Open Access Article
Open Access ArticleThe morphology and structural features of self-aggregating hexapeptides with antibiofilm formation activity†
Dongru
Chen‡
 a,
Tingyu
Wang‡
a,
Yiyi
Huang
b,
Yucong
Chen
b,
Huancai
Lin
*b and
Liping
Wu
*a
a,
Tingyu
Wang‡
a,
Yiyi
Huang
b,
Yucong
Chen
b,
Huancai
Lin
*b and
Liping
Wu
*a
aDepartment of Orthodontics, Hospital of Stomatology, Guanghua School of Stomatology, Sun Yat-sen University, Guangdong Provincial Key Laboratory of Stomatology, Guangzhou, Guangdong, China. E-mail: wulping@mail.sysu.edu.cn
bDepartment of Preventive Dentistry, Hospital of Stomatology, Guanghua School of Stomatology, Sun Yat-sen University, Guangdong Provincial Key Laboratory of Stomatology, Guangzhou, Guangdong, China. E-mail: linhc@mail.sysu.edu.cn
First published on 2nd June 2023
Abstract
Though self-aggregating peptides all aggregate into fibrils, they have been found to exert different roles against microbes, and the reasons remain unknown. In this study, we aimed to screen more self-aggregating hexapeptides with the highest fibrillation abilities using combined computational simulation methods and to study their effects against microbes (antimicrobial effect and antibiofilm-formation effect). Thereafter, we further explored the structural features of the self-aggregating hexapeptides leading to the different functions. By combining the use of three amyloid prediction software packages, we screened out 25 hexapeptides. Among them, 12 hexapeptides had antibiofilm activities (7 were stable), which were named as antibiofilm self-aggregating hexapeptides (ASAPs), while the remaining 13 hexapeptides had no effects on microbes. The ASAPs aggregated into fibril agglutinating bacteria, with the fibrils mainly divided into two classical morphologies, namely “twisted filaments” and “flat striated ribbons”, while the non-ASAPs aggregated into long bundles composed of numerous nanofibrils, or into amorphous polymers. Secondary structure analysis showed that the ASAP fibrils had predominantly β-sheets, and almost had no α-helices. In conclusion, our study revealed that the two types of fibrils aggregated by self-aggregating hexapeptides, i.e., “twisted filaments” and “flat striated ribbons”, were correlated with the agglutination function and the subsequent antibiofilm-formation activity, revealing a structure-function relationship of self-aggregating peptides.
Introduction
The concept of self-aggregating peptides or self-assembling peptides is broader than that of amyloid peptides. Self-aggregating peptides refer to peptides that can aggregate spontaneously into well-ordered supramolecular nanostructures due to intramolecular or intermolecular interactions, which can explain the aggregation of natural peptides into amyloid fibrils.1 For a protein/peptide aggregate to be considered as an amyloid, it must contain a high amount of β-sheets.2 Amyloids were previously regarded as the hallmark of more than 20 human disorders of unrelated etiologies, such as Alzheimer's disease, Parkinson's disease, and type II diabetes.3 However, recent studies have revealed that amyloids are used by nature in multifaceted and ingenious ways, exerting many functional roles, including as structures for microbial biofilm formation, for the storage of peptide hormones in mammalian secretory granules, and as information carriers.4 Moreover, synthetic self-aggregating peptides have been explored as antimicrobial and anticancer agents, as biomaterials for cell adhesion, and in tissue engineering applications, showing great application potential in many fields.5–8Previously, we innovatively found that some hexapeptides derived from the C123 truncated protein of Streptococcus mutans (S. mutans) could suppress microbial biofilm formation by aggregating into amyloid fibril agglutinating microbes, without bactericidal effects.9 Besides, the derivatives of hexapeptides are not limited to the C123 protein, indicating more potential hexapeptides possessing antibiofilm-formation abilities.9 The effect of self-aggregating peptide agglutinating microbes was also reported in human α-defensin 6 and Aβ, while most self-aggregating peptides were discovered to exert bactericidal functions.10–15 However, in our previous study, no self-aggregating hexapeptides with bactericidal function were found.9 The reasons for the various functions of self-aggregating peptides against microbes remained unknown. The hidden world of amyloid biology has suddenly snapped into an atomic-level focus in recent years, revealing over 80 fibrils.16 Though self-aggregating peptides with different sequences all aggregate into fibrils, they might yield distinct fibril polymorphs and structures.17 Increasing evidence has revealed a link between the fibrils structure of self-aggregating peptides and their function.16,18 It was shown that fibrils formed in vitro by Aβ40 and Aβ42 were polymorphic, which correlated with different AD phenotypes.19 The self-aggregating hexapeptides, LFKFFK derived from PSMα3, IIKVIK derived from PSMα1, and IIKIIK derived from PSMα4, all aggregated into fibrils, but only LFKFFK elicited antimicrobial activities. Structure analysis revealed that LFKFFK formed reversible fibrils and atypical amyloid structures, in which each β-strand was at an angle of ∼50° from the fibril axis, while IIKVIK and IIKIIK formed highly stable steric-zipper architectures, in which each β-strand was at an angle close to 90°.12 However, no consensus has been reached on this opinion.
In this study, we aimed to screen more self-aggregating hexapeptides with the highest fibrillation ability, to explore their effects on microbes, and to discover the possible structure and function relationship. We anticipated that our insights into the structure and function relationship of self-aggregating peptides would shed light on their structural interpretation, and therefore aid discovering more functional self-aggregating hexapeptides with antibiofilm-formation effects.
Materials and methods
Computational simulation of hexapeptides
ZipperDB (https://services.mbi.ucla.edu/zipperdb) is a frequently used amyloid hexapeptide prediction database with data from the analysis of over 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 putative protein sequences for segments with high fibrillation propensity. Hexapeptides with Rosetta energies equal to or below −23 kcal mol−1 are deemed to have high fibrillation propensity, whereby the lower the Rosetta energy, the higher the fibrillation propensity. In this case, we contacted the ZipperDB server and obtained the hexapeptides with the highest fibrillation propensity.20 We synthesized the first 10 hexapeptides, and examined their bactericidal activities and antibiofilm-formation activities on Streptococcus mutans ATCC 700610TM (S. mutans) at a concentration of 0.05 mg mL−1. However, only the first self-aggregating hexapeptide (LFWLVF) exhibited antibiofilm-formation activity, and no hexapeptide with bactericidal activity was found (Table S1, ESI†). The above screening method showed limitations, mainly because the screening efficiency was very low. To improve the screening efficiency, we combined another two amyloid prediction software systems: TANGO21 and Waltz.22 Tango and Waltz were used to verify the fibrillation propensity of the hexapeptides predicted by ZipperDB. If the fibrillation propensity of a hexapeptide was also verified by both Tango and Waltz, the hexapeptide was selected. We synthesized the first 25 hexapeptides selected according to their Rosetta energies.
000 putative protein sequences for segments with high fibrillation propensity. Hexapeptides with Rosetta energies equal to or below −23 kcal mol−1 are deemed to have high fibrillation propensity, whereby the lower the Rosetta energy, the higher the fibrillation propensity. In this case, we contacted the ZipperDB server and obtained the hexapeptides with the highest fibrillation propensity.20 We synthesized the first 10 hexapeptides, and examined their bactericidal activities and antibiofilm-formation activities on Streptococcus mutans ATCC 700610TM (S. mutans) at a concentration of 0.05 mg mL−1. However, only the first self-aggregating hexapeptide (LFWLVF) exhibited antibiofilm-formation activity, and no hexapeptide with bactericidal activity was found (Table S1, ESI†). The above screening method showed limitations, mainly because the screening efficiency was very low. To improve the screening efficiency, we combined another two amyloid prediction software systems: TANGO21 and Waltz.22 Tango and Waltz were used to verify the fibrillation propensity of the hexapeptides predicted by ZipperDB. If the fibrillation propensity of a hexapeptide was also verified by both Tango and Waltz, the hexapeptide was selected. We synthesized the first 25 hexapeptides selected according to their Rosetta energies.
Peptide synthesis, storage, and fibrils preparation
All the hexapeptides were synthesized at >95% purity by Genscript (Nanjing, China) following a solid-phase peptide synthesis protocol. All the hexapeptides were not modified by N-terminal acetylation and C-terminal amidation. Lyophilized hexapeptide powder was stored or diluted in 5% dimethylsulfoxide (DMSO) to a concentration of 1 mg mL−1 at −20 °C.For fibrils preparation, hexapeptides (0.5 mg mL−1) were agitated at 700 rpm, pH 7, 37 °C for 4 days to ensure that the hexapeptide monomers with self-aggregating ability aggregated thoroughly.
Microbial stains and growth conditions
First, Gram-positive bacteria S. mutans were incubated statically in brain heart infusion broth medium (BHI) at 37 °C for 18 h under anaerobic conditions (85% N2, 10% H2, 5% CO2) for planktonic bacteria growth to the plateau stage, and in BHI + 1% sucrose for 24 h for biofilm formation. Gram-positive bacteria Staphylococcus aureus (S. aureus) ATCC 6538p and Gram-negative Escherichia coli (E. coli) ATCC 8739 were incubated in tryptic soy broth at 37 °C for 18 h for planktonic bacteria growth to the plateau stage.Microbe proliferation and crystal violet assays
To test the antimicrobial activity, 106 CFU mL−1 planktonic microbes were incubated in Eppendorf tubes with the 25 hexapeptides at a concentration of 0.05 mg mL−1, respectively, or with the same concentration of DMSO for the control group. After 24 h cultivation, the planktonic microbe solution was added to 96-well plates after being well mixed. The value of OD600 for planktonic microbe proliferation was detected using a spectrophotometer.To test the antibiofilm-formation activity, crystal violet assays were performed. Here, 106 CFU mL−1 planktonic S. mutans was added into the medium with 25 of the 0.05 mg mL−1 hexapeptides, respectively, and then incubated in 96-well plates for 24 h. Then, the 96-well plate was washed twice with ddH2O, fixed with absolute methanol for 15 min, air-dried for at least 15 min, stained with 0.1% crystal violet for 15 min, and washed with ddH2O until the wells of the control group were clean. The absorbance was measured using a spectrophotometer for obtaining the OD600. All the experiments were replicated three times.
Transmission electron microscopy (TEM) assay
A 10 μL aggregated fibrils solution was deposited on carbon-coated nickel grids for 2 min, and negative staining was performed using 3% phosphotungstic acid for 1 min. Observations were made using a Hitachi TEM system.Atomic force microscopy (AFM) assay
A 10 μL aggregated fibrils solution was deposited on the surface of freshly cleaved mica and left to dry in air at room temperature overnight. Images were obtained using a NanoScope IIIa Multimode control system (Dimension Fastscan, Bruker) operating in the tapping mode using 100 mm long silicon nitride cantilevers with a resonance frequency of 70 kHz. Images were captured by NanoScope Analysis Software.Circular dichroism (CD) assay
The hexapeptides monomers solution was dissolved by ddH2O to a concentration of 0.5 mg mL−1, and the monomers solution was agitated for 4 days at 37 °C for fibrillation. The CD spectra were recorded from 197–260![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nm using a Chirascan-plus CD spectrometer (Applied Photophysics, Leatherhead, England). Measurements were obtained with a bandwidth of 1.0
nm using a Chirascan-plus CD spectrometer (Applied Photophysics, Leatherhead, England). Measurements were obtained with a bandwidth of 1.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nm in 1.0
nm in 1.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nm steps and 0.5
nm steps and 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) s time per point at 37 °C. CDToolX was used to manage the CD spectroscopic data and the secondary structure was analyzed by CONTINLL.23
s time per point at 37 °C. CDToolX was used to manage the CD spectroscopic data and the secondary structure was analyzed by CONTINLL.23
Fourier-transform infrared spectroscopy (FTIR) assay
The aggregated fibrils solution was centrifuged at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 20 min to collect the pellets and the supernatant was discarded. Subsequently, the pellets were freeze-dried overnight to get fibrils powder. The powder was thoroughly mixed with anhydrous potassium bromide, and then pressure was applied at 1333.22 Pa cm−2 for 15 min to get the potassium bromide tablet. Data were recorded using an FT-Raman Spectrometer Nicolet NXR 9650 in the spectral region of 4000–200 cm−1. Each spectrum was processed and analyzed by PeakFit v4.12 (Systat Software Inc., USA). After correction with a linear baseline, information of the peaks was obtained through Gaussians smoothing and numerical fitting for the spectra of the amide I band (1600–1700 cm−1). As a rule of thumb, a peak in the 1600–1640 cm−1 or 1670–1680 cm−1 region is indicative of β-sheets, 1650–1660 cm−1 indicates α-helix, and 1640–1650 cm−1 random coils (Table S2, ESI†).24,25
000 g for 20 min to collect the pellets and the supernatant was discarded. Subsequently, the pellets were freeze-dried overnight to get fibrils powder. The powder was thoroughly mixed with anhydrous potassium bromide, and then pressure was applied at 1333.22 Pa cm−2 for 15 min to get the potassium bromide tablet. Data were recorded using an FT-Raman Spectrometer Nicolet NXR 9650 in the spectral region of 4000–200 cm−1. Each spectrum was processed and analyzed by PeakFit v4.12 (Systat Software Inc., USA). After correction with a linear baseline, information of the peaks was obtained through Gaussians smoothing and numerical fitting for the spectra of the amide I band (1600–1700 cm−1). As a rule of thumb, a peak in the 1600–1640 cm−1 or 1670–1680 cm−1 region is indicative of β-sheets, 1650–1660 cm−1 indicates α-helix, and 1640–1650 cm−1 random coils (Table S2, ESI†).24,25
Results
Simulations of the hexapeptides using computational methods
The screened 25 hexapeptides were named H1 to H25 (Table 1), respectively. By analyzing the primary sequences of the 25 hexapeptides superficially, we found that all the predicted hexapeptides side-chains contained asparaginate (A), isoleucine (I), or valine (V), which are common in self-aggregating proteins.| Name | Sequence | Gravy | Isoelectric point (pI) | Rosetta energy |
|---|---|---|---|---|
| H1 | AIIIII | 4.05 | 5.57 | −32.7 |
| H2 | AIIVII | 4 | 5.57 | −32.4 |
| H3 | AIIIIV | 4 | 5.57 | −32.1 |
| H4 | ALIIII | 3.93 | 5.57 | −31.6 |
| H5 | GIVVII | 3.58 | 5.52 | −31.5 |
| H6 | VVVIII | 4.35 | 5.49 | −31.3 |
| H7 | VIVIII | 4.35 | 5.49 | −31.3 |
| H8 | AIIIIY | 3.08 | 5.57 | −31.3 |
| H9 | AIIVIL | 3.88 | 5.57 | −31.3 |
| H10 | AIIIIA | 3.6 | 5.57 | −31.2 |
| H11 | VIVVII | 4.35 | 5.49 | −31.1 |
| H12 | VVVVII | 4.3 | 5.49 | −31.1 |
| H13 | AVIIIF | 3.72 | 5.57 | −31.1 |
| H14 | AVIVIM | 3.52 | 5.57 | −31.1 |
| H15 | GVVIIV | 3.53 | 5.52 | −31.1 |
| H16 | GIVIIV | 3.58 | 5.52 | −31.1 |
| H17 | AVIVIA | 3.5 | 5.57 | −31 |
| H18 | AVILII | 3.88 | 5.57 | −31 |
| H19 | AYIVII | 3.03 | 5.57 | −31 |
| H20 | AAIIII | 3.6 | 5.57 | −31 |
| H21 | AIILII | 3.93 | 5.57 | −30.9 |
| H22 | AIIVIA | 3.55 | 5.57 | −30.9 |
| H23 | AVIVIF | 3.67 | 5.57 | −30.9 |
| H24 | GVVVIV | 3.48 | 5.52 | −30.9 |
| H25 | GLVIII | 3.52 | 5.52 | −30.8 |
Effect of the hexapeptides monomers on bacteria
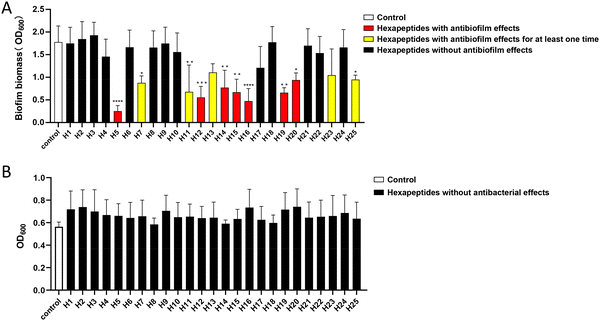
The antibacterial and antibiofilm-formation effects of the above 25 hexapeptides monomers were examined on S. mutans. It was shown that seven hexapeptides (H5, H12, H14, H15, H16, H19, and H20) always exhibited efficient antibiofilm activity against S. mutans in our multiple replication studies (p < 0.05), while five hexapeptides (H7, H11, H13, H23, and H25) showed antibiofilm activity in at least one experiment, but were not stable in all the multiple replication studies (n = 3), and thus they were excluded from our following experiments. The remaining 13 hexapeptides (H1, H2, H3, H4, H6, H8, H9, H10, H17, H18, H21, H22, and H24) had no antibiofilm activities (Fig. 1A). It should be noted that no hexapeptides exhibited bactericidal effects, and no significant differences in the planktonic proliferation values were observed between the control group and hexapeptides group (Fig. 1B). The results indicated that the combined screening method greatly improved the successful screening efficiency, and we successfully found more hexapeptides with antibiofilm-formation effects.In vitro aggregated morphologies of the hexapeptides
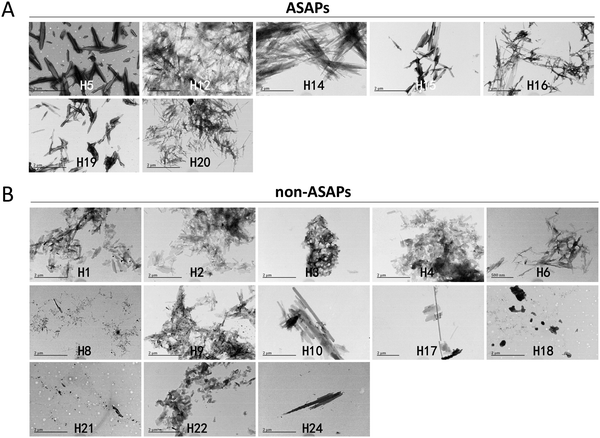
After agitating for 4 days, most hexapeptides aggregated into fibrils, but with different morphologies when observed under TEM. The fibrils formed by the antibiofilm-formation self-aggregating hexapeptides (H5, H12, H14, H15, H16, H19, and H20) seemed to be more rigid, less erratic, and longer (Fig. 2A), and we named these hexapeptides as antibiofilm self-aggregating hexapeptides (ASAPs), while the fibrils formed by non-ASAPs were in quite a small amount, or in a large amount with shorter fragments (Fig. 2B).ASAPs formed fibrils agglutinating microbes
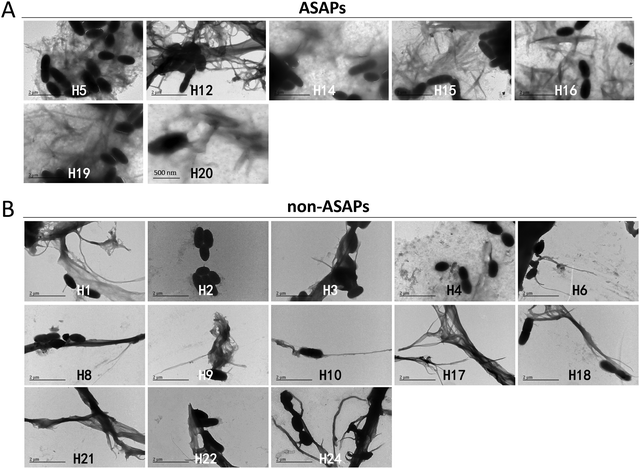
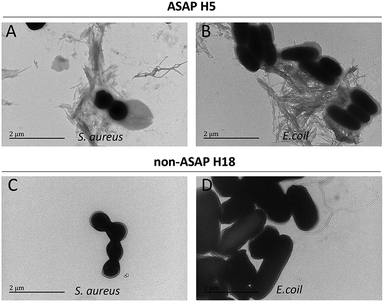
The effects of hexapeptides against microbes were further studied by TEM. It was seen that all the ASAPs (H5, H12, H14, H15, H16, H19, and H20) aggregated into fibrils agglutating S. mutans. These fibrils formed a network-like structure wrapping S. mutans, and the fibrils around S. mutans were in a large number (Fig. 3A). Though the non-ASAPs also showed fibrils occasionally (e.g., H1, H6, H8, H9, H10, H17, H18, H21, H22, and H24) and sometimes a few bacteria were attached to the fibrils, the fibrils were mostly in long bundles composed of numerous nanofibrils (Fig. 3B), which were quite different in their morphologies compared to that of the ASAPs. Besides, the morphologies of the fibrils formed by the non-ASAPs in the presence of S. mutans were different from those without S. mutans.To examine the effects of ASAPs on other strains of bacteria, we selected two classical model strains, S. aureus and E. coli. We also selected H5 and H18 as representative hexapeptide for ASAPs and non-ASAPs groups, respectively. The TEM results also verified that ASAP H5 aggregated into fibrils agglutinating S. aureus and E. coli (Fig. 4A and B), indicating that the agglutination effects were broad-spectrum. The non-ASAP H18 had no effect on S. aureus and E. coli.
Aggregated morphologies revealed by AFM
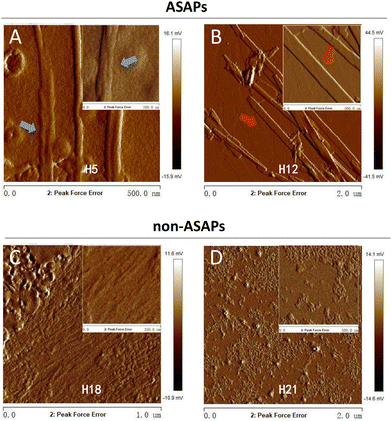
A particular powerful approach to probe the key physical properties of fibrillar structures is AFM. Since the fibrils morphologies of ASAP H5, H15, and H19 were similar, as seen in Fig. 2, and the same as H12, H14, H16, and H20, thus we selected H5 and H12 as representative hexapeptides to explore their fine structures, and H18 and H21 as representative hexapeptides for the non-ASAP groups. In addition, H18 and 21 were randomly selected as representatives of the non-ASAPs. The AFM images showed that both H5 and H12 formed typical fibrils after 4 days agitating. The fibrils formed by H5 were “twisted filaments” (Fig. 5A), while the fibrils formed by H12 were “flat striated ribbons” (Fig. 5B). The non-ASAP H18 formed bundles of numerous soft nanofibrils (Fig. 5C), similar with that seen in Fig. 3. Another non-ASAP H21 formed an amorphous polymer of small particles (Fig. 5D). The AFM results further verified the diversities of the fibrils morphologies observed by TEM.Secondary structures features
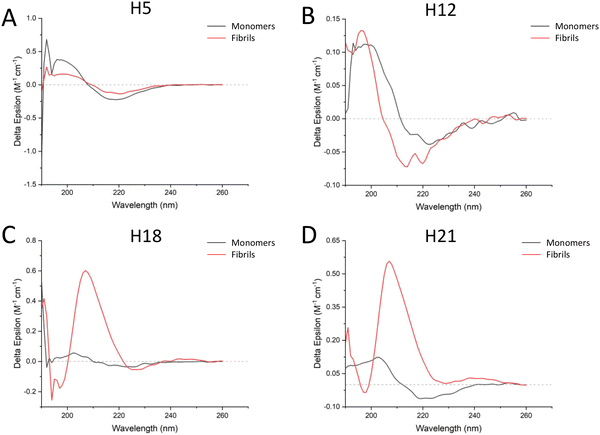
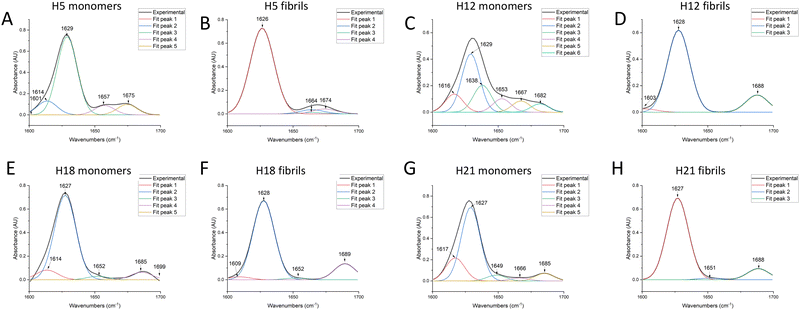
The above results indicated that the fibrils formed by the ASAPs had distinguished morphologies compared to that formed by the non-ASAPs. To further investigate their structural features, CD and FTIR were performed. As shown in Fig. 6A and B, ASAPs H5 and H12 kept predominantly a β-sheet-rich secondary structure from monomers to fibrils, which displayed a positive peak around 200 nm and negative peaks around 220 nm. However, the non-ASAPs H18 fibrils and H21 fibrils had a high positive peak around 210 nm (Fig. 6C and D), which might indicate amorphous aggregates were also formed besides fibrils. When analyzed by the CONTIN/LL programs, both the monomers and fibrils of H5 and H12 yielded high β-sheet contents (H5 monomers: 43.7%, H5 fibrils: 42.4%, H12 monomers: 42.3%, H12 fibrils: 42.6%), and a very small content of helix structures (Table S3, ESI†).FTIR worked best with concentrated solutions, solids, and films and could resolve with accuracy the β-sheet composition.26 A good correlation between the peptide structure and amide band frequency was confirmed by scientific calculations and experimental studies. The FTIR results showed that both ASAPs (H5 and H12) monomers and non-ASAPs (H18 and H21) monomers exhibited peak maxima in the region of 1620–1630 cm−1, indicating the predominant presence of β-sheets, and more than one weak peak indicated the presence of β-turns, α-helices, and random coils (Fig. 7A, C, E, and G). After fibrillation, the H5 fibrils displayed intense peaks in the region of 1620–1630 cm−1 and weak peaks around 1670 cm−1, which were assigned to parallel β-sheets (Fig. 7B), while the H12 fibrils displayed intense peaks in the region of 1620–1630 cm−1 and weak peaks around 1688 cm−1, which were assigned to antiparallel β-sheets (Fig. 7D). The Non-ASAPs H18 and H21 fibrils both exhibited intense peaks in the region of 1620–1630 cm−1 and weak peaks around 1688 cm−1, also indicating β-sheets; in addition, weak peaks at 1652 and 1651 cm−1 in the H18 fibrils and H21 fibrils, respectively, indicated the presence of α-helices (Fig. 7F and H). In order to obtain a deeper insight into the secondary structures of hexapeptides, the corresponding secondary structure in the amide I region was quantified according to the relative assignments of these bands. In general, the β-sheets were a dominant structure for both the monomers and fibrils of ASAPs (H5 and H12) and non-ASAPs (H18 and H21). The proportion of β-sheets increased from 91.7% (monomers) to 94.2% (fibrils) for H5, and from 75.8% (monomers) to 83.2% (fibrils) for H12, while it decreased from 89.5% (monomers) to 82.2% (fibrils) in H18 and increased from 75.8% (monomers) to 83.2% (fibrils) in non-ASAPs H21. It was a remarkable result that the fibrils of H5 and H12 contained no α-helices.
Discussion
By combining the use of three amyloid prediction software systems, we successfully screened more self-aggregating hexapeptides with antibiofilm-formation activity, improving the screen efficiency, and the method could be adopted in future screening. These ASAPs aggregated into typical fibrils agglutinating microbes, and thus prevented bacteria from adhering to surfaces. The fibrils formed by ASAPs were noted in a large amount with distinguished morphologies, mainly as twisted filaments or flat striated ribbons, while the non-ASAPs fibrils were in amorphous polymers or bundles of numerous soft nanofibrils. The two types of twisted filaments and flat striated ribbons were the classical morphologies for many different self-aggregating peptides fibrillation states.27,28 In a saline environment, β-endorphin primarily fibrillizes into flat striated ribbons, while in a saline-free environment, β-endorphin fibrillizes into twisted filaments. By using 3D solid-state nuclear magnetic resonance (NMR) spectroscopy to assess their structural differences at the atomic level, Seuring et al.28 found the spectral fingerprints and chemical shifts were very similar for both fibril types, indicating that the monomer structure and molecular interfaces were almost the same. However, experimental and theoretical considerations attributed the mesoscopic difference in the twisted fibrils versus flat ribbons to electrostatic intermolecular repulsions. Our structural analysis by CD and FTIR of the two fibril types also indicated similar β-sheet contents. Besides, our study correlated the two fibrils structures with their microbes agglutination activities, and their subsequent antimicrobial formation activities.The agglutination functions of self-aggregating peptides is appealing, as these could help prevent the spread of infection and facilitate bacterial clearance by the immune cells of the host. However, very few synthetic self-aggregating peptides have been found to exert agglutination functions without bacteriacidal effects up to now.9,10,29,30 As far as we know, self-aggregating hexapeptides are the shortest peptides with simple microbes agglutination functions and antibiofilm-formation activities. In our opinion, there might be numerous amyloid peptides with microbes agglutination activities that have not been revealed yet. Previously, Chu et al.15 demonstrated that human α-defensin 6, an important defensin secreted in the intestinal tract, underwent ordered self-assembly to form amyloid fibrils and nanonets that could surround and entangle bacteria. Importantly, HD6 only entangled bacteria and inhibited their adhesion, without bactericidal effects. Fibrils aggregated by self-aggregating peptides could also agglutinate viruses and inhibit virus entry into cells. Various infectious pathogens had been detected in the amyloids of neurodegeneration patients’ brains, which might indicate the agglutination effects of amyloid peptides against pathogens.7,31 The membranes formed by β-lactoglobulin (BLG) amyloid fibrils coated with iron oxyhydroxide nanoparticles (Fe NPs) were found to be able to eliminate viruses effectively from water by retaining and inactivating a broad range of viruses, including SARS-CoV-2, H1N1 (the influenza A virus strain responsible for the swine flu pandemic in 2009), and enterovirus 71 (a non-enveloped virus resistant to harsh conditions).32 Hence, self-aggregating peptides with broad-spectrum agglutination ability could be further explored for their application value in many aspects.
A notable finding is that no self-aggregating hexapeptide with bacteriacidal activity were found; however, most native and engineered self-aggregating proteins/peptides have been found to display functional roles against microbes, including a number of antimicrobial peptides (AMPs) or host defense peptides.30 By analyzing the structural features of peptides, Landau's group found both amphibian antimicrobial peptide uperin 3.5 and human LL-37 active core (residues 17–29) assembled into helical fibrils, and they both exerted antimicrobial effects.13,18 The crystal structure of LL-37 fibrils show alternating hydrophobic and polar (positively charged) zigzagged belts on their surface, suggesting interactions with and disruption of the negatively charged lipid bilayers, e.g., bacterial membranes.18 Prasad et al.33 used sodium dodecyl sulfate (SDS) micelles to simulate the cell membrane of bacteria in order to investigate the secondary structure transitions of U3 peptides during the antibacterial process. Their results indicated that SDS micelles promoted the stabilization of the α-helical structure of U3 peptides by promoting residue–residue hydrogen bonds within the peptide. In addition, the β-sheet dimers decreased and α-helical content increased after attachment to SDS micelles. The α-helical forms of U3 peptides adopted amphipathic confirmations, which enable insertion of peptides into the hydrophobic core of micelles. Sequence 2–12 (EFVAKLFKFFK, named UP) of PSMα3 was reported to play an important role in cross-α nanofibrils formation of a hydrophobic interface between PSMα3 monomers, and UP also had antibacterial effects. Aside from the cross-α fibrils being correlated with antibacterial effects, atypical cross-β fibrils, not classical cross-β fibrils, with antimicrobial effect were also reported in LFKFFK.12 However, in our study, all the self-aggregating hexapeptides fibrils yielded high β-sheet contents, and no α-helical contents. Therefore, we speculated that self-aggregating peptides with agglutination effects should be features of both their structure and morphology, and there is a structure–function relationship for self-aggregating peptides.
In conclusion, we successfully screened out more self-aggregating hexapeptides with microbes agglutination effects and antibiofilm-formation activity by combining the use of three amyloid prediction software systems. Also, the two types of fibrils, i.e., twisted filaments and flat striated ribbons, were correlated with the agglutination function and antibiofilm-formation activity. This study overall indicates that diversifications in fibril structures aggregated by self-aggregating hexapeptides might be associated with different functions, revealing a structure–function relationship of fibrils formed by self-aggregating hexapeptides, which is an important research area to investigate further in the future.
Study limitations
One distinct limitation was that not all the selected hexapeptides could fibrillate into fibrils actually, because the screening method was based on computational simulation, and it should be examined in in vitro experiments. Second, the predicted Rosetta energy of hexapeptide could not reflect its antibiofilm effectiveness, thus the screened self-aggregating hexapeptides in this study might not be the ones with the greatest antibiofilm-formation abilities. Third, though combining the use of three prediction methods significantly improved the screening efficiency, some self-aggregating hexapeptides might be excluded by this method. For example, the first hexapeptide LFWLVF with the lowest Rosetta energy predicted by ZipperDB was verified to have antibiofilm effects, but it was not predicted by Waltz, and thus it was excluded. Last but not least, the included 25 hexapeptides had similar sequences, so that studies of the diversities in sequence were limited. In future, more self-aggregating hexapeptides with various sequences need to be explored.Author contributions
Dongru Chen contributed to the conceptualization, methodology, investigation, formal analysis, resources, funding acquisition, validation, writing – original draft, and writing – review & editing. Tingyu Wang contributed to the methodology, investigation, formal analysis, writing – original draft, and writing – review & editing. Yiyi Huang contributed to the investigation, formal analysis, and writing – review & editing. Yucong Chen contributed to the methodology, investigation, formal analysis, and writing – review & editing. Huancai Lin and Liping Wu contributed to the conceptualization, project administration, and writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was funded by the National Natural Science Foundation of China (no. 82204253) and Guangdong Basic and Applied Basic Research Foundation (no. 2019A1515110847).References
- J. M. Lehn, Toward complex matter: supramolecular chemistry and self-organization, Proc. Natl. Acad. Sci. U. S. A., 2002, 99(8), 4763–4768 CrossRef CAS PubMed.
- S. Paul, K. Kumari and S. Paul, Molecular Insight into the Effects of Enhanced Hydrophobicity on Amyloid-like Aggregation, J. Phys. Chem. B, 2020, 124(45), 10048–10061 CrossRef CAS PubMed.
- F. Chiti and C. M. Dobson, Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade, Annu. Rev. Biochem., 2017, 86, 27–68 CrossRef CAS PubMed.
- D. Otzen and R. Riek, Functional Amyloids, Cold Spring Harbor Perspect. Biol., 2019, 11, 12 Search PubMed.
- L. Schnaider, S. Brahmachari and N. W. Schmidt, et al., Self-assembling dipeptide antibacterial nanostructures with membrane disrupting activity, Nat. Commun., 2017, 8(1), 1365 CrossRef PubMed.
- R. Gallardo, M. Ramakers and F. De Smet, et al., De novo design of a biologically active amyloid, Science, 2016, 354, 6313 CrossRef PubMed.
- K. Bourgade, G. Dupuis and E. H. Frost, et al., Anti-Viral Properties of Amyloid-beta Peptides, J. Alzheimer's Dis., 2016, 54(3), 859–878 CAS.
- S. Das, R. S. Jacob and K. Patel, et al., Amyloid Fibrils: Versatile Biomaterials for Cell Adhesion and Tissue Engineering Applications, Biomacromolecules, 2018, 19(6), 1826–1839 CrossRef CAS PubMed.
- D. Chen, J. Li and T. Pan, et al., The broad-spectrum antibiofilm activity of amyloid-forming hexapeptides, Microb. Biotechnol., 2021, 14(2), 656–667 CrossRef CAS PubMed.
- D. K. Kumar, S. H. Choi and K. J. Washicosky, et al., Amyloid-beta peptide protects against microbial infection in mouse and worm models of Alzheimer's disease, Sci. Transl. Med., 2016, 8(340), 340r–372r Search PubMed.
- Z. Wei, S. Wu and J. Xia, et al., Enhanced Antibacterial Activity of Hen Egg-White Lysozyme against Staphylococcus aureus and Escherichia coli due to Protein Fibrillation, Biomacromolecules, 2021, 22(2), 890–897 CrossRef CAS PubMed.
- N. Salinas, J. P. Colletier and A. Moshe, et al., Extreme amyloid polymorphism in Staphylococcus aureus virulent PSMalpha peptides, Nat. Commun., 2018, 9(1), 3512 CrossRef PubMed.
- N. Salinas, E. Tayeb-Fligelman and M. D. Sammito, et al., The amphibian antimicrobial peptide uperin 3.5 is a cross-alpha/cross-beta chameleon functional amyloid, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, 3 CrossRef PubMed.
- W. Tu, K. Xue and S. Lou, et al., Self-assembly of virulent amyloid-derived peptides into nanoantibacterials, Nanoscale, 2021, 13(21), 9864–9872 RSC.
- H. Chu, M. Pazgier and G. Jung, et al., Human alpha-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets, Science, 2012, 337(6093), 477–481 CrossRef CAS PubMed.
- M. R. Sawaya, M. P. Hughes and J. A. Rodriguez, et al., The expanding amyloid family: Structure, stability, function, and pathogenesis, Cell, 2021, 184(19), 4857–4873 CrossRef CAS PubMed.
- R. Tycko, Physical and structural basis for polymorphism in amyloid fibrils, Protein Sci., 2014, 23(11), 1528–1539 CrossRef CAS PubMed.
- Y. Engelberg and M. Landau, The Human LL-37(17-29) antimicrobial peptide reveals a functional supramolecular structure, Nat. Commun., 2020, 11(1), 3894 CrossRef PubMed.
- W. Qiang, W. M. Yau and J. X. Lu, et al., Structural variation in amyloid-beta fibrils from Alzheimer's disease clinical subtypes, Nature, 2017, 541(7636), 217–221 CrossRef CAS PubMed.
- L. Goldschmidt, P. K. Teng and R. Riek, et al., Identifying the amylome, proteins capable of forming amyloid-like fibrils, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(8), 3487–3492 CrossRef CAS PubMed.
- X. Lu, C. R. Brickson and R. M. Murphy, TANGO-Inspired Design of Anti-Amyloid Cyclic Peptides, ACS Chem. Neurosci., 2016, 7(9), 1264–1274 CrossRef CAS PubMed.
- N. Louros, K. Konstantoulea and M. De Vleeschouwer, et al., WALTZ-DB 2.0: an updated database containing structural information of experimentally determined amyloid-forming peptides, Nucleic Acids Res., 2020, 48(D1), D389–D393 CrossRef CAS PubMed.
- A. J. Miles and B. A. Wallace, CDtoolX, a downloadable software package for processing and analyses of circular dichroism spectroscopic data, Protein Sci., 2018, 27(9), 1717–1722 CrossRef CAS PubMed.
- A. Dong, P. Huang and W. S. Caughey, Protein secondary structures in water from second-derivative amide I infrared spectra, Biochemistry, 1990, 29(13), 3303–3308 CrossRef CAS PubMed.
- P. I. Haris and D. Chapman, The conformational analysis of peptides using Fourier transform IR spectroscopy, Biopolymers, 1995, 37(4), 251–263 CrossRef CAS PubMed.
- J. M. Ruysschaert and V. Raussens, ATR-FTIR Analysis of Amyloid Proteins, Methods Mol. Biol., 2018, 1777, 69–81 CrossRef CAS PubMed.
- K. Arima, Ultrastructural characteristics of tau filaments in tauopathies: immuno-electron microscopic demonstration of tau filaments in tauopathies, Neuropathology, 2006, 26(5), 475–483 CrossRef PubMed.
- C. Seuring, J. Verasdonck and P. Ringler, et al., Amyloid Fibril Polymorphism: Almost Identical on the Atomic Level, Mesoscopically Very Different, J. Phys. Chem. B, 2017, 121(8), 1783–1792 CrossRef CAS PubMed.
- S. Voth, M. Gwin and C. M. Francis, et al., Virulent Pseudomonas aeruginosa infection converts antimicrobial amyloids into cytotoxic prions, FASEB J., 2020, 34(7), 9156–9179 CrossRef CAS PubMed.
- D. Chen, X. Liu and Y. Chen, et al., Amyloid peptides with antimicrobial and/or microbial agglutination activity, Appl. Microbiol. Biotechnol., 2022, 106(23), 7711–7720 CrossRef CAS PubMed.
- M. A. Wozniak, A. P. Mee and R. F. Itzhaki, Herpes simplex virus type 1 DNA is located within Alzheimer's disease amyloid plaques, J. Pathol., 2009, 217(1), 131–138 CrossRef CAS PubMed.
- A. Palika, A. Armanious and A. Rahimi, et al., An antiviral trap made of protein nanofibrils and iron oxyhydroxide nanoparticles, Nat. Nanotechnol., 2021, 16(8), 918–925 CrossRef CAS PubMed.
- A. K. Prasad, C. Tiwari and S. Ray, et al., Secondary Structure Transitions for a Family of Amyloidogenic, Antimicrobial Uperin 3 Peptides in Contact with Sodium Dodecyl Sulfate, ChemPlusChem, 2022, 87(1), e202100408 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ma00014a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |