Open Access Article
Open Access ArticleTonicity-induced cargo loading into extracellular vesicles†
Chaeeun
Lee‡
ab,
Sumit
Kumar‡
ab,
Juhee
Park
b,
Yongjun
Choi
 b,
Elizabeth Maria
Clarissa
ab and
Yoon-Kyoung
Cho
b,
Elizabeth Maria
Clarissa
ab and
Yoon-Kyoung
Cho
 *ab
*ab
aDepartment of Biomedical Engineering, Ulsan National Institute of Science and Technology (UNIST), Ulsan 44919, Korea. E-mail: ykcho@unist.ac.kr
bCenter for Soft and Living Matter, Institute for Basic Science (IBS), Ulsan 44919, Korea
First published on 28th February 2024
Abstract
The current challenge in using extracellular vesicles (EVs) as drug delivery vehicles is to precisely control their membrane permeability, specifically in the ability to switch between permeable and impermeable states without compromising their integrity and functionality. Here, we introduce a rapid, efficient, and gentle loading method for EVs based on tonicity control (TC) using a lab-on-a-disc platform. In this technique, a hypotonic solution was used for temporarily permeabilizing a membrane (“on” state), allowing the influx of molecules into EVs. The subsequent isotonic washing led to an impermeable membrane (“off” state). This loading cycle enables the loading of different cargos into EVs, such as doxorubicin hydrochloride (Dox), ssDNA, and miRNA. The TC approach was shown to be more effective than traditional methods such as sonication or extrusion, with loading yields that were 4.3-fold and 7.2-fold greater, respectively. Finally, the intracellular assessments of miRNA-497-loaded EVs and doxorubicin-loaded EVs confirmed the superior performance of TC-prepared formulations and demonstrated the impact of encapsulation heterogeneity on the therapeutic outcome, signifying potential opportunities for developing novel exosome-based therapeutic systems for clinical applications.
Introduction
Extracellular vesicles (EVs) inherit diverse cellular contents, playing a pivotal role in facilitating intercellular communication.1–4 Substantial endeavors have been dedicated to unraveling the significance of EVs in various diseases, alongside exploring their potential applications in diagnostics and therapeutics.5–8 Different from synthetic nanocarriers, EVs possess the unique ability to traverse diverse physiological barriers, including the formidable blood–brain barrier, rendering them invaluable for the treatment of neurological disorders.3,9–15Moreover, EVs are recognized for orchestrating intricate signalling cascades that influence a spectrum of physiological and pathological processes, including metastasis.16–18 This sophisticated cargo transfer occurs through an array of endocytic pathways, enabling EVs to mediate intricate cellular responses. EVs transfer their cargo to target cells through various endocytic pathways for cargo release, and these pathways may coexist and not always be mutually exclusive for the internalization of the same set of EVs,19,20 avoiding lysosomal degradation,21,22 and increasing their effectiveness as therapeutic delivery vehicles, presenting a promising candidate for various biomedical applications. Capitalizing on the advantages of EVs as nanocarriers, they have been harnessed for drug encapsulation, exemplified by instances such as curcumin (NCT01294072), siRNA against KrasG12D (NCT03608631), and antigen (NCT01159288).23–25
To successfully utilize EVs as nanocarriers, loading cargo molecules into EVs has been demonstrated as an effective means of preserving them in bio-fluids.26–28 It is critical to shield drugs from components in body fluids, especially in the bloodstream, until they reach the intended target; encapsulation within EVs creates a protective barrier, preventing interactions with immune cells, serum proteins, and nucleases that could otherwise render the cargo ineffective or lead to its premature clearance from circulation.29
However, current loading methods, such as extrusion, electroporation, and sonication, can damage27,30–32 the original membrane structure of EVs, which is important for cellular uptake and drug delivery.1,33,34 Although treating with chemicals, such as saponin, can be utilized as an alternative to the use of external forces, the residue left within the EVs can cause toxicity issues depending on the specific chemical compositions, its concentration, and the intended treatment time.27,35,36 Similarly, the freeze–thaw cycle can damage native EVs by causing lipid damage,30 protein denaturation, and precipitation of their cargo.37–40 Therefore, it is necessary to develop an efficient material-loading method to enhance the membrane permeability of EVs without causing such damage. Several attempts have been made to enhance drug loading into EVs by optimizing conventional techniques,41,42 or by introducing new techniques such as nanoporation43 or the pH gradient method.44 Nevertheless, in certain cases, these methods entail multiple steps, including dehydration and prolonged adjustments to buffer conditions. In this study, instead of using harsh stimuli or chemicals, we employed hypotonic shocks to load external cargo into EVs.
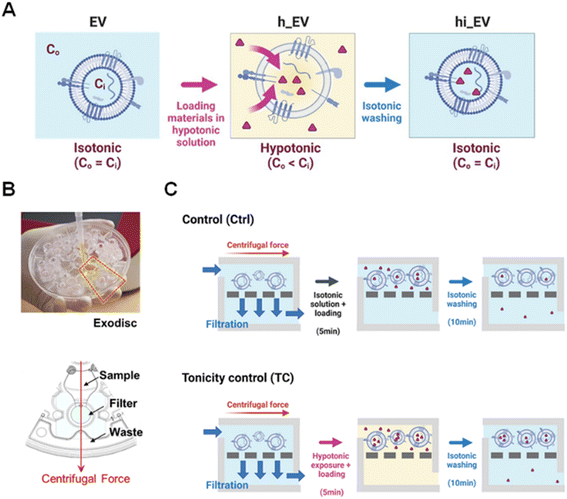
Hypotonic environments have been used to transport materials into cells,45–47 tissues,48 and unilamellar vesicles49,50 through osmotic-pressure-induced swelling, which decreases the lipid density and affects membrane permeability.51,52 Unlike cells, nano-sized EVs can tolerate greater changes in tonicity without rupturing.50,53 To enhance their use as nanoreactors for drug delivery or other applications, we propose a feasible approach involving the application of a hypotonic treatment to rapidly permeabilize EV membranes for efficient loading of external cargo without requiring harsh stimuli or chemicals (Fig. 1). Our approach, called “tonicity control (TC)”, utilizes Exodisc, a lab-on-a-disc platform developed for EV separation using centrifugal tangential flow filtration.54 This enables multiple steps of hypotonic treatment, facilitating the influx of diverse cargo molecules into EVs. The TC method was applied to load the chemotherapeutic drug (Dox), different sizes of dextrans, and single-stranded DNA (ssDNA) as cargo models to evaluate the loading performance for EVs. Finally, we demonstrated that miRNA-497 and Dox-encapsulating EVs could penetrate tissue spheroids and reduce their size by supplying the drug, demonstrating the potential of this method for targeted drug delivery.
Experimental section
EV isolation using Exodisc
HEK 293T cells and A549 cells were cultured at 37 °C with 5% CO2. Dulbecco's modified Eagle's medium (DMEM) (Gibco) was used for the cell culture medium after supplementing with 5% (v/v) of fetal bovine serum (FBS) and 1% (v/v) of antibiotic-antimycotic solution (Gibco). In detail, 2 × 106 cells were seeded in, and the medium was changed to the exo-free medium, containing 5% (v/v) of exosome depleted FBS after 1 day. The cell culture supernatant was collected after 2 days and used for EV separation. Before the separation, the sample was subjected to the following preparation steps to remove cellular debris; 300 g for 10 min and 2000 g for 10 min followed by filtration using a 0.2 μm pore syringe filter (Pall Corporation, NY, USA) and separation by Exodisc using the ExoDiscovery system (Labspinner, South Korea) at 3000 rpm followed by two washing steps with PBS.Controlling tonicity and material loading
Following the EV separation, a hypotonic solution (1 mL) containing the loading material was added to the sample chamber of the Exodisc and rotated for 5 min at 3000 rpm. As the loading materials, we used Dox-HCl (5 μM, 1 mL), FITC-dextran (5 μM, 1 mL), and FAM-labelled ssDNA (1 μM, 1 mL). Following the hypotonic exposure, the EVs were washed twice with 1 mL of PBS to restore them to the isotonic solution and remove any unloaded materials. Fluorescence signals were measured using a TECAN plate reader (F200, Tecan, Switzerland), and the loading amount was quantified using a standard curve.Dox loading into EVs by extrusion and sonication
For each method, we started from the same amount of the pre-filtered cell culture supernatant (3 mL). The EVs were isolated using Exodisc, as described previously.54 The experimental conditions were chosen as commonly used in previous studies. In the extrusion method, the EVs were extruded through a 0.2 μm pore syringe filter (Pall Corporation, USA) 20 times.55 In the case of the sonication method, the EVs were sonicated for six cycles, with the “on mode” lasting 30 s and the “off mode” lasting 150 s according to a previous report.56 After loading, extrusion, or sonication, the free drugs were washed using Exodisc under the same conditions.Calcein–cobalt assay for vesicle permeability test
Following EV separation, calcein-AM (1 μM; final concentration) was added to the filter chamber and incubated at room temperature for 30 min. Free calcein-AM was washed twice with PBS. Next, the EVs were exposed to hypotonic, isotonic, or hypotonic to isotonic solution and eluted from the disc. The EVs were transferred to a 96-well black plate, and CoCl2·6H2O solution was added (4 mm; final working concentration). The F200 plate reader (Tecan, Switzerland) was used to measure the intensity in the wavelength range of 485–535 nm.Western blotting
Firstly, EVs were lysed with RIPA buffer (Millipore, USA) and protease inhibitor (Thermo Fisher, USA). Following lysis, lysed EVs were boiled at 95 °C for 10 min with SDS-PAGE loading buffer (Biosesang, South Korea) and run on an 8% Tris-glycine SDS-PAGE gel. The gel was subsequently transferred to polyvinylidene fluoride (PVDF) membranes (Bio-Rad). The membranes were blocked with 5% skim milk at RT for 1 h, and they were incubated overnight at 4 °C with primary antibodies using CD9, CD81, and TSG101 (BD bioscience, USA). After 1 day, the membranes were washed three times with TBST buffer and incubated with secondary horse radish peroxidase (HRP)-conjugated antibody at room temperature for 1 h. Chemiluminescent detection was performed using the Pierce™ ECL Western Blotting Substrate (Thermo Fisher,) and the result was obtained using Azure C600 equipment (Azure Biosystems, USA).Indirect enzyme-linked immunosorbent assay (ELISA)
The EV samples were diluted in PBS using the same dilution factor for all conditions, and 50 μL of the diluted EV samples were incubated in a 96-well plate overnight at 4 °C. After the incubation, the remaining samples were removed, and 1% BSA was added to block nonspecific interactions, followed by incubation at RT for 1 h. A washing step was performed using 0.1% BSA, and primary antibodies (CD9, CD63, and CD81) were added to the respective designated wells and incubated at RT for 1 h, followed by another washing step. Horseradish peroxidase (HRP)-conjugated antibodies were loaded into each well at RT for 1 h, and another washing step was performed before adding the TMB solution. After 15 min, the reaction was stopped by adding a stop solution (2 N H2SO4) to each well. The absorbance was measured at 450 nm using an Infinite M200 plate reader (Tecan, Switzerland).mRNA expression of EVs
To confirm mRNA expression, the total RNA from EVs and hi_EVs were extracted using the miRNeasy kit (Qiagen). cDNA was synthesized using the SuperScript VILO cDNA synthesis kit (Thermo Fisher Scientific). qRT-PCR was performed as follows: 50 °C for 2 min, 95 °C for 10 min; 40 cycles of 94 °C for 10 s and 60 °C for 20 s. Alix (Hs00994346_m1) and GAPDH(Hs99999905_m1) were purchased from Thermo Fisher Scientific. The primers and probes for CD9, CD63, and CD81 were synthesized by Macrogen (South Korea). CD9 forward primer 5′-GGCTTCCTCTTGGTGATATTCG-3′, probe 5′-TCCTGGACTTCCTTAATCACCTCATCCT-3′, and reverse primer 5′-GGCTCATCCTTGGTTTTCAG-3′; CD63 forward primer 5′-AACGAGAAGGCGATCCATAAG-3′, probe 5′-CCTCGACAAAAGCAATTCCAAGGGC-3′, and reverse primer 5′-GCAGGCAAAGACAATTCCC-3′; CD81 forward primer 5′-AGATCGCCAAGGATGTGAAG-3′, probe 5′-AGCAGTCAAGCGTCTCGTGGAAG-3′, and reverse primer 5′-AGGTGGTCAAAGCAGTCAG-3′.miRNA loading measurement and cell proliferation assay
MiR-497 mimics were purchased from Bioneer (South Korea), and the sequences were as follows: 5′-CAG CAG CAC ACU GUG GUU UGU-3′. After loading into EVs, miRNAs were isolated using the miRNeasy Micro Kit (Qiagen) according to the manufacturer's protocol. cDNA was synthesized using the TaqMan Advanced miRNA cDNA synthesis kit, according to the manufacturer's protocol. To quantify the miRNA amount, qRT-PCR was conducted for 40 cycles on a QuantStudio 6 Flex machine, and the loading amount was calculated using a standard curve. Subsequently, 4 × 109 A549 cells are seeded in a 96 well plate and cultured at 37 °C with 5% CO2. After 24 h, miR-497 loaded EVs were applied to the cells with 5% of exo-free FBS in the same cell culture medium described above. After treatment of the miR-497 loaded EVs, images were captured using a JuLI™ Stage automated imaging system (NanoEntek Inc., Seoul, Korea). The proliferation rate was analyzed by the JuLI™ STAT software.Spheroid culture and imaging after Dox-EV treatment
A total of 5 × 104 A549 cells were seeded in ultra-low attachment (ULA) 96-well microplates (Corning) and centrifuged at 1000 rpm for 3 min. The spheroids were cultured with Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with 5% (v/v) of FBS and 1% (v/v) of antibiotic–antimycotic solution (Gibco). After 48 h, the cell culture medium was replaced with the Dox-EV containing medium, followed by washing with PBS and imaging for 48 h. Live imaging of the A549 spheroids was performed using a 3D laser scanning confocal microscope (LSM 980) with a 10× objective lens and transmitted bright field imaging. The diameter of the spheroid was measured after performing max intensity projection on the 3D scanned spheroid image. The ZEN software (Zeiss, Germany) was used for microscopy operation and image processing, and ImageJ software was used to measure the spheroid size. Regarding the fluorescence imaging of Dox-EV in spheroids, 4 × 104 A549 cells were seeded with the same conditions, and equal amounts of Dox and Dox-EV (3 μM) are treated. After 48 h, spheroids are washed twice with PBS and fixed with 4% of paraformaldehyde and measured. The images are generated by max intensity projection of Z-stacks through the THUNDER imaging system (Leica) using 10× magnification.Characterization of extracellular vesicles using NTA and BCA
A Nanosight NS 500 system (Malvern Instruments, UK) was used to measure the particle concentration of the EVs. The EV samples were diluted with pre-filtered PBS according to the manufacturer's recommended concentration range (25–100 particles per frame). To ensure consistency, identical settings were used for all measurements. The protein concentration of EVs was quantified using a bicinchoninic acid (BCA) assay kit (Thermo Fisher Scientific) following the manufacturer's protocol.Dynamic light scattering (DLS)
The size of EVs was measured using a Zetasizer Nano ZSP (Malvern Instruments, UK). The EV solution (109 particles per mL) were transferred to a microcuvette (ZEN0040; Malvern Instruments, UK), and the particle size was determined using the Zetasizer software according to the manufacturer's protocol. To measure the swollen EV in solutions with different tonicities, all the EV cases were fixed with 4% paraformaldehyde after hypotonic exposure to maintain the structure and washed with deionized water (DI) to remove the impact of salt during the measurement.Transmission electron microscopy (TEM) imaging
EVs were fixed with 4% paraformaldehyde followed by washing with DI, and the samples were dropped on a pre-cleaned parafilm. Formvar/carbon film grids, coated with poly-L-lysine (PLL), were flipped onto a drop of EVs on the parafilm. After flipping back, the grids were covered with lids and incubated at RT for 1 h. UranyLess EM Stain solution (EMS) was dropped on the grid over 1 min and removed by flipping on a drop of DI. The remaining solution was absorbed using a paper and dried. Imaging was performed using a transmission electron microscope (JEM-2100; JEOL, Tokyo, Japan).Cell imaging
After treating the A549 cells with Dox–EVs for 24 h, the samples were fixed with 4% paraformaldehyde (PFA) for 20 min, followed by gentle washing with PBS twice. The fixed cells were permeabilized with 0.1% Triton-X and washed. The nuclei of the A549 cells were stained with Hoechst for 10 min and washed. Cell imaging was performed using a confocal microscope (LSM 980) equipped with a 60× oil-immersion objective lens. Microscopic operation and imaging were conducted using the ZEN software (Zeiss, Wetzlar, Germany).Cell internalization assay
EVs were labelled with 1 μm 1,1-dioctadecyl-3,3,3,3-tetramethylindodicarbocyanine (DiD) dye on the disc, and the remaining dye was washed out. A total of 105 A549 cells were seeded in each well of the 12-well plate and incubated at 37 °C for 24 h. Next, 100 μL of DiD-labeled EVs was added to each well and incubated for 2 h. The cells were then collected using trypsin–EDTA, washed twice with PBS, and centrifuged at 1800 rpm for 5 min at 4 °C. The collected cells were analyzed by flow cytometry, and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events were counted for each sample. The mean fluorescence intensity was normalized to those of the control.
000 events were counted for each sample. The mean fluorescence intensity was normalized to those of the control.
Cell viability test
The cell viability was assessed using the MTT assay (V13154, Invitrogen, Carlsbad, CA, USA). A total of 104 A549 cells were seeded in a 96-well plate and cultured with the medium containing 5% CO2 at 37 °C. After one day, the culture medium was removed, and the cells were washed with PBS. Subsequently, Dox-loaded EVs produced by each method and the EVs without Dox (negative control) were added to each well (final volume of 100 μL) in the culture medium. After 48 h of incubation, MTT stock solution (12 mm, 10 μL) was added to each well, and the plate was incubated for 4 h. The SDS-HCl solution was then added, and the absorbance was measured using a TECAN plate reader (M200, Tecan, Männedorf, Switzerland) at an excitation wavelength of 480 nm and emission wavelengths of 595 and 535 nm for another 4 h.Results and discussion
Tonicity control and rapid tonicity change using Exodisc
We utilized the Exodisc, a centrifugal microfluidic platform equipped with anodic aluminum oxide membrane filters featuring 20 nm of pore sizes, to isolate EVs from the HEK293T cell-culture supernatant (CCS) in a highly efficient and label-free manner.54 This technique allowed us to rapidly enrich EVs, and not only the EV separation step, but cargo loading, and washing, was performed on the same disc without sample transfer steps, leading to minimal particle loss (Fig. 1). The efficient washing capability of Exodisc effectively removed unloaded materials, and its rapid solution exchange feature enabled a quick exchange of solutions within 5 min (Video S1†). To characterize the effect of the hypotonic treatment, we collected two types of EV samples: h_EV, which was exposed to only hypotonic solution, and hi-EV, which was exposed to hypotonic solution followed by isotonic washing (Fig. 1).Enhancing EV permeability and cargo loading through hypotonic treatment
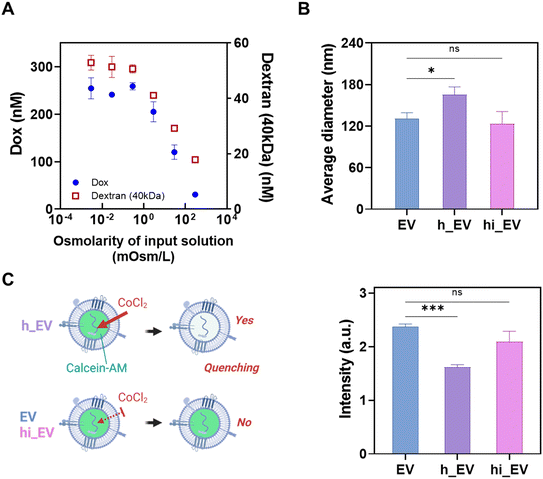
Through variations in the osmolarity of the hypotonic solution, we identified the optimal conditions at 296 μOsm L−1, allowing for maximum loading of representative materials like Dox and FITC-dextran (40 kDa) while minimizing hypotonic stress (Fig. 2A). After EV isolation using the Exodisc, we introduced a hypotonic solution containing these loading materials, subsequently followed by an isotonic solution to remove free loading molecules. We employed this optimized condition for subsequent experiments, demonstrating the potential of leveraging hypotonic shocks to enhance EV permeability and streamline cargo loading.A dynamic light scattering (DLS) analysis revealed that exposure to the hypotonic solution (296 μOsm L−1) resulted in a significant increment of 26.8 ± 15% in EV size due to osmotic pressure-induced swelling and temporary change in their membrane permeability (h_EV). After returning to isotonic conditions (hi_EV), the EVs showed no significant change in their size from the original sample (EV) size, as shown in Fig. 2B.
Furthermore, a cobalt–calcein assay was used to investigate the membrane permeability of EVs during the hypotonic–isotonic treatment. EVs were incubated with a calcein-AM dye, (2′,7′-([bis(carboxymethyl)amino]methyl) fluorescein), and free calcein-AM was washed away with Exodisc. Subsequently, cobalt chloride was added, which decreased the fluorescence intensity owing to the formation of a complex between the cobalt Co2+ ions and the fluorophore calcein, resulting in the quenching of calcein fluorescence (excitation/emission: 494/517 nm). Fig. 2C shows that EVs quench to a greater extent when placed in the hypotonic solution (−31.6 ± 1.9%), implying an increase in their membrane permeability, which facilitated the entry of cobalt chloride across the EV membrane. However, when cobalt chloride was introduced after the EVs were restored to an isotonic solution, quenching was not considerable (−11.7 ± 14%), indicating a decrease in the membrane permeability of the EVs.
In addition, we quantified Dox concentrations inside and outside EVs using Exodisc. Following the loading and two subsequent washing steps, we measured the Dox concentrations in solutions collected from the top of the filter (representing total Dox) and the flow-through samples (representing Dox outside of EVs). Utilizing the NTA data, we approximated an average radius (r) of 65.3 nm, a total of 5 × 109 EVs (NEV), and a total volume of 100 μL of EV solution on the filter (Vtotal). Subsequently, the initial Dox concentration inside EVs after the loading step was estimated to be 18.8 ± 3.47 mM, with the measured external concentration of 3.96 μM. Approximately 56.3% of the Dox permeated the EV membrane during loading. After the first washing step, the internal Dox concentration decreased to 4.7 ± 0.3 mM, indicating cargo leakage. Subsequent washing resulted in an internal Dox concentration of 3.8 ± 0.3 mM within the EVs, while the measured external concentration was 0.02 μM.
Hypo-osmotic exposure facilitates cargo transport from outside to inside EVs via osmotic pressure differences. Various factors such as membrane permeability of water and cargo, vesicle size, and osmolarity differences play key roles in this mechanism.57 The cargo loading process induced by hypotonic conditions can be understood through theoretical analysis using the Kedem–Katchalsky (K–K) formalism. The hypothesis and the calculation parameters are presented in Fig. S1 and Table S1,† respectively. Detailed results can be found in Fig. S2,† with a comprehensive discussion in the ESI† section.
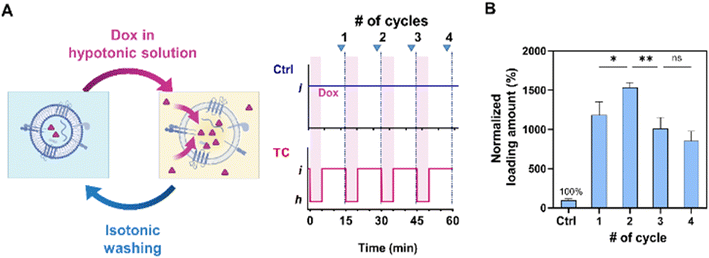
Improved EV cargo loading by osmotic cycles
In our subsequent experiment, we conducted multiple osmotic cycles on EVs to emulate the loading process and determine their maximum cargo loading capacity (Fig. 3A). We compared these results with those obtained using the control (Ctrl) method, which involved applying the same Dox concentration and washing steps without subjecting the EVs to hypotonic shocks (Fig. 1C). The loading amount remained relatively constant across time intervals ranging from 5 to 30 min. Consequently, we fixed the duration of each step to the minimum required time of 5 min for solution filtration.Notably, the highest loading amount was achieved after subjecting the EVs to two cycles of hypotonic shocks, resulting in a 29.4% increase compared to the single-cycle case (Fig. 3B). However, the loading amount showed a decrease after the third cycle, possibly attributed to the cumulative effects of washing steps and particle loss. On average, the two-cycle method resulted in loading amounts approximately 15.4 times higher than those of the Ctrl method. Consequently, we employed the two-cycle TC method to maximize the cargo loading for further analysis.
Preservation of EV characteristics after tonicity control-induced osmotic cycles
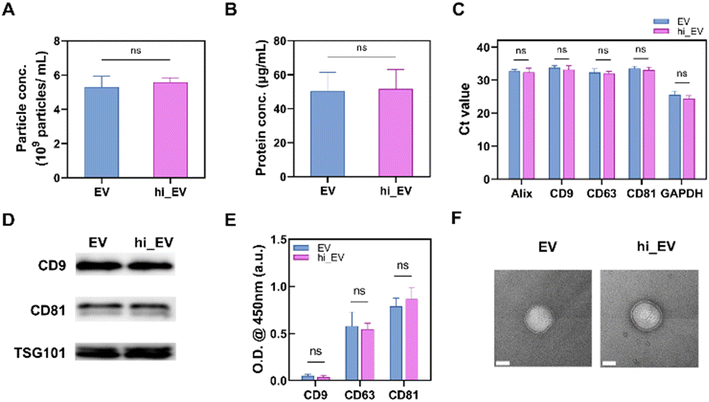
To use EVs for cargo delivery without affecting their structure and functionality, it is crucial to preserve their innate features, such as their small size, lipid bilayer encapsulation, and bioactive surface proteins. We compared the EVs and hi_EVs to test whether the TC process harmed their structure or functionality. We found that the particle numbers (∼5 × 109 particles per mL) and protein concentration (∼50 μg mL−1) were not significantly different between EVs and hi_EVs after the TC treatment (Fig. 4A and B). Additionally, representative mRNA markers, such as Alix, CD9, CD63, CD81, and GAPDH, did not significantly differ between EVs and hi_EVs, indicating that the hypotonic treatment did not affect the inner components of the EVs (Fig. 4C). No significant differences in the EV-specific proteins were observed between EVs and hi_EVs by Western blot (Fig. 4D) or enzyme-linked immunosorbent assay (ELISA) (Fig. 4E). In addition, we confirmed these results using transmission electron microscopy (TEM) images, which demonstrated that the double-layer and membrane of the EVs remained intact after the TC treatment (Fig. 4F). Overall, these findings indicated that the TC process does not significantly alter the characteristics of EVs, further highlighting their potential as a drug delivery system.TC method for exogenous cargo loading into EVs
We conducted experiments with different loading materials of varying sizes and shapes to further evaluate the effectiveness of the TC method in loading different types of exogenous cargos into EVs. For FITC-dextran of three different sizes (3–5, 10, and 40 kDa), the TC technique resulted in a loading amount 2.2 to 3.6 times higher than the control method (Ctrl), which did not involve hypotonic shock application (Fig. 5A). We also investigated short-length nucleic acids for loading by using FAM-labeled ssDNA (10, 30, and 50 mer). The loading amount increased in the TC case, whereas the nucleic acids could not be loaded using the Ctrl method (Fig. 5B). The loading amounts were determined from the calibration curves shown in Fig. S3.† For the investigated cargos, such as dextran (3–40 kDa) and ssDNA (10–50 mer), the background signal, which was the fluorescence intensity after washing, was minimal. Larger molecules with different shapes, such as Y-shaped DNA composed of three strands of 30 mers and a tetrahedral structure composed of four strands of DNA molecules (55 bp, 79 bp), could not be completely washed away using Exodisc with filters having 20 nm pores (data not shown); thus, the loading amount could not be determined. Taken together, these findings indicate that the TC technique is adaptable and has the potential to increase the cargo delivery capacity.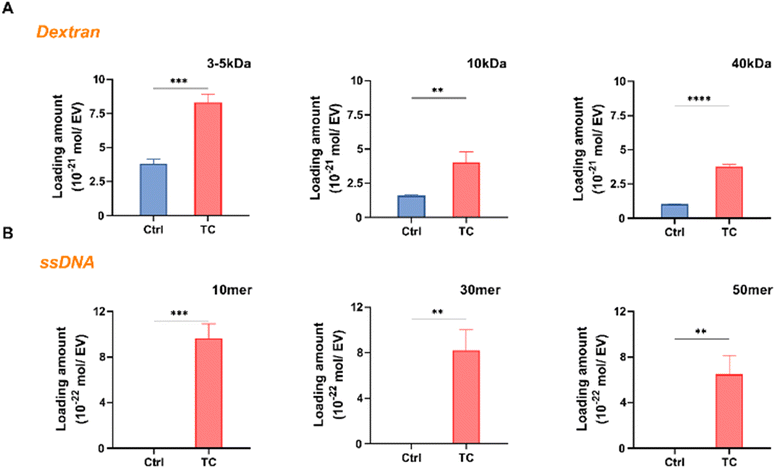 | ||
| Fig. 5 TC method is used for loading different cargo materials. (A) Loading of FITC-dextran (3–5, 10, and 40 kDa) into EVs by using the TC vs. Ctrl method (****p < 0.0001, ***p < 0.001, **p < 0.01). The Ctrl method is without hypotonic treatment (Fig. S1†); (B) loading of FAM-labeled ssDNA (10, 30, and 50 mer) into EVs using the TC method compared with the Ctrl (**p < 0.01), data represent mean ± s.d. of n = 3 independent experiments. | ||
Enhanced miRNA loading and therapeutic efficacy of EVs through the TC method
To determine whether the TC method affected the cellular uptake of EVs, we performed a cell internalization assay using cancer cells. Both EVs and hi_EVs were fluorescently labeled with the dye, 1,1-dioctadecyl-3,3,3,3-tetramethylindodicarbocyanine (DiD), and used to treat lung cancer cells (A549). After 24 h, we observed the cells under a confocal microscope and found no significant difference in internalization between the EVs and hi_EVs (Fig. S4A†). To further quantify the internalization, we used flow cytometry and calculated the cell internalization efficiency by comparing the mean fluorescence intensities of hi_EV and EVs, as previously reported.30 Our analysis showed no significant difference in the cell internalization efficiency between hi_EVs and EVs (Fig. S4B and C†). These results indicate that the TC method does not affect the ability of EVs to be taken up by the cells and support the potential use of TC-loaded EVs as a drug delivery system.To demonstrate the therapeutic implications of our technique, we selected miRNA-497 (miR-497) for loading into EVs using the TC method, as it is known to have an inhibitory effect on the proliferation of lung cancer cells.58,59 As shown in Fig. S5A,† our findings reveal a substantial increase in the quantity of miR-497 loaded into EVs with the TC method compared with the Ctrl method, as confirmed by the real-time PCR analysis. Next, we evaluated the efficacy of the miR-497-loaded EVs in reducing the proliferation of A549 cells. As shown in Fig. S5B,† both the miR-497 loaded EVs exhibit reduced proliferation compared with the negative control group, which only received EVs without miR-497. The group treated with miR-497-loaded EVs produced using the TC method showed a significant difference in inhibiting cell proliferation compared with the Ctrl method after 72 h, indicating that the higher loading quantity achieved with the TC method contributed to its superior efficacy. These results suggest that our TC technique has significant potential for loading therapeutic molecules, such as miRNAs, into EVs thereby enhancing their cargo delivery applications.
Comparison of loading methods for EVs in drug delivery applications
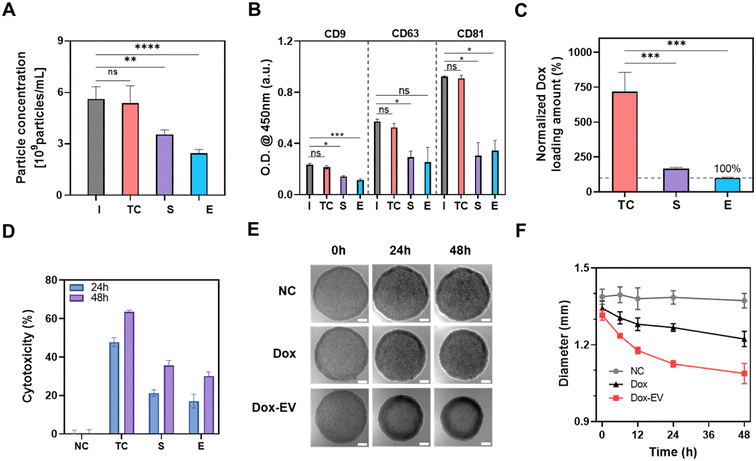
The loading of materials into EVs is a crucial step in drug delivery applications. However, conventional techniques such as sonication and extrusion have potential to induce membrane damage in EVs due to external forces or strong stimulation.27,30 Here, we conducted a comparative analysis between the TC method and traditional loading methods, including sonication and extrusion, employing experimental conditions commonly used in previous reports.55,56Using the same initial volume of the conditioned media, we found no significant particle loss in the case of the TC method, whereas particle losses were 34.9 ± 4.8% and 57.0 ± 1.6%, respectively, in the case of sonication and extrusion (Fig. 6A). Additionally, we evaluated the expressions of EV tetraspanin markers, such as CD9, CD63, and CD81, after each loading process. Compared with the TC method, the other methods exhibited a decrease in the signal for all the markers (Fig. 6B), which could be attributed to damage or particle loss during the loading procedure.
When the Dox loading amount was investigated, the TC approach produced a loading amount 4.3 ± 0.82 and 7.2 ± 1.3 times higher than the sonication and extrusion methods, respectively (Fig. 6C). For cellular treatment, we prepared Dox-loaded EVs using the same volume of the cell culture medium with different loading methods, such as sonication, extrusion, and the TC method, and then treated them with A549 cells. After 48 h, Dox-EV generated using the TC method had a higher anti-tumor effect (52.1 ± 1.3%) than that generated using the other loading methods (31.1 ± 1.6% for sonication and 25.7 ± 2.5% for extrusion) (Fig. 6D). These findings suggest that the TC process did not result in particle loss or surface protein degradation and that it produced better results and cell internalization than the conventional loading methods (Fig. S6†).
The stability of EVs after each process was tested using sandwich ELISA. EVs were captured by CD81 antibody, and the signal was detected by CD9 antibody (Fig. S7†). Following the TC processes, the CD9/CD81 ELISA results exhibited a trend similar to the control (Ctrl). However, sonication and extrusion resulted in significantly lower signals than the Ctrl. Across all cases, there was a notable decrease in signal after 5 days. After 21 days of storage at 4 °C, both Ctrl and TC samples showed a signal decrease of 51.0 ± 1.70% and 53.0% ± 4.3%, respectively, compared to day 1. The difference in signal between TC and Ctrl was not statistically significant. However, after sonication and extrusion, the signal was substantially decreased by 76.9 ± 3.52% and 88.7 ± 2.56%, respectively, compared to the initial signal (Fig. S7B†). These signal reductions suggest the potential damage caused by sonication or extrusion methods.
To evaluate EV damage after each loading method, we conducted a sandwich ELISA using tetraspanin markers (CD81 as capture antibody and CD9 as detection antibody). This was done with the same initial volume of CCS (Fig. S8A†) and EV samples adjusted to the same particle concentration after various loading processes (Fig. S8B†). In both scenarios, no significant differences were observed between the Ctrl and TC methods.
When using the same input CCS amount, both sonication and extrusion processes resulted in a signal decrease of 38.9 ± 1.6% and 50.2 ± 1.9% (Fig. S8A†). These optical density (O.D.) reductions corresponded to an effective particle concentration loss of 65.2 ± 2.7% and 84.2 ± 3.2% after sonication and extrusion, respectively.
Furthermore, when maintaining the same particle concentration after each loading process, sonication and extrusion demonstrated a signal reduction of 17.8 ± 3.1% and 29.2 ± 3.0%, as depicted in Fig. S8B.† These decreases corresponded to an effective particle concentration decrease of 29.9 ± 5.2% and 49.2 ± 5.0% after sonication and extrusion, respectively, while there was no loss in the TC method. Taken all together, the reduction in ELISA signal observed in sonication or extrusion methods can be attributed to both surface damage and particle loss.
When we used Dox-EVs obtained from the experiments using the same initial volume of CCS (Fig. S9A†), the TC method displayed 63.4 ± 1.0% cytotoxicity, while sonication showed 35.6 ± 2.5% and extrusion showed 30.0 ± 2.2% cytotoxicity. The difference is attributed to both surface damage and particle loss.
When equal particle concentrations of EVs obtained from different loading processes were administered to A549 cells, the TC method exhibited 64.5 ± 2.1% cytotoxicity, showing better results compared to sonication (44.6 ± 0.97%) and extrusion (42.5 ± 7.81%) (Fig. S9B†).
To test the effectiveness of EVs engineered using the TC method in delivering cargo to 3D cancer cell environments, we treated A549 cell spheroids with Dox-EV and an equal amount of unencapsulated Dox (Fig. 6E). Compared with the negative control (NC), in which only the same concentration of EVs without Dox was used, both the Dox and Dox-EV treatments reduced the spheroid size after 48 h of incubation; Dox-EV-treated spheroids showed better results than the Dox-treated spheroids (Fig. 6F). On average, the size of Dox-EV-treated spheroids decreased by 18.1 ± 3.0%, whereas that of the Dox-treated spheroids decreased by only 9.9 ± 1.5%, despite receiving the same amount of Dox.
When equal amounts of Dox and Dox-EV (3 μM) were administered, Dox-EV prepared using the TC method displayed a 2.2 times higher mean intensity within the spheroid (Fig. S10†) compared to unencapsulated Dox. These results indicate that Dox-EV produced via the TC method exhibits superior retention within the spheroid compared to the unencapsulated form of Dox. It suggests that the uptake of Dox encapsulated within EVs by the cells is notably more efficient than that of unencapsulated drugs. Additionally, it supports the idea that EVs possess enhanced particle retention within spheroids due to their cellular origin and inherent properties, consistent with previous reports.60,61
Conclusions
The study introduced an innovative method for loading cargo molecules into EVs using tonicity control-induced osmotic cycles. This method involves a controlled membrane permeabilization of EVs using a hypotonic solution, allowing the influx of molecules into EVs, followed by isotonic washing to restore the membrane integrity, while preserving the surface markers, concentration, and shape of the EVs. The loading amount of different cargo molecules, such as Dox, dextran, ssDNA, and miRNA-497, could be enhanced. Compared with conventional loading methods, such as sonication and extrusion, the TC method is more efficient in terms of particle retention and cargo delivery. Intracellular assessments of miRNA-497-loaded EVs and Dox-loaded EVs showed promising results, suggesting their potential for developing innovative EV-based therapeutic systems.Author contributions
The manuscript was written with the contributions from all the authors. All the authors approved the final version of the manuscript.Conflicts of interest
UNIST has filed patents on Exodisc, with YKC as an inventor, licensed to LabSpinner, Inc., where both JP and YKC are stockholders. The other authors declare no conflicts of interest.Acknowledgements
This work was supported by the Center for Soft and Living Matter, Institute for Basic Science [IBS-R020-D1] by the Korean Government, and the BK21 program through the National Research Foundation (NRF), funded by the Ministry of Education of Korea.References
- I. K. Herrmann, M. J. A. Wood and G. Fuhrmann, Nat. Nanotechnol., 2021, 16, 748–759 CrossRef CAS PubMed.
- S. E. L. Andaloussi, I. Mager, X. O. Breakefield and M. J. Wood, Nat. Rev. Drug Discovery, 2013, 12, 347–357 CrossRef PubMed.
- M. Mathieu, L. Martin-Jaular, G. Lavieu and C. Théry, Nat. Cell Biol., 2019, 21, 9–17 CrossRef CAS PubMed.
- R. Kalluri and V. S. LeBleu, Science, 2020, 367, eaau6977 CrossRef CAS PubMed.
- S. Kumar, M. Karmacharya, I. J. Michael, Y. Choi, J. Kim, I. Kim and Y.-K. Cho, Nat. Catal., 2021, 4, 763–774 CrossRef CAS.
- S. Kumar, M. Karmacharya and Y.-K. Cho, Small, 2023, 19, 2202962 CrossRef CAS PubMed.
- M. Karmacharya, S. Kumar and Y.-K. Cho, J. Funct. Biomater., 2023, 14, 117 CrossRef CAS PubMed.
- S. Kumar, J.-A. Han, I. J. Michael, D. Ki, V. Sunkara, J. Park, S. Gautam, H. K. Ha, L. Zhang and Y.-K. Cho, Adv. Funct. Mater., 2019, 29, 1902669 CrossRef.
- K. O'Brien, K. Breyne, S. Ughetto, L. C. Laurent and X. O. Breakefield, Nat. Rev. Mol. Cell Biol., 2020, 21, 585–606 CrossRef.
- R. Xu, A. Rai, M. Chen, W. Suwakulsiri, D. W. Greening and R. J. Simpson, Nat. Rev. Clin. Oncol., 2018, 15, 617–638 CrossRef CAS PubMed.
- I. Keklikoglou, C. Cianciaruso, E. Güç, M. L. Squadrito, L. M. Spring, S. Tazzyman, L. Lambein, A. Poissonnier, G. B. Ferraro, C. Baer, A. Cassará, A. Guichard, M. L. Iruela-Arispe, C. E. Lewis, L. M. Coussens, A. Bardia, R. K. Jain, J. W. Pollard and M. De Palma, Nat. Cell Biol., 2019, 21, 190–202 CrossRef CAS PubMed.
- S. El Andaloussi, I. Mäger, X. O. Breakefield and M. J. A. Wood, Nat. Rev. Drug Discovery, 2013, 12, 347–357 CrossRef CAS PubMed.
- C. Li, S. Qin, Y. Wen, W. Zhao, Y. Huang and J. Liu, J. Controlled Release, 2022, 349, 902–916 CrossRef CAS.
- S. Saeedi, S. Israel, C. Nagy and G. Turecki, Transl. Psychiatry, 2019, 9, 122 CrossRef.
- Y. Liang, Z. Iqbal, J. Lu, J. Wang, H. Zhang, X. Chen, L. Duan and J. Xia, Mol. Ther., 2023, 31, 1207–1224 CrossRef CAS.
- W. Tian, S. Liu and B. Li, BioMed Res. Int., 2019, 2019, 4649705 Search PubMed.
- J. Dai, Y. Su, S. Zhong, L. Cong, B. Liu, J. Yang, Y. Tao, Z. He, C. Chen and Y. Jiang, Signal Transduction Targeted Ther., 2020, 5, 145 CrossRef CAS.
- L. Zhang and D. Yu, Biochim. Biophys. Acta, Rev. Cancer, 2019, 1871, 455–468 CrossRef CAS.
- L. A. Mulcahy, R. C. Pink and D. R. Carter, J. Extracell. Vesicles, 2014, 3, 24641 CrossRef.
- A. Nasiri Kenari, L. Cheng and A. F. Hill, Methods, 2020, 177, 103–113 CrossRef CAS.
- G. van Niel, G. D'Angelo and G. Raposo, Nat. Rev. Mol. Cell Biol., 2018, 19, 213–228 CrossRef CAS.
- M. Mathieu, L. Martin-Jaular, G. Lavieu and C. Thery, Nat. Cell Biol., 2019, 21, 9–17 CrossRef CAS PubMed.
- T. Yong, Z. Wei, L. Gan and X. Yang, Adv. Mater., 2022, 34, e2201054 CrossRef PubMed.
- L. Cheng and A. F. Hill, Nat. Rev. Drug Discovery, 2022, 21, 379–399 CrossRef CAS PubMed.
- R. Tenchov, J. M. Sasso, X. Wang, W. S. Liaw, C. A. Chen and Q. A. Zhou, ACS Nano, 2022, 16, 17802–17846 CrossRef CAS PubMed.
- M. Zhang, X. Zang, M. Wang, Z. Li, M. Qiao, H. Hu and D. Chen, J. Mater. Chem. B, 2019, 7, 2421–2433 RSC.
- M. J. Haney, N. L. Klyachko, Y. Zhao, R. Gupta, E. G. Plotnikova, Z. He, T. Patel, A. Piroyan, M. Sokolsky, A. V. Kabanov and E. V. Batrakova, J. Controlled Release, 2015, 207, 18–30 CrossRef CAS PubMed.
- K. O'Brien, S. Ughetto, S. Mahjoum, A. V. Nair and X. O. Breakefield, Cell Rep., 2022, 39, 110651 CrossRef.
- L. Cheng and A. F. Hill, Nat. Rev. Drug Discovery, 2022, 21, 379–399 CrossRef CAS PubMed.
- S. Le Saux, H. Aarrass, J. Lai-Kee-Him, P. Bron, J. Armengaud, G. Miotello, J. Bertrand-Michel, E. Dubois, S. George, O. Faklaris, J. M. Devoisselle, P. Legrand, J. Chopineau and M. Morille, Biomaterials, 2020, 231, 119675 CrossRef CAS PubMed.
- K. B. Johnsen, J. M. Gudbergsson, M. N. Skov, G. Christiansen, L. Gurevich, T. Moos and M. J. C. Duroux, Cytotechnology, 2016, 68, 2125–2138 CrossRef CAS PubMed.
- J. Roerig and M. Schulz-Siegmund, Small, 2023, e2301763, DOI:10.1002/smll.202301763.
- L. Zhao, C. Gu, Y. Gan, L. Shao, H. Chen and H. Zhu, J. Controlled Release, 2020, 318, 1–15 CrossRef CAS PubMed.
- Y. Yang, Y. Hong, E. Cho, G. B. Kim and I. S. Kim, J. Extracell. Vesicles, 2018, 7, 1440131 CrossRef PubMed.
- F. Susa, T. Limongi, B. Dumontel, V. Vighetto and V. Cauda, Cancers, 2019, 11, 1979 CrossRef CAS PubMed.
- C. Chen, Y. Li, Q. Wang, N. Cai, L. Wu and X. Yan, Anal. Bioanal. Chem., 2023, 415, 1287–1298 CrossRef CAS PubMed.
- H. Ding, Y. Cai, L. Gao, M. Liang, B. Miao, H. Wu, Y. Liu, N. Xie, A. Tang, K. Fan, X. Yan and G. Nie, Nano Lett., 2019, 19, 203–209 CrossRef CAS PubMed.
- C.-S. Lee, J. Fan, H. S. Hwang, S. Kim, C. Chen, M. Kang, T. Aghaloo, A. W. James and M. Lee, Nano Lett., 2023, 23, 1202–1210 CrossRef CAS PubMed.
- K. B. Johnsen, J. M. Gudbergsson, M. N. Skov, G. Christiansen, L. Gurevich, T. Moos and M. Duroux, Cytotechnology, 2016, 68, 2125–2138 CrossRef CAS.
- D. S. S. M. Uppu, Y. Min, I. Kim, S. Kumar, J. Park and Y.-K. Cho, ACS Appl. Mater. Interfaces, 2021, 13, 29313–29324 CrossRef CAS.
- M. A. C. Pomatto, B. Bussolati, S. D'Antico, S. Ghiotto, C. Tetta, M. F. Brizzi and G. Camussi, Mol. Ther.--Methods Clin. Dev., 2019, 13, 133–144 CrossRef CAS.
- C. Chen, Y. Li, Q. Wang, N. Cai, L. Wu and X. Yan, Anal. Bioanal. Chem., 2023, 415, 1287–1298 CrossRef CAS.
- R. Hao, Z. Yu, J. Du, S. Hu, C. Yuan, H. Guo, Y. Zhang and H. Yang, Small, 2021, 17, e2102150 CrossRef.
- A. Jeyaram, T. N. Lamichhane, S. Wang, L. Zou, E. Dahal, S. M. Kronstadt, D. Levy, B. Parajuli, D. R. Knudsen, W. Chao and S. M. Jay, Mol. Ther., 2019, 28(3), 975–985 CrossRef PubMed.
- A. Jaiswal, C. H. Hoerth, A. M. Zuniga Pereira and H. Lorenz, Sci. Rep., 2019, 9, 12911 CrossRef PubMed.
- Y. H. Lee and C. A. Peng, Biochem. Biophys. Res. Commun., 2009, 390, 1367–1371 CrossRef CAS PubMed.
- I. Khmelinskii and V. I. Makarov, Sci. Rep., 2020, 10, 8395 CrossRef CAS PubMed.
- J. L. Lemoine, R. Farley and L. Huang, Gene Ther., 2005, 12, 1275–1282 CrossRef CAS PubMed.
- N. Groulx, F. Boudreault, S. N. Orlov and R. Grygorczyk, J. Membr. Biol., 2006, 214, 43–56 CrossRef CAS PubMed.
- S. U. Alam Shibly, C. Ghatak, M. A. Sayem Karal, M. Moniruzzaman and M. Yamazaki, Biophys. J., 2016, 111, 2190–2201 CrossRef CAS PubMed.
- C.-M. Lin, D. T. Wu, H.-K. Tsao and Y.-J. Sheng, Soft Matter, 2012, 8(22), 6139–6150 RSC.
- D. Kumar, A. Gayen and M. Chandra, Lab Chip, 2023, 9, 2471–2481 CAS.
- C. C. Logisz and J. S. Hovis, Biochim. Biophys. Acta, 2005, 1717, 104–108 CrossRef CAS PubMed.
- H. K. Woo, V. Sunkara, J. Park, T. H. Kim, J. R. Han, C. J. Kim, H. I. Choi, Y. K. Kim and Y. K. Cho, ACS Nano, 2017, 11, 1360–1370 CrossRef CAS PubMed.
- H. Zeng, S. Guo, X. Ren, Z. Wu, S. Liu and X. Yao, Cells, 2023, 12(10), 1416 CrossRef CAS PubMed.
- H. Lee, H. Park, G. J. Noh and E. S. Lee, Carbohydr. Polym., 2018, 202, 323–333 CrossRef CAS.
- H. Y. Elmoazzen, J. A. Elliott and L. E. McGann, Biophys. J., 2009, 96, 2559–2571 CrossRef CAS.
- K. Jeong, Y. J. Yu, J. Y. You, W. J. Rhee and J. A. Kim, Lab Chip, 2020, 20, 548–557 RSC.
- Q. Huang, H. Li, X. Dai, D. Zhao, B. Guan and W. Xia, Mol. Med. Rep., 2019, 20, 3959–3967 CAS.
- Y. Jiang, Z. Lyu, B. Ralahy, J. Liu, T. Roussel, L. Ding, J. Tang, A. Kosta, S. Giorgio, R. Tomasini, X. J. Liang, N. Dusetti, J. Iovanna and L. Peng, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2215308120 CrossRef CAS.
- V. D. Nguyen, H. Y. Kim, Y. H. Choi, J. O. Park and E. Choi, Drug Delivery, 2022, 29, 2621–2631 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3lc00830d |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |