Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Osteogenic potential of a 3D printed silver nanoparticle-based electroactive scaffold for bone tissue engineering using human Wharton's jelly mesenchymal stem cells†
Mira
Mira
a,
Arie
Wibowo
 *bc,
Gusti Umindya Nur
Tajalla
d,
Glen
Cooper
e,
Paulo Jorge Da Silva
Bartolo
f and
Anggraini
Barlian
*cg
*bc,
Gusti Umindya Nur
Tajalla
d,
Glen
Cooper
e,
Paulo Jorge Da Silva
Bartolo
f and
Anggraini
Barlian
*cg
aMaster of Biotechnology, School of Life Sciences and Technology, Institut Teknologi Bandung, Jl. Ganesha 10, Bandung 40132, West Java, Indonesia
bMaterials Science and Engineering Research Group, Faculty of Mechanical and Aerospace Engineering, Institut Teknologi Bandung, Jl. Ganesha 10, Bandung 40132, West Java, Indonesia. E-mail: ariewibowo@itb.ac.id
cResearch Center for Nanoscience and Nanotechnology, Institut Teknologi Bandung, Jl. Ganesha 10, Bandung 40132, West Java, Indonesia
dMaterials and Metallurgy Engineering, Institut Teknologi Kalimantan, Jl. Soekarno Hatta 15, Balikpapan 76127, East Kalimantan, Indonesia
eSchool of Engineering, University of Manchester, Manchester M13 9PL, UK
fThe Singapore Centre for 3D Printing (SC3DP), Nanyang Technological University, 50 Nanyang Avenue, Block N3.1-B2C-03, Singapore 639798, Singapore
gSchool of Life Sciences and Technology, Institut Teknologi Bandung, Jl. Ganesha 10, Bandung 40132, West Java, Indonesia. E-mail: aang@sith.itb.ac.id
First published on 17th November 2023
Abstract
This study aims to perform biological assessments of an electroactive and anti-infection scaffold based on polycaprolactone/0.5 wt% silver nanoparticles (PCL/AgNPs) that was fabricated using a green synthesis approach followed by a 3D printing method without utilization of any toxic solvents, which has not been explored previously. For this purpose, human Wharton's jelly mesenchymal stem cells (hWJ-MSCs) were used as a cell source to explore the biocompatibility and the ability to induce the osteogenesis process on the fabricated PCL and PCL/AgNPs scaffolds. Scanning electron microscopy (SEM), confocal microscopy and an alamar blue assay up to day 14 revealed that the PCL/AgNPs scaffolds have better cell attachment, penetration and proliferation than the PCL scaffolds. A gene expression study up to day 21 using the reverse transcription-quantitative polymerase chain reaction (RT-qPCR) showed that the PCL/AgNPs scaffolds have better osteogenic differentiation at the gene level than the PCL scaffolds. This is indicated by the 2–3 fold greater expression of runt-related transcription factor 2 (RUNX2), collagen type I alpha 1 chain (COL1A1), and osteopontin (OPN) than the PCL scaffold. A protein expression study up to day 21 using immunocytochemistry and detection of alkaline phosphatase (ALP) revealed that the PCL/AgNPs scaffolds have better osteogenic differentiation at the protein level than the PCL scaffolds. This is shown by the observed collagen type I and osteopontin protein, and ALP activity at day 21 of PCL/AgNPs scaffolds (768 U L−1) which is 1.3 times higher than that of the PCL scaffolds (578 U L−1). These biological assessments showed that the combination of a green synthesis approach to prepare AgNPs and solvent-free 3D printing methods to fabricate the PCL/AgNPs scaffolds led to better biocompatibility and ability to induce the osteogenesis process, which is attractive for bone tissue engineering and regenerative medicine applications.
1. Introduction
Osteoporosis is a degenerative disease that causes bone weakening and leads to bone fractures, particularly in adult women and elderly people.1 This disease is known as a silent killer that gradually decreases the patient's quality of life and affects 200 million women worldwide.1,2 To date, most of the patients rely on natural bone healing processes to recover from bone fracture. However, elderly people might not fully recover from bone fracture because their healing rate is significantly slower than younger people.3 Thus, it is necessary to create bone scaffolds that could provide not only mechanical support, but also accelerate the bone healing process.Utilization of external stimuli together with stimuli-responsive scaffolds could solve this problem because many external stimuli, such as magnetic,4 mechanical,5 temperature,6,7 and electrical stimulation,7–9 are able to improve the bone tissue regeneration rate. Among them, electrical stimulation is an attractive option because it has been demonstrated that sending signals across cell membranes with −10 to −90 mV would encourage cells to travel to the site of injury,10 and also boost cardiac,11 nerve,12,13 and bone tissue regeneration.14,15 Therefore, electroactive scaffolds have emerged as a promising solution for bone therapies because they could deliver electrical stimulation directly to cells to improve incorporation, growth, and formation of new bone tissue.9,16
Electroactive scaffolds can be prepared by combining non-conductive biocompatible and biodegradable polymers, such as polycaprolactone (PCL), with conductive fillers, such as carbon-based,17,18 conductive polymeric-based,9,19 or metallic-based nanoparticles.20,21 This approach shows great potential for developing scaffolds that can effectively stimulate cells, promote tissue growth, and enhance healing. Previously, 3D printed electroactive scaffolds based on PCL/silver nanoparticles (AgNPs) were investigated by incorporation of 0.5 wt% of AgNPs into PCL.22 Compared to the PCL scaffold alone, the PCL/AgNPs scaffold exhibited a six-fold increase in compressive strength (3.88 ± 0.42 MPa), greater hydrophilicity (contact angle of 76.8 ± 1.7°), and conductivity (2.3 ± 0.5 × 10−3 S cm−1).22 These results show the potency of the electroactive PCL/AgNPs scaffolds for bone tissue engineering and regenerative medicine applications. Moreover, the addition of AgNPs demonstrated anti-bacterial properties against Staphylococcus aureus (decreased by 99.5%) which is beneficial for anti-infection for the implant.22 This scaffold's characteristic is crucial to ensure the success of the bone implant because infection is the main cause of implant failure in orthopedic surgery.23,24
In addition to the appropriate mechanical properties, the scaffold must be biocompatible with cells and able to integrate with patient tissues without causing an immune response or cytotoxicity. The scaffold must also be able to provide attachment, growth, proliferation, penetration, and cell differentiation.25 Previously, Lu et al. showed that the presence of exosomes and AgNPs on the surface of a PCL scaffold could boost osteogenesis differentiation of human bone marrow mesenchymal stem cells (hBMSC).26 However, the PCL scaffold was prepared by electrospinning after dissolving PCL in organic solvent (1,1,1,3,3,3-hexafluoro-2-propanol/HFP), which is considered as toxic with an LC50 (rat) of 1974 ppm/4 h.27 In this context, 3D-printed PCL/AgNPs scaffolds are attractive for biomedical application because they were prepared by a green synthesis and solvent-free manufacturing approach. Despite its favorable solvent-free process, mechanical and electrical conductivity properties, biocompatibility and ability to induce osteogenesis of 3D-printed PCL/AgNPs scaffolds were not known. Therefore, this research aims to determine the biocompatibility and osteogenesis induction capabilities of a solvent free 3D-printed PCL/AgNPs scaffold.
For this purpose, not only a scaffold with appropriate characteristics is needed for tissue growth and development, but also a suitable cell source is important to facilitate the formation of the necessary tissue.28 In this research, human Wharton's jelly MSCs (hWJ-MSCs) were used as precursor cells in the initiation of bone regeneration. hWJ-MSCs were chosen over other MSCs due to their numerous advantages, including abundant sources, a non-invasive collection method, a high cell yield, their phenotype and stem cell properties persisting even after long-term culture, allowing mass production of cells normally required for regenerative medicine, excellent proliferative potential, and a low risk of rejection when transplanted.29–32 Evaluation of the biocompatibility of the scaffold and its ability to support cell growth and differentiation will provide valuable insights into the potential of PCL/AgNPs scaffolds for bone tissue applications.
2. Materials and methods
2.1. Materials
AgNPs were prepared using a facile green synthesis approach without the use of toxic chemicals, instead using an extract of Cilembu sweet potatoes, as described in our previous report.22 PCL (CAPA 6500, Mw of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, glass transition temperature of ∼60 °C, melting point of 58–60 °C, and density of 1.146 g mL−1 at 25 °C) were brought from Perstorp, Warrington, UK. Umbilical cord samples were provided by Dr Hasan Sadikin Central General Hospital. Dulbecco's Modified Eagle Medium (DMEM)-high glucose (11995065), DMEM-low glucose (11885084), Fetal Bovine Serum (FBS, 10270106), Antibiotic–Antimycotic (ABAM, 15240062), 0.25% trypsin–EDTA (25200056), and StemPro™ Differentiation Kit (A1007001, A1007101, and A1007201) were purchased from Gibco, Thermo Fisher Scientific, Massachusetts, USA. Phosphate Buffered Saline (PBS, P4417-50TAB) and Collagenase Type I (SA C1.28-100MG) were obtained from Sigma-Aldrich, Merck KGaA, Missouri, USA. 4′,6-Diamidino-2-phenylindole/DAPI D1306, Anti-Collagen Type I Antibody, and Alexa Fluor 647 were supplied by from Invitrogen, Thermo Fisher Scientific, Massachusetts, USA. Anti-Osteopontin Antibody ab8448 (Abcam, Cambridge, UK), Hexamethyldisilazane (HMDS, Electron Miscroscopy Science 16700), Surface Marker Analysis Kit for MSCs (BD Biosciences, Erembodegem, Belgium), Quick-RNA™ MiniPrep Plus Kit R1057 (Zymo Research, California, USA), SensiFAST SYBR Lo-ROX One-Step Kit BIO-74005 (Bioline, London, UK), 100 bp DNA Ladder DL007 (Geneaid Biotech Ltd, New Taipei City, Taiwan), and Alkaline Phosphatase Kit (Reiged Diagnostics, Gaziemir, Turkey).
000, glass transition temperature of ∼60 °C, melting point of 58–60 °C, and density of 1.146 g mL−1 at 25 °C) were brought from Perstorp, Warrington, UK. Umbilical cord samples were provided by Dr Hasan Sadikin Central General Hospital. Dulbecco's Modified Eagle Medium (DMEM)-high glucose (11995065), DMEM-low glucose (11885084), Fetal Bovine Serum (FBS, 10270106), Antibiotic–Antimycotic (ABAM, 15240062), 0.25% trypsin–EDTA (25200056), and StemPro™ Differentiation Kit (A1007001, A1007101, and A1007201) were purchased from Gibco, Thermo Fisher Scientific, Massachusetts, USA. Phosphate Buffered Saline (PBS, P4417-50TAB) and Collagenase Type I (SA C1.28-100MG) were obtained from Sigma-Aldrich, Merck KGaA, Missouri, USA. 4′,6-Diamidino-2-phenylindole/DAPI D1306, Anti-Collagen Type I Antibody, and Alexa Fluor 647 were supplied by from Invitrogen, Thermo Fisher Scientific, Massachusetts, USA. Anti-Osteopontin Antibody ab8448 (Abcam, Cambridge, UK), Hexamethyldisilazane (HMDS, Electron Miscroscopy Science 16700), Surface Marker Analysis Kit for MSCs (BD Biosciences, Erembodegem, Belgium), Quick-RNA™ MiniPrep Plus Kit R1057 (Zymo Research, California, USA), SensiFAST SYBR Lo-ROX One-Step Kit BIO-74005 (Bioline, London, UK), 100 bp DNA Ladder DL007 (Geneaid Biotech Ltd, New Taipei City, Taiwan), and Alkaline Phosphatase Kit (Reiged Diagnostics, Gaziemir, Turkey).
2.2. Research ethics
Human Wharton's jelly mesenchymal stem cells (hWJ-MSCs) that were used in this study have received ethical approval from the Research Ethics Commission of Padjadjaran University, no. 432/UN6.KEP/EC/2021.2.3. Fabrication of 3D printed PCL/AgNPs scaffolds
The 3D printed PCL/AgNPs scaffolds were fabricated according to our previous report.22 In general, a solvent-free approach was used to prepare the homogeneous PCL/AgNPs blends by adding 0.5 wt% of AgNPs powder to the melted PCL (at ±80 °C) and physically stirring to obtain a uniform blend (namely physical melt blending method). After cooling, the PCL/AgNPs blend was sliced into tiny pieces to accommodate loading into the 3D printer's hopper. Scaffolds were then fabricated by 3D-printing using a 3D Discovery printer (regenHU, Villaz-St-Pierre, Switzerland). The scaffolds were designed using computer-aided design software model construction (BioCAD, regenHU, Villaz-St-Pierre, Switzerland) with dimensions of 20 mm × 20 mm × 3 mm, a fiber distance of 1 mm, a slice width of 280 μm, and a 0°/90° lay-up profile. The scaffolds were then produced employing a 5 mm s−1 deposition velocity, a melting temperature of 90 °C, an extrusion pressure of 6 bar, and a screw rotation velocity of 15 rpm with a printing nozzle diameter of 0.5 mm.2.4. Isolation, subculture and characterizations of the hWJ-MSCs
Umbilical cord samples were taken aseptically from Dr Hasan Sadikin Central General Hospital after obtaining approval from the donor. The sample was then cleaned in 70% alcohol, followed by PBS 1× containing 1% ABAM. The umbilical cord was cut along a 1 cm length. The sample is cut vertically so that the arteries and veins are visible and can be removed. The sample is washed until it is completely clean of blood. Samples were cut into small pieces with a size of less than 5 mm.Then, the cells were isolated from human Wharton's jelly (hWJ) using enzymatic methods. Briefly, the hWJ samples were digested with 0.2% collagenase enzyme in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio with pellets, then digested with 0.25% trypsin–EDTA enzyme in a 1
1 ratio with pellets, then digested with 0.25% trypsin–EDTA enzyme in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio with pellets. The collagenase enzyme was used to break down the triple helix domain of collagen so that cells are separated from other cells without damaging the cell membrane.33,34 While the trypsin enzyme was used to hydrolyze the peptide bonds between the amino acids lysine and arginine into simpler compounds, thereby optimizing cell separation.35
1 ratio with pellets. The collagenase enzyme was used to break down the triple helix domain of collagen so that cells are separated from other cells without damaging the cell membrane.33,34 While the trypsin enzyme was used to hydrolyze the peptide bonds between the amino acids lysine and arginine into simpler compounds, thereby optimizing cell separation.35
The digested cell samples were put into a T25 flask. The flask was filled with 1 mL of complete medium (DMEM + 10% FBS + 1% ABAM) and incubated at 37 °C with 5% CO2. After 24 hours, 1 mL of complete medium was added to the flask. Subculture is carried out until it reaches passage 4 for further testing.36,37
To ensure that the cells isolated from human Wharton's jelly were truly hWJ-MSCs cells and meet the requirements of the International Society for Cell Therapy (ISCT) as hWJ-MSCs, characterization was carried out. Cells were characterized using an inverted microscope to determine the attachment of cells to the substrate and the morphology of the isolated cells, analyzing cell surface marker expression using flow cytometry (BD FACSCanto II, BD Biosciences, Erembodegem, Belgium), and differential staining to determine multilineage differentiation potential.38
Analysis of cell surface marker expression was carried out by resuspending the cells with 3 mL of PBS. 100 μL of cell suspension was transferred into each prepared 10 mL tube, then the following were added: (1) 10 μL PE mouse anti-human CD44 (51-9007656) into the first tube; (2) 10 μL hMSC positive cocktail (51-9007663) and 10 μL PE hMSC negative cocktail (51-9007661) into the second tube; (3) 10 μL hMSC isotype control positive cocktail (51-9007664) and 10 μL PE hMSC isotype control negative cocktail (51-9007662) into the third tube; and (4) 10 μL propidium iodide staining solution (51-66211E) into the fourth tube; while the fifth tube was not given any antibodies (only cell suspension). After incubation for 30 minutes in the dark, samples were analyzed using flow cytometry (BD FACSCanto II).38
The multipotency test begins with cell harvesting using 0.25% trypsin–EDTA. A total of 2 × 104 cells were placed into each well on a 12-well plate. Then the cells were induced using differentiation medium for 14 days. After 14 days, the cells were fixed using 4% formalin and incubated for 30 minutes. Next, cells were rinsed with PBS 1 × 3 times. Then, the cells were stained using: (1) oil red o (0.3% in 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 isopropanol
1 isopropanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) dH2O) (adipogenic differentiation) for 30 minutes; (2) alcian blue (1% in 0.1 N HCl) (chondrogenic differentiation) for 30 minutes; and alizarin red (2% in dH2O, pH 4.2) (osteogenic differentiation) for 30 minutes. Next, the cells were rinsed using distilled water three times and observed using an inverted microscope.38
dH2O) (adipogenic differentiation) for 30 minutes; (2) alcian blue (1% in 0.1 N HCl) (chondrogenic differentiation) for 30 minutes; and alizarin red (2% in dH2O, pH 4.2) (osteogenic differentiation) for 30 minutes. Next, the cells were rinsed using distilled water three times and observed using an inverted microscope.38
2.5. Cell seeding on a scaffold and their characterization
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per scaffold) in the middle of the scaffold. The scaffolds were then incubated for 2 hours under standard cell culture incubator conditions to allow cell attachment before adding complete medium to obtain a final volume of 800 μL. To minimize cell migration and attachment to the underlying tissue culture plastic, an untreated well plate was utilized.19
000 cells per scaffold) in the middle of the scaffold. The scaffolds were then incubated for 2 hours under standard cell culture incubator conditions to allow cell attachment before adding complete medium to obtain a final volume of 800 μL. To minimize cell migration and attachment to the underlying tissue culture plastic, an untreated well plate was utilized.19
2.6. Gene and protein expression during the osteogenesis process
| Gene | Forward primer | Reverse primer |
|---|---|---|
| RUNX2 | TCGCCTCACAAACAACCACA | CTGGTAGTGACCTGCGGAGA |
| COL1A1 | TGGAAAGAATGGAGATGATGGG | ACTGAAACCTCTGTGTCCCT |
| OPN | GTGCCATACCAGTTAAACAG | CATCTACATCATCAGAGTCGT |
| GAPDH | ATCAAGAAGGTGGTGAAGCAG | TCGTTGTCATACCAGGAAATGAG |
2.6.2.1. Protein analysis of collagen type I and osteopontin. Protein analysis of collagen type I and osteopontin with immunocytochemistry was carried out on days 7, 14, and 21. 1 × 105 cells of hWJ-MSCs were grown on the PCL scaffold and the PCL/AgNPs scaffold under standard culture conditions. At each time point, cells grown on the scaffold were washed with PBS twice. Cells were fixed with methanol-DMEM (50%, 60%, 70%, 80%, and 90%), then fixed with 100% methanol and 50% methanol-PBS, and then cells were washed with PBS three times. Cells were permeabilized with PBST for 20 minutes at room temperature and washed with PBS three times. Then, 3% BSA in PBST was added to the sample, which was incubated for 60 minutes at room temperature. 50 μL of primary antibody (anti-collagen type I; anti-osteopontin) was added to the sample and incubated overnight in a 37 °C water bath, then cells were washed with PBS three times. 50 μL of secondary antibody (Alexa Fluor 647) was added to the sample which was then incubated for 2 hours at room temperature. The cells were then washed with PBS 3 times. After that, 50 μL of DAPI was added to the middle of the scaffold and incubated for 10 minutes at room temperature in the dark. Cells were washed with PBS three times. Then the scaffolds were carefully transferred into a confocal dish and observed using a confocal microscope (FV1200 Laser Scanning Microscopes, Olympus, Tokyo, Japan).40
2.6.2.2. Detection of alkaline phosphatase. hWJ-MSCs with a density of 1 × 105 cells were grown on the PCL scaffold and the PCL/AgNPs scaffold under standard culture conditions. Alkaline phosphatase detections were carried out on days 7, 14, and 21 respectively. The detection of alkaline phosphatase was done using procedures according to the Alkaline Phosphatase Kit protocol.44
2.7. Statistical analysis
All quantitative biological experiments were performed at least in triplicate (n ≥ 3), which is commonly used as the minimum number of experiments in biological research.45–47 All data were expressed as mean ± standard deviation and statistical analysis was evaluated using SPSS 20.0. Comparisons of groups for cell proliferation, gene expression analysis using RT-qPCR and detection of alkaline phosphatase were achieved using one-way analysis of variance (ANOVA) and Mann Whitney. Significance levels were set at p < 0.05.19 All statistical analysis results are presented in the ESI† (Tables S1–S44).3. Results and discussion
3.1. Stem cell characterization of the isolated cells from human Wharton's jelly
One of the important issues in cell therapy is the variety of stem cells used as cell sources, stem cells that have no significant difference in growth rate or differentiation from the target tissue, have high potential, and are easy to produce.48 This study employed human Wharton's jelly mesenchymal stem cells (hWJ-MSCs) as progenitor cells to initiate the process of bone repair.After isolation of stem cells from human Wharton's jelly (hWJ), it is important to perform stem cell characterization to ensure that the isolated cells are the stem cells from hWJ (hWJ-MSCs) and meet the International Society for Cell Therapy (ISCT) requirements as hWJ-MSCs. There are several criteria for mesenchymal stem cells according to the International Society for Cell Therapy (ISCT), namely: (1) plastic-adherent, (2) ≥95% of the MSCs population should express CD105, CD73, and CD90 but lack expression (≤2% positive) of CD45, CD34, CD14, CD11b, CD79a, or CD19 and HLA class II, and (3) under standard in vitro differentiating conditions, the cells must be able to differentiate into osteoblasts, adipocytes, and chondroblasts.49
In this study, characterization using an inverted microscope was performed to determine the plastic adherence and morphology of the isolated cells. Microscopical observation revealed that the isolated cells from hWJ were successfully attached to the plastic substrate and have an elongated shape, which is fibroblast-like morphology (Fig. 1). This result is consistent with a previous study in which it was discovered that the isolated stem cells from WJ have fibroblast-like morphology.50
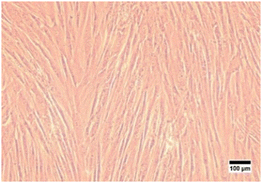 | ||
| Fig. 1 Cells isolated from Wharton's jelly were observed with an inverted microscope at 100× magnification. | ||
Then, stem cell characterization using flow cytometry was implemented to identify surface marker expression of the isolated cells. As shown in Fig. 2, the isolated cells highly expressed the CD 73 marker (95.8%; Fig. 2(a)), the CD 90 marker (92.2%; Fig. 2(b)), and the CD 105 marker (89.7%; Fig. 2(c)), but the isolated cells did not express the marker lin-negative (0%; Fig. 2(d)). Even though ISCT mentioned that ≥95% of MSCs should express CD105, there are several studies demonstrating that expression of the CD105 marker by MSCs could be varied, such as 88.1%,39 90.76%,36 92.94%,51 and 94.54%,52 depending on the isolated methods, cell source, culture time in vitro, and differentiation status.36,53 Thus, the isolated cells still comply with the ISCT's criteria even though the cell population that emits CD105 does not reach 95%.
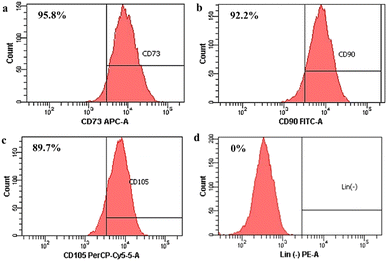 | ||
| Fig. 2 Surface marker identification of human Wharton's jelly mesenchymal stem cells (hWJ-MSCs). (a) CD73. (b) CD90. (c) CD105. (d) Lin (−) negative marker. | ||
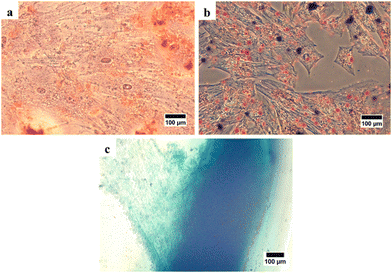
After that, stem cell characterization using differential staining was performed to determine the multilineage differentiation potential of the isolated cells. After treating with the StemPro Osteogenesis Differentiation Kit and staining with alizarin red, the cells were stained red implying the presence of calcium deposits as an indication that the cells had differentiated into osteocytes (Fig. 3(a)). After treating with the StemPro Adipogenesis Differentiation Kit and staining with oil red O, the cells were stained red suggesting the presence of lipid droplets as the indication, that the cells had differentiated into adipocytes (Fig. 3(b)). In addition, after treating with the StemPro Chondrogenesis Differentiation Kit and staining with alcian blue, the cells were stained blue, implying that there were glycosaminoglycans and glycoproteins as components of the extracellular matrix of chondrocytes (Fig. 3(c)).38 These results showed that the isolated cells are fulfilling the criteria of MSCs from ISCT.
3.2. Morphology and dimension of 3D printed PCL/AgNPs scaffolds
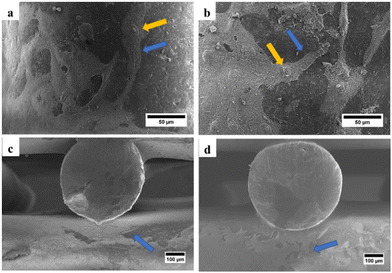
SEM was used to examine the morphology of the scaffold, determine the pore size and fiber diameter of the scaffold, and observe the morphology of the hWJ-MSCs grown on the scaffold. SEM observations showed that the fabricated scaffolds are well-aligned scaffolds with identical circulated pores and shapes (Fig. 4). Also, the PCL/AgNPs scaffolds had a larger fiber diameter and smaller pore size than the PCL scaffolds (Table 2). This result is consistent with our previous work that the addition of AgNPs on the PCL scaffold could increase the fiber diameter and reduce the pore size of the scaffold.22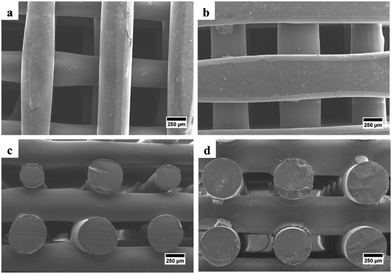 | ||
| Fig. 4 SEM images of the fabricated scaffolds that were taken from the top-view: (a) PCL and (b) PCL/AgNPs; and the side-view: (c) PCL and (d) PCL/AgNPs. | ||
| Scaffold sample | Fiber diameter (μm) | Pore size (μm) |
|---|---|---|
| PCL | 474 ± 93 | 522 ± 51 |
| PCL/AgNPs | 534 ± 18 | 405 ± 22 |
In the creation of scaffolds, pore size and pore interconnectivity are crucial factors for cell adhesion, migration, and growth.28 A cellular capsule will form around the edges of the scaffold if the pores are too tiny, restricting cell migration. This can then prevent debris from being removed and nutrients from diffusing through the construct, leading to necrotic areas. In contrast, if the pores are too big, the surface area decreases, which in turn reduces cell adhesion.28 Appropriate pore diameters for bone formation were determined to be greater than 300 μm in order to allow for adequate vascularisation of the material and prevent hypoxic conditions in the inner regions.54,55 The results of Krieghoff et al. (2019) showed that scaffolds with pores ranging from 300 to 500 μm produced the best results because they were capable of producing collagen, hydroxyapatite deposition, and bone mineral maturation.56 Research conducted by Sicchieri et al. (2012) also shows that PLGA-CaP scaffolds promoted bone formation, and an increase in the number of blood vessels was observed within pores ranging from 470 to 580 μm.57
3.3. Attachment, penetration and proliferation of the hWJ-MSCs on the scaffold
The area covered with the white substances on the PCL/AgNPs scaffolds (Fig. 5(b) and (d)) is larger than that on the PCL scaffolds (Fig. 5(a) and (c)). This suggested that the number of hWJ-MSCs attached to the PCL/AgNPs scaffold was greater than that of the PCL scaffold. Mammalian cells rely on their connections with other cells, the extracellular matrix (ECM), and material substrates to control vital functions such as intra- and intercellular communication, apoptosis, morphology, function, and differentiation.58 To produce functional and morphologically accurate cell populations in tissue culture, scaffolds must imitate the biological environment from which the particular cell type was derived.59 Previously, we found that the 3D-printed PCL scaffold has a contact angle of 80.6 ± 2.0°, and the addition of 0.5 wt% AgNPs which causes a slight decrease in the scaffold's contact angle (76.8 ± 1.7°).22 Given that scaffolds with a lower contact angle are more preferable for cell attachment due to their hydrophilic nature, the addition of 0.5 wt% AgNPs to PCL scaffolds improves cell attachment to the scaffold.22,60
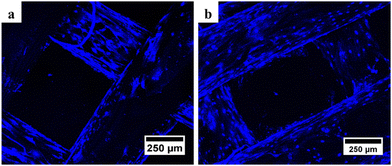 | ||
| Fig. 6 Confocal microscopy results of (a) PCL and (b) PCL/AgNPs scaffolds showed the presence of blue oval particles indicating the presence of the nucleus after DAPI staining. | ||
It is noteworthy that the number of nuclei at the bottom surface of the PCL/AgNPs scaffolds (Fig. 6(b)) is higher than the PCL scaffolds (Fig. 6(a)). These results imply that the PCL/AgNPs scaffolds are more favorable for hWJ-MSCs growth than PCL scaffolds. The pore size must be large enough to facilitate cell migration, nutrient transport, and waste removal. In contrast, if the pores are too large, the surface area decreases, which in turn limits cell adhesion.28
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 682 cells and the number of cells increased to 88
682 cells and the number of cells increased to 88![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 431 cells on day 14, while in the PCL scaffold there were 41
431 cells on day 14, while in the PCL scaffold there were 41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 874 cells attached on the first day and the number increased to 79
874 cells attached on the first day and the number increased to 79![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 618 cells on day 14 (Table S1, ESI†). These results suggest that the PCL/AgNPs scaffold could support the proliferation of hWJ-MSCs better than the PCL scaffold, which is consistent with other reports that silver nanoparticles (AgNPs) were able to promote the proliferation of MSCs.63 Zhang et al., stated that although PCL has many advantages, it also has several disadvantages, such as poor cell adhesion, low bioactivity, and hydrophobicity.64 Therefore, the PCL scaffold should add other components, such as AgNPs as a filler in this study, to obtain all the desired scaffold properties.
618 cells on day 14 (Table S1, ESI†). These results suggest that the PCL/AgNPs scaffold could support the proliferation of hWJ-MSCs better than the PCL scaffold, which is consistent with other reports that silver nanoparticles (AgNPs) were able to promote the proliferation of MSCs.63 Zhang et al., stated that although PCL has many advantages, it also has several disadvantages, such as poor cell adhesion, low bioactivity, and hydrophobicity.64 Therefore, the PCL scaffold should add other components, such as AgNPs as a filler in this study, to obtain all the desired scaffold properties.
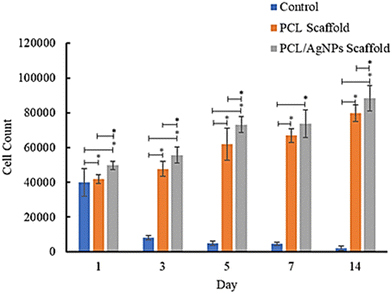 | ||
| Fig. 7 Results of analysis of the proliferative ability of hWJ-MSCs grown on the control, PCL and PCL/AgNPs scaffolds. | ||
In this study, the well plates that were used are non-treated polystyrene to prevent cell migration and adhesion to the underlying tissue culture plastic. Since the treatment group utilized non-treated well plates, the control group (cells grown on well plates without scaffold) also utilized non-treated well plates. The surfaces of non-treated polystyrene are composed primarily of hydrophobic phenyl groups, which are not normally found in the body and are detrimental to cell anchorage.59 Consequently, the number of cells in the control group decreased from day to day.
3.4. Gene and protein expression during the osteogenesis process of the hWJ-MSCs on the scaffold
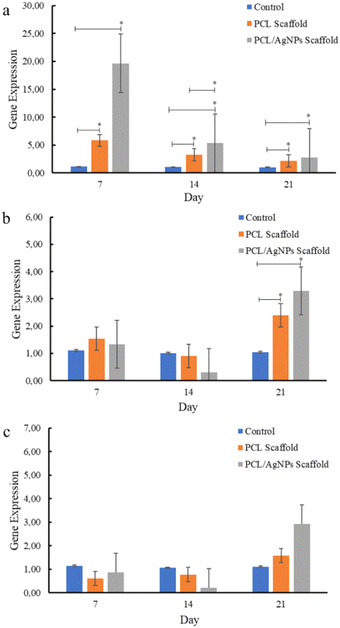 | ||
| Fig. 8 Expression of osteogenesis marker genes in hWJ-MSCs that were grown on the control, PCL and PCL/AgNPs scaffold: (a) RUNX2, (b) COL1A1, and (c) OPN. | ||
PCL/AgNPs scaffolds upregulated RUNX2 gene expression threefold more than the PCL scaffolds on day 7. The PCL/AgNPs scaffolds also significantly upregulated RUNX2 gene expression higher than the PCL scaffolds on day 14. Therefore, it can be concluded that the PCL/AgNP scaffolds induce hWJ-MSCs osteogenesis more effectively than the PCL scaffolds (Fig. 8). This result was in line with a current study that showed silver nanoparticles (AgNPs) promoted the osteogenesis of MSCs.63 On day 21, RUNX2, COL1A1, and OPN genes were upregulated in hWJ-MSCs samples grown on the PCL scaffolds and the PCL/AgNPs scaffolds (Fig. 8). The PCL/AgNPs scaffolds upregulated OPN gene expression twofold more than the PCL scaffolds. These findings suggest that on day 21 of treatment, the hWJ-MSCs had reached the mature osteoblast stage. Amarasekara et al. showed that mature osteoblasts secrete numerous osteoblastogenic markers, including OPN, while ALP and COL1A1 remain expressed.67 The results of RT-qPCR showed that both scaffolds caused the RUNX2 gene to be expressed, which in turn caused the COL1A1 and OPN genes to be expressed. This led to more osteogenic differentiation.8
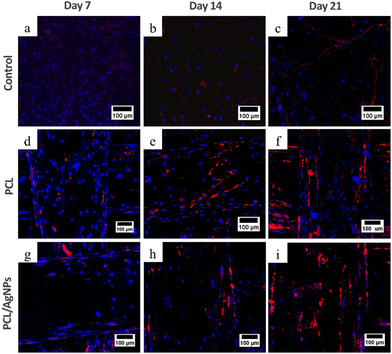
3.4.2.1. Expression of collagen type I protein. Analysis of collagen type I protein and osteopontin protein using the immunocytochemistry method was performed to confirm the RT-qPCR results. Immunocytochemistry results showed that collagen type I was observed on days 7, 14, and 21 in hWJ-MSCs grown on the PCL scaffold and PCL/AgNPs scaffold (Fig. 9). This is in accordance with the statement by Amarasekara et al. that collagen type I is expressed in the pre-osteoblast, immature-osteoblast, and mature-osteoblast stages.67
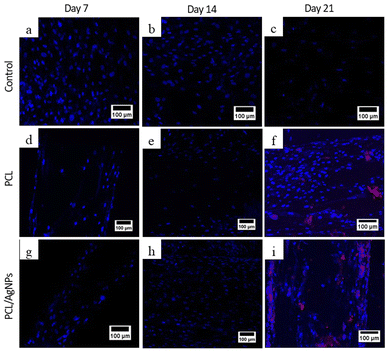
3.4.2.2. Expression of osteopontin protein. To verify the RT-qPCR results, immunocytochemistry analysis of the osteopontin protein was performed. On day 21, osteopontin protein was detected by immunocytochemistry in hWJ-MSCs grown on the PCL scaffolds and the PCL/AgNPs scaffolds (Fig. 10). This is consistent with the assertion made by Stein & Lian (1993) that osteopontin protein achieves peak levels of expression during mineralization (mature osteoblast stage).66
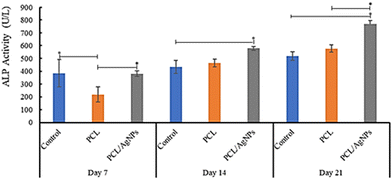
3.4.2.3. Detection of alkaline phosphatase protein. The activity of alkaline phosphatase (ALP) is considered to be a crucial indicator of early osteogenic differentiation.69 The ALP activity of the hWJ-MSCs grown on the PCL scaffold at days 7, 14, and 21 are 218 U L−1, 464 U L−1, and 578 U L−1 respectively (Fig. 11 and Table S40, ESI†). This increment trend of ALP activity on the PCL scaffold was also observed by Abazari et al.,69 and is in accordance with the statement by Amarasekara et al. that the activity of ALP is still low in the pre-osteoblast stage but will increase in the immature-osteoblast and mature-osteoblast stages.67 Interestingly, ALP activity of the hWJ-MSCs grown on the PCL/AgNPs scaffold (381 U L−1, 578 U L−1, and 768 U L−1 at days 7, 14, and 21 respectively; Fig. 11 and Table S44, ESI†) was considerably higher than the PCL scaffold. The results suggested that the PCL/AgNPs scaffold induces osteogenic differentiation of hWJ-MSCs better than the PCL scaffold.
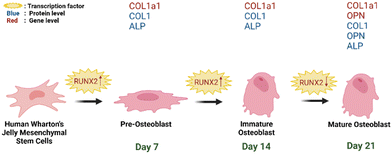 | ||
| Fig. 12 Schematic representation of hWJ-MSCs osteogenesis grown on PCL/AgNPs scaffolds (created with https://BioRender.com). | ||
On day 14 of the hWJ-MSCs treatment, it had entered the immature osteoblast stage (Fig. 12). According to Amarasekara et al. (2021), immature osteoblasts secrete collagen type 1 as the major constituent of the ECM and express ALP to mature the ECM.67 This is consistent with the findings of research indicating that the RUNX2 gene was upregulated on day 14 in hWJ-MSCs samples grown on the PCL scaffolds and the PCL/AgNPs scaffolds (Fig. 8). The PCL/AgNPs scaffolds upregulated RUNX2 gene expression twofold more than the PCL scaffolds. On day 14, collagen type I protein was detected in hWJ-MSCs samples grown on the PCL scaffolds and the PCL/AgNPs scaffolds, as determined by ICC (Fig. 9). The activity of ALP was also observed in hWJ-MSC samples grown on the PCL scaffolds and the PCL/AgNPs scaffolds (Fig. 10). Whereas, the ALP activity was greater in hWJ-MSCs grown on the PCL/AgNPs scaffolds than in hWJ-MSCs grown on the PCL scaffolds (Fig. 10).
On day 21 of treatment, hWJ-MSCs had reached the mature osteoblast stage. According to Amarasekara et al., mature osteoblasts secrete numerous osteoblastogenic markers, such as osteopontin, while ALP and Col-I continue to be expressed.67 Osteopontin increases bone formation and mineralization.67 This is consistent with the findings of research indicating that RUNX2, COL1A1, and OPN genes were upregulated in hWJ-MSC samples grown on the PCL scaffold and the PCL/AgNPs scaffold on day 21 (Fig. 8). The PCL/AgNPs scaffolds upregulated OPN gene expression twofold more than the PCL scaffolds. The RT-qPCR results were strengthened by ICC data, which showed that on day 21, collagen type I and Osteopontin were observed in hWJ-MSCs samples grown on the PCL scaffolds and on the PCL/AgNPs scaffolds (Fig. 9). hWJ-MSCs grown on the PCL/AgNPs scaffolds had higher ALP activity than hWJ-MSCs grown on the PCL scaffolds (Fig. 10).
In this study, it was observed that the osteogenesis process was divided into pre-osteoblast (day 7), immature osteoblast (day 14), and mature osteoblast (day 21). If this treatment is continued, the hWJ-MSCs will enter the osteocyte stage. Therefore, the osteogenesis process in 3D-printed PCL/AgNPs scaffolds in mRNA and protein levels, is in line with previous research. Thus, an illustration to summarize the osteogenesis process that occurs in hWJ-MSCs grown on an electroactive scaffold based on silver nanoparticles is shown in Fig. 12.
The presence of AgNPs can indirectly improve osteogenesis in the PCL/AgNPs scaffold due to their ability to enhance hydrophilicity of the scaffold that led to adhesion increment of hWJ-MSCs to the PCL/AgNPs scaffold.28,60 Strong adhesion is required for osteogenic differentiation of MSCs, because it will activate FAK, ERK1/2 and RUNX2 respectively.70 Therefore, we found the PCL/AgNPs scaffold upregulated RUNX2 gene expression 2–3 times higher than the PCL scaffold. Similar phenomena were also observed by Lu et al. when using AgNPs and human bone marrow MSCs (hBMSCs)-derived exosomes (MSCs-exo) to modify the surface of the PCL scaffold.26 They found that AgNPs with a particle size of 10 nm could enhance cell spreading of hBMSCs on the surface of the Ag hybrid PCL scaffold and elevate osteogenic differentiation of MSCs, which is indicated by a significant increment of upregulated osteogenic-related gene expression (including RUNX2).26
Even though the exact direct role of AgNPs on osteogenesis improvement of MSCs remains unclear, several researchers found interesting results. Greulic et al.,71 and Jung et al.,72 found that AgNPs could enhance osteogenesis of MCSs by penetrating the cells (human MSCs), followed by integrating with DNA and activating the expression of genes such as HIF-1 and IL-8. Zhang et al., found that the addition of AgNPs at low concentration could effectively promote mMSC differentiation toward osteoblasts, which are indicated by higher calcium deposition and alkaline phosphate activity, and upregulated Cbfa1 gene expression at day 14. Furthermore, AgNPs could stimulate TGF-β/BMP signaling in a mouse model, causing cells to undergo chondrogenic and osteogenic differentiation, encouraging fracture callus development and filling fracture voids.63
4. Conclusions
Based on the results of this study, it can be concluded that silver nanoparticle-based electroactive scaffolds have better biocompatibility than PCL scaffolds. This is evidenced by the fact that the PCL/AgNPs scaffolds can promote attachment, penetration, and proliferation of hWJ-MSCs better than the PCL scaffolds. The PCL scaffolds and PCL/AgNPs scaffolds were able to induce hWJ-MSCs osteogenesis. On day 7 of the hWJ-MSCs treatment, they had entered the pre-osteoblast stage, as indicated by the expression of RUNX2, COL1A1, and ALP activity. On day 14 of the hWJ-MSCs treatment, they had entered the immature osteoblast stage, as evidenced by the expression of RUNX2, collagen type 1 being observed as the major constituent of the ECM, and the ALP activity that acted to mature the ECM. On day 21 of treatment, hWJ-MSCs had reached the mature osteoblast stage, as demonstrated by the expression of RUNX2, COL1A1, OPN, and ALP activity. However, the 3D-printed PCL/AgNPs scaffolds induced hWJ-MSC osteogenesis better than the PCL scaffolds. Which is indicated by the 2–3 fold greater expression of RUNX2, COL1A1, and OPN compared with the PCL scaffolds. ALP activity at day 21 of the PCL/AgNPs scaffolds (768 U L−1) was also 1.3 times higher than that of the PCL scaffolds (578 U L−1). These findings demonstrated that the 3D printed electroactive scaffolds based on silver nanoparticles can enhance osteogenesis of MCSs at their low concentration and is an attractive scaffold for bone tissue engineering and regenerative medicine applications. Despite the promising results, the current work was only based on an in vitro study using hWJ-MSCs, which is the limitation of this study. Thus, future work is recommended using an in vivo model to provide insights into the scaffold's effectiveness in a more physiologically relevant environment and to prove the stability of mature osteoblasts and differentiation towards mature mineralized osteocytes.Author contributions
Mira Mira (M. M.): writing – original draft, editing, data acquisition, data analysis. Arie Wibowo (A. W.): resources, supervision, conceptualization, data analysis, writing – original draft, writing – review. Gusti Umindya Nur Tajalla (G. U. N. T.): data acquisition, writing – review. Glen Cooper (G. C.): conceptualization, writing – review. Paulo Jorge Da Silva Bartolo (P. J. D. B.): conceptualization, writing – review. Anggraini Barlian (A. B.): supervision, resources, conceptualization, methodology, data analysis, writing – review. All authors have given approval to the submitted version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to acknowledge the ITB Research Fund 2023 scheme from the Institut Teknologi Bandung (PN-6-02-2023). The authors would like to thank Dr Hasan Sadikin Central General Hospital for providing the umbilical cord samples and the Research Center for Nanoscience and Nanotechnology, Institut Teknologi Bandung for allowing us to utilize their characterization facilities. The authors would also like to thank Fitria Dwi Ayuningtyas, PhD, Rizka Musdalifah Amsar, Salindri Prawitasari, and Safira Meidina Nursatya for their invaluable assistance.References
- S. R. Cummings and L. J. Melton, Lancet, 2002, 359, 1761–1767 CrossRef PubMed
.
- Y. Shen, X. Huang, J. Wu, X. lin, X. Zhou, Z. Zhu, X. Pan, J. Xu, J. Qiao, T. Zhang, L. Ye, H. Jiang, Y. Ren and P.-F. Shan, Front. Endocrinol., 2022, 13, 882241 CrossRef PubMed
.
- J. T. Lin and J. M. Lane, Clin. Orthop. Relat. Res., 2004, 425, 126–134 CrossRef
.
- A. C. Manjua, J. M. Cabral, C. A. Portugal and F. C. Ferreira, Sci. Technol. Adv. Mater., 2021, 22, 461–480 CrossRef CAS PubMed
.
- P. Augat, M. Hollensteiner and C. von Rüden, Injury, 2021, 52, S78–S83 CrossRef PubMed
.
- X. Hu, J. He, X. Yong, J. Lu, J. Xiao, Y. Liao, Q. Li and C. Xiong, Colloids Surf., B, 2020, 195, 111218 CrossRef CAS PubMed
.
- I. J. Budiarso, N. D. W. Rini, A. Tsalsabila, M. D. Birowosuto and A. Wibowo, ACS Biomater. Sci. Eng., 2023, 9, 3084–3115 CrossRef CAS PubMed
.
- L. Leppik, H. Zhihua, S. Mobini, V. T. Parameswaran, M. Eischen-Loges, A. Slavici, J. Helbing, L. Pindur, K. M. C. Oliveira, M. B. Bhavsar, L. Hudak, D. Henrich and J. H. Barker, Sci. Rep., 2018, 8, 6307 CrossRef PubMed
.
- M. A. Marsudi, R. T. Ariski, A. Wibowo, G. Cooper, A. Barlian, R. Rachmantyo and P. J. D. S. Bartolo, Int. J. Mol. Sci., 2021, 22, 11543 CrossRef CAS PubMed
.
- C. Ning, Z. Zhou, G. Tan, Y. Zhu and C. Mao, Prog. Polym. Sci., 2018, 81, 144–162 CrossRef CAS PubMed
.
- H. Esmaeili, A. Patino-Guerrero, M. Hasany, O. M. Ansari, A. Memic, A. Dolatshahi-Pirouz and M. Nikkhah, Acta Biomater., 2022, 139, 118–140 CrossRef CAS PubMed
.
- A. R. Mutepfa, J. G. Hardy and C. F. Adams, Front. Med. Technol., 2022, 4, 693438 CrossRef PubMed
.
- H. Cheng, Y. Huang, H. Yue and Y. Fan, Stem Cells Int., 2021, 2021, 6697574 Search PubMed
.
- L. Cui, J. Zhang, J. Zou, X. Yang, H. Guo, H. Tian, P. Zhang, Y. Wang, N. Zhang, X. Zhuang, Z. Li and J. Ding, Biomaterials, 2020, 230, 119617 CrossRef CAS PubMed
.
- B. C. Heng, Y. Bai, X. Li, L. W. Lim, W. Li, Z. Gae, X. Zhang and X. Deng, Adv. Sci., 2023, 10, 2204502 CrossRef CAS PubMed
.
- A. Oryan, S. Alidadi, A. Moshiri and N. Maffulli, J. Orthop. Surg. Res., 2014, 9, 18 CrossRef PubMed
.
- A. M. Martins, G. Eng, S. G. Caridade, J. F. Mano, R. L. Reis and G. Vunjak-Novakovic, Biomacromolecules, 2014, 15, 635–643 CrossRef CAS PubMed
.
- C. A. Pineda, K. Srirussamee, P. Palma, V. M. Fuenzalida, S. H. Cartmell and H. Palza, Nanomaterials, 2020, 10, 428 CrossRef PubMed
.
- A. Wibowo, C. Vyas, G. Cooper, F. Qulub, R. Suratman, A. I. Mahyuddin, T. Dirgantara and P. Bartolo, Materials, 2020, 13, 512 CrossRef CAS PubMed
.
- Y. Gao, A. M. Hassanbhai, J. Lim, L. Wang and C. Xu, RSC Adv., 2017, 7, 10051–10056 RSC
.
- S. Ghaziof, S. Shojaei, M. Mehdikhani, M. Khodaei and M. J. Nodoushan, J. Mech. Behav. Biomed. Mater., 2022, 132, 105271 CrossRef CAS PubMed
.
- A. Wibowo, G. U. N. Tajalla, M. A. Marsudi, G. Cooper, L. A. T. W. Asri, F. Liu, H. Ardy and P. D. S. B. Bartolo, Molecules, 2021, 26, 2042 CrossRef CAS PubMed
.
- D. Campoccia, L. Montanaro and C. R. Arciola, Biomaterials, 2006, 27, 2331–2339 CrossRef CAS PubMed
.
- M. Ribeiro, F. J. Monteiro and M. P. Ferraz, Biomatter, 2012, 2, 176–194 CrossRef PubMed
.
- C. M. Murphy, M. G. Haugh and F. J. O’Brien, Biomaterials, 2010, 31, 461–466 CrossRef CAS PubMed
.
- H. Lu, Y. Zhang, S. Xiong, Y. Zhou, L. Xiao, Y. Ma, Y. Xiao and X. Wang, Front. Chem., 2021, 9, 699802 CrossRef CAS PubMed
.
- National Technical Information Service, OTS0571423.
- C. M. Murphy, F. J. O’Brien, D. G. Little and A. Schindeler, Eur. Cells Mater., 2013, 26, 120–132 CrossRef CAS PubMed
.
- H. S. Wang, S. C. Hung, S. T. Peng, C. C. Huang, H. M. Wei, Y. J. Guo, Y. S. Fu, M. C. Lai and C. C. Chen, Stem Cells, 2004, 22, 1330–1337 CrossRef PubMed
.
- S. Tipnis, C. Viswanathan and S. Majumdar, Immunol. Cell Biol., 2010, 88, 795–806 CrossRef PubMed
.
- J. Lim, Z. R. M. Razi, J. X. Law, A. M. Nawi, R. Idrus, T. G. Chin, M. Mustangin and M. W. Ng, J. Tissue Eng. Regen. Med., 2018, 15, 75–87 CrossRef CAS PubMed
.
- A. S. Ansari, M. D. Yazid, N. Q. A. V. Sainik, R. A. Razali, A. B. Saim and R. Idrus, Stem Cells Int., 2018, 2018, 2406462 Search PubMed
.
- S. M. Daboor, S. M. Budge, A. E. Ghaly, S. L. Brook and D. Dave, Am. J. Biochem. Biotechnol., 2010, 6, 239–263 CrossRef CAS
.
- H. Alipour, A. Raz, S. Zakeri and N. D. Djadid, Asian Pac. J. Trop. Biomed., 2016, 6, 975–981 CrossRef CAS
.
- E. Sulistyowati, D. Salirawati and Amanatie, J. P. Saintek, 2016, 21, 107–119 Search PubMed
.
- W. Widowati, R. F. Gunanegara, R. Rizal, W. S. Widodo, A. Amalia, S. H. B. Wibowo, K. Handono, M. Marlina, I. N. E. Lister and L. Chiuman, J. Phys. Conf. Ser., 2019, 1374, 012024 CrossRef CAS
.
- D. Bharti, S. B. Shivakumar, J. K. Park, I. Ullah, R. B. Subbarao, J. S. Park, S. L. Lee, B. W. Park and G. J. Rho, Cell Tissue Res., 2018, 372, 51–65 CrossRef CAS PubMed
.
- N. Beeravolu, C. McKee, A. Alamri, S. Mikhael, C. Brown, M. Perez-Cruet and G. R. Chaudhry, J. Visualized Exp., 2017, 122, e55224 Search PubMed
.
- N. Vanawati, A. Barlian, Y. Tabata, H. Judawisastra and I. Wibowo, Future Sci. OA, 2022, 8, FSO810 CrossRef CAS PubMed
.
- R. M. Amsar, A. Barlian, H. Judawisastra, U. A. Wibowo and K. Karina, Future Sci. OA, 2021, 7, FSO734 CrossRef CAS PubMed
.
- E. M. Czekanska, Methods Mol. Biol., 2011, 740, 27–32 CrossRef CAS PubMed
.
- N. Ninan, B. Joseph, R. M. Visalakshan, R. Bright, C. Denoual, P. Zilm, Y. B. Dalvi, P. V. Priya, A. Mathew, Y. Grohens, N. Kalarikkal, K. Vasilev and S. Thomas, Mater. Adv., 2021, 2, 6620–6630 RSC
.
- J. Kim and T. Adachi, Front. Bioeng. Biotechnol., 2019, 7, 288 CrossRef PubMed
.
- L. Liu, P. Li, G. Zhou, M. Wang, X. Jia, M. Liu, X. niu, W. Song, H. Liu and Y. Fan, J. Biomed. Nanotechnol., 2013, 9, 1532–1539 CrossRef CAS PubMed
.
- L. Gachkar, D. Yadegari, M. B. Rezaei, M. Taghizadeh, S. A. Astaneh and I. Rasooli, Food Chem., 2007, 102, 898–904 CrossRef CAS
.
- D. L. Buckley, K. Raina, N. Darricarrere, J. Hines, J. L. Gustafson, I. E. Smith, A. H. Miah, J. D. Harling and C. M. Crews, ACS Chem. Biol., 2015, 10, 1831–1837 CrossRef CAS PubMed
.
- J. L. Chen, T. W. J. Steele and D. C. Stuckey, Biotechnol. Bioeng., 2018, 115, 351–358 CrossRef CAS PubMed
.
- F. S. Hosseini, F. Soleimanifar, A. Ardeshirylajimi, S. Vakilian, M. Mossahebi-Mohammadi, S. E. Enderami, A. Khojasteh and S. Z. Karizi, Artif. Cells Nanomed. Biotechnol., 2019, 47, 300–307 CrossRef CAS PubMed
.
- M. Dominici, K. L. Blanc, I. Mueller, I. Slaper-Cortenbach, F. C. Marini, D. S. Krause, R. J. Deans, A. Keating, D. J. Prockop and E. M. Horwitc, Cytotherapy, 2006, 8, 315–317 CrossRef CAS PubMed
.
- S. Borys-Wojcik, M. Brazert, M. Jankowski, K. Ozegowska, B. Chermula, H. Piotrowska-Kempisty, D. Bukowska, P. Antosik, L. Pawelczyk, M. Nowicki, M. Jaseta and B. Kempisty, J. Biol. Regul. Homeost. Agents, 2019, 33, 119–123 CAS
.
- M. Borhani-Haghighi, T. Talaei-Khozani, M. Ayatollahi and Z. Vojdani, Iran. J. Med. Sci., 2015, 40, 143–151 Search PubMed
.
- M. A. A. Hussein, H. A. M. Hussein, A. A. Thabet, K. M. Selim, M. A. Dawood, A. M. El-Adly, A. A. Wardany, A. Sobhy, S. Magdeldin, A. Osama, A. M. Anwar, M. Abdel-Wahab, H. Askar, E. K. Bakhiet, S. Sultan, A. A. Ezzat, U. A. Raouf and M. M. Afifi, Cells, 2022, 11, 1408 CrossRef CAS PubMed
.
- P. Anderson, A. B. Carrillo-Gálvez, A. García-Pérez, M. Cobo and F. Martín, PLoS One, 2013, 8(10), e376979 Search PubMed
.
- V. Karageorgiou and D. Kaplan, Biomaterials, 2005, 26, 5474–5491 CrossRef CAS PubMed
.
- A. Penk, Y. Förster, H. A. Scheidt, A. Nimptsch, M. C. Hacker, M. Schulz-Siegmund, P. Ahnert, J. Schiller, S. Rammelt and D. Huster, Magn. Reson. Med., 2013, 70, 925–935 CrossRef CAS PubMed
.
- J. Krieghoff, A.-K. Picke, J. Salbach-Hirsch, S. Rother, C. Heinemann, R. Bernhard, C. Kascholke, S. Möller, M. Rauner, M. Schnabelrauch, V. Hintze, D. Scharnweber, M. Schulz-Siegmund, M. C. Hacker, L. C. Hofbauer and C. Hofbauer, Biomater. Res., 2019, 23, 26 CrossRef CAS PubMed
.
- L. G. Sicchieri, G. E. Crippa, P. T. de Oliveira, M. M. Beloti and A. L. Rosa, J. Tissue Eng. Regener. Med., 2012, 6, 155–162 CrossRef CAS PubMed
.
- A. A. Khalili and M. R. Ahmad, Int. J. Mol. Sci., 2015, 16, 18149–18184 CrossRef CAS PubMed
.
- M. J. Lerman, J. Lembong, S. Muramoto, G. Gillen and J. P. Fisher, Tissue Eng., Part B, 2018, 24, 359–372 CrossRef CAS PubMed
.
- Y.-L. Qiu, X. Chen, Y.-L. Hou, Y.-J. Hou, S.-B. Tian, Y.-H. Chen, L. Yu, M.-H. Nie and X.-Q. Liu, Mol. Med. Rep., 2019, 19, 4043–4056 CAS
.
- S. Ji and M. Guvendiren, Micromachines, 2020, 11, 31 CrossRef PubMed
.
- J. Ge, L. Guo, S. Wang, Y. Zhang, T. Cai, R. C. H. Zhao and Y. Wu, Stem Cell Rev. Rep., 2014, 10, 295–303 CrossRef CAS PubMed
.
- R. Zhang, P. Lee, V. C. H. Lui, Y. Chen, X. Liu, C. N. Lok, M. To, K. W. K. Yeung and K. K. Y. Wong, Nanomedicine, 2015, 11, 1949–1959 CrossRef CAS PubMed
.
- Q. Zhang, S. Lv, J. Lu, S. Jiang and L. Lin, Int. J. Biol. Macromol., 2015, 76, 94–101 CrossRef CAS PubMed
.
- R. J. Miron and Y. F. Zhang, J. Dent. Res., 2012, 91, 736–744 CrossRef CAS PubMed
.
- G. S. Stein and J. B. Lian, Endocr. Rev., 1993, 14, 424–442 CrossRef CAS PubMed
.
- D. S. Amarasekara, S. Kim and J. Rho, Int. J. Mol. Sci., 2021, 22, 2851 CrossRef CAS PubMed
.
- H. C. Tenenbaum, Bone Miner., 1987, 3, 13–26 CAS
.
- M. F. Abazari, Z. Hosseini, S. Z. Karizi, S. Norouzi, M. A. Faskhoudi, E. Saburi, S. E. Enderami, A. Ardeshirylajimi and H. Mohajerani, Gene, 2020, 740, 144534 CrossRef CAS PubMed
.
- H. Donnelly, M. Salmeron-Sanchez and M. J. Dalby, J. R. Soc., Interface, 2018, 15, 20180388 CrossRef PubMed
.
- C. Greulich, S. Kittler, M. Epple, G. Muhr and M. Koller, Langenbecks Arch. Surg., 2009, 394, 495–502 CrossRef CAS PubMed
.
- S. K. Jung, J. H. Kim, H. J. Kim, Y. H. Ji, J. H. Kim and S. W. Son, J. Invest. Dermatol., 2014, 134, 3003–3007 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ma00332a |
| This journal is © The Royal Society of Chemistry 2023 |