Angio-Net: deep learning-based label-free detection and morphometric analysis of in vitro angiogenesis†
Suryong
Kim‡
a,
Jungseub
Lee‡
a,
Jihoon
Ko‡
 b,
Seonghyuk
Park
a,
Seung-Ryeol
Lee
a,
Youngtaek
Kim
b,
Seonghyuk
Park
a,
Seung-Ryeol
Lee
a,
Youngtaek
Kim
 a,
Taeseung
Lee
a,
Sunbeen
Choi
a,
Taeseung
Lee
a,
Sunbeen
Choi
 a,
Jiho
Kim
a,
Wonbae
Kim
a,
Yoojin
Chung
c,
Oh-Heum
Kwon
d and
Noo Li
Jeon
a,
Jiho
Kim
a,
Wonbae
Kim
a,
Yoojin
Chung
c,
Oh-Heum
Kwon
d and
Noo Li
Jeon
 *ae
*ae
aDepartment of Mechanical Engineering, Seoul National University, Seoul, 08826, Republic of Korea. E-mail: njeon@snu.ac.kr
bDepartment of BioNano Technology, Gachon University, Gyeonggi 13120, Republic of Korea
cDivision of Computer Engineering, Hankuk University of Foreign Studies, Yongin, 17035, Republic of Korea
dDepartment of IT convergence and Applications Engineering, Pukyong National University, Busan, 48513, Republic of Korea
eInstitute of Advanced Machines and Design, Seoul National University, Seoul, 08826, Republic of Korea
First published on 9th January 2024
Abstract
Despite significant advancements in three-dimensional (3D) cell culture technology and the acquisition of extensive data, there is an ongoing need for more effective and dependable data analysis methods. These concerns arise from the continued reliance on manual quantification techniques. In this study, we introduce a microphysiological system (MPS) that seamlessly integrates 3D cell culture to acquire large-scale imaging data and employs deep learning-based virtual staining for quantitative angiogenesis analysis. We utilize a standardized microfluidic device to obtain comprehensive angiogenesis data. Introducing Angio-Net, a novel solution that replaces conventional immunocytochemistry, we convert brightfield images into label-free virtual fluorescence images through the fusion of SegNet and cGAN. Moreover, we develop a tool capable of extracting morphological blood vessel features and automating their measurement, facilitating precise quantitative analysis. This integrated system proves to be invaluable for evaluating drug efficacy, including the assessment of anticancer drugs on targets such as the tumor microenvironment. Additionally, its unique ability to enable live cell imaging without the need for cell fixation promises to broaden the horizons of pharmaceutical and biological research. Our study pioneers a powerful approach to high-throughput angiogenesis analysis, marking a significant advancement in MPS.
Introduction
Advances in three-dimensional (3D) cell culture models have dramatically reshaped the landscape of in vitro research, providing spatiotemporal data on the human body microenvironment.1–5 Despite the wealth of data being generated by researchers, persistent concerns regarding the reliability and standardization of these datasets act as barriers to their seamless adoption into real-world preclinical and clinical applications.6–8Traditionally, the acquisition of image data from 3D cell culture models has been incorporated with labor-intensive processes such as immunofluorescence staining and subsequent microscopic imaging.9,10 This method not only imposes economic burdens but also necessitates cell fixation, imposing limitations on data collection. Moreover, the inefficiency of staining in thick tissues mandates the use of powerful lasers, introducing undesirable noise into the imaging process.
In navigating these challenges, the advent of machine learning techniques has spurred notable advancements in image analysis. This paradigm shift ushers in fresh opportunities to overcome the constraints inherent in traditional methodologies, presenting a more seamless and standardized approach to analyzing data derived from 3D cell culture models. Notably, recent strides in deep neural networks have found application in image enhancement for immunocytochemistry research and medical imaging. Techniques like convolutional neural networks (CNNs) and generative adversarial networks (GANs) have been deployed in diverse biomedical contexts,11,12 encompassing super-resolution microscopy,13,14 tumor segmentation in magnetic resonance (MR) images,15,16 and virtual histological staining.2,17,18 Moreover, these techniques apply 3D cell culture organ chip and 3D organoid such as OrganoID19 (a deep learning application for 3d organoid dynamics tracking and analysis), detection and tracking organoid dynamics,20 and evaluation of cell morphology21,22 and cell–cell interactions23,24 in MPS. Despite these strides, the broader integration of deep learning technology in the realm of 3D cell culture remains somewhat limited, partly due to the challenges associated with obtaining a substantial volume of high-quality images for effective training.25–28
In this research, we introduce Angio-Net, an innovative label-free fluorescence image reconstruction technique employing a segmentation architecture based on GANs. Using a large collection of high-quality images obtained from an advanced high-throughput microfluidic cell culture platform, we focus on demonstrating the virtual staining of blood vessels—an organ known for its complex structure in the human body. The proficient machine learning architecture, powered by a substantial dataset, skillfully converts brightfield images into synthetic fluorescence representations, highlighting its potential usefulness in various disease models and for evaluating the effectiveness of drugs.
Results
Integration of microphysiological system-based data acquisition with machine learning-based analysis
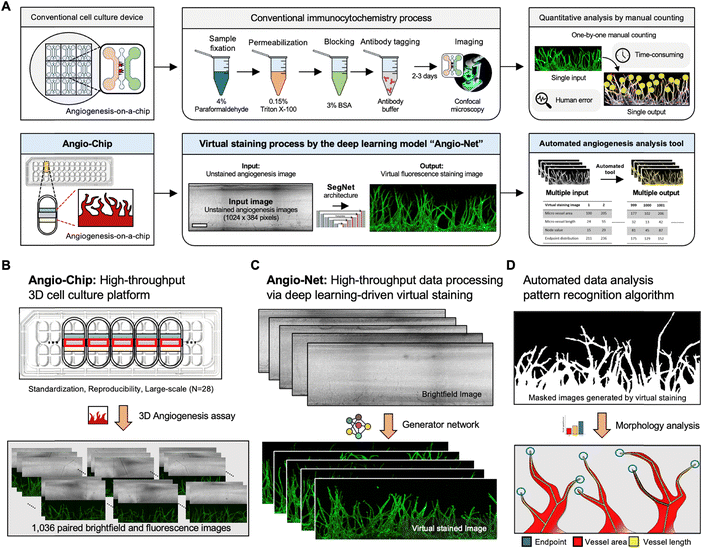
Conventional fluorescent staining process for 3D cell culture models typically encompasses several steps, such as cell fixation, membrane permeabilization, blocking, and fluorescent antibody tagging. Moreover, wide-field fluorescent microscopy faces limitations in detecting 3D tissue structures exceeding 100 mm in height. In contrast, our approach leverages a neural network architecture and a virtual fluorescent staining process, eliminating the need for conventional staining or confocal microscopy. Specifically, our deep learning model is designed to transform transmitted (brightfield) microscopy images into fluorescent images, accomplishing this process in a matter of milliseconds per image (as shown in Fig. 1A).Typically, machine learning-based image analysis necessitates hundreds of paired brightfield and fluorescence images, posing a challenge for acquisition. To address this, we developed Angio-Net, capable of achieving high-throughput experiments through deep learning-based virtual staining of large-scale data obtained from an injection-molded 3D cell culture platform (Angio-Chip). This single standardized microfluidic chip can accommodate up to 28 samples, offering the potential for expanded experimental capacity. Fig. 1B illustrates the internal structure of each well, comprising three microchannels: the center channel, lower channel, and upper channel. Briefly, we begin by injecting acellular fibrinogen solution into the central channel along with thrombin to initiate polymerization. Subsequently, in the upper channel, we introduce lung fibroblast, which secretes angiogenesis-inducing factors, and combine it with the fibrin polymer within the central channel. Finally, endothelial cells are introduced into the lower channel to encourage their adhesion to the fibrin polymer wall within the central channel. In conjunction with this configuration, the generation of hydrostatic pressure due to the volume difference in culture medium propels endothelial proliferation and migration, ultimately resulting in the observation of angiogenic sprouts (for more details, refer to the Materials and methods section).
We obtained angiogenesis image data from the Angio-Chip using confocal microscopy, resulting in 1036 paired, Z-stacked images (brightfield and fluorescent), each measuring 512 × 512 pixels. A single unit captures three images, which are stitched together to form a continuous image, resized to 1024 × 384 pixels for effective machine learning processing. The dataset was divided into training and test sets, encompassing 828 and 208 pairs, respectively.
Establishment in neural network architecture for image conversion
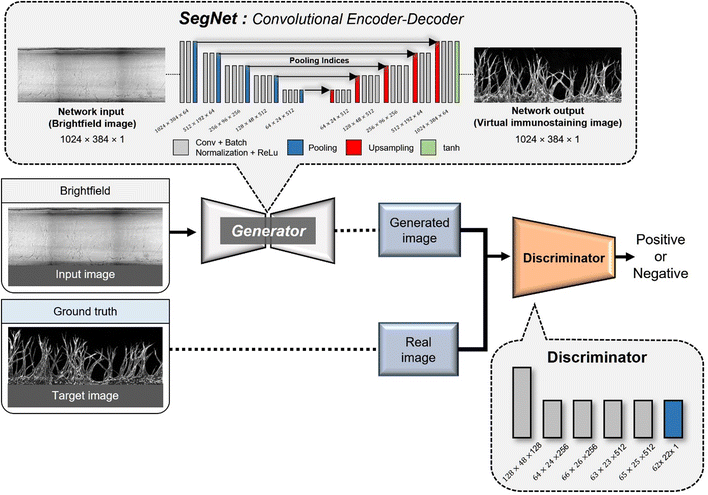
An encoder–decoder network represents a neural network architecture characterized by the symmetrical connection of an encoder and a decoder (Fig. 2). In this architecture, the encoder is tasked with encoding the input into a specific state, while the decoder generates the output based on this state. Encoder–decoder networks find extensive utility in image conversion tasks, where the input image is transformed into another image sharing the same fundamental structure. Typically, the encoder comprises multiple convolutional layers, mirroring the structure of the encoder. As the input image traverses the encoder's successive layers, it undergoes downsampling, which is later reversed during the decoding phase.In many image conversion scenarios, the input and output images share significant commonalities, necessitating the transmission of this shared information to the output layer via shortcuts in the network architecture. To achieve this, network designs that incorporate skip connections into the encoder–decoder network are used. Notable examples include U-Net and SegNet, both of which introduce skip connections that enable the output layer to receive both global information from the encoded state and local information from the skipped connection. In the case of U-Net, the output of a corresponding layer is relayed through the skip connection, while SegNet transmits index information from the max-pooling operation.
GANs are a neural network architecture grounded in Minmax game theory. It operates by simultaneously optimizing a generative model and an adversarial discriminative model to refine perceptual-level loss functions. GANs have found widespread application in medical image processing, demonstrating effectiveness in tasks such as image super-resolution reconstruction and brightfield holography. Conditional GAN (cGAN) extends the capabilities of the generative and discriminative models by incorporating additional information into the GAN framework. The objective function of cGAN can be expressed as follows:
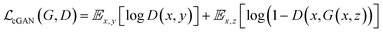 | (1) |
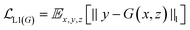 | (2) |
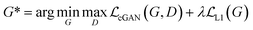 | (3) |
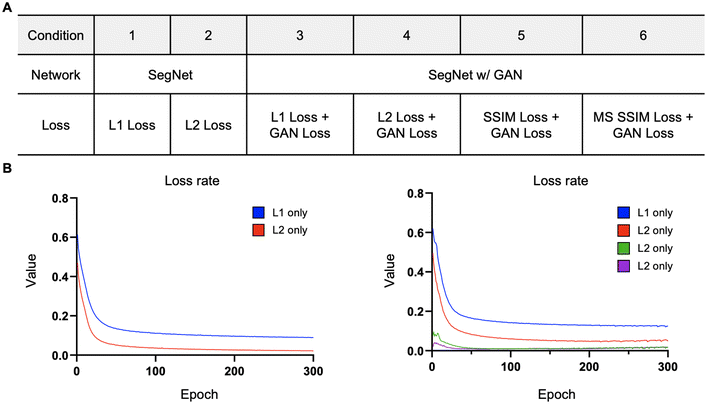
Assessing loss functions in generating virtual stained images from complex vascular networks
The loss function primarily consists of GAN loss (LcGAN), indicating the discriminator's loss, and image loss (Limage), which measures the disparity between the target and generated images. The selection of loss function plays a crucial role in neural network design. Traditional L2-norm, widely used, tends to produce blurry images. Pix2pix, opting for the L1 loss function, demonstrated sharper image outcomes compared to L2. Besides L1 and L2, metrics addressing perceptional image quality exist, such as structural similarity index (SSIM) and multiscale structural similarity index (MS-SSIM). SSIM gauges perceived image quality by assessing structural changes rather than absolute error like mean squared error (MSE) or peak signal-to-noise ratio (PSNR). As the human visual system excels at deriving structural information, SSIM calculates structural information, SSIM calculates structural differences by averaging them over constant-sized windows in the images. For windows x and y, SSIM is defined as follows: μx and μy are average pixel values, σ2x and σ2y are pixel value variances, σxy is covariance, and c1 and c2 are constants to prevent division by zero errors.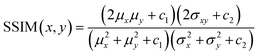 | (4) |
| L = wGAN × LGAN + wL1 × LL1 + wL2 × LL2 + wSSIM × LSSIM + wMS-SSIM × LMS-SSIM | (5) |
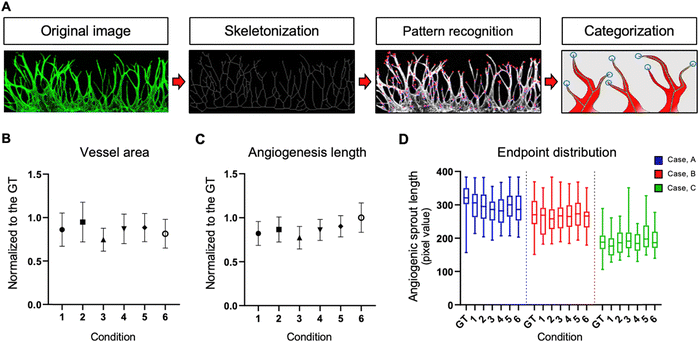
Comparative analysis of vascular morphology in virtual staining
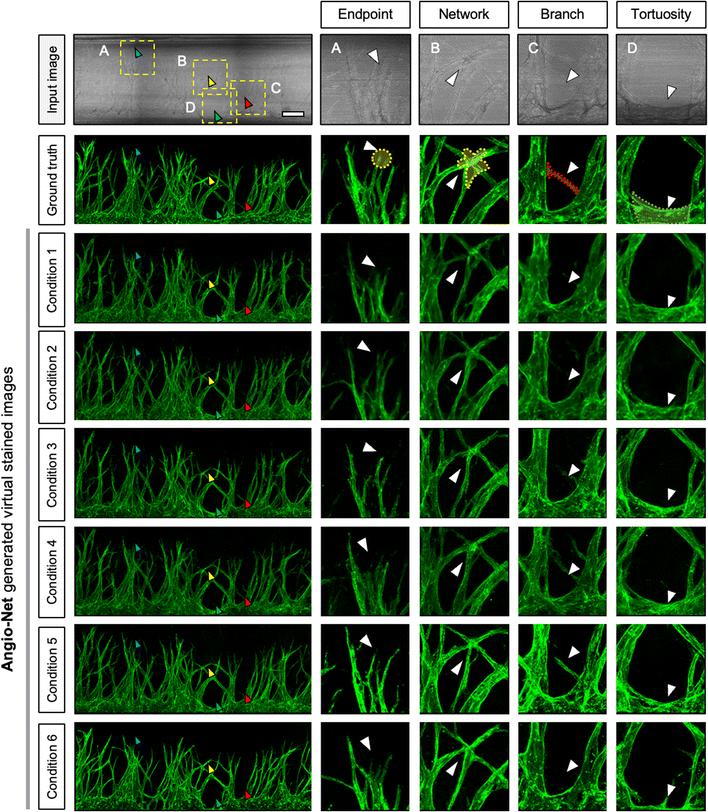
We present the distinctions between the input image, the ground truth (GT), and the virtual image under six loss conditions (Fig. 4). The image highlights four key regions—endpoint, network, branch, and tortuosity—revealing significant differences. Endpoint signifies the termination point of angiogenic sprouts (Fig. 4A), while network denotes the intersection point of vascular networks (Fig. 4B). Branch represents the line connecting two vessels (Fig. 4C), and tortuosity (Fig. 4D) is the primary part of the vascular networks.In certain instances, the number of endpoints in the virtual fluorescent images may appear less distinct than in the input image. This discrepancy arises because the endpoints in the input image are typically thinner than the main vessel. As endpoints traverse the network, information on their numbers diminishes, causing a deviation from the GT image, as indicated by the white arrow in Fig. 4A.
Despite these challenges, the virtual image successfully reproduces the morphology of blood vessels (Fig. 4B and C). While the virtual blood vessel is accurately rendered when the input image vessel is thick, details are lost when the vessel in the input is thin. Minimal error is observed in the main vessel, with only the minute vessel detail being lost, insignificantly affecting quantitative analysis. However, the area disappears entirely due to the neural network's struggle to learn adequately from the input image's brightness, resulting in a lower overall area value compared to the actual area (Fig. 4D).
Comparing virtual immunostaining images to GT images, macroscopic morphology remains similar, but discrepancies emerge at the local scale. Conditions 1 to 6 display virtual immunostaining images with varying levels of detail. Condition 1 and 2 present a general vascular morphology, but the vessel outlines and interiors appear blurred, with faint luminance discoloration. Background and endpoint details remain hazy, making clear distinction challenging. Condition 3 and 4 feature a brighter interior and a slightly more defined outline than condition 1 and 2, but endpoint blurring persists.
Condition 5 and 6, however, closely resemble the GT, exhibiting a distinct outline and a bright interior. Endpoints exhibit a well-defined morphology and are easily countable. Among the conditions, Condition 5 represents the optimal choice for loss, demonstrating vessel morphology nearly identical to the GT.
Developing an automated angiogenesis analysis algorithm based on virtual staining
Conventional 3D cell culture models offer detailed structural insights. However, challenges arise with thicker tissue samples, impacting staining efficiency. To overcome this limitation, high-powered lasers are often employed, but this can introduce noise, making it challenging to achieve noise-free results. That is where Angio-Net comes in as a solution. It allows for the visual refinement of objects through virtual staining, beginning with a refined brightfield image. In this study, 208 test images underwent processing, and a quantification algorithm was applied to determine the results (Fig. 5A). These results were then normalized against GT values. Fig. 5B to D illustrate the normalized distribution of virtual immunostaining images for each loss condition. The X-axis represents loss conditions (L1, L2, L1 + GAN, L2 + GAN, SSIM + GAN, and MS-SSIM + GAN), while the Y-axis represents normalized values, with 1.0 being the GT standard. The average area values for each condition ranges from 0.74 to 0.95, with values for all conditions ranging from 0.60 to 1.20 (Fig. 5B). Differences in area levels are attributed to variations in brightness between the internal area of the vessel and the GT (Fig. 4). The range of length values is from 0.65 to 1.18, with a mean ranging from 0.77 to 1.00 (Fig. 5C). Notably, the variation in endpoint values exceeds that of the other two parameters.To accurately assess the quantitative trends in virtual staining images, a novel reference point—the endpoint sprout distribution—was introduced. This metric served to quantify the actual degree of blood vessel growth, exhibiting relatively consistent values even with variations in the number of endpoints. The endpoint sprout distribution was classified into three cases—A, B, and C—based on sprout length and area size (Fig. 5D).
Case A, with an average endpoint sprout value of 318.55 for the GT, had the highest among the three cases. When normalized to GT, the average values for the six conditions ranged from 88.44 to 94.01%. Case B, with an average GT of 269.92, displayed a distribution across six conditions ranging from 95.21 to 101.48% when normalized to GT. Case C, with the lowest average endpoint sprout value of 188.44 for GT, exhibited a distribution across six conditions ranging from 94.10 to 110.23%. All images of cases A, B, and C with GT and under six conditions are presented in Fig. S2.† The results of the endpoint sprout inclination serve as a metric for comparing GT with other conditions, providing a quantified average sprout distribution value across the entire test set.
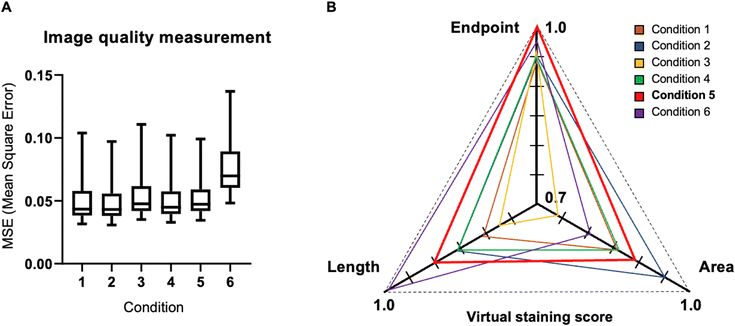
Quantitative evaluation of loss functions in virtual staining
The accuracy of loss functions was assessed through image quality metrics, employing L2 loss for each test set pair and comprehensive quantification data encompassing area, length, and endpoint distribution. MSE was adopted as the image quality evaluation metric, reflecting the difference between virtual images and GT images (Fig. 6A). The average MSE value was 0.049, 0.048, 0.053, 0.050, 0.052, and 0.072 across condition 1 to 6, with conditions 1 to 5 exhibiting similar MSE values between 0.048 to 0.052.Analyzing the average quantification data for area, length, and number, revealed that condition 5 demonstrated the highest accuracy. It achieved normalized values of 0.887, 0.903, and 1.015 for area, length, and number, respectively, closely approximating the GT value of 1.000. In contrast, condition 1 recorded values of 0.863, 0.822, and 0.945, while condition 2 exhibited values of 0.951, 0.867, and 0.959. Condition 3 reported values of 0.748, 0.774, and 0.975; condition 4 showed values of 0.872, 0.862, and 0.951; and condition 6 presented values of 0.816, 1.002, and 0.974. Generally, these values were lower compared to condition 5, signifying lower accuracy. Condition 5 emerged as the most accurate among the six conditions for area, length, and number measurements, boasting the highest values across all three components. Conversely, the other conditions generally exhibited lower values, indicating diminished accuracy.
Application of Angio-Net to evaluate anti-angiogenic drug efficacy
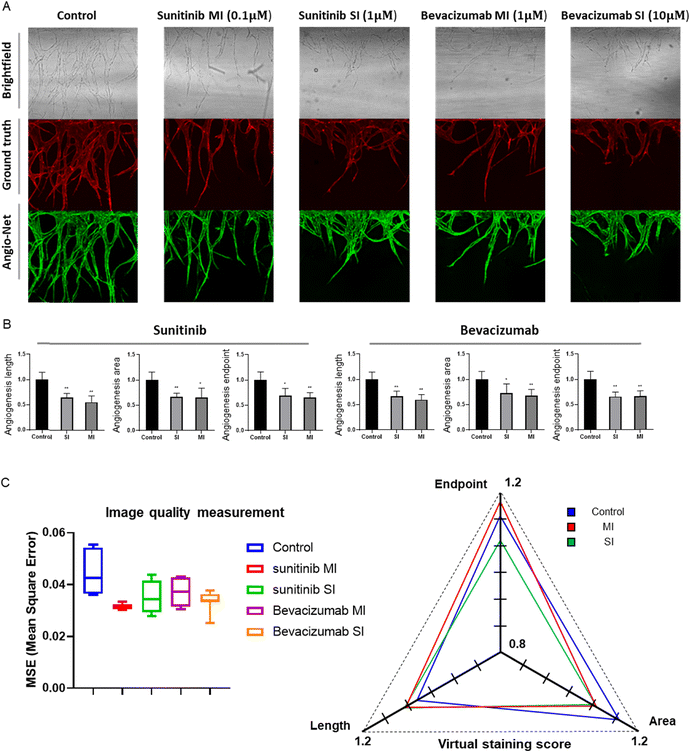
To expeditiously and accurately assess drug efficacy, we conducted an evaluation of angiogenesis inhibitors using the pre-trained Angio-Net system. Clinical-grade anti-angiogenic drugs, namely sunitinib (VEGF inhibitor) and bevacizumab (anti-VEGF monoclonal antibody), were administered to the Angio-Chip. We have only trained Angio-Net with vessels that had sprouted to 0.6 mm or larger. Our objective was to determine if we could accurately capture morphological features, even in vessels inhibited to 50–70% compared to drug negative controlled group by angiogenesis inhibiters, and observe morphological changes. For efficient evaluation of the reconstructed images, we categorized the drug-treated experimental groups into 1) the control group, 2) the moderate inhibition group (50–65%), and 3) the significant inhibition group (65–80%). Following reference guidelines, we selected drug concentrations of 0.1 μM and 1 μM (sunitinib) and 1 μM and 10 μM (bevacizumab) for ‘moderate inhibition group (MI)’ and ‘significant inhibition group (SI)’ and 0.1% DMSO for negative control group.A series of images displays brightfield, actual immunostained fluorescent, and virtually stained angiogenesis images from Angio-Net. While there is a remarkable resemblance in vascular patterns between the control and drug-treated groups, closer inspection reveals occasional undetected small vessels and false-positive features (Fig. 7A). A detailed evaluation of virtually stained images for each condition reveals significant differences compared to the control group, demonstrating Angio-Net's effectiveness in assessing anti-angiogenic drug effects on angiogenic sprouting with sunitinib and bevacizumab (Fig. 7B). In the quantification under sunitinib conditions, length values of approximately 0.75 and 0.55 relative to the control were observed, while the area had values of 0.74 and 0.65. Finally, the endpoint was found to have values of 0.78 and 0.65. It is evident that as the drug concentration increased, the quantitative values of angiogenesis consistently decreased. Similarly, in the quantification under bevacizumab conditions, length values of approximately 0.66 and 0.59 relative to the control were observed, while the area had values of 0.72 and 0.67. Finally, the endpoint was found to have values of 0.66 and 0.65. It is quantitatively confirmed that as the drug concentration increased, the quantitative values of angiogenesis consistently decreased or reached saturation. Therefore, when observing the differences in values according to the drug concentrations, we conclude that we have appropriately selected the experimental group originally intended to exhibit inhibition in the range of approximately 50% to 80%.
We virtually stained these inhibited vessels using Angio-Net and evaluated the mean squared error (MSE) and scores in the virtual staining (Fig. 7C). The left graph represents the results of calculating the MSE between virtual stained images and immunostained images, while the right graph displays the vessel feature values (length, endpoint, area) as ratios of virtual stained/immunostained values. Despite no further network training and the use of pre-trained weights for image transformation, it is evident that the MSE remains nearly identical across the control group, MI group, and SI Group. Additionally, in this Angio-Net scoring graph, while most values are slightly higher than 1 (ranging from 1.05 to 1.17), it demonstrates that there is no significant difference in scores between the control group and the inhibited group. Particularly noteworthy is the angiogenic length, which is found to be predictably accurate across all groups within 5% error rates.
In our comprehensive analysis, we extended the scope of our study to encompass a broader range of drug concentrations, thereby enabling a detailed investigation of the resultant morphological variations in angiogenesis images. This expanded analysis is illustrated in Fig. S3,† where we applied virtual staining techniques to brightfield images of blood vessels treated with sunitinib (at concentrations of 10 and 100 μM), wortmannin (50 μM), and Paclitaxel (50 μM). When comparing the GT images to the virtually stained images under high concentration conditions, such as those treated with high dose of sunitinib, the virtual stained images that were highly like the GT. Furthermore, even in more extreme scenarios where vascular structures were extensively disrupted by treatments like wortmannin and paclitaxel, virtual stained images were like the GT images (Fig. S3†). We anticipate improved accuracy with a extensive dataset encompassing images from various drug treatment groups.
Discussion
Advances in organoid and MPS have shown promise in shedding light on uncharted territory. While a wealth of information is being generated by researchers, issues such as reliability and standardization of this data still leave doubts over its adoption in clinical and preclinical domains.6 The data analysis process that 3D cell culture models undergo is prone to errors due to various factors that affect the image generation and analysis process. To address this issue, machine learning techniques are being applied to perform morphological analysis of cells or organoids with relatively simple structures.29 In this study, we successfully performed machine learning-based morphological analysis of complex blood vessels with tortuous and side branches using Angio-Chip, which develops robust and reproducible angiogenesis models, to generate large-scale image data, and Angio-Net, which trained on this data set. Through our image analysis, we assessed biochemical changes at the tissue level upon drug administration by analyzing vascular area, length, and tip cell count as evaluation metrics. Notably, several anti-angiogenic drugs including bevacizumab and sunitinib, employ a mechanism of action centered on inhibiting VEGFR activation, thereby disrupting endothelial cell proliferation, migration, and network formation. To decipher these intricate biochemical transformations, we evaluated cellular proliferation based on vascular area, quantified cell migration through tip cell count, and measured vascular length to comprehensively assess network formation.30–32When analyzed by conventional fluorescence microscopy, thicker samples reduce staining efficiency and rely on more intense lasers to achieve data acquisition, a process that introduces unnecessary noise and makes it difficult to acquire data in the desired area. Also, as shown in Fig. S4,† brightfield images itself pose challenges, featuring extraneous features such as shadows, channel traces, and migrated lung fibroblasts intermingled with blood vessels. The intricacies of distinguishing between vessels and artifacts underscore the necessity for a sophisticated discrimination method, emphasizing the pivotal role of advanced AI assistance. Virtual staining is a technique that can free us from these problems. By eliminating the need for immunocytochemistry, which requires several steps including cell fixation, samples can be preserved and examination time can be reduced. In addition, real-time analysis can potentially be performed by coloring the object as well as the brightfield. This includes automated analysis algorithms that measure the morphological characteristics of angiogenesis without human intervention, leading to significant time savings and error reduction by establishing standardized protocols.
Our study developed a high-throughput analysis process that uses widely adopted removal, skeletonization, and binarization techniques. The algorithm analyzes the number of endpoints, length, and area of angiogenesis, which are standard metrics for vascular morphology analysis. We validated the algorithm's results with image generated by deep learning. A deep learning architecture with six objective functions generated 208 pairs of test images. Qualitative evaluation showed that most images in the six conditions retained their overall morphology and branching structure, even though with some loss of detail compared to the GT images. The models without GAN loss weighting showed significant differences from the actual fluorescence images, while the models with GAN loss weighting were indistinguishable from the actual stained image, especially for the SSIM and MS-SSIM loss models (Fig. 4).
Traditional manual counting methods and commonly used angiogenesis assessment tools (e.g., AngioTool33,34) are labor-intensive and subjective, requiring different parameters for each image. In addition, these tools are also limited to planar 2D blood vessels, making it difficult to evaluate 3D blood vessel data. Angio-Net provides specialized analysis tools for 3D blood vessels, from imaging to quantification. The system's method can replace immunocytochemical processes, allowing for non-destructive real-time imaging, which is useful for high-throughput screening and creating visible microenvironments. Additionally, non-destructive real-time imaging enables end-users to continuously track the growth of 3D vascular networks while collecting quantitative data.
Angio-Net has demonstrated as an invaluable tool for rapidly validating the efficacy of anti-angiogenic drugs. However, when processing images with excessively high drug concentrations, there is a risk of inducing a “ghosting” effect,35 generating vessel-like contours that do not actually present. Previous studies on a similar platform36–38 revealed that sunitinib (0.1 μM and 1 μM) and bevacizumab (1 μM and 10 μM) resulted in a 30–70% reduction in tip cells, length, and area compared to control conditions. In Fig. 7C, the analysis of actual fluorescent staining images aligns with the anticipated 50% reduction. While the virtual staining data in this experiment show slightly increased values (10–15%) over immunostained data, it is crucial to note that the network's weights were not trained on drug experiment data but solely on normal blood vessels. The ghosting effect also count for increased end point and vessel area in our drug test experiments. This is attributed to the lack of diversity in the training data, and we anticipate that it can be controlled by further training with various drug-treated datasets. This result serves as evidence of its potential when applied to images of blood vessels under diverse conditions.
Recently, various research groups employing in vitro platforms have embraced the integration of AI for visual analysis and classification. However, the majority of these studies have been constrained to relatively simple structures, such as contour extraction of organoids or virtual staining limited to single cell level.25,39 Notably, there has been a lack of effective analytical tools for dissecting complex tissues, such as vascular networks or tumor microenvironments, using deep learning. Our contribution lies in pioneering the training of complex vascular tissue obtained through confocal microscopy. This work enables the transformation of brightfield images into virtual stained images. We implemented our network structure based on the well-established pix2pix, a conditional Generative Adversarial Network widely used and validated in conventional image transformations. To achieve fine pixel-level accuracy, we employed SegNet as the generator, aiming for an accessible research approach. Through this study, we anticipate opening a new avenue in the field of image transformation and analysis within Microphysiological Systems (MPS) using deep learning. This work lays the foundation for virtual staining in diverse tissues, holding the potential to contribute significantly to the advancement of MPS research. Furthermore, we have developed an algorithm for extracting quantitative morphological features from these transformed images. While the virtual stained image may not provide values as precise as the immunostained image, it serves as a valuable tool for decision-making and evaluation before immunostaining by presenting an image closely resembling the real one.
Conclusions
This study aimed to advance high-throughput analysis by introducing the innovative Angio-Net system. This system enables the acquisition of large-scale bright-field images of objects without requiring fluorescent staining. Leveraging deep learning-based virtual staining, these images are characterized, mimicking the process of traditional methods. The algorithmic measurement tools are optimized for automated morphological analysis, eliminating the need for manual intervention. The feasibility and efficacy of this approach have been successfully demonstrated, particularly in its application to the intricate structure of 3D blood vessels. The proposed model holds the potential to instill reliability and standardization in data analysis across various domains of cell culture research.Materials and methods
Design and fabrication of microfluidic devices
The Angio-Chip is designed with a well layout that aligns with the standard 384-well plate, featuring a 4.5 mm pitch between wells in the column and a 9 mm pitch in the row. Utilizing a common slide glass size of 3′′ by 1′′, each Angio-Chip accommodates 28 individual wells for cell culture. The chip's compatibility with automated machinery and microscopes facilitates high-throughput angiogenesis experiments. The well structure, as illustrated in Fig. 1C, comprises three distinct channels. These include the central channel, the lower channel, and the upper channel. The angiogenic sprouting process primarily occurs within the central channel, characterized by dimensions of 3 mm in width, 1 mm in length, and a depth of 100 μm. The Angio-Chip was produced using injection molding technique, specifically with polystyrene (PS) injection molding (R&D Factory, Korea). The aluminum alloy mold core underwent machining, which included processing and polishing. During the injection process, a clamping force of 130 tons was applied, with a maximum injection pressure of 55 bar, a cycle time of 15 s, and a nozzle temperature of 220 °C. The device was assembled by securely attaching a film substrate to the injection molded PS microfluidic body. The design of the alloy mold core was created using Solidworks software (Dassault System).Cell preparation
Human umbilical vein endothelial cells (HUVECs; Lonza, Switzerland) were cultured in endothelial growth medium 2 (EGM-2; Lonza) at passage numbers 4 to 5 for the experiments. Lung fibroblasts (LFs; Lonza) were cultured in fibroblast growth medium 2 (FGM-2; Lonza) at passage numbers 5 to 6 for the experiments. Cells were incubated at 37 °C with 5.0% CO2 for 2–3 days before seeding onto the Angio-Chip. Prior to seeding, HUVECs and LFs were detached from the culture dish using 0.25% trypsin–EDTA (HyClone, USA). The cells were subsequently re-suspended in bovine fibrinogen solutions at the concentrations tailored to each experimental model.Hydrogel and cell seeding
Prior to cell seeding, each device underwent a plasma surface treatment at 70 W for three minutes to promote surface hydrophilicity (Femto Science, Korea). In the central channel, 1 μl of acellular bovine fibrinogen solution (Sigma, USA) with a concentration of 2.5 mg ml−1 was introduced. The upper channel was filled with 3 μl of fibrinogen embedded with LFs at a cell concentration of 6.0 million cells per ml. The fibrinogen hydrogel in the central and upper channel was mixed with a 2.0% of bovine thrombin solution (0.5 U ml−1, Sigma) and allowed to undergo polymerization. Subsequently, after hydrogel polymerization, the lower channel was seeded with 3 μl of HUVECs diluted in the culture medium to a concentration of 3.0 million cells per ml. The devices, following cell seeding, were tilted until HUVECs were completely adhered to the central acellular fibrin hydrogel interface. Each media reservoir was filled with 100 μl of the growth medium after 15 minutes, and the growth medium was changed daily. To induce shear stress and interstitial flow, all medium from the lower reservoir was removed, and 100 μl of medium was added solely into the upper reservoir.40 In the angiogenesis inhibitor treatment experiments, we treated sunitinib at concentrations of 1 μM and 0.1 μM, and bevacizumab at 10 μM and 1 μM in 0.1% dimethyl sulfoxide (DMSO) solution respectively. All treatments were conducted on culture day 3, followed by fixation and staining on day 5.Immunocytochemistry
The samples within the device were fixed with 4.0% (w/v) paraformaldehyde (Biosesang, Korea) in PBS (Gibco, USA) for 15 minutes, followed by permeabilization through immersion in 0.15% Triton X-100 (Sigma) for 20 minutes. Subsequently, the samples were treated with 3.0% BSA (Sigma) for a hour. To achieve endothelial cell-specific staining, 488 fluorescein-labeled Ulex Europaeus agglutinin I (Vector, UK) was utilized, prepared at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 ratio of dye in BSA and incubated for 12 hours at 4.0 °C.
500 ratio of dye in BSA and incubated for 12 hours at 4.0 °C.
Imaging and data acquisition
Imaging was conducted with a confocal microscope (Nikon Ti-2, Japan) to capture both slice and Z-stackable images of angiogenesis, enabling the creation of paired brightfield and fluorescent images. For efficient data management and high-speed acquisition in a well-plate format, high-throughput imaging software (Nikon High Content NIS-Elements Package, Japan) was used. Subsequently, the confocal images were analyzed using Fiji (https://www.fiji.sc), an open-access software. The confocal 3D images were converted to 2D images through Z-projection and then cropped to a defined region of interest.Image preprocessing and quantification algorithm
To enhance data quantification accuracy, preprocessing of fluorescence angiogenesis images was necessary due to inherent noise caused by variation in brightness, contrast difference, and tiny particles. Fig. S1† depicts the complete image quantification process. The initial step involved noise reduction through image blurring, including methods such as averaging filtering, median filtering, and Gaussian filtering. Specifically, Gaussian filtering, implemented using the Python OpenCV library, was chosen as it effectively preserved the blood vessel contours' values, essential for maintaining the original blood vessel shape. Gaussian filtering primarily aimed to improve value uniformity in fluorescent angiogenesis images, which exhibited non-uniform values. Subsequently, an appropriate binary threshold was applied to isolate the actual vessel area, compensating value spread from blurring and removing low fluorescence noise. Even after Gaussian filtering and binary thresholding, the angiogenesis images still contained multiple black and white blobs due to value non-uniformity within the angiogenesis area and external factors.To address this, the OpenCV algorithm, with the Find-contour library, was utilized to remove small islands or particle blobs below a specified area threshold. Furthermore, a skeletonization algorithm was applied to extract a 1 pixel-wide skeleton from the binary blood vessel image.41 This angiogenesis skeleton image facilitated the determination of the total number of vessels and angiogenesis endpoints. To ensure accurate endpoint counting, our algorithm calculated the average distances of points from the baseline in the top 20% of endpoints, setting 50% of this average as the reference point for prioritizing the endpoints that higher growth rates. This analytical algorithm enabled the automated quantification of various parameter trends in angiogenesis images. The resulting quantified data were plotted using PRISM (GraphPad Prism 9).
Angio-Net network architecture
In Angio-Net, we utilize a network structure based on the pix2pix model to transform unstained images into their stained counterparts. Our network is configured with a SegNet architecture instead of U-Net. Both SegNet and U-Net share a common encoder–decoder structure comprising three distinct paths: (i) the first path, known as the contracting path, involves a series of convolutional layers and max-pooling layers. It serves the purpose of capturing the overall context information of the image while progressively reducing its size through downsampling; (ii) the second path is the expansion path, which mirrors the structure of the contracting path. It aims to up-sample the previously down-sampled image back to its original dimensions while traversing through the contracting path; (iii) the third path, known as the skip connection path and indicated by the red arrows connecting the contracting and expansive paths, facilitates the transfer of information from corresponding layers in the contracting path to the corresponding layer in the expansive path. This one-to-one correspondence between the layers ensures that local information is retained when performing upsampling.42,43The primary difference between U-Net and SegNet lies in the information conveyed through the connection path. In U-Net, the output of the corresponding layer in the downsampling path is combined with the output of the previous layer in the upsampling path through the connection path.43 In contrast, SegNet transfers the index information from the max-pooling operation performed in the corresponding layer of the downsampling path to the corresponding layer in the upsampling path. The upsampling in the expansive path is accomplished by inversely applying the index information.
We made minimal modifications to input and output layer sizes to accommodate our data. Furthermore, we leveraged the encoder layers of SegNet, which are consistent with the well-known VGG16 network, and performed transfer learning by applying the pre-trained weights from the VGG16 network.44 To serve as the activation function for the final layer, we employed the hyperbolic tangent function (tan![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) h). To enhance the discriminator's capabilities, we implemented the PatchedGAN technique. PatchedGAN involves dividing the image into smaller patches, evaluating the authenticity of each patch, and subsequently averaging the discriminant values.45
h). To enhance the discriminator's capabilities, we implemented the PatchedGAN technique. PatchedGAN involves dividing the image into smaller patches, evaluating the authenticity of each patch, and subsequently averaging the discriminant values.45
Author contributions
The work was conceived and designed by NLJ. The figures were designed by SK, JL, and JK, and they also contributed to the writing of the manuscript. Technical and material support were provided by SP, SL, and YK. The work was supervised by YC, OK and NLJ. All authors reviewed and contributed to the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2021R1A3B1077481; NRF-2020R1A2C1006331; RS-2023-00253722; NRF-2022M3A9B6082680).References
- S. Breslin and L. O'Driscoll, Drug Discovery Today, 2013, 18, 240–249 CrossRef CAS PubMed.
- K. Ben-Yehuda, S. K. Mirsky, M. Levi, I. Barnea, I. Meshulach, S. Kontente, D. Benvaish, R. Cur-Cycowicz, Y. N. Nygate and N. T. Shaked, Adv. Intell. Syst., 2022, 4, 2100200 CrossRef.
- E. Berthier, E. W. K. Young and D. Beebe, Lab Chip, 2012, 12, 1224–1237 RSC.
- M. Chung, S. Lee, B. J. Lee, K. Son, N. L. Jeon and J. H. Kim, Adv. Healthcare Mater., 2018, 7, 1700028 CrossRef PubMed.
- S. Jalili-Firoozinezhad, F. S. Gazzaniga, E. L. Calamari, D. M. Camacho, C. W. Fadel, A. Bein, B. Swenor, B. Nestor, M. J. Cronce, A. Tovaglieri, O. Levy, K. E. Gregory, D. T. Breault, J. M. S. Cabral, D. L. Kasper, R. Novak and D. E. Ingber, Nat. Biomed. Eng., 2019, 3, 583–583 CrossRef CAS PubMed.
- C. M. Leung, P. de Haan, K. Ronaldson-Bouchard, G. A. Kim, J. Ko, H. S. Rho, Z. Chen, P. Habibovic, N. Li Jeon, S. Takayama, M. L. Shuler, G. Vunjak-Novakovic, O. Frey, E. Verpoorte and Y. C. Toh, Nat. Rev. Methods Primers, 2022, 2, 33 CrossRef CAS.
- P. Vulto and J. Joore, Nat. Rev. Drug Discovery, 2021, 20, 961–962 CrossRef CAS PubMed.
- Y. Lee, J. W. Choi, J. Yu, D. Park, J. Ha, K. Son, S. Lee, M. Chung, H. Y. Kim and N. L. Jeon, Lab Chip, 2018, 18, 2433–2440 RSC.
- B. N. Ondatje, S. Sances, M. J. Workman and C. N. Svendsen, Lab Chip, 2022, 22, 4246–4255 RSC.
- S. Peel, A. M. Corrigan, B. Ehrhardt, K. J. Jang, P. Caetano-Pinto, M. Boeckeler, J. E. Rubins, K. Kodella, D. B. Petropolis, J. Ronxhi, G. Kulkarni, A. J. Foster, D. Williams, G. A. Hamilton and L. Ewart, Lab Chip, 2019, 19, 410–421 RSC.
- I. J. Goodfellow, J. Pouget-Abadie, M. Mirza, B. Xu, D. Warde-Farley, S. Ozair, A. Courville and Y. Bengio, Adv. Neural Inf. Process. Syst., 2014, 27, 2672–2680 Search PubMed.
- P. Isola, J. Y. Zhu, T. H. Zhou and A. A. Efros, Proc. CVPR IEEE, 2017, pp. 5967–5976, DOI:10.1109/Cvpr.2017.632.
- M. Weigert, U. Schmidt, T. Boothe, A. Muller, A. Dibrov, A. Jain, B. Wilhelm, D. Schmidt, C. Broaddus, S. Culley, M. Rocha-Martins, F. Segovia-Miranda, C. Norden, R. Henriques, M. Zerial, M. Solimena, J. Rink, P. Tomancak, L. Royer, F. Jug and E. W. Myers, Nat. Methods, 2018, 15, 1090–1097 CrossRef CAS PubMed.
- W. Ouyang, A. Aristov, M. Lelek, X. Hao and C. Zimmer, Nat. Biotechnol., 2018, 36, 460–468 CrossRef CAS PubMed.
- F. Borrelli, J. Behal, A. Cohen, L. Miccio, P. Memmolo, I. Kurelac, A. Capozzoli, C. Curcio, A. Liseno, V. Bianco, N. T. Shaked and P. Ferraro, APL Bioeng., 2023, 7, 026110 CrossRef CAS PubMed.
- M. Mittal, L. M. Goyal, S. Kaur, I. Kaur, A. Verma and D. J. Hemanth, Appl. Soft Comput., 2019, 78, 346–354 CrossRef.
- Y. Rivenson, H. D. Wang, Z. S. Wei, K. de Haan, Y. B. Zhang, Y. C. Wu, H. Gunaydin, J. E. Zuckerman, T. Chong, A. E. Sisk, L. M. Westbrook, W. D. Wallace and A. Ozcan, Nat. Biomed. Eng., 2019, 3, 466–477 CrossRef CAS PubMed.
- Y. Rivenson, T. R. Liu, Z. S. Wei, Y. Zhang, K. de Haan and A. Ozcan, Light: Sci. Appl., 2019, 8, 23 CrossRef PubMed.
- J. M. Matthews, B. Schuster, S. S. Kashaf, P. Liu, R. Ben-Yishay, D. Ishay-Ronen, E. Izumchenko, L. Shen, C. R. Weber, M. Bielski, S. S. Kupfer, M. Bilgic, A. Rzhetsky and S. Tay, PLoS Comput. Biol., 2022, 18, e1010584 CrossRef CAS PubMed.
- X. S. Bian, G. Li, C. Wang, W. Q. Liu, X. H. Lin, Z. X. Chen, M. C. Cheung and X. B. A. Luo, Comput. Biol. Med., 2021, 134, 104490 CrossRef PubMed.
- A. Mencattini, D. Di Giuseppe, M. C. Comes, P. Casti, F. Corsi, F. R. Bertani, L. Ghibelli, L. Businaro, C. Di Natale, M. C. Parrini and E. Martinelli, Sci. Rep., 2020, 10, 7653 CrossRef CAS PubMed.
- W. Lee, B. Yoon, J. Lee, S. Jung, Y. S. Oh, J. Ko and N. L. Jeon, BioChip J., 2023, 17, 357–368 CrossRef CAS.
- M. C. Comes, J. Filippi, A. Mencattini, P. Casti, G. Cerrato, A. Sauvat, E. Vacchelli, A. De Ninno, D. Di Giuseppe, M. D'Orazio, F. Mattei, G. Schiavoni, L. Businaro, C. Di Natale, G. Kroemer and E. Martinelli, Neural. Comput. Appl., 2021, 33, 3671–3689 CrossRef.
- S. Park, J. Newton, T. Hidjir and E. W. K. Young, Lab Chip, 2023, 23, 3671–3682 RSC.
- E. M. Christiansen, S. J. Yang, D. M. Ando, A. Javaherian, G. Skibinski, S. Lipnick, E. Mount, A. O'Neil, K. Shah, A. K. Lee, P. Goyal, W. Fedus, R. Poplin, A. Esteva, M. Berndl, L. L. Rubin, P. Nelson and S. Finkbeiner, Cell, 2018, 173, 792–803 CrossRef CAS PubMed.
- Z. Z. Chen, N. Ma, X. W. Sun, Q. W. Li, Y. Zeng, F. Chen, S. Q. Sun, J. Xu, J. Zhang, H. Ye, J. J. Ge, Z. Zhang, X. R. Cui, K. Leong, Y. Chen and Z. Z. Gu, Biomaterials, 2021, 272, 120770 CrossRef CAS PubMed.
- V. Anagnostidis, B. Sherlock, J. Metz, P. Mair, F. Hollfelder and F. Gielen, Lab Chip, 2020, 20, 889–900 RSC.
- T. Tran, O. H. Kwon, K. R. Kwon, S. H. Lee and K. W. Kang, 2018 IEEE International Conference on Electronics and Communication Engineering (ICECE 2018), 2018, pp. 13–16 Search PubMed.
- J. J. Metzger, C. Pereda, A. Adhikari, T. Haremaki, S. Galgoczi, E. D. Siggia, A. H. Brivanlou and F. Etoc, Cells Rep. Methods, 2022, 2, 100297 CrossRef CAS PubMed.
- R. A. Brekken, J. P. Overholser, V. A. Stastny, J. Waltenberger, J. D. Minna and P. E. Thorpe, Cancer Res., 2000, 60, 5117–5124 CAS.
- A. R. Quesada, R. Muñoz-Chápuli and M. A. Medina, Med. Res. Rev., 2006, 26, 483–530 CrossRef CAS PubMed.
- S. Hyung, J. Ko, Y. J. Heo, S. M. Blum, S. T. Kim, S. H. Park, J. O. Park, W. K. Kang, H. Y. Lim, S. J. Klempner and J. Lee, Sci. Adv., 2023, 9, eadk1098 CrossRef CAS PubMed.
- N. Popovic, S. Vujosevic and T. Popovic, Sci. Rep., 2019, 9, 16340 CrossRef PubMed.
- I. S. Zaitoun, C. M. Wintheiser, N. Jamali, S. J. Wang, A. Suscha, S. R. Darjatmoko, K. Schleck, B. A. Hanna, V. Lindner, N. Sheibani and C. M. Sorenson, Sci. Rep., 2019, 9, 9700 CrossRef PubMed.
- J. Y. Zhu, T. Park, P. Isola and A. A. Efros, IEEE I Conf. Comp. Vis., 2017, pp. 2242–2251, DOI:10.1109/Iccv.2017.244.
- S. Kim, J. Ko, S. R. Lee, D. Park, S. Park and N. L. Jeon, Biotechnol. Bioeng., 2021, 118, 2524–2535 CrossRef CAS PubMed.
- J. Ko, J. Ahn, S. Kim, Y. Lee, J. Lee, D. Park and N. L. Jeon, Lab Chip, 2019, 19, 2822–2833 RSC.
- V. van Duinen, W. Stam, E. Mulder, F. Famili, A. Reijerkerk, P. Vulto, T. Hankemeier and A. J. van Zonneveld, Int. J. Mol. Sci., 2020, 21, 4804 CrossRef CAS PubMed.
- L. Hradecka, D. Wiesner, J. Sumbal, Z. S. Koledova and M. Maska, IEEE Trans. Med. Imaging, 2023, 42, 281–290 Search PubMed.
- J. Yu, S. Lee, J. Song, S. R. Lee, S. Kim, H. Choi, H. B. Kang, Y. C. Hwang, Y. K. Hong and N. L. Jeon, Nano Convergence, 2022, 9, 16 CrossRef CAS PubMed.
- T. Y. Zhang and C. Y. Suen, Commun. ACM, 1984, 27, 236–239 CrossRef.
- V. Badrinarayanan, A. Kendall and R. Cipolla, IEEE Trans. Pattern Anal. Mach. Intell., 2017, 39, 2481–2495 Search PubMed.
- O. Ronneberger, P. Fischer and T. Brox, Lect. Notes Comput. Sci., 2015, 9351, 234–241 Search PubMed.
- K. Simonyan and A. Zisserman, arXiv, 2014, preprint, arXiv:1409.1556, DOI:10.48550/arXiv.1409.1556.
- S. G. Wang, K. Miao, S. Y. Li and Q. An, Electronics, 2022, 11, 124 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3lc00935a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |