples form almost identical pattern as the intact AgNC@hpC12. Exceptions are T1–T7 and T2–T8 pairs for which the resemblance is minimal.
The change in the fluorescence pattern of the variants suggests that cytosines in positions 6 and 12 do not participate in the formation of the “red” AgNC@hpC12. We propose that C6 and C12 do not make any contact with the cluster and positioned far from making such contacts possible. The rest of the cytosines are critically involved in stabilization of AgNC@hpC12 either through direct or indirect bonding with cytosines. Direct bonding might take place via Ag to N3 heteroatom of cytosine, while indirect bonding might take place via water bridges between Ag or other N heteroatoms of cytosines and O. Both types of bonding are feasible and have been demonstrated to be involved in cluster stabilization via X-ray crystallography.8,9
3.3 EDS studies of Ag/DNA stoichiometry in the nanocluster
Additional confirmation of AgNCs size was obtained using the EDS analysis. EDS was performed to quantify the number of silver atoms bound to the hpC12 template. The samples were purified to remove any unbound silver from solution and were deposited onto a silicon wafer for analysis (Fig. 3). The ratio of the relative atomic percentages of the Ag and P in the EDS spectrum were used for evaluating the stoichiometric ratio of silver per hpC12. Multiple readings were collected and averaged to provide that each AgNC@hpC12 binds an average of 10.3 ± 0.6 silver atoms averaged over 13 independent measurements, Table S1†. We conclude, therefore, that the number of cytosines in the loop does not correspond to the number of silver atoms in the bound AgNCs. The EDS measurements indicated that on average there are ∼10 atoms of silver per 12 cytosines in each AgNC@hpC12. This cluster size is consistent with previously reported data on similar sized hairpin loop AgNC@DNAs.35 We have recently used various loop sizes between C7–C13 for modulating antibacterial activity of AgNC@DNA26 and, interestingly, all the studied loop sizes C7–C13 contained ∼10–11 silver atoms in the final AgNC composition.263.4 Model of AgNC@hpC12
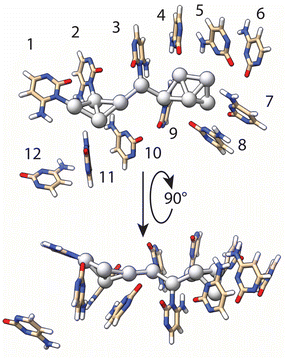
Using the results of T-scanning to examine the importance of each single cytosine in the formation of the AgNC, we have built a model of AgNC@hpC12 intended to reflect Ag–C interactions within the loop in order to use it further for DFT computations. Both the fluorescence pattern and intensity of the fluorescence remained unchanged for the T6 and T12 variants (Fig. 2F and L) as compared with the intact AgNC@hpC12 (Fig. 1B). Therefore, we inferred that cytosines C6 and C12 do not directly participate in the cluster coordination. It is apparent from the inspection of the C → T replacement in Fig. 2 that positions 1, 2, 3, and 9 produce the largest variation in the fluorescence. The fluorescence pattern now resembles C11 loop (Fig. S3†) – the size with one less cytosine further corroborating the importance of the cytosines at these four positions. The “blue” shifts of the main red peak for the T4, T5, T7, T8, T10, and T11 variants (Fig. 2) also suggest that contacts of cytosines in positions 4, 5, 7, 8, 10, and 11 are important for cluster coordination but perhaps not as critical as 1, 2, 3, 9. Therefore, we ranked the importance of the cytosine bases in stabilization of AgNCs based on the effect they produced in T-scanning and grouped them into the following three groups: group I (C1, C2, C3, and C9) > group II (C4, C5, C7, C8, C10, and C11) > group III (C6 and C12). This ranking was also utilized in geometry optimization of the AgNC@hpC12 described below. Additionally, SEM-EDS measurements identified that 10 silver atoms are bound to the hpC12. Cytosines of the loop bind and concentrate silver ions before the reduction step. In the first stage of the theoretical modeling, we chose to coordinate each silver atom with group I and group II cytosines, C1–C5 and C7–C11, constituting an elongated chain of the complexed ions ligated with the N3 heteroatoms of cytosines. This assumption is reasonable based on well-established affinity of Ag+ to N3 of cytosines. The model at this step resembles a zig-zag type of structure with silver atoms facing inside the C12 loop structure. The reduction step induces dispersing and clustering of the silver by competing between the process of the DNA chelation and silver agglomeration.50 Partial reduction of the Ag10 construct reorganizes the DNA host as well as the shape of the cluster itself. Next, for the geometry optimization, we imposed the following restrictions: (i) silver atoms near C1, C2, C3, C9 remained directly linked to N3 heteroatom of cytosines with Ag–N distance kept between 2.0–2.1 Å; (ii) binding of the silver atoms near C4, C5, C7, C8, C10, C11 nucleobases was relaxed to allow for more positional freedom in the optimization step; and (iii) neighboring silver atoms were forced to maintain metal–metal bonds (2.7–2.8 Å), matching the Ag–Ag distances observed in crystal structures of AgNCs.8 This initial structure was then subjected to multiple runs of geometry optimization using the Universal Force Field (UFF) at the Molecular Mechanics level of theory. Final refinement of the structure was performed by fixing the geometry of DNA and allowing the optimization of the embedded cluster structure with a single run using DFT level of theory (see details in ESI†). The final optimized model structure of the full AgNC@hpC12 is shown in Fig. S5.†Fig. 4 shows the final geometry of the cluster and all 12 cytosines of the loop while the sugar-phosphate backbone was removed for clarity. The structure of Ag10NC resembles very well the “Big Dipper” structure obtained for Ag8NC using X-ray crystallography.9 At this stage of optimization, the silver atoms maintained multiple contacts with the cytosines at the beginning of the loop anchoring the cluster. The loop forms a framed pocket of cytosines for the core of the AgNCs to be stably accommodated. Pseudo-trapezoidal Ag5 moiety is located within the pocket of the loop surrounded by the cytosines C4, C5, C6, C7, and C8 with C6 rotated out and making no evident contact with any of the silver atoms. The distance between Ag and N3 of the cytosines in the pocket is relaxed during optimization to larger distances ranging between 2.88 to 3.53 Å. The shortest distance, 2.88 Å, was observed between C5 and the corresponding Ag atom. Interestingly, the replacement of C5 → T5 also resulted in the noticeable appearance of an “orange” peak with significant intensity suggesting more critical role of C5 as compared to other members of the second group of cytosines (group II). Two other remarkably close distances are between the tip of the cluster and carbonyl O2 atoms of the C7 and C8 cytosines in the loop. These two bonds, 2.54 Å and 3.05 Å for C7 and C8 respectively, were not imposed but were formed during the geometry optimization. Such distances were observed in crystal structure of AgNC@DNA for Ag–O2 stabilizing bonds.8 Furthermore, at 2.54 Å, this distance is close to a typical Ag–O coordination bond observed in metal–organic frameworks (MOF).51 Both C7 and C8 have common signature when replaced with thymine (Fig. 2G and H), they both exhibit clear green peaks at λEXC/λEM = 420/535 nm suggesting similar effect on optical properties of Ag10 when Ag–O2 interaction is destabilized. Recently obtained crystal structures of AgNC@DNA show critical involvement of water molecules and an extensive network of hydrogen bonds between O2, N7, and silver in stabilization of the cluster.8 Although these interactions are not obvious in our model (Fig. 4), a more careful inspection suggests that NH2 groups and O2 carbonyl atoms are positioned favorably to form such networks protruding into the minor and major grooves. We can assume, thus, that our model structure comes very close to the expected real structure, although verification of its exact correctness might require further studies involving mass-spectrometry and X-ray crystallography in the future. The very small size of AgNC@DNA in the range of only a few silver atoms presents a difficult task in studying the exact structures of the nanoclusters. A limited number of studies have shown resolved structures of AgNCs including X-ray crystallography8,9 and computational reports.34 Further studies are expected to uncover more details of structural control and reproducibility, making it possible to regulate properties of the nanoclusters both wanted (fluorescence wavelength and intensity) and unwanted (stability).3.5 Optical transitions of the Ag10NC@hpC12
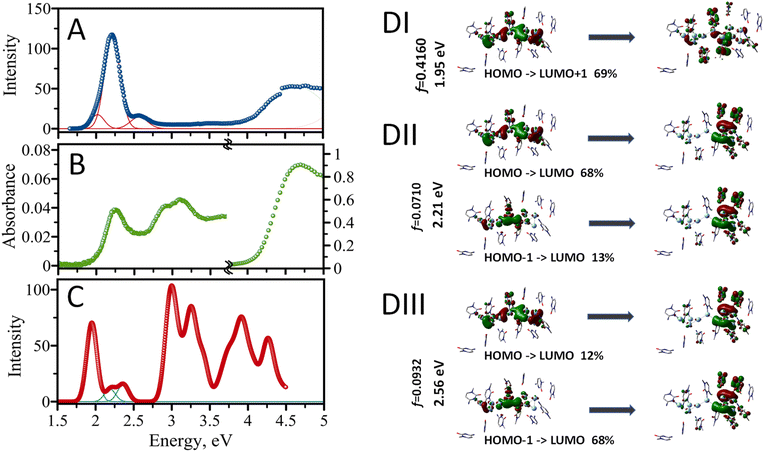
The experimental excitation spectrum of the AgNC@hpC12 collected in both visible and UV ranges is shown in Fig. 5A. Several obvious peaks in the spectrum spanning the UV-vis range indicate that multiple optical transitions are allowed for the nanocluster. The UV range, 4–5 eV, is an almost continuous spectrum of excitations without any obvious maxima where many distinct transitions are possible (Fig. 5A). Previously, UV excitation for AgNCs was in part assigned to an efficient energy transfer from DNA to AgNC.18 Next, there is a very small, almost undetectable, peak with the maximum at 3.54 eV at the border of UV and visible ranges (350 nm). The visible range is dominated by one intense peak with the maximum at E = 2.21 eV. This peak corresponds to the “red” RI emission peak observed in the EEM at λEXC/λEM = 560/635 nm. The other obvious feature is a shoulder to the right of the main peak at higher energy values. If fitted with the Gaussian function, this shoulder peaks at 2.56 eV, which corresponds to λEXC/λEM = 485/525 nm band in the EEM. A clear asymmetric character of the red peak's excitation shows a longer tail towards the lower energy values on the left side of the 2.21 eV maximum. Such appearance is associated with red edge emission shift (REES) which was previously observed in several cases for AgNCs.19,20,26 REES usually emerges when the motion of solvent molecules is restricted and is not capable of providing fast enough relaxation of excited fluorophore to the lowest vibrational state, such as low temperature or high viscosity.45 This observation suggests complexity of the AgNC's environment inside the hpC12 loop which may cause certain unusual optical behavior of the nanoclusters such as better stability, shifts of the maxima, etc.19 The absorption spectrum shown in Fig. 5B closely resembles the experimental excitation spectrum (Fig. 5A). The spectrum's UV portion is dominated by DNA absorption with a maximum at E = 4.7 eV (∼260 nm). The absorption due to DNA is large, and it was necessary to zoom into the visible region ∼100× times to highlight the characteristics of the spectrum in the visible part (1.5–4.5 eV). The visible part features a prominent peak at E = 2.21 eV. This peak coincides with the main peak in the excitation spectrum and corresponds to the excitation of the main fluorescence transition for the AgNC@hpC12. Next, there are obvious peaks in the range between 2.7 and 3.5 eV, most likely corresponding to the absorption by silver and silver nanostructures.52 Although these peaks are not obvious in the excitation spectrum, they become apparent in the computed spectrum as shown in Fig. 5C.The structural model of AgNC@hpC12 created with the help of T-scanning results and geometry optimization allowed us to evaluate the absorption spectrum using the TD-DFT approach.52 A partial positive charge is expected for the AgNC upon reduction with NaBH4, and it was previously proposed that a positive charge is critical for the existence of the AgNC@DNA.53 Therefore, we begin by placing the smallest possible positive charge, p = +2, on the entire optimized structure of AgNC@hpC12, and increased it by an increment of 2 (p = +4, p = +6, and p = +8). Odd overall positive charges, p = +1, +3, +5, +7, and +9, lead to a triplet electronic configuration and thus were discarded from consideration as such transition would be spin forbidden. Simple considerations using a superatom model suggest that the Ag10NCs would be stable if they contained 2, 8, 18, 20, … electrons on their shared orbitals.54 Since each Ag atom can contribute one electron (from [Kr]4d105s1 configuration), the combination of 8Ag0 and 2Ag+ in the cluster (Ag102+) results in a singlet state with closed-shell electronic configuration.55 For the calculation of the absorption spectrum, the sugar-phosphate backbone was removed, and only cytosine bases of the hpC12 were considered (Fig. 4). Ninety-six singlet excited states were then calculated using TD-DFT with M06-2X/LANL2DZ combination of the functional/basis set. ONIOM procedures in Gaussian 16 allowed to treat the AgNC and the cytosine bases separately as first set of atoms (high-level layer) and second set of atoms (low-level layer). We assign a +2 charge for the Ag10NC leaving cytosine bases uncharged. The calculated absorption energies confirm that many different transitions are allowed for a single nanocluster, including several transitions in the visible and multiple in UV region (Fig. 5C). The first main peak is observed at 1.95 eV (635 nm), and two more transitions are obvious in the visible range at 2.21 eV (560 nm) and 2.36 eV (525 nm), which blend into a continuous shoulder of the main peak at spectral broadening of σ = 0.09 eV (Fig. 5B). Many closely positioned transitions then follow these three peaks at higher energies and in the UV region resembling the experimental excitation spectrum with a multitude of UV peaks.
The calculated energy of the main first transition, 1.95 eV, is not the same as the experimentally observed 2.21 eV for the maximum of excitation wavelengths. This difference of the underestimate, ΔE = 0.26 eV, is quite acceptable considering the possible structural deviations of the nanocluster. Overestimates or underestimates of the calculated transitions as compared with experimental absorption values are often reported in the literature and associated with the choice of an exchange functional.56–58 Our choice of M06-2X functional was justified by previously reported vertical transition energy values close to the experimental ones for AgNCs34 and other complex molecules.58 Next, we have compared the computed results using two other common functionals, B3LYP and B3PW91, and found that the use of these functional underestimates the energy of transitions even more than the chosen M06-2X (ESI Table 1†). Additionally, we used water with Solvation Model based on Density (SMD) for our computations, while it may not be the best fit for the dielectric environment of AgNC inside the hpC12 loop. Restrictive diffusion and complex interplay of the interactions between Ag and nucleobases may result in less polar environment shifting, thus, the actual spectrum to lower energy values (hyperchromic effect) as observed previously for solvents of various polarities59 or water/methanol mixtures.60 Modeling SMD solvent as benzene for our AgNC@hpC12 results in a shift the maximum of the first transition to 1.92 eV, cyclohexane to 1.91 eV, acetonitrile to 1.86 eV. These factors can also affect the quantum yield of the emitting species.61 Another support for our conclusion that the calculated spectrum is red-shifted altogether is the energy of the second prominent transition. If the entire calculated spectrum is shifted by 0.3 eV to lower energy values, computed peaks coincide very well with the experimental excitation ones matching 2.25 eV (550 nm – “RII”) and 2.56 eV (485 nm – “O”) as indicated in Fig. 6C.
Ag108+ (2Ag0 and 8Ag+) is another expected stable configuration according to the superatom model with 2 electrons filling 2S orbital. 2 (S), 8 (2S + 6P), 18 (2S + 6P + 10D), … “magic number” rule applies to symmetric spherical or near-spherical metal nanoclusters where degeneracy of P and D orbitals are expected.32 Although we have focused primarily on the superatom model assumption for symmetric near-spherical metal nanoclusters, we acknowledge that our Ag10NC@hpC12 model suggests an elongated shape of the nanocluster with several elements including a zig-zag handle and a clustered head. The structural asymmetry of the Ag10NC@hpC12 cluster will most likely remove the degeneracy of the higher level, P and D, orbitals. Without this degeneracy requiring 6ē to completely fill the P shell of Ag10NC, magic numbers 2 and 8 are no longer needed for stability. For example, intermediate Ag104+ (6Ag0 and 4Ag+) and Ag106+ (4Ag0 and 6Ag+) configurations will also yield stable singlet states. It is worth noting that experimentally observed “magic numbers” for DNA-templated AgNCs are N0 = 6 and N0 = 4.21N0 = 4 emits primarily in “green” and N0 = 6 emits in “red”, with both structures containing various numbers of silver ions in their composition.21 Interestingly, all charged species of the Ag10NC@hpC12 cluster (Ag102+, Ag104+, Ag106+, Ag108+) have similar positions of major bands in the computed absorption spectrum (ESI Table 2†), suggesting a real possibility for a “magic” arrangement of silver atoms, Ag104+, with NAg0 = 6 as the core and NAg+ = 4. The visible absorption spectra for all the charged species show the presence of the three peaks observed for Ag102+, albeit with different relative intensities as indicated by the oscillator strengths of the corresponding transitions between ground and excited states. We notice that the increase in overall positive charge of the Ag10@hpC12 results in a slight shift to higher energy values for all three observed transitions: RI, RII, and O (ESI Table 2†).
Silver belongs to group 11 metals with the outermost valence 5s electrons being extremely delocalizable. The electron delocalization and complexation with DNA ligands are expected to result in uneven charge distribution. It is becoming apparent that not all the silver in a cluster might be actively participating in the MO building. Some comprise a neutral core, some comprise a positively charged shell. It has been recently suggested that the core of the “luminescent” cluster may be different from the size of the full AgNC.34 Further visualization of molecular orbitals involved in the visible transitions confirmed the highly delocalized properties of the AgNC@hpC12. Fig. 5D (I–III) shows molecular orbitals involved in the three identified visible transitions. It is evident from Fig. 5D, all of the MOs involved have a delocalized nature. For example, the transition at 1.95 eV is primarily HOMO to LUMO+1 transition. HOMO's electron density is smeared over all the silver atoms of the AgNC and even spills over to nucleobases. Small contributions can be observed on cytosines 1, 2, 3, 4, 5, 7, 8, 9, 10, and 11. The excited state LUMO+1 also has delocalized density but with larger involvement of the cytosines, particularly in positions 1, 2, 3, 7, 9, and 10. This observation coincides well with our previous statement that C1, C2, C3, and C9 are critically involved in the optical properties of AgNCs, and replacing them with thymines will surely alter the distribution of electron density and thus optical transitions, as indicated by T-scans (Fig. 2A–C, and 2I).
3.6 Biological activity of AgNC@hpC12
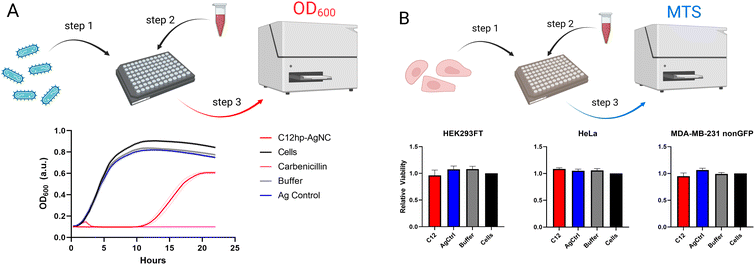
AgNCs as supra-atomic structures have been reported to possess a variety of interesting properties besides obvious optical beneficial behavior. We have previously reported that AgNCs encapsulated in hairpin loops exhibit antibacterial activity while remaining non-toxic to human cell lines.26 Four different sizes of the loops (C7, C9, C11, C13) resulted in varied bioactivity of AgNCs.26 We expected the AgNC@hpC12 to be also quite bioactive and capable of significantly slowing down the growth of bacteria.To assess the differences in bacterial growth between treatments, the time it takes each culture to reach half its maximal optical density is calculated, along with its 95% confidence interval using a non-linear fit of the data. The AgNC@hpC12, at a concentration of 3 μM, was able to significantly slow the growth of K12 E. coli in liquid cultures (Fig. 6A). There was a minimal difference between the buffer treated cells, Ag control-treated cells, and the untreated bacteria culture, which all reached half their maximum growth after approximately 4.3 hours. The E. coli treated with the AgNC@hpC12, however, did not reach half of their maximum growth until 15.2 ± 0.2 hours had passed. From the growth curves, it is also apparent that the treatment with AgNC@hpC12 lowered the maximum amount of bacterial growth in the culture, as the untreated cells reached an OD600 of approximately 0.9 while the AgNC@hpC12 treated bacteria were only able to achieve an OD600 of 0.6 over 22 hours. This same concentration of AgNC@hpC12, 3 μM, was found to not decrease the viability of three different mammalian cell lines that were tested (Fig. 6B). These results are consistent with that of our previously reported work using similar AgNC@DNA hairpin structures.26 The non-toxic nature of AgNC@DNAs is critical to their potential advancement in biomedical applications.
Although it is evident that AgNCs@hpC12 possess antibacterial activity, it is not obvious what the mechanism of such activity is. So far, various mechanisms have been proposed, including oxidative release of Ag+ similar to AgNPs.62 We have previously outlined several possible explanations of such useful bioactivity.26 The ability of AgNC@DNA to act as antibacterial agent might be related to the composition of AgNCs which have both silver atoms (Ag0) and silver ions (Ag+). Further oxidation can change the ratio of Ag+/Ag0 modulating the antibacterial activity of AgNC@DNA. Another possibility is related to AgNCs' reactivity towards oxygen. AgNCs react with oxygen which is apparent when AgNCs age. Molecular oxygen, O2, is inert and exists in the ground stable triplet state. Metal nanoclusters with closed shell configurations, forming singlet spin states, are expected to be inert towards molecular oxygen.63 For such a reaction to proceed – a spin accommodation is required.63 We hypothesize that AgNC@hpC12 is capable of such a spin accommodation and thus gets oxidized with molecular oxygen (aging) and H2O2 which promotes AgNC@hpC12 to a “dark” state (Fig. S6†). It has been previously reported that nanostructured silver can generate large amounts of intracellular reactive oxygen species contributing to the antibacterial activity of AgNCs.64 Both theoretical32 and experimental21 studies indicate that certain shape, composition, and charge states of AgNC@DNA can form optically “dark” states with high-spin multiplicity. We have computed several excited triplet states for the model of AgNC@hpC12. Indeed, several triplet states have energies comparable to the energies of visible transitions. Fig. S7† shows a comparative diagram of three main singlet state transitions and three computed triplet state transitions found in the same visible region. Each singlet excited state of AgNC@hpC12 appears to have a corresponding triplet excited state suggesting the possibility of intersystem crossing from *SN to *TN. Therefore, it is reasonable to propose that the singlet excited state of AgNCs may interact with abundant triplet oxygen stimulating the transition of triplet to singlet oxygen, 3O2 → 1O2, while converting from 1(AgNC) to 3(AgNC) via an accelerated intersystem crossing. Efficient intersystem crossing with the help of molecular oxygen has been recently demonstrated,65 and molecular oxygen is known to accelerate S → T transition in fluorophores.66 In reaction with triplet molecular oxygen, the generation of singlet 1O2* is allowed whenever the energy gap between S → T exceeds 0.98 eV.65 This amount of energy is required for the conversion from 3O2 to 1O2,67 and has been observed experimentally in optical spectra of oxygen.68,69 It is tempting to hypothesize that *S3 → *T1 intersystem crossing shown in Fig. S5† with energy gap of 0.96 eV is close enough to 0.98 eV (ΔE ≈ 0.02 eV), considering computational error, and that such transition can be accelerated according to the following reaction:
| *S3(Ag10NC) + 3O2 = *T1(Ag10NC) + 1O2 |
Since singlet oxygen is far more reactive as compared to triplet oxygen, this can explain why AgNC@DNA is toxic to bacteria.26 While it is a reasonable hypothesis, more details will be needed to further clarify the role of ISC in ROS formation and the contribution of this pathway to cell viability.
4 Conclusions
Structural, optical, and antibacterial properties were evaluated both experimentally and computationally for AgNCs formed using C12 hairpin loop DNA structure as a template. Both experimental and computational results indicate that the hpC12 loop templates the formation of a 10-atom AgNC within the loop. Not all the cytosines in the loop participate equally in stabilizing the AgNCs. At least three different roles of cytosines were identified. Four cytosines are critically involved in stabilization of the cluster and its optical properties, six cytosines are important for stabilization but not critical for its optical properties, and two cytosines don't participate in the formation of AgNCs. Geometrically optimized structure of Ag10NCs@hpC12 in the shape of “Big Dipper” suggests that the two parts of the nanocluster handle and pseudo-trapezoidal head might contribute differently to optical and biological properties of AgNCs. Molecular orbital visualization shows almost equal electron density distribution spread over all silver atoms with small contributions from DNA bases. The degree of contribution from the DNA bases matches very well the roles of cytosines identified in optical experimental measurements. Computational analysis suggests the richness of possible electronic transitions, which may contribute to a variety of colors observed for AgNC with well-defined peaks in the red, orange, and green parts of the visible spectrum. We observe reasonable agreement of computational results with experimental suggesting the close match of the optimized geometry to the expected one. The high antibacterial activity of Ag10NC@hpC12 identified in this study is most likely associated with the charged character of AgNC and its ability to interact with environmental oxygen generating harmful to bacteria singlet oxygen species.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Research reported in this publication was in part supported by the National Science Foundation, Division of Material Research: Award Numbers DMR-2203946 (to K. A. A.) and DMR-2204027 (to A. V. K.); by ACS PRF # 58498-UNI5 (to A. V. K.); and by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R35GM139587 (to K. A. A.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors would like to thank Dr Ryan Riskowski (UNO) for allowing us to use the Duetta Spectrometer. AFM scans were obtained with the use of MultiMode Nanoscope IV system at the Nanoimaging Core Facility, UNMC, and we thank Dr Alexander Lushnikov (NCF-UNMC) for his assistance in sample preparation for AFM imaging. Also, we would like to thank Alora Dunnavant (UNC Charlotte) for her help with preparations for the bacterial growth assays.References
- A. Gonzàlez-Rosell, C. Cerretani, P. Mastracco, T. Vosch and S. M. Copp, Nanoscale Adv., 2021, 3(5), 1230–1260 RSC.
- C. M. Ritchie, K. R. Johnsen, J. R. Kiser, Y. Antoku, R. M. Dickson and J. T. Petty, J. Phys. Chem. C, 2007, 111(1), 175–181 CrossRef CAS PubMed.
- J. T. Petty, J. Zheng, N. V. Hud and R. M. Dickson, J. Am. Chem. Soc., 2004, 126(16), 5207–5212 CrossRef CAS PubMed.
- S. Y. New, S. T. Lee and X. D. Su, Nanoscale, 2016, 8(41), 17729–17746 RSC.
- M. Chandler, O. Shevchenko, J. L. Vivero-Escoto, C. D. Striplin and K. A. Afonin, J. Chem. Educ., 2020, 97(7), 1992–1996 CrossRef CAS.
- P. R. O'Neill, E. G. Gwinn and D. K. Fygenson, J. Phys. Chem. C, 2011, 115(49), 24061–24066 CrossRef.
- T.-H. Lee, J. I. Gonzalez, J. Zheng and R. M. Dickson, Acc. Chem. Res., 2005, 38(7), 534–541 CrossRef CAS PubMed.
- C. Cerretani, H. Kanazawa, T. Vosch and J. Kondo, Angew. Chem., Int. Ed., 2019, 58(48), 17153–17157 CrossRef CAS PubMed.
- D. J. E. Huard, A. Demissie, D. Kim, D. Lewis, R. M. Dickson, J. T. Petty and R. L. Lieberman, J. Am. Chem. Soc., 2019, 141(29), 11465–11470 CrossRef CAS PubMed.
- L. Yourston, P. Dhoqina, N. Marshall, R. Mahmud, E. Kuether and A. V. Krasnoslobodtsev, Processes, 2021, 9(10), 1699 CrossRef CAS.
- P. Shah, A. Rørvig-Lund, S. B. Chaabane, P. W. Thulstrup, H. G. Kjaergaard, E. Fron, J. Hofkens, S. W. Yang and T. Vosch, ACS Nano, 2012, 6(10), 8803–8814 CrossRef CAS PubMed.
- L. E. Yourston and A. V. Krasnoslobodtsev, Molecules, 2020, 25(13), 3026 CrossRef CAS PubMed.
- S. Choi, J. Yu, S. A. Patel, Y. L. Tzeng and R. M. Dickson, Photochem. Photobiol. Sci., 2010, 10(1), 109–115 CrossRef PubMed.
- J. M. Obliosca, C. Liu, R. A. Batson, M. C. Babin, J. H. Werner and H.-C. Yeh, Biosensors, 2013, 3(2), 185–200 CrossRef CAS PubMed.
- H.-C. Yeh, J. Sharma, J. J. Han, J. S. Martinez and J. H. Werner, Nano Lett., 2010, 10(8), 3106–3110 CrossRef CAS PubMed.
- W. Guo, J. Yuan, Q. Dong and E. Wang, J. Am. Chem. Soc., 2010, 132(3), 932–934 CrossRef CAS PubMed.
- M. Ganguly, C. Bradsher, P. Goodwin and J. T. Petty, J. Phys. Chem. C, 2015, 119(49), 27829–27837 CrossRef CAS PubMed.
- I. L. Volkov, Z. V. Reveguk, P. Y. Serdobintsev, R. R. Ramazanov and A. I. Kononov, Nucleic Acids Res., 2018, 46(7), 3543–3551 CrossRef CAS PubMed.
- L. Yourston, L. Rolband, C. West, A. Lushnikov, K. A. Afonin and A. V. Krasnoslobodtsev, Nanoscale, 2020, 12(30), 16189–16200 RSC.
- L. E. Yourston, A. Y. Lushnikov, O. A. Shevchenko, K. A. Afonin and A. V. Krasnoslobodtsev, Nanomaterials, 2019, 9(4), 613 CrossRef CAS PubMed.
- S. M. Copp, D. Schultz, S. Swasey, J. Pavlovich, M. Debord, A. Chiu, K. Olsson and E. Gwinn, J. Phys. Chem. Lett., 2014, 5(6), 959–963 CrossRef CAS PubMed.
- S. M. Copp and A. Gonzàlez-Rosell, Nanoscale, 2021, 13(8), 4602–4613 RSC.
- M. Chandler, O. Shevchenko, J. L. Vivero-Escoto, C. D. Striplin and K. A. Afonin, J. Chem. Educ., 2020, 97(7), 1992–1996 CrossRef CAS.
- K. A. Afonin, D. Schultz, L. Jaeger, E. Gwinn and B. A. Shapiro, Methods Mol. Biol., 2015, 1297, 59–66 CrossRef CAS PubMed.
- P. R. O'Neill, L. R. Velazquez, D. G. Dunn, E. G. Gwinn and D. K. Fygenson, J. Phys. Chem. C, 2009, 113(11), 4229–4233 CrossRef.
- L. Rolband, L. Yourston, M. Chandler, D. Beasock, L. Danai, S. Kozlov, N. Marshall, O. Shevchenko, A. V. Krasnoslobodtsev and K. A. Afonin, Molecules, 2021, 26(13), 4045 CrossRef CAS PubMed.
- C. M. Ritchie, K. R. Johnsen, J. R. Kiser, Y. Antoku, R. M. Dickson and J. T. Petty, J. Phys. Chem. C, 2007, 111(1), 175–181 CrossRef CAS PubMed.
- T. Li, N. He, J. Wang, S. Li, Y. Deng and Z. Wang, RSC Adv., 2016, 6(27), 22839–22844 RSC.
- J. Zhu, L. Zhang, Y. Teng, B. Lou, X. Jia, X. Gu and E. Wang, Nanoscale, 2015, 7(31), 13224–13229 RSC.
- G. Tao, Y. Chen, R. Lin, J. Zhou, X. Pei, F. Liu and N. Li, Nano Res., 2018, 11(4), 2237–2247 CrossRef CAS.
- Y. Guo, F. Shen, Y. Cheng, H. Yu, Y. Xie, W. Yao, R. Pei, H. Qian and H.-W. Li, J. Phys. Chem. B, 2020, 124(9), 1592–1601 CAS.
- K. L. D. M. Weerawardene, H. Häkkinen and C. M. Aikens, Annu. Rev. Phys. Chem., 2018, 69(1), 205–229 CrossRef CAS PubMed.
- S. M. Swasey, N. Karimova, C. M. Aikens, D. E. Schultz, A. J. Simon and E. G. Gwinn, ACS Nano, 2014, 8(7), 6883–6892 CrossRef CAS PubMed.
- Z. V. Reveguk, V. A. Pomogaev, M. A. Kapitonova, A. A. Buglak and A. I. Kononov, J. Phys. Chem. C, 2021, 125(6), 3542–3552 CrossRef CAS.
- Y. Zhang, C. He, J. T. Petty and B. Kohler, J. Phys. Chem. Lett., 2020, 11(21), 8958–8963 CrossRef CAS PubMed.
- A. V. Krasnoslobodtsev, in Supramolecular Nanotechnology, 2023, pp. 823–844, DOI:10.1002/9783527834044.ch30.
- E. Gwinn, D. Schultz, S. M. Copp and S. Swasey, Nanomaterials, 2015, 5(1), 180–207 CrossRef PubMed.
- V. Packirisamy and P. Pandurangan, BioTechniques, 2022, 74(1), 51–62 CrossRef PubMed.
- D. Fredrick, L. Yourston and A. V. Krasnoslobodtsev, Luminescence, 2023, 1 DOI:10.1002/bio.4471.
- D. Lyu, J. Li, X. Wang, W. Guo and E. Wang, Anal. Chem., 2019, 91(3), 2050–2057 CrossRef CAS PubMed.
- M. Yang, X. Chen, Y. Su, H. Liu, H. Zhang, X. Li and W. Xu, Front. Chem., 2020, 8, 601621 CrossRef CAS PubMed.
- S. Javani, R. Lorca, A. Latorre, C. Flors, A. L. Cortajarena and A. Somoza, ACS Appl. Mater. Interfaces, 2016, 8(16), 10147–10154 CrossRef CAS PubMed.
- S. M. Swasey, L. E. Leal, O. Lopez-Acevedo, J. Pavlovich and E. G. Gwinn, Sci. Rep., 2015, 5(1), 10163 CrossRef CAS PubMed.
- H. Day, C. Huguin and Z. Waller, Chem. Commun., 2013, 49, 7696–7698 RSC.
- J. R. Lakowicz and S. Keating-Nakamoto, Biochemistry, 1984, 23(13), 3013–3021 CrossRef CAS PubMed.
- L. E. Yourston and A. V. Krasnoslobodtsev, Molecules, 2020, 25(13), 3026 CrossRef CAS PubMed.
- L. Rolband, L. Yourston, M. Chandler, D. Beasock, L. Danai, S. Kozlov, N. Marshall, O. Shevchenko, A. V. Krasnoslobodtsev and K. A. Afonin, Molecules, 2021, 26(13), 4045 CrossRef CAS PubMed.
- D. Schultz, K. Gardner, S. S. R. Oemrawsingh, N. Markešević, K. Olsson, M. Debord, D. Bouwmeester and E. Gwinn, Adv. Mater., 2013, 25(20), 2797–2803 CrossRef CAS PubMed.
- J. T. Petty, M. Ganguly, A. I. Yunus, C. He, P. M. Goodwin, Y.-H. Lu and R. M. Dickson, J. Phys. Chem. C, 2018, 122(49), 28382–28392 CrossRef CAS.
- J. T. Petty, D. Lewis, S. Carnahan, D. Kim and C. Couch, J. Phys. Chem. B, 2022, 126(21), 3822–3830 CrossRef CAS PubMed.
- M. J. Alhilaly, R.-W. Huang, R. Naphade, B. Alamer, M. N. Hedhili, A.-H. Emwas, P. Maity, J. Yin, A. Shkurenko, O. F. Mohammed, M. Eddaoudi and O. M. Bakr, J. Am. Chem. Soc., 2019, 141(24), 9585–9592 CrossRef CAS PubMed.
- A. Singh, S. Jha, G. Srivastava, P. Sarkar and P. Gogoi, Int. J. Sci. Technol. Res., 2013, 2, 153–157 Search PubMed.
- J. T. Petty, O. O. Sergev, M. Ganguly, I. J. Rankine, D. M. Chevrier and P. Zhang, J. Am. Chem. Soc., 2016, 138(10), 3469–3477 CrossRef CAS PubMed.
- H. Häkkinen, Adv. Phys.: X, 2016, 1(3), 467–491 Search PubMed.
- A. Gonzàlez-Rosell, R. Guha, C. Cerretani, V. Rück, M. B. Liisberg, B. B. Katz, T. Vosch and S. M. Copp, J. Phys. Chem. Lett., 2022, 8305–8311, DOI:10.1021/acs.jpclett.2c02207.
- F. Rabilloud, Eur. Phys. J. D, 2013, 67(1), 18 CrossRef.
- G. U. Kuda-Singappulige and C. M. Aikens, J. Phys. Chem. C, 2021, 125(45), 24996–25006 CrossRef CAS.
- M. Kowalczyk, N. Chen and S. J. Jang, ACS Omega, 2019, 4(3), 5758–5767 CrossRef CAS PubMed.
- Z. Lei, X.-K. Wan, S.-F. Yuan, J.-Q. Wang and Q.-M. Wang, Dalton Trans., 2017, 46(11), 3427–3434 RSC.
- I. Díez, M. Pusa, S. Kulmala, H. Jiang, A. Walther, A. S. Goldmann, A. H. E. Müller, O. Ikkala and R. H. A. Ras, Angew. Chem., Int. Ed., 2009, 48(12), 2122–2125 CrossRef PubMed.
- A. Marini, A. Muñoz-Losa, A. Biancardi and B. Mennucci, J. Phys. Chem. B, 2010, 114(51), 17128–17135 CrossRef CAS PubMed.
- X. Tian, X. Jiang, C. Welch, T. R. Croley, T.-Y. Wong, C. Chen, S. Fan, Y. Chong, R. Li, C. Ge, C. Chen and J.-J. Yin, ACS Appl. Mater. Interfaces, 2018, 10(10), 8443–8450 CrossRef CAS PubMed.
- A. C. Reber, S. N. Khanna, P. J. Roach, W. H. Woodward and A. W. Castleman, J. Am. Chem. Soc., 2007, 129(51), 16098–16101 CrossRef CAS PubMed.
- L. S. de Melo, A. S. Gomes, S. Saska, K. Nigoghossian, Y. Messaddeq, S. J. Ribeiro and R. E. de Araujo, J. Fluoresc., 2012, 22(6), 1633–1638 CrossRef PubMed.
- T. Suenobu, I. Arahori, K.-i. Nakayama, T. Suzuki, R. Katoh and T. Nakagawa, J. Phys. Chem. A, 2020, 124(1), 46–55 CrossRef CAS PubMed.
- J. R. Lakowicz and G. Weber, Biochemistry, 1973, 12(21), 4161–4170 CrossRef CAS PubMed.
- C. Schweitzer and R. Schmidt, Chem. Rev., 2003, 103(5), 1685–1758 CrossRef CAS PubMed.
- R. M. Badger, A. C. Wright and R. F. Whitlock, J. Chem. Phys., 1965, 43(12), 4345–4350 CrossRef CAS.
- A. S. Kozlov, O. N. Egorova, O. I. Medvedkov and A. A. Krasnovsky, Opt. Lett., 2021, 46(3), 556–559 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3na00092c |
| This journal is © The Royal Society of Chemistry 2023 |