Open Access Article
Open Access ArticlePsoralidin–cucurbit[7]uril complex with improved solubility to tackle human colorectal cancer: experimental and computational study†
Fortuna
Ponte‡
a,
Nada K.
Sedky‡
 b,
Iten M.
Fawzy
b,
Iten M.
Fawzy
 c,
Fatma
Mokhtar
c,
Fatma
Mokhtar
 d,
Emilia
Sicilia
d,
Emilia
Sicilia
 *a and
Sherif Ashraf
Fahmy
*a and
Sherif Ashraf
Fahmy
 *e
*e
aDepartment of Chemistry and Chemical Technologies, University of Calabria, Arcavacata di Rende 87036, Italy. E-mail: emilia.sicilia@unical.it
bDepartment of Biochemistry, School of Life and Medical Sciences, University of Hertfordshire Hosted by Global Academic Foundation, R5 New Garden City, New Administrative Capital, Cairo, Egypt
cDepartment of Pharmaceutical Chemistry, Faculty of Pharmacy, Future University in Egypt, 11835 Cairo, Egypt
dDepartment of Pharmacognosy, Faculty of Pharmacy, El Saleheya El Gadida University, 23 El Saleheya El Gadida, 44813, Sharkia, Egypt
eDepartment of Chemistry, School of Life and Medical Sciences, University of Hertfordshire Hosted by Global Academic Foundation, R5 New Garden City, New Capital, Cairo 11835, Egypt. E-mail: sheriffahmy@aucegypt.edu; Tel: +201222613344
First published on 17th October 2023
Abstract
Nowdays, natural compounds are extensively studied for the prevention and treatment of various types of cancer due to their remarkable healing properties. In this field, encapsulating such natural anticancer agents into different delivery systems is a promising strategy for improving their therapeutic efficacy, selectivity, uptake into target tumor cells, and reducing adverse side effects. In this study, the supramolecular host–guest complexation of one of the promising bioactive compounds, Psoralidin (Ps), with cucurbit[7]uril (CB7) has been experimentally and computationally investigated. The Ps@CB7 complex has been characterized using 1H NMR and UV spectroscopy, phase solubility method, and DFT calculations. The phase solubility study suggests the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry for the formed complexes and reveals the enhancement of the Ps solubility upon complexation. The stability constant of the host–guest complex has been computed to be 2.9 × 104 M−1, which corresponds to a complexation-free energy of −6.0 kcal mol−1. Morphology and dissolution studies have been performed. Ps, CB-7, and Ps@CB7 cytotoxicity was tested against colon cancer cells (HT-29). This was followed by apoptotic assay and cell cycle investigations for both the free drug (Ps) and the formulated complex (Ps@CB7). The obtained results clearly indicate that the guest molecule forms a stable complex with the CB7 host. Calculations show that non-covalent van der Waals interactions play the most decisive role in forming a stable adduct between the host and the guest. The formed Ps@CB7 exhibits a 9-fold enhanced cytotoxic activity against HT-29 colorectal cells compared to Ps. Apoptotic assay findings also revealed a remarkably higher percent cell population among the early apoptosis, late apoptosis, and necrosis quartiles in HT-29 cells treated with Ps@CB7 than their comparable, which were treated with Ps only (P-value ≤ 0.001). Ps@CB7 superseded the effects of Ps alone in arresting the cells at the G2 phase and reducing the number of cells undergoing DNA synthesis in the S phase (P-value ≤ 0.01). In conclusion, the novel Ps@CB7 complex would be an effective supramolecular system in the fight against cancer.
1 stoichiometry for the formed complexes and reveals the enhancement of the Ps solubility upon complexation. The stability constant of the host–guest complex has been computed to be 2.9 × 104 M−1, which corresponds to a complexation-free energy of −6.0 kcal mol−1. Morphology and dissolution studies have been performed. Ps, CB-7, and Ps@CB7 cytotoxicity was tested against colon cancer cells (HT-29). This was followed by apoptotic assay and cell cycle investigations for both the free drug (Ps) and the formulated complex (Ps@CB7). The obtained results clearly indicate that the guest molecule forms a stable complex with the CB7 host. Calculations show that non-covalent van der Waals interactions play the most decisive role in forming a stable adduct between the host and the guest. The formed Ps@CB7 exhibits a 9-fold enhanced cytotoxic activity against HT-29 colorectal cells compared to Ps. Apoptotic assay findings also revealed a remarkably higher percent cell population among the early apoptosis, late apoptosis, and necrosis quartiles in HT-29 cells treated with Ps@CB7 than their comparable, which were treated with Ps only (P-value ≤ 0.001). Ps@CB7 superseded the effects of Ps alone in arresting the cells at the G2 phase and reducing the number of cells undergoing DNA synthesis in the S phase (P-value ≤ 0.01). In conclusion, the novel Ps@CB7 complex would be an effective supramolecular system in the fight against cancer.
Introduction
Colorectal cancer has been reported as the second leading cause of death after lung cancer, with about 2.5 million diagnosed cases.1 Currently, chemotherapy, radiotherapy, or tumorectomy are extensively used alone or in combination to treat different types of tumors.2 Nonetheless, these treatment options are challenging owing to the systemic side effects due to the non-selectivity effects on healthy cells, modest pharmacokinetics, high probabilities of cancer recurrence and multiple-drug resistance (MDR).2,3 Moreover, high doses for long periods are required to be administered to achieve the desired effects, leading to more toxic reactions.2,3Nature is prosperous with thousands of natural therapeutic phytochemicals that are well-accredited for their anticancer activities. In addition, natural compounds have several advantages, including lower side effects and the ability to overcome MDR, which have given them a superior preference over conventional chemotherapeutics.4,5 Furthermore, natural compounds have been co-administered with chemotherapeutics to achieve synergistic and protective effects. Thus, phytochemicals have been proposed as potential alternatives for synthetic anticancers owing to their abundant content of biologically active metabolites, such as coumarins, flavonoids, phenolic compounds, saponins, isocyanins, and others. These therapeutic metabolites have shown considerable anticancer effects through different mechanisms.4–10
Psoralidin (Ps) is a phenolic coumarin isolated from Psoralea corylifolia L. seeds which are cultivated in several regions in Asia and Europe. Several reports studied the anticancer activities of Ps, whose structure is presented in Scheme 1, against several tumors, such as lung, esophageal, breast, and colon cancers.11–13 The anticancer activities of Ps are attributed to its ability to stimulate apoptotic pathways, generate DNa anomalies, and inhibit the proliferation of cancer cells.11,12 Albeit its hopeful anticancer effects, the clinical translation of PS has been hindered by its water insolubility, poor targeting affinities, high therapeutic concentrations needed, and poor biodistribution.11 Thus, it is inspiring to innovate an unorthodox approach to address the obstacles that prevent the clinical use of Ps in cancer therapy.
Recent advances in drug delivery have enabled the transport of several medicinal phytochemicals, overcoming their shortcomings and improving their hydrophilicity, bioavailability and hence, their therapeutic effectiveness.14–17
Drug delivery systems based on supramolecular systems have recently gained enormous attention for their ability to host several therapeutically active molecules inside their cavities. Supramolecules, such as calix[n]arenes, cyclodextrins, cucurbit[n]urils, and pillar[n]arenes, have been extensively used as potential host molecules for different drug molecules owing to their extraordinary physicochemical and dynamic structural features. Such features have permitted the accommodation of several bioactive compounds inside their cavities, increasing their water solubility, improving their bioavailability, and eventually, escorting the payloads selectively to the intended sites of action.18–25
Captivatingly, cucurbit[n]urils have shown considerable benefits, such as the facile methods of fabrication, the ability to be decorated with different functional groups, and their potential to form host–guest complexes with various molecules. This presents a promising approach for carrying phytochemicals to the targeted organs while reducing their off-target effects.18,19
To this end, this study utilized cucurbit[7]uril (CB7) to host Ps via host–guest complexation forming Ps@CB7. Several characterization tests have been performed to assess the obtained system features, such as 1H NMR, UV spectrophotometry, phase solubility study, morphological properties, and dissolution study. Moreover, computational investigations have been conducted to characterize the host–guest supramolecular adducts that can be formed by Ps and CB7. Biological investigations were also performed to test the potentiated effects of Ps complexation with CB7 compared to free Ps treatment on colon cancer cells (HT-29). In vitro studies included the assessment of cell viability, apoptosis, and cell cycle kinetics following the treatment of HT-29 cells with Ps and Ps@CB7. To the best of our knowledge, no previous studies involved the complexation of Ps with CB7 intending to overcome its shortcomings in cancer therapy.
2. Materials and methods
2.1. Materials
Cucurbit[7] uril, Psoralidin, and all other chemicals have been purchased from Sigma-Aldrich (St. Louis, MO, USA). Dulbecco's modified Eagle's medium (DMEM), Fetal bovine serum, and phosphate-buffer saline (PBS) were obtained from Lonza, Basel, Switzerland. Penicillin–Streptomycin Solution was purchased from Corning (Life Science, USA). Annexin V-FITC/PI Apoptosis Detection Kit was obtained from Elabscience (Wuhan, China). HT-29 cell line was purchased from ATCC (Manassas, VA, USA).2.2. Host–guest complex formation
The complexation of Ps@CB7 has been prepared in a molar ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in an aqueous solution at 24 °C.
1 in an aqueous solution at 24 °C.
2.3. Instrumentation
UV spectrophotometric measurements have been conducted on a UV-Vis spectrophotometry (Peak instruments T-9200, USA) at 357 nm. Proton (1H) NMR spectra have been recorded using NMR Bruker™ spectrometer (Bremen, Germany).2.4. Phase solubility study
The Ps@CB7 complex has been analyzed by the phase solubility study, which has been carried out using Higuchi and Connors's method, with modifications.21,26 In brief, an excess amount of Ps has been added to aqueous solutions comprising various concentrations of CB7 (0–14 mM). Then, solutions were kept at 24 °C, shaken for 24 h, and left until equilibrium had been reached. Then, the solutions were filtered through a 0.45 μm membrane filter and measured at 357 nm using a UV-Vis spectrophotometer (Peak instruments T-9200, USA).The complex's stability constant (K) has been calculated using eqn (1).9
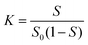 | (1) |
Then, Gibb's rule (eqn (2)) was used to estimate the free energy of the formed Ps@CB7 complex, as follows:
ΔG° = −RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) k k | (2) |
2.5. Morphological features of Ps@CB7
The morphological features and size range of the formed complex have been studied using a transmission electron microscope (JEOL-JEM 2100, Musashino, Akishima, Tokyo, Japan), operating at 160 kV.2.6. In vitro dissolution study
In vitro dissolution study has been conducted for free Ps and Ps@CB7 complex, as previously detailed, with some modifications.27 The dissolution assay was carried out employing a shaking incubator (Jeio Tech SI-300, Seoul, Korea), rotating at 120 rpm with a temperature kept at 37 °C ± 0.2 °C. In brief, 1 mL of free Ps or Ps@CB7 complex has been placed in a dialysis sac inserted into a dissolving container containing 150 mL of PBS adjusted at pH 7.4. Samples (1 mL) have been withdrawn at pre-determined time slots and restored with an equal volume of fresh buffer medium. Then, samples were centrifuged and quantified using UV-vis spectrophotometry at a wavelength of 357 nm. The release % of Ps has been determined utilizing eqn (3)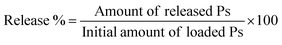 | (3) |
2.7. Cell culture
HT-29 cell lines have been purchased from the American type of culture collection (ATCC, Wesel, Germany). DMEM media has been used to culture the cells. A mixture of penicillin–streptomycin, along with 10% of heat-inactivated fetal bovine serum, have been used to supplement the culture media. After that, cells were kept in a humidified, 5% (v/v) CO2 incubator at 37 °C.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells) were collected by trypsinization and washed twice with ice-cold PBS (pH 7.4). After that, 0.5 mL of Annexin V-FITC/PI was added to the cells and incubated for 30 min at room temperature in the dark. After incubation, cells were injected in ACEA Novocyte™ flow cytometer (ACEA Biosciences Inc., San Diego, CA, USA). The cells have been evaluated for FITC and PI fluorescent signals using FL1 (λex/em 488/530 nm) and FL2 (λex/em 535/617 nm) signal detectors, respectively. A number of 12
000 cells) were collected by trypsinization and washed twice with ice-cold PBS (pH 7.4). After that, 0.5 mL of Annexin V-FITC/PI was added to the cells and incubated for 30 min at room temperature in the dark. After incubation, cells were injected in ACEA Novocyte™ flow cytometer (ACEA Biosciences Inc., San Diego, CA, USA). The cells have been evaluated for FITC and PI fluorescent signals using FL1 (λex/em 488/530 nm) and FL2 (λex/em 535/617 nm) signal detectors, respectively. A number of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events have been obtained per sample, and positive FITC and/or PI cells have been computed using ACEA NovoExpress™ software (ACEA Biosciences Inc., San Diego, CA, USA).
000 events have been obtained per sample, and positive FITC and/or PI cells have been computed using ACEA NovoExpress™ software (ACEA Biosciences Inc., San Diego, CA, USA).
2.8. Statistical analysis
Statistical analysis has been performed using GraphPad Prism software version (GraphPad, San Diego, CA, USA). All experiments have been performed in triplicate, and standard deviations (SD) have been calculated. A one-way analysis of variance (ANOVA) test has been used for multiple group analysis, followed by the post hoc Tukey–Kramer test to compare between two individual groups. The threshold of significance is denoted by (*); p-value < 0.05, (**) = P < 0.01, (***); and p-value < 0.0001. Meanwhile, ns refers to non-significant.2.9. Computational details
All the calculations presented in this paper have been carried out with the Gaussian 16 suite of programs29 employing the DFT method.The geometries of the investigated host–guest complexes have been fully optimized in water media employing the B97-D exchange and correlation functional, including Grimme dispersion corrections.30 Geometry optimizations have been performed using the 6-311G(d,p) basis set for all the atoms of the studied systems. The impact of solvation effects has been considered to carry out geometry optimization using the continuum solvation model based on density (SMD).31 whereas seven explicit water molecules in close proximity to the border of the host cavity have been added. In order to confirm the minimum character of the intercepted structures, vibrational frequency analysis has been carried out at the same level of theory, and frequency calculations within the harmonic approximation have been used to calculate the Gibbs free energies for the formation of host–guest complexes between cucurbit[7]uril (CB7) cavity and Psoralidin (Ps).
The complexation free energies in solution ΔGsol, in implicit water, have been calculated as the difference between the free energy in a solvent of the host–guest complex (GHG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) complexsol) and the free energies of the separated species in the solvent (GHsol and GGsol):
complexsol) and the free energies of the separated species in the solvent (GHsol and GGsol):
ΔGH–G![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) complexsol = GH–G complexsol = GH–G![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) complexsol − (GHsol + GGsol) complexsol − (GHsol + GGsol) |
Non-covalent interactions (NCI) in the formed clusters have been investigated by using the method proposed by Johnson et al.,33 RDG (Reduced Density Gradient) analysis, using the Multiwfn 3.8 software34 and VMD visualization program.35 The generated colored RDG maps allow precisely locating the interaction regions where the blue, green, and red regions denote the presence of H-bond and van der Waals interactions and steric effect, respectively.
3. Results & discussion
3.1. Spectrophotometric analysis of the interaction between Ps and Cb7
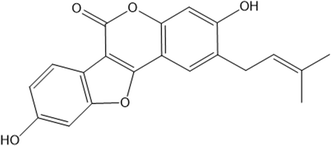
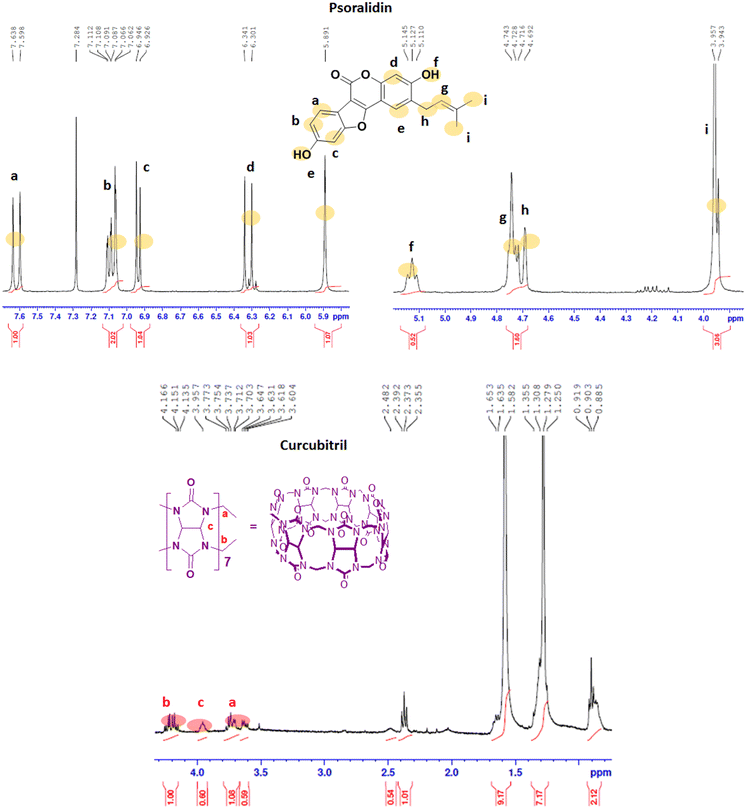
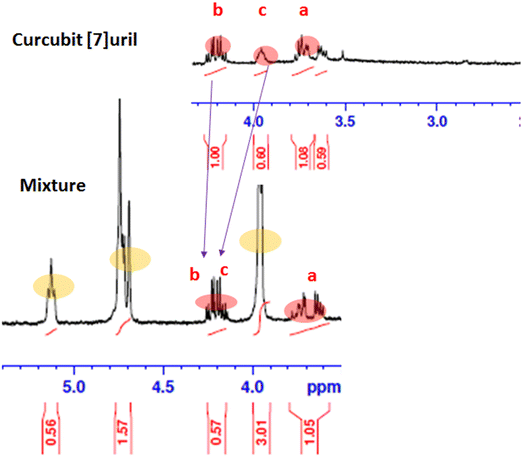
The 1H NMR spectra of the Ps, CB7, and Ps@CB7 are presented in Fig. 1 and 2, in which TMS has been used as an internal standard and chemical shifts have been expressed as ppm. 1H-NMR (400 MHz, CDCl2) has been detailed in Fig. 1, where Ps showed aromatic hydrogens at δ 7.63–7.59 (d, 1H, ArH), 7.11–7.06 (m, 1H, ArH), 6.94–6.92 (d, 1H, ArH), 6.34–6.30 (d, 1H, ArH) and 5.89 (s, 1H, ArH), while the aliphatic hydrogens appeared at δ 4.74–4.72 (t, 1H, methylene CH), 4.71–4.69 (d, 2H, CH2) and the two methyl groups appeared at δ 3.95 and 3.94 as two singlets (s, 3H, CH3). The hydroxy group has proved to be at δ 5.1 since the exchangeable peak disappeared in the performed D2O supplied in the supplementary. On the other hand, the 1H NMR (400 MHz, CDCl2) of CB7 showed no peaks in the aromatic region, and only aliphatic hydrogens appeared at δ 4.16–4.13 (b) for the CH2 protons and δ 3.71–3.60 (a) for the other CH2 protons. Meanwhile, δ 3.95–3.73 (c) was shown for CH protons.The 1H-NMR (400 MHz, CDCl2) of the mixture approved the existence of peaks corresponding to both Ps and CB7, as shown in Fig. 2, revealing the complexation of Ps with CB7 via the downfield shift evident for both (b) and (c) peaks of CFb7. Due to the complexation, –CH2 protons (b) have been shifted downfield from δ 4.16–4.13 to δ 4.25–4.21, while CH protons downshifted from δ 3.95–3.73 to δ 4.19–4.13. This could be explained by the formation of hydrogen bonds between Ps and Cb7 since the crowding of hydrogens displayed by hydrogen bonds always deshields hydrogens to the downfield.
The UV spectra attained from the aqueous solutions comprising a fixed concentration of Ps guest molecule (0.24 mM) and varying concentrations of CB7 (0–0.27 mM) are illustrated in Fig. 3.
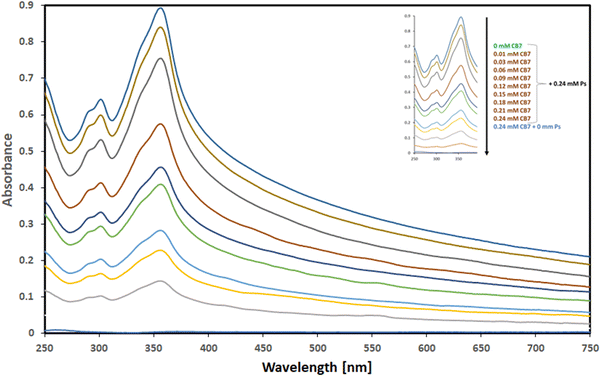 | ||
| Fig. 3 UV absorption spectra of Ps in the presence of increasing concentrations of CB7 (ranging from 0.00–0.24 mM as marked in the insert). | ||
The spectrum of CB7, in the absence of Ps, shows no absorbance in the range of 250–750 nm, while the spectrum of Ps, on the other hand, shows strong UV absorbance with a distinctive absorption maximum at 357 nm in the absence of CB7. The spectra of the mixtures also show a hypochromic shift, as evident by the observed decrease in absorbance of Ps with increasing concentrations of CB7 until a very weak absorbance of Ps has been observed when the concentration of CB7 has been equal to that of Ps (0.24 mM). Throughout this hypochromic trend of decreased absorbance, the observed absorption spectrum of Ps has been shown to diminish with increasing concentrations of CB7 used while the Ps concentration has been kept fixed. This proposed that Ps has been being complexed with CB7 and suggested the formation of a complex between Ps guest molecule and CB7 host. This is in line with previous studies showing a similar trend upon the complexation of various guest molecules with different host supramolecules.20,22,24,36–38
3.2. Phase solubility study
The phase solubility method has been used to evaluate the aqueous solubility of Ps upon its complexation with CB7 at 25 °C, as demonstrated in Table 1 and Fig. S1 (ESI†). In addition, the phase solubility method has been employed to estimate the stoichiometry, stability constant, and complexation-free energy of the Ps@CB7 complex.| Equation | R 2 | Stability constant (Ks, M−1) | ΔG° (kcal mol1) | PSD type | |
|---|---|---|---|---|---|
| Ps@CB7 | Y = 0.9043x − 0.1183 | 0.994 | 2.9 × 104 | −6.0 | AL |
The solubility of Ps increased linearly as a function of CB7, indicating a linear correlation with an R2 value of 0.994. Based on previous reports, the observed phase solubility diagram (PSD) is classified as AL-type, suggesting a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex formation between Ps and CB7 (Ps@CB7).9,21,39 Additionally, the solubility of Ps has been dramatically increased by about twenty-four times higher than its intrinsic solubility upon complexation with CB7. The calculated complex stability constant (Ks) and complexation free energy have been 2.9 × 104 M−1 and −6.0 kcal mol−1, respectively, as presented in Table 1, indicating the outstanding stability of the developed complex. The obtained Ks value is within the previously reported stability constants range (0.02 × 103–14 × 105) M− 1, envisioned for delivering various biologically active guest molecules.20,22–24,40–42
1 complex formation between Ps and CB7 (Ps@CB7).9,21,39 Additionally, the solubility of Ps has been dramatically increased by about twenty-four times higher than its intrinsic solubility upon complexation with CB7. The calculated complex stability constant (Ks) and complexation free energy have been 2.9 × 104 M−1 and −6.0 kcal mol−1, respectively, as presented in Table 1, indicating the outstanding stability of the developed complex. The obtained Ks value is within the previously reported stability constants range (0.02 × 103–14 × 105) M− 1, envisioned for delivering various biologically active guest molecules.20,22–24,40–42
3.3. Morphology and dissolution study
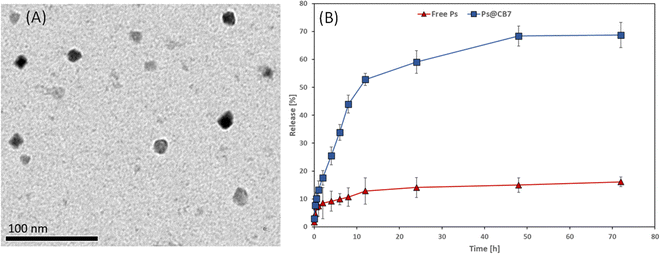
The morphological features and size range of the developed Ps@CB7 have been studied using TEM, as presented in Fig. 4A. The TEM analysis revealed the formation of spherical particles in the nanoscale range of 12–15 nm, which agrees very well with previous work reporting the design of host–guest complexes using cucurbiturils as host molecules.43Dissolution profiles of free Ps and Ps@CB7 complex are depicted in Fig. 4B. The dissolution rate of Ps has been found to increase in the CB7 complex, as evidenced by increased time-dependent release % in the case of Ps@CB7, as compared to free Ps. The Ps exhibited a high release rate from the Ps@CB7 (69%) within 72 h, as compared to free Ps (16%). Thus, the designed complex demonstrated a faster dissolution rate than free Ps. These findings align with the phase solubility study results and support the ability of the CB7 to improve the aqueous solubility and bioavailability of the hydrophobic guest molecules via the formation of host–guest complexes.
3.4. Cell viability assay
Ps has precedently demonstrated potent antiproliferative activity against different types of tumors, including gastric, colon, breast cancer, and liver cancer cells.44 CB7 is a host supramolecule that may alter the efficacy and safety of anticancer agents by forming complexes with them. For instance, CB7 improved the antitumor activity of carboplatin and oxaliplatin, platinum-based anticancer drugs.45The anticancer activity of Ps against HT-29 and HCT-116 colon cancer cells has been previously established.46 Several mechanisms have been suggested. For example, Ps has been reported to induce apoptosis and inhibit tumor cell proliferation in the colon47 and esophageal cancers.48 Ps treatment caused a significant decrease in the gene expression of the forskolin-induced corticotrophin-releasing factor.49 Ps treatment also hindered the activity of the protein tyrosine phosphatase 1B.50
In the current study, human colorectal adenocarcinoma cells (HT-29) have been treated for 48 h with 10 different concentrations (ranging from 0.01–300 μg mL−1) of blank CB7, Ps, and Ps@CB7. SRB assay has been then applied to determine the cellular viability of HT-29 following each of the investigated treatments. Our findings showed that Ps had an IC50 value of 76.88 μg mL−1 on HT-29 cells, which improved to 8.19 μg mL−1 when complexed with Cucurbit[7]uril host molecule (Ps@CB7) as demonstrated by Fig. S2 and Table 1 (ESI†). Accordingly, Ps@CB7 resulted in a 9-fold increase in cytotoxic activity as compared to free Ps treatment. This may be attributed to the improved solubility and bioavailability of Ps upon its complexation with CB7.51 Furthermore, CB7 host molecule has previously demonstrated potential use in drug delivery, providing physical and chemical stability and improving hydrophilicity for various therapeutic molecules.52
A similar strategy has been reported previously where a stable host–guest complex was designed between oxaliplatin and CB7 with potent and anticancer activity and lesser side effects. CB7 caused a significant decrease in the protein binding of oxaliplatin and a remarkable reduction in the side effects.51 This reinforces the substantial enhancement in the biological properties due to drug complexation with CB7.
To warrant the safety of CB7 as a vehicle encapsulating the examined drug (Ps), the cytotoxicity of blank CB7 has also been tested on HT-29. Our findings showed that the IC50 of CB7 against HT-29 cellular has been 211.8 μg mL−1 (i.e., >100 μg mL−1), Fig. S2 and Table 2 (ESI†). As such, the complex is rendered a safe vehicle for carrying Ps. The SRB assay findings drove potential interest to evaluate further the effects of Ps and Ps@CB7 on apoptosis and cell cycle kinetics.
| Treatment | IC50 on HT-29 (μg mL−1) |
|---|---|
| Cucurbit[7]uril (CB7) | 211.8 |
| Psoralidin (Ps) | 76.88 |
| Psoralidin/Cucurbit[7] uril complex (Ps@CB7) | 8.19 |
3.5. Apoptosis
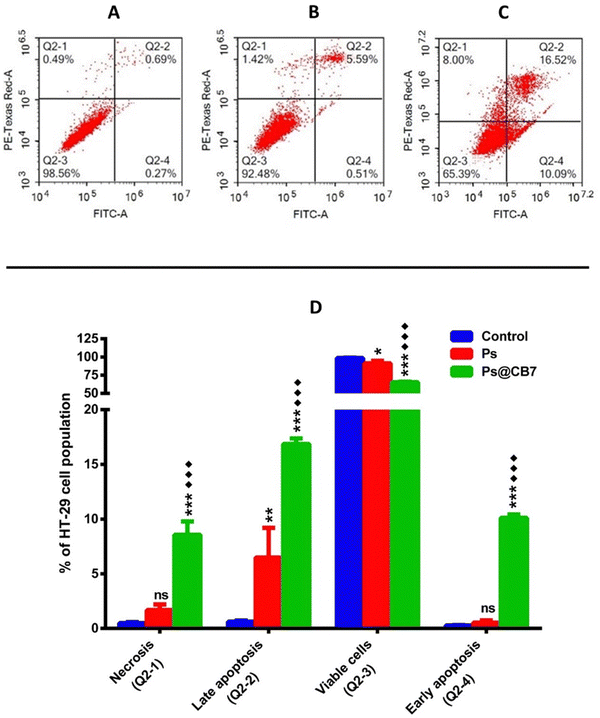
Colon cancer (HT-29) cells have been treated with the corresponding IC50 concentrations of Ps and Ps@CB7 for 48 h. Nontreated HT-29 cells have been deployed as the negative control, and results are displayed in Table 3 and Fig. 5. Ps has been shown to significantly increase the percentage of cells undergoing late apoptosis while causing only a slight increase in the percentage of cells undergoing early apoptosis. Cells pre-incubated with Ps demonstrated a 10-fold increase in the late apoptosis quartile compared to the control (untreated). Moreover, Ps treatment resulted in a noticeable decrease in the viable cells quartile (∼7.3% less than the control). Similar previous observations have been recorded upon exposure to Ps for a shorter period (8 h), where it caused a dose-dependent increase in apoptosis, especially the early apoptosis quartile among both types of colon cancer cells, HT-29 and HCT-116.46 Previous work also indicated that exposure to Ps for 48 h resulted in apoptosis of esophageal carcinoma cells (Eca9706) in a dose-dependent manner.48 As such, the apoptotic effects of Ps on cancer cells are highly ascertained.| Apoptotic stage | Percent HT-29 cell population | ||
|---|---|---|---|
| Control | Ps | Ps@CB7 | |
| The reported percent is the average of triplicates ± standard deviation. The symbols (*) and (♦) refer to statistical significance from control and Ps, respectively. The existence of any symbol once corresponds to statistical significance at P-value ≤ 0.05. The duplication of any symbol indicates statistical a P-value ≤ 0.01, and the repetition of any symbol thrice refer to values ≤ 0.001. | |||
| Necrosis (Q2-1, AV/PI+) | 0.487 ± 0.095 | 1.677 ± 0.524 | ***♦♦♦ 8.56 ± 1.22 |
| Late apoptosis (Q2-2, AV+/PI+) | 0.617 ± 0.119 | 6.483 ± 2.722** | ***♦♦♦ 16.86 ± 0.515 |
| Viable cells (Q2-3, AV−/PI−) | 98.63 ± 0.237 | 91.30 ± 3.441* | ***♦♦♦ 65.13 ± 0.860 |
| Early apoptosis (Q2-4, AV+/PI−) | 0.273 ± 0.035 | 0.543 ± 0.202 | ***♦♦♦ 10.12 ± 0.291 |
As anticipated, Ps@CB7 has been much more potent at inducing apoptosis. Treatment with Ps@CB7 reduced the percentage of cells in the viable quartile by 33.5% and 26% compared to untreated (control) and free Ps-treated groups, respectively. Such findings came to assert and provide further evidence for the cytotoxicity assay. Ps@CB7 caused a remarkable increase in both early and late apoptosis quartiles by 37 and 27-fold compared to untreated cells. Ps@CB7 also significantly increased- 19-and 3-folds in early and late apoptosis compared to Ps sole treatment. Furthermore, Ps@CB7 complex exerted an additional effect by significantly promoting necrosis. Ps@CB7 complex could induce necrosis at 18 and 5 times higher than untreated and pure Ps treatment groups. Consequently, Ps@CB7 has been more efficient at reducing the number of HT-29 cancer cells by activating both apoptotic and necrotic pathways. A similar pattern was described upon complexation of photosensitizers with CB7; simply, the complex possessed higher photodynamic, apoptotic, and necrotic effects on neuroblastoma cells than pure photosensitizer treatment, owing to the production of more significant amounts of reactive oxygen species (ROS).53
3.6. Cell cycle analysis
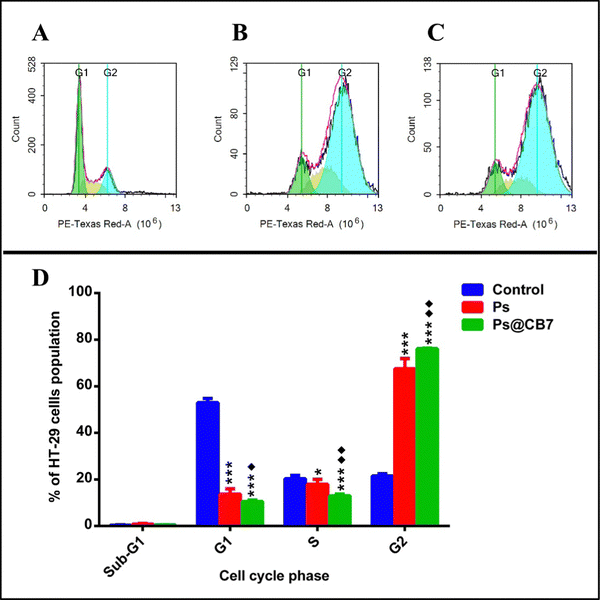
DNA flow cytometry analysis has been implemented to study the cell cycle kinetics of HT-29 cells after 48 h exposure to Ps and Ps@CB7 treatments. Untreated HT-29 cells have been employed as the control group. The assay aimed to determine the specific cell cycle phase mostly affected by Ps and Ps@CB7 treatments.G2 is a gap phase that follows the S phase (DNA synthesis phase) and prepares the cell to continue with mitosis (M-phase) and cell division. Thus, the sequestration of cells at the G2 phase and preventing their passage to the next phase (mitosis) is a successful strategy to inhibit uncontrolled cell division and cancer progression.54 Our findings suggested that Ps could trap HT-29 cells in the G2 phase and cause cycle arrest at G2/M of the cell cycle. A 3-fold increase in the frequency of cells at the G2 phase has been observed upon exposure to Ps treatment as compared to the control group (Table 4 and Fig. 6). Likewise, Li and colleagues found that Ps treatment resulted in G2/M cell cycle arrest of liver cancer cells (HepG2).55
| HT-29 cell cycle phases | Control (untreated) | Ps (76.88 μg mL−1) | Ps@CB7 (8.19 μg mL−1) |
|---|---|---|---|
| The displayed data are the mean of triplicate individual runs ± standard deviation (SD). Freq stands for the frequency of cells at the different phases of the cell cycle. | |||
| Freq Sub-G1 | 0.36 ± 0.181 | 0.87 ± 0.218 | 0.447 ± 0.085 |
| Freq G1 | 52.9 ± 1.839 | *** 13.7 ± 2.243 | ***♦ 10.52 ± 0.581 |
| Freq S | 20.33 ± 1.466 | * 17.92 ± 2.144 | ***♦♦ 12.94 ± 0.735 |
| Freq G2 | 21.54 ± 0.8997 | *** 67.51 ± 4.396 | ***♦♦ 76.09 ± 0.268 |
Nevertheless, the effect of Ps treatment on human lung cancer A549 cells has been somewhat different, where it induced cell cycle arrest at the G1 phase and caused a significant increase in the cell population at the G1 stage.56 S-phase is considered of utmost importance because it is the limiting step towards cell division. This is because the genetic material is copied or duplicated at this phase in preparation for cell division.54 The current study also noted that Ps decreased the frequency of cells in the S-phase by ∼12% and, thus, proved successful at reducing the number of cells undergoing DNA synthesis.
Ps@CB7 treatment surpassed Ps effects and resulted in more potentiated actions. Ps@CB7 induced a 3.5-fold increase in cells at the G2 phase compared to the control group. Moreover, Ps@CB7 has been highly distinguished at reducing the number of cells undergoing DNA synthesis in the S-phase by 36% less than the control group. Consequently, CB7 complex augmented the biological effects of Ps, where Ps@CB7 complex displayed much better biological properties than Ps, improved its cytotoxic activity apoptotic effects, and caused significant interference with the different phases of the cell cycle.
3.7. QM investigation of the host–guest interaction between Ps and CB7
Quantum mechanical calculations, especially when the DFT approach is used, are considered a powerful tool for the investigation of supramolecular interactions established between drugs and host molecules such as calix[n]arenes (CXs), cyclodextrins (CDs) and cucurbiturils (CBs). Several computational studies have been carried out during the past few years involving the encapsulation of several drug molecules such as carboplatin,57 paracetamol and ibuprofen,58 vemurafenib,59 pyrazinamide and isoniazid60 into the cavity of CB. To the best of our knowledge, no previous computational studies reported the encapsulation of coumestan derivatives using CB7.In this work, psoralidin (Ps), a coumestan derivative having an isopentenyl substituent in the C-2 position of coumestrol, has been chosen as a guest molecule.
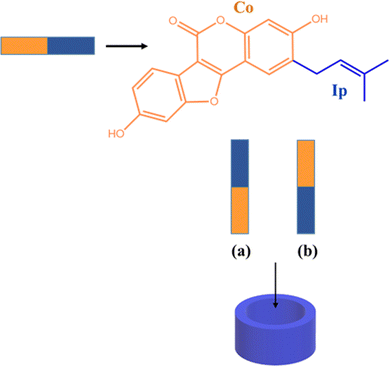
The interaction of the CB7 cavity with Ps molecule has been studied by considering different inclusion modes. Owing to the symmetrical structure of the CB7 cavity, the guest insertion only from the upper rim of the macrocycle has been considered. Several initial positions of the guest with respect to the host have been examined, placing the guest very far and close to the host. All reasonable modes of inclusion were computationally examined converging to two structures reported in Scheme 2.
In particular, Ps can assume two different orientations with respect to the host macrocycle. The arrangement in which the coumestrol moiety (Co) of Ps points towards the inside of the cavity and the isopentenyl group (Ip) points outside the cavity is labeled as A, while the label B indicates the disposition in which the unsaturated chain of Ps, Ip, points towards the interior of the cavity and the Co group is outwardly oriented.
All the reliable examined geometries describing the inclusion modes have been optimized, adopting the B97-D DFT functional in an implicit aqueous solution. The use of the B97-D functional, able to take into account non-covalent interactions, whose inclusion is critical for the proper reproduction of the properties of the investigated systems, is supported by its successful use for previous host–guest system investigations.20,23,24,40,61,62
Seven explicit water molecules around the host cavity have been added. These seven water molecules have been added to avoid that, since the impact of the solvent is taken into consideration adopting an implicit approach, weak interactions established with the peripheral carbonyl oxygens on the rim could block the inclusion of the guest inside the host cavity. Fig. 7 shows the optimized geometries of the corresponding host–guest complexes between CB7 host and Ps guest in A and B arrangements. Amongst the adopted distinct spatial arrangements of the Ps with respect to the CB7 molecule, the one that should favor the inclusion is identified as “perpendicular” with respect to the host portal. Therefore, optimized geometries have been obtained starting from such perpendicular arrangments for both A and B conformations. Despite these precautions, the drug does not enter the cavity and establishes weak interactions with the macromolecule at its border.
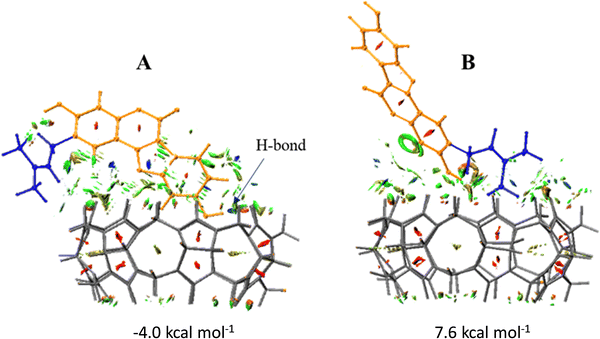
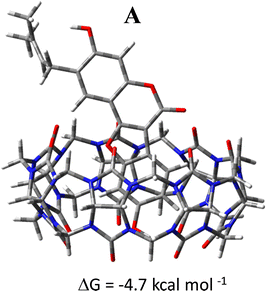
The graphical visualizations of non-covalent interactions obtained using the RDG analysis of non-covalent interactions are reported for each supramolecular aggregate. The energies of complexation have been calculated in solution, including the effect of basis set superposition error (BSSE) and entropy change corrections, and are reported in Fig. 7.
The DFT results suggest that the complexation process of Ps and CB7 is thermodynamically favorable in only one of the two investigated conformations, named A, supporting a good propensity of the macrocycle to maximize interactions with this guest. For the most stable A conformation host–guest adduct, the calculated complexation-free energy is −4.0 kcal mol−1. Instead, the adduct in B conformation is not stabilized as the complexation-free energy of 7.6 kcal mol−1, which implies lower adaptability of the host cavity when the Ps is rotated with the unsaturated Ip chain pointing inside it. Furthermore, the complexation of the Ps molecule induces, in both supramolecular complexes, small structural variations in the CB7 cavity.
From the analysis of the colored RDG map, in both configurations, it can be deduced that intramolecular interactions inside the host cavity are dominated by steric repulsion. At the same time, the prevailing color green indicates that the interactions in the intermolecular region are of van der Waals type. Precisely, in A conformation, the system is stabilized by weak interactions. A single hydrogen bond is formed, marked in blue and indicated with an arrow, between the phenolic OH group of Co and a carbonyl oxygen atom of the macrocyclic host. 1H-NMR experiments have evidenced the formation of such an H-bond.
Furthermore, a representation of the host–guest A and B arrangements, in which a model of interpenetrating spheres of van der Waals radii has been used for the guest, is reported in Fig. S3 of the ESI.† It appears that, although the Ps guest should be included into the CB7 host cavity, the rigidity of its structure does not allow, when both terminal parts of the molecule interact with the rim atoms, that further interactions are established with the internal regions of the cavity driving the inclusion.
Additionally, the encapsulation of the guest has been forced including manually the Ps guest into the CB7 host cavity avoiding interactions at the border to be established hampering the encapsulation. The optimized geometrical structure of the obtained host–guest inclusion complex in conformation A is reported in Fig. 8. The complexation free energy has been calculated in solution, including the effect of the basis set superposition error (BSSE) and entropy change correction, and is reported in the same Fig. 8. The value of the calculated free energy change of the inclusion complex formed by including manually the guest inside the host is −4.7 kcal mol−1 compatible with the value estimated experimentally.
4. Conclusions
The present work provides a comprehensive study for forming supramolecular host–guest complexes between cucurbit[7]uril and Psoralidin, a natural molecule with multiple therapeutic effects proven in preclinical studies.The phase solubility studies show that Ps and CB7 form a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 host–guest inclusion complex. CB7 binds to Ps molecule in an aqueous solution with good affinity (K = 2.9 × 104 M−1 that corresponds to a complexation-free energy of −6.0 kcal mol−1). According to DFT calculations, the dominating intermolecular interactions in the most stable adduct are of van der Waals type, with additional contributions coming from the interaction of the phenolic OH group of the coumestrol with the CB7 guest cavity. As a result, the complex Ps@CB7 in its conformation with the coumestrol moiety pointing towards the host cavity, is formed in an aqueous solution. The calculated complexation free energy of −4.0 kcal mol−1 is in very good agreement with the experimental value. The guest can be also placed manually inside the cavity, bypassing the interactions that prevent the encapsulation, forming an inclusion complex, whose complexation fre energy is calculated to be −4.7 kcal mol−1. This value, if it is assumed that the complex can be forced to be encapsulated, fits well the estimated experimental value.
1 host–guest inclusion complex. CB7 binds to Ps molecule in an aqueous solution with good affinity (K = 2.9 × 104 M−1 that corresponds to a complexation-free energy of −6.0 kcal mol−1). According to DFT calculations, the dominating intermolecular interactions in the most stable adduct are of van der Waals type, with additional contributions coming from the interaction of the phenolic OH group of the coumestrol with the CB7 guest cavity. As a result, the complex Ps@CB7 in its conformation with the coumestrol moiety pointing towards the host cavity, is formed in an aqueous solution. The calculated complexation free energy of −4.0 kcal mol−1 is in very good agreement with the experimental value. The guest can be also placed manually inside the cavity, bypassing the interactions that prevent the encapsulation, forming an inclusion complex, whose complexation fre energy is calculated to be −4.7 kcal mol−1. This value, if it is assumed that the complex can be forced to be encapsulated, fits well the estimated experimental value.
The IC50 on human colorectal adenocarcinoma cells (HT-29) of Ps@CB7 complex is 8.19 μg mL−1 compared to free Ps, (76.88 μg mL−1). The ability of CB7 to strengthen the cytotoxic and apoptotic effects of Ps was confirmed through the apoptosis assay and cell cycle analysis. Ps@CB7 treatment resulted in a significant increase in the percent cell population among both apoptotic and necrotic quartiles as compared to free Ps.
Ps@CB7 also showed distinguished ability at trapping HT-29 cells in the G2 phase of the cell cycle and markedly decreasing the number of cells undergoing DNA synthesis in the S-phase.
It is clear that the host–guest complex between Ps and CB7 enhances the bioavailability, solubility, and stability of the natural drug and improves its desired pharmacological activities.
Data availability
Data is contained within the article.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work has been funded by the RSC Research Fund grant (ID: R22-3870733873) from the Royal Society of Chemistry to Dr Sherif Ashraf Fahmy and by the Italian Association for Cancer Research, AIRC (ID: 25578) to Dr Fortuna Ponte.References
- H. Sung, et al., Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries, Ca-Cancer J. Clin., 2021, 71(3), 209–249 CrossRef PubMed.
- A. Yadav, et al., Advances in delivery of chemotherapeutic agents for cancer treatment, AAPS PharmSciTech, 2022, 23, 1–14 CrossRef PubMed.
- I. Ritacco, et al., Hydrolysis in acidic environment and degradation of satraplatin: a joint experimental and theoretical investigation, Inorg. Chem., 2017, 56(10), 6013–6026 CrossRef CAS PubMed.
- C. C. Patiño-Morales, et al., Antitumor effects of natural compounds derived from Allium sativum on neuroblastoma: an overview, Antioxidants, 2021, 11(1), 48 CrossRef PubMed.
- M. Kubczak, A. Szustka and M. Rogalińska, Molecular targets of natural compounds with anti-cancer properties, Int. J. Mol. Sci., 2021, 22(24), 13659 CrossRef CAS PubMed.
- S. A. Fahmy, et al., PEGylated chitosan nanoparticles encapsulating ascorbic acid and oxaliplatin exhibit dramatic apoptotic effects against breast cancer cells, Pharmaceutics, 2022, 14(2), 407 CrossRef CAS PubMed.
- H. M. E.-S. Azzazy, et al., Essential Oils Extracted from Boswellia sacra Oleo Gum Resin Loaded into PLGA–PCL Nanoparticles: Enhanced Cytotoxic and Apoptotic Effects against Breast Cancer Cells, ACS Omega, 2022, 8(1), 1017–1025 CrossRef PubMed.
- H. M. E.-S. Azzazy, et al., Peganum harmala Alkaloids and Tannic Acid Encapsulated in PAMAM Dendrimers: Improved Anticancer Activities as Compared to Doxorubicin, ACS Appl. Polym. Mater., 2022, 4(10), 7228–7239 CrossRef CAS.
- S. A. Fahmy, et al., Enhanced antioxidant, antiviral, and anticancer activities of the extract of fermented Egyptian rice bran complexed with hydroxypropyl-β-cyclodextrin, ACS Omega, 2022, 7(23), 19545–19554 CrossRef CAS PubMed.
- K. A. Mohamad, R. N. El-Naga and S. A. Wahdan, Neuroprotective effects of indole-3-carbinol on the rotenone rat model of Parkinson's disease: Impact of the SIRT1-AMPK signaling pathway, Toxicol. Appl. Pharmacol., 2022, 435, 115853 CrossRef CAS PubMed.
- J. Sharifi-Rad, et al., Pharmacological activities of psoralidin: a comprehensive review of the molecular mechanisms of action, Front. Pharmacol., 2020, 11, 571459 CrossRef CAS PubMed.
- Z. Xin, et al., Mechanisms explaining the efficacy of psoralidin in cancer and osteoporosis, a review, Pharmacol. Res., 2019, 147, 104334 CrossRef CAS PubMed.
- R. A. Youness, et al., Oral Delivery of Psoralidin by Mucoadhesive Surface-Modified Bilosomes Showed Boosted Apoptotic and Necrotic Effects against Breast and Lung Cancer Cells, Polymers, 2023, 15(6), 1464 CrossRef CAS PubMed.
- O. A. A. Alabrahim and H. M. E.-S. Azzazy, Polymeric nanoparticles for dopamine and levodopa replacement in Parkinson's disease, Nanoscale Adv., 2022, 4(24), 5233–5244 RSC.
- L. A. Gnann, et al., Hematological and hepatic effects of vascular epidermal growth factor (VEGF) used to stimulate hair growth in an animal model, BMC Dermatol., 2013, 13(1), 1–5 CrossRef PubMed.
- H. Park, A. Otte and K. Park, Evolution of drug delivery systems: From 1950 to 2020 and beyond, J. Controlled Release, 2022, 342, 53–65 CrossRef CAS PubMed.
- K. Paunovska, D. Loughrey and J. E. Dahlman, Drug delivery systems for RNA therapeutics, Nat. Rev. Genet., 2022, 23(5), 265–280 CrossRef CAS PubMed.
- O. A. A. Alabrahim, S. A. Fahmy and H. M. E.-S. Azzazy, Stimuli-Responsive Cucurbit [n] uril-Based Supramolecular Nanocarriers for Delivery of Chemotherapeutics, ACS Appl. Nano Mater., 2023, 6(5), 3139–3158 CrossRef CAS.
- S. A. Fahmy, et al., Chemotherapy based on supramolecular chemistry: A promising strategy in cancer therapy, Pharmaceutics, 2019, 11(6), 292 CrossRef CAS PubMed.
- S. A. Fahmy, et al., Synthesis, characterization and host-guest complexation of asplatin: improved in vitro cytotoxicity and biocompatibility as compared to cisplatin, Pharmaceuticals, 2022, 15(2), 259 CrossRef CAS PubMed.
- S. A. Fahmy, et al., PLGA/PEG nanoparticles loaded with cyclodextrin-Peganum harmala alkaloid complex and ascorbic acid with promising antimicrobial activities, Pharmaceutics, 2022, 14(1), 142 CrossRef CAS PubMed.
- S. A. Fahmy, et al., Experimental and Computational Investigations of Carboplatin Supramolecular Complexes, ACS Omega, 2020, 5(48), 31456–31466 CrossRef CAS PubMed.
- S. A. Fahmy, et al., Betaine host–guest complexation with a calixarene receptor: Enhanced in vitro anticancer effect, RSC Adv., 2021, 11(40), 24673–24680 RSC.
- S. A. Fahmy, et al., Host-Guest Complexation of Oxaliplatin and Para-Sulfonatocalix [n] Arenes for Potential Use in Cancer Therapy, Molecules, 2020, 25(24), 5926 CrossRef CAS PubMed.
- S. A. Fahmy, et al., Peganum harmala alkaloids self-assembled supramolecular nanocapsules with enhanced antioxidant and cytotoxic activities, ACS Omega, 2021, 6(18), 11954–11963 CrossRef CAS PubMed.
- T. Higuchi, Phase-solubility techniques, Adv. Anal. Chem. Instrum., 1965, 4, 117–212 CAS.
- P. Arya and N. Raghav, In-vitro studies of Curcumin-β-cyclodextrin inclusion complex as sustained release system, J. Mol. Struct., 2021, 1228, 129774 CrossRef CAS.
- S. Ioniţă, et al., Resveratrol encapsulation and release from pristine and functionalized mesoporous silica carriers, Pharmaceutics, 2022, 14(1), 203 CrossRef PubMed.
- M. E. Frisch, et al., Gaussian 16, revision C. 01, 2016, Gaussian, Inc., Wallingford CT Search PubMed.
- S. Grimme, Semiempirical GGA-type density functional constructed with a long-range dispersion correction, J. Comput. Chem., 2006, 27(15), 1787 CrossRef CAS PubMed.
- A. V. Marenich, C. J. Cramer and D. G. Truhlar, Universal solvation model based on solute electron density and on a continuum model of the solvent defined by the bulk dielectric constant and atomic surface tensions, J. Phys. Chem. B, 2009, 113(18), 6378 CrossRef CAS PubMed.
- S. F. Boys and F. Bernardi, The calculation of small molecular interactions by the differences of separate total energies. Some procedures with reduced errors, Mol. Phys., 1970, 19(4), 553–566 CrossRef CAS.
- E. R. Johnson, et al., Revealing non-covalent interactions, J. Am. Chem. Soc., 2010, 132(18), 6498–6506 CrossRef CAS PubMed.
- T. Lu and F. Chen, Multiwfn: A multifunctional wavefunction analyzer, J. Comput. Chem., 2012, 33(5), 580–592 CrossRef CAS PubMed.
- H. William, VMD-visual molecular dynamics, J. Mol. Graphics, 1996, 14, 33–38 CrossRef PubMed.
- H. Zhang, et al., Host–guest interactions of 6-benzyladenine with normal and modified cucurbituril: 1H NMR, UV absorption spectroscopy and phase solubility methods, Supramol. Chem., 2011, 23(7), 527–532 CrossRef CAS.
- S. A. Ahmed, et al., Inclusion of a coumarin derivative inside the macrocyclic hosts: A spectroscopic, thermodynamic and theoretical investigation, J. Mol. Liq., 2018, 264, 550–562 CrossRef CAS.
- S. A. Fahmy, et al., Stimuli-Responsive Amphiphilic Pillar [n] arene Nanovesicles for Targeted Delivery of Cancer Drugs, ACS Omega, 2021, 6(40), 25876–25883 CrossRef CAS PubMed.
- Y. Huang, et al., Solubility enhancement of kinetin through host–guest interactions with cucurbiturils, J. Inclusion Phenom. Macrocyclic Chem., 2008, 61, 171–177 CrossRef CAS.
- S. A. Fahmy, et al., Investigation of the host-guest complexation between 4-sulfocalix [4] arene and nedaplatin for potential use in drug delivery, Spectrochim. Acta, Part A, 2018, 193, 528–536 CrossRef CAS PubMed.
- A. Praetorius, et al., Design of a fluorescent dye for indicator displacement from cucurbiturils: A macrocycle-responsive fluorescent switch operating through ap K a shift, Org. Lett., 2008, 10(18), 4089–4092 CrossRef CAS PubMed.
- N. J. Wheate, et al., Side-on binding of p-sulphonatocalix [4] arene to the dinuclear platinum complex trans-[{PtCl (NH3) 2} 2μ-dpzm] 2+ and its implications for anticancer drug delivery, J. Inorg. Biochem., 2009, 103(3), 448–454 CrossRef CAS PubMed.
- B. Al Tbakhi, et al., Cinnamaldehyde–cucurbituril complex: Investigation of loading efficiency and its role in enhancing cinnamaldehyde in vitro anti-tumor activity, RSC Adv., 2022, 12(12), 7540–7549 RSC.
- J. Li, et al., Psoralidin inhibits the proliferation of human liver cancer cells by triggering cell cycle arrest, apoptosis and autophagy and inhibits tumor growth in vivo, J BUON, 2019, 24(5), 1950–1955 Search PubMed.
- E. Pashkina, et al., The Effect of Cucurbit[7]uril on the Antitumor and Immunomodulating Properties of Oxaliplatin and Carboplatin, Int. J. Mol. Sci., 2021, 22, 14 CrossRef PubMed.
- C. Sun, et al., Psoralidin, a natural compound from Psoralea corylifolia, induces oxidative damage mediated apoptosis in colon cancer cells, J. Biochem. Mol. Toxicol., 2022, 36(7), e23051 CrossRef CAS PubMed.
- Z. Jin, et al., Differential effect of psoralidin in enhancing apoptosis of colon cancer cells via nuclear factor-κB and B-cell lymphoma-2/B-cell lymphoma-2-associated X protein signaling pathways, Oncol. Lett., 2016, 11(1), 267–272 CrossRef CAS PubMed.
- Z. Jin, et al., Psoralidin inhibits proliferation and enhances apoptosis of human esophageal carcinoma cells via NF-κB and PI3K/Akt signaling pathways, Oncol. Lett., 2016, 12(2), 971–976 CrossRef CAS PubMed.
- Y. Chen, et al., Transcriptional regulation of corticotrophin releasing factor gene by furocoumarins isolated from seeds of Psoralea corylifolia, Life Sci., 2008, 82(21–22), 1117 CrossRef CAS PubMed.
- L. Ren, et al., New compounds from the seeds of Psoralea corylifolia with their protein tyrosine phosphatase 1B inhibitory activity, J. Asian Nat. Prod. Res., 2020, 22(8), 732–737 CrossRef CAS PubMed.
- Y. J. Jeon, et al., Novel molecular drug carrier: encapsulation of oxaliplatin in cucurbit[7]uril and its effects on stability and reactivity of the drug, Org. Biomol. Chem., 2005, 3(11), 2122 RSC.
- J. A. Plumb, et al., Cucurbit [7] uril encapsulated cisplatin overcomes cisplatin resistance via a pharmacokinetic effect, Metallomics, 2012, 4(6), 561–567 CrossRef CAS PubMed.
- X. Li, et al., Cucurbit [7] uril enhances photosensitization of porphyrins in neuroblastoma cells, Photodiagn. Photodyn. Ther., 2019, 25, 364–368 CrossRef CAS PubMed.
- G. H. Williams and K. Stoeber, The cell cycle and cancer, J. Pathol., 2012, 226(2), 352–364 CrossRef CAS PubMed.
- J. Li, et al., Psoralidin inhibits the proliferation of human liver cancer cells by triggering cell cycle arrest, apoptosis and autophagy and inhibits tumor growth in vivo, J BUON, 2019, 24(5), 1950 Search PubMed.
- W. Hao, et al., Psoralidin induces autophagy through ROS generation which inhibits the proliferation of human lung cancer A549 cells, PeerJ, 2014, 2, e555 CrossRef PubMed.
- E. Pashkina, et al., The effect of cucurbit [7] uril on the antitumor and immunomodulating properties of oxaliplatin and carboplatin, Int. J. Mol. Sci., 2021, 22(14), 7337 CrossRef CAS PubMed.
- P. G. de Lima, R. G. Viegas and O. V. de Oliveira, Computational studies of the encapsulation of ibuprofen and paracetamol into cucurbit [7] uril, Comput. Theor. Chem., 2021, 1206, 113465 CrossRef.
- A. D. Bani-Yaseen, The supramolecular host-guest complexation of Vemurafenib with β-cyclodextrin and cucurbit [7] uril as drug photoprotecting systems: A DFT/TD-DFT study, Comput. Theor. Chem., 2020, 1191, 113026 CrossRef CAS.
- M. Cheriet, et al., A DFT study of inclusion complexes of the antituberculosis drugs pyrazinamide and isoniazid with cucurbit [7] uril, J. Inclusion Phenom. Macrocyclic Chem., 2017, 89(1–2), 127–136 CrossRef CAS.
- A. Bouhadiba, S. Rahali, Y. Belhocine, H. Allal, L. Nouar and M. Rahim, Structural and energetic investigation on the host/guest inclusion process of benzyl isothiocyanate into β-cyclodextrin using dispersion-corrected DFT calculations, Carbohydr. Res., 2020, 491, 107980, DOI:10.1016/j.carres.2020.107980.
- R. Sure and S. Grimme, Comprehensive Benchmark of Association (Free) Energies of Realistic Host-Guest Complexes, J. Chem. Theory Comput., 2015, 11(8), 3785, DOI:10.1021/acs.jctc.5b00296.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ma00521f |
| ‡ Both authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |