Open Access Article
Open Access ArticleA vibrating capillary for ultrasound rotation manipulation of zebrafish larvae†
Zhiyuan
Zhang
 a,
Yilin
Cao
a,
Sara
Caviglia
b,
Prajwal
Agrawal
a,
Yilin
Cao
a,
Sara
Caviglia
b,
Prajwal
Agrawal
 a,
Stephan C. F.
Neuhauss
b and
Daniel
Ahmed
*a
a,
Stephan C. F.
Neuhauss
b and
Daniel
Ahmed
*a
aAcoustic Robotics Systems Laboratory, Institute of Robotics and Intelligent Systems, Department of Mechanical and Process Engineering, ETH Zurich, Säumerstrasse 4, CH-8803 Zurich, Switzerland. E-mail: dahmed@ethz.ch
bNeuhauss Laboratory, Department of Molecular Life Sciences, University of Zurich, Winterthurerstrasse 190, CH-8057 Zurich, Switzerland
First published on 29th December 2023
Abstract
Multifunctional micromanipulation systems have garnered significant attention due to the growing interest in biological and medical research involving model organisms like zebrafish (Danio rerio). Here, we report a novel acoustofluidic rotational micromanipulation system that offers rapid trapping, high-speed rotation, multi-angle imaging, and 3D model reconstruction of zebrafish larvae. An ultrasound-activated oscillatory glass capillary is used to trap and rotate a zebrafish larva. Simulation and experimental results demonstrate that both the vibrating mode and geometric placement of the capillary contribute to the developed polarized vortices along the long axis of the capillary. Given its capacities for easy-to-operate, stable rotation, avoiding overheating, and high-throughput manipulation, our system poses the potential to accelerate zebrafish-directed biomedical research.
Introduction
Zebrafish exhibit significant physiological and genetic parallels to humans, sharing common functional features with us and other mammals.1–4 They have been pivotal in genetic screenings to identify mutant phenotypes mirroring human clinical conditions such as heart disease, various anemias, cancer, and neurological disorders. As such, zebrafish are also a crucial model for probing complex brain disorders. Zebrafish larval models serve as high-throughput and cost-effective alternatives in drug screening processes and are frequently used to elucidate the molecular mechanisms that underlie human diseases and identify treatments.5–10Rotational manipulation of zebrafish larvae is a crucial capability to extract valuable anatomical information and achieve multidimensional analysis. As zebrafish become more central to research, the shortcomings of traditional handling platforms are evident, especially for tasks requiring high-throughput manipulation. The current practices—using agarose/gel11,12 for mounting and fixation—are not only labor-intensive but also pose a risk of injury or death to the delicate larvae. Optical tweezers have been introduced to manipulate micro-objects13,14 and organs15,16 inside the body of a zebrafish larva, offering high precision but limited by a small operational area and shallow penetration depth, restricting their ability to handle the entire fish. Magnetic fields have also been utilized to rotate embryos by pre-injecting magnetic particles into the chorion, however, this method will inherently become ineffective when the embryos hatch from the chorion around two days post fertilization (dpf).17 Managing the rotation of the slender body of a larva remains a significant challenge. To address this challenge, numerous automated zebrafish screening platforms have been created;18–20 yet, these methods often restrict the larva to an enclosed space, hindering direct-contact and three-dimensional downstream treatments like micro-injection, mechanical analysis, and surface examination. Moreover, these platforms are inadequate to operate multiple zebrafish simultaneously.21–23 Therefore, developing a multifunctional manipulation and analysis system remains key to accelerating zebrafish-directed biomedical research.
In contrast, ultrasound is an attractive modality to manipulate micro-objects with distinct advantages of biocompatibility, deeper penetration, no requirement for special sample properties (conductivity, transparency, and more), and the ability to generate large forces.24–38 Currently, ultrasound-driven manipulation, in terms of trapping and rotating, has been achieved for microbeads,39–43 cells,44–50 and plant seeds,51,52 by using surface acoustic waves and microbubbles. However, most of these subjects have nearly spherical structures with good symmetry and are ∼5–100 μm in size; only a few work to date has been conducted on model organisms with slender bodies like C. elegans (∼600–1000 μm)53,54 and zebrafish (∼1–5 mm).55–58 Such methods are also strictly limited to rotational instability, bubble collapse, size mismatch, the requirement for a confined operation space, overheating from dissipated acoustic energy, complex instruments, time-consuming, and expensive fabrication. There is still a need for a biologically safe, easy-to-fabricate, easy-to-operate, and high-efficient manipulation method for small model organisms.
Here, we designed an acoustofluidic micromanipulation system for rapid trapping, high-speed rotation, multi-angle imaging, and 3D model reconstruction of zebrafish larvae. The vibrating capillary produces asymmetric acoustic forces and polarized streaming vortices to effectively trap and rotate the zebrafish larva. As a new mechanism, our zebrafish rotating method gives its capacities for ultrafast actuation responses, stable rotation, high-throughput manipulation, safe operation at moderate levels of acoustic pressure, avoiding overheating, easy-to-operate, and portable setup.
We then expanded our research to examine the flow dynamics within the open liquid channel, exploring the effects of varying capillary vibrations, capillary positioning, and the channel's cross-sectional geometry. Our experimental findings, alongside simulations, confirmed the underlying rotational mechanism and offered a deeper understanding of how channel shape influences flow patterns. By tuning the ultrasound excitation voltage, we demonstrated the precisely controlled manipulation of zebrafish larvae. Finally, to showcase its applicability in biological studies, we successfully executed rotational manipulation on fluorescently labeled zebrafish larvae enabling detailed cellular examination, and conducted 3D reconstructions to investigate their morphological characteristics. Given the importance of animal-on-chip technologies for studying small model animals, our method is expected to become a pivotal tool with diverse applications across the biological and engineering disciplines.
Rotational mechanism
Polarized vortices induced by a vibrating glass capillary
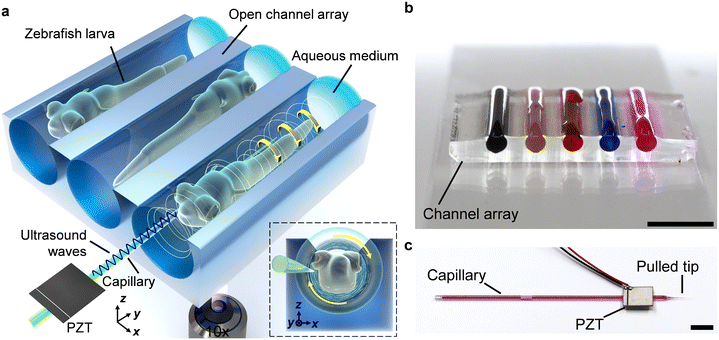
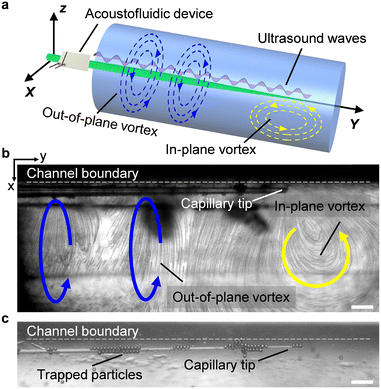
The acoustofluidic micromanipulation system consists of an acoustofluidic device and an open fluidic channel array, illustrated in Fig. 1 and Video S1.† For the acoustofluidic device, a piezoelectric transducer (PZT) is bonded on a glass capillary, which possesses a pre-pulled sharp tip (Fig. 1c). During experiments, the capillary is horizontally aligned and is positioned close to the channel boundary with its tip submerged into one of the cylindrical liquid channels (Fig. S1, ESI†). When the transducer is excited with a high-frequency square wave signal, ultrasound waves generate and propagate through the capillary, causing it to periodically vibrate. We recorded and analyzed the vibration amplitude of the capillary tip in both air and liquid (Fig. S2, ESI†). When keeping the ultrasound excitation voltage constant, the vibrating amplitude in liquid is weaker than that in air. Due to the acoustofluidic interaction, the vibrating capillary generates polarized acoustic streaming in the surrounding liquid that features the out-of-plane vortex along the long axis of the capillary and the in-plane vortex around the capillary tip, as shown in Fig. 2.Microparticles placed in an acoustofluidic field predominantly experience both the acoustic radiation force (ARF) and streaming force.59 Based on the gradient of the Gor'kov potential,60 the time-averaged ARF acting on a single isolated incompressible particle can be expressed as
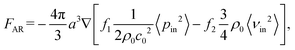 | (1) |
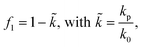 | (2) |
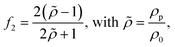 | (3) |
The force arising from acoustic streaming can be estimated by the Stokes' drag
| FAS = 6πηau, | (4) |
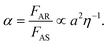 | (5) |
Cross-section flow profile analysis
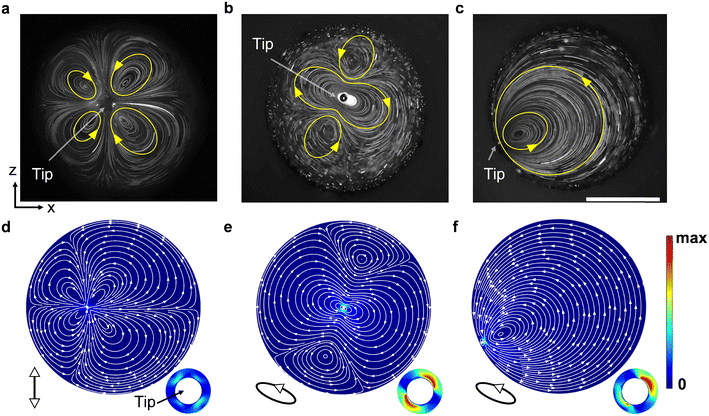
To gain further insights into the rotational mechanism, we visualized the flow profile through the cross-sectional perspective, (i.e., from the xz plane). The acoustofluidic device was held vertically and the tip of the capillary was submerged into the liquid in a cylindrical channel with a consistent inner diameter (1.14 mm) as the open liquid channel used above. The flow profile was observed and recorded from beneath via an inverted microscope, as shown in Fig. 3. We then analyzed the generated acoustic streaming by ImageJ.61Fig. 4 shows that when the capillary is excited with different ultrasound excitation frequencies or placed at different positions in the channel, various flow profiles can be developed in the channel. Firstly, as the tip of the capillary was sited near the channel center, we observed two different flow profiles. When the ultrasound excitation frequency was 240 kHz (the natural frequency of the utilized transducer), four in-plane vortices were generated around the tip (Fig. 4a and Video S3†); on the other hand, at 185 kHz, three vortices were observed, with one larger vortex rotating around the tip (Fig. 4b). Accordingly, we hypothesize that the variable flow profile is due to the distinct vibrating modes of the capillary tip induced by different ultrasound excitation frequencies: linear vibration in the normal direction and elliptical vibration. Based on theories described in ref. 48 different vibrating modes are caused by complex vibrational coupling between the transducer and capillary. When the exciting frequency of the transducer is far from the natural frequency of the capillary, the transducer prompts the glass capillary to vibrate along the normal direction, and the entire device is linear vibration. However, if the frequency is close to the capillary's natural frequency, the resonance of the entire acoustofluidic device increases significantly, resulting in an elliptical vibration with the oscillation amplitude in the normal direction greater than that in the tangential direction.
To verify the hypothesis, we developed a numerical simulation model to estimate the natural frequency of the utilized glass capillary (see Numerical simulation for configuration details). Simulations of a 50 mm-length capillary without pulling show that its first-order to eight-order natural frequencies are in the range of 453.65 Hz to 52.331 kHz, which is totally out of the operational frequency range in this work. Thus, with the operation frequencies used in this work, the vibration of the bulk capillary body is negligible in simulations. Then, we simplified the model of the capillary tip without the bulky body (Fig. S3, ESI†). Simulations show that the used excitation frequency, 185 kHz, is close to the 5th-order natural frequency of the capillary tip, verifying the proposed hypothesis.
Next, by keeping the ultrasound excitation frequency at 185 kHz and moving the tip closer to the channel boundary, we observed that those vortices near the boundary decrease in size, and a single vortex becomes dominant across the whole channel (Fig. 4c). This phenomenon is attributable to the no-slip condition of the channel boundary induces asymmetric fluid body forces. We further developed a two-dimensional simulation model to verify this hypothesis (see Numerical simulation for configuration details). In the model, the diameter of the liquid domain was set as 1.14 mm and the tip of the capillary was defined as a circle with a diameter of 20 μm. Fig. 4d–f show that the simulated streamlines coincide with the experimental results. The simulations additionally show that by adjusting the direction of capillary vibration and the distance between the capillary and the boundary, a single polarized vortex in the channel can be generated with multiple vibrating modes (Fig. S4, ESI†), suggesting a good compliance.
Then, to optimize the channel design, we developed different channel types with various cross-section shapes in the simulation model, namely square-, triangle-, and droplet-shaped channels (with similar dimensions to experimental channels). We found that by placing the vibrating capillary near the channel boundary, a single polarized vortex can be developed in all the simulated channels (Fig. S5, ESI†). We further calculated the streaming velocity of the liquid in the middle of each channel along the z-axis. As depicted in Fig. S5e,† the streaming velocity starts from zero and rapidly increases to maximum over a short z-distance (0–∼3 μm). This is because the capillary boundary is also set as a no-slip wall. As the distance from the capillary increases, the streaming velocity decreases to nearly zero, with a large slope (i.e., over ∼3 μm to ∼150 μm). It is given that acoustic energy attenuates as propagation distance increases. However, the streaming velocity has a small but notable increase at an intermediate distance (∼150–∼200 μm), which is expected from the dampening effect of the bulk liquid. Finally, the streaming velocity decreases gradually to zero (~200–∼1100 μm). The streaming velocity gradient implies the asymmetrical body force in the liquid along the z-axis. As the channel with a circular cross-section featured streaming velocity at a stable amplitude over a large area and by considering the size of zebrafish larvae, the complexity of channel fabrication, and the observation view, we selected the circular cross-section to demonstrate the manipulation of zebrafish larvae.
Results and discussion
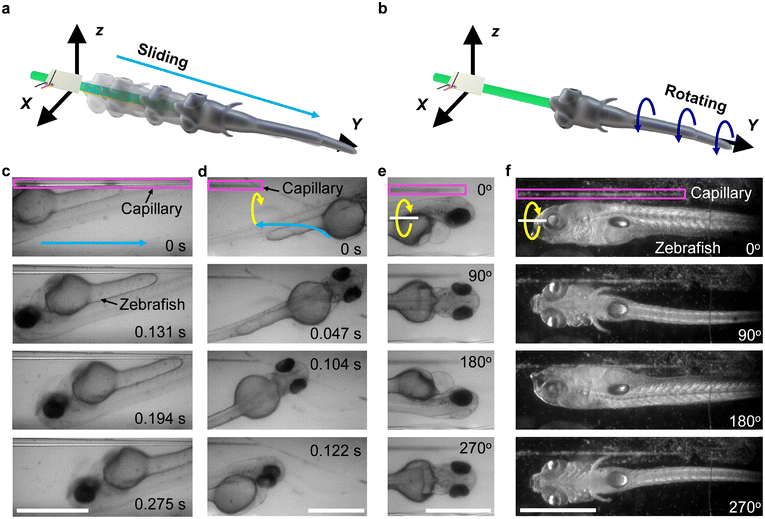
With the designed acoustofluidic manipulation system, we demonstrated the trapping and out-of-plane rotation on living zebrafish larvae, as illustrated in Fig. 5a and b. Firstly, a 4 dpf zebrafish larva anesthetized with sedatives was placed with a pipette into an open channel filled with deionized water. Upon exciting the transducer with the ultrasound excitation frequency of 185 kHz and excitation voltage of 12 VPP, the zebrafish larva was trapped around the capillary tip, as shown in Fig. 5c (where a zebrafish larva slides along the capillary to the tip) and Fig. 5d (where a larva initially in front of the capillary tip was trapped toward it) (Video S4†). The larva can be trapped in 0.1–1 s based on the applied ultrasound excitation voltage. During the process of trapping, the larva began out-of-plane rotation as demonstrated in Fig. 5e and Video S5.† Optical images of the larva from multiple view angles can thus be readily captured.This finding suggests that both the vortices in the liquid (shown in Fig. 2b) contribute to the trapping and rotation of the zebrafish larva; to be specific, the in-plane vortex traps the larva while the out-of-plane vortex rotates it. The calculation of acoustic forces acting on a zebrafish larva is discussed in the ESI† Note. We further demonstrated that an 8 dpf zebrafish larva, which has a bigger swimming bladder and is larger than the 4 dpf larva, was also continuously rotated (Fig. 5f). More than 40 zebrafish larvae of 2–10 dpf with lengths of 700–1200 μm and diameters of 200–300 μm were used to test the stability and repeatability of the system. They were arbitrarily placed into the microchannel and all of them showed stable rotation independent of the orientation, size, and age.
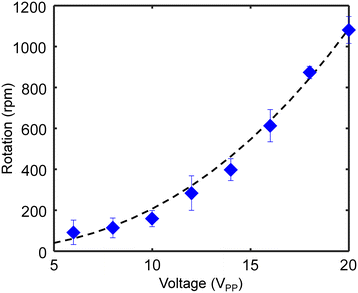
In the experimental domain, the highest Reynolds number was achieved around the capillary tip and was estimated as 50.16 based on Re = ρvsD/μ, where ρ is the density of deionized water, 1000 kg m−3; D is the characteristic diameter of the channel, 1140 μm; vs is the highest measured velocity of the fluid, 44 mm s−1; and μ is the fluid viscosity, 10−3 Pa s. The flow of the rotational system can be assumed to be laminar. We observed that in this low Reynolds number regime, the rotating zebrafish larva comes to a stop instantaneously when the ultrasound field is switched off. Based on ref. 51, the relationship between the drag torque T acting on the zebrafish, the acoustic pressure amplitude P, and the excitation voltage V is T ∝ P2 ∝ V2. Therefore, the rotational velocity of the zebrafish larva is tunable by changing the ultrasound excitation voltage applied to the transducer. As shown in Fig. 6, effective rotation can be performed at a low voltage, ∼5 VPP. The rotational velocity increases quadratically with the excitation voltage (where the power function fitting curve has a slope of 2.4); the highest rotational velocity achieved here was ∼1080 revolutions per minute (rpm) with an excitation voltage of 20 VPP and excitation frequency of 185 kHz. Notably, the rotational performance and efficiency are affected by multiple factors, including the diameter of the capillary tip, the position of the transducer, and the amount of liquid.
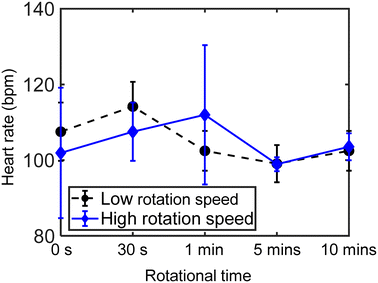
During experiments, we measured the temperature around the zebrafish larva. As depicted in Fig. S6,† the temperature of the liquid did not show a noticeable change before and during acoustofluidic actuation, which is conducive to avoiding overheating that may hurt the biological specimen. We also tested the viability of the rotated zebrafish larvae, as shown in Fig. 7. The larvae that were continuously rotated with a moderate rotational speed for 30 seconds, 1 minute, 5 minutes, and 10 minutes, respectively, had a similar heart rate (90–130 bpm) and blood flow to those grown in E3 medium, implying that the moderate ultrasound treatment and rotation did not harm them. In addition, however, if the ultrasound excitation voltage is ≥20 VPP, the lava is at risk of being crushed by large shear forces, as shown in Video S6.† In addition, we operated the zebrafish larvae without a sedative, and they can also be trapped and rotated (Video S6†), implying that a sedative is not necessary for the rotational manipulation. However, without a sedative, the larvae do have the ability and consciousness to escape from the vortex. We envision this phenomenon may open up possibilities for studying stress responses in zebrafish.
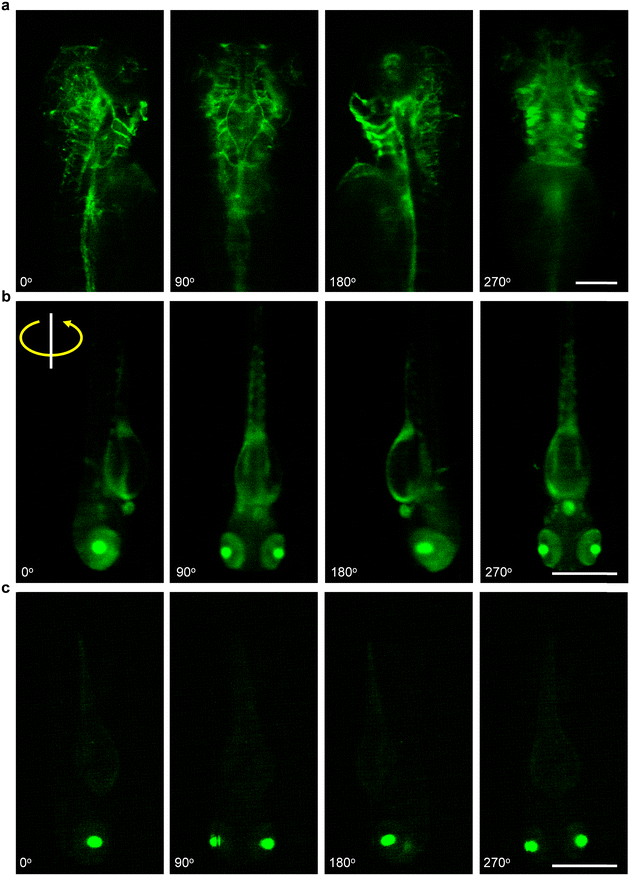
To further demonstrate the potential of our acoustofluidic device for zebrafish-based biological applications, we performed rotation manipulation on three types of fluorescently labeled zebrafish larvae (Video S7†). As shown in Fig. 8a, through rotating manipulation, we visualized the distribution of blood vessels through the kdrl:GFP transgenic zebrafish larva from multiple view angles. Fig. 8b shows the prox1a > Lyn-Citrine; cmlc2:GFP transgenic zebrafish larva with its heart, eyes, brain, and spinal cord fluorescently labeled. During rotation, we can observe the entire profile of the heart and its beating. Notably, also distinct neuronal subpopulations in the retina are visualizable. Fig. 8c shows the rotational image sequences of the fluorescently labeled eyes of an alpha-crystallin:YFP transgenic zebrafish larva. We further studied the 3D distribution of the brain endothelial cells and distinct neurons of the kdrl:GFP zebrafish larva. As shown in Fig. 9, the larva was precisely rotated to the target angles in turn and at each pose, we adjusted the focus of the microscope from bottom to up to observe the intricate vasculature distribution in the zebrafish brain slice by slice. We believe our method is promising and useful for zebrafish morphological studies, organ analysis, tissue investigations, and interrogations at the cellular level.
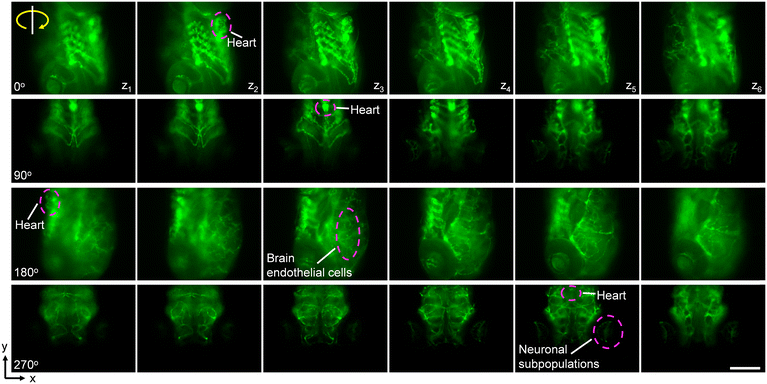 | ||
| Fig. 9 3D visualization of the fluorescently labeled blood vasculature and neurons. The label of zi denotes the focus plane of the microscope from bottom to up. Scale bar, 100 μm. | ||
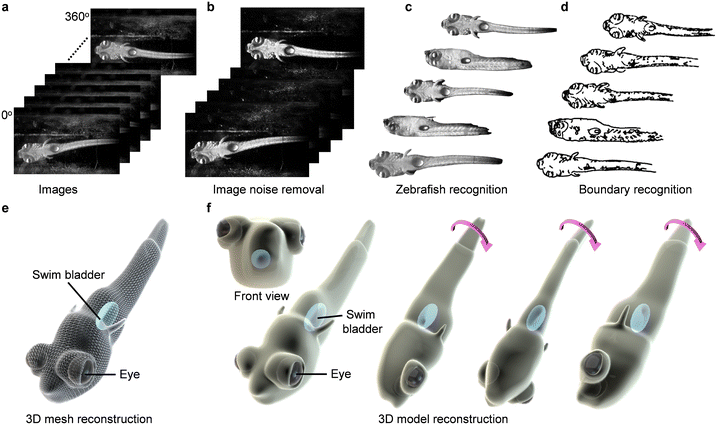
The controllable out-of-plane rotation of zebrafish larvae empowers exciting 3D reconstruction from the captured multi-view 2D image sequences. We first collected images of the zebrafish larva from different view angles during several rotation cycles. Imaging preprocesses, including removing noise, enhancing contrast, and adjusting brightness and sharpness were performed to improve the image quality. Subsequently, a feature-based alignment algorithm62,63 was used to align images by identifying common features, such as corners and edges. The aligned images are then used to reconstruct a point cloud of the larva using a process called stereo matching.64 The point cloud is further converted into a mesh by connecting the points in a triangular pattern. By averaging the positions of the points in the mesh, the mesh is smoothed to remove sharp edges or artifacts. A hole-filling process is then used to fill the holes in the mesh.65Fig. 10 shows such an example rendering from the 3D mesh with a visualized ellipsoidal swim bladder inside the larva (Video S8†). 3D reconstruction of other internal organs (heart, liver, bone, etc.) of zebrafish larvae is challenging and suffers from problems such as being off the axis of rotation, small size, occluded by other objects, and out of focus at certain viewing angles. Increasing the camera's frame rate, reducing the rotation speed, and optimizing geometric parameters of the channel can reveal more detailed information about the larvae.
Currently, confocal imaging and light-sheet imaging can perform 3D imaging and 3D reconstruction with a high resolution. However, the focus range of confocal imaging is narrow, which makes it difficult to scan the whole structure with a single view and requires a long operation time. Light-sheet imaging is much faster than confocal imaging by using a plane of light instead of a point. But this method is also difficult for high-throughput imaging. The 3D reconstruction through these machines could also have imaging distortion, for example, a sphere may be reconstructed as an ellipsoid. These machines are hard to integrate into other systems for comprehensive analysis. Additionally, these machines are expensive and not accessible to all labs. By contrast, our system offers the following advantages: (i) our platform allows fast 3D imaging of the whole zebrafish larva, which could be captured within milliseconds; (ii) our platform could enable high-throughput imaging by simultaneously rotating multiple zebrafish larvae; (iii) our system is miniaturized, portable, and adaptable for integration with other operation systems, for example, microfluidic devices and microinjection probes; (iv) our system has a high cost-effective. For a single chip, the cost is around 10 dollars.
Conclusions
An accurate system for positioning and rotating zebrafish larvae is a powerful and fundamental tool for further biomedical manipulation of this vertebrate model organism. In this work, we designed an on-chip acoustofluidic micromanipulation system. The acoustofluidic device induces asymmetrical forces into the surrounding liquid. By controlling the polarized vortices thus generated, we demonstrated fast trapping and high-speed rotation of zebrafish larvae around the capillary tip. Simulations of cross-sectional flow profiles were consistent with the experimental results, which verified the mechanism of the polarized vortex, and provided insights into the effects of channel geometry. Compared to existing methods, our acoustofluidic system provides a fast and inexpensive approach to manipulating and observing small living organisms without complex sample preparation and setup. The open channel allows further sophisticated contact manipulation. Since acoustic energy is not directly leaked into the liquid, overheating during operation is avoided. Most importantly, our portable manipulation system allows array expansion for high-throughput manipulation. It is expected that the sequence operation can be developed to operate larvae one by one; the channel and capillary can also be facilely copied to parallelly operate multiple larvae in one batch. A high rotational speed can accelerate the imaging and analysis process, which also has the potential for high-throughput manipulation. Additionally, it could become a new tool to capture the dynamic response of zebrafish larvae, for example, the blood flow, heart beating, drug metabolism, neuron response, and so on. With its superior performance and general applicability, we demonstrated the 3D visualization of fluorescently labeled neurons and vasculature distribution of zebrafish larvae. We further performed 3D reconstruction of the larvae, suggesting our acoustofluidic micromanipulation system will be a valuable tool for diverse studies in model organisms. In the future, we will explore more zebrafish-based biomedical applications of the system, including 3D mechanical characterization, surface detection, micro-injection, drug screening, Immune response, neural linking, and more; and optimize the channel properties for precise manipulation of small organisms on different scales.To characterize the resolution of our technique, we observed the blood cells (with a diameter of ∼8 μm) of the rotating zebrafish. As shown in Fig. S7,† with the 10× objective, a high-speed camera (3030 frames per second), and a lower rotational velocity, 0–100 rpm, the blood cells in the larva's vessels are clearly observed without a fluorescent label. Based on the experiment results, we believe our technique is valuable for cell-level zebrafish studies. The imaging resolution could be further improved using ultra-high-speed and ultra-high-sensitive cameras.
Materials and method
Device design and fabrication
For the acoustofluidic device, glass capillaries with a diameter of 1.14 mm were first pulled using a micropipette puller. The tip of the pulled capillary is then broken manually with a diameter of 20 ± 8 μm for a stable flow field. A rectangular piezoelectric transducer with dimensions 7.0 × 8.0 × 0.2 mm (Steminc, USA) was bonded using a two-component epoxy (UHU Plus Sofortfest) to the pulled capillary at 5 mm from the end of the tip. To produce the open channel, polydimethylsiloxane (PDMS) resin (mixed with a curing agent at a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio) was poured into a chassis that was 3D printed out of black photopolymer resin (Form3, FormLabs), and then a glass capillary was submerged ∼80 percent into the mixture from the top. After curing, the PDMS substrate was lifted away. The cross-sectional area of the microchannel is 80% of a circle with a diameter of 1.14 mm, and the channel length is 8 mm. For rotation operations, the PDMS channel was first bonded onto a glass slide by oxygen plasma treatment. Then, to ensure precise positioning of the capillary in the channel, a 3D-printed casing was used to mount the acoustofluidic device on the glass slide. The casing allows the fast replacement of the capillary between channels. Channels for the cross-section flow profile study were fabricated by the same method. During experiments, the transducer is excited with a high-frequency square wave signal generated by a function generator (0–25 MHz, 0–10 Vpp, Tektronix AFG1022, Tektronix, Inc., USA) and an amplifier (0–60 Vpp, 15× amplification, High Wave 3.2v, Digitum-Elektronik).
1 ratio) was poured into a chassis that was 3D printed out of black photopolymer resin (Form3, FormLabs), and then a glass capillary was submerged ∼80 percent into the mixture from the top. After curing, the PDMS substrate was lifted away. The cross-sectional area of the microchannel is 80% of a circle with a diameter of 1.14 mm, and the channel length is 8 mm. For rotation operations, the PDMS channel was first bonded onto a glass slide by oxygen plasma treatment. Then, to ensure precise positioning of the capillary in the channel, a 3D-printed casing was used to mount the acoustofluidic device on the glass slide. The casing allows the fast replacement of the capillary between channels. Channels for the cross-section flow profile study were fabricated by the same method. During experiments, the transducer is excited with a high-frequency square wave signal generated by a function generator (0–25 MHz, 0–10 Vpp, Tektronix AFG1022, Tektronix, Inc., USA) and an amplifier (0–60 Vpp, 15× amplification, High Wave 3.2v, Digitum-Elektronik).
Zebrafish larvae preparation
Zebrafish larvae (2–10 dpf) were used to show the rotation operation. Zebrafish strain were kept in a 14 h/10 h light/dark cycle under standard conditions. To avoid unwanted movement during experiments, zebrafish larvae were paralyzed with a sedative (3-amino-benzoic acid ethyl ester (MS-222, tricaine, MESAB; Sigma Aldrich) at 0.01% from pH 7.0 bicarbonate buffered stock solution) in an E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, and 0.33 mM MgSO4) at 26 °C. The following fluorescent lines were used: tg(kdrl:EGFP)s843,66TgBAC(prox1a:KALTA4, 4xUAS-E1B:TagRFP)nim5, Tg(5xUAS-E1B:LY-Citrine, cryaa:Citrine)nim23,67Tg(hsp70l:GFP-RAB11a, cmlc2:GFP)pd1031.68 Three day-old larvae were pre-screened for the presence of transgenes of interest using a fluorescence stereo-microscope (Olympus).Numerical simulation
Numerical simulations were conducted using the commercial COMSOL Multiphysics software (v5.6, Burlington, MA). For the natural frequency study, the material of the capillary tip is solid glass (a built-in material in COMSOL). The inner and outer diameters of the capillary are 1.14 mm and 0.2 mm, respectively. The length of the capillary tip is 8 mm, and the minimum diameter of the tip is 20 μm. For the cross-section flow profile study, we performed ultrasound vibration of the capillary with the “thermoviscous acoustics, frequency domain” physical field. The “laminar flow” physical field was used to denote the liquid domain. The “no-slip” condition was used to set boundaries. The physics-controlled mesh was utilized. Other parameters and configuration used in numerical simulations are listed in Table S1.†Imaging and data analysis
The rotation system was mounted on an inverted microscope (Axiovert 200 M, Zeiss). Videos were recorded with a high-speed camera (Chronos 1.4, Kron Technologies) and then analyzed in ImageJ.Live subject statement
All experiments were conducted in accordance with local authorities (Zürich Switzerland: Kantonales Veterinäramt TV4206).Author contributions
D. A. conceived and supervised the project. Z. Z. and Y. C. performed all the experiments, data analysis, and numerical simulation. S. N. and S. C. contributed to the cultivation and fluorescent label of zebrafish larvae. P. A. and Z. Z. performed the 3D reconstruction of zebrafish larvae. Z. Z., D. A., and S. N. contributed to the scientific discussion. All authors wrote the manuscript.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This project has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme grant agreement No. 853309 (SONOBOTS) and the Swiss National Science Foundation (SNSF) under the SNSF Project funding MINT 2022 No. 213058 and the SNSF Spark funding 2023 No. 221285. Z. Z. acknowledges funding from the China Scholarship Council (No. 202006210065). P. A. acknowledges funding from SNSF BRIDGE under the BRIDGE Proof of Concept Funding 2023 No. 218795. Z. Z., Y. C., P. A., and D. A. thank Adriana L. Hotz and Martin Walther for the zebrafish cultivation and preparation; Jan Durrer for the experimental setup; Mahmoud Medany, Guru Aathavan, Shaolin Kataria, and Shenyang Zhuang for the zebrafish 3D reconstruction.References
- M. B. Orger and G. G. De Polavieja, Annu. Rev. Neurosci., 2017, 40, 125 CrossRef CAS PubMed.
- R. Dahm and R. Geisler, Mar. Biotechnol., 2006, 8, 329 CrossRef CAS PubMed.
- K. Liu, C. Petree, T. Requena, P. Varshney and G. K. Varshney, Front. Cell Dev. Biol., 2019, 7, 1 CrossRef PubMed.
- H. W. Detrich, M. Westerfield and L. I. Zon, Methods Cell Biol., 1999, 59, 3 CrossRef CAS PubMed.
- C. A. MacRae and R. T. Peterson, Nat. Rev. Drug Discovery, 2015, 14, 721 CrossRef CAS PubMed.
- G. Kari, U. Rodeck and A. P. Dicker, Clin. Pharmacol. Ther., 2007, 82, 70 CrossRef CAS PubMed.
- H. Feitsma and E. Cuppen, Mol. Cancer Res., 2008, 6, 685 CrossRef CAS PubMed.
- D. G. Howe, Y. M. Bradford, A. Eagle, D. Fashena, K. Frazer, P. Kalita, P. Mani, R. Martin, S. T. Moxon, H. Paddock, C. Pich, S. Ramachandran, L. Ruzicka, K. Schaper, X. Shao, A. Singer, S. Toro, C. Van Slyke and M. Westerfield, Nucleic Acids Res., 2017, 45, D758 CrossRef CAS PubMed.
- E. E. Patton, L. I. Zon and D. M. Langenau, Nat. Rev. Drug Discovery, 2021, 20, 611 CrossRef CAS PubMed.
- S. Cassar, I. Adatto, J. L. Freeman, J. T. Gamse, I. Iturria, C. Lawrence, A. Muriana, R. T. Peterson, S. Van Cruchten and L. I. Zon, Chem. Res. Toxicol., 2020, 33, 95 Search PubMed.
- S. C. Baraban, M. T. Dinday, P. A. Castro, S. Chege, S. Guyenet and M. R. Taylor, Epilepsia, 2007, 48, 1151 CrossRef PubMed.
- Y. Geng and R. T. Peterson, Zebrafish, 2021, 18, 376 CrossRef CAS PubMed.
- L. Lin, X. Peng, X. Wei, Z. Mao, C. Xie and Y. Zheng, ACS Nano, 2017, 11, 3147 CrossRef CAS PubMed.
- J. Li, P. S. Kollipara, Y. Liu, K. Yao, Y. Liu and Y. Zheng, ACS Nano, 2022, 16, 6 Search PubMed.
- I. A. Favre-Bulle, A. B. Stilgoe, H. Rubinsztein-Dunlop and E. K. Scott, Nat. Commun., 2017, 8, 630 CrossRef PubMed.
- P. Yu, Y. Liu, Q. Zhao, Z. Wang, Y. M. Li and L. Gong, Appl. Phys. Express, 2020, 13, 032008 CrossRef CAS.
- F. Berndt, G. Shah, R. M. Power, J. Brugués and J. Huisken, Nat. Commun., 2018, 9, 1 CrossRef CAS PubMed.
- M. F. Yanik, C. B. Rohde and C. Pardo-Martin, Annu. Rev. Biomed. Eng., 2011, 13, 1 CrossRef PubMed.
- C. Pardo-Martin, T. Y. Chang, B. K. Koo, C. L. Gilleland, S. C. Wasserman and M. F. Yanik, Nat. Methods, 2010, 7, 634 CrossRef CAS PubMed.
- Y. Guo, W. J. Veneman, H. P. Spaink and F. J. Verbeek, Biomed. Opt. Express, 2017, 8, 2611 CrossRef PubMed.
- S. G. Hong, P. Lee, S. C. Baraban and L. P. Lee, Sci. Rep., 2016, 6, 1 CrossRef PubMed.
- F. Yang, Z. Chen, J. Pan, X. Li, J. Feng and H. Yang, Biomicrofluidics, 2011, 5, 024115 CrossRef PubMed.
- N. M. Fuad, J. Kaslin and D. Wlodkowic, Biomicrofluidics, 2017, 11, 05110 CrossRef.
- V. M. Jooss, J. S. Bolten, J. Huwyler and D. Ahmed, Sci. Adv., 2022, 8, 1 Search PubMed.
- P. Agrawal, Z. Zhang, Z. Ghorbanikharaji, Z. Shi and D. Ahmed, Acoustic field techniques for cell characterization in health monitoring, Elsevier Inc., 2023 Search PubMed.
- J. Rufo, F. Cai, J. Friend, M. Wiklund and T. J. Huang, Nat. Rev. Methods Primers, 2022, 2, 1 CrossRef.
- A. Ozcelik, J. Rufo, F. Guo, Y. Gu, P. Li, J. Lata and T. J. Huang, Nat. Methods, 2018, 15, 1021 CrossRef CAS PubMed.
- C. Dillinger, N. Nama and D. Ahmed, Nat. Commun., 2021, 12, 1–11 CrossRef PubMed.
- Z. Zhang, Z. Shi and D. Ahmed, arXiv, 2023, preprint, DOI:10.48550/arXiv.2307.05754.
- Y. Deng, A. Paskert, Z. Zhang, R. Wittkowski and D. Ahmed, Sci. Adv., 2023, 9, eadh526 Search PubMed.
- A. D. C. Fonseca, C. Glück, J. Droux, Y. Ferry, C. Frei, S. Wegener, B. Weber, M. E. Amki and D. Ahmed, Nat. Commun., 2023, 14, 5889 CrossRef PubMed.
- Z. Zhang, A. Sukhov, J. Harting, P. Malgaretti and D. Ahmed, Nat. Commun., 2022, 13, 7347 CrossRef PubMed.
- J. Janiak, Y. Li, Y. Ferry, A. A. Doinikov and D. Ahmed, Nat. Commun., 2023, 14, 4705 CrossRef CAS PubMed.
- J. Rufo, F. Cai, J. Friend, M. Wiklund and T. J. Huang, Nat. Rev. Methods Primers, 2022, 2, 30 CrossRef CAS.
- P. Zhang, H. Bachman, A. Ozcelik and T. J. Huang, Annu. Rev. Anal. Chem., 2020, 13, 17–43 CrossRef CAS PubMed.
- Z. Tian, S. Yang, P. Huang, Z. Wang, P. Zhang, Y. Gu, H. Bachman, C. Chen, M. Wu and T. J. Huang, Sci. Adv., 2019, 5, 5 Search PubMed.
- Q. Zhou, V. Sariola, K. Latifi and V. Liimatainen, Nat. Commun., 2016, 7, 12764 CrossRef CAS PubMed.
- A. Kopitca, K. Latifi and Q. Zhou, Sci. Adv., 2021, 7, 39 Search PubMed.
- X. Bai, S. Bin, D. Yuguo, Z. Wei, F. Yanmin, C. Yuanyuan, Z. Deyuan, A. Fumihito and F. Lin, Sens. Actuators, A, 2020, 315, 112340 CrossRef.
- L. Feng, B. Song, Y. Chen, S. Liang, Y. Dai, Q. Zhou, D. Chen, X. Bai, Y. Feng, Y. Jiang, D. Zhang and F. Arai, Biomicrofluidics, 2019, 13, 064103 CrossRef PubMed.
- E. Kim, M. Kojima, L. Xiaoming, T. Hattori, K. Kamiyama, Y. Mae and T. Arai, ROBOMECH J., 2017, 4, 5 CrossRef.
- Y. Li, X. Liu, Q. Huang, A. T. Ohta and T. Arai, Lab Chip, 2021, 21, 1016 RSC.
- X. Liu, Y. Li, L. Li, M. Kojima, Q. Shi, Q. Huang, T. Fukuda and T. Arai, IEEE/ASME Trans. Mechatron., 2022, 3164263 Search PubMed.
- A. Ozcelik, N. Nama, P. H. Huang, M. Kaynak, M. R. McReynolds, W. Hanna-Rose and T. J. Huang, Small, 2016, 12, 5120 CrossRef CAS PubMed.
- Y. Zhou, J. Liu, J. Yan, S. Guo and T. Li, Small, 2021, 17, 1 Search PubMed.
- Q. Tang, F. Liang, L. Huang, P. Zhao and W. Wang, Biomed. Microdevices, 2020, 22, 1 CrossRef PubMed.
- I. Bernard, A. A. Doinikov, P. Marmottant, D. Rabaud, C. Poulain and P. Thibault, Lab Chip, 2017, 17, 2470 RSC.
- X. Liu, Q. Shi, Y. Lin, M. Kojima, Y. Mae, T. Fukuda, Q. Huang and T. Arai, Small, 2019, 15, 1 CAS.
- C. Dai, Y. Sun, L. Xin, Z. Zhang, G. Shan, T. Wang, K. Zhang, X. Wang, L. T. Chu and C. Ru, IEEE Robot. Autom. Lett., 2020, 5, 339 Search PubMed.
- N. F. Läubli, M. S. Gerlt, A. Wüthrich, R. T. M. Lewis, N. Shamsudhin, U. Kutay, D. Ahmed, J. Dual and B. J. Nelson, Anal. Chem., 2021, 93, 9760 CrossRef PubMed.
- N. F. Läubli, N. Shamsudhin, H. Vogler, G. Munglani, U. Grossniklaus, D. Ahmed and B. J. Nelson, Small Methods, 2019, 3, 1 Search PubMed.
- N. F. Läubli, J. T. Burri, J. Marquard, H. Vogler, G. Mosca, N. Vertti-Quintero, N. Shamsudhin, A. deMello, U. Grossniklaus, D. Ahmed and B. J. Nelson, Nat. Commun., 2021, 12, 2583 CrossRef PubMed.
- J. Zhang, S. Yang, C. Chen, J. H. Hartman, P. H. Huang, L. Wang, Z. Tian, P. Zhang, D. Faulkenberry, J. N. Meyer and T. J. Huang, Lab Chip, 2019, 19, 984 RSC.
- D. Ahmed, A. Ozcelik, N. Bojanala, N. Nama, A. Upadhyay, Y. Chen, W. Hanna-Rose and T. J. Huang, Nat. Commun., 2016, 7, 11085 CrossRef CAS PubMed.
- J. Durrer, P. Agrawal, A. Ozgul, S. C. F. Neuhauss, N. Nama and D. Ahmed, Nat. Commun., 2022, 13, 6370 CrossRef CAS PubMed.
- C. Chen, Y. Gu, J. Philippe, P. Zhang, H. Bachman, J. Zhang, J. Mai, J. Rufo, J. F. Rawls, E. E. Davis, N. Katsanis and T. J. Huang, Nat. Commun., 2021, 12, 1118 CrossRef CAS PubMed.
- Z. Chen, X. Liu, X. Tang, Y. Li, D. Liu, Y. Li, Q. Huang and T. Arai, IEEE Robot. Autom. Lett., 2022, 7, 4 Search PubMed.
- Z. Zhang, L. K. Allegrini, N. Yanagisawa, Y. Deng, S. C. F. Neuhauss and D. Ahmed, IEEE Robot. Autom. Lett., 2023, 8, 5 Search PubMed.
- D. L. Miller, J. Acoust. Soc. Am., 1988, 84, 1378 CrossRef CAS PubMed.
- G. Go, J. Han, J. Zhen, S. Zheng, A. Yoo, M. J. Jeon, J. O. Park and S. Park, Adv. Healthcare Mater., 2017, 6, 1 Search PubMed.
- C. A. Schneider, W. S. Rasband and K. W. Eliceiri, Nat. Methods, 2012, 9, 671 CrossRef CAS PubMed.
- F. Liang, J. Zhu, H. Chai, Y. Feng, P. Zhao, S. Liu, Y. Yang, L. Lin, L. Cao and W. Wang, Small Methods, 2023, 7, 2201492 CrossRef CAS PubMed.
- C. A. Brassey and J. D. Gardiner, R. Soc. Open Sci., 2015, 2, 150302 CrossRef PubMed.
- L. Huang, Y. Feng, F. Liang, P. Zhao and W. Wang, Biomicrofluidics, 2021, 15, 014106 CrossRef CAS PubMed.
- L. Huang, P. Zhao and W. Wang, Lab Chip, 2018, 18, 2359 RSC.
- D. Beis, T. Bartman, S. W. Jin, I. C. Scott, L. A. D'Amico, E. A. Ober, H. Verkade, J. Frantsve, H. A. Field, A. Wehman, H. Baier, A. Tallafuss, L. Bally-Cuif, J. N. Chen, D. Y. R. Stainier and B. Jungblut, Development, 2005, 132, 4193–4204 CrossRef CAS PubMed.
- J. Cayuso, A. Dzementsei, J. C. Fischer, G. Karemore, S. Caviglia, J. Bartholdson, G. J. Wright and E. A. Ober, Dev. Cell, 2016, 39, 316–328 CrossRef CAS PubMed.
- A. L. Alvers, S. Ryan, P. J. Scherz, J. Huisken and M. Bagnat, Development, 2014, 141, 1110–1119 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3lc00817g |
| This journal is © The Royal Society of Chemistry 2024 |