Open Access Article
Open Access ArticleLabel-free cell classification in holographic flow cytometry through an unbiased learning strategy
Gioele
Ciaparrone
a,
Daniele
Pirone
b,
Pierpaolo
Fiore
 a,
Lu
Xin
c,
Wen
Xiao
c,
Xiaoping
Li
d,
Francesco
Bardozzo
ab,
Vittorio
Bianco
a,
Lu
Xin
c,
Wen
Xiao
c,
Xiaoping
Li
d,
Francesco
Bardozzo
ab,
Vittorio
Bianco
 b,
Lisa
Miccio
b,
Feng
Pan
b,
Lisa
Miccio
b,
Feng
Pan
 *c,
Pasquale
Memmolo
*c,
Pasquale
Memmolo
 *b,
Roberto
Tagliaferri
*ab and
Pietro
Ferraro
*b,
Roberto
Tagliaferri
*ab and
Pietro
Ferraro
 b
b
aNeurone Lab, Department of Management and Innovation Systems (DISA-MIS), University of Salerno, Fisciano, Italy. E-mail: robtag@unisa.it
bCNR – Institute of Applied Sciences and Intelligent Systems “Eduardo Caianiello”, Pozzuoli, Italy. E-mail: pasquale.memmolo@isasi.cnr.it
cKey Laboratory of Precision Opto-Mechatronics Technology of Ministry of Education, School of Instrumentation Science & Optoelectronics Engineering, Beihang University, 100191 Beijing, China. E-mail: panfeng@buaa.edu.cn
dDepartment of Obstetrics and Gynecology, Peking University People's Hospital, Beijing 100044, China
First published on 9th January 2024
Abstract
Nowadays, label-free imaging flow cytometry at the single-cell level is considered the stepforward lab-on-a-chip technology to address challenges in clinical diagnostics, biology, life sciences and healthcare. In this framework, digital holography in microscopy promises to be a powerful imaging modality thanks to its multi-refocusing and label-free quantitative phase imaging capabilities, along with the encoding of the highest information content within the imaged samples. Moreover, the recent achievements of new data analysis tools for cell classification based on deep/machine learning, combined with holographic imaging, are urging these systems toward the effective implementation of point of care devices. However, the generalization capabilities of learning-based models may be limited from biases caused by data obtained from other holographic imaging settings and/or different processing approaches. In this paper, we propose a combination of a Mask R-CNN to detect the cells, a convolutional auto-encoder, used to the image feature extraction and operating on unlabelled data, thus overcoming the bias due to data coming from different experimental settings, and a feedforward neural network for single cell classification, that operates on the above extracted features. We demonstrate the proposed approach in the challenging classification task related to the identification of drug-resistant endometrial cancer cells.
Introduction
Imaging flow cytometry (IFC) is the current state of the art technology for single-cell analysis,1 exploited for powerful clinical applications, both diagnostic2,3 and therapeutical.4,5 In IFC systems, thousands of cells are individually analysed in very short times as they are illuminated by a light source while they are flowing in a microfluidic channel.6 The high-throughput property allows collecting large amounts of data, thus providing a multi-scale characterization of a heterogenous cell line.7 In fact, average parameters can be measured, but, at the same time, the intra-species variability is not lost since each single cell is regarded as a separate entity. Therefore, a technological effort is underway aimed at the miniaturization of the IFCs towards on-chip systems, cheap and simple to fabricate.8 The conventional IFC benefits from the use of fluorescent dyes with the aim of staining in different ways the several subcellular structures, thus making an intra-cellular analysis possible. However, the same exogenous markers play a destructive role, since they interfere with the cell (photodamaging) and alter the imaging (photobleaching), with the risk of recording an untrue signal. Besides, staining protocols can be expensive and time-consuming, and they often require trained operators. More importantly, in addition to the 2D morphological information, no quantitative measurement can be extracted from the recorded images about the cell biophysical properties. These drawbacks reduce the IFC potential in understanding more complex phenomena and point out the need to integrate into IFCs a label-free and quantitative imaging module. Among several technologies, digital holography (DH) in microscopy is a remarkable candidate as it exploits the light interference principle to image a biological specimen, thus allowing quantitative phase-contrast information without using exogenous agents.9,10 The simpler and cheaper sample preparation, along with the unique capability of numerical refocusing, makes the combination of DH imaging to conventional IFC systems a natural evolution,11,12 further exploitable for on-chip implementations.13,14 By using an appropriate numerical processing, the reconstruction of a quantitative phase map (QPM) from the recorded digital hologram can be obtained.15 Unlike fluorescence imaging, in a QPM the cell biophysical information is recorded, as the cell refractive index (RI) is coupled to the cell morphology in the form of a 2D image, thus explaining the increasing applications of quantitative phase imaging (QPI) in biomedicine.16 The most fascinating evolution of QPI is phase-contrast tomography (PCT), which consists in mapping the 3D RI spatial distribution at the single-cell level,17 and which has been also reproduced under flow cytometry conditions.18,19 Therefore, the capability of a Holographic-IFC (HIFC) to provide a quantitative biophysical cell fingerprint suggests the enormous potential in combining this technology with artificial intelligence (AI) for label-free single-cell phenotyping. While in the 3D PCT case, the use of machine learning20,21 and deep learning22 is starting to be tested, in the 2D QPI case the learning approaches have been widely investigated,23–27 also in a flow cytometry environment.28–35 The standard workflow consists of creating a training dataset, usually obtained by acquiring digital holograms of single cells in the HIFC system under investigation. Then, QPMs are retrieved and used to train classifiers exploiting the paradigms of feature selection or convolutional neural networks. The assessment of the learned classification model is made by using testing datasets with additional QPMs obtained by using the same acquisition setting and processing pipeline. However, it is demonstrated that this protocol suffers from generalization robustness with respect to data coming from other imaging systems or processed in a different way,36i.e., the training stage can be consider biased by the employed recording and processing settings. In addition, heavy and long computations to process a digital hologram to recover the corresponding QPM may represent a further bottleneck for on-chip applications.37 In this paper, we propose a strategy to overcome unwanted biases mainly related to the intrinsic and unpredictable chances within the experimental protocol, e.g., sample preparation by different users, recording in different days, imaging system calibration, etc. We design a learning scheme based on the combination of a convolutional neural network for single cell hologram detection, followed by a convolutional autoencoder operating on unlabelled data and a shallow neural network for cell classification. We demonstrate that the adopted solution is robust with respect to different HIFC acquisition biases, mainly related to the recorded image brightness changes, different levels of speckle noise and fringe orientation variations, thanks to the use of the unsupervised autoencoder for the image feature extraction. It overcomes possible biases caused by different holographic reconstruction methodologies since the learning model is trained with unprocessed digital holograms. In fact, a digital hologram encodes compact and dense information about the light–cell interaction in the form of a 2D intensity map, hence including the phase delay retrieved in the corresponding QPM. Actually, learning approaches fed by the raw holographic diffraction patterns are expected to provide even better results in terms of using QPMs, as recently demonstrated.38,39Here we employ the proposed methodology in detecting drug resistant and non-drug resistant cells within the same Endometrial Cancer (EC) cell line, recorded in a HIFC system. EC is the sixth most diagnosed cancer in women worldwide40,41 and the incidence of EC has increased in many countries.42 To date, the best therapeutical approach is chemotherapy, which mainly exploits platinum-based drugs. Among them, cisplatin is believed to effectively affect EC proliferation and apoptosis.43 Unfortunately, the recurrent or advanced disease that develops in a minority of patients reduces the therapeutical efficacy because of chemoresistance.44,45 Hence, in such cases an accurate evaluation of the drug sensitivity of the patient can allow planning a more effective individualized chemotherapy. A promising solution to this challenging clinical issue can be found in the well assessed discovery about distinctive morphological changes in drug resistant cancer cells.46–50 Then, single-cell analysis becomes the key-technique to fully exploit this property. In summary, the results reported in this paper demonstrate that the combination among raw hologram inference, the high-throughput and single-cell analysis provided by HIFC, and the fast, automatic and accurate results obtained by the proposed unbiased learning approach pushes towards the realization of lab-on-chip devices for aiding medical decision making.51 Then, the label-free and non-destructive DH properties become fundamental to allow downstream more in-depth studies on drug resistant cells after a real-time AI-based cell identification.52–54
Materials and methods
Sample preparation
Two kinds of human EC cells, namely ISK cells and ISK-CisR cells, were considered in this study. ISK is a well-differentiated adenocarcinoma, oestrogen receptor α (ERα)(+), ERβ(+), and progesterone receptor (PR)(+) cell line derived from the American Type Culture Collection and stored in the Obstetrics and Gyneacology Laboratory of Peking University People's Hospital. The drug-resistant characteristics of ISK-CisR cells were induced by exposing ISK cells to cisplatin for 10 months. Both cell types were cultured and grown in DMEM/F12 (Dulbecco's Modified Eagle's Medium/Ham's F-12 50/50 Mix, CORNING, USA) supplemented with L-glutamine, 15 mM HEPES and 10% fetal bovine serum (Gibco 10099-141, Australia). Then, cells were seeded onto a 100 mm glass Willco dish at a density of 13.7 × 106 cells for 24 hours with 5% CO2 and 37 °C. Finally, the cells were collected from wells using trypsin digestion, washed in phosphate-buffered saline (PBS) to dilute to half the concentration (about 5 × 106), and then suspended in 500 μL of 1× binding buffer (10 mM HEPES, 140 mM NaCl, 2.5 mM CaCl2, pH 7.4) with 10 μL of annexin conjugate and PI.HIFC system design and implementation
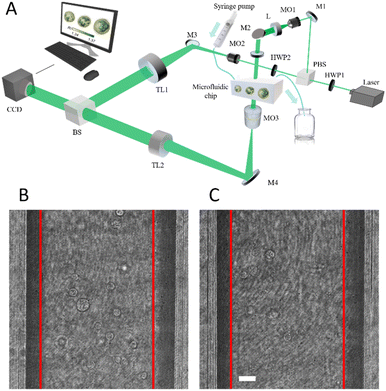
We built up a HIFC system to acquire holographic images of flowing cells, as shown in Fig. 1A. The recording system is equipped with a Mach–Zehnder imaging interferometer in off-axis configuration. A 100 mW coherent laser at 532 nm is used as the light source employing a single-mode fiber. The plane wave is produced by a collimating lens (L) and split into the object beam and the reference beam by a polarizing beam splitter (PBS). In the object arm we used a 20× microscope objective (MO) with a numerical aperture of 0.50 and a tube lens (TL) to amplify the object and output plane wave. Such an object wave is then combined with the reference one in a beam splitter (BS) at a small angle and the digital holograms of the object are recorded by a 1024 × 1024 CCD camera having 5.5 × 5.5 μm2 pixel sizes (PointGrey, Canada) operating at 20 fps. The cell solution is flowing in a microfluidic PMMA channel with 100 μm in width and depth 58.5 mm in length (microfluidic-chipshop, Fluidic 144, Germany) driven by a syringe pump system with an injection rate of 5 μl min−1.Dataset construction
In order to create the dataset for solving the above-mentioned drug-resistant classification problem, 9 holographic video sequences per class were recorded, corresponding to 9316 and 8789 holographic frames containing respectively the ISK and ISK-CisR cells flowing in the microfluidic channel, as shown in Fig. 1B and C. Among them, we randomly select 8 videos (4 for ISK and 4 for ISK-CisR) to be used for validating and testing the performance of the entire classification scheme. In this way, it can be ensured that the learning system is not influenced by the bias induced in the Mask R-CNN and concurrently in the classification learning system. Two possible strategies were tested, with the aim of understanding how the holographic diffraction pattern biases can influence the classifier's performances. The first one consists in taking all the diffraction information by selecting a bounding box around the unfocused cell. The second one is based on the segmentation of the cell with respect to its background by means of a binary mask, thus reducing the diffraction content. In order to perform a fast and automatic selection of the region of interests (ROIs) containing the single cells used for feeding the classifier, a Mask R-CNN55 with ResNet-101 (ref. 56) backbone was trained to extract both bounding boxes and cell masks from each holographic frame. The detection network was trained on a subset of 4 videos (2 per class), while 2 videos (1 per class) were used as a validation set for hyperparameter tuning. Other 4 videos (2 per class) were instead used as an independent test set. More precisely, the Computer Vision Annotation Tool (CVAT)57 was used to label 2 different datasets made of the same 120 × 120 ROIs, i.e. a bounding box (bbox) dataset and a mask dataset. In this way, a total of 71![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 528 cells were labelled, i.e., 44
528 cells were labelled, i.e., 44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 456 cells belonging to the ISK class and 27
456 cells belonging to the ISK class and 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 072 cells belonging to the ISK-CisR class. In addition, 14
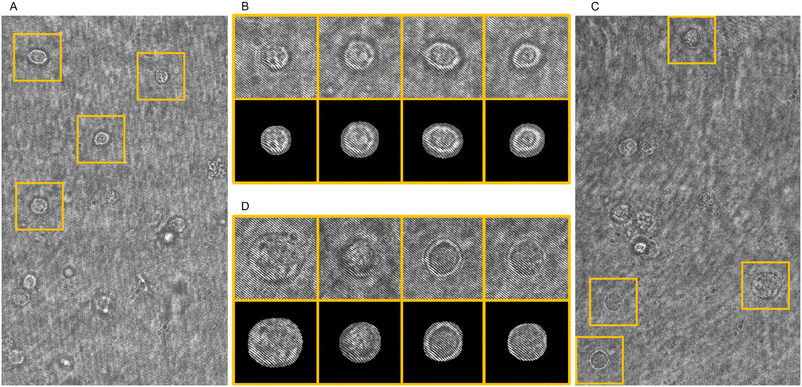
072 cells belonging to the ISK-CisR class. In addition, 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 890 dubious objects were identified. They were considered false positives during the training step and manually removed because we wanted the model to extract only real cells, while they were disregarded at evaluation time for computing the Average Precision (AP). AP is used to evaluate the precision (the ratio of correctly identified positives to the total identified positives) across varying thresholds of recall (the ratio of correctly identified positives to the total actual positives). AP is essentially an average measure of precision at different levels of recall, providing an overall performance metric for the precision–recall curve. We used transfer learning by initializing the convolutional neural network (CNN) with weights trained on the COCO object detection dataset.58 Frames were cropped to 1024 × 625 by removing the channel's borders, as displayed in Fig. 1B and C. The Mask R-CNN was trained for 25 epochs using Stochastic Gradient Descent with momentum, with a learning rate of 10−3, a momentum of 0.9, and 4 images per batch. We chose the best epoch using the standard AP metric computed at Intersection over Union IoU = 0.5 on the validation set. Finally, two different test analyses are needed to ultimately evaluate both learning systems. The output of the Mask R-CNN is reported in Fig. 2A and C, along with some bounding boxes and cell masks displayed as examples in Fig. 2B and D about the ISK and ISK-CisR cells, respectively. Then, for the cell classification experiments, we created
890 dubious objects were identified. They were considered false positives during the training step and manually removed because we wanted the model to extract only real cells, while they were disregarded at evaluation time for computing the Average Precision (AP). AP is used to evaluate the precision (the ratio of correctly identified positives to the total identified positives) across varying thresholds of recall (the ratio of correctly identified positives to the total actual positives). AP is essentially an average measure of precision at different levels of recall, providing an overall performance metric for the precision–recall curve. We used transfer learning by initializing the convolutional neural network (CNN) with weights trained on the COCO object detection dataset.58 Frames were cropped to 1024 × 625 by removing the channel's borders, as displayed in Fig. 1B and C. The Mask R-CNN was trained for 25 epochs using Stochastic Gradient Descent with momentum, with a learning rate of 10−3, a momentum of 0.9, and 4 images per batch. We chose the best epoch using the standard AP metric computed at Intersection over Union IoU = 0.5 on the validation set. Finally, two different test analyses are needed to ultimately evaluate both learning systems. The output of the Mask R-CNN is reported in Fig. 2A and C, along with some bounding boxes and cell masks displayed as examples in Fig. 2B and D about the ISK and ISK-CisR cells, respectively. Then, for the cell classification experiments, we created
• a training set made of the 10 videos (5 per class) used to train, validate, and test the cell detector;
• a validation set and a test set each made of 4 videos (2 per class) not used to train, validate, and test the cell detector.
The training set included 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 236 cells (10
236 cells (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 881 ISK and 6355 ISK-CisR), the validation set included 8842 cells (5843 ISK and 2999 ISK-CisR), while the test set included 9126 cells (6171 ISK and 2955 ISK-CisR). Notice that the validation and test sets are created from the 8 videos set aside previously and never seen by the network during the training stage.
881 ISK and 6355 ISK-CisR), the validation set included 8842 cells (5843 ISK and 2999 ISK-CisR), while the test set included 9126 cells (6171 ISK and 2955 ISK-CisR). Notice that the validation and test sets are created from the 8 videos set aside previously and never seen by the network during the training stage.
CNN classification
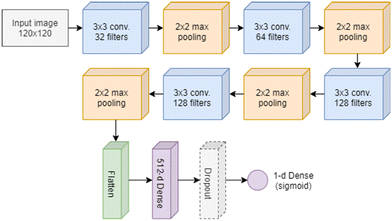
The CNN model used to discriminate ISK and ISK-CisR cells is made of two main sections, as sketched in Fig. 3. The former, which includes the convolution and pooling layers, extracts features from the images, while the latter (i.e., the fully-connected head) classifies them. The input layer takes 120 × 120 images normalized in the [0, 1] interval. It is followed by 4 blocks, each made of a 3 × 3 convolution layer with different number of filters (32, 64, 128, and 128) and a max pooling layer with 2 × 2 size, in order to reduce the number of parameters (i.e., the computational complexity) and to prevent over-fitting. All convolutional layers use Rectified Linear Unit (ReLu) as activation function. The classifier head has two fully-connected (dense) layers (512-d and 1-d) and a dropout layer again to prevent over-fitting.In particular, the final 1-d fully connected layer predicts the probability that a cell belongs to one of the two classes by using a sigmoid activation function. We trained the network using a binary cross-entropy loss, the RMSprop optimization algorithm59 with the learning rate set to 10−4, and a batch size of 32. We used an early stopping strategy in order to stop the training process when no improvement on the validation loss was obtained for at least 5 consecutive epochs.
Results
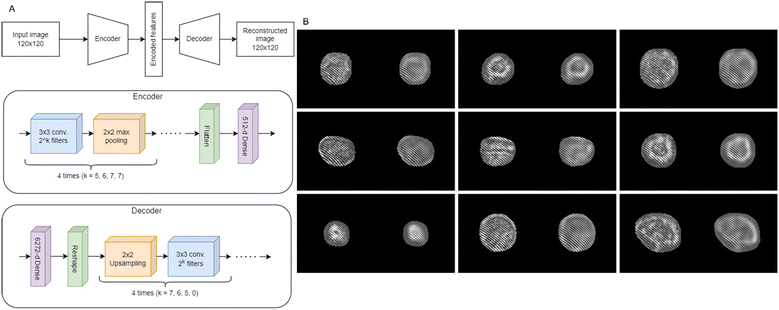
We firstly evaluate the performance of the Mask-RCNN by using 4 test videos. The network obtained 95.9%, 95.7%, 97.8%, and 94.4% AP in the bounding box detection task, with a corresponding miss rate between 5.6% and 2.1%, and 96.0%, 95.7%, 97.9%, and 94.4% AP in the cell mask prediction task with a corresponding miss rate between 5.6% and 2.2%. The abovementioned miss rate refers to the proportion of actual positive cases (cells, in this context) that the system fails to detect. It is equivalent to the false negative rate, or 1 – recall. The first two scores refer to the 2 ISK videos, while the last two scores refer to the 2 ISK-CisR videos. The same CNN classification architecture and training procedure were used for identifying ISK and ISK-CisR cells in both the mask and bbox datasets, and 99.93% and 100% accuracies were reached, respectively. Notice that the presence of the diffraction content included in the bbox case seems to push up the classification performance. Moreover, the almost equal accuracies can be explained by referring to Fig. 2, in the case of cells recorded very close to their focus plane, since their sharpness appears very low as in the numerical refocusing. However, high classification performances with simple CNN architectures, as in our case, are typically achieved when the classification problem is relatively easy to address. Definitely, by considering that the bbox strategy does not introduce any significant performance's gain, it is more effective to consider the mask detection method only for evaluating the overall performance of the proposed unbiased learning strategy due to its ability to extract only the smallest region containing the cell, thus reducing the number of informative pixels and becoming more suitable for the subsequent steps. As claimed in the introduction, the aim of this study is to test an unbiased learning strategy to identify drug-resistant EC cells, namely ISK-CisR, with respect to the background cell line, i.e., ISK. Essentially, the identification problem can be seen as a phenotyping of a cell sub-class within the same cell line, that is usually a hard issue. Therefore, one can expect that the CNN has learned intrinsic biases of the training dataset.The main causes of biases in HIFC data are related to different recording and processing settings. To avoid the latter, we implement a classifier that works on raw hologram images, thus completely skipping the processing step. Instead, there are several intrinsic recorded video properties that could be sources of bias and their removal can be a hard procedure. For example, the holographic sequences used for creating the training sets were recorded in different days, showing variation in frame brightness. In this case, the brightness may be removed by rescaling the intensity of all the holographic videos to the same average value. In addition, although training sets contain only suitable cells, the network classifies also non-cell objects as belonging to one of the two possible classes. Thus, the network may be using more than just intrinsic cell properties to classify input images. In order to extract actual cell features less likely to be biased by intrinsic video properties, we employed the convolutional autoencoder60–62 sketched in Fig. 4A. The autoencoder is a type of encoder/decoder neural network that allows for the extraction of a compact representation capable of providing a reasonably accurate reconstruction of the input, while eliminating various types of noise and perturbations at multiple levels of abstraction, such as in microscopy63 or, for example, in smoke removal from soft tissues in surgical videos,64etc. We trained the network using the Adam optimizer65 with a learning rate of 0.01, a batch size of 32 images, and the mean squared error as loss function. We used the same training, validation, and test sets as the classification experiment. We only performed the experiments on the mask dataset, in order to remove any information outside the cell boundaries that could induce a further bias. We used an early stopping strategy to stop training when the validation loss did not decrease by at least 10−7 for 30 consecutive epochs. A 512-d bottleneck produced a suitable cell reconstruction, shown as an example in Fig. 4B. It is clear that the network is able to preserve the main informative content at low-frequency, while the fine details at high-frequency are instead mostly lost. We thus expect that this approach should discard the residual experimental bias due to its unsupervised nature. Therefore, we trained a shallow fully-connected neural network with 2 hidden layers with 32 neurons each to perform the classification task according to the 512 features extracted by the CNN autoencoder. We used binary cross-entropy loss, Adam optimizer with 0.01 learning rate, and early stopping with patience of 30 epochs, and we obtained a test set accuracy of 92.26 ± 0.67% computed by averaging the results obtained by repeating 10 times the training of the shallow network, in order to take into account the random weight initialization. Of course, while always remaining high, the performances of the unsupervised approach are lower than the supervised one, but with the advantage of having removed possible biases due to the experiment that could reduce the classification robustness.
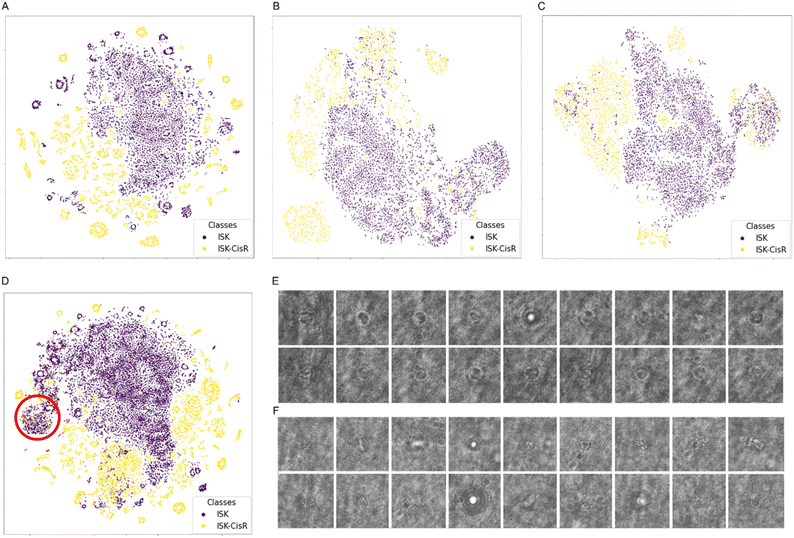
By using tSNE,66 we projected the 512 unsupervised features of the training, validation, and test sets, as well as those of the entire dataset, in order to produce the human-readable 2D plots reported in Fig. 5A–D, respectively. As further confirmation of the advantages in exploiting the holographic diffraction patterns for solving classification problems, it is evident that the two classes are well separated even by using the autoencoder features.
However, a major overlap area can be observed, as highlighted in red in Fig. 5D. As visible in Fig. 5E and F, manual inspection showed that points in this area correspond to a high number of non-cell objects, resulting from detection errors of the Mask R-CNN. Therefore, we filtered the dataset by removing this cluster, thus obtaining a cleaner dataset including 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 064 cells in the training set (10
064 cells in the training set (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 709 ISK, 6355 ISK-CisR), 7779 cells in the validation set (4894 ISK, 2885 ISK-CisR), and 8046 cells in the test set (5411 ISK, 2635 ISK-CisR). Then, we trained the shallow network using the 512-d autoencoder features of the new clean dataset. As reported by the confusion matrix in Table 1, the accuracy on the test set rose to 96.68 ± 0.61%. In order to assess the robustness of the CNN model with respect to the brightness changes, we both decreased or increased the image pixel values of the test set by 50%. In the first case, the network obtained 99.65% and 99.41% accuracies on the mask and bbox test sets, respectively, while in the second case it obtained 99.44% and 100% accuracy on the mask and bbox test sets, respectively. The almost unchanged performances point out that the network is able to generalize with respect to the brightness of the DH experiment.
709 ISK, 6355 ISK-CisR), 7779 cells in the validation set (4894 ISK, 2885 ISK-CisR), and 8046 cells in the test set (5411 ISK, 2635 ISK-CisR). Then, we trained the shallow network using the 512-d autoencoder features of the new clean dataset. As reported by the confusion matrix in Table 1, the accuracy on the test set rose to 96.68 ± 0.61%. In order to assess the robustness of the CNN model with respect to the brightness changes, we both decreased or increased the image pixel values of the test set by 50%. In the first case, the network obtained 99.65% and 99.41% accuracies on the mask and bbox test sets, respectively, while in the second case it obtained 99.44% and 100% accuracy on the mask and bbox test sets, respectively. The almost unchanged performances point out that the network is able to generalize with respect to the brightness of the DH experiment.
| Predicted | |||
|---|---|---|---|
| ISK | ISK-CisR | ||
| 7 | ISK | 96.76 ± 0.84% | 3.24 ± 0.84% |
| ISK-CisR | 3.49 ± 0.99% | 96.51 ± 0.99% | |
CNN validation in the presence of biases never seen by the trained model
In order to evaluate the effectiveness and the robustness of the proposed method with respect to further sources of biases never seen by the trained CNN model, two test sets have been created numerically for simulating the presence of higher image noise level and interference fringe variation. In general, the noise level of the holographic images as well as the direction, orientation and frequency of the interference fringe patterns can remarkably change when the digital hologram recording is made by different imaging systems. In addition, noise increasing and/or fringe pattern variation can be observed on the same optical setup if, for example, unwanted misalignment occurs. To test and validate the proposed unbiased learning method in the above cases, we randomly selected 1000 images per class from the test set used in the previous section and we simulated the biases on them. In Table 2 the resulting subset of 2000 images is named “Originally Recorded”. In particular, we create three new test sets by adding speckle noise numerically with three different standard deviations of noise, namely σ = {0.01, 0.03, 0.05}. Moreover, we simulate the fringe orientation variation in the ranges [−3, +3], [−6, +6], [−10, +10] around the recorded fringe direction. In this way, we create 6 new datasets and used them for testing the performance of the proposed CNN model.| Dataset | Accuracy (%) |
|---|---|
| Originally recorded | 99.70 |
| Speckle noise with σ = 0.01 | 99.60 |
| Speckle noise with σ = 0.03 | 99.75 |
| Speckle noise with σ = 0.05 | 99.70 |
| Fringe rotation in the range [−3, +3] | 99.15 |
| Fringe rotation in the range [−6, +6] | 98.40 |
| Fringe rotation in the range [−10, +10] | 94.35 |
Fig. 6 reports an example of an ISK cell (Fig. 6A) and the related simulated biases, i.e., the image with increased noise level obtained by adding the speckle noise with σ = 0.05 (Fig. 6B) and a fringe rotation of 10 degrees (Fig. 6C). The evaluation of the performance of the CNN model is made by calculating the average accuracy in classification, as reported in Table 2.
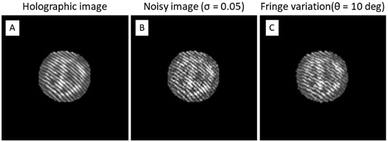 | ||
| Fig. 6 Simulation of experimental biases. A) Originally recorded ISK cell. In B a speckle noise is added to the image in A. In C a rotation of the fringe pattern is simulated on the image in A. | ||
As expected, the convolutional autoencoder is highly robust with respect to the increased noise level. In fact, even with more noisy images, the classification performance remains almost the same. Instead, the fringe variation can cause a remarkable image distortion if compared to the originally recorded one (as shown in Fig. 6A and C) but the proposed CNN model still provides a very good average accuracy. This result confirms that the convolutional autoencoder is capable of strongly mitigating different types of “noise” (i.e., biases) under specific conditions and with several experimental configurations. Finally, we consider the merged dataset obtained from the cases “Speckle noise with σ = 0.05” and “Fringe rotation in the range [−10, +10]”, to simulate multiple biases simultaneously, achieving an average accuracy of 92.84%.
Conclusions
Currently, deep learning for image classification from flow cytometry data is one of the most interesting strategies for cell phenotyping. The possibility to perform cell identification in label-free modality has been established by using a HIFC system. However, the numerical processing needed to recover quantitative phase images from recorded holograms, to be further used to train and infer the deep CNN, is often cumbersome due to the computation cost. Here we investigate the possibility of using raw holographic images as input of a CNN, thus completely skipping the processing step. Although this solution was already demonstrated in previous papers, the generalization capabilities of such models may be limited by intrinsic biases within the recording stage, thus limiting their exploitation only for data coming from the same imaging system and acquired exactly with the same setting. In this paper, we propose a learning pipeline able to counteract experimental and numerical processing biases, thus making the CNN inference robust and independent from the origin of datasets. In particular, we used a convolutional autoencoder, whose aim is to remove noise, and mitigate the biases effect, assuming them as a generic “noise”. The reported results show that the proposed solution is able to effectively overcome the effect of different experimental biases, related to image brightness, noise level and fringe orientation. We successfully apply the proposed methodology for the identification of drug resistant and non-drug resistant cells within the same EC cell line, thus demonstrating the effectiveness of the unbiased learning strategy. Of course, a generalized unbiased classification framework needs to be further investigated to include other sources of biases, also considering data collected with other HIFC systems and/or data recording settings. This would be obtained only by remarkably increasing the training set by adding data from different recording systems. Therefore, based on the achieved proof-of-concept results, this strategy is found to be promising and encouraging for further investigations in other applications of cytopathology studies. We anticipate that a fully automatic pipeline is one of the main goals to be aimed at. In fact, the miss rate of the Mask R-CNN needs to be pushed down, also avoiding the necessity of manually removing false positive predictions. Finally, further types of bias need to be considered and, in case of low performance of the current CNN model, other more complex models and strategies might be used.Author contributions
G. C., D. P. and F. B. were in charge of the data analysis/numerical results and wrote the original draft; P. Fi. was in charge of the data analysis/numerical results in the revised draft; L. X., W. X. and X. L. took care of biological samples and performed the experiment; V. B. and L. M. revised the original draft; P. M. supervised, wrote and revised the original draft; F. P., R. T. and P. Fe. revised the original draft, conceptualized and supervised the research.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by project PRIN 2017, MORphological biomarkers For Early diagnosis in Oncology (MORFEO) Prot. 2017N7R2CJ funded by the Italian Ministry of University & Research.References
- T. Blasi, H. Hennig, H. D. Summers, F. J. Theis, J. Cerveira, J. O. Patterson, D. Davies, A. Filby, A. E. Carpenter and P. Rees, Nat. Commun., 2016, 7, 10256 CrossRef CAS PubMed.
- M. Doan, I. Vorobjev, P. Rees, A. Filby, O. Wolkenhauer, A. E. Goldfeld, J. Lieberman, N. Barteneva and A. E. Carpenter, Trends Biotechnol., 2018, 36, 649–652 CrossRef CAS PubMed.
- L. Miccio, F. Cimmino, I. Kurelac, M. M. Villone, V. Bianco, P. Memmolo, F. Merola, M. Mugnano, M. Capasso, A. Iolascon, P. L. Maffettone and P. Ferraro, View, 2020, 1, 20200034 CrossRef.
- C. Probst, Y. Zeng and R.-R. Zhu, J. Pharm. Sci., 2017, 106, 1952–1960 CrossRef CAS PubMed.
- J. Hazin, G. Moldenhauer, P. Altevogt and N. R. Brady, J. Immunol. Methods, 2015, 423, 70–77 CrossRef CAS PubMed.
- Y. G. Yuanyuan Han, Y. Gu, A. Ce Zhang and Yu.-H. Lo, Lab Chip, 2016, 16, 4639–4647 RSC.
- N. S. Barteneva, E. Fasler-Kan and I. A. Vorobjev, J. Histochem. Cytochem., 2012, 60(10), 723–733 CrossRef CAS PubMed.
- M. I. Lapsley, L. Wang and T. J. Huang, Biomarkers Med., 2013, 7, 75–78 CrossRef CAS PubMed.
- P. Ferraro, A. Wax and Z. Zalevsky, Coherent Light Microscopy, Springer Series in Surface Sciences, Springer, 2011, vol. 46 Search PubMed.
- M. K. Kim, SPIE Rev., 2010, 1, 018005 Search PubMed.
- J. Min, B. Yao, V. Trendafilova, S. Ketelhut, L. Kastl, B. Greve and B. Kemper, J. Biophotonics, 2019, 12(9), e201900085 CrossRef PubMed.
- N. A. Turko and N. T. Shaked, Biomed. Opt. Express, 2020, 11, 6649–6658 CrossRef CAS PubMed.
- S. Seo, T. W. Su, D. K. Tseng, A. Erlinger and A. Ozcan, Lab Chip, 2009, 9, 777–787 RSC.
- F. Merola, P. Memmolo, L. Miccio, V. Bianco, M. Paturzo and P. Ferraro, Proc. IEEE Inst. Electr. Electron Eng., 2015, 103(2), 192–204 CAS.
- M. K. Kim, SPIE Rev., 2010, 1, 018005 Search PubMed.
- Y. K. Park, C. Depeursinge and G. Popescu, Nat. Photonics, 2018, 12, 578–589 CrossRef CAS.
- W. Choi, C. Fang-Yen, K. Badizadegan, S. Oh, N. Lue, R. R. Dasari and M. S. Feld, Nat. Methods, 2007, 4, 717–719 CrossRef CAS PubMed.
- F. Merola, P. Memmolo, L. Miccio, R. Savoia, M. Mugnano, A. Fontana, G. D'Ippolito, A. Sardo, A. Iolascon, A. Gambale and P. Ferraro, Light: Sci. Appl., 2017, 6, e16241 CrossRef CAS PubMed.
- D. Pirone, J. Lim, F. Merola, L. Miccio, M. Mugnano, V. Bianco, F. Cimmino, F. Visconte, A. Montella, M. Capasso, A. Iolascon, P. Memmolo, D. Psaltis and P. Ferraro, Nat. Photonics, 2022, 16, 851–859 CrossRef CAS PubMed.
- J. Yoon, Y. Jo, M.-H. Kim, K. Kim, S. Y. Lee, S.-J. Kang and Y. K. Park, Sci. Rep., 2017, 7, 6654 CrossRef PubMed.
- D. Pirone, L. Xin, V. Bianco, L. Miccio, W. Xiao, L. Che, X. Li, P. Memmolo, F. Pan and P. Ferraro, Sens. Actuators, B, 2023, 375, 132963 CrossRef CAS.
- D. Ryu, J. Kim, D. Lim, H.-S. Min, I. Y. Yoo, D. Cho and Y. K. Park, BME Front., 2021, 2021, 9893804 CrossRef PubMed.
- Y. Jo, H. Cho, S. Y. Lee, G. Choi, G. Kim, H.-S. Min and Y. K. Park, IEEE J. Sel. Top. Quantum Electron., 2018, 25(1), 1–14 Search PubMed.
- C. X. Chen, H. S. Park, H. Price and A. Wax, Front. Phys., 2021, 9, 759142 CrossRef.
- P. Memmolo, G. Aprea, V. Bianco, R. Russo, I. Andolfo, M. Mugnano, F. Merola, L. Miccio, A. Iolascon and P. Ferraro, Biosens. Bioelectron., 2022, 201, 113945 CrossRef CAS PubMed.
- G. Kim, Y. Jo, H. Cho, H.-S. Min and Y. K. Park, Biosens. Bioelectron., 2019, 123, 69–76 CrossRef CAS PubMed.
- Y. H. Lin, K. Y. K. Liao and K. B. Sung, J. Biomed. Opt., 2020, 25(11), 116502 Search PubMed.
- Y. Li, B. Cornelis, A. Dusa, G. Vanmeerbeeck, D. Vercruysse, E. Sohn, K. Blaszkiewicz, D. Prodanov, P. Schelkens and L. Lagae, Comput. Biol. Med., 2018, 96, 147–156 CrossRef PubMed.
- M. Ugele, M. Weniger, M. Stanzel, M. Bassler, S. W. Krause, O. Friedrich, O. Hayden and L. Richter, Adv. Sci., 2018, 5(12), 1800761 CrossRef PubMed.
- M. Ugele, M. Weniger, M. Leidenberger, Y. Huang, M. Bassler, O. Friedrich and L. Richter, Lab Chip, 2018, 18(12), 1704–1712 RSC.
- D. Roitshtain, L. Wolbromsky, E. Bal, H. Greenspan, L. L. Satterwhite and N. T. Shaked, Cytometry, Part A, 2017, 91(5), 482–493 CrossRef CAS PubMed.
- D. K. Singh, C. C. Ahrens, W. Li and S. A. Vanapalli, Lab Chip, 2017, 17(17), 2920–2932 RSC.
- N. Nissim, M. Dudaie, I. Barnea and N. T. Shaked, Cytometry, Part A, 2021, 99(5), 511–523 CrossRef PubMed.
- M. Rubin, O. Stein, N. A. Turko, Y. Nygate, D. Roitshtain, L. Karako, I. Barnea, R. Giryes and N. T. Shaked, Med. Image Anal., 2019, 57, 176–185 CrossRef PubMed.
- L. Xin, W. Xiao, L. Che, J. Liu, L. Miccio, V. Bianco, P. Memmolo, P. Ferraro, X. Li and F. Pan, ACS Omega, 2021, 6(46), 31046–31057 CrossRef CAS PubMed.
- H. Chen, L. Huang, T. Liu and A. Ozcan, Light: Sci. Appl., 2022, 11, 254 CrossRef CAS PubMed.
- D. Pirone, D. Sirico, L. Miccio, V. Bianco, M. Mugnano, P. Ferraro and P. Memmolo, Lab Chip, 2022, 22, 793–804 RSC.
- M. Delli Priscoli, P. Memmolo, G. Ciaparrone, V. Bianco, F. Merola, L. Miccio, F. Bardozzo, D. Pirone, M. Mugnano, F. Cimmino, M. Capasso, A. Iolascon, P. Ferraro and R. Tagliaferri, IEEE J. Sel. Top. Quantum Electron., 2021, 27(5), 1–9 Search PubMed.
- Y. Zhu, C. H. Yeung and E. Y. Lam, JPhys Photonics, 2021, 3, 024013 CrossRef.
- F. Bray, J. Ferlay, I. Soerjomataram, R. L. Siegel, L. A. Torre and A. Jemal, Ca-Cancer J. Clin., 2018, 68, 394–424 CrossRef PubMed.
- M. T. Doherty, O. B. Sanni, H. G. Coleman, C. R. Cardwell, W. G. McCluggage, D. Quinn, J. Wylie and U. C. McMenamin, PLoS One, 2020, 15(4), e0232231 CrossRef CAS PubMed.
- J. Lortet-Tieulent, J. Ferlay, F. Bray and A. Jemal, J. Natl. Cancer Inst., 2018, 110(4), 354–361 CrossRef PubMed.
- D. Cella, H. Huang, H. D. Homesley, A. Montag, R. Salani, K. D. Geest, R. Lee and N. M. Spirtos, Gynecol. Oncol., 2010, 119, 538–542 CrossRef CAS PubMed.
- Y. Xiao, L. Jin, C. Deng, Y. Guan, E. Kalogera, U. Ray, P. Thirusangu, J. Staub, S. S. Bhattacharya, H. Xu, X. Fang and V. Shridhar, Oncogene, 2021, 40(8), 1409–1424 CrossRef CAS PubMed.
- T. Yao, R. Cao, W. Xiao, F. Pan and X. Li, J. Biophotonics, 2019, 12(7), e201800443 CrossRef PubMed.
- T. S. Gerashchenko, M. V. Zavyalova, E. V. Denisov, N. V. Krakhmal, D. N. Pautova, N. V. Litviakov, S. V. Vtorushin, N. V. Cherdyntseva and V. M. Perelmuter, Acta Nat., 2017, 9, 56–67 CrossRef CAS.
- A. Pasqualato, A. Palombo, A. Cucina, M. A. Mariggiò, L. Galli, D. Passaro and M. Bizzarri, Exp. Cell Res., 2012, 318(7), 835–846 CrossRef CAS PubMed.
- Y. Kanintronkul, R. Worayuthakarn, N. Thasana, P. Winayanuwattikun, K. Pattanapanyasat, R. Surarit and J. Svasti, Anticancer Res., 2011, 31(3), 921–927 CAS.
- N. A. Puspita and A. Bedford, Int. J. Health Sci., 2017, 5(1), 8–14 Search PubMed.
- R. Domura, R. Sasaki, Y. Ishikawa and M. Okamoto, J. Funct. Biomater., 2017, 8(2), 18 CrossRef PubMed.
- A. Isozaki, J. Harmon, Y. Zhou, S. Li, Y. Nakagawa, M. Hayashi, H. Mikami, C. Lei and K. Goda, Lab Chip, 2020, 20(17), 3074–3090 RSC.
- Y. Li, A. Mahjoubfar, C. L. Chen, K. R. Niazi, L. Pei and B. Jalali, Sci. Rep., 2019, 9, 11088 CrossRef PubMed.
- A. Isozaki, H. Mikami, K. Hiramatsu, S. Sakuma, Y. Kasai, T. Iino, T. Yamano, A. Yasumoto, Y. Oguchi, N. Suzuki, Y. Shirasaki, T. Endo, T. Ito, K. Hiraki, M. Yamada, S. Matsusaka, T. Hayakawa, H. Fukuzawa, Y. Yatomi, F. Arai, D. Di Carlo, A. Nakagawa, Y. Hoshino, Y. Hosokawa, S. Uemura, T. Sugimura, Y. Ozeki, N. Nitta and K. Goda, Nat. Protoc., 2019, 14(8), 2370–2415 CrossRef CAS PubMed.
- Y. J. Heo, D. Lee, J. Kang, K. Lee and W. K. Chung, Sci. Rep., 2017, 7, 11651 CrossRef PubMed.
- K. He, G. Gkioxari, P. Dollar and R. Girshick, in Proceedings of the IEEE International Conference on Computer Vision (ICCV), 2017, pp. 2961–2969 Search PubMed.
- K. He, X. Zhang, S. Ren and J. Sun, in Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp. 770–778 Search PubMed.
- “openvinotoolkit/cvat: Powerful and efficient Computer Vision Annotation Tool (CVAT).” https://github.com/openvinotoolkit/cvat (accessed Jul. 23, 2021).
- T.-Y. Lin, M. Maire, S. Belongie, J. Hays, P. Perona, D. Ramanan, P. Dollár and C. Lawrence Zitnick, in European conference on computer vision, 2014, pp. 740–755 Search PubMed.
- G. Hinton, “Neural Networks for Machine Learning. Lecture 6a: Overview of mini-batch gradient descent,” Coursera. https://www.cs.toronto.edu/~hinton/coursera/lecture6/lec6.pdf (accessed Jul. 29, 2021).
- P. Baldi, in Proceedings of ICML Workshop on Unsupervised and Transfer Learning, PMLR, 2012, vol. 27, pp. 37–49 Search PubMed.
- I. Goodfellow, Y. Bengio and A. Courville, Deep Learning, MIT Press, Cambridge, MA, USA, 2016 Search PubMed.
- W. Liu, Z. Wang, X. Liu, N. Zeng, Y. Liu and F. E. Alsaadi, Neurocomputing, 2017, 234, 11–26 CrossRef.
- F. Bardozzo, T. Collins, A. Forgione, A. Hostettler and R. Tagliaferri, Med. Image Anal., 2022, 77, 102380 CrossRef PubMed.
- F. H. G. Zuluaga, F. Bardozzo, J. I. R. Patino and R. Tagliaferri, in 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), 2021, pp. 3483–3486 Search PubMed.
- D. P. Kingma and J. Ba, arXiv, 2014, preprint, arXiv:1412.6980, DOI:10.48550/arXiv.1412.6980.
- L. van der Maaten and G. Hinton, J. Mach. Learn. Res., 2008, 9, 2579–2605 Search PubMed.
| This journal is © The Royal Society of Chemistry 2024 |