Open Access Article
Open Access ArticleThe complete change in bile acids and steroids in systematic metabolomics applied to the intrahepatic cholestasis of pregnancy†
Hualin
Xu‡
a,
Yupin
Xu‡
b,
Guoqiang
Zhao
a,
Xukun
Fu
c,
Jian
Zhao
a,
Huaqian
Wang
a,
Yuliang
Cai
a and
Hongmei
Lin
 *a
*a
aDepartment of Obstetrics and Gynecology, Shaoxing Maternity and Child Health Care Hospital, Shaoxing 312000, Zhejiang Province, China. E-mail: linhongmei@sxfby.com
bSchool of Medicine, Shaoxing University, Shaoxing 312000, Zhejiang Province, China
cDepartment of Medical Record, Shaoxing Maternity and Child Health Care Hospital, Shaoxing 312000, Zhejiang Province, China
First published on 31st March 2023
Abstract
Intrahepatic cholestasis of pregnancy (ICP) is a pregnancy-specific hepatobiliary disease, leading to an abnormal increase in total bile acid in the blood of pregnant women. To systematically explore the similarities and differences in metabolites and metabolic pathways among three types of biological samples from ICP women, a study of 18 ICP and 6 healthy (as a normal control) pregnant women was performed to investigate their clinical information and biochemical features. Based on validated LC–MS/MS methods 1–5 for hydrophilic and hydrophobic metabolites (molecular weight <2000 Dalton), an untargeted-metabolomic strategy was applied to 24 pregnant women to determine the metabolites from 22 serum, 15 placental and 22 urine samples. Then 1137 metabolites from serum, 876 metabolites from placental tissue and 311 metabolites from urine with a coefficient of variation <30% in the pooled quality control samples were found. Furthermore, orthogonal partial least squares–discriminate analysis (OPLS–DA), correlation analysis, chemical enrichment analysis and metabolic pathway analysis were carried out by a bioinformatics process. On the OPLS–DA model analysis, the metabolites in urine were better than those in serum or placental tissue to reflect the metabolic changes of ICP disease. Some metabolites were significantly changed in serum (n = 71), placental tissue (n = 46) and urine (n = 36), such as bile acids, triacylglycerols, lysoPCs, and steroids. Primary bile acid biosynthesis was the main metabolic pathway in ICP disease, and taurine and hypotaurine metabolism and sphingolipid metabolism were also found. More specifically, bile acids increased and steroids decreased in the serum, placental and urine samples. For complex metabolic diseases such as ICP disease, untargeted-metabolomic analysis of multiple biological samples could provide a systematic understanding of the changes in metabolic types and pathways.
1. Introduction
Women have a physiological systematic adaptation for fetal growth to prompt better conditions during pregnancy.1 These changes require more energy to help the body adjust better, which makes higher demands on liver metabolism.2 Intrahepatic cholestasis of pregnancy (ICP) is a pregnancy-specific hepatobiliary disease with an incidence of 0.32% in China.3 In European and American countries, its incidence is 1–2% of pregnant women.4 ICP disease has common clinical characteristics, including pruritus, jaundice, and elevated total bile acids (TBA). Specifically, an increased TBA level above 10 μmol L−1 serves as the basis for diagnosis of this disease.Although TBA is the main diagnostic basis for ICP disease, when using it, it is impossible to distinguish ICP women with mild pruritus from normal pregnant women.5 Therefore, the relationship between bile acid profile and ICP disease has been intensively studied. In 2007, LC–MS/MS was used for quantitation of 15 bile acids to analyze bile acid profiles in the serum of ICP women.6 There was then a further discovery that concentrations of lithocholic acid in serum showed a dramatic decrease during ursodeoxycholic acid treatment.7 Severe ICP disease increased serum levels of taurochenodeoxycholic acid, tauroursodeoxycholic acid, glycocholic acid, taurocholic acid, and glycochenodeoxycholic acid.8
With the development of omics, researchers were not limited only to the study of bile acid profiles when proteomics was applied to the study in 20139 and placental gene-expression profiles in ICP women were also studied in 2014.10 Urinary and serum microRNA profiling detection in ICP as a non-invasive biomarker found its potential.11,12 For lipid profiles, decreasing lipoprotein lipase in serum and placental tissues was possibly related to ICP by participating in abnormal lipid metabolism.13 Gene, transcript, protein and metabolite move around tightly in life, and metabolites are very sensitive to a disease microenvironment. Metabolomics, an important means of translational medical research, has been used to study pathogenesis and biomarkers. The metabolomics result can reflect metabolite changes in specific pathophysiological conditions, providing a new research method for disease diagnosis.14 Recent studies of ICP disease with targeted metabolomics have mostly focused on bile acids from a single type of biological sample, such as serum samples15,16 or urine.17 Untargeted metabolomics in ICP disease has also analyzed the composition of urine.18 However, there have been few metabolomic studies comparing different types of samples for ICP disease.
In this study, we used untargeted metabolomics with validated LC–MS/MS methods 1–5 for hydrophilic and hydrophobic metabolites to detect metabolites in serum (n = 1143), placental tissue (n = 877) and urine (n = 311) from 18 ICP women and 6 healthy women. Based on OPLS–DA analysis, correlation analysis, chemical enrichment analysis and metabolic pathway analysis, a systematic understanding of the changes in metabolic types and pathways in ICP disease has been established.
2. Experimental
2.1 Chemical reagents
LC/MS-grade methanol, acetonitrile, isopropanol, formic acid, ammonium acetate and ammonium hydroxide were purchased from Fisher Scientific (Fair Lawn, USA). LC/MS-grade ammonium bicarbonate and LC-grade methyl-tert-butyl-ether (MTBE) were purchased from Sigma-Aldrich Co. (St. Louis, USA). Isotopic internal standards were purchased from Cambridge Isotope Laboratories Inc. (Tewksbury, USA) and Toronto Research Chemicals (Toronto, Canada). A lipid mixture of SPLASH LIPIDOMIX isotopic internal standard was purchased from Avanti Polar Lipids (Alabama, USA). Ultrapure water (18.2 MΩ cm) was prepared with an in-house Milli-Q Purified Water system (Merck KGaA, Darmstadt, Germany).2.2 Biological sample collection and pretreatment
From September 2020 to November 2021, 24 pregnant women were enrolled in the study and their clinical data were collected from the medical records in the Shaoxing Maternity and Child Health Care Hospital (Shaoxing, Zhejiang Province, China). Biological samples were collected from ICP/healthy women at the end of their pregnancy. As far as possible, their blood, urine and placental tissue were collected at the same time point in this systematic study. In the study, participants met the following inclusion criteria: married women were aged 20–45 years without any severe systemic diseases (including the heart, brain, liver, kidney, and hematopoietic system), or any history of drug abuse. The ICP group consisted of pregnant women who were diagnosed with ICP disease in an early stage of their pregnancy. They were also given virological tests (including hepatitis virus, Epstein–Barr virus and cytomegalovirus) and given an organic morphological examination by liver ultrasound technology (results are given in the File named “ultrasound image”). After being diagnosed with ICP disease, pregnant women in the ICP group were treated with 15 mg (kg−1 d−1) UDCA. The control group consisted of healthy pregnant women without any pregnancy complications (threatened abortion, gestational diabetes mellitus, hypertensive disorder, placenta previa, intrahepatic cholestasis of pregnancy, oligohydramnios, intrauterine growth restriction, etc.). The study protocol was established according to the ethical guidelines of the Helsinki Declaration and was approved by the Human Ethics Committee of Shaoxing Maternity and Child Health Care Hospital (No. 2018050). All study participants were adults and their written informed consent was obtained.The sample preparation procedures used in this research were adopted from previous reports.19,20 Pooled quality control samples for serum, tissue and urine were prepared, respectively, to monitor analytical performance.
Serum: in short, each serum sample was prepared using two independent methods. Firstly, 150 μL aliquots of serum were deproteinized using 600 μL of methanol–acetonitrile solution (1/1, v/v) fortified with multiple isotope-labeled compounds as extraction internal standards (lysine-d4 1.0 μg mL−1, hippuric acid-d5 1.0 μg mL−1, stearic acid-d35 1.0 μg mL−1, chenodesoxycholic acid-d4 1.0 μg mL−1, palmitic acid-13C16 0.2 μg mL−1, AcCa(12:0)-d9 0.2 μg mL−1, AcCa(18:0)-d3 0.2 μg mL−1). Aliquots of each resultant supernatant were transferred into polypropylene tubes and dried down in a CentriVap concentrator to harvest hydrophilic metabolite extracts (Labconco Corporation, Kansas City, USA). Prior to LC–MS metabolomic analysis, each extract was reconstituted with 100 μL of methanol–water mixture (1/1, v/v). Secondly, another set of 50 μL aliquots of serum samples were subjected to liquid–liquid partition to extract the lipidome using methanol and MTBE. Aliquots of the upper layer extracts were pipetted into polypropylene tubes and dried down under vacuum in a CentriVap concentrator to harvest hydrophobic lipidome extracts, which were re-dissolved using 150 μL of acetonitrile–isopropanol mixture (1/1, v/v) before being analyzed with an LC–MS based lipidomic method.
Placental tissue: briefly, 20.0 ± 1.0 mg tissue samples were weighed after thawing at ambient temperature and 600 μL of methanol–water solution (hippuric acid-d5 1.0 μg mL−1, chlorophenylalanine 1.0 μg mL−1) was added prior to homogenizing with a high-throughput tissue grinder SCIENTZ-24 (Scientz, Ningbo, China). Then 300 μL aliquots of homogenates were subjected to liquid–liquid partition to extract lipids (upper layer) and polar metabolites (lower layer), simultaneously, using 250 μL of water and 900 μL of MTBE. Aliquots of each extract were transferred into polypropylene tubes and dried in a CentriVap concentrator. Prior to LC–MS analysis, 80 μL of methanol–water solution (1/1, v/v) and 80 μL of acetonitrile–isopropanol mixture (1/1, v/v) were employed to reconstitute the polar metabolite extracts for metabolomic analysis and lipids for lipidomic analysis, respectively.
Urine: in short, 100 μL aliquots of urine samples were deproteinized using 400 μL of the same methanol–acetonitrile solution as that used for serum preparation. Aliquots of each resultant supernatant were transferred into polypropylene tubes and dried down in a CentriVap concentrator to harvest hydrophilic extracts, which were reconstituted with 80 μL of methanol–water solution (1/1, v/v) prior to LC–MS metabolomic analysis.
2.3 Metabolomic and lipidomic analysis
We adopted identical metabolomic and lipidomic analytical platforms to those reported in previous publications.21 In short, serum and tissue samples were submitted to a meta-Phenotyper™ high-coverage and precision metabolomic platform, in which five analytical methods based on ultra-high performance liquid chromatography-high resolution mass spectrometry were employed to achieve comprehensive detection of metabolites and lipids. Metabolomic analyses were conducted on an Ultimate™ 3000 UHPLC system coupled to a Q Exactive™ quadrupole Orbitrap mass spectrometer (Thermo Scientific, San Jose, USA). The first fraction of hydrophilic extract was profiled on an ACE C18-PFP column (Advanced Chromatography Technologies Ltd, Aberdeen, Scotland) and detected with positive electrospray mode. The second fraction was measured on an Acquity HSS C18 column (Waters Corporation, Milford, USA) column, and finally detected with negative electrospray mode. The third hydrophilic fraction was analyzed using hydrophilic interaction chromatography on an Acquity BEH Amide column (Waters Corporation, Milford, USA) and detected with negative electrospray ionization mode. Lipidomic analyses were performed on the same analytical instrument which was operated in positive/negative polarity switching mode for lipid detection, and an Accucore C30 core–shell column (Thermo Scientific, Bellefonte, USA) was employed for chromatographic separation of lipids. For all the analytical methods above, the tuning parameters of the mass spectrometer were set as follows: HESI ion source, sheath gas flow rate 50 arb, aux gas 15 arb, capillary temperature 325 °C, heater temperature 400 °C, S-lens RF level 50%, spray voltage 3.5 kV and 3.0 kV for positive and negative ionization, respectively. Next, 70–1000 m/z (metabolomic analyses) and 300–2000 m/z (lipidomic analyses) full scan mass spectra data under 70000 FWHM resolution as well as Top 7 or Top 10 full-scan data-dependent MS/MS spectra data were acquired with XCalibur software (Thermo Scientific, San Jose, USA). The primary settings of MS1 spectrum data acquisition included: microscans 1, AGC target 3e6, and maximum injection time 200 ms, For MS2 data acquisition, the parameters were set as follows: precursor isolation window 1.0 Da, normalized collision energy 30, resolution of MS2 spectra 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 FWHM, AGC target 1e5, maximum IT 50 ms. In addition, 3 pooled quality control samples were inserted into each analytical sequence during data acquisition.
500 FWHM, AGC target 1e5, maximum IT 50 ms. In addition, 3 pooled quality control samples were inserted into each analytical sequence during data acquisition.
2.4 Raw data process and statistics
Clinical parameters were collected from the hospital system in Shaoxing Maternity and Child Health Care Hospital. Univariate analysis including the Shapiro–Wilk test and the Mann–Whitney U-test were conducted with SPSS20.0 (IBM, USA).Raw spectral data for polar metabolites was processed by Compound Discoverer 3.3 software (Thermo Scientific, San Jose, USA) for peak extraction and structural annotation through searching against a local HMDB metabolite database,22 and a local proprietary iPhenome™ SMOL high resolution MS/MS spectrum library created using authentic standards as well as the online mzCloud library (www.mzcloud.org). Multiple chemical information, including exact mass of precursor, isotopic pattern fit score, MS/MS spectra similarity and retention time, was involved in metabolite structural annotation. Meanwhile, untargeted lipidomic data process, including peak picking and lipid identification, was executed with Lipid Search software (Thermo Scientific, San Jose, USA), through which the acquired MS/MS spectra were searched against the in silico predicted spectra of diverse endogenous lipid classes. The proposed identification results were further manually checked one-by-one based on peak shape, adduct ion behavior, fragmentation pattern, and chromatographic behavior to strictly eliminate false positives. The peak areas were extracted as relative quantitative information for annotated metabolites and lipids with TraceFinder software (Thermo Scientific, San Jose, USA). RSD% < 50% in the quality control samples was used as a filtering criterion to guarantee that only valid metabolites in the bio-samples analyzed herein were involved in further processing. Finally, the resultant data matrices from all the measurements were merged and trimmed before statistical analysis. The chemical identification results were finally annotated with classification criteria proposed by MSI (Metabolomics Standardization Initiative).23 Multivariate analyses, including principal component analysis and orthogonal partial least square–discriminant analysis, were conducted with SIMCA-P software (Sartorius Umetrics, Germany), and other univariate analyses, including independent sample t test, FDR multiple test correction, ChemRICH chemical enrichment analysis24 and data visualization, were conducted with a proprietary cloud computing platform, IPOS (http://82.157.20.231:3838/ipos/ (http://82.157.20.231:3838/ipos/)) based on R. Metabolic enrichment analysis, and pathway analysis was conducted on the MetaboAnalyst website.25
3. Results
3.1 Clinical information
Comparisons of the clinical characteristics of participants between the intrahepatic cholestasis of pregnancy (ICP) and normal control (NC) groups are presented in Table 1 and Table S1 (ESI†). The measurement data were compared statistically, showing that the serum levels of total bile acid (TBA) and direct bilirubin (DB) were higher and the serum levels of albumin/globulin (A/G), Ca, Mg were significantly lower in the ICP group (P value < 0.01). It is worth mentioning that the apolipoprotein A (ApoA) level in the ICP group had decreased (P value < 0.05). There were no differences in age, body mass index (BMI), gestational period or other clinical parameters.| Group | ICP (n = 18) | NC (n = 6) | P value |
|---|---|---|---|
| Abbreviations in the table: SD means standard deviation; BMI means body mass index; TBA means total bile acid. The P value from the Mann–Whitney U-test to compare the clinical parameters of ICP and NC groups. P value < 0.05 indicates significance for statistical analysis. | |||
| Age (year), Mean ± SD | 28.78 ± 5.83 | 29.83 ± 5.00 | 0.422 |
| BMI (kg m−2), Mean ± SD | 25.15 ± 4.29 | 24.67 ± 1.66 | 0.739 |
| Alcohol history (%) | 0/18 | 0/6 | |
| Smoking history (%) | 0/18 | 0/6 | |
| Twin pregnancy (%) | 2/18 | 0/6 | |
| Assisted reproductive technology (%) | 1/18 | 0/6 | |
| Gestational period (week), mean ± SD | 37.71 ± 1.99 | 39.33 ± 1.37 | 0.054 |
| Gestational number (1/2/3/4) | 10/3/2/3 | 3/2/1/0 | |
| Delivery number (1/2) | 15/3 | 6/0 | |
| TBA (μmol L−1), mean ± SD | 27.63 ± 21.11 | 3.52 ± 1.86 | 0.001 |
| Adverse pregnancy outcome (%) | 1/18 | 0/6 | |
3.2 Method validation
In accordance with the international academic community, the strategy of Pooled Quality Control was used for quality control (QC) in the study.19,22,25,26Principal component analysis (PCA) is an unsupervised method commonly used in multivariate statistics, reflecting the most real and original mathematical structure of multidimensional data. Comparing QC samples with all analysis samples, results showing the QC samples were closely clustered together are shown in Fig. S2 (ESI†). This means that the quality of the detected data is reliable. On the basis of the PCA results, the time series characteristics of the first principal component (PC1) scores from three QC samples were studied. The low level of PC1 variation meant that the analytical quality remains stable in Fig. S3 (ESI†). According to Fig. S4 (ESI†), more than 95% of metabolites had RSD% < 30%, which means the values of metabolites were also stable. Spearman correlation analysis was performed among QC1-3 data matrices. As expected theoretically, the correlation coefficient between QC1 and QC3 was the smallest. However, QC1 and QC3 samples are highly correlated in Fig. S5 (ESI†). High correlation indicates the high data quality of the acquired untargeted metabolomic data. The CV% of each MS response value (representing a metabolite) in the sample and the QC group was calculated, and the results are shown in Fig. S6 (ESI†). It can be seen that the CV% of metabolites in the QC group was far less than that in the sample group, which means that the methods in the study can accurately measure the differences in metabolic levels between different samples. In addition, RSD% values of isotopic internal standards were less than 15% in each sample.
3.3 Metabolic profile and the OPLS–DA model analysis
There were 1143 metabolites in serum samples (n = 22), 877 metabolites in placental tissue samples (n = 15), and 311 metabolites in urine samples (n = 22), detected with the UPLC-HRMS platform, most of them (1137, 876 and 311 metabolites in serum, tissue and urine, respectively) showed acceptable quantitative precision in the QC samples with RSD < 30%. The composition and proportions of these metabolites varied between serum, placental tissue and urine samples (Fig. S1, ESI†). There was an attempt to use the unsupervised PCA score plots of metabolic phenotypes to discriminate the ICP/NC groups, but the outcome was not effective (Fig. S7, ESI†). Therefore, the OPLS–DA model was adopted in following data analysis.The results of the OPLS–DA classification of untargeted metabolomic data from serum samples are shown in detail in Fig. 1(a), R2Y = 0.894, Q2 = 0.400. The result of the S-plot is given in Fig. S8a (ESI†), where metabolites are colored according to their chemical classes. The robustness of this OPLS–DA model was evaluated by 999 times permutation, as shown in Fig. S9a (ESI†). The intercept of Q2 was −0.239 and its slope coefficient was positive, showing that the model has no overfitting problem. In conclusion, the OPLS–DA model was relatively robust and it had general prediction ability.
The results of the OPLS–DA classification of untargeted metabolomic data from placenta tissue samples are shown in detail in Fig. 2(a), R2Y = 0.802, Q2 = 0.289. The result of the S-plot is given in Fig. S8b (ESI†) where metabolites are colored according to their chemical classes. The robustness of this OPLS–DA model was evaluated by 999 times permutation, as shown in Fig. S9b (ESI†). The intercept of Q2 was −0.385 and its slope coefficient was positive, showing that the model has no overfitting problem. In conclusion, although the results of 999 times permutation showed that the model had no overfitting problem, the explanatory and predictive ability of the OPLS–DA model were not ideal because the value of the cumulative Q2 was too small, suggesting that the difference between the ICP/NC groups was very slight.
The results of the OPLS–DA classification of untargeted metabolomic data from urine samples are shown in detail in Fig. 3(a), R2Y = 0.851, Q2 = 0.621. The result of the S-plot is given in Fig. S8c (ESI†), where metabolites are colored according to their chemical classes. The robustness of this OPLS–DA model was evaluated by 999 times permutation, as shown in Fig. S9c (ESI†). The intercept of Q2 was −0.239 and its slope coefficient was positive, showing that the model has no overfitting problem. In conclusion, the OPLS–DA model was robust when its prediction ability and accuracy were good.
Through systematic analysis of metabolites from serum, placental tissue and urine samples of ICP patients, from the mathematical point of view, the metabolites in urine were better than those in serum or placental tissue at reflecting the metabolic changes in ICP patients. Of course, the changes in serum were more significant than those in placental tissue.
3.4 Altered metabolic pathways and significant biomarkers
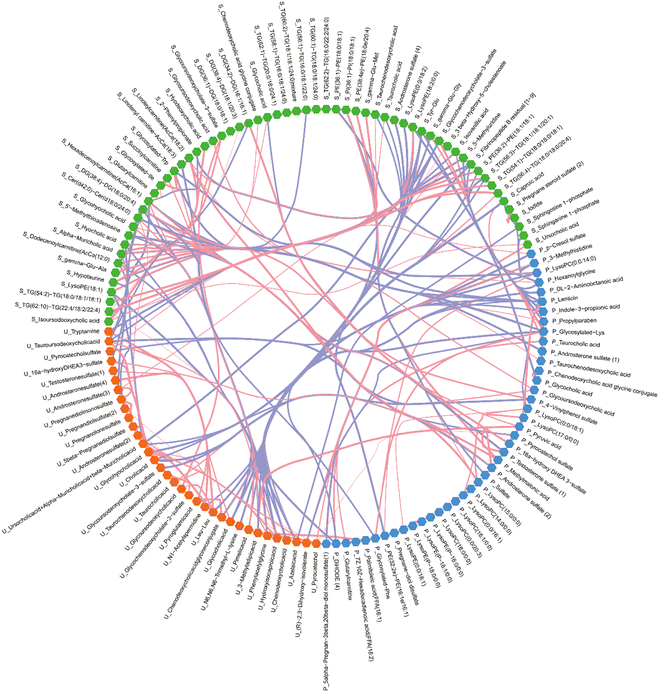
In this systematic study, using the untargeted metabolomic technique, the chemical structures of 1143 metabolites from serum samples, 877 metabolites from placental samples and 311 metabolites from urine samples were putatively annotated. Fig. S10 (ESI†) shows the chemical structure classification of differential metabolites detected systematically between the ICP/NC groups in the study. A significance level with p value < 0.05 of independent sample t tests as well as RSD% < 30% in QC samples were used to screen significantly differential metabolites between the ICP/NC groups (results are in the Excel file named “MS data (P value screening)”). Based on these significant data, the heatmaps show in detail the differences in the contents of different metabolites between the ICP/NC groups (Fig. 4). To systematically understand the correlation between the influence of ICP disease and the change in important metabolic contents between serum, urine and placental tissue, a correlation network diagram was drawn to show the complex relationships through data visualization. In Fig. 5, there are two major trends: a positive correlation among serum metabolites by themselves and a negative correlation between metabolites from serum and urine. Moreover, there was also a weak negative correlation between metabolites from urine and placenta. In ICP disease, the state of blood and placenta tended to be very similar, while urine was different because the body fluids had undergone a more complex process of metabolism.There were 71 metabolites with significant changes in the metabolic level from serum samples, and the main types were bile acids (n = 14), triacylglycerols (n = 11), acylcarnitines (n = 6) and peptides (n = 5). The proportion of different metabolic types from the ICP women is shown in Fig. 1(c). As to the change in metabolic types, diacylglycerol decreased while bile acids and sphingoid bases increased. Remarkably, three main pathways of primary bile acid biosynthesis, taurine and hypotaurine metabolism and sphingolipid metabolism can be found in Fig. 1(d), which are associated with the matching metabolites and their changing trend shown in Fig. 1(b).
There were 46 metabolites with significant changes in the metabolic level from placenta samples, and the main types were lysoPCs (n = 9), steroids (n = 6), bile acids (n = 5) and amino acids (n = 5). The proportions of different metabolic types from the ICP women are shown in Fig. 2(c). More specifically, bile acids and LysoPCs increased while benzenoids and steroids decreased. The only main pathway in Fig. 2(d) is primary bile acid biosynthesis. Although taurine and hypotaurine metabolism and sulfur metabolism did not show a strong relationship with metabolic change in Fig. 2(b), these two pathways had latent possibilities.
Compared with the NC group, the metabolic state of some metabolites (n = 36) in urine samples from the ICP group had significant changes, such as bile acids (n = 13), steroids (n = 9) and amino acids (n = 6). In addition, steroids decreased and bile acids increased in the urine from ICP women (Fig. 3(c)). The only main pathway in Fig. 3(d) is primary bile acid biosynthesis, and the changed metabolites are marked in Fig. 3(b).
With systematic analysis of metabolites from three types of sample, primary bile acid biosynthesis was the main pathway of change. There were 17 bile metabolites closely related to ICP disease from the three kinds of sample, which could be divided into two categories: (a) free bile acids, including ursocholic acid + α-muricholic acid + β-muricholic acid, cholic acid, chenodeoxycholic acid, hyodeoxycholic acid, ursocholic acid, hyocholic acid, isoursodeoxycholic acid, and α-muricholic acid; (b) conjugated bile acids, including glycohyocholic acid, glycoursodeoxycholate-3-sulfate, tauroursodeoxycholic acid, glycochenodeoxycholate-3-sulfate, chenodeoxycholic acid glycine conjugate, glycocholic acid, glycoursodeoxycholic acid, taurochenodesoxycholic acid, and taurocholic acid. From a systematic point of view, the changes in chenodeoxycholic acid glycine conjugate, glycocholic acid, glycoursodeoxycholic acid, taurochenodesoxycholic acid, and taurocholic acid took place in serum, placenta and urine together. More importantly, they were all conjugated bile acids. Apart from glycoursodeoxycholic acid, the other four bile acids were all bile acids expressed highly in human blood, and they increased in all samples from the ICP group, reflecting the overall trend of total bile acids. 3β-hydroxy-5-cholestenoate was involved in the primary bile acid biosynthesis pathway, belonging to the second pathway of bile acid synthesis (Fig. 1(d)). Unlike the above trend of conjugated bile acids, 3β-hydroxy-5-cholestenoate decreased in serum from the ICP group (Fig. 1(b)).
4. Discussion
This study presents a systematic investigation of clinical and biochemical features in ICP women by a comparison of ICP patients with normal pregnant women. In clinical practice, the diagnosis of ICP is mostly based on the concentration of TBA in serum, but the mechanism of this metabolic disease has not been thoroughly studied. Untargeted metabolomics is a useful method to explore complex diseases.27In this study, three OPLS–DA models were built for metabolites from serum (Fig. 1(a)), placental tissue (Fig. 2(a)) and urine (Fig. 3(a)). Compared with the other two types of sample, the OPLS–DA model of urine samples was robust and its prediction ability was accurate. Urine samples are more stable and simpler in composition than blood or tissue samples, and are more suitable as test samples. In 2017, a study of urine samples from ICP women had the same result for the changes in bile acids.18 But it is worth mentioning that there are no coincident metabolic indexes from amino acid metabolism and steroid hormones. In particular, steroids such as pregnanolone sulfate, pregnandiol sulfate, 16a-hydroxy DHEA 3-sulfate, pregnanediol monosulfate, 5beta-pregnanediol sulfate, androsterone sulfate, and testosterone sulfate decreased in our results (results are given in the Excel file named “MS data (P value screening)”), but steroids such as testosterone glucuronide, 18-oxocortisol, 11-oxo-androsterone glucuronide, estrone glucuronide, estriol-3-glucuronide, and cortolone-3-glucuronide increased in Ma L's results.18 Untargeted metabolomics used on biological samples has the significant advantage of an expanded range of metabolic classifications, but the coverage of metabolites will be incomplete because of their complexity.28
The differences in metabolite classes among different types of sample has been systematically compared in this study. Bile acids, steroids, organic acids and amino acids were main metabolites among the serum, placental and urine samples affected by ICP disease (results are given in the Excel file named “MS data (P value screening)”). Essential nutrients for pregnancy, such as amino acids, are metabolized differently depending on the state of the woman.1 Most amino acids showed an upward trend, but lenticin and 3-methylhistidine from placental tissue and phenylacetylglycine from urine showed a downward trend in our results. Similarly, organic acids did not show a consistent trend in different types of sample. But it is an important finding that bile acids increased and steroids decreased in serum, placental and urine samples. The abnormal change in bile acids and sulfated steroids is generally thought to be caused by ICP disease.29 Endogenous steroids may play a key role in impaired biliary lipid secretion.30 More specifically, the two classes of compound (bile acids and steroids) do not share the same transport mechanisms for excretion, but their changes are compatible with a specific defect in the biliary excretion of certain steroid sulfates.31 There is still a controversial issue whether the abnormal metabolic profiles of steroid sulfates are due to a change in progesterone metabolism or to impaired biliary secretion.
As the metabolism of basic nutrients increases during pregnancy, the liver is required to provide more energy.32 The change in bile acids is influenced in two ways: physiological self-regulation33 and ICP disease.6,8,15–17 There is no doubt that primary bile acid biosynthesis plays an important role in ICP disease.34 In the meantime, taurine and hypotaurine metabolism alleviates the liver damage as an antioxidative mechanism.35 Sphingolipid metabolism is found in our results, which has significant signaling and regulatory roles within cells.36 In recent years, researchers have also found a link between sphingolipids as modulators of liver regeneration and ICP disease.37
Although this study showed the differences between metabolites from serum, placental tissue and urine and their altered metabolic pathways in ICP disease, further exploration of the mechanism is lacking because of the limitation of current experimental conditions. In subsequent studies, we will explore potential changes in proteases based on the metabolic pathways identified and expand the sample size.
5. Conclusion
This study demonstrates that metabolomics can expand the range of exploration of metabolites associated with human disease. For complex metabolic diseases such as ICP disease, untargeted-metabolomic analysis of multiple biological samples can provide a systematic understanding that primary bile acid biosynthesis is active, bile acids increased and steroids decreased in serum, placental and urine samples, which promotes a theoretical basis for researching the mechanism of ICP disease.Data availability
All the data and material can be available. The data used to support the findings of this study are available from the corresponding author upon request.Author contributions
Hongmei Lin, Hualin Xu and Yupin Xu designed experiments; Hualin Xu and Yupin Xu analyzed results and wrote the manuscript; Guoqiang Zhao and Jian Zhao carried out experiments; Xukun Fu collected clinical information. Huaqian Wang and Yuliang Cai collected and kept the biological samples. All authors have read and approved the final manuscript.Conflicts of interest
The authors declare that they have no competing interests.Acknowledgements
We thanks for the 2019 Medical and Health Research Projects in Zhejiang Province (No. 2019KY718) for financial support. We thank Dr Wu Zeming (Iphenome, Dalian) for the technical support and Prof. Zhang Jianhua (Shaoxing University, Shaoxing) for the support of his lab.References
- J. C. King, Physiology of pregnancy and nutrient metabolism, Am. J. Clin. Nutr., 2000, 71(5), 1218S–1225S CrossRef CAS PubMed.
- B. Armistead, E. Johnson and R. VanderKamp, et al., Placental regulation of energy homeostasis during human pregnancy, Endocrinology, 2020, 161(7), bqaa076 CrossRef PubMed.
- D. D. Smith and K. M. Rood, Intrahepatic cholestasis of pregnancy, Clin. Obstet. Gynecol., 2020, 63(1), 134–151 CrossRef PubMed.
- J. Piechota and W. Jelski, Intrahepatic cholestasis in pregnancy: review of the literature, J. Clin. Med., 2020, 9(5), 1361 CrossRef CAS PubMed.
- M. Martinefski, M. Contin and S. Lucangioli, et al., In search of an accurate evaluation of intrahepatic cholestasis of pregnancy, Scientifica, 2012, 2012 Search PubMed.
- L. Ye, S. Liu and M. Wang, et al., High-performance liquid chromatography–tandem mass spectrometry for the analysis of bile acid profiles in serum of women with intrahepatic cholestasis of pregnancy, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2007, 860(1), 10–17 CrossRef CAS PubMed.
- S. E. Lucangioli, G. Castaño and M. D. Contin, et al., Lithocholic acid as a biomarker of intrahepatic cholestasis of pregnancy during ursodeoxycholic acid treatment, Ann. Clin. Biochem., 2009, 46(1), 44–49 CrossRef CAS PubMed.
- J. Chen, W. Deng and J. Wang, et al., Primary bile acids as potential biomarkers for the clinical grading of intrahepatic cholestasis of pregnancy, Int. J. Gynecol. Obstet., 2013, 122(1), 5–8 CrossRef CAS PubMed.
- T. Zhang, Y. Guo and X. Guo, et al., Comparative proteomics analysis of placenta from pregnant women with intrahepatic cholestasis of pregnancy, PLoS One, 2013, 8(12), e83281 CrossRef PubMed.
- Q. L. Du, Y. D. Pan and Y. H. Zhang, et al., Placental gene-expression profiles of intrahepatic cholestasis of pregnancy reveal involvement of multiple molecular pathways in blood vessel formation and inflammation, BMC Med. Genomics, 2014, 7(1), 1–11 CrossRef PubMed.
- L. Ma, X. Q. Zhang and D. X. Zhou, et al., Feasibility of urinary microRNA profiling detection in intrahepatic cholestasis of pregnancy and its potential as a non-invasive biomarker, Sci. Rep., 2016, 6(1), 1–7 CrossRef PubMed.
- P. Zou, L. Luo and C. Zhao, et al., The serum microRNA profile of intrahepatic cholestasis of pregnancy: identification of novel noninvasive biomarkers, Cell. Physiol. Biochem., 2018, 51(3), 1480–1488 CrossRef CAS PubMed.
- Z. M. Hao, Y. F. Ye and Y. K. Zhang, et al., Lipoprotein lipase and lipid profiles in plasma and placenta from normal pregnancies compared with patients with intrahepatic cholestasis of pregnancy, Eur. J. Obstet. Gynecol. Reprod. Biol., 2016, 203, 279–285 CrossRef CAS PubMed.
- G. A. N. Gowda, S. Zhang and H. Gu, et al., Metabolomics-based methods for early disease diagnostics. Expert review of molecular diagnostics, Expert Rev. Mol. Diagn., 2008, 8(5), 617–633 CrossRef CAS PubMed.
- Y. Cui, B. Xu and X. Zhang, et al., Diagnostic and therapeutic profiles of serum bile acids in women with intrahepatic cholestasis of pregnancy-a pseudo-targeted metabolomics study, Clin. Chim. Acta, 2018, 483, 135–141 CrossRef CAS PubMed.
- Y. He, X. Zhang and Y. Shao, et al., Recognition of asymptomatic hypercholanemia of pregnancy: Different clinical features, fetal outcomes and bile acids metabolism from intrahepatic cholestasis of pregnancy, Biochim. Biophys. Acta, Mol. Basis Dis., 2022, 1868(1), 166269 CrossRef CAS PubMed.
- Y. Li, X. Zhang and J. Chen, et al., Targeted metabolomics of sulfated bile acids in urine for the diagnosis and grading of intrahepatic cholestasis of pregnancy, Genes Dis., 2018, 5(4), 358–366 CrossRef CAS PubMed.
- L. Ma, X. Zhang and F. Pan, et al., Urinary metabolomic analysis of intrahepatic cholestasis of pregnancy based on high performance liquid chromatography/mass spectrometry, Clin. Chim. Acta, 2017, 471, 292–297 CrossRef CAS PubMed.
- B. Du, D. Ding and C. Ma, et al., Locust density shapes energy metabolism and oxidative stress resulting in divergence of flight traits, Proc. Natl. Acad. Sci. U. S. A., 2022, 119(1), e2115753118 CrossRef CAS PubMed.
- Y. Zhi, Y. Sun and Y. Jiao, et al., HR-MS Based Untargeted Lipidomics Reveals Characteristic Lipid Signatures of Wilson's Disease, Front. Pharmacol., 2021, 12 Search PubMed.
- L. Jia, J. Yang and M. Zhu, et al., A metabolite panel that differentiates Alzheimer's disease from other dementia types, Alzheimer's Dementia, 2022, 18(7), 1345–1356 CrossRef CAS PubMed.
- D. S. Wishart, A. C. Guo and E. Oler, et al., HMDB 5.0: the human metabolome database for 2022, Nucleic Acids Res., 2022, 50(D1), D622–D631 CrossRef CAS PubMed.
- L. W. Sumner, A. Amberg and D. Barrett, et al., Proposed minimum reporting standards for chemical analysis, Metabolomics, 2007, 3(3), 211–221 CrossRef CAS PubMed.
- D. K. Barupal and O. Fiehn, Chemical Similarity Enrichment Analysis (ChemRICH) as alternative to biochemical pathway mapping for metabolomic datasets, Sci. Rep., 2017, 7(1), 1–11 CrossRef CAS PubMed.
- J. Chong, D. S. Wishart and J. Xia, Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis, Curr. Protoc. Bioinformatics, 2019, 68(1), e86 Search PubMed.
- E. Ryan and G. E. Reid, Chemical derivatization and ultrahigh resolution and accurate mass spectrometry strategies for “shotgun” lipidome analysis, Acc. Chem. Res., 2016, 49(9), 1596–1604 CrossRef CAS PubMed.
- A. D. Kennedy, B. M. Wittmann and A. M. Evans, et al., Metabolomics in the clinic: A review of the shared and unique features of untargeted metabolomics for clinical research and clinical testing, J. Mass Spectrom., 2018, 53(11), 1143–1154 CrossRef CAS PubMed.
- W. J. Nash and W. B. Dunn, From mass to metabolite in human untargeted metabolomics: Recent advances in annotation of metabolites applying liquid chromatography-mass spectrometry data, TrAC, Trends Anal. Chem., 2019, 120, 115324 CrossRef.
- L. J. Meng, Steroids and bile acids in intrahepatic cholestasis of pregnancy: effects of treatment with ursodeoxycholic acid, J. Hepatol., 1997, 27(6), 1029–1040 CrossRef CAS PubMed.
- A. Pařízek, M. Hill and M. Dušková, et al., A comprehensive evaluation of steroid metabolism in women with intrahepatic cholestasis of pregnancy, PLoS One, 2016, 11(8), e0159203 CrossRef PubMed.
- J. Sjövall, Fifty years with bile acids and steroids in health and disease, Lipids, 2004, 39(8), 703–722 CrossRef PubMed.
- L. Rui, Energy metabolism in the liver, Compr. Physiol., 2014, 4(1), 177 Search PubMed.
- B. Zhu, P. Yin and Z. Ma, et al., Characteristics of bile acids metabolism profile in the second and third trimesters of normal pregnancy, Metabolism, 2019, 95, 77–83 CrossRef CAS PubMed.
- Y. Kiriyama and H. Nochi, The biosynthesis, signaling, and neurological functions of bile acids, Biomolecules, 2019, 9(6), 232 CrossRef CAS PubMed.
- T. Mizota, T. Hishiki and M. Shinoda, et al., The hypotaurine-taurine pathway as an antioxidative mechanism in patients with acute liver failure, J. Clin. Biochem. Nutr., 2021, 21–50 Search PubMed.
- C. R. Gault, L. M. Obeid and Y. A. Hannun, An overview of sphingolipid metabolism: from synthesis to breakdown, Adv Exp Med Biol., 2010, 1–23 CAS.
- A. Mikucka-Niczyporuk, P. Pierzynski and A. Lemancewicz, et al., Role of sphingolipids in the pathogenesis of intrahepatic cholestasis, Prostaglandins Other Lipid Mediators, 2020, 147, 106399 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2mo00305h |
| ‡ These authors have contributed equally to this work and share first authorship. |
| This journal is © The Royal Society of Chemistry 2023 |