Open Access Article
Open Access ArticleUnderstanding the relationships between solubility, stability, and activity of silicatein†
Toriana N.
Vigil
 a,
Mary-Jean C.
Rowson
b,
Abigail J.
Frost
a and
Bryan W.
Berger
a,
Mary-Jean C.
Rowson
b,
Abigail J.
Frost
a and
Bryan W.
Berger
 *a
*a
aDepartment of Chemical Engineering, University of Virginia, Charlottesville, Virginia, USA. E-mail: bryan.berger@virginia.edu
bDepartment of Biomedical Engineering, University of Virginia, Charlottesville, Virginia, USA
First published on 15th December 2022
Abstract
Silicatein is an enzyme that mineralizes environmental precursors to patterned nanomaterials and is found naturally orchestrating the complex and beautiful exoskeletons of marine sponges. To harness this activity for nanomaterial biomanufacturing, enzyme solubility and stability have been widely studied. We address the enzyme's solubility challenge via protein fusion tags: enhanced green fluorescent protein (eGFP), monomeric superfolder GFP (msGFP2), and trigger factor (TF). All three silicatein fusion proteins form oligomers to varying degrees, that are partially modulated by disulfide bridges. Biomineralization activity was assessed with silica and nanoceria, showing comparable yields for eGFP-silicatein and TF-silicatein, as well as identical composition of mineralized products regardless of disulfide bridge reduction, shown via XRD characterization of silicatein's nanocrystalline product. This implies that solubility has only minor effects on silicatein activity and that continued improvement in this area is currently inessential. Furthermore, these results suggest that silicatein biomineralization activity is inherent to the enzyme itself. Thus, future studies should be aimed at understanding silicatein's kinetic mechanisms.
Introduction
Nanomaterials are increasingly studied and used in nearly every sector, ranging from medicine to renewable energy and catalysis. Accordingly, the need for a sustainable and inexpensive source of nanomaterials is paramount. Biomineralization is a green, natural process that is becoming a promising strategy for the production of nanoparticles via synthetic biology.1 This biological approach for the synthesis of inorganic nanoparticles leverages biomineralization protein activity at mild temperatures and with green solvents, in contrast to the high temperatures and toxic reagents required to perform traditional chemical synthesis techniques.2,3 Furthermore, the mild reaction conditions required for biomineralization are considerably more cost-effective than current nanomaterial synthesis.4Silicatein is a biomineralization protein originally identified as a key player in the generation of the silica exoskeleton in marine sponges.5 The particular role of silicatein has been uniquely classified as “direct” biomineralization protein, encompassing both the initiation and subsequent patterning of mineralization.6,7 Practically, this concept is manifested as the production of mineralized, crystalline nanoparticles.8 Previous studies have illustrated a purported evolutionary relaxation of substrate specificity for silicatein, which has been shown to mineralize titanium, gallium, barium,9 ceria, zirconia,7 and other metals.8,10 This substrate flexibility effectively broadens the scope of nanomaterials that can be made via silicatein biomineralization, widening the variety of nanoparticles that can be made with this green approach.
Despite the many attractive attributes of silicatein, the potential to harness its biomineralization capabilities remains limited, as a result of conflicting theories and reports surrounding the mechanism of action. Due to sequence homology with cathepsin L,5,11–13 a catalytic triad mechanism involving S26, H165, and N185 is most commonly proposed, despite being recently disputed in the work of Povarova, et al. (2018), who argue in favor of a biotemplating mechanism.13 The variety of conflicting results are likely due to poor enzyme/substrate solubility.12–16 Wild-type silicatein has extremely low solubility, with recent work reporting maximum production of 8 nmol L−1 cell culture in optimal buffer conditions.16
To address this challenge, and facilitate a reactor-based nanoparticle synthesis process, the field has focused on identifying a soluble version of silicatein. Studies by Dakhili, et al. (2017) and Oguri, et al. (2021) explore the addition of several different solubility tags to silicatein, highlighting minor solubility improvements with the addition of Trigger Factor and ProS2, respectively.15,17 These studies note that cleavage of the solubility tag results in massive aggregation and precipitation of wild-type silicatein.17,18 In their recent work, Oguri, et al. suggest enzyme aggregation may not be inherently detrimental to activity, referencing previous work by Murr and Morse that utilizes hydropathy modelling to highlight silicatein's natural propensity for aggregation leading to the complex superstructures found in vivo.19 In fact, some biomineralization processes are mediated entirely by protein aggregation, often called amyloid-like aggregation. Nacre protein PFMG1 is an example, where-in calcium biomineralization is dependent on protein aggregation.17 In these cases, aggregation ultimately results in phase separation, leading to focused crystalline nucleation and growth.18 Although protein aggregation is key for biomineralization in many cases – thus far, silicatein biomineralization has been shown to be catalytic and independent of protein aggregation.7,11–16 Furthermore, for materials synthesis a soluble silicatein would be instrumental for use with a simple reactor system.
To date, TF-silicatein is the most soluble silicatein fusion, and can be produced at approximately 1 μmol L−1 of cell culture, approximately 100× more than the wild-type.16 In this work, we examine the role of protein fusion partners, oligomerization, and disulfide bridges with respect to biomineralization activity by comparison of our own eGFP-silicatein, msGFP2-silicatein, and the widely used TF-silicatein, each based on silicatein from P. ficiformis. Furthermore, we perform biomineralization assays with ceria ammonium nitrate alongside the more commonly used tetraethyl-ortho-silicate and show that use of the water-soluble precursor ceria ammonium nitrate is comparable to the native substrate. With this in mind, ceria biomineralization may be valuable for further studies examining protein structure and function.
Results and discussion
Protein fusion partner
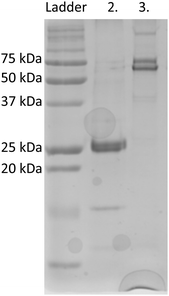
eGFP was chosen as a fusion partner for recombinant silicatein in order to enhance solubility and to provide a visible representation of protein in solution, as shown previously for other difficult-to-solubilize proteins.19,20 eGFP-silicatein expression and purification did not result in anticipated 52 kDa band in purified fraction, but with an intense 26 kDa band, as shown in Fig. 1. This 26 kDa band was sent to University of Virginia's Biomolecular Analysis Facility for analysis via ESI MS/MS. 30 E coli proteins were identified from unique peptide sequences present, six of which were chaperone proteins known to be associated with complex protein folding (Table S1, ESI†). Protein mass, sequence coverage, and unique properties were used to identify the 26 kDa band as E coli chaperone DnaK. Additional induced overexpression of DnaK led to increased band intensity on an SDS PAGE gel, supporting this identification (Fig. S1A, ESI†). DnaK is a “holdase” chaperone that is known to prevent the formation of protein aggregates.21–23 Expression of unstable proteins can lead to overexpression of DnaK due to instability or disruption of folding equilibrium.24 The overexpression of DnaK here suggests that eGFP-silicatein is a very unstable protein with significant, prolonged hydrophobic regions. Additional induced overexpression of DnaK did not result in further improvement of eGFP-silicatein expression (Fig. S1B and C, ESI†). Furthermore, it is likely the high levels of DnaK resulted in quenching of eGFP fluorescence, which is consistent with previous observations by Martinez-Alonso, et al.25An engineered monomeric GFP (msGFP2)26 was also selected as a fusion partner for silicatein, and subsequently accompanied by endogenous overexpression and fluorescence quenching of DnaK. No significant expression or solubility differences were observed between eGFP-silicatein (230 nmol L−1) and msGFP2-silicatein (720 nmol L−1). TF-silicatein expression was consistent with previous reports,16 with yields of approximately 1 μmol protein per liter culture (Fig. 1 and Table S2 ESI†).
Oligomerization
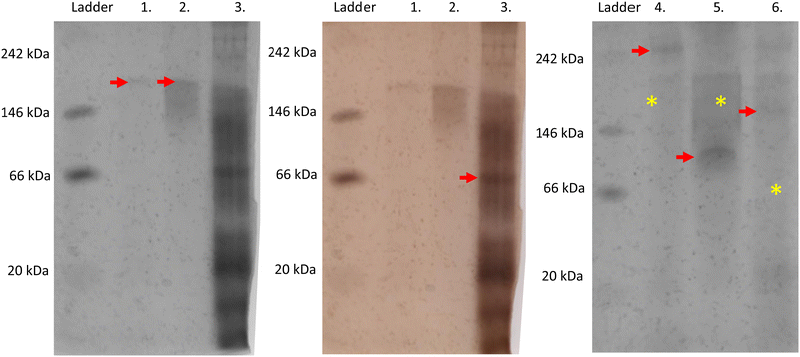
On native PAGE, eGFP-silicatein and msGFP2-silicatein each appear to form a high molecular weight complex greater than 150 kDa, whereas TF-silicatein is at the anticipated MW of 75 kDa (Fig. 2). Although TF-silicatein displays a variety of breakdown products in Fig. 2, the 75 kDa band was identified from MW shown in Fig. 1 (Fig. 1, lane 3). Lane 1 in Fig. 3 shows a typical example of eGFP-silicatein solution on native PAGE, with several bands evident after Coomassie staining (10 kDa, 26 kDa, 70 kDa). However, it should be noted that since these proteins were not denatured prior to gel electrophoresis, they travel according to their folded, compact structure at a different apparent molecular weight rather than denatured where they would migrate according to their true molecular weight. To identify protein biomineralization activity, eGFP-silicatein in native PAGE was incubated in 2 mM ceria ammonium nitrate for 12 hours, then imaged at 460 nm, which is consistent with optical activity of cerium.27,28 Signal under these conditions is indicative of protein interaction with ceria, consistent with biomineralization activity. This illustrates that catalytic activity of eGFP-silicatein is associated with the 150 kDa band (Fig. S3 and S2, ESI†), which interestingly does not appear via Coomassie staining (Lane 1 Fig. 3). This suggests that eGFP-silicatein forms a 150 kDa catalytically active oligomer that is present in small amounts, but still contributes significant catalytic activity. Notably, TF-silicatein did not show the same activity on native PAGE (Fig. S3, ESI†). The occurrence of the >150 kDa bands suggests eGFP-silicatein and msGFP2-silicatein are forming oligomers in solution. eGFP is known to form multimers, however, oligomerization with msGFP2-silicatein suggests that this behavior is silicatein-driven, rather than GFP-driven. TF-silicatein appears at the expected molecular weight of 75 kDa, thus not oligomerizing. In the case of TF-silicatein, the tendency for aggregation is diminished by other forces, with one possibility being the notable steric hindrance of TF as described by Ullers, et al. and Hoffmann, et al.21,22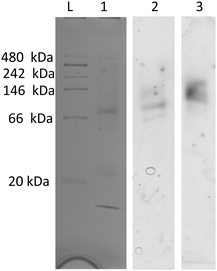 | ||
| Fig. 3 Native PAGE for eGFP-silicatein, (1) stained with Coomassie blue, (2) incubated in water bath, (3) incubated in CAN bath. Imaged at 460 nm for ceria detection. Additional information regarding the processing of this gel can be found in ESI,†Fig. 2. | ||
Disulfide bridges
Silicatein aggregation is credited to the large, exposed hydrophobic patches of the protein, with intermolecular disulfide bridges providing a stabilizing effect.29,30 Recombinant silicatein has four cysteine residues, two of which form a bridge with each other (C34, C76), and an additional two that are available to interact with other molecules (C43, C145), according to previous work by Gorlich, et al. 202031 (Fig. 4). Free cysteines C43 and C145 may form intermolecular disulfide bridges with other eGFP-silicatein molecules, or other nearby molecules such as chaperone DnaK. Native PAGE shows that for eGFP-silicatein and TF-silicatein, thiol reduction with β-mercaptoethanol and blocking with acrylamide resulted in larger MW bands at approximately 250 and 150 kDa respectively (Fig. 2). This suggests that without disulfide stabilization, additional aggregation occurs, forming larger high molecular weight complexes. For TF-silicatein, which did not display oligomerization or aggregation behavior prior to reduction, this suggests the intramolecular disulfide bridge between C34 and C76 plays a key role in stabilizing the monomer.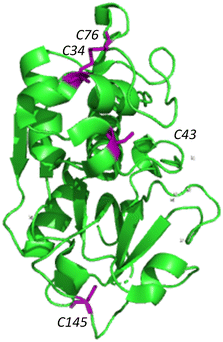 | ||
| Fig. 4 Ribbon structure of silicatein (6ZQ3)30 with cysteine residues highlighted in purple. Figure made with Pymol. | ||
In contrast, msGFP2-silicatein appears at approximately 100 kDa when reduced (Fig. 2), approximately 50 kDa less than previously shown in native conditions. This suggests that in a native state, free cysteines C43 and C145 likely facilitate intermolecular disulfide bridges with multiple msGFP2-silicatein molecules. Nonetheless, reduced msGFP2-silicatein still appears to be dimerizing (monomer MW 48 kDa), implying that other intermolecular forces contribute to silicatein aggregation behavior. This result also indicates that fusion partner may have a limited impact on oligomerization and in some instances is able to reduce silicatein aggregation.
Biomineralization activity
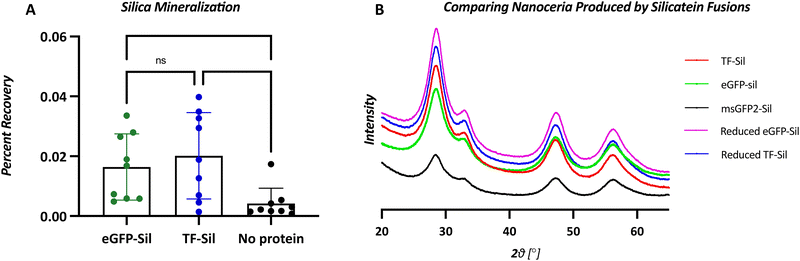
Although biomineralization was assessed with the commonly used precursor TEOS, assays were also performed with water-soluble precursor ceria ammonium nitrate to avoid any confounding factors due to TEOS insolubility and auto-hydrolysis.32 In a side-by-side comparison, eGFP-silicatein and TF-silicatein display similar biomineralization activity with TEOS (Fig. 5). Notably, this reaction yields approximately 0.02% recovery of silicon from precursor to product over a period of 24 hours, consistent with other works.15,29 As by Curran et al., additional biomineralization assays were conducted with ceria ammonium nitrate (CAN) to yield nanoscopic ceric oxide. These experiments yielded visible precipitates after 24 hours, suggesting greater conversion than with native substrate silica (Table S3 and Fig. 4, ESI†). This result suggests that slow kinetic activity of silicatein may be due to a lack of evolutionary pressure, an abundance of available precursors, and minimal needs for quick mineralization for silicatein within marine sponges.Nanoceria biomineralization products were assessed with x-ray diffraction, and no significant differences were observed between nanoceria made by eGFP-silicatein, msGFP2-silicatein, and TF-silicatein (Fig. 5). Furthermore, no differences were observed in nanoceria made by reduced eGFP-silicatein and TF-silicatein (Fig. 5). Therefore, fusion partner, degree of oligomerization, and disulfide bridges are irrelevant to the quality of biomineralization product. This suggests that silicatein activity is intrinsic to the protein itself, without strong influence from the immediate microenvironment in an oligomeric or fused state. Furthermore, these results show that oligomerization and aggregation of silicatein is not inherently detrimental to enzyme activity, and suggests that current eGFP-silicatein, msGFP2-silicatein, TF-silicatein, and others are sufficiently soluble and stable for further studies.
Conclusions
Increasing the solubility of monomeric silicatein has been a major focus in improving biomineralization in an effort to enable greener, safer nanoparticle synthesis. In this work we show that TF-silicatein can reliably be expressed and purified in higher amounts (1 μmol L−1) than eGFP-silicatien and msGFP2-silicatein. Through investigation of protein structure in native conditions, we show that eGFP-silicatein and msGFP2-silicatein form oligomeric structures, and that silicatein fusion proteins have comparable mineralization activity with commonly used precursor TEOS. Furthermore, we probe the role of oligomerization in relation to enzyme activity, and show that fusion proteins that are thiol reduced and blocked produce nanoceria of the same crystalline quality as native oligomeric proteins. Since biomineralization occurred regardless of oligomerization state, these findings support a catalytic mechanism. Furthermore, these results imply that fusion protein, and thus solubility, has only minimal effects on activity and highlight that silicatein aggregation is not inherently detrimental to biomineralization activity. Said differently, silicatein biomineralization is intrinsic to the enzyme, and thus fusions that seek to influence solubility do not improve biomineralization; rather, strategies focused on improving intrinsic activity of silicatein are likely to improve overall biomineralization activity. Therefore, for greener, safer nanoparticle synthesis with silicatein future research should focus on intrinsic properties of the enzyme or strategic approaches that capitalize on silicatein's propensity for oligomerization.Materials and methods
Recombinant protein expression in Escherichia coli
A codon-optimized DNA sequence for enhanced green fluorescent protein (eGFP) linked to truncated silicatein (from Curran, et al.) was cloned into a pet28a+ plasmid for over-expression in BL21 E. coli.7 BL21 E.coli cells containing the cloned eGFP silicatein pet28a + plasmid were grown in 300 mL Terrific Broth until they reached an optical density of 0.6–0.8. Protein expression was induced with Isopropyl β-D-1-thiogalactopyranoside (IPTG) followed by incubation at 20 °C overnight.Protein purification
Cells were recovered from solution via centrifugation at 3000 × g for 10 minutes and resuspended in lysis buffer (5% v/v 99% glycerol, 36 mM Tris HCl, 20 mM Tris Base, 300 mM sodium chloride, 5 mM imidazole). This suspension was then subject to microtip sonication using a Q500 QSonica sonicator with alternating 20 second on/off cycles for one hour, then transferred to centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 15 minutes. The cell pellet was discarded, and the supernatant containing protein was filtered with a 0.4 then 0.2 μm syringe filter. Immobilized metal affinity chromatography with Chelating Sepharose fast flow resin (ThermoFisher) and nickel chloride was performed with various imidazole elutions. Purified protein was dialyzed with 10
000 × g for 15 minutes. The cell pellet was discarded, and the supernatant containing protein was filtered with a 0.4 then 0.2 μm syringe filter. Immobilized metal affinity chromatography with Chelating Sepharose fast flow resin (ThermoFisher) and nickel chloride was performed with various imidazole elutions. Purified protein was dialyzed with 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 MWCO snakeskin dialysis tubing in deionized water. A280 protein concentrations were measured with a NanoDrop 1000 spectrophotometer.
000 MWCO snakeskin dialysis tubing in deionized water. A280 protein concentrations were measured with a NanoDrop 1000 spectrophotometer.
Native PAGE
An 8% Native PAGE resolving gel was loaded with Native Mark Unstained Protein Standard ladder (ThermoFisher) followed by protein samples (at least 50 μmol L−1). For oligomerization experiments: gel was stained with FASTsilver (G Biosciences). For biomineralization assay: the gel was run at 110 V for approximately 2 hours, then was cut into three separate pieces and transferred into either a Coomassie blue staining solution, 0.1 M Tris buffer, or 2 mM ceria ammonium nitrate bath for 12 hours incubation. The Coomassie blue gel was imaged colorimetrically, while buffer control and 2 mM CAN gels were imaged at 460 nm excitation to detect nanoceria. All gels were imaged using an Amersham Imager 680, exposure times were adjusted as needed. Graphical explanation is shown in ESI.†Thiol reduction and blocking
β-mercaptoethanol and acrylamide were added to 1 μm silicatein fusion protein to final concentrations of 10 mM and 20 mM, respectively. Solution was incubated for 1 hour at room temperature, followed by dialysis (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 MWCO) overnight.
000 MWCO) overnight.
Biomineralization assays
Protein function was evaluated via combination of 2 μm silicatein fusion protein, 2 mM precursor, 100 mM Tris, and DI water. Protein concentrations were adjusted to be consistent across runs using A280 measurements as described above in ‘protein purification’. Two controls of the biomineralization assay were performed to properly evaluate silicatein function - 2 μm silicatein fusion protein without precursor, as well as 2 mM precursor without silicatein fusion protein. Solutions were placed on an orbital mixer and incubated at room temperature for 24 hours. Nanoparticles were recovered via centrifugation at 21000 × g for 30 min in an Eppendorf 5424 centrifuge.Silicomolybdate assay
Silica recovery was measured with the silicomolybdate assay adapted from Povarova, et al. 2018 with extinction coefficient as derived by Coradin, et al. 2004.33,34Nanoceria recovery
Biomineralization samples were centrifuged at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 30 minutes. The supernatant was discarded, and pellets were washed with ethanol, then left to dry overnight. Precipitate mass (mg) was measured with Mettler-Toledo XPR-56 micro-analytical balance with anti-static capability.
000 × g for 30 minutes. The supernatant was discarded, and pellets were washed with ethanol, then left to dry overnight. Precipitate mass (mg) was measured with Mettler-Toledo XPR-56 micro-analytical balance with anti-static capability.
X-ray diffractions
Nanoceria was characterized with a Bruker ApexII Duo diffractometer with a Cu 1.54 angstrom source and at room-temperature. Sample spectra were compared to standards listed in the Inorganic Crystal Structure Database.Author contributions
TNV did conceptualization, data curation, formal analysis, investigation, methodology, validation, visualization, writing – original draft, and writing – review and editing. MCR did investigation, formal analysis, validation, writing – original draft. AJF did investigation and validation. BWB did conceptualization, formal analysis, funding acquisition, investigation, methodology, project administration, resources, supervision, validation, writing – review and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This material is based on work supported by the National Science Foundation under Grant No. 1727-166. This work used ESI MS/MS, from Thermo Electron Orbitrap Exploris 480 in the Biomolecular Analysis Facility which is supported by the University of Virginia School of Medicine. X-ray diffraction was made possible by the NSF-MRI program (CHE-2018870) and Dr Diane Dickie at the UVa XRD Core.References
- T. V. Avramenko, Y. N. Shkryl, G. N. Veremeichik and V. P. Bulgakov, Biomanufacturing of nanocrystals using protein biocatalysts, J. Nanoparticle Res., 2020, 22, 5 CrossRef.
- O. M. Sinatra, L. Pan and J. Bakr, Methods of synthesizing monodisperse colloidal quantum dots, Mater. Matters, 2017, 12, 3–7 Search PubMed.
- R. L. Roberts, E. J. Karadagh, L. R. Wang, L. Malmstadt and N. Brutchey, Continuous flow methods of fabricating catalytically active metal nanoparticles, ACS Appl. Mater. Interfaces, 2019, 11, 27479–27502 CrossRef PubMed.
- P. Pandy and G. Jain, Assessing the nanotechnology on the grounds of costs, benefits, and risks, Beni-Suef Univ. J. Basic Appl. Sci., 2020, 9, 63 CrossRef.
- K. Shimizu, J. Cha, G. D. Stucky and D. E. Morse, Silicatein α: Cathepsin L-like protein in sponge biosilica, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 6234–6238 CrossRef CAS PubMed.
- S. Mann, Biomineralization: Principles and Concepts in Bioinorganic Materials Chemistry, Oxford University Press, 2001 Search PubMed.
- C. D. Curran, L. Lu, Y. Jia, C. J. Kiely, B. W. Berger and S. McIntosh, Direct Single-Enzyme Biomineralization of Catalytically Active Ceria and Ceria-Zirconia Nanocrystals, ACS Nano, 2017, 11, 3337–3346 CrossRef CAS PubMed.
- W. Andre, R. Tahir, M. N. Natalio and F. Tremel, Bioinspired synthesis of multifunctional inorganic and bio-organic hybrid materials, FEBS J., 2012, 279, 1737–1749 CrossRef PubMed.
- R. L. Brutchey and D. E. Morse, Silicatein and the translation of its molecular mechanism of biosilicification into low temperature nanomaterial synthesis, Chem. Rev., 2008, 108, 4915–4934 CrossRef CAS PubMed.
- D. Otzen, The Role of Proteins in Biosilification, Scientifica, 2012, 867562 Search PubMed.
- J. N. Cha, K. Shimizu, Y. Zhou, S. C. Christiansen, B. F. Chmelka, G. D. Stucky and D. E. Morse, Silicatein filaments and subunits from a marine sponge direct the polymerization of silica and silicones in vitro, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 361–365 CrossRef CAS PubMed.
- C. F. Fairhead, M. Johnson, K. A. Kowatz, T. McMahon, S. A. Carter, L. G. Oke, M. Liu, H. Naismith and J. H. van der Walle, Crystal structure and silica condensing activities of silicatein a-cathepsin L chimeras, Chem. Commun., 2008, 1765–1767 RSC.
- N. V. Povarova, N. A. Barinov, M. S. Baranov, N. M. Markina, A. M. Varizhuk, G. E. Pozmogova, D. V. Klinov, V. B. Kozhemyako and K. A. Lukyanov, Efficient silica synthesis from tetra(glycerol)orthosilicate with cathepsin- and silicatein-like proteins, Sci. Rep., 2018, 8, 1–9 CAS.
- Y. Zhou, K. Shimizu, J. N. Cha, G. D. Stucky and D. E. Morse, Efficient Catalysis of Polysiloxane Synthesis by Silicateinα Requires Specific Hydroxy and Imidazole Functionalities, Angew. Chem., Int. Ed., 1999, 15, 779–782 CrossRef.
- L. S. Dakhili, S. Y. T. Caslin, S. A. Faponle, A. S. Quayle, P. de Visser and S. P. Wong, Recombinant silicateins as model biocatalysts in organosiloxane chemistry, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E5285–E5291 Search PubMed.
- E. I. Sparkes, R. A. Kettles, C. S. Egedeuzu, N. L. Stephenson, S. A. Caslin, S. Y. T. Dakhili and L. S. Wong, Improved production and biophysical analysis of recombinant silicatein-α, Biomolecules, 2020, 10, 1–17 CrossRef PubMed.
- J. S. Perovic, I. Mandal and T. Evans, A pearl protein self-assembles to form protein complexes that amplify mineralization, Biochemistry, 2013, 52, 5696–5702 CrossRef PubMed.
- M. Tarczewska, A. Bielak, K. Zoglowek, A. Soltys, K. Dobryszycki, P. Ozyhar and A. Rozycka, The role of intrinsically disordered proteins in liquid-liquid phase separation during calcium carbonate biomineralization, Biomolecules, 2022, 12, 1266 CrossRef PubMed.
- T. C. Waldo, G. S. Standish, B. M. Berendzen and J. Terwilliger, Rapid protein-folding assay using green fluorescent protein, Nat. Biotechnol., 1999, 17, 891–895 CrossRef PubMed.
- I. M. Weber, E. Guth and C. Weiss, GFP Facilitates Native Purification of Recombinant Perlucin Derivatives and Delays the Precipitation of Calcium Carbonate, PLoS ONE2, 2012, 7, 46653 CrossRef PubMed.
- P. Ullers, R. S. Ang, D. Schwager, F. Georgopoulos and C. Genevaux, Trigger Factor can antagonize both SecB and DnaK/DnaJ chaperone functions in Escherichia coli, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 3101–3106 CrossRef PubMed.
- G. Hoffmann, A. Bukau and B. Kramer, Structure and function of the molecular chaperone Trigger Factor, Biochim. Biophys. Act., 2010, 1803, 650–661 CrossRef PubMed.
- F. U. Teter, S. A. Houry, W. A. Ang, D. Tradler, T. Rockabrand, D. Fischer, G. Blum, P. Georgopoulos and C. Hartl, Polypeptide flux through bacterial Hsp70, Cell, 1999, 97, 755–765 CrossRef PubMed.
- I. Christensen, S. Ramisch and S. Andre, DnaK response to expression of protein mutants is dependent on translation rate and stability, Commun. Biol., 2022, 5, 597 CrossRef PubMed.
- A. Martinez-Alonso, M. Vera and A. Villaverde, Role of the chaperone DnaK in protein solubility and conformational quality in inclusion body-forming Escherichia coli cells, FEMS Microbiol. Lett., 2007, 273, 187–195 CrossRef PubMed.
- B. S. Valbuena, F. M. Fitzgerald, I. Strack, R. L. Andruska, N. Smith and L. Glick, A photostable monomeric superfolder green fluorescent protein, Traffic, 2020, 8, 534–544 CrossRef PubMed.
- S. Inerbaev, T. M. Karakoti, A. S. Kuchibhatla, S. V. N. T. Kumar, A. Masunov and A. E. Seal, Aqueous medium induced optical transitions in cerium oxide nanoparticles, Phys. Chem. Chem. Phys., 2015, 17, 6217–6221 RSC.
- H. Masui, T. Fujiwara, K. Machida, K. Adachi, G. Sakata and T. Mori, Characterization of Cerium(IV) Oxide Ultrafine Particles Prepared Using Reversed Micelles, Chem. Mater., 1997, 9, 2197–2204 CrossRef.
- S. Oguri, H. Nakashima, K. Godigamuwa, K. Okamoto, J. Takeda, Y. Okazaki, F. Sakono and M. Kawasaki, Solubilization and aggregation control of silica-polymerizing enzyme fused with a removable soluble protein, J. Biosci. Bioeng., 2021, 133, 222–228 CrossRef PubMed.
- D. E. M. Meredith and M. Murr, Fractal intermediates in the self-assembly of silicatein filaments, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 11657–11662 CrossRef PubMed.
- I. Gorlich, S. Samuel, A. J. Best, R. J. Seidel, R. Vacelet, J. Leonarski, F. K. Tomizaki, T. Rellinghaus, B. Pohl and D. Zlotnikov, Natural hybrid silica/protein superstructure at atomic resolution, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 31088–31093 CrossRef PubMed.
- C. J. Brinker, Hydrolysis and condensation of silicates: effects on structure, J. Non. Cryst. Solids, 1988, 100, 31–50 CrossRef CAS.
- K. A. Povarova, N. V. Markina, N. M. Baranov, M. S. Barinov, N. A. Klinov, D. V. Kozhemyako and V. B. Lukyanov, A water-soluble precursor for efficient silica polymerization by silicateins, Biochem. Biophys. Res. Commun., 2018, 495, 2066–2070 CrossRef PubMed.
- J. Coradin, T. Eglin and D. Livage, The silicomolybdic acid spectrophotometric method and its application to silicate/biopolymer interaction studies, Spectroscopy, 2004, 18, 567–576 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ma00938b |
| This journal is © The Royal Society of Chemistry 2023 |