Open Access Article
Open Access ArticleUltrasensitive low-probe-concentration PANC-1 and MCF-7 cancer cell sensors enabled by combined 2D-material-polymer-phage frameworks†
Denise
Lee
 a,
Sophia Shuwn-Yi
Chan
a,
Sophia Shuwn-Yi
Chan
 a,
J Shamita
Naikar
a,
J Shamita
Naikar
 b,
Maria Prisca
Meivita
b,
Maria Prisca
Meivita
 a,
Wey-Chyi
Teoh
b,
Natasa
Bajalovic
*a and
Desmond K.
Loke
a,
Wey-Chyi
Teoh
b,
Natasa
Bajalovic
*a and
Desmond K.
Loke
 *ab
*ab
aDepartment of Science, Mathematics and Technology, Singapore University of Technology and Design, Singapore 487372, Singapore. E-mail: natasa_bajalovic@sutd.edu.sg; desmond_loke@sutd.edu.sg
bOffice of Innovation, Changi General Hospital, Singapore, 529889, Singapore
First published on 5th December 2022
Abstract
Unique biophysical properties of cancer cells can provide essential knowledge that can aid in the diagnosis of disease, clinical trial designs and drug developments. The collective response of cancer cells is of significant interest to the research community. However, sensing cells in a medium population is extremely challenging. Here, we controlled the electrical conduction of cancer cells by using combined 2D-material-polymer-phage frameworks via current flow effects. We then developed an AC-pulse sensor system based on MoS2/PEG/M13 nanoprobes for accurate detection of clinically relevant cancer cells such as PANC-1 cells and MCF-7 cells. The detection limit can reach ∼15 cells per µL, which is below an average of ∼45 cells per µL for existing sensing methods with medium cell populations. Moreover, excellent cell viability was achieved in PANC-1 cells for nanoprobe concentrations of up to 50 vol%, which surpassed those used in detection experiments. Furthermore, a nanoprobe concentration of ∼10 vol%/7.1 nM was achieved, below an average of ∼195 nM used for state-of-the-art sensing methods with medium cell populations. The combination of a previously unreported exotic sensing material and an AC-pulse strategy represents an approach for unlocking the ultrasensitive detection of cancer cells and provides a promising avenue for early cancer diagnosis, staging and monitoring.
Introduction
Nanotechnology is an exciting area of research involving nanomaterials with sizes in the 1–900 nm range. It has an ever-increasing number of applications that cover a wide range of industries, such as medicine and life sciences. A promising application is its utility in biosensors. For instance, medical researchers have leveraged the benefits of nanotechnology in areas ranging from early detection of Alzheimer's disease to diabetes management to early cancer screening, including Covid-19 drug discovery.1–3 Nano-scale sensors are utilized in cases requiring greater precision and continuity.4–7 Moreover, they play an important role in the development of the emerging initiative bio-digital twins.8,9 Cancer is the second leading cause of death worldwide, accounting for 10 million deaths in 2020.10 The cancer burden continues to grow globally. Nevertheless, survival rates for many types of cancers are improving, thanks to accessible early detection.Recently, characterizing electrical signatures of cancer cell populations has generated substantial interest in the scientific community.11–16 The unique biophysical properties of each cancer cell type provide essential knowledge that could aid in the diagnosis of diseases, clinical trial designs and drug development.14 The electrical responses of cancer cell populations are of particular interest, because cancer cells exhibit a collective behavior in terms of cell migration and invasion.17–19 However, sensing medium cell populations is extremely challenging because traditional analytical methodologies can be labor-intensive and the compositions of cell monolayers tend to be different. Furthermore, the impedance of cell population is highly sensitive to minute changes in the microenvironment, such as the cell media, pH, temperature and air composition.20–23
Using alternating current (AC) pulses can reduce the noise generated from minute changes in microenvironments of the cell layers, since the impedimetric measurements resulting from an AC pulse from 10 to 100 kHz are largely dominated by the biological cells and the changes within them.16,24 This is based on the β-dispersion phenomenon, a theory originally hypothesized by Schwan et al. The β-dispersion theory describes how biological cell membranes at low frequencies have a significantly larger impedance compared to cell membranes at high frequencies.25,26
Nanomaterials are promising candidates for detecting cancer cells. The utilization of nanomaterials such as nanosheets, nanotubes and nanodots in cytosensors is becoming increasingly popular, since the quantitative or qualitative detection of cells has wide-ranging applications, such as cancer diagnostics,27–30 cell monitoring,31 drug development, and other applications.32–34 With properties that are significantly different from those of the bulk, the addition of nanomaterials enhances the detection sensitivity of these sensors. For example, leukemia cytosensors using gold nanoparticles have been developed by Zheng et al., which utilize the difference in the expression levels of death receptors on leukemia cells.35 Moreover, Tang et al. were able to harness the unique properties of Pt@AgNFs, which were immobilized on a substrate to create circulating-tumor-cell cytosensors.36
Molybdenum disulfide (MoS2) has emerged as a leading contender for enhancing cancer cell detection due to its unique electrical properties.37–39 A two-dimensional (2D) MoS2 nanosheet comprises a few S–Mo–S monolayers assembled via van der Waals (vdW) forces between the layers.37 However, MoS2 can be utilized in the biomedical field in its early stages due to the limitations imposed by the cytotoxicity of the material and the decreased stability of MoS2 in aqueous solutions for some applications.40 For instance, MoS2 nanosheets may aggregate in water due to its strong hydrophobic nature and increased vdW attractions between the layers.28,41,42
Herein, we show that by stimulating and altering current flow effects, we can control the electrical conduction of cancer cells via combined 2D-material-polymer-phage frameworks. We then developed an AC-pulse sensor system by using MoS2/polyethylene glycol (PEG)/M13 nanoprobes for targeted pancreatic cancer cell (PANC-1) and breast cancer cell (MCF-7) detection. This method achieved a low limit of detection (LOD) of ∼15 cells per µL, which is below an average of ∼45 cells per µL for existing sensing methods using medium cell population. This allows cancer detection to be performed with a better accuracy for enhancing diagnosis reliability. Additionally, excellent cell viability was achieved in PANC-1 cells at nanoprobe concentrations of up to 50 vol%, which exceeds those utilized in sensing experiments. Furthermore, a nanoprobe concentration of ∼10 vol%/7.1 nM was attained, below an average probe concentration of ∼195 nM for current sensing methods with medium cell populations. And, this enables the use of a smaller amount of material for saving diagnosis costs. This proposed methodology for cancer cell detection holds intriguing potential for developing multiplexed lab-on-chip platforms, which can be further applied for clinical purposes such as cancer diagnosis, as well as broad applications in analyte-based sensing.
Methods
Cell lines and cell culture
The PANC-1 cell line was purchased from American Type Culture Collection (ATCC). The PANC-1 cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Nacalai Tasque) supplemented with 10% fetal bovine serum (FBS, Thermo Fisher), 1% L-glutamine (Sigma Aldrich) and 1% penicillin–streptomycin (Fisher Scientific). To elucidate the selectivity of the nanoprobe-type cancer cell system, the MCF-7 and MCF-10A cells were also utilized. The MCF-7 cells were cultured in phenol-red-containing DMEM (Nacalai Tesque), supplemented with 7.5% FBS (Thermo Fisher) and 2 mM of L-glutamine (Sigma Aldrich). The healthy breast epithelial cell line MCF-10A was cultured in DMEM supplemented with Ham's F12 (DMEM/F12, Gibco) containing 10% FBS, 20 ng mL−1 epidermal growth factor (EGF, Gibco), 0.5 µg mL−1 hydrocortisone (Sigma Aldrich) and 10 µg mL−1 insulin (Sigma-Aldrich). The cells were incubated at 37 °C in a humidified incubator in an atmosphere of 5% CO2.M13 phage propagation
Escherichia coli (E. coli) bacteriophage M13 15669-B1 (M13 phage) was purchased from ATCC. M13 phage was revived according to the phage recovery and propagation protocol (ATCC). 5-Alpha F’Iq competent E. coli (high efficiency E. coli) purchased from New England Biolabs (NEB) was utilized as the host cell for M13 phage propagation. An overnight culture (O.C.) of E. coli was prepared with tetracycline (TET, Sigma Aldrich) and left to incubate on a shaker at 37 °C for 4–6 h at 90 rpm until the mixture showed a cloudy color with an optical absorbance of 0.4 (OD600). A 48 h incubation was then performed with the O.C. and Lennox L broth base (LB Broth, Sigma Aldrich). Similarly, a new culture (N.C.) was prepared and left to incubate at 37 °C for 48 h on a shaker at 90 rpm. First and second precipitations were achieved according to the M13-amplification-protocol (NEB). Concentration of the M13 phage was measured using a µDrop plate (Thermo Fisher Scientific Inc.).Molecular probe conjugation
Molecular probe conjugation was performed under sterile conditions. MoS2 (92 mg L−1) in DI water was purchased from 2D Semiconductors Inc. The MoS2 solution was sonicated prior to molecular probe conjugation. LA-PEG-NHS was purchased from Nanocs Inc. and reconstituted with DI water. The LA-PEG-NHS solution was added to a sterile glass bottle containing MoS2 in DI water. The mixture was left to incubate on a shaker at 25 °C for 48 h at 120 rpm. The filtered M13 phage was added to the mixture and was then left to incubate on a shaker at 25 °C for 48 h at 120 rpm.Material characterization
Atomic force microscopy (AFM) was performed using a Bruker Dimension Icon (Bruker Cooperation) system with a scanning area of 1.5 µm × 1.5 µm. Probe samples were drop casted on Si substrates prior to imaging. Raw AFM data were analyzed with NanoScope analysis software and ImageJ software. Fourier-transform infrared (FTIR) analysis of the probe was performed using Nicolet IS10, US (Thermo Fisher Scientific Inc.) with the wavenumber in the 400–4000 cm−1 range. Transmission electron microscopy (TEM) was carried out using FEI Talos F200 (Thermo Fisher Scientific Inc.). Probe samples were negatively dyed prior to TEM imaging. UV-vis absorbance spectroscopy was performed with a Variskan Lux multimode microplate reader (Thermo Fisher Scientific Inc.).Cell viability studies
The indium tin oxide (ITO) subsystems were sterilized with 100% ethanol for 20 min prior to exposure to ultra-violet (UV) irradiation for 20 min. The subsystem comprised two left and right ITO electrodes on a glass substrate (Latech) with a cloning cylinder (C3983-50EA, Sigma Aldrich) secured using a silicone adhesive. The size of the gap between the electrodes was chosen to be ∼0.1 mm, and the thickness of the electrode was fixed at 650 nm. PANC-1 cells were plated in each ITO subsystem at a seeding density of 5 × 103 cells per well and the cells were left to incubate for 24 h. Different concentrations of MoS2 or MoS2/PEG/M13 nanoprobes were added to each subsystem and the cells with the MoS2 or MoS2/PEG/M13 nanoprobes were incubated for 24–48 h at 37 °C in a humidified incubator in an atmosphere of 5% CO2. The cytotoxicity of MoS2 was examined using WST-1 assays at 24 h and 48 h after adding MoS2. Similarly, by utilizing WST-1 assays at 24 h after inserting the nanoprobes, the cytotoxicity of MoS2/PEG/M13 nanoprobes was evaluated. The absorbance was measured at a wavelength of 450 nm.Electrical characterization
The ITO subsystems were sterilized with 100% ethanol for 20 min prior to exposure to UV irradiation for 20 min. The PANC-1/MCF-7/MCF-10A cells were plated in each subsystem at different seeding densities (1 × 103–7 × 103 cells per well) and the cells were left to incubate for 24 h. 10 vol% MoS2/PEG/M13 nanoprobes was added to each subsystem. The cells with MoS2/PEG/M13 nanoprobes were then incubated for 24 h at 37 °C in a humidified incubator in an atmosphere of 5% CO2. Cell impedance was measured using an arbitrary waveform generator (Tektronix Inc.) and a digital oscilloscope (Tektronix Inc.). Different sensing voltages in the 0.2–0.7 V range were utilized, and the pulse length was fixed at 1 ms.Results
Methodology of the MoS2/PEG/M13 sensor framework
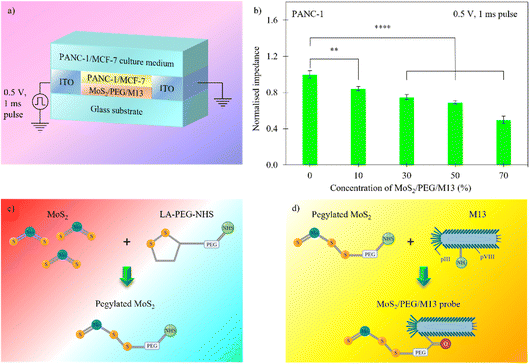
The strategy adopted for the highly sensitive detection of cells on the MoS2/PEG/M13 sensor system is illustrated in Fig. 1. A glass substrate was utilized as the starting material, on which two 650 nm-thick left and right ITO electrodes were deposited for external circuitry connection (Fig. 1(a)). The distance between the electrodes was chosen to be ∼100 µm. The PANC-1/MCF-7 cell layer was then plated. Finally, the MoS2/PEG/M13 nanoprobes were added to complete the system structure. Fig. 1(b) shows the variation of normalized system impedance for different probe concentrations. The systems exhibit a decreasing normalized impedance (from 1 to 0.5) with an increase in nanoprobe concentrations (between 0 and 70%). Thus, the electrical conductivity of the sensor system can be controlled by varying nanoprobe concentrations. The M13 phage acts as the cell targeting agent, while MoS2 nanosheets play the role of an impedance contrast agent. PEG comprises lipoic acid (LA) and N-hydroxysuccinimide (NHS) ester branches, which can be harnessed to create a conjugated MoS2/M13 system to link MoS2 nanosheets to the M13 phage.43 The LA-terminal of the PEG linker chemically binds to the MoS2 nanosheet via disulfide binding (Fig. 1(c)).44–46 As shown in Fig. 1(d), through amine reactions, the pegylated MoS2 nanosheets then attach to the M13 phage.47 Furthermore, experiments have demonstrated that the pegylation process enhances the stability of MoS2 nanosheets dispersed in water.40,48M13 is a cylindrical, ∼880 nm long and ∼6.6 nm wide in diameter, nonlytic bacteriophage composed of proteins.47 The uptake of M13 phage by cells has been attributed to different pathways of endocytosis.49 Moreover, experiments have revealed that the M13 phage shows preferential binding to the membranes of epithelial cells, and is internalized mainly through clathrin-mediated-endocytosis.50 When left at room temperature, MoS2 nanosheets undergo spontaneous desulfurization, creating defect sites at the edges of nanosheets.51 This results in surface electron accumulation, which is substantially larger compared to that in the bulk.51 The electrical conduction of MoS2 nanosheets is also stronger than that of bulk materials. Moreover, experiments have demonstrated that surface functionalization of MoS2 with LA enhances the conduction of MoS2 nanosheets.52 When LA conjugation is utilized, the current flow in MoS2 nanosheets increases significantly due to enhanced electron mobility. This can facilitate the electrical conductivity of MoS2/PEG/M13 and improve the sensitivity of nanoprobe sensor systems. Experiments have also revealed that the M13 phage assembles well on cell surfaces.49,53 Moreover, high electrical conductance in cancer cells incubated with MoS2 is disclosed in recent studies.54,55 Based on these findings and to achieve strong assembly interactions and high electrical conductivity, we chose to utilize the M13 phage and MoS2 in this work.
Preparation of the MoS2/PEG/M13 nanoprobe
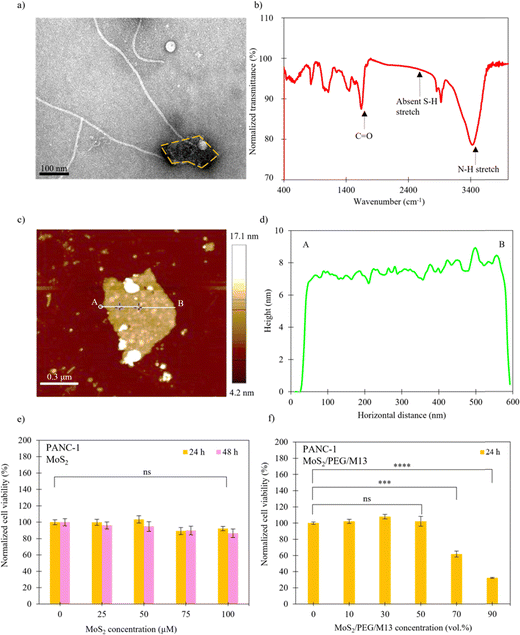
Liquid-phase exfoliation is an effective method for preparing few-layer MoS2 nanosheets by breaking weak vdW forces between the layers.56,57 We obtained few-layer MoS2 nanosheets using sonication liquid exfoliation. PEG was then added. The PEG links MoS2 nanosheets through covalent disulfide bonds between sulphur atoms on LA terminals and exposed sulphur atoms on MoS2 nanosheets.58 Finally, the M13 phage was added to complete the MoS2/PEG/M13-nanoprobe-structure. The PEGylated MoS2 binds to the M13 phage in an NHS ester-amine reaction to form an amide bond between PEG and M13 phage.43The chemical composition and morphology of the prepared samples were systematically investigated. Fig. 2(a) shows the transmission electron microscopy (TEM) image of the MoS2/PEG/M13 nanoprobe, in which thin 2D nanosheets and 1D wire-like structures were observed. To investigate the crystal structure and quality of the nanoprobe, the Fourier-transform infrared (FTIR) spectra of the MoS2/PEG/M13 nanoprobe sample were obtained (Fig. 2(b)). A broad peak at ∼3400 cm−1 was observed in the FTIR spectra of the MoS2/PEG/M13 nanoprobe sample. This corresponds to an amide bond stretching (N–H) between PEG and M13 phage. Another peak at ∼1640 cm−1 corresponds to a carbonyl stretch (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) associated with amide bonds. Although amide bonds are present in all biological molecules, including the M13 phage, these bonds are vital in determining the success of probe conjugation. Additionally, the lack of a peak in the 2500–2600 cm−1 range, corresponding to a thiol stretch (S–H), indicates that all LA terminals on the PEG linker are covalently bound to the available MoS2 nanosheets. Additional peaks observed at 607, 875, 1112, 1427 and 1627 cm−1 are characteristic of MoS2,45,59,60 while the peaks at 2800–3000 cm−1 correspond to the presence of PEG.61,62 These results indicate that both MoS2 and PEG are present in the nanoprobes. The atomic force microscopy (AFM) topology of the nanosheet of the MoS2/PEG/M13 nanoprobe is shown in Fig. 2(c). The thickness of the nanosheet was determined by cross-sectional analysis (Fig. 2(c), line A–B). The measured height of ∼7 nm shown in Fig. 2(d) corresponds to MoS2 nanosheets with ∼10 triple layers.63
O) associated with amide bonds. Although amide bonds are present in all biological molecules, including the M13 phage, these bonds are vital in determining the success of probe conjugation. Additionally, the lack of a peak in the 2500–2600 cm−1 range, corresponding to a thiol stretch (S–H), indicates that all LA terminals on the PEG linker are covalently bound to the available MoS2 nanosheets. Additional peaks observed at 607, 875, 1112, 1427 and 1627 cm−1 are characteristic of MoS2,45,59,60 while the peaks at 2800–3000 cm−1 correspond to the presence of PEG.61,62 These results indicate that both MoS2 and PEG are present in the nanoprobes. The atomic force microscopy (AFM) topology of the nanosheet of the MoS2/PEG/M13 nanoprobe is shown in Fig. 2(c). The thickness of the nanosheet was determined by cross-sectional analysis (Fig. 2(c), line A–B). The measured height of ∼7 nm shown in Fig. 2(d) corresponds to MoS2 nanosheets with ∼10 triple layers.63
Ultraviolet-visible (UV-vis) absorbance spectroscopy was further performed to investigate the presence of the M13 phage and MoS2 in the conjugated MoS2/PEG/M13 nanoprobe system (Fig. S1, ESI†). Fig. S1a (ESI†) shows that both MoS2 and MoS2/PEG/M13 samples exhibit similar spectral features. However, the absorbance intensity of the MoS2/PEG/M13 sample is smaller compared to that of the MoS2 sample. The absorbance intensity may correlate to the concentration of MoS2. As MoS2 is hydrophobic, the conjugation process can result in an increased dispersion of MoS2, resulting in a decrease in the absorbance intensity of the MoS2/PEG/M13 sample. A signature peak at ∼250 nm was observed for both M13 and MoS2/PEG/M13 samples, similar to those reported in the literature.44,64,65 This indicates that the conjugation process does not alter spectral characteristics of the M13 phage.
The viability of PANC-1 cells with pure MoS2 was investigated at different material concentrations using WST-1 cell viability assay after 24 h and 48 h incubation periods (Fig. 2(e)). The cells with 25–100 µM MoS2 disclose similar cell viability to that of the control (cells only/0 µM MoS2), indicating that MoS2 has negligible cytotoxic effects. Additionally, we investigated the viability of PANC-1 cells with MoS2/PEG/M13 nanoprobes 24 h after incubation (Fig. 2(f)). The cells with 10–50 vol% MoS2/PEG/M13 nanoprobes exhibit a similar cell viability of ∼100% compared to that of the control (0 vol% MoS2/PEG/M13 nanoprobes), whereas a decreased cell viability of ∼60% compared to that of the control was shown by cells with ∼70 vol% MoS2/PEG/M13 nanoprobes. For bare MoS2, small/large material concentrations can maintain high cell viability, as shown in experiments. However, for the MoS2/PEG/M13 nanoprobe, the charge level of the nanoprobe can reach a value that induces cell death when nanoprobes are incubated with cells.32 However, the concentration of the nanoprobe is not small enough to prevent cell death at high nanoprobe concentrations, leading to low cell viability.
Cell sensing performance of MoS2/PEG/M13 nanoprobes
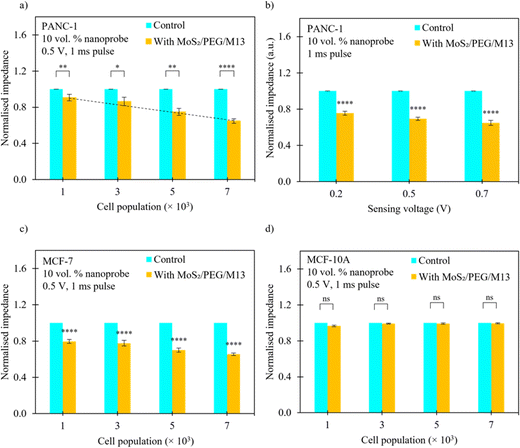
We measured and calculated the normalized impedance of the PANC-1/MCF-7 cell systems with MoS2/PEG/M13 nanoprobes for different cell populations/sensing voltages. The impedance of the system was normalized to that of the control (cells only). The PANC-1 cell system with 10 vol% MoS2/PEG/M13 nanoprobes showed ∼16% decrease in impedance compared to that of the control, whereas ∼50% smaller impedance than that of the control was observed for the PANC-1 system with 70 vol% MoS2/PEG/M13 nanoprobes (Fig. 1(b)). To increase the conductance, and at the same time, maintain excellent cell viability, and also minimal nanoprobe concentrations, we chose to utilize the PANC-1 cell system with 10 vol% MoS2/PEG/M13 nanoprobes. Utilizing the PANC-1 cell system with 10 vol% MoS2/PEG/M13 nanoprobes, we observed a decrease in the normalized impedance (the normalized impedance is defined as the ratio of the impedance of the cell system with MoS2/PEG/M13 nanoprobes to the impedance of a pure cell system) for increasing cell populations (Fig. 3(a)). When 3 × 103 and 5 × 103 cells were utilized, we observed a corresponding ∼14% and ∼25% decrease in the normalized impedance. The largest percentage decrease in normalized impedance was observed for 7 × 103 cells (above 30%).To investigate the LOD of the PANC-1 cell system with 10 vol% MoS2/PEG/M13 nanoprobes, we performed a linear regression analysis based on the slope of the linear plot of the normalized impedance as a function of the cell population (gradient of the dotted line in Fig. 3a) (s), and the standard deviation (σ) of the normalized impedance:
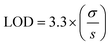 | (1) |
The calculated LOD was found to be ∼15 cells per µL 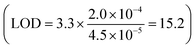 , below the baseline of ∼50 cells per µL for state-of-the-art sensing methods66–70 with medium cell populations (Fig. S2, ESI†). Moreover, the concentration of the MoS2/PEG/M13 nanoprobe utilized for the LOD analysis was ∼10 vol%/7.1 nM, which is lower than the average probe concentration of ∼195 nM for existing sensing methods71–73 with medium cell populations (Fig. S3, ESI†). Additionally, the normalized impedance of the PANC-1 cell system with 10 vol% MoS2/PEG/M13 nanoprobes decreases (from 0.7 to 0.6) with an increase in sensing voltage (0.2–0.7 V) (Fig. 3(b)). These results indicate that the electrical conductance of nanoprobe-based cell systems can be tuned for sensitive cancer cell detection.
, below the baseline of ∼50 cells per µL for state-of-the-art sensing methods66–70 with medium cell populations (Fig. S2, ESI†). Moreover, the concentration of the MoS2/PEG/M13 nanoprobe utilized for the LOD analysis was ∼10 vol%/7.1 nM, which is lower than the average probe concentration of ∼195 nM for existing sensing methods71–73 with medium cell populations (Fig. S3, ESI†). Additionally, the normalized impedance of the PANC-1 cell system with 10 vol% MoS2/PEG/M13 nanoprobes decreases (from 0.7 to 0.6) with an increase in sensing voltage (0.2–0.7 V) (Fig. 3(b)). These results indicate that the electrical conductance of nanoprobe-based cell systems can be tuned for sensitive cancer cell detection.
In cancer cell sensing studies, the ability to distinguish cancer cells from normal cells is an important aspect. Experiments have demonstrated that nanomaterial-based platforms disclose the ability to distinguish cancer cells from normal cells for different types of organs or between different cell types. The MoS2-based framework with cervical and lung cancer blood samples reveals a different relative impedance as compared with that of the framework with healthy cervical and lung blood samples.74 A lower impedance is exhibited in a gold nanoparticle-based system with HeLa cells compared to the system with human embryo skin fibroblast cells (HESF cells).75 The graphene-based platform with HeLa cells also shows a smaller impedance than that of the platform with NIH3T3 cells.76 Furthermore, we compared the normalized impedance of the MCF-7 cell system with 10 vol% MoS2/PEG/M13 nanoprobes with that of the MCF-10A cell system with 10 vol% MoS2/PEG/M13 nanoprobes, albeit utilizing different cell lines (Fig. 3(c) and (d)). When the MCF-7 cell system with MoS2/PEG/M13 nanoprobes for moderate cell populations (i.e., 3 × 103 and 5 × 103 cells) was examined, the normalized impedance (0.7–0.8) was found to be smaller relative to that of the MCF-10A cell system with MoS2/PEG/M13 nanoprobes (∼1.0), indicating that MCF-7 cells are more sensitive/selective to the MoS2/PEG/M13 nanoprobe than MCF-10A cells in terms of sensing ability.
Discussion
Electrical-based sensing is a popular approach for detecting cancer cells, since cell types have unique electrical signatures that can be harnessed for disease diagnosis.16,77–80 In disease diagnosis, cell counts are clinically relevant, where their utilization has been adopted as a predictor of patient prognosis, such as lymphocyte counts in patients with lung cancer,81 monocyte counts in malignant-pleural-mesothelioma,82 and white-blood-cell counts for other types of cancers.83–85 In this work, the systems with MoS2/PEG/M13 nanoprobes exhibit a low LOD of ∼15 cells per µL, below the reference of ∼50 cells per µL for current sensing methods with medium cell populations. This was achieved by enhancing current flow through the cells, and at the same time, maintaining excellent cell viability.Moreover, achieving rapid responses in sensor systems is critical for providing key information that could aid in patient management. Traditional sensors may exhibit a detection time in the 1–24 h range.68,86 In this work, the impedance could be obtained using millisecond pulses, enabling real-time monitoring of cancer cells. Additionally, the setup is facile and most of the components in the system are reusable, which can save diagnosis-related costs.
In the future, work can be performed to utilize the multiplexed potential of the M13 phage to match the complexity of cancer as a disease. The high density of available functionalization sites at the pIII and pVIII protein sites has not been fully utilized to broaden the capabilities of MoS2/PEG/M13 nanoprobes for improved targeting and detection.47 Other nanomaterials/phages could also be utilized to optimize cancer cell sensor performance. Experiments have shown that biological systems with other nanomaterials, viz., metal nanoparticles and nanowires, reveal a low LOD.87,88 Additionally, biological platforms with other phages, e.g., the Listeria phage and T7 bacteriophage, which disclose a low LOD, have been demonstrated in recent studies.89,90 These experiments elucidate that the nanomaterial/phage-based interaction facilitated an increase in sensitivity. Application of the MoS2/PEG/M13 nanoprobes can be further explored in animal studies and clinical trials, with different types of cancers. Oncologists may rely on traditional morphological or histological characteristics of tumor tissues for disease diagnosis, which cannot be determined affirmatively with a single type of analysis. Tumor development is an ongoing biological process,91 due to which the validity of these analyses is short-lived.92 Multi-parametric testing using the MoS2/PEG/M13 nanoprobes open a path to enhance the sensitivity and speed of diagnosis for improving patient well-being.
In recent years, there has been a gradual shift in understanding the developmental behavior of cancer. A growing number of studies have suggested that the predominant mode of cancer invasion is through collective cell migration,17 where cells move as clusters, strands or sheets. Collective cell migration has become a hallmark of cancer metastasis in many types of tumor,19 implying that traditional single-cell analyses may no longer be sufficient for diagnosing and understanding the cancer-cell behavior accurately.93 Furthermore, the bioelectrical properties of cancer cells have been demonstrated to affect not just its own behavior, but also the behavior of neighboring cells.94 With the growing association between bioelectrical properties and collective cell behaviors demonstrated in cancer cells, impedimetric cell-population-based methods of detection can provide valuable information in understanding cancer cell development.
Conclusion
In this work, we demonstrated an ultra-sensitive and low-probe-concentration MoS2/PEG/M13-nanoprobe-based AC-pulse sensor system for detecting PANC-1 and MCF-7 cells. This is achieved through combined 2D-material-polymer-phage frameworks that alter the electrical conduction of cancer cells. The MoS2/PEG/M13-nanoprobe-based PANC-1 cell system has an LOD of ∼15 cells per µL, below the reference value of ∼50 cells per µL for current sensing methods using medium cell population. In addition, excellent cell viability was achieved in PANC-1 cells with a MoS2/PEG/M13 nanoprobe concentration up to 50 vol%, which exceeds the concentrations utilized in detection experiments. Furthermore, a nanoprobe concentration of ∼10 vol%/7.1 nM was achieved, below an average of ∼195 nM used for existing sensing methods with medium cell populations. As a result, the proposed system represents the first methodology reported using MoS2/PEG/M13 nanoprobes for clinically relevant cancer cell detection and constitutes an excellent opportunity for the development of lab-on-chip platforms.Data availability
The authors declare that the data supporting the finding of this study are available within the article and the ESI.† Other data are available from the corresponding authors upon reasonable request.Conflicts of interest
The authors declare no competing interests.Acknowledgements
We thank K. G. Lim, L. T. Ng, Y. L. Koh, and A. Huzaifah for important discussions. The authors acknowledge support from the Singapore University of Technology and Design (SKI 2021 01 04, SKI 2021 02 03, and SKI 2021 02 17), Changi General Hospital (Singapore) (CGH-SUTD-HTIF2019-001), the Ministry of Education (Singapore) (MOE-T2EP50220-0022), SUTD-Zhejiang-University (SUTD-ZJU (VP) 201903), and the Agency of Science Technology and Research (Singapore) (A20G9b0135) grant programs. D. K. L. acknowledges support from the Massachusetts Institute of Technology-SUTD International Design Centre and National Supercomputing Centre, Singapore (15001618). D. L. acknowledges support from the SUTD President Graduate Scholarship.References
- X.-J. Liang, C. Chen, Y. Zhao, L. Jia and P. Wang, Biopharmaceutics and therapeutic potential of engineered nanomaterials, Curr. Drug Metab., 2008, 9(8), 697–709, DOI:10.2174/138920008786049230.
- L. Li, A. M. Belcher and D. K. Loke, Simulating selective binding of a biological template to a nanoscale architecture: A core concept of a clamp-based binding-pocket-favored N-terminal-domain assembly, Nanoscale, 2020, 12(47), 24214–24227, 10.1039/D0NR07320B.
- M. Abd Elkodous, G. S. El-Sayyad, I. Y. Abdelrahman, H. S. El-Bastawisy, A. E. Mohamed, F. M. Mosallam, H. A. Nasser, M. Gobara, A. Baraka, M. A. Elsayed and A. I. El-Batal, Therapeutic and diagnostic potential of nanomaterials for enhanced biomedical applications, Colloids Surf., B, 2019, 180, 411–428, DOI:10.1016/j.colsurfb.2019.05.008.
- S. Barua, H. S. Dutta, S. Gogoi, R. Devi and R. Khan, Nanostructured MoS2-based advanced biosensors: A review, ACS Appl. Nano Mater., 2018, 1(1), 2–25, DOI:10.1021/acsanm.7b00157.
- D. Loke, J. M. Skelton, T. C. Chong and S. R. Elliott, Design of a nanoscale, CMOS-integrable, thermal-guiding structure for boolean-logic and neuromorphic computation, ACS Appl. Mater. Interfaces, 2016, 8(50), 34530–34536, DOI:10.1021/acsami.6b10667.
- D. K. Loke, G. J. Clausen, J. F. Ohmura, T. C. Chong and A. M. Belcher, Biological-templating of a segregating binary alloy for nanowire-like phase-change materials and memory, ACS Appl. Nano Mater., 2018, 1(12), 6556–6562, DOI:10.1021/acsanm.8b01508.
- M. Gheorghiu, A short review on cell-based biosensing: Challenges and breakthroughs in biomedical analysis, J. Biomed. Res., 2021, 35(4), 255, DOI:10.7555/JBR.34.20200128.
- C. L. Gargalo; S. C. de Las Heras; M. N. Jones; I. Udugama; S. S. Mansouri; U. Krühne and K. V. Gernaey, Towards the Development of Digital Twins for the Bio-Manufacturing Industry, in Advances in biochemical engineering/biotechnology, 2020, vol. 176, pp.1–34 DOI:10.1007/10_2020_142.
- K. Bruynseels, F. Santoni de Sio and J. van den Hoven, Digital twins in health care: Ethical implications of an emerging engineering paradigm, Front. Genet., 2018, 9, 1–11, DOI:10.3389/fgene.2018.00031.
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries, Ca-Cancer J. Clin., 2021, 71(3), 209–249, DOI:10.3322/caac.21660.
- P. R. C. Gascoyne, S. Shim, J. Noshari, F. F. Becker and K. Stemke-Hale, Correlations between the dielectric properties and exterior morphology of cells revealed by dielectrophoretic field-flow fractionation, Electrophoresis, 2013, 34(7), 1042–1050, DOI:10.1002/elps.201200496.
- G. Qiao, W. Duan, C. Chatwin, A. Sinclair and W. Wang, Electrical properties of breast cancer cells from impedance measurement of cell suspensions, J. Phys.: Conf. Ser., 2010, 224(1), 9–13, DOI:10.1088/1742-6596/224/1/012081.
- D. Lee, S. S. Chan, N. Aksic, N. Bajalovic and D. K. Loke, Ultralong-time recovery and low-voltage electroporation for biological cell monitoring enabled by a microsized multipulse framework, ACS Omega, 2021, 6(51), 35325–35333, DOI:10.1021/acsomega.1c04257.
- D. Lee, J. S. Naikar, S. S. Chan, M. P. Meivita, L. Li, Y. S. Tan, N. Bajalovic and D. K. Loke, Ultralong recovery time in nanosecond electroporation systems enabled by orientational-disordering processes, Nanoscale, 2022, 14, 7934–7942 RSC.
- G. Qiao, W. Wang, W. Duan, F. Zheng, A. J. Sinclair and C. R. Chatwin, Bioimpedance analysis for the characterization of breast cancer cells in suspension, IEEE Trans. Biomed. Eng., 2012, 59(8), 2321–2329, DOI:10.1109/TBME.2012.2202904.
- M. Al Ahmad, Z. Al Natour, F. Mustafa and T. A. Rizvi, Electrical characterization of normal and cancer cells, IEEE Access, 2018, 6, 25979–25986, DOI:10.1109/ACCESS.2018.2830883.
- J. A. Park, L. Atia, J. A. Mitchel, J. J. Fredberg and J. P. Butler, Collective migration and cell jamming in asthma, cancer and development, J. Cell Sci., 2016, 129(18), 3375–3383, DOI:10.1242/jcs.187922.
- P. Friedl, Y. Hegerfeldt and M. Tusch, Collective cell migration in morphogenesis and cancer, Int. J. Dev. Biol., 2004, 48(5–6), 441–449, DOI:10.1387/ijdb.041821pf.
- P. Friedl and D. Gilmour, Collective cell migration in morphogenesis, regeneration and cancer, Nat. Rev. Mol. Cell Biol., 2009, 10(7), 445–457, DOI:10.1038/nrm2720.
- J. Dermol, O. N. Pakhomova, A. G. Pakhomov and D. Miklavěiě, Cell electrosensitization exists only in certain electroporation buffers, PLoS One, 2016, 11(7), 1–19, DOI:10.1371/journal.pone.0159434.
- A. Mansoorifar, A. Koklu, S. Ma, G. V. Raj and A. Beskok, Electrical impedance measurements of biological cells in response to external stimuli, Anal. Chem., 2018, 90(7), 4320–4327, DOI:10.1021/acs.analchem.7b05392.
- D. A. McRae and M. A. Esrick, Monitoring Temperature and Tissue-Dependent Changes via Electrical Impedance Trajectories during Therapy, in Thermal Treatment of Tissue with Image Guidance, ed. T. P. Ryan and T. Z. Wong, 1999, vol. 3594, pp. 129–138 DOI:10.1117/12.348731.
- C. Brosseau and E. Sabri, Resistor-capacitor modeling of the cell membrane: A multiphysics analysis, J. Appl. Phys., 2021, 129, 011101, DOI:10.1063/5.0033608.
- S. Arndt, J. Seebach, K. Psathaki, H. J. Galla, J. Wegener, S. Arndt, J. Seebach, K. Psathaki, H. J. Galla and J. Wegener, Bioelectrical impedance assay to monitor changes in cell shape during apoptosis, Biosens. Bioelectron., 2004, 19(6), 583–594, DOI:10.1016/S0956-5663(03)00269-0.
- K. Cheung, S. Gawad and P. Renaud, Impedance spectroscopy flow cytometry: On-chip label-free cell differentiation, Cytometry, Part A, 2005, 65(2), 124–132, DOI:10.1002/cyto.a.20141.
- H. P. Schwan Electrical Properties of Tissues and Cell Suspensions: Mechanisms and Models. In Proceedings of 16th Annual International Conference of the IEEE Engineering in Medicine and Biology Society; IEEE, 2002, pp. A70–A71 DOI:10.1109/IEMBS.1994.412155.
- J. Chen, C. Liu, D. Hu, F. Wang, H. Wu, X. Gong, X. Liu, L. Song, Z. Sheng and H. Zheng, Single-layer MoS2 nanosheets with amplified photoacoustic effect for highly sensitive photoacoustic imaging of orthotopic brain tumors, Adv. Funct. Mater., 2016, 26(47), 8715–8725, DOI:10.1002/adfm.201603758.
- R. Anbazhagan, Y.-A. Su, H.-C. Tsai and R.-J. Jeng, MoS2–Gd chelate magnetic nanomaterials with core–shell structure used as contrast agents in in vivo magnetic resonance imaging, ACS Appl. Mater. Interfaces, 2016, 8(3), 1827–1835, DOI:10.1021/ACSAMI.5B09722.
- N. Xia, Y. Huang, Z. Cui, S. Liu, D. Deng, L. Liu and J. Wang, Impedimetric biosensor for assay of caspase-3 activity and evaluation of cell apoptosis using self-assembled biotin-phenylalanine network as signal enhancer, Sens. Actuators, B, 2020, 320, 128436, DOI:10.1016/j.snb.2020.128436.
- T. H. Hui, Z. L. Zhou, H. W. Fong, R. K. C. Ngan, T. Y. Lee, J. S. K. Au, A. H. W. Ngan, T. T. C. Yip and Y. Lin, Characterizing the malignancy and drug resistance of cancer cells from their membrane resealing response, Nat. Publ. Gr., 2016, 1–8, DOI:10.1038/srep26692.
- J. H. Lee, H. K. Choi, L. Yang, S. T. D. Chueng, J. W. Choi and K. B. Lee, Nondestructive real-time monitoring of enhanced stem cell differentiation using a graphene-Au HYBRID NANOELECTRODE Array, Adv. Mater., 2018, 30(39), 1–8, DOI:10.1002/adma.201802762.
- Z. Kou, X. Wang, R. Yuan, H. Chen, Q. Zhi, L. Gao, B. Wang, Z. Guo, X. Xue, W. Cao and L. Guo, A Promising gene delivery system developed from PEGylated MoS2 nanosheets for gene therapy, Nanoscale Res. Lett., 2014, 9(1), 1–9, DOI:10.1186/1556-276X-9-587.
- B. He, T. J. Morrow and C. D. Keating, Nanowire sensors for multiplexed detection of biomolecules, Curr. Opin. Chem. Biol., 2008, 12(5), 522–528, DOI:10.1016/j.cbpa.2008.08.027.
- X. Ren, Y. Han, Y. Xu, T. Liu, M. Cui, L. Xia, H. Li, Y. Gu and P. Wang, Diversified strategies based on nanoscale metal-organic frameworks for cancer therapy: The leap from monofunctional to versatile, Coord. Chem. Rev., 2021, 431, 213676, DOI:10.1016/j.ccr.2020.213676.
- T. Zheng, J. J. Fu, L. Hu, F. Qiu, M. Hu, J. J. Zhu, Z. C. Hua and H. Wang, Nanoarchitectured electrochemical cytosensors for selective detection of leukemia cells and quantitative evaluation of death receptor expression on cell surfaces, Anal. Chem., 2013, 85(11), 5609–5616, DOI:10.1021/ac400994p.
- S. Tang, H. Shen, Y. Hao, Z. Huang, Y. Tao, Y. Peng, Y. Guo, G. Xie and W. Feng, A Novel cytosensor based on Pt@Ag nanoflowers and AuNPs/acetylene black for ultrasensitive and highly specific detection of circulating tumor cells, Biosens. Bioelectron., 2018, 104, 72–78, DOI:10.1016/j.bios.2018.01.001.
- J. Zhou, Y. Zhao, J. Bao, D. Huo, H. Fa, X. Shen and C. Hou, One-step electrodeposition of Au–Pt bimetallic nanoparticles on MoS2 nanoflowers for hydrogen peroxide enzyme-free electrochemical sensor, Electrochim. Acta, 2017, 250, 152–158, DOI:10.1016/j.electacta.2017.08.044.
- B. Dou, J. Yang, R. Yuan and Y. Xiang, Trimetallic hybrid nanoflower-decorated MoS2 nanosheet sensor for direct in situ monitoring of H2O2 secreted from live cancer cells, Anal. Chem., 2018, 90(9), 5945–5950, DOI:10.1021/acs.analchem.8b00894.
- M. Govindasamy, V. Mani, S. M. Chen, R. Karthik, K. Manibalan and R. Umamaheswari, MoS2 flowers grown on graphene/carbon nanotubes: A versatile substrate for electrochemical determination of hydrogen peroxide, Int. J. Electrochem. Sci., 2016, 11(4), 2954–2961, DOI:10.20964/110402954.
- T. Liu, C. Wang, W. Cui, H. Gong, C. Liang, X. Shi, Z. Li, B. Sun and Z. Liu, Combined photothermal and photodynamic therapy delivered by PEGylated MoS2 nanosheets, Nanoscale, 2014, 6(19), 11219–11225, 10.1039/c4nr03753g.
- Y. Wang, Y. Du, J. Deng and Z. Wang, Friction reduction of water based lubricant with highly dispersed functional MoS2 nanosheets, Colloids Surf., A, 2019, 562, 321–328, DOI:10.1016/J.COLSURFA.2018.11.047.
- S. Jatav, K. P. Furlan, J. Liu and E. H. Hill, Heterostructured monolayer MoS2 nanoparticles toward water-dispersible catalysts, ACS Appl. Mater. Interfaces, 2020, 12(17), 19813–19822, DOI:10.1021/ACSAMI.0C02246.
- E. Grelet and R. Rana, From soft to hard rod behavior in liquid crystalline suspensions of sterically stabilized colloidal filamentous particles, Soft Matter, 2016, 12(20), 4621–4627, 10.1039/c6sm00527f.
- W. Feng, L. Chen, M. Qin, X. Zhou, Q. Zhang, Y. Miao, K. Qiu, Y. Zhang and C. He, Flower-like PEGylated MoS2 nanoflakes for near-infrared photothermal cancer therapy, Sci. Rep., 2015, 5, 1–13, DOI:10.1038/srep17422.
- T. Liu, C. Wang, X. Gu, H. Gong, L. Cheng, X. Shi, L. Feng, B. Sun and Z. Liu, Drug delivery with PEGylated MoS2 nano-sheets for combined photothermal and chemotherapy of cancer, Adv. Mater., 2014, 26(21), 3433–3440, DOI:10.1002/adma.201305256.
- F. Yin, T. Anderson, N. Panwar, K. Zhang, S. C. Tjin, B. K. Ng, H. S. Yoon, J. Qu and K. T. Yong, Functionalized Mos2 nanosheets as multi-gene delivery vehicles for in vivo pancreatic cancer therapy, Nanotheranostics, 2018, 2(4), 371–386, DOI:10.7150/ntno.27308.
- W. J. Chung, D. Y. Lee and S. Y. Yoo, Chemical modulation of M13 bacteriophage and its functional opportunities for nanomedicine, Int. J. Nanomed., 2014, 9(1), 5825–5836, DOI:10.2147/IJN.S73883.
- H. Zhao, Y. Li, B. Tan, Y. Zhang, X. Chen and X. Quan, PEGylated molybdenum dichalcogenide (PEG-MoS2) nanosheets with enhanced peroxidase-like activity for the colorimetric detection of H2O2, New J. Chem., 2017, 41(14), 6700–6708, 10.1039/c7nj00899f.
- M. Zaczek, A. Górski, A. Skaradzińska, M. Łusiak-Szelachowska and B. Weber-D’browska, Phage penetration of eukaryotic cells: Practical implications, Future Virol., 2019, 14(11), 745–760, DOI:10.2217/fvl-2019-0110.
- Y. Tian, M. Wu, X. Liu, Z. Liu, Q. Zhou, Z. Niu and Y. Huang, Probing the endocytic pathways of the filamentous bacteriophage in live cells using ratiometric PH fluorescent indicator, Adv. Healthcare Mater., 2015, 4(3), 413–419, DOI:10.1002/adhm.201400508.
- M. D. Siao, W. C. Shen, R. S. Chen, Z. W. Chang, M. C. Shih, Y. P. Chiu and C.-M. Cheng, Two-dimensional electronic transport and surface electron accumulation in MoS2, Nat. Commun., 2018, 9(1), 1–12, DOI:10.1038/s41467-018-03824-6.
- C. Nie, B. Zhang, Y. Gao, M. Yin, X. Yi, C. Zhao, Y. Zhang, L. Luo and S. Wang, Thickness-dependent enhancement of electronic mobility of MoS2 transistors via surface functionalization, J. Phys. Chem. C, 2020, 124(31), 16943–16950, DOI:10.1021/ACS.JPCC.0C02657.
- K. Mohan and G. A. Weiss, Engineering chemically modified viruses for prostate cancer cell recognition, Mol. BioSyst., 2015, 11(12), 3264–3272, 10.1039/C5MB00511F.
- S. S. Y. Y. Chan, D. Lee, M. P. Meivita, L. Li, Y. S. Tan, N. Bajalovic and D. K. Loke, Ultrasensitive two-dimensional material-based MCF-7 cancer cell sensor driven by perturbation processes, Nanoscale Adv., 2021, 3(24), 6974–6983, 10.1039/D1NA00614B.
- X. Zhu, X. Ji, N. Kong, Y. Chen, M. Mahmoudi, X. Xu, L. Ding, W. Tao, T. Cai, Y. Li, T. Gan, A. Barrett, Z. Bharwani, H. Chen and O. C. Farokhzad, Intracellular mechanistic understanding of 2D MoS2 nanosheets for anti-exocytosis-enhanced synergistic cancer therapy, ACS Nano, 2018, 12(3), 2922–2938, DOI:10.1021/acsnano.8b00516.
- H. Zhao, Y. Li, B. Tan, Y. Zhang, X. Chen and X. Quan, PEGylated molybdenum dichalcogenide (PEG-MoS2) nanosheets with enhanced peroxidase-like activity for the colorimetric detection of H2O2, New J. Chem., 2017, 41(14), 6700–6708, 10.1039/c7nj00899f.
- G. Oudeng, M. Au, J. Shi, C. Wen and M. Yang, One-step in situ detection of MiRNA-21 expression in single cancer cells based on biofunctionalized MoS2 nanosheets, ACS Appl. Mater. Interfaces, 2018, 10(1), 350–360, DOI:10.1021/acsami.7b18102.
- J. Kim, H. Kim and W. J. Kim, Single-layered MoS2-PEI-PEG nanocomposite-mediated gene delivery controlled by photo and redox stimuli, Small, 2016, 12(9), 1184–1192, DOI:10.1002/smll.201501655.
- T. Wang, W. Liu and J. Tian, Preparation and characterization of gold/poly(vinyl alcohol)/MoS2 intercalation nanocomposite, J. Mater. Sci.: Mater. Electron., 2004, 15(7), 435–438, DOI:10.1023/B:JMSE.0000031597.00716.2E.
- F. L. Pua, C. H. Chia, S. Zakari, T. K. Liew, M. A. Yarmo and N. M. Huang, Preparation of transition metal sulfide nanoparticles via hydrothermal route, Sains Malays., 2010, 39(2), 243–248 Search PubMed.
- K. Shameli, M. B. Ahmad, S. D. Jazayeri, S. Sedaghat, P. Shabanzadeh, H. Jahangirian, M. Mahdavi and Y. Abdollahi, Synthesis and characterization of polyethylene glycol mediated silver nanoparticles by the green method, Int. J. Mol. Sci., 2012, 13(6), 6639, DOI:10.3390/IJMS13066639.
- L. Saravanan and S. Subramanian, Surface chemical studies on the competitive adsorption of poly(ethylene glycol) and ammonium poly(methacrylate) onto alumina, J. Colloid Interface Sci., 2005, 284(2), 363–377, DOI:10.1016/J.JCIS.2004.08.188.
- M.-W. Lin, I. I. Kravchenko, J. Fowlkes, X. Li, A. A. Puretzky, C. M. Rouleau, D. B. Geohegan and K. Xiao, Thickness-dependent charge transport in few-layer MoS2 field-effect transistors, Nanotechnology, 2016, 27(16), 165203, DOI:10.1088/0957-4484/27/16/165203.
- A. Casadevall and L. A. Day, DNA packing in the filamentous viruses Fd, Xf, Pf1 and Pf3, Nucleic Acids Res., 1982, 10(7), 2467, DOI:10.1093/NAR/10.7.2467.
- P. Passaretti, I. Khan, T. R. Dafforn and P. Goldberg Oppenheimer, Improvements in the production of purified M13 bacteriophage bio-nanoparticle, Sci. Rep., 2020, 10(1), 1–9, DOI:10.1038/s41598-020-75205-3.
- D. Garcia, I. Ghansah, J. LeBlanc and M. J. Butte, Counting cells with a low-cost integrated microfluidics-waveguide sensor, Biomicrofluidics, 2012, 6(1), 014115, DOI:10.1063/1.3689857.
- J. B. M. Rocha Neto, A. C. Soares, R. A. Bataglioli, O. Carr, C. A. R. Costa, O. N. Oliveira, M. M. Beppu and H. F. Carvalho, Polysaccharide multilayer films in sensors for detecting prostate tumor cells based on hyaluronan-CD44 interactions, Cells, 2020, 9(6), 1563, DOI:10.3390/cells9061563.
- E. Sharon, E. Golub, A. Niazov-Elkan, D. Balogh and I. Willner, Analysis of telomerase by the telomeric hemin/G-quadruplex-controlled aggregation of Au nanoparticles in the presence of cysteine, Anal. Chem., 2014, 86(6), 3153–3158, DOI:10.1021/ac5000152.
- X. Cheng, Y. S. Liu, D. Irimia, U. Demirci, L. Yang, L. Zamir, W. R. Rodríguez, M. Toner and R. Bashir, Cell detection and counting through cell lysate impedance spectroscopy in microfluidic devices, Lab Chip, 2007, 7(6), 746–755, 10.1039/b705082h.
- Y. F. Chen, H. W. Wu, Y. H. Hong and H. Y. Lee, 40 GHz RF biosensor based on microwave coplanar waveguide transmission line for cancer cells (HepG2) dielectric characterization, Biosens. Bioelectron., 2014, 61, 417–421, DOI:10.1016/j.bios.2014.05.060.
- X. Zhang, K. Xiao, L. Cheng, H. Chen, B. Liu, S. Zhang and J. Kong, Visual and highly sensitive detection of cancer cells by a colorimetric aptasensor based on cell-triggered cyclic enzymatic signal amplification, Anal. Chem., 2014, 86(11), 5567–5572, DOI:10.1021/AC501068K.
- C. Pan, M. Guo, Z. Nie, X. Xiao and S. Yao, Aptamer-based electrochemical sensor for label-free recognition and detection of cancer cells, Electroanalysis, 2009, 21(11), 1321–1326, DOI:10.1002/elan.200804563.
- L. Liu, R. J. Lv, J. K. Leung, Q. Zou, Y. Wang, F. Li, W. Liang, S. Feng and M. Y. Wu, A Near-infrared biothiol-specific fluorescent probe for cancer cell recognition, Analyst, 2019, 144(16), 4750–4756, 10.1039/c9an00795d.
- Y. Chen, J. Peng, Y. Lai, B. Wu, L. Sun and J. Weng, Ultrasensitive label-free detection of circulating tumor cells using conductivity matching of two-dimensional semiconductor with cancer cell, Biosens. Bioelectron., 2019, 142, 111520, DOI:10.1016/j.bios.2019.111520.
- R. Wang, J. Di, J. Ma and Z. Ma, Highly sensitive detection of cancer cells by electrochemical impedance spectroscopy, Electrochim. Acta, 2012, 61, 179–184, DOI:10.1016/j.electacta.2011.11.112.
- L. Feng, Y. Chen, J. Ren and X. Qu, A graphene functionalized electrochemical aptasensor for selective label-free detection of cancer cells, Biomaterials, 2011, 32(11), 2930–2937, DOI:10.1016/j.biomaterials.2011.01.002.
- S. S. Y. Chan, Y. Sing Tan, K.-X. Wu, C. Cheung and K. D. Loke, Ultra-high signal detection of human embryonic stem cells driven by two-dimensional materials, ACS Appl. Bio Mater., 2018, 1(2), 210–215, DOI:10.1021/acsabm.8b00085.
- A. A. Marino, D. M. Morris, M. A. Schwalke, I. G. Iliev and S. Rogers, Electrical potential measurements in human breast cancer and benign lesions, Tumor Biol., 1994, 15(3), 147–152, DOI:10.1159/000217885.
- I. R. Suhito, K. M. Koo and T. H. Kim, Recent advances in electrochemical sensors for the detection of biomolecules and whole cells, Biomedicines, 2021, 9(1), 1–20, DOI:10.3390/biomedicines9010015.
- Y. Cho, H. S. Kim, A. B. Frazier, Z. G. Chen, D. M. Shin and A. Han, Whole-cell impedance analysis for highly and poorly metastatic cancer cells, J. Microelectromech. Syst., 2009, 18(4), 808–817, DOI:10.1109/JMEMS.2009.2021821.
- O. Cho, Y.-T. Oh, M. Chun, O. K. Noh and H.-W. Lee, Radiation-related lymphopenia as a new prognostic factor in limited-stage small cell lung cancer, Tumor Biol., 2015, 37(1), 971–978, DOI:10.1007/S13277-015-3888-Y.
- B. M. Burt, S. J. Rodig, T. R. Tilleman, A. W. Elbardissi, R. Bueno and D. J. Sugarbaker, Circulating and tumor-infiltrating myeloid cells predict survival in human pleural mesothelioma, Cancer, 2011, 117(22), 5234–5244, DOI:10.1002/CNCR.26143.
- S. H. Jee, J. Y. Park, H.-S. Kim, T. Y. Lee and J. M. Samet, White blood cell count and risk for all-cause, cardiovascular, and cancer mortality in a cohort of Koreans, Am. J. Epidemiol., 2005, 162(11), 1062–1069, DOI:10.1093/AJE/KWI326.
- J. Trujillo-Santos, P. D. Micco, M. Iannuzzo, R. Lecumberri, R. Guijarro, O. Madridano and M. Monreal, Investigators, for the R. Elevated white blood cell count and outcome in cancer patients with venous thromboembolism, Thromb. Haemostasis, 2017, 100(05), 905–911, DOI:10.1160/TH08-05-0339.
- A. Shankar, J. J. Wang, E. Rochtchina, M. C. Yu, R. Kefford and P. Mitchell, Association between circulating white blood cell count and cancer mortality: A population-based cohort study, Arch. Intern. Med., 2006, 166(2), 188–194, DOI:10.1001/ARCHINTE.166.2.188.
- Y. Xiao, V. Pavlov, T. Niazov, A. Dishon, M. Kotler and I. Willner, Catalytic beacons for the detection of DNA and telomerase activity, J. Am. Chem. Soc., 2004, 126(24), 7430–7431, DOI:10.1021/ja031875r.
- D. Ghosh, Y. Lee, S. Thomas, A. G. Kohli, D. S. Yun, A. M. Belcher and K. A. Kelly, M13-templated magnetic nanoparticles for targeted in vivo imaging of prostate cancer, Nat. Nanotechnol., 2012, 7(10), 677–682, DOI:10.1038/nnano.2012.146.
- L. Ceppi, N. M. Bardhan, Y. Na, A. Siegel, N. Rajan, R. Fruscio, M. G. Del Carmen, A. M. Belcher and M. J. Birrer, Real-time single-walled carbon nanotube-based fluorescence imaging improves survival after debulking surgery in an ovarian cancer model, ACS Nano, 2019, 13(5), 5356–5365, DOI:10.1021/acsnano.8b09829.
- J. W. Kretzer, M. Schmelcher and M. J. Loessner, Ultrasensitive and fast diagnostics of viable listeria cells by CBD magnetic separation combined with A511::LuxAB detection, Viruses, 2018, 10(11), 626, DOI:10.3390/v10110626.
- T. C. Hinkley, S. Singh, S. Garing, A. L. M. Le Ny, K. P. Nichols, J. E. Peters, J. N. Talbert and S. R. Nugen, A phage-based assay for the rapid, quantitative, and single CFU visualization of E. coli (ECOR #13) in drinking water, Sci. Rep., 2018, 8(1), 1–8, DOI:10.1038/s41598-018-33097-4.
- M. Ferrari, Cancer nanotechnology: Opportunities and challenges, Nat. Rev. Cancer, 2005, 5(3), 161–171, DOI:10.1038/nrc1566.
- A. Rasooly and J. Jacobson, Development of biosensors for cancer clinical testing, Biosens. Bioelectron., 2006, 21(10), 1851–1858, DOI:10.1016/j.bios.2006.01.003.
- T. S. Deisboeck and I. D. Couzin, Collective behavior in cancer cell populations, BioEssays, 2009, 31(2), 190–197, DOI:10.1002/bies.200800084.
- M. Levin, Molecular bioelectricity: How endogenous voltage potentials control cell behavior and instruct pattern regulation in vivo, Mol. Biol. Cell, 2014, 25(24), 3835, DOI:10.1091/MBC.E13-12-0708.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ma00669c |
| This journal is © The Royal Society of Chemistry 2023 |