Open Access Article
Open Access ArticleAdvancing in situ single-cell microbiological analysis through a microwell droplet array with a gradual open sidewall†
Jie
Wang‡
a,
Lin
Du‡
 b,
Yuwei
Han
c,
Dawei
Zhang
b,
Yuwei
Han
c,
Dawei
Zhang
 *a and
Dalei
Jing
*b
*a and
Dalei
Jing
*b
aSchool of Optical-Electrical and Computer Engineering, University of Shanghai for Science and Technology, Shanghai, China 200093. E-mail: dwzhang@usst.edu.cn
bSchool of Mechanical Engineering, University of Shanghai for Science and Technology, Shanghai, China 200093. E-mail: jingdalei@usst.edu.cn
cState Key Laboratory of Genetic Engineering, School of Life Science, Fudan University, Shanghai, China 200433
First published on 6th November 2023
Abstract
The utilization of microfluidic analysis technology has resulted in the advancement of fast pathogenic bacteria detection, which can accurately provide information on biochemical reactions in a single cell and enhance detection efficiency. Nevertheless, the achievement of rapid and effective in situ detection of single-bacteria arrays remains a challenge due to the complexity of bacterial populations and low Reynolds coefficient fluid, resulting in insufficient diffusion. We develop microwell droplet array chips from the lateral hydrodynamic wetting approach to address this issue. The sidewall of the microwell gradually opens which aids in advancing the liquid–air interface and facilitates the impregnation of the solid microwells, preserving the Wenzel state and assisting in resisting the liquid force to separation from the drop. The feasibility of preparing cell arrays and identifying them inside the microwells was demonstrated through the simulated streamlined distribution of gradual and traditional microwells with different sizes. The water-based ink diffusion experiment examined the relationship between diffusion efficiency and flow velocity, as well as the position of the microwell relative to the channel. It showed that the smaller gradual microwell still has a good diffusion efficiency rate at a flow velocity of 2.1 μL min−1 and that the infiltration state is easier to adjust. With this platform, we successfully isolated a mixed population containing E. coli and S. aureus, obtained single-bacteria arrays, and performed Gram assays after in situ propagation. After 20 hours of culture, single bacteria reproduced demonstrating the capability of this platform to isolate, cultivate, and detect pathogenic bacteria.
Introduction
Infectious diseases pose a significant risk to human health, and the spread of pathogenic microorganisms can lead to widespread disease outbreaks. Patients with pre-existing chronic conditions or weakened immune systems are particularly vulnerable to infections caused by pathogenic bacteria.1,2 The development of precise and effective pathogen detection technologies is essential for the successful diagnosis and treatment of infectious illnesses since existing detection methods are sometimes time-consuming and difficult.3,4 The single-cell analysis (SCA) technique based on microfluidics provides detailed information about intracellular substances and biochemical reactions. This makes it an invaluable tool for obtaining individual differences among pathogenic microorganisms at the single-cell level, thus facilitating a more comprehensive understanding of complex life processes.5–7 In the field of microbial physiology, cell phenotyping and histological analysis, the SCA technique has played a crucial role in improved pathogen detection tools and processes depending on the advantage in pathogen isolation and detection systems in recent years.8–10The preferred method for bacterial single-cell analysis in SCA technology is droplet microfluidics, which is effective manipulation on a chip and enables multiple analyses of individual droplets for ultra-high throughput acquisition of single cells.11–14 Among them, microwell droplet arrays have inherent advantages over continuous flow droplets in that they allow for in situ detection by fixed physical microwells, which create a more stable microenvironment, prevent sample contamination, and allow for in situ culture and colony analysis of single bacterial cells through cell attachment and diffusion.15–19 The capture of a single bacterium in a microwell to create a single bacterial array has led to major improvements in the creation and manufacture of bioapplications. With the aid of supporting instruments, it has been demonstrated that the morphological form and hydrodynamic properties of microwells permit the capture of a single bacterium.20,21 However, due to the complexity of the microbial community and the low diffusion efficiency of low Reynolds number laminar flow, the droplet array used for single-cell analysis has certain limitations when it comes to effectively isolating and culturing pathogenic bacteria. These limitations include inadequate representation of cells and the inability to verify the activity of individual bacterial cells.22 With lateral contact flow23 and wetting transition criteria,24 it is possible to achieve single-cell separation inside the microwell by using a physical barrier to separate the target from the general flora in transitional methods of surface modification or valve control.25,26 The energy barrier created at the liquid–air interface of a hydrophobic surface during single-cell separation controls the wetting transition from the Cassie to the Wenzel state, which is usually metastable within the constraints of a thin microstructure. Filling the voids of the rough surface morphology with liquid can result in a reversible change to the unstable transition state, causing non-intimate contact between solid and liquid and the introduction of air.27–29 This significantly decreased the ability to identify pathogenic bacteria by separating them into microwells and staining them. As a result, it is necessary to create microwell droplet array chips for extensive pathogenic bacteriology research that are easy to use and affordable and limit droplet fusion while preserving analytes in tiny volumes.30,31
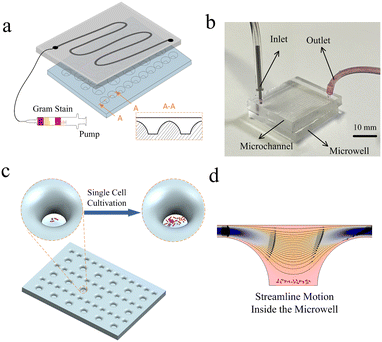
In this study, we develop a microwell droplet array chip that expedites wetting transitions into stable droplet arrays in microwells, enabling more effective in situ detection of single-bacteria arrays. By using the lateral hydrodynamic approach, we were able to capture individual bacterial cells and provide ample growth space inside the microwell arrays. Our design employs a gradual opening trend, which is inspired by the surface form of a lotus leaf, to enhance the movement of the liquid–air interface and promote the pre-embedding of bacterial cells to the bottom of the well. Unlike previous methods, we focused on the stability of the liquid–air interface rather than surface modification or valve control. The channel above the microwell provides a confined environment and allows for the diversion of reagents. Through our investigations of microwell and channel dimensions, relative positions, and flow rate variations, we have discovered a high diffusion efficiency within the system. Our chip has been successfully used for single-cell isolation and in situ propagation culture experiments of mixed pathogenic bacteria. The microwell droplet array chip makes it simple to speed up wetting transitions for in situ detection of single bacteria, as demonstrated in Fig. 1. A conceptual representation of the channels and microwells can be seen in Fig. 1(a). Four different Gram reagents were sequentially pumped onto the chip after it had dried to stain the bacteria. The cross-section of the microwells is shown in the diagram in A-A. The assembled device is shown in Fig. 1(b). The gradually opening shaped microwells are displayed at the bottom, and the vertical sidewall microchannels are shown at the top. Fig. 1(c) shows a 3D schematic of single bacterium isolation and in situ cultivation in a microwell. The simulated streamlined motion of the reagents as they pass is shown in Fig. 1(d).
Experimental
Materials and equipment
Wet etching is used to create a glass-positive mould (Dongcheng Microfluidics Co., Ltd., Zhenjiang, China) for microwell array chip preparation. An SU-8 (2025, Kayaku Advanced Materials, Inc., MA) positive mould with a 50 μm width and a 25 μm thickness for a channel is created on silicon wafers (MJB4, SUSS Micro-Tec., Germany) by using the UV photolithography technique (see Fig. S1 and S2 in the ESI† for more details about the preparation process). The polydimethylsiloxane (PDMS) chips with microwells and channels are made of a Sylgard prepolymer and curing agent (184, Sigma-Aldrich, MO) in accordance with 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 from the glass positive mould and SU-8 positive mould, respectively. The inlet and outlet PDMS chips are punched with a 0.7 mm punch (iBiochip, China). With plasma surface treatment equipment (PDC-32G-2, Harrick Plasma, USA), two PDMS chips with microwells and channels are bonded together to obtain the microwell droplet array chip. A single disposable medical sterile 1 mL syringe and a syringe pump (PHD2000, Harvard Apparatus) are used to load various amounts of reagents into the chip. A charge-coupled device (CCD) camera (DP73, Olympus, Japan) is used to monitor the isolation and in situ propagation of single bacterial cells in the microwell arrays under an Olympus IX71 microscope.
1 from the glass positive mould and SU-8 positive mould, respectively. The inlet and outlet PDMS chips are punched with a 0.7 mm punch (iBiochip, China). With plasma surface treatment equipment (PDC-32G-2, Harrick Plasma, USA), two PDMS chips with microwells and channels are bonded together to obtain the microwell droplet array chip. A single disposable medical sterile 1 mL syringe and a syringe pump (PHD2000, Harvard Apparatus) are used to load various amounts of reagents into the chip. A charge-coupled device (CCD) camera (DP73, Olympus, Japan) is used to monitor the isolation and in situ propagation of single bacterial cells in the microwell arrays under an Olympus IX71 microscope.
Bioreagents and pathogenic bacteria
Bacterial single-cell isolation and in situ propagation validation were carried out using S. aureus (29213, ATCC, VA) and E. coli (25922, ATCC, VA). The bacteria are measured and expanded quickly in a Luria-Bertani broth medium (HBO128, Qingdao Haibo, China) and blood agar plates (Bkmam, China) at 37 °C. A Gram Stain Kit (G1060, Solarbio, China) is used to stain for the identification of colonized bacteria. The concentration of the bacterial solution is figured out by contrasting the turbidity of the bacterial solution with that of a Mackenzie turbidimetric tube. At enough concentration of 1.5 × 103 copies per mL in accordance with the McFarland Equivalence Turbidity Standard, we carried out single-cell isolation tests for S. aureus. The bacteria are first purified by overnight incubation on blood agar plates, and the colonized bacteria are then transferred into a Luria-Bertani broth medium to obtain an appropriate concentration of the bacterial solution with a Mackenzie turbidimetric tube. A mixed flora solution was made in the same way. A droplet array is created by loading the solution onto a microwell array chip. It is incubated for in situ growth for 20 hours at constant humidity and standard atmosphere. We pre-embedded the bacteria into the bottom of the microwell and applied the Gram stain. Unlike regular staining, the channel is vented with air after each staining to “blow away” the staining chemical and prevent harm to bacteria caused by washing. Finally, the chips can be sent for observation.Simulation model of microwells
The multiphysics field simulation program (COMSOL Multiphysics 5.6) microfluidic module is used to model the flow in microwells. The entire system is sealed and the slow laminar flow within the channel can be analysed for bacteria due to the presence of a drainage channel above the microwell. The sidewall expansion type microwell is formed by two 1/4-like circular arcs. The depth of the narrow channel above the microwell is 25 μm. We investigated how the geometry of the microwell affects the flow pattern. Simulations were performed to observe the effect of flow lines generated near individual microwells on the distribution of bacteria. The working fluid is assumed to be water (a homogeneous, incompressible Newtonian fluid; density 998.2 kg m−3, dynamic viscosity 0.001 kg m−1 s−1) and the flow is assumed to be laminar steady flow. Based on the Navier–Stokes momentum conservation equation, the steady-state velocity field and streamline in the microwell are solved. No-slip conditions are used on the walls of channels and microwells. The inlet solution is an aqueous solution, and the inflow velocity is 0.1 m s−1.Results and discussion
Transition principle for droplets and diffusion
The hydrophobic effect refers to the phenomenon caused by the reduction of the solid–liquid contact area due to air pockets trapped in the rough, porous microstructure. When the droplet is stationary on an ideal solid surface, Young obtained the equilibrium equation based on the principle of mechanical equilibrium32σsl + σla![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θY = σsa θY = σsa |
P + P∞ − Pa − σ ∇·![[n with combining right harpoon above (vector)]](https://www.rsc.org/images/entities/i_char_006e_20d1.gif) = 0 = 0 |
![[n with combining right harpoon above (vector)]](https://www.rsc.org/images/entities/i_char_006e_20d1.gif) is the interface unit normal vector. The critical pressure mostly is obtained by force balance analysis and depends on the homogeneity of the meniscus slope near the well walls. When its shape becomes more circular, the resistance of the pores to hydrostatic pressure increases.
is the interface unit normal vector. The critical pressure mostly is obtained by force balance analysis and depends on the homogeneity of the meniscus slope near the well walls. When its shape becomes more circular, the resistance of the pores to hydrostatic pressure increases.
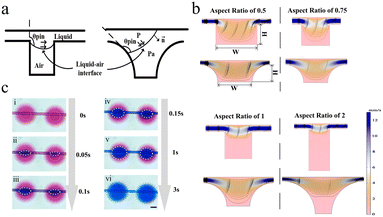
The movement of the forward liquid–air interface is enhanced by the gradual unfolding design of the sidewalls of the microwell, the bottom of the microwell is in contact with the liquid–air interface, and the liquid exhibiting the Cassie state on the hydrophobic surface is transformed into the Wenzel state. Although the Wenzel state is at a lower energy state, the energy potential barrier needs to be crossed for the system to change the permeability state. Ref. 41 discussed the effect of substrate surface topography on wettability, which was inspired from here. We changed the sidewall of the microwell from a vertical type structure to a 1/4 rounded type structure to enhance the transformation process of the depinning. The transition process of the two sidewall structures off the depinning is shown in Fig. 2a, where θpin is the contact angle of the liquid–air interface on the microwell sidewall, and the contact line is shifted when θpin is greater than the intrinsic contact angle of the material. In the Cassie state, the liquid–air interface is suspended between the vertical sidewalls (as shown on the left in Fig. 2a). The interface is curved due to the Laplace pressure inside the liquid. If the suspended interface does not remain fixed to the top of the sidewall, then it will be moved down into the microwell and wet on the surface. If the contact angle formed at the liquid–air interface is greater than the advance angle at that point, it will lead to depinning and the system will become the Wenzel state (as shown on the right in Fig. 2a). In contrast to vertical sidewalls, the pressure–volume energy of the liquid is modified by the gradually expanding sidewall design. The transition caused by depinning can also be considered as a process in which the work done by pressure is greater than the energy potential generated by the surface energy associated with the wetting roughness.
Fig. 2b is the steady-state flow velocity field and streamlined profile of the rectangular and gradient expansion microwells. We found that the wet state was affected by the aspect ratio of the microwell. The smaller the aspect ratio of the microwell, the easier it is to be wet. However, when the aspect ratio reaches 0.5 and below, although the microwell can be wetted, the bottom has a strong streamlined distribution, which means that the flowing liquid will affect the internal state of the microwell. Through the comparison of the streamlined distribution and velocity field, it is found that the gradient expansion microwells are more conducive to the entry of fluid. Due to the surface energy of the liquid and the pinning effect, the liquid is confined by the rectangular microwell and flows directly through the channel.42 The streamlines with gradient expansion microwells43,44 of the aspect ratio of 1 have a relatively uniform slope, which indicates smooth velocity changes within the microwells and that resistance to water pressure is enhanced at the bottom. The bottom of the gradient extension microwells is not disturbed by any strong flow and therefore does not encounter damage to bacteria caused by the high shear forces that can occur in lateral flow. We used microwells with an aspect ratio of 1 to perform diffusion tests in which blue ink aqueous solution was substituted for the red, as shown in Fig. 2c, to confirm the diffusion effect in this manner. Fig. 2c, i represents the state inside the system when the blue ink has not yet diffused. The white dotted line in Fig. 2c, ii–vi indicates the border between the two solutions, and it shows how quickly the blue liquid entered the microwells during the first 0.1 s of the experiment. The subsequent stage of 3 s begins with the liquid progressively diffusing, and the blue liquid fills the well until the red liquid completely fills the pores (as illustrated in Fig. 2c, v and vi).
Study of the staining procedure of Gram stain
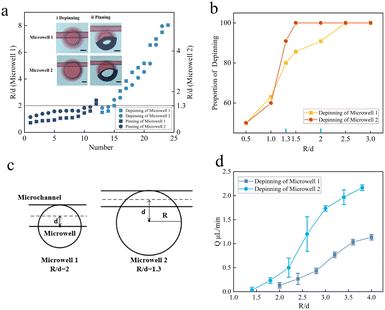
To utilize microwell arrays in single-bacteria assays, we investigated the impact of the alignment, intake flow rate, and diffusion efficiency of the two chips on pinning and depinning. The PDMS chip with microwells could produce droplets of approximately 5 nL and 30 nL for small and large wells, respectively. The depth and breadth of a PDMS chip with channels were 25 μm and 50 μm, respectively. After bonding the two chips, we evaluated the relationship between microwells and channels (R is the radius of the microwell, and d for the distance between its centre and the centreline of the channel flow channel). In Fig. 3a, we observed that depinning occurs when R/d is larger for both microwell 1 and microwell 2. For microwell 1, the liquid exits the channel and enters the microwell when R/d is equal to 2. When the liquid–air interface touches the bottom of the wall, the microwell quickly enters the Wenzel wetting condition. The liquid–air contact is essentially fixed to the edge of the microwell as R/d slowly drops. As a result of the closed air inside the microstructure, the microwell is currently in the Cassie state, preventing liquid from entering. The liquid is depinning from the channel into the microwell for microwell 2 when R/d is equal to 1.3. The liquid is essentially trapped at the edge of the microwell as R/d steadily drops, preventing it from entering the interior of the microwell. This demonstrates that in the case of tiny R/d, the pinning of the liquid–air interface at the edge of the microwell results in the system being in the Cassie state, which is the non-wetting condition. As seen in Fig. 3b, depinning is more effective when the R/d is higher (i.e., the distance between the centreline of the channel and the centre of the circle is closer). When R/d is 2, the depinning efficiency for microwell 1 reaches 90.9%, and when it is increased to 2.5, it achieves 100%. This phenomenon was also seen in microwell 2, and when R/d was higher than 1.3, the depinning efficiency was 100%. The value of R/d at 100% depinning efficiency is hence referred to as a unique position. As seen in Fig. 3c, the location is currently tangent to both the upper border of the flow channel and the microwell. When R/d decreases, the channel is closer to the microwell centre from that particular point, and when it increases, the channel is farther from the microwell centre.The condition involving depinning and pinning is influenced by the chips' alignment as well as the pace at which liquid flows through the channel. We looked at the impact that different intake flow rates had on depinning. The flow rate range of microwell 2 is higher than that of microwell 1, even when the flow rate exceeds 2.1 μL min−1, depinning may be generated at a rate within the ranges of the error bars, as illustrated in Fig. 3d. The greatest flow rate of depinning of microwell 1 is just 1.1 μL min−1, though.
The alignment of the chips and the rate of liquid flow through the channel both have an impact on the depinning and pinning state. We investigated the effects of various intake flow rates on depinning. The relationship between the flow rate at which it is capable of depinning and the relative positions of the microwell and microchannel are shown in Fig. 3d. In microwell 2, depinning may happen when the rate surpasses 2.1 μL min−1. However, the maximum flow rate of depinning for microwell 1 is only 1.1 μL min−1. The infiltration conditions within the microwell will change regardless of whether microwell 1 or microwell 2 is within a broad range of set flow rates, which decreases the reliance of the system on the flow rate, can simplify operations, and can speed up the process of biological detection.
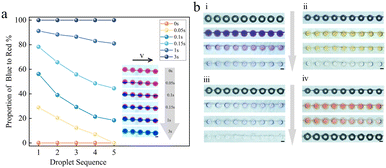
To confirm that the microwell array chip is useful for Gram stain, we next investigated the diffusion ability of the two-phase liquid. We are all aware that different reagents and sample solutions at the microscale often flow laminarly with a low Reynolds coefficient, and the mixtures are frequently inadequate, which restricts the use of microfluidic technology. Our goal was to create a microfluidic system that could identify bacteria quickly using the Gram stain. Numerous substances, such as violet dyes, potassium iodide, and detergents, are used as Gram stain reagents. A crucial stage in the Gram stain procedure is washing. The cells will be treated with various staining agents at different stages of the staining procedure, and the unabsorbed agents must be removed by proper washing processes to guarantee that the final staining result is accurate. To produce the optimum Gram stain effect, it is specifically required to gently rinse with water after Gram stain to remove extra staining reagents and lingering detergent from the cell surface. To explore the diffusion replacement impact of the two-phase liquid, we first successively injected red/blue ink into the chip, as shown in Fig. 4a. The timing begins when the first droplet changes color, and within 3 seconds, all 5 droplets in the observation area shift from entirely red to completely blue, demonstrating that the microwell has undergone a complete solution diffusion replacement. This diffusion replacement takes around 9 seconds to complete on a 45-droplet array. Software called ImageJ was used to calculate the area of the blue/red portion of the five drops. The portion of the blue ink made up 28.9% of the red ink at time 0.05 s, and by the time 0.15 s, it made up 78.3%, an increase of 49.4% from the 0.05 s period. The portion of the blue ink to the red ink is 91.2% at 1 second, and even after the blue area reaches 100% at 3 s, it only grows by 8.8%. Without a change in pump speed, there is quite minimal change in the area ratio of the two inks in the image within 1–3 seconds, showing that post-diffusion is still occurring. The three-dimensional structure and the volume of the microwell, which is relatively large compared to the channel, are to blame for the extended diffusion substitution.
Then, as shown in Fig. 4b, the four reagent solutions included in the Gram kit were put through diffusion staining tests. The screenshots of crystal violet, iodine solution, 95% ethanol, and safranin solution flowing into the chip, respectively, are shown in Fig. 4b. The dyeing reagent solution entry microwells, the clear solution entry microwells, and the microwells following water washing are all shown in the screenshot as four rows of microwell arrays. See Video in the ESI† for further information on the dynamic staining and washing procedure.
Diffusion stain studies were then conducted using the four reagent solutions included in the Gram kit. Screenshots of crystal violet, iodine solution, 95% ethanol, and safranin solution, respectively, are presented in Fig. 4b, i–iv. Each screenshot shows four rows of microwell arrays that are empty, filled with the staining reagent solution, filled with a water solution, and finished washing. The results demonstrate that the Gram stain reagent solution can also effectively penetrate the microwell and diffuse, suggesting that the system has strong stability and a high efficiency of diffusion and can be used to observe the identification of bacteria as Gram-positive or Gram-negative.
Single bacteria array and in situ detection
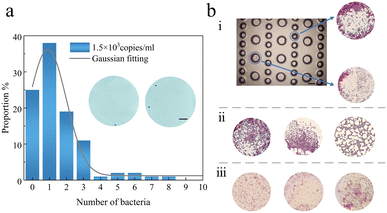
Then, we grew the extracted bacteria in situ, created a single bacterial array using the chip, and ultimately performed Gram stain to confirm the viability for biological applications. As is well known, the Gram stain procedure must penetrate the bacteria in the four reagent solutions for a few seconds to distinguish between positive and negative bacteria. There is a definite sequence to the reagent solutions that cannot be changed or skipped. As the chip for the bacterial array, we employed a PDMS chip with two kinds of microwells, and the channel leading to the reagent solution was 50 μm wide. In order for the bacteria to enter the microwells, droplets of the bacterial solution are first formed on the PDMS chip. We let the bacteria return to atmospheric pressure after turning off the pumping machinery. When the bacteria in the microwells are to be identified by Gram stain, the chip is put in an alcohol lamp to increase the temperature and let the solution evaporate, leaving the bacteria fixed in the microwells. A period of aqueous solution and air was necessary in the chip after each set of staining solutions colored the bacteria in order to isolate the effects of subsequent staining reagent solutions. Air must be passed between the two portions of solution in the capillary tube in order to prevent the fusion of each sequence of Gram-stained and aqueous solutions. Several sets of dye sequences were passed through for testing at the bacterial solution density of 1.5 × 103 copies per ml. The population distribution in Fig. 5a reveals that just one bacterium was present in around 38% of the microwells. The bacteria are more likely to be pre-buried towards the border of the microwell due to the lateral flow process in the microwell, which is also a major benefit for analyzing the development of adherent cells.To undertake cellular phenotyping, cell–cell interaction research, and omics analysis, single-cell array in situ analysis is a crucial step. We combined S. aureus and E. coli solutions to create a mixed bacterial solution and then loaded it onto the microwell array based on studies on the production of single-cell arrays. After 20 hours, Gram stain allowed the bacteria in the microwells to be identified. The bacteria in the L-B medium solution reproduced freely and independently. The bacteria in the microwells are stained as indicated in Fig. 5b, which depicts the outcome of in situ propagation. After propagation is finished, Fig. 5b(i) presents a partial top view of the microwell array and the mixture of E. coli and S. aureus that has been placed inside the microwells. Fig. 5b(ii) and (iii) show that only S. aureus and E. coli are present in the microwell, respectively. For further stained images of single bacteria and mixed bacteria, see Fig. S3 in the ESI.†
The build-up of metabolites and chemicals released by single cells or their offspring can occur more quickly when grown in arrays of microwells than when cultivated in bulk cultures. Numerous biological processes, including sample diagnostics, microbial genetic analysis, and antibacterial drug screening, depend on colony analysis at the single-cell level. The chips we developed enable cell-to-cell communication, the transport of some signalling molecules, such as peptides, that may contribute to cell growth, and can also be used to explain the pairing of taxa interactions or signal dependencies with other organisms.
Conclusions
In conclusion, we introduce, validate, and utilize microwell droplet array chips based on the lateral hydrodynamic wetting approach to in situ isolate and cultivate single-celled bacteria. The dynamics of bacterial populations, the stimulating impact at the gene level, and the clinical drug diagnostic analysis may all benefit greatly from the employment of SCA technology in the identification of harmful bacteria. The negative mould of the lotus leaf is where the concept of the sidewall gradually opening came from. This design is advantageous for controlling the wetting state of the liquid because it can overcome the energy barrier between the Cassie and Wenzel states. Due to the great diffusion efficiency of the liquid in the microwell array caused by pinning, it has several uses in amplification, sequencing, and metabolic processes. More notably, the chip can analyse at the single-cell level using just the lateral hydrodynamic wetting approach and can trap a variety of bacteria in a specific region without the need for an external force field, which is a significant benefit for the omics research of cell co-culture.Data availability
The data that support the findings of this study are available from the corresponding authors upon reasonable request.Author contributions
J. Wang and L. Du: conceptualisation, data curation, formal analysis, investigation, methodology, verification, visualisation, writing – original draft preparation. Y. Han: formal analysis, investigation, conceptualisation, methodology, supervision, visualisation. D. Jing: conceptualisation, data curation, investigation, methodology, project administration, supervision, verification, and writing – review and editing. D. Zhang: conceptualisation, methodology, funding acquisition, project administration, supervision.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Dr Enqi Wu and Mr Ming Yang for helping with the design parts of the experiment. This work was supported by the National Natural Science Foundation of China under Grant No. 62104148, No. 62274039 and No. 12272236.References
- G. Xing, W. Zhang, N. Li, Q. Pu and J.-M. Lin, Chin. Chem. Lett., 2022, 33, 1743–1751 CrossRef CAS.
- Z. Xu, J. Wang, Z. Jia, Y.-X. Wu, N. Gan and S. Yu, Analyst, 2023, 148, 1093–1101 RSC.
- C. Mao, C. Xue, X. Wang, S. He, L. Wu and X. Yan, Talanta, 2020, 217, 121020 CrossRef CAS PubMed.
- L. A. Harris, S. Beik, P. M. M. Ozawa, L. Jimenez and A. M. Weaver, Curr. Opin. Syst. Biol., 2019, 17, 24–34 CrossRef PubMed.
- X. Xu, J. Wang, L. Wu, J. Guo, Y. Song, T. Tian, W. Wang, Z. Zhu and C. Yang, Small, 2020, 16, 1903905 CrossRef CAS PubMed.
- S. K. Govers, J. Mortier, A. Adam and A. Aertsen, PLoS Biol., 2018, 16, 2003853 CrossRef PubMed.
- C. Brasko, K. Smith, C. Molnar, N. Farago, L. Hegedus, A. Balind, T. Balassa, A. Szkalisity, F. Sukosd, K. Kocsis, B. Balint, L. Paavolainen, M.-Z. Enyedi, I. Nagy, L.-G. Puskas, L. Haracska, G. Tamas and P. Horvath, Nat. Commun., 2018, 9, 226 CrossRef PubMed.
- J. Shrestha, S. R. Bazaz, L. Ding, S. Vasilescu, S. Idrees, B. Söderström, P.-M. Hansbro, M. Ghadiri and M. E. Warkiani, Lab Chip, 2022, 23, 146–156 RSC.
- C. H. Lin, Y. H. Hsiao, H. C. Chang, C. F. Yeh, C. K. He, E. M. Saln, C. Chen, L. M. Chiu and C. H. Hsu, Lab Chip, 2015, 15, 2928–2938 RSC.
- J. Zhang, J. Xue, N. Luo, F. Chen, B. Chen and Y. Zhao, Lab Chip, 2023, 23, 1066–1079 RSC.
- L. Cong, J. Wang, X. Li, Y. Tian, S. Xu, C. Liang, W. Xu, W. Wang and S. Xu, Anal. Chem., 2022, 94, 10375–10383 CrossRef CAS PubMed.
- F. Mi, C. Hu, Y. Wang, L. Wang, F. Peng, P. F. Geng and M. Guan, Anal. Bioanal. Chem., 2022, 414, 2883–2902 CrossRef CAS PubMed.
- X. Xu, L. Cai, S. Liang, Q. Zhang, S. Lin, M. Li, Q. Yang, C. Li, Z. Han and C. Yang, Lab Chip, 2023, 23, 1169–1191 RSC.
- Y. Suo, W. Yin, W. Wu, W. Cao, Q. Zhu and Y. Mu, Analyst, 2022, 147, 3305–3314 RSC.
- Y. Zhang, I. Naguro and A.-E. Herr, Angew. Chem., 2019, 131, 14067–14072 CrossRef.
- X. L. Guo, Y. Wei, Q. Lou, Y. Zhu and Q. Fang, Small, 2022, 18, 2107992 CrossRef PubMed.
- Y. Huang, Z. Gao, C. Ma, Y. Sun, Y. Huang, C. Jia, J. Zhao and S. Feng, Analyst, 2023, 148, 2758–2766 RSC.
- J. Shemesh, T. B. Arye, J. Avesar, J. H. Kang, A. Fine, M. Super, A. Meller, D. E. Ingber and S. Levenberg, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 11293–11298 CrossRef CAS PubMed.
- M. Azizi, M. Zaferani, B. Dogan, S. Zhang, K. W. Simpson and A. Abbaspourrad, Anal. Chem., 2018, 90, 14137–14144 CrossRef CAS PubMed.
- Z. Gao, J. Yi, J. Zhao, H. Gu, H. Zhou and H. Xu, Sens. Actuators, B, 2020, 324, 128716 CrossRef CAS.
- L. Du, H. Liu and J. Zhou, Microsyst. Nanoeng., 2020, 6, 33 CrossRef CAS PubMed.
- E. Pope, C. Cartmell, B. Haltli, A. Ahmadi and R.-G. Kerr, Front. Microbiol., 2022, 13, 958660 CrossRef PubMed.
- R. L. Lai and N. T. Huang, Microfluid. Nanofluid., 2019, 23, 121 CrossRef.
- A. Tuteja, W. Choi, M. Ma, J. M. Mabry, S. A. Mazzella, G. C. Rutledge and R. E. Cohen, Science, 2007, 318, 1618–1622 CrossRef CAS PubMed.
- A. Karimi, S. Yazdi and A.-M. Ardekani, Biomicrofluidics, 2013, 7, 21501 CrossRef CAS PubMed.
- V. Narayanamurthy, S. Nagarajan, F. Samsuri and T. M. Sridhar, Anal. Methods, 2017, 9, 3751–3772 RSC.
- N. A. Patankar, Langmuir, 2010, 26, 8941–8945 CrossRef CAS PubMed.
- H. Kusumaatmaja, M.-L. Blow, A. Dupuis and J.-M. Yeomans, Europhys. Lett., 2008, 81, 36003 CrossRef.
- R. Malinowski, I.-P. Parkin and G. Volpe, Chem. Soc. Rev., 2020, 49, 7879–7892 RSC.
- Z. Gao, J. Yi, J. Zhao, H. Gu, H. Zhou and H. Xu, Sens. Actuators, B, 2020, 324, 128716 CrossRef CAS.
- D. C. Duffy, Lab Chip, 2023, 23, 818–847 RSC.
- Y. Thomas, Philos. Trans. R. Soc. London, 1805, 95, 65–87 CrossRef.
- R. N. Wenzel, Ind. Eng. Chem., 1936, 28, 988–994 CrossRef CAS.
- A. B. D. Cassie and S. Baxter, Trans. Faraday Soc., 1944, 40, 546–551 RSC.
- Y. Xue, S. Chu, P. Lv and H. Duan, Langmuir, 2012, 28, 9440–9450 CrossRef CAS PubMed.
- W. Ren, Langmuir, 2014, 30, 2879–2885 CrossRef CAS PubMed.
- Q. S. Zheng, Y. Yu and Z. H. Zhao, Langmuir, 2005, 21, 12207–12212 CrossRef CAS PubMed.
- M. C. Salvadori, M. Cattani, M. R. S. Oliveira, F. S. Teixeira and I. G. Brown, J. Appl. Phys., 2010, 108, 024908 CrossRef.
- B. Emami, A. A. Hemeda and M. M. Amrei, Phys. Fluids, 2013, 25, 89–109 Search PubMed.
- B. Emami, H. V. Tafreshi, M. Gad-el-Hak and G. C. Tepper, Appl. Phys. Lett., 2012, 100, 13104 CrossRef.
- G. Whyman and E. Bormashenko, Langmuir, 2011, 27, 8171–8176 CrossRef CAS PubMed.
- V. H. Lieu, T. A. House and D. T. Schwartz, Anal. Chem., 2012, 84, 1963–1968 CrossRef CAS PubMed.
- L. Du, Y. X. Li and X. L. Zhang, ACS Appl. Mater. Interfaces, 2023, 15, 17413–17420 CrossRef CAS PubMed.
- L. Du, A. Riaud and J. Zhou, IEEE Trans. Nanotechnol., 2020, 19, 102–106 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3lc00590a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |