Open Access Article
Open Access ArticleA smart nanopaper sensor for optical diagnosis of Helicobacter pylori infection†
Zeinab Asghari
Adib
a,
Amir Reza
Sharifi
a,
Mohammad Ali
Kiani
a,
Hossein
Yousefi
b,
Daniel
Horák
 c,
Uliana
Kostiv
c,
Ali
Nabavi-Rad
d,
Abbas
Yadegar
d,
Mohammad
Yaghoubi-Avini
e and
Hamed
Golmohammadi
c,
Uliana
Kostiv
c,
Ali
Nabavi-Rad
d,
Abbas
Yadegar
d,
Mohammad
Yaghoubi-Avini
e and
Hamed
Golmohammadi
 *af
*af
aNanosensor Bioplatforms Laboratory, Chemistry and Chemical Engineering Research Center of Iran, 14335-186, Tehran, Iran. E-mail: hamed.golmohammadi@imtek.uni-freiburg.de; golmohammadi@ccerci.ac.ir
bLaboratory of Sustainable Nanomaterials, Department of Wood Engineering and Technology, Gorgan University of Agricultural Sciences and Natural Resources, Gorgan, 4913815739, Iran
cInstitute of Macromolecular Chemistry, Academy of Sciences of the Czech Republic, Heyrovského nám. 2, 162 06 Prague 6, Czech Republic
dFoodborne and Waterborne Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran
eDepartments of Microbiology and Microbial Biotechnology, Faculty of Life Sciences and Biotechnology, Shahid Beheshti University G.C., Tehran, Iran
fDepartment of Microsystems Engineering (IMTEK), University of Freiburg, 79110 Freiburg, Germany
First published on 2nd October 2023
Abstract
Despite a decline in its prevalence, Helicobacter pylori (H. pylori), as the most common causative agent of chronic gastrointestinal diseases, still infects more than half of the world's population. Although effective, current H. pylori infection diagnostic approaches entail some drawbacks and hurdles, including requiring endoscopy and multiple biopsies, harmful chemicals, and expensive and bulky apparatus. Herein, to tackle some of state-of-the-art limitations, we have developed a biocompatible nanosensor for diagnosis of H. pylori infection. The developed optical sensor comprises NaYF4:Yb,Er,Lu@PEG-NH2 upconversion nanoparticles, red cabbage-extracted anthocyanins, and iron oxide nanoparticles embedded within chitin nanopaper. The enzymatic reaction of H. pylori urease enzyme with urea, which leads to pH changes, and consequently changes in the pH-dependent fluorescence/color profile of the developed sensor via an inner filter effect mechanism, was utilized as a sensing strategy for H. pylori infection diagnosis. A portable 3D-printed smartphone-based imaging platform was also fabricated for monitoring the developed sensor. We believe that our developed smart nanosensor can not only potentially be exploited as an ingestible sensor for non-invasive diagnosis of H. pylori infection, its therapeutic monitoring, and gastrointestinal pH monitoring, but also provide fresh insights into the development of a variety of innovative ingestible/swallowable sensors for non-invasive gastrointestinal diagnostics.
Introduction
Bacterial infections have remained as one of the prevailing and critical global health concerns as they still lead to numerous costly outbreaks along with increasing mortality annually worldwide, chiefly owing to the lack of rapid, reliable, and cost-effective sensing methods/tools for their timely diagnosis.1,2 Among infectious bacteria, Helicobacter pylori (H. pylori) is known as one of the most prevalent and perilous bacterial pathogens, infecting over 50% of the world's population.3,4H. pylori is a fastidious Gram-negative spiral-shaped microaerophilic human pathogenic bacterium that colonizes the gastric mucosa and can survive for a prolonged time in the stomach.5–7H. pylori produces several enzymes such as urease, oxidase, and catalase, of which the urease plays a crucial role in the survival of the bacterium in gastric media by neutralizing acids in the immediate environment through the hydrolysis of urea into ammonia (NH3) and carbon dioxide (CO2).6,8H. pylori infection is involved in various gastric-duodenal diseases and is responsible for 60–100% of peptic and duodenal ulcers, 80% of chronic atrophic gastritis, and 90% of mucosa-associated lymphoid tissue lymphoma.9 Remarkably, gastric cancer, which is categorized as the sixth commonly diagnosed cancer and the fourth leading factor of cancer death in 2020, is mainly caused by the chronic infection of H. pylori.10,11 Regrettably, a large number of studies conducted, especially in recent years, have underscored the antibiotic-resistant H. pylori strains.12,13 Given the importance of H. pylori infection diagnosis, particularly at early stages, and its therapeutic monitoring, to date, several invasive and non-invasive diagnostic methods/tests have been developed for its infection detection.14,15 Invasive tests (ITs) such as a rapid urease test, histology, culture, and polymerase chain reaction (PCR) are based on the direct detection of the H. pylori organism in gastric biopsies, which are obtained through endoscopy. ITs involve timely procedures along with several restrictions and are used as routine tests for H. pylori detection, the most important of which are the need for sophisticated apparatus and professional labor to run the tests and analyze the results.16,17 On the flip side, serology, a stool antigen test (SAT), a urea breath test (UBT), and a H. pylori saliva antigen test (HPS) are categorized as non-invasive tests (NITs) for H. pylori infection detection. The UBT and SAT are among the most commonly used NITs for H. pylori infection detection. The UBT, which relies on the H. pylori urease activity, measures the amount of 13CO2 or 14CO2 in exhaled breath of patients before and after ingestion of 13C or 14C-labeled urea, while the SAT detects H. pylori-associated antigens in stool samples utilizing an enzyme immunoassay. Although these diagnostic tests provide non-invasive approaches for H. pylori infection detection, they suffer from major hurdles, including poor specificity, sensitivity, and reproducibility, and the use of relatively expensive laboratory equipment as well as harmful and isotopically labeled chemicals.17–20
To remedy the above-mentioned impediments to ITs and NITs, during the past few years, biosensors have opened a new avenue for the detection of H. pylori biomarkers, mainly utilizing functional nucleic acids, particularly DNAzymes and DNA aptamers as the recognition elements.21–23 Notwithstanding the high sensitivity, selectivity, and specificity revealed by these biosensors, they still suffer from major shortcomings such as low stability and applicability for in vivo testing that hinder their further applications in real media.24,25
Herein, aiming to tackle some of the aforementioned obstacles we benefit from the concept of ingestible sensors as a new emerging technology at the cutting edge of biosensing to propose a swallowable nanosensor for non-invasive smart diagnosis of H. pylori infection. In recent decades, ingestible sensors have arisen as a fascinating and efficient sensing technology for gastrointestinal (GI) tract health monitoring through which different biological parameters such as enzymes, metabolites, hormones, electrolytes, and microbial communities can be monitored and measured in the digestive system. This technology is built upon the capability of sensors to non-invasively pass along the gut lumen and access the GI tract organs, while providing valuable information about the health status of the GI tract.26–31
In the current work, for the first time, a nanosensor has been fabricated via embedding an anthocyanin pigment (extracted from red cabbage) and NaYF4:Yb,Er,Lu@PEG-NH2 upconversion nanoparticles (UCNPs) within magnetic chitin nanofiber (ChNF) paper and employed in combination with smartphone technology for smart diagnosis of H. pylori infection. The sensing strategy for H. pylori infection diagnosis relies on the enzymatic reaction of H. pylori urease with urea and production of ammonia through an inner filter effect (IFE) mechanism that leads to variations in the pH-dependent fluorescence/color profile of the fabricated sensor. In order to utilize our fabricated sensor for in vitro monitoring of pH/H. pylori, we have also developed a portable smartphone-based imaging platform laser setup (SIS). Regarding the non-toxicity, ease of fabrication, and high efficiency of our developed smart sensor for pH/H. pylori monitoring, it is envisioned that it can also be potentially inspiring for the development of innovative ingestible/swallowable sensors amenable to diagnosing and monitoring GI tract diseases and parameters.
Experimental section
Materials and equipment
In all experiments, Milli-Q ultrapure water was used. Hydrochloric acid (37%), sodium hydroxide, urea, ammonia solution (25%), ethanol, pepsin, FeCl3·6H2O and FeCl2·4H2O were purchased from Merck, Germany. Normal saline solution was purchased from a local drugstore in Iran. For the production of ChNF paper, industrial hydrochloric acid, sodium hydroxide, hydrogen peroxide, and acetic acid were purchased from Mojalali Co., Iran, and used without further purification.For the synthesis of UCNPs, YCl3, YbCl3, ErCl3·6H2O, LuCl3, NH4F, sodium alendronate, dichloromethane, trifluoroacetic acid, triisopropylsilane, and oleic acid were purchased from Sigma-Aldrich (St. Luis, MO, USA). Sodium hydroxide was obtained from Lach-Ner (Neratovice, Czech Republic) and α-amino-ω-Boc-amino PEG (H2N-PEG-NH-Boc; 5 kDa) was purchased from Rapp Polymere (Tübingen, Germany). The solvents and cellulose dialysis membranes were purchased from Lachema (Brno, Czech Republic) and Spectrum Europe (Breda, Netherlands), respectively.
The UCNP emission spectrum was recorded using an Edinburgh Instruments FS5 spectrofluorometer (Edinburgh, UK) coupled with a CW 980 nm laser diode as an excitation source. The digital images were captured using a Motorola Edge 20 Pro smartphone with the Pro mode. Tecnai Spirit G2 transmission electron microscopy (TEM; FEI; Brno, Czech Republic) and dynamic light scattering (DLS; Scatteroscope I; Qudix, Inc., South Korea) analyses were performed to analyze the shape and size distribution of NaYF4:Yb,Er,Lu nanoparticles. UV-vis absorption spectra were recorded using a spectrophotometer model DR5000TM (Hach, Canada). pH measurements were conducted using a consort C1010 pH meter, United Kingdom. The 3D printing process was carried out using a prusa mark3 slicer, the Czech Republic.
Production of ChNF paper (chitin nanopaper)
As schematically illustrated in Fig. 1A, briefly, the shrimp's shell (Litopenaeus vannamei) was treated first with HCl acid solution (5 wt%) so as to eliminate minerals and further refluxed twice with NaOH solution (5 wt%) for protein elimination. Second, the H2O2 solution (0.1 wt%) was utilized to eliminate pigments. Next, a chicken grinder was used to ground the white shells to attain chitin powder. The obtained powder was further converted to 1 wt% chitin suspension and passed through a super disk grinder (MKCA6-3; Masuko Sangyo Co., Ltd) for mechanical nanofibrillation. The chitin suspension was converted to the highly viscous wet gel of ChNF after grinding. The well-dispersed ChNF suspension (0.2 wt%) was filtered by vacuuming followed by hot pressing at 100 °C and 2 MPa for 1 h. The mentioned process led to the formation of a transparent ChNF paper with a thickness and a density of ∼40 μm and 1.1 g cm−1, respectively. Full details of the characterization of the ChNF paper is provided in the ESI.†![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 32
32
Synthesis of UCNPs
The UCNPs were synthesized in a three-step procedure as follows:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v; 15 mL each) using centrifugation (3460 rcf) and redispersion and transferred in water.
1 v/v; 15 mL each) using centrifugation (3460 rcf) and redispersion and transferred in water.
Extraction of red cabbage anthocyanins (RCAs)
Approximately, 450 g of fresh red cabbage leaves were chopped into small pieces. Afterward, 240 mL of an ethanol/water solution (7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 v/v) was added to the chopped leaves and the mixture pH was adjusted to 2.0 using HCl (1.0 M). The mixture was then stored at 4 °C for 24 h, while preventing from light. After this, the extract was filtered followed by centrifugation at 4000 rpm for 15 min. The extracted anthocyanin solution was stored at 4 °C in the dark place for further usage.35
3 v/v) was added to the chopped leaves and the mixture pH was adjusted to 2.0 using HCl (1.0 M). The mixture was then stored at 4 °C for 24 h, while preventing from light. After this, the extract was filtered followed by centrifugation at 4000 rpm for 15 min. The extracted anthocyanin solution was stored at 4 °C in the dark place for further usage.35
Synthesis of Fe3O4 magnetic NPs
Fe3O4 Magnetic NPs were synthesized using a co-precipitation method. Firstly, a mixture of FeCl3·6H2O and FeCl2·H2O at a molar ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was prepared in water. Then, 30 mL of concentrated ammonia solution (25%) was added to the prepared solution under vigorous stirring. The mixture was further stirred for 1 h. The resulting Fe3O4 NPs were then separated using a strong magnet and washed several times with water and placed at ambient temperature to be dried.36
1 was prepared in water. Then, 30 mL of concentrated ammonia solution (25%) was added to the prepared solution under vigorous stirring. The mixture was further stirred for 1 h. The resulting Fe3O4 NPs were then separated using a strong magnet and washed several times with water and placed at ambient temperature to be dried.36
Design and fabrication of magnetic nanopaper-based sensors
First, a concentrated suspension of Fe3O4 NPs was prepared in water and one side of circular pieces of ChNF paper (6 mm diameter, cut using a hole puncher) was then casted using the as-prepared suspension in such a way that a crescent shape was created. The ChNF papers were then placed at ambient temperature for ∼1 h to be dried. Next, a mixture of the synthesized UCNPs (6 mg mL−1) and RCAs (0.56 mM) was prepared with a volume ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25. 8 μL of the as-prepared mixture was further dropped on another side of the ChNF paper and allowed for ∼30 min to be dried at ambient temperature. Then, the ChNF paper pieces were rinsed with water to remove any non-immobilized reagents. Fig. 3A shows the schematic procedure of nanopaper-based sensor fabrication.
25. 8 μL of the as-prepared mixture was further dropped on another side of the ChNF paper and allowed for ∼30 min to be dried at ambient temperature. Then, the ChNF paper pieces were rinsed with water to remove any non-immobilized reagents. Fig. 3A shows the schematic procedure of nanopaper-based sensor fabrication.
Design and fabrication of a portable smartphone-based imaging platform laser setup (SIS)
In order to design a portable smartphone-based imaging platform equipped with a laser setup, the SolidWorks 2020 program was used. The main compartments of the fabricated imaging platform are composed of an infrared laser diode module (980 nm, input power supply: AC 100–240 V, 1.0 A, 50/60 Hz, output power: 400 mW, class: IIIa and IIIb, beam diameter at aperture: 2.0 mm × 3.5 mm, dimension: D16 mm × L71 mm, Berlin Lasers, China), an optical ultra-violet (UV)/near-infrared (NIR) cut-off filter (ID ∼ 50 mm, Iran), and also a glass slide/strip, Iran.Self-developed android app (IMA)
An android-based mobile app was programmed in Java to analyze the color intensity of the optical sensors in RGB and grayscale color spaces. The app enables the condition in which any saved images in the smartphone can be imported to that. The imported images can be divided into multiple segments for easier color analysis. A pixel-scale input was also coded to cut a specific area of the probes’ images, in square, rectangle, or circular shapes without any quality loss. There is no limitation to the number of imported or cut images, however the better the computation capability of the smartphone, the faster the analysis. The developed app autonomously calculates the final concentration of the desired analytes based on the predefined regression equations and color space of each probe. Eventually, the final concentration of each analyte can be stored and transferred via any internet-based service such as email services, instant messengers, and cloud services.Analytical experiments
Gastric juice simulation
For gastric juice simulations, 0.4 g of pepsin was dissolved in 5 mL of HCl (0.1 M), and the mixture was then added to 100 mL of saline solution (0.9% w/v). Eventually, the solution pH was adjusted to the desired pH with HCl/NaOH (1 M).37In vitro detection of pH
To conduct pH detection by using the developed sensor, it was immersed in 3 mL of simulated gastric juice with different pHs ranging from 1.5 to 4.5 and after 10 s its fluorescence/color profile was measured with SIS and subsequently quantified by using IMA.Bacterial strains and culture
H. pylori clinical strain BY-1 was obtained from the H. pylori biobank of Helicobacter Research Laboratory in the Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran. The isolate was cultured on Brucella agar plates containing 5% sheep blood, 10% fetal calf serum (FCS), the Campylobacter-selective supplement (vancomycin 2.0 mg L−1, polymyxin B 0.05 mg L−1, and trimethoprim 1.0 mg L−1), and amphotericin B (2.5 mg L−1) for 2–3 days at 37 °C under microaerophilic conditions (5% O2, 10% CO2, and 85% N2) in a CO2 incubator (Innova® CO-170; New Brunswick Scientific, USA). Suspensions of H. pylori in normal saline were prepared at a density of 9 × 108 CFU mL−1 and immediately used in the following experiments.38In vitro detection of H. pylori
For in vitro H. pylori detection using the developed sensor, it was immersed in 3 mL of simulated gastric juice spiked with H. pylori with a total concentration of 108 CFU mL−1. Subsequently, the fluorescence/color profile of the developed sensor was imaged using SIS before and 2–3 min after adding 2 μL of urea (1.6 M).Results and discussion
Nanosensor for the diagnosis of H. pylori infection
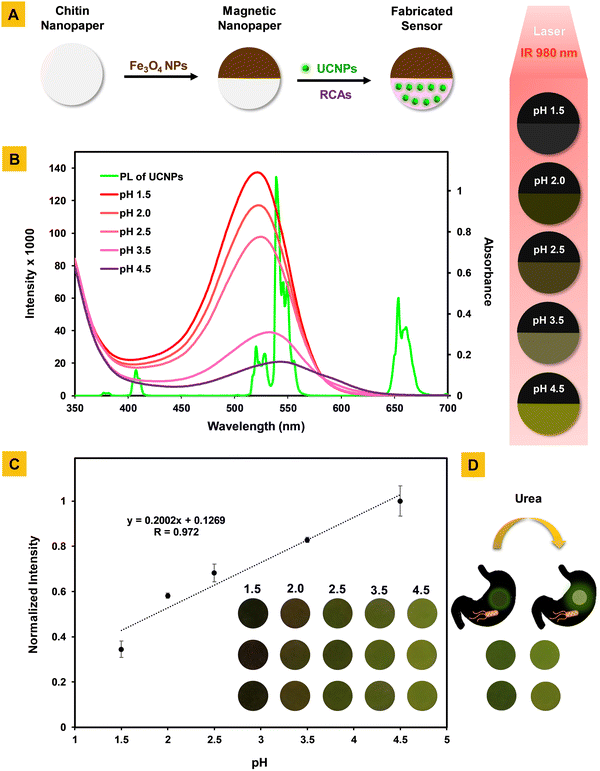
The developed nanosensor for the diagnosis of H. pylori infection comprises UCNPs, RCAs, and Fe3O4 NPs immobilized within the 3D scaffold of ChNF paper.In this work, NaYF4:Yb,Er,Lu@PEG-NH2 nanoparticles were prepared using a high-temperature co-precipitation method. The mean diameter and height of the synthesized sphero-cylindrical shape nanoparticles were 55 nm and 41 nm, respectively (Fig. 1E). They were uniform in size and are of great importance for achieving reproducible results in different applications. Also, the luminescence spectrum of the particles showed characteristic green and red emissions (Fig. 1D) of Er3+ attributed to the 2H11/2 → 2I15/2 (409 nm), 4S3/2 → 2I15/2 (525 nm), 2H9/2 → 4I15/2 (542 nm), and 4F9/2 → 2I15/2 (656 nm) transitions.
It is noteworthy that the reason for introducing a PEG protective coating on the particle surface was to prevent dissolution, reduce the possible toxicity, and maintain the luminescence of the NaYF4:Yb,Er,Lu nanoparticles. We have already evaluated the in vitro cytotoxicity of the synthesized UCNPs using primary fibroblasts and cervical carcinoma HeLa cell lines, proving that they do not show significant toxicity even at high concentrations.34
A portable smartphone-based imaging platform laser setup for optical monitoring
As shown in Fig. 2A, a 3D-printed dark chamber (181 mm × 129 mm × 10 mm), with explicit places for an adjustable gradient structure equipped with an infrared laser diode module, as an excitation source for UCNPs embedded in the fabricated fluorescent sensors, an adjustable movable sample holder structure equipped with a glass-based strip/slide with defined zones for the sample placement, a circular chamber containing an optical (UV)/NIR cut-off filter and a hole for the smartphone camera was first fabricated and the mentioned compartments were then fixed in their predetermined places. In order to take the fluorescence image of the fabricated ChNF paper-based sensors, they are placed within the dark chamber through a side strip hole, and the setup is subsequently integrated with the smartphone's camera. Furthermore, we used our developed android app (IMA) in order to quantify our nanosensor response. Fig. 2B demonstrates the different steps from imaging to analyzing and data sharing enabled by IMA. Apart from cost-effectiveness (Table S1, ESI†), ease of fabrication and use, and on-body monitoring capability compared to reported methods for the diagnosis of H. pylori infection, our fabricated smartphone-based imaging platform also benefits from fixed imaging conditions. It is noteworthy that one of the main drawbacks that significantly affects the accuracy and reliability of imaging-based optical sensors is the dependence of the analytical results obtained on the ambient light and the distance and angle of imaging, all of which have been well considered in the design of our developed imaging module.Recommended sensing strategy for H. pylori infection diagnosis
The sensing strategy for the diagnosis of H. pylori infection using the developed sensor relies on the enzymatic reaction of H. pylori urease with urea and production of ammonia. The produced ammonia in turn via a mechanism called the “inner filter effect” (IFE) leads to variations in the pH-dependent color/fluorescence profile of the developed sensor. The IFE phenomenon refers to the absorption of the excitation and/or emission light of a fluorophore using an appropriate absorber, which results in the fluorescence quenching of the fluorophore due to the spectral overlap between the absorption (excitation) and/or emission bands of the fluorophore with the absorption band of the absorber.53–56 In this work, as shown in Fig. 3B, the photoluminescence (PL) of the developed sensor is dynamically quenched in the pH range of 4.5 to 1.5 as a result of the relative overlap between the different absorption spectra of RCAs and the PL spectrum of UCNPs in the spectral region of 510–560 nm. Given the pH-dependency of RCAs and the attributed color/absorbance variations in different pHs, the PL of the developed sensor would be specific at each pH level. Accordingly, since the addition of urea to gastric juice by interacting with bacteria's urease can lead to ammonia production and subsequent pH change, the developed sensor is capable of diagnosing the H. pylori infection in a fast manner.Analytical performance and applicability of the developed sensor for pH/H. pylori detection in simulated gastric juice: an in vitro study
In this work, the pH differentiation before and after the addition of urea is measured by SIS and considered as an output signal that is in relationship with the presence of H. pylori in media. Primarily, by considering the optimal concentrations of sensing elements (UCNPs and RCAs), the analytical characteristics of our sensor were assessed in simulated gastric juice. The calibration curve was plotted and showed a linear relationship in the pH range of 1.5–4.5 with a correlation coefficient of r = 0.972. The regression equation for pH was also y = 0.2002x + 0.1269, where y is the optical analytical signal (normalized RGB value) and x is the pH of the solution (Fig. 3C). Also, the relative standard deviation for seven replicate measurements of pH = 1.5 was 1.47, which confirms the reproducibility of our developed sensor. It should be highlighted that the fluorescence/color profile of our developed sensor in different pHs are in the range of dark to bright green, which can be easily screened even by the naked eye (Fig. 3C, inset image). What's more, by using the analytical characteristics of our pH-responsive sensor, it was employed for the diagnosis of H. pylori infection according to the recommended sensing strategy. The fluorescence/color profile of our sensor, as shown in Fig. 3D, well indicates that in the presence of H. pylori and urea, the pH of simulated gastric juice increases and reaches ∼4, confirming its high efficiency for diagnosis of H. pylori infection.Conclusions
Considering its high prevalence as one of the main causes of most digestive diseases, such as gastroduodenal ulcers, chronic active gastritis, and gastric cancer, the early diagnosis of H. pylori infection is of great significance. In this work, we have developed a nanopaper-based sensor for smart non-invasively diagnosis of H. pylori infection. The sensing elements consist of UCNPs and RCAs immobilized in the 3D scaffold of a magnetic nanopaper. The sensing strategy is based on the resulting pH differentiation before and after the addition of urea, which originates from the interaction of urea with H. pylori's urease and production of ammonia. The resulting pH variations affected the color/fluorescence profile of our sensor, which was monitored using a smartphone-based laser imaging platform.While somewhat effective, existing ITs for H. pylori infection diagnosis have several disadvantages, such as demanding endoscopy and multiple biopsies, costly and bulky apparatus, timely procedures, and professional labor to run and analyze the tests.57,58 On the other hand, current NITs suffer from poor specificity, sensitivity, and reproducibility, and the use of relatively expensive laboratory equipment as well as harmful and isotopically labeled chemicals.59,60 Meanwhile, due to the use of non-toxic materials (ChNF paper, RCAs, UCNPs and iron oxide NPs)34,61 in the fabrication of our developed sensor, as well as the ease of fabrication and use, the relatively low price (Table S1, ESI†), the use of smartphone technology, and on-body monitoring capability, our sensing platform can potentially be exploited as an ingestible/wearable sensor for smart non-invasively diagnosis of H. pylori infection.
Furthermore, considering the high performance of our optical sensing platform for measuring pH in the range of 1.5 to 4.5, which was also validated by the in vitro study in simulated gastric juice, it can be utilized as an explicit sensor for pH monitoring of the GI tract to diagnose several pH-related upper GI diseases including hiatal hernia, gastritis, ulcers, gastro-esophageal reflux disease (GERD), etc.62–66 In addition, our developed sensor by detecting gastric H. pylori infection during the treatment process can help specialists in its therapeutic monitoring.
A schematic overview of our proposed workflow on how our developed sensing platform can be employed as an ingestible/wearable sensor for on-body and non-invasive monitoring of gastric pH/H. pylori infection is shown in Scheme 1.
 | ||
| Scheme 1 A schematic overview of our proposed workflow on how to use our developed sensing platform for on-body and non-invasive monitoring of the gastric pH/H. pylori infection. | ||
Altogether, owing to the results obtained from our developed sensor, we believe that such smart nanosensors can be potentially applicable for monitoring proteins, enzymes, hormones, metabolites, and microbial agents in gut lumen as well, while inspiring the design and fabrication of innovative swallowable/ingestible sensors for non-invasive diagnosis of a variety of GI disorders/diseases.
Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
All authors gratefully acknowledge support from the Chemistry & Chemical Engineering Research Centre of Iran (Tehran, Iran), the University of Gorgan (Gorgan, Iran), the Shahid Beheshti University of Medical Sciences (Tehran, Iran), and the Czech Science Foundation (no. 21-04420S). H. G. would like to thank to the Alexander von Humboldt-Stiftung/Foundation for their financial support under the Georg Förster Research Fellowship Program.Notes and references
- J. Chen, S. M. Andler, J. M. Goddard, S. R. Nugen and V. M. Rotello, Chem. Soc. Rev., 2017, 46, 1272–1283 RSC.
- M. S. Mannoor, H. Tao, J. D. Clayton, A. Sengupta, D. L. Kaplan, R. R. Naik, N. Verma, F. G. Omenetto and M. C. McAlpine, Nat. Commun., 2012, 3, 763 CrossRef PubMed.
- A. K. Miller and S. M. Williams, Genes Immunity, 2021, 22, 218–226 CrossRef PubMed.
- J. Khatoon, R. P. Rai and K. N. Prasad, World J Gastrointest. Oncol., 2016, 8, 147 CrossRef PubMed.
- P. Knezevic, V. A. Sabo, N. Simin, M. Lesjak and N. Mimica-Dukic, J. Pharm. Biomed. Anal., 2018, 152, 271–278 CrossRef CAS PubMed.
- K. Saxena, N. Chauhan and U. Jain, Anal. Biochem., 2021, 630, 114325 CrossRef CAS PubMed.
- I. A. Charitos, D. D’Agostino, S. Topi and L. Bottalico, Gastroenterology Insights, 2021, 12, 111–135 CrossRef.
- C.-Y. Kao, B.-S. Sheu and J.-J. Wu, Biomedical J, 2016, 39, 14–23 CrossRef PubMed.
- K. S. Bailey, H. E. Brown, V. Lekic, K. Pradeep, J. L. Merchant and R. B. Harris, Helicobacter, 2023, 28, e12954 CrossRef PubMed.
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, Ca-Cancer J. Clin., 2021, 71, 209–249 CrossRef PubMed.
- S. Salvatori, I. Marafini, F. Laudisi, G. Monteleone and C. Stolfi, Int. J. Mol. Sci., 2023, 24, 2895 CrossRef CAS PubMed.
- R. Ghotaslou, H. E. Leylabadlo and Y. M. Asl, World J. Methodol., 2015, 5, 164 CrossRef PubMed.
- E. Tacconelli, E. Carrara, A. Savoldi, S. Harbarth, M. Mendelson, D. L. Monnet, C. Pulcini, G. Kahlmeter, J. Kluytmans and Y. Carmeli, Lancet Infect. Dis., 2018, 18, 318–327 CrossRef PubMed.
- N. S. Atkinson and B. Braden, Dig. Dis. Sci., 2016, 61, 19–24 CrossRef CAS PubMed.
- A. I. Cardos, A. Maghiar, D. C. Zaha, O. Pop, L. Fritea, F. Miere and S. Cavalu, Diagnostics, 2022, 12, 508 CrossRef CAS PubMed.
- A. Talebi Bezmin Abadi, J. Pathog., 2018, 2018, 9064952 Search PubMed.
- R. Nosrati, B. Golichenari, A. Nezami, S. M. Taghdisi, B. Karimi, M. Ramezani, K. Abnous and S. A. M. Shaegh, TrAC, Trends Anal. Chem., 2017, 97, 428–444 CrossRef CAS.
- E. Garza-González, G. I. Perez-Perez, H. J. Maldonado-Garza and F. J. Bosques-Padilla, World J. Gastroenterol., 2014, 20, 1438 CrossRef PubMed.
- D. Y. Graham and M. Miftahussurur, J. Adv. Res., 2018, 13, 51–57 CrossRef CAS PubMed.
- T. Shimoyama, World J. Gastroenterol., 2013, 19, 8188 CrossRef PubMed.
- E. M. McConnell, D. Morrison, M. A. R. Rincon, B. J. Salena and Y. Li, TrAC, Trends Anal. Chem., 2020, 124, 115785 CrossRef CAS.
- M. M. Ali, M. Wolfe, K. Tram, J. Gu, C. D. Filipe, Y. Li and J. D. Brennan, Angew. Chem., 2019, 131, 10012–10016 CrossRef.
- Z. Liu and X. Su, Biosens. Bioelectron., 2017, 87, 66–72 CrossRef CAS PubMed.
- H. Yoo, H. Jo and S. S. Oh, Mater. Adv., 2020, 1, 2663–2687 RSC.
- E. M. McConnell, I. Cozma, Q. Mou, J. D. Brennan, Y. Lu and Y. Li, Chem. Soc. Rev., 2021, 50, 8954–8994 RSC.
- K. Kalantar-Zadeh, N. Ha, J. Z. Ou and K. J. Berean, ACS Sens., 2017, 2, 468–483 CrossRef CAS PubMed.
- L. A. Beardslee, G. E. Banis, S. Chu, S. Liu, A. A. Chapin, J. M. Stine, P. J. Pasricha and R. Ghodssi, ACS Sens., 2020, 5, 891–910 CrossRef CAS PubMed.
- E. De la Paz, N. H. Maganti, A. Trifonov, I. Jeerapan, K. Mahato, L. Yin, T. Sonsa-Ard, N. Ma, W. Jung and R. Burns, Nat. Commun., 2022, 13, 7405 CrossRef CAS PubMed.
- M. Mimee, P. Nadeau, A. Hayward, S. Carim, S. Flanagan, L. Jerger, J. Collins, S. McDonnell, R. Swartwout and R. J. Citorik, Science, 2018, 360, 915–918 CrossRef CAS PubMed.
- K. Kalantar-Zadeh, K. J. Berean, N. Ha, A. F. Chrimes, K. Xu, D. Grando, J. Z. Ou, N. Pillai, J. L. Campbell and R. Brkljača, Nat. Electron., 2018, 1, 79–87 CrossRef.
- C. J. Bettinger, Angew. Chem., Int. Ed., 2018, 57, 16946–16958 CrossRef CAS PubMed.
- T. Naghdi, H. Golmohammadi, H. Yousefi, M. Hosseinifard, U. Kostiv, D. Horak and A. Merkoci, ACS Appl. Mater. Interfaces, 2020, 12, 15538–15552 CrossRef CAS PubMed.
- U. Kostiv, Z. K. Farka, M. J. Mickert, H. H. Gorris, N. Velychkivska, O. Pop-Georgievski, M. J. Pastucha, E. K. Odstrčilíková, P. Skládal and D. Horák, Biomacromolecules, 2020, 21, 4502–4513 CrossRef CAS PubMed.
- U. Kostiv, V. Lobaz, J. Kučka, P. Švec, O. Sedláček, M. Hrubý, O. Janoušková, P. Francová, V. Kolářová and L. Šefc, Nanoscale, 2017, 9, 16680–16688 RSC.
- S. Pourjavaher, H. Almasi, S. Meshkini, S. Pirsa and E. Parandi, Carbohydr. Polym., 2017, 156, 193–201 CrossRef CAS PubMed.
- S. A. Jadhav and S. V. Patil, Front. Mater. Sci., 2014, 8, 193–198 CrossRef.
- S. Malek, M. K. Pirouzifard, A. Yadegar, S. M. V. Bonab, B. Jannat and M. Moslemi, Appl. Food Biotechnol., 2022, 9, 321–331 CAS.
- A. Nabavi-Rad, S. Jamshidizadeh, M. Azizi, A. Yadegar, K. Robinson, T. M. Monaghan and M. R. Zali, Front. Cell. Infect. Microbiol., 2023, 13, 529 Search PubMed.
- L. Bai, L. Liu, M. Esquivel, B. L. Tardy, S. Huan, X. Niu, S. Liu, G. Yang, Y. Fan and O. J. Rojas, Chem. Rev., 2022, 122, 11604–11674 CrossRef CAS PubMed.
- T. Naghdi, H. Yousefi, A. R. Sharifi and H. Golmohammadi, Comprehensive analytical chemistry, Elsevier, 2020, vol. 89, pp. 257–312 Search PubMed.
- A. R. Sharifi, T. Naghdi, H. Yousefi, M. A. Kiani and H. Golmohammadi, ACS Sustainable Chem. Eng., 2022, 16995–17026 CrossRef CAS.
- E. M. Chan, Chem. Soc. Rev., 2015, 44, 1653–1679 RSC.
- S. Wen, J. Zhou, K. Zheng, A. Bednarkiewicz, X. Liu and D. Jin, Nat. Commun., 2018, 9, 2415 CrossRef PubMed.
- F. Wang, D. Banerjee, Y. Liu, X. Chen and X. Liu, Analyst, 2010, 135, 1839–1854 RSC.
- S. Ghosh, Y.-F. Chang, D.-M. Yang and S. Chattopadhyay, Biosens. Bioelectron., 2020, 155, 112115 CrossRef CAS PubMed.
- C. Ding, S. Cheng, C. Zhang, Y. Xiong, M. Ye and Y. Xian, Anal. Chem., 2019, 91, 7181–7188 CrossRef CAS PubMed.
- H. Chen, J. Yu, J. Zhang, K. Sun, Z. Ding, Y. Jiang, Q. Hu, C. Wu and D. T. Chiu, Angew. Chem., 2021, 133, 19480–19485 CrossRef.
- T. Naghdi, S. Faham, T. Mahmoudi, N. Pourreza, R. Ghavami and H. Golmohammadi, ACS Sens., 2020, 5, 3770–3805 CrossRef CAS PubMed.
- S. Fang, Z. Guan, C. Su, W. Zhang, J. Zhu, Y. Zheng, H. Li, P. Zhao and X. Liu, Food Control, 2022, 138, 109018 CrossRef CAS.
- N. Ghareaghajlou, S. Hallaj-Nezhadi and Z. Ghasempour, Food Chem., 2021, 365, 130482 CrossRef CAS PubMed.
- G. Hrazdina, The flavonoids: advances in research, 1982, 135–188 Search PubMed.
- R. E. Wrolstad, Color and pigment analyses in fruit products, Oregon State University, 1993 Search PubMed.
- M. Shafizadeh, S. Abbasi-Moayed, Z. Hamzei, A. Keshavarz, S. Yousefi, M. R. Hormozi-Nezhad and H. Golmohammadi, Biosensors Bioelectronics: X, 2022, 11, 100150 CAS.
- H.-R. Nan, Y.-H. Liu, W.-J. Gong, H.-B. Peng, Y.-Q. Wang, Z.-B. Zhang and X.-H. Cao, Anal. Methods, 2022, 14, 532–540 RSC.
- K. Ning, Y. Fu, J. Wu, Y. Sun, K. Liu, K. Ye, J. Liu, Y. Wu and J. Liang, Anal. Methods, 2023, 15, 79–86 RSC.
- E. Sheng, Y. Lu, Y. Tan, Y. Xiao, Z. Li and Z. Dai, Anal. Chem., 2020, 92, 4364–4370 CrossRef CAS PubMed.
- L. Chen, X. Li, T. Zou, T. Wang, X. Cui, Y. Chen, C. Zhang and S. Zhao, Analyst, 2019, 144, 4086–4092 RSC.
- K. Saxena, A. Kumar, N. Chauhan, M. Khanuja, B. D. Malhotra and U. Jain, ACS Omega, 2022, 7, 32292–32301 CrossRef CAS PubMed.
- Y. Zilberman and S. R. Sonkusale, Biosens. Bioelectron., 2015, 67, 465–471 CrossRef CAS PubMed.
- H. Wu, L. Gu, X. Ma, X. Tian, S. Fan, M. Qin, J. Lu, M. Lyu and S. Wang, ACS Omega, 2021, 6, 3771–3779 CrossRef CAS PubMed.
- U. Kostiv, H. Engstová, B. Krajnik, M. Šlouf, V. Proks, A. Podhorodecki, P. Ježek and D. Horák, Front. Chem., 2020, 8, 497 CrossRef CAS PubMed.
- J. E. Richter and J. H. Rubenstein, Gastroenterology, 2018, 154, 267–276 CrossRef PubMed.
- J. Gray and M. Shiner, Gut, 1967, 8, 74 CrossRef PubMed.
- P.-J. Lu, P.-I. Hsu, C.-H. Chen, M. Hsiao, W.-C. Chang, H.-H. Tseng, K.-H. Lin, S.-K. Chuah and H.-C. Chen, World J. Gastroenterol., 2010, 16, 5496 CrossRef PubMed.
- A. Tenca, S. Massironi, D. Pugliese, D. Consonni, A. Mauro, F. Cavalcoli, M. Franchina, M. Spampatti, D. Conte and R. Penagini, Neurogastroenterol. Motil., 2016, 28, 274–280 CrossRef CAS PubMed.
- Y. Ning, S. Cheng, J.-X. Wang, Y.-W. Liu, W. Feng, F. Li and J.-L. Zhang, Chem. Sci., 2019, 10, 4227–4235 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ma00612c |
| This journal is © The Royal Society of Chemistry 2023 |