Open Access Article
Open Access ArticleLocal transplantation of GMSC-derived exosomes to promote vascularized diabetic wound healing by regulating the Wnt/β-catenin pathways†
Ziwei
Liu‡
 abc,
Shuo
Yang‡
abc,
Shuo
Yang‡
 b,
Xiaoming
Li‡
d,
Situo
Wang
abc,
Tong
Zhang
b,
Na
Huo
b,
Ruixin
Duan
e,
Quan
Shi
*b,
Jianjun
Zhang
b,
Xiaoming
Li‡
d,
Situo
Wang
abc,
Tong
Zhang
b,
Na
Huo
b,
Ruixin
Duan
e,
Quan
Shi
*b,
Jianjun
Zhang
 *d and
Juan
Xu
*b
*d and
Juan
Xu
*b
aMedical School of Chinese PLA, Beijing, 100853, China
bDepartment of Stomatology, The First Medical Center, Chinese PLA General Hospital, Beijing, 100853, China. E-mail: newxj@hotmail.com; shiquan3333@sina.cn
cOrthopedic Laboratory of PLA General Hospital, Beijing, 100853, China
dCollege of Chemical Engineering, Beijing University of Chemical Technology, Beijing, 100029, China. E-mail: zhangjj@mail.buct.edu.cn
eDepartment of Stomatology, The People's Hospital of Anyang City, Henan 455000, China
First published on 8th December 2022
Abstract
With the increasing number of diabetic patients, chronic wound healing remains a great challenge in clinical medicine. As one of the main components secreted by stem cells, the exosome is considered to be a promising candidate for promoting chronic wound healing. Here, gingival mesenchymal stem cell (GMSC)-derived exosomes (GMSC-Exo) were isolated and demonstrated to promote the proliferation, migration, and tube formation of human umbilical vein endothelial cells (HUVECs) by regulating the Wnt/β-catenin signaling pathway in a diabetic-mimicking high glucose environment. In order to deliver GMSCs-Exo to the target site and prolong their local retention, porous microspheres consisting of poly-lactic-co-glycolic acid (PLGA), amphiphilic block copolymer (PLLA-PEG-PLLA), nano-hydroxyapatite (nHAP), and poly-ε-L-lysine (EPL) coating were fabricated through a double emulsion method and following surface treatment, hereafter referred to as PHE microspheres. PHE microspheres loaded with GMSCs-Exo were implanted into the full-thickness skin wound of a diabetic mouse model, resulting in significant vascularized wound healing when compared to a control group only injected with GMSCs-Exo suspension or filled with PHE microspheres. These findings indicated that the GMSCs-Exo-loaded porous microspheres could efficiently treat diabetic wounds and have promising potential for future clinical translations.
Introduction
Skin, as the first protective barrier between the body and the external environment, can effectively prevent damage caused by external irritating factors and can self-repair after injury.1 Under normal circumstances, the process of skin wound healing will be quickly activated and then enter the four healing stages spontaneously: hemostasis, inflammation, proliferation, and remodeling.2 For chronic wounds caused by diabetes, however, the wound healing process can be delayed or may even fail.3 Diabetes mellitus (DM), characterized by long-term high blood sugar levels, is a global disease that can eventually lead to disability and even death. With narrow blood vessels, patients with diabetes always lack sufficient blood circulation, which causes an insufficient supply of oxygen and nutrients, therefore also having a passive effect on skin wound repair.4,5 Since delayed wound healing not only impairs the life quality of patients, but also increases the financial burden to society,6 many researchers are striving to find novel and efficient ways to overcome this complicated clinical problem of wound healing.In the past two decades, mesenchymal stem cells (MSCs) have been widely studied in the regenerative medicine field, owing to their self-renewal potential, multilineage potency, and immunoregulatory capacity.7 For skin wound repair, MSCs can accelerate the healing process by promoting the proliferation and migration of tissue cells, facilitating the formation of blood vessels,8,9 and increasing the rate of re-epithelialization.10 Gingival mesenchymal stem cells (GMSCs), a kind of oral MSCs separated and isolated from human gingival tissue, are considered as an ideal source of MSCs because of their high genomic stability, rich source, easy accessibility, and fast proliferation rate.7,11 Shi et al. proved that GMSCs-Exo could effectively promote skin wound healing in a diabetic rat model by enhancing angiogenesis, and promoting the re-epithelialization, deposition, and remodeling of the extracellular matrix (ECM).12 Although the therapeutic effect of stem cells on promoting skin wound repair has already been affirmed to some extent, there are also certain problems in stem-cell therapy. For example, it may present both problems of storage and transportation, or incur risks of induced tumorigenesis and deformity.13 Hence, a strong research trend is to find a substitute that can not only have the effect of stem-cell therapy, but also avoid the above-mentioned risks.
Exosome, a kind of small extracellular vesicle with a diameter between 30 and 150 nm secreted by living cells, is the main component of cell paracrine.14 It is composed of a phospholipid bilayer and inner cargos, such as DNA, mRNAs, miRNAs, proteins, and lipids.15 As a communicator, the exosome can release its cargo to target cells and regulate biological events, like proliferation, migration, angiogenesis, apoptosis, and immunomodulatory reactions, via multiple signal pathways,6 and may become a promising alternative to stem cells in wound repair and skin regeneration.16 Hence, we hypothesized that the local transplantation of exosomes derived from GMSCs (GMSCs-Exo) with a comparable regenerative potential to GMSCs may promote diabetic wound healing.
At present, the common methods for applying exosomes are intravenous injection and local injection, both of which can suffer from rapid exosomes clearance and secondary injury.17 To solve these problems, the development of novel biomaterials capable of loading and transplanting exosomes has attracted much attention. Bioresorbable porous microspheres made from biodegradable aliphatic polyester (e.g., poly-L-lactide (PLLA) and poly-lactic-co-glycolic acid (PLGA)) is considered a promising biomaterial suitable for minimally invasive drug delivery and tissue regeneration.18 With surface-affinity coatings, these microspheres have exhibited a desired absorption rate and sustained release of exosomes.19
Herein, we first investigated the underlying mechanism by figuring out the function of HUVECs treated with GMSCs-Exo in vitro, and then explored the role of the Wnt/β-catenin signaling pathway for GMSCs-Exo-induced angiogenesis. To locally transplant GMSC-derived exosomes, a porous microsphere matrix was fabricated from PLGA and an amphiphilic block copolymer composed of a hydrophobic PLLA block and hydrophilic poly (ethyl glycol) (PEG) block through a double emulsion method, followed by alkali treatment. In the meantime, nano-hydroxyapatite (nHAP) was incorporated to neutralize the acidic environment generated by the degradation of the aliphatic polyester. Poly-ε-L-lysine (EPL), a natural cationic polypeptide that has shown biocompatibility and inherent antibacterial activity,20 was then introduced in the surface of porous microspheres to adsorb the exosomes. The final PLGA/nHAP/EPL microspheres (denoted as PHE) could be applied as an excellent carrier for GMSCs-Exo, thereby promoting the healing of chronic wounds in a diabetic mouse model.
Materials and methods
Isolation and characterization of the GMSCs
Adipogenic differentiation: GMSCs were seeded by the same general steps above. When the cells had reached 100% confluence, the original medium was changed for adipogenic differentiation medium (Cyagen) to continue the culture. After 21 days, the wells were also washed twice with PBS and fixed with 4% paraformaldehyde for 20 min at room temperature, and the formation of lipid-laden fat cells was detected by staining with 1 mL of 0.5% (w/v) Oil Red O (Sigma) per well for 10 min.
Chondrogenic differentiation: 1 × 106 GMSCs at the third passage were suspended in a 15 mL centrifuge tube at 1000g for 5 min to form cell pellets. The cell pellets were then cultured for 28 days in chondrogenic induction medium (Cyagen), which was changed every 3 days. After the induction, the pellets were collected and fixed in 4% paraformaldehyde at 4 °C for 24 h, and then paraffin-embedded and sectioned for Alcian blue (Cyagen) staining for 30 min.
Isolation and characterization of the GMSCs-Exo
The typical ultracentrifugation method was used to extract and collect the exosomes from the supernatant of the GMSCs culture. When the GMSCs (P3–P5) had achieved a confluence of 80–85%, the medium was replaced by fresh DMEM/F12 containing 10% exosome-free FBS, and then the cells were further cultures for 48 h. The supernatant was collected and centrifuged at 500g for 10 min, 2000g for 20 min, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 min sequentially to eliminate any dead cells and cell debris. Clear supernatants were passed through 0.22 μm filters and transferred into 15 mL Amicon Ultra-30kd ultracentrifugation tubes, for ultracentrifugation at 100
000g for 30 min sequentially to eliminate any dead cells and cell debris. Clear supernatants were passed through 0.22 μm filters and transferred into 15 mL Amicon Ultra-30kd ultracentrifugation tubes, for ultracentrifugation at 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 60 min twice. The final precipitates from the supernatant were resuspended with PBS and stored at −80 °C.
000g for 60 min twice. The final precipitates from the supernatant were resuspended with PBS and stored at −80 °C.
Transmission electron microscopy (TEM, HITACHI) was utilized to observe the morphology of the GMSCs-Exo. Nanoparticle tracking analysis (NTA, IZON) was used to measure the size of the exosomes and analyze the size distribution. The concentration of GMSCs-Exo was determined by the BCA protein assay kit (Servicebio), while the exosomal markers CD63 and Tsg101 were detected by western blot analysis.
Effect of GMSCs-Exo on HUVECs under a high glucose condition in vitro
Effect of GMSCs-Exo on the Wnt/β-catenin signaling pathway
After determining the effects of the exosomes on the function of HUVECs, we further explored whether these effects were achieved by activating the Wnt/β-catenin signaling pathway. The western blot method was used to detect the expression changes of β-catenin and the downstream proteins related to cell proliferation and migration. HUVECs were cultured in normal medium as a control group and three experimental groups: HUVECs cultured in HG medium, HG medium + GMSCs-Exo, and HG medium + GMSCs-Exo + ICG-001. Then we extracted β-catenin protein from the nucleus and chose Histone H3 as an internal reference. While N-cadherin and cyclin D3 were extracted from the whole cell, GADPH was used as an internal reference. Their expression level under different treatments was determined by western blot analysis. At the same time, we also used the CCK-8 assay, transwell assay, and tube formation assay to evaluate the function of the HUVECs after treatment with GMSCs-Exo and ICG-001, detecting the changes of function under different conditions.Preparation and characterization of PHE
First, nHAP was generated by a high-gravity method and PLGA/nHAP microspheres were prepared by an emulsion-solvent evaporation method according to previous works.21–23 Subsequently, we dispersed the microspheres in 40 mL of 0.2 mol per L NaOH solution and obtained hydrolyzed PLGA/nHAP porous microspheres. Then, EPL was added into the buffer containing the hydrolyzed PLGA/nHAP porous microspheres. Finally, after washing with 1 mol per L NaCl solution three times and washing with water for another five times, the PHE porous microspheres were freeze-dried and stored for later use.Scanning electron microscopy (SEM) was used to observe the internal morphology of the microsphere, and the types of surface elements were determined by element mapping. Zeta potential detection was performed to detect the exosomes-loading capacity of the material. In addition, antibacterial experiments were used to prove that PHE has good antibacterial ability. The DIO-labeled GMSCs-Exo were loaded onto PHE porous microspheres and observed under a confocal microscope.
GMSCs-Exo combined with PHE accelerated wound repair in diabetic mice model
After 2 weeks adaptive feeding, all the animals were given a high-fat and high-sugar feed for another 2 weeks. The rats' weight and blood glucose were recorded every three days. The day before the experiment, the mice were fasted for 12 h, and the preoperative body weight and fasting blood glucose were measured and recorded. Mice with fasting blood glucose >16.7 mmol L−1 were selected for the experimental group. Then, 2 mice were randomly selected to be sacrificed, and the pancreatic tissue was excised to serve as the experimental group showing the pancreatic tissue structure of the diabetic mice, and the remaining mice were used for the subsequent experiments for modeling diabetic skin damage.
We divided the 40 experimental animals into 4 groups, each group included 10 animals: control group (G1) – the wound was rinsed by PBS and covered by a gauze infiltrated with PBS; exosome group (G2) – 4 points were selected around each wound, and 25 μL suspension of 100 μg/100 μL exosomes and PBS was injected at each point, and then a gauze infiltrated with PBS was placed above; PHE group (G3) – a skin dressing without exosomes was applied to the wound, and then 3 mg of the material needed by each mouse was dissolved in 100 μL of PBS, and then a gauze infiltrated with PBS was placed above; PHE@Exo group (G4) – the material loaded with the exosomes was applied to the wound, and also the 3 mg of the material needed by each mouse was dissolved in 100 μg/100 μL exosomes and PBS suspension, and then a gauze infiltrated with PBS was placed above.
Before the surgery, the mice were anesthetized by an intraperitoneal injection of pentobarbital sodium (50 mg kg−1). After successful anesthesia, the hair on the back of the mice was first removed with a shaver, and then a depilatory cream was applied topically for further hair removal. After skin preparation, a circular full-thickness skin wound with a diameter of 1 cm was made in the center of the back of each mouse. Then we dealt with the wound according to the groupings above, respectively, and changed the skin dressing every 3 days. The wounds were recorded and measured to evaluate the wound-closure rates; furthermore, on the 7th day and 14th day, 5 mice of each group were killed and their newly formed skin tissue was excised to make pathological slices for further study of the wound healing.
Image analysis and statistical analysis
Images in this article were analyzed by ImageJ software (NIH). Data were analyzed by SPSS Statistics 26 software (IBM SPSS Statistics Inc), and were presented as the mean ± SD from at least three separate experiments. T-Test was used to compare differences between two groups. One-way ANOVA, followed by LSD multiple comparison tests were used to analyze the differences in more than two groups. Graphs were performed in GraphPad Prism 9.0.0 (GraphPad Software Inc). Student's p < 0.05 was considered statistically significant.Results and discussion
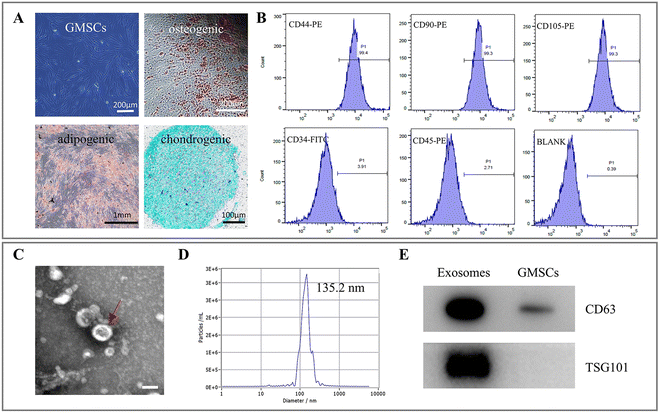
Characterization of the GMSCs and GMSCs-derived exosomes
GMSCs were successfully isolated and cultured as previously described.24 Compared with the MSCs of other tissue origin, GMSCs possess some unique properties. Despite their relatively high proliferative rate25 and stable morphology,26 GMSCs also have the potential to transdifferentiate into ectodermal and endodermal cell lineages, such as neural cells,27 keratinocytes,28 and endothelial cells.29 As shown in Fig. 1A, GMSCs at the third passage showed a normal spindle-shaped morphology. The results of the osteogenic-, adipogenic-, and chondrogenic-induced differentiation experiments demonstrated the multi-potency of the GMSCs. The flow cytometry assay revealed that the GMSCs positively expressed the surface markers of MSCs (CD44, CD90, and CD105), and negatively expressed the surface markers of hematopoietic stem cells (CD34 and CD45) (Fig. 1B).GMSCs-Exo was successfully extracted by ultracentrifugation. Observed by TEM, GMSCs-Exo presented a spherical structure (Fig. 1C), with a mean diameter of 135.2 ± 44.3 nm as analyzed by NTA (Fig. 1D). In addition, the western blot assay confirmed that GMSCs-Exo expressed the surface markers CD63 and Tsg101 (Fig. 1E, raw data: Fig. S1†).
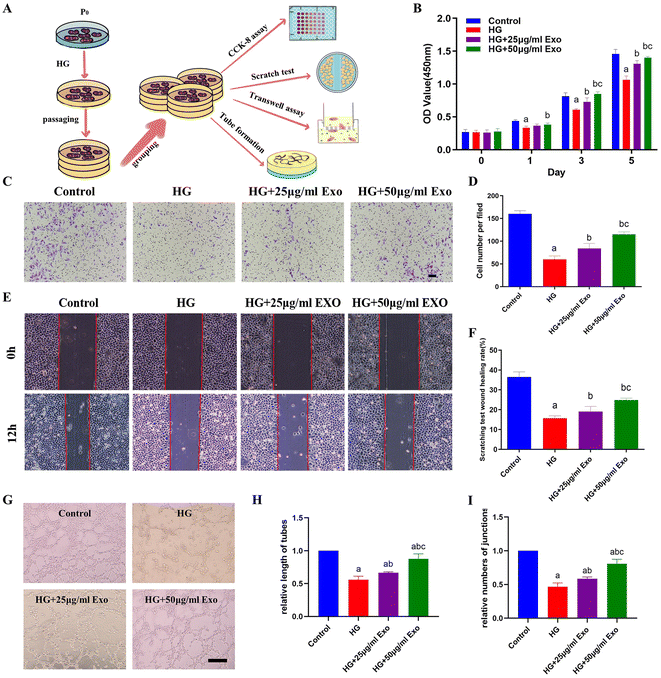
GMSCs-Exo promoted HUVECs proliferation, migration, and tube formation under a high glucose condition in vitro
It is well-acknowledged that the high glucose environment in diabetic patients can cause problems in keratinization30 and angiogenesis,31 thereby hindering the process of wound healing.31 Researchers found that exosomes could apparently improve the biological function of HUVECs, including their proliferation, migration, and tube formation after damage by high glucose in vitro.32 Here, in order to mimic the high glucose microenvironment in diabetic patients, HUVECs were also cultured in HG medium for further evaluation. First, DIO-labeled GMSCs-Exo were co-cultured with HUVECs cultured in HG medium for 24 h in vitro. Abundant green fluorescence (from the GMSCs-Exo) could be observed around the blue-labeled nucleus of the HUVECs, indicating the successful uptake of exosomes by the HUVECs (Fig. S2†).High glucose could inhibit the proliferation of HUVECs compared to normal conditions. In the presence of GMSCs-Exo, however, HUVECs proliferation was promoted, and the highest OD values were obtained in the HG+50 μg mL−1 GMSCs-Exo group (Fig. 2B). Transwell migration assay demonstrated that the migration of HUVECs could be inhibited by a high glucose condition at 24 h, but could be increased in the presence of exosomes as compared to the HG group (Fig. 2C and D). Moreover, the scratch closure test results also showed similar outcomes, whereby GMSCs-Exo could promote HUVECs migration in a dose-dependent manner (Fig. 2E and F).
In the tube formation assay, a fewer number of junctions and shorter length of tubes were presented in the HG group compared with the control group, which indicated that the high glucose condition may weaken the tube formation ability of HUVECs. The 50 μg mL−1 GMSCs-Exo formed significantly more tubular and cord-like structures on the matrigel with higher numbers of junctions and length of tubes compared to the control group, followed by 25 μg mL−1 GMSCs-Exo (Fig. 2G–I).
GMSCs-exo activated the Wnt/β-catenin pathway to promote wound healing
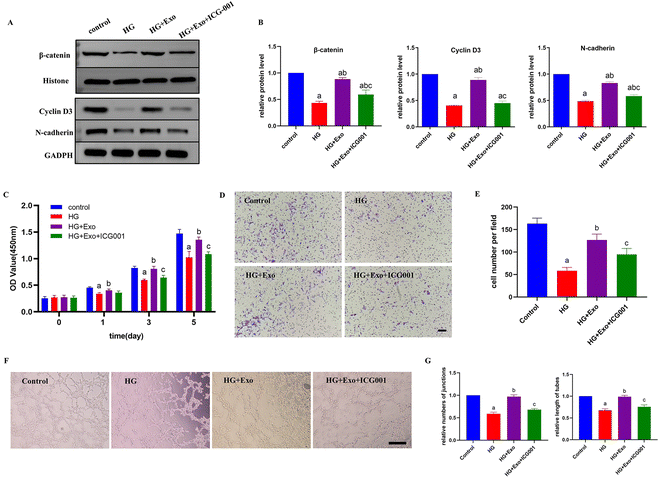
The Wnt/β-catenin signaling pathway is a highly conservative pathway in the process of evolution. In the classic Wnt pathway, β-catenin is the central promoter.33 The application of MSCs-Exo could not only regulate the proliferation, migration, and collagen secretion of skin cells through the Wnt/β-catenin signaling pathway, but also affect the function of vascular endothelial cells and thus affect angiogenesis at the wound site. Zhang et al. proved that exosomes derived from human umbilical cord mesenchymal stem cells could promote β-catenin nuclear transfer through Wnt4 and promote skin cell proliferation and migration.34 Another study pointed out that one of the regulatory mechanisms of this pathway was to promote the angiogenesis of endothelial cells, and this effect could be blocked by the inhibitors β-catenin or Wnt4.35Here, ICG-001, a competitive antagonist, was used to block the Wnt/β-catenin signal transition pathway via combining with its specific receptor.35 The results of western blotting showed that the expressions of β-catenin, cyclin D3, and N-cadherin were upregulated with the presence of GMSCs-Exo and decreased after adding ICG-001, indicating that GMSCs-Exo could activate the Wnt/β-catenin signaling pathway and the inhibitor of β-catenin had the opposite effect (Fig. 3A and B; raw data: Fig. S3†). To make the results more convincing, further research was done on the functions of the HUVECs. CCK-8 assay (Fig. 3C) and transwell assay (Fig. 3D and E) showed that the HUVECs treated with HG medium supplemented with GMSCs-Exo had stronger proliferative and migratory capacities compared with the cells treated merely with HG medium. However, HUVECs in the HG+Exo+ICG-001 group had a significant diminishment in these abilities compared to in the HG+Exo group. Likewise, the tube formation assay showed that the HG+Exo group formed significantly more tubular and cord-like structures on the matrigel with higher numbers of junctions and length of tubes compared to the HG group, and this function could also be inhibited after adding ICG-001, which was associated with wound healing dysfunction (Fig. 3F and G).
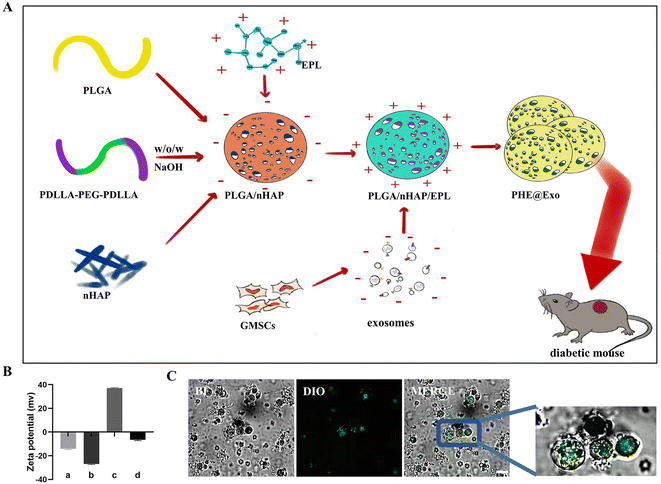
Fabrication and characterization of the PHE porous microspheres
PHE porous microspheres were fabricated through a double emulsion method, alkali treatment, and following EPL coating (Fig. 4A). As shown in Fig. S4,† the PHE showed a porous microsphere structure under SEM. P, N, and O elements could be detected on the surface of the PHE porous microspheres, indicating that nHAP and EPL were successfully combined with PLGA. The zeta potential detection showed that the average potential of PLGA-nHAP was −14.1 mV, and the average potential of the EPL-loaded material surface was 37 mV. After reloading the exosomes, the average potential became −6.45 mV. This demonstrated that the PLGA-nHAP exhibited a positive potential after EPL coating, and therefore could adsorb more negatively charged exosomes (Fig. 4B). Correspondingly, laser confocal microscopy also showed that green-labeled exosomes were successfully loaded on PHE (Fig. 4C). In addition, in the presence of EPL, the PHE porous microspheres exhibited an obvious antibacterial ability, which would be beneficial for diabetic patients who are prone to wound infection (Fig. S5†).In the current PHE porous microspheres, PLGA, as a kind of synthetic polymer with good biocompatibility and biodegradability, has been widely used in the field of tissue regeneration.36,37 During the evaporation of the organic solvent, the PLLA-PEG-PLLA copolymer enables the formation of a porous architecture, which was confirmed in our previous work.38 Nanoscale nHAP can neutralize the acidic environment caused by PLGA degradation products because of its alkalinity, thereby effectively preventing the denaturation of exosomes and creating a more favorable environment for wound healing.39 Poly-ε-L-lysine (EPL) is a natural cationic polypeptide produced from Streptomyces albulus that is rich in cations and has a strong antimicrobial property on skin wounds of diabetic patients.40 Hence, PHE porous microspheres could be expected to be applied as an excellent carrier for GMSCs-Exo, thereby promoting the healing of diabetic wounds.
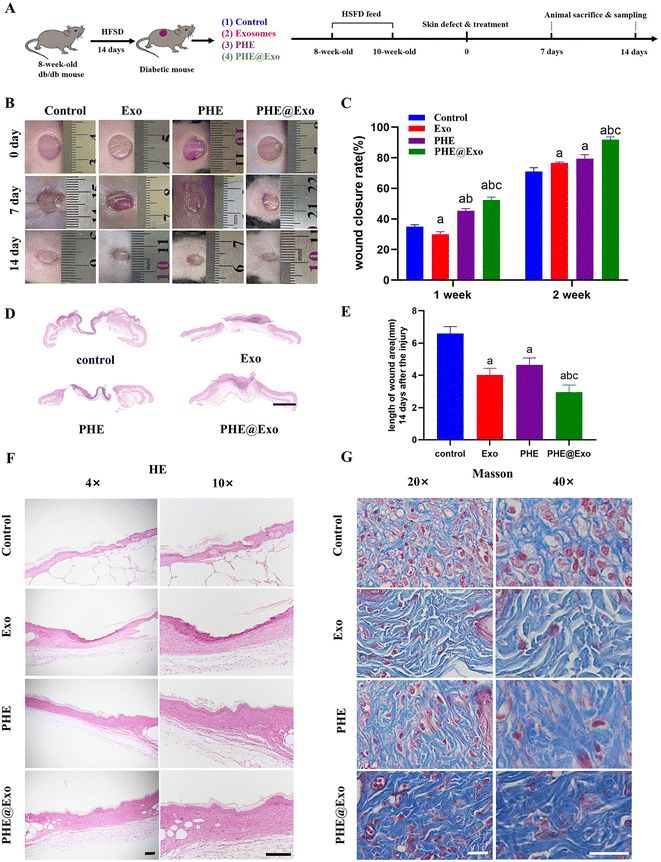
Diabetic wound healing examinations in a db/db mice model
Db/db is a kind of mouse with spontaneous type 2 diabetes mellitus (T2DM), which has insulin resistance and exhibits spontaneous hyperglycemia in an early stage of T2DM, and a deficiency of insulin secretion in the late stage of diabetes,41 and therefore it can be used to establish a stable type 2 diabetic skin model.42 The entire surgical procedure of the animal experiments is roughly as shown in Fig. 5A. After being fed with high-fat and high-sugar foods for 2 weeks, a total of 42 mice with fasting blood glucose >16.7 mmol L−1 (19.81 ± 2.79 mmol L−1) were regarded as the induced diabetic mice models, and the induction success rate was 89.4%. Those mice showed symptoms of polydipsia, polyphagia, polyuria, and weight loss, and developed obvious obesity within 1 week. Compared with normal mice, the H&E staining of the pancreatic tissue in the diabetic mice presented clear damage of the functional structure (Fig. S6†).We compared the wound healing of each group at days 0, 7, and 14 after surgery, and performed a quantitative analysis of the wound-closure rates (Fig. 5B and C). The results showed that the wound sizes in all the groups decreased with time. More specifically, PHE and PHE@Exo showed a faster healing speed at both 1 week and 2 weeks post-surgery compared to the control; but Exo showed a lower closure rate at 1 week and a higher closure rate at 2 weeks compared to the control. PHE@Exo showed a significant better healing effect than in the other three groups, while Exo and PHE showed no significant distinction at 2 weeks post-surgery.
H&E staining showed the re-epithelium length of the skin defects at 2 weeks post-surgery in the four groups (Fig. 5D and E). PHE@Exo presented the longest neo-epithelium and was significantly different from the other three groups. Exo, PHE, and PHE@Exo also showed a significant difference compared to the control; however, there were no difference in the length of the neo-epithelium between the Exo and PHE groups.
In addition, the thickness of the neo-epithelium was also observed by H&E staining (Fig. 5F). The neo-epithelial layer of PHE@Exo was significantly thicker than in the other three groups, while Exo and PHE also had a thicker neo-epithelial layer than the control (Fig. S7†). Masson trichrome staining demonstrated there was more extensive collagen deposition in PHE@Exo, and the defect sites in PHE@Exo were almost fully covered by neo-epithelium with more mature and wavy collagen fibers. Exo and PHE also showed a thicker and better arranged collagen than the control (Fig. 5G). These results indicated that GMSCs-Exo could promote the formation and arrangement of collagen, and PHE@Exo had a superior ECM remodeling ability than the other three groups.
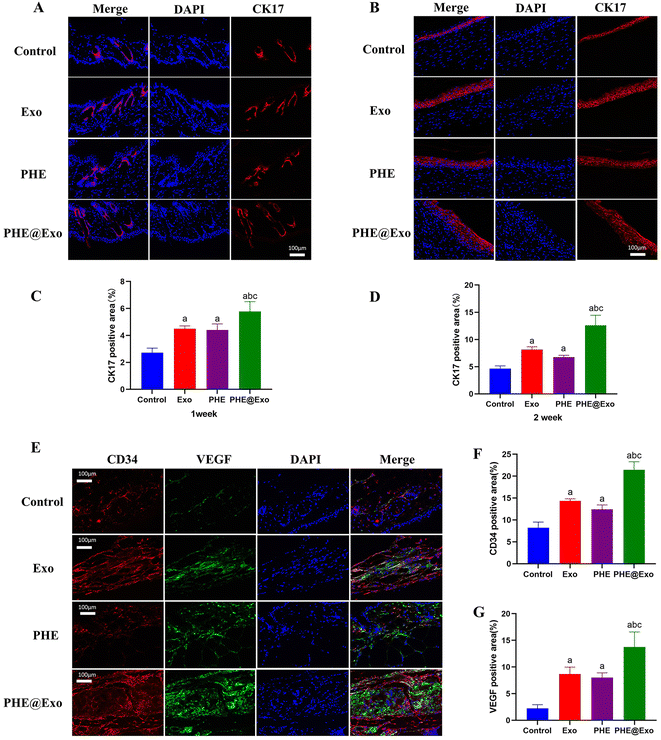
Immunofluorescence staining showed that the expression of CK17 in the PHE@Exo group was significantly higher than that in the other three groups a week after the operation (Fig. 6A and C). Meanwhile, the injection group had a higher-level expression than the control group, indicating that GMSCs-Exo could effectively promote keratinization. Two weeks after the operation, immunofluorescence staining showed that a layer of intact neo-epithelial structure was formed at the injury site, which was mainly composed of collagen fibers, lacking glands and adipose tissue. In addition, the epithelial tissue generated in the PHE@Exo group was the thickest, which was significantly different from the other three groups (Fig. 6B and D).
On the other hand, we used red, green, and blue fluorescence to stain CD34, VEGF, and DAPI, respectively, to evaluate the angiogenic capacity of the four groups. It could be seen that the PHE@Exo group had the highest expression of CD34 and VEGF, and the strongest angiogenesis ability, which was significantly different from the other three groups (Fig. 6E–G).
Our research found that GMSCs-Exo combined with PHE accelerated diabetic wound healing in vivo by reepithelization, collagen deposition, and ECM remodeling. It was also found that GMSCs-Exo reversed the inhibition of HUVECs proliferation, migration, and tube formation in vitro induced by a high glucose environment, probably attributed to their activating the Wnt/β-catenin pathway (Fig. 7). However, a key problem for the healing of chronic wounds lies in the fact that it is difficult to progress from the inflammatory phase to the proliferative phase. Therefore, how GMSCs-Exo modulate inflammatory responses will be another focus of our attention in future research.
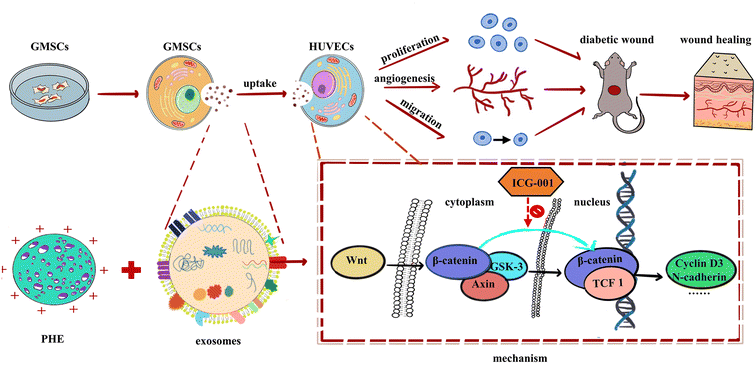 | ||
| Fig. 7 Schematic diagram showing GMSCs-Exo accelerating diabetic wound repair through enhancing angiogenesis by activation of the Wnt/β-catenin signaling pathway. | ||
Conclusion
In conclusion, we successfully isolated GMSCs-Exo and proved that such exosomes can promote the proliferation, migration, and tube formation of HUVECs in a high glucose environment through a series of in vitro experiments. Further, we demonstrated that such angiogenetic enhancement was achieved by activating the Wnt/β-catenin signaling pathway, and could be inhibited by ICG-001. Finally, PHE microspheres, a kind of novel porous microscaffolds with excellent biocompatibility, were prepared to load and deliver GMSCs-Exo for the first time. GMSCs-Exo-loaded PHE could significantly accelerate diabetic mice wound healing in vivo by enhancing re-epithelization, collagen deposition, and angiogenesis, thereby demonstrating great promise for diabetic patients for future clinical chronic wound treatment.Author contributions
J. Xu, Q. Shi, and J. Zhang provided the lab resource and funding, supervised project; Z. Liu, S. Yang, X. Li and R. Duan performed the experimental work and data analysis; Z. Liu, X. Li and S. Wang wrote the manuscript and plotted the schemes; T. Zhang and N. Huo revised and edited the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Siyu Liu for providing a photographic guidance. This work was financially supported by Beijing Natural Science Foundation (7212091), National Natural Science Foundation of China (81901034), Military Medical Science Foundation (21QNPY104), and Military Health Care Special Project (19BJZ22).Notes and references
- J. M. J. E. Proksch, Giorn. Ital. Dermatol. Venereol., 2009, 144, 689–700 Search PubMed.
- P. H. Wang, B. S. Huang, H. C. Horng, C. C. YehandY and J. Chen, J. Chin. Med. Assoc., 2018, 81, 94–101 CrossRef PubMed.
- M. Changand and T. T. Nguyen, Acc. Chem. Res., 2021, 54, 1080–1093 CrossRef PubMed.
- S. Hamed, C. L. Bennett, C. Demiot, Y. Ullmann and L. T. A. Desmoulière, Wound Repair Regen., 2014, 22, 23–33 CrossRef PubMed.
- K. Wicks, T. TorbicaandK and A. Mace, Semin. Immunol., 2014, 26, 341–353 CrossRef CAS PubMed.
- N. B. Vu, H. T. Nguyen, R. Palumbo, R. Pellicano, S. FagooneeandP and V. Pham, Minerva Med., 2021, 112, 384–400 Search PubMed.
- D. Kim, A. E. Lee, Q. Xu, Q. ZhangandA and D. Le, Front. Immunol., 2021, 12, 667221 CrossRef CAS PubMed.
- R. Tutuianu, A. M. Rosca, D. M. Iacomi and M. S. I. Titorencu, Int. J. Mol. Sci., 2021, 22, 6239 CrossRef CAS PubMed.
- S. Schlosser, C. Dennler, R. Schweizer, D. Eberli, J. V. Stein, V. Enzmann, P. Giovanoli, D. ErniandJ and A. Plock, Microvasc. Res., 2012, 83, 267–275 CrossRef CAS PubMed.
- G. Luo, W. Cheng, W. He, X. Wang, J. Tan, M. Fitzgerald and X. L. J. Wu, Wound Repair Regen., 2010, 18, 506–513 CrossRef PubMed.
- L. Fonticoli, Y. Della Rocca, T. S. Rajan, G. Murmura, O. Trubiani, S. Oliva, J. Pizzicannella and G. D. M. F. Diomede, Int. J. Mol. Sci., 2022, 23, 4135 CrossRef CAS PubMed.
- Q. Shi, Z. Qian, D. Liu, J. Sun, X. Wang, H. Liu and J. X. X. Guo, Front. Physiol., 2017, 8, 904 CrossRef PubMed.
- A. Trounsonand and C. McDonald, Cell Stem Cell, 2015, 17, 11–22 CrossRef PubMed.
- H. S. Joo, J. H. Suh, H. J. Lee, E. S. Bangand and J. M. Lee, Int. J. Mol. Sci., 2020, 21, 727 CrossRef CAS PubMed.
- I. S. Kang, J. Suh, M. N. Lee, C. Lee, J. Jin, C. Lee, Y. I. Yang, Y. JangandG and T. Oh, BMB Rep., 2020, 53, 118–123 CrossRef CAS PubMed.
- Y. Zhang, Y. Pan, Y. Liu, X. Li, L. Tang, M. Duan and J. L. G. Zhang, Stem Cell Res. Ther., 2021, 12, 434 CrossRef CAS PubMed.
- B. Safari, M. Aghazadeh and S. D. L. Roshangar, Eur. J. Pharm. Biopharm., 2022, 171, 50–59 CrossRef CAS PubMed.
- Z. Yuan, Z. Wan, P. Wei, X. Lu, J. Mao, Q. Cai and X. Z. X. Yang, Adv. Healthcare Mater., 2020, 9, e2000211 CrossRef PubMed.
- Y. Gao, Z. Yuan, X. Yuan, Z. Wan, Y. Yu, Q. Zhan, Y. Zhao, J. Han, J. Huang and C. X. Q. Cai, Bioact. Mater., 2022, 14, 377–388 CrossRef CAS PubMed.
- L. Zhou, Y. Xi, M. Chen, W. Niu, M. Wang and P. X. M. B. Lei, Nanoscale, 2018, 10, 17304–17317 RSC.
- V. Sokolova, K. Kostka, K. T. Shalumon, O. Prymak and J. P. C. M. Epple, J. Mater. Sci., 2020, 31, 102 CAS.
- P. Wei, W. Jing, Z. Yuan, Y. Huang, B. Guan, W. Zhang, X. Zhang, J. Mao, Q. Cai and D. C. X. Yang, ACS Appl. Mater. Interfaces, 2019, 11, 30596–30609 CrossRef CAS PubMed.
- P. F. Wei, Z. Y. Yuan, W. Jing, B. B. Guan, Z. H. Liu, X. Zhang, J. P. Mao, D. F. Chen, Q. CaiandX and P. Yang, Biomater. Sci., 2018, 7, 272–286 RSC.
- Q. C. Xu, Z. G. Wang, Q. X. Ji, X. B. Yu, X. Y. Xu, C. Q. Yuan, J. Dengand and P. S. Yang, Int. J. Clin. Exp. Pathol., 2014, 7, 4922–4929 Search PubMed.
- Q. Sun, H. Nakata, M. Yamamoto and S. K. S. Kuroda, J. Cell. Mol. Med., 2019, 23, 7592–7601 CrossRef CAS PubMed.
- G. B. Tomar, R. K. Srivastava, N. Gupta, A. P. Barhanpurkar, S. T. Pote, H. M. Jhaveri, G. C. MishraandM and R. Wani, Biochem. Biophys. Res. Commun., 2010, 393, 377–383 CrossRef CAS PubMed.
- D. Li, X. Y. Zou, I. El-Ayachi, L. O. Romero, Z. Yu, A. Iglesias-Linares, J. F. Cordero-MoralesandG and T. Huang, Stem Cell Rev. Rep., 2019, 15, 67–81 CrossRef CAS PubMed.
- D. Murugan Girija, M. Kalachaveedu, S. Ranga Raoand and R. Subbarayan, J. Cell. Physiol., 2018, 233, 8450–8457 CrossRef CAS PubMed.
- X. Liu, J. Wang, F. Dong and H. L. Y. Hou, Dev., Growth Differ., 2016, 58, 702–713 CrossRef CAS PubMed.
- T. Ma, B. Fu, X. Yang and Y. X. M. Pan, J. Cell. Biochem., 2019, 120, 10847–10854 CrossRef CAS PubMed.
- H. Wu, F. Li, W. Shao and J. G. D. Ling, ACS Cent. Sci., 2019, 5, 477–485 CrossRef CAS PubMed.
- Y. Hu, R. Tao, L. Chen, Y. Xiong, H. Xue, L. Hu, C. Yan, X. Xie, Z. Lin, A. C. Panayi and B. M. G. Liu, J. Nanobiotechnol., 2021, 19, 150 CrossRef CAS PubMed.
- L. B. S. Scholpp, Cell. Mol. Life Sci., 2018, 75, 785–795 CrossRef PubMed.
- B. Zhang, X. Wu, X. Zhang, Y. Sun, Y. Yan, H. Shi, Y. Zhu, L. Wu, Z. Pan, W. Zhu and H. Q. W. Xu, Stem Cells Transl. Med., 2015, 4, 513–522 CrossRef CAS PubMed.
- B. Zhang, M. Wang, A. Gong, X. Zhang, X. Wu, Y. Zhu, H. Shi, L. Wu, W. Zhu and H. Q. W. Xu, Stem Cells, 2015, 33, 2158–2168 CrossRef CAS PubMed.
- Z. Yuan, P. Wei, Y. Huang, W. Zhang, F. Chen, X. Zhang, J. Mao, D. Chen and Q. C. X. Yang, Acta Biomater., 2019, 85, 294–309 CrossRef CAS PubMed.
- L. Zhang, Z. Wan, Z. Yuan, J. Yang, Y. Zhang, Q. Cai and J. H. Y. Zhao, J. Mater. Chem. B, 2022, 10, 3344–3356 RSC.
- P. Wei, Z. Yuan, Q. Cai and J. M. X. Yang, Macromol. Rapid Commun., 2018, 39, e1800062 CrossRef PubMed.
- H. Shen, X. Hu, F. Yang and J. B. S. Wang, Acta Biomater., 2010, 6, 455–465 CrossRef CAS PubMed.
- C. Wang, M. Wang, T. Xu, X. Zhang, C. Lin, W. Gao, H. Xu and B. L. C. Mao, Theranostics, 2019, 9, 65–76 CrossRef CAS PubMed.
- C. W. Cheng, V. Villani, R. Buono, M. Wei, S. Kumar, O. H. Yilmaz, P. Cohen, J. B. Sneddon, L. Perinand and V. D. Longo, Cell, 2017, 168, 775–788 CrossRef CAS PubMed.
- P. S. Hinton, K. Shankar, L. M. EatonandR and S. Rector, Metab. Clin. Exp., 2015, 64, 905–916 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2na00762b |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |