Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Scalable mesenchymal stem cell enrichment from bone marrow aspirate using deterministic lateral displacement (DLD) microfluidic sorting†
Nicholas
Tan Kwan Zen‡
 ab,
Kerwin Kwek
Zeming‡
ab,
Kerwin Kwek
Zeming‡
 a,
Kim Leng
Teo
d,
Mavis
Loberas
ef,
Jialing
Lee
d,
Chin Ren
Goh
a,
Da Hou
Yang
a,
Steve
Oh
d,
James
Hui Hoi Po
ef,
Simon M.
Cool
a,
Kim Leng
Teo
d,
Mavis
Loberas
ef,
Jialing
Lee
d,
Chin Ren
Goh
a,
Da Hou
Yang
a,
Steve
Oh
d,
James
Hui Hoi Po
ef,
Simon M.
Cool
 fgh,
Han Wei
Hou
fgh,
Han Wei
Hou
 *abc and
Jongyoon
Han
*abc and
Jongyoon
Han
 *aij
*aij
aCritical Analytics for Manufacturing of Personalized Medicine, Singapore-MIT Alliance for Research and Technology (SMART), 138602, Singapore
bSchool of Mechanical and Aerospace Engineering, Nanyang Technological University, 639798, Singapore
cLee Kong Chian School of Medicine, Nanyang Technological University, 308232, Singapore
dBioprocessing Technology Institute, Agency for Science, Technology and Research (A*STAR), 138668, Singapore
eNUS Tissue Engineering Program, Life Sciences Institute, National University of Singapore, 117510, Singapore
fDepartment of Orthopaedic Surgery, Yong Loo Lin School of Medicine, National University Health System, National University of Singapore, 119288, Singapore
gInstitute of Molecular and Cell Biology, Agency for Science, Technology and Research (A*STAR), 138668, Singapore
hSchool of Chemical Engineering, University of Queensland, Brisbane, 4072, Australia
iDepartment of Biological Engineering, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, USA
jDepartment of Electrical Engineering and Computer Science, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, USA. E-mail: jyhan@mit.edu
First published on 6th September 2023
Abstract
The growing interest in regenerative medicine has opened new avenues for novel cell therapies using stem cells. Bone marrow aspirate (BMA) is an important source of stromal mesenchymal stem cells (MSCs). Conventional MSC harvesting from BMA relies on archaic centrifugation methods, often leading to poor yield due to osmotic stress, high centrifugation force, convoluted workflow, and long experimental time (∼2–3 hours). To address these issues, we have developed a scalable microfluidic technology based on deterministic lateral displacement (DLD) for MSC isolation. This passive, label-free cell sorting method capitalizes on the morphological differences between MSCs and blood cells (platelets and RBCs) for effective separation using an inverted L-shaped pillar array. To improve throughput, we developed a novel multi-chip DLD system that can process 2.5 mL of raw BMA in 20 ± 5 minutes, achieving a 2-fold increase in MSC recovery compared to centrifugation methods. Taken together, we envision that the developed DLD platform will enable fast and efficient isolation of MSCs from BMA for effective downstream cell therapy in clinical settings.
I. Introduction
The advent of regenerative medicine marks a new paradigm shift in medical treatment through therapeutic cell delivery by autologous or allogenic means to replace/regenerate damaged cells, thereby creating personalized medicine.1,2 The discovery of stromal mesenchymal stem cells (MSCs) has led to the pursuit of pioneering novel therapies due to their tri-lineage differentiation potential into mesodermal cell types in vitro (adipocytes, chondrocytes, and osteocytes).3 This unique multilineage differentiation has spearheaded MSC therapies to treat health conditions such as neurological disorders,4,5 cardiac ischemia,6 diabetes,7,8 and cartilage diseases.9 Apart from their unique differentiation capabilities, MSCs are also known to exhibit immunomodulatory properties,10 enabling expanded cell therapeutic applications. These cells can be found in various biological samples such as adipose tissues,11 umbilical cord blood12 and salivary glands.13 A traditional MSC harvesting method is aspiration from human bone marrow (BMA)3,14–16 as it has a relatively higher MSC quantity compared to other tissues. Despite the abundance of MSCs in the bone marrow, they only constitute 0.01% and 0.005% of the bone marrow mononucleated cell (BMMNC) population for males and females, respectively.17 This advocates a critical need for more effective and efficient MSC extraction from biological samples.Advancements in cell sorting methods exploit cells' biophysical and biochemical properties. Current cell sorting techniques such as fluorescence-activated cell sorting (FACS) and magnetic-activated cell sorting (MACS) leverage antibody labeling targeted to cell surface markers.18,19 These techniques can reproduce extremely high sorting resolution but are limited by the high cost of antibodies, bulky machinery, and laborious sample preparation. They also require technical expertise and high operating pressure, which may influence MSC consistency and cell integrity.20 Besides, it has been challenging to determine MSC-specific cell surface markers.21 Antibody binding on cells may also affect their native biological or signalling pathways,22 thus unsuitable for downstream therapeutic purposes.
The conventional MSC isolation technique from BMA adopts a label-free density-based centrifugation23–25 method known as Ficoll density gradient centrifugation (DGC). Through Ficoll DGC, the mononuclear cell (MNC) fraction contains a heterogeneous cell population consisting mainly of monocytes, lymphocytes, hematopoietic stem cells (HSCs), and MSCs. Despite its wide usage, Ficoll DGC has the poorest yield amongst different MSC isolation techniques, including RBC lysis, immunomagnetic depletion, and simple filtration.26–28 It was shown that Ficoll DGC typically suffers from significant cell loss amongst human progenitor cells (30% for HSCs and 55% for MSCs).26 MSCs can aggregate to form large clusters, congregating in the polymorphonuclear (PMN) fraction during centrifugation.29 Apart from the modest cell recovery, the workflow for Ficoll DGC (∼2 to 3 h) suffers from poor repeatability due to inconsistent preparation protocols, clinical personnel training, and increased exposure to contamination.
Microfluidic technologies have generated substantial advancement in cell sorting technologies over the years.20,30 Cell sorting using microfluidics often leverages the intrinsic biophysical properties of cells, rendering the techniques label-free.31 They can be classified into two groups: active and passive methods. Active methods exploit externally applied fields or forces such as acoustophoresis,32 electrokinetics,33 and magnetophoresis34 to enhance separation. In contrast, passive methods rely on inherent hydrodynamic interactions between the channel structures and cells for separation, optimized by manipulating channel geometries.20 Passive methods include inertial focusing,35 deterministic lateral displacement (DLD),36 and hydrodynamic filtration.37 These microfluidic techniques are often easy to operate and integrate and are scalable into clinical sample processing workflows. To our knowledge, no work has been done on directly isolating MSCs from whole undiluted BMA using microfluidic techniques.
Previous studies addressing the separation of MSCs have relied mostly on the inertial focusing technique.38–40 However, these investigations focus on size-based sorting for separating different MSC subpopulations from a pure culture population of MSCs. A significant constraint posed by this technique is the low sample concentration required for effective sorting to occur, limiting the use in high cell concentration samples such as blood or bone marrow samples. Conversely, DLD has demonstrated proficiency in processing samples with high concentrations,41–43 making it more suitable for clinical applications with the absence of preprocessing steps or reagents. Therefore, this study aims to determine the potential of employing DLD technology for direct BMA processing as an alternative to Ficoll DGC, the preferred method for isolating MSCs. We demonstrate a simple user-friendly workflow for BMA processing using a benchtop microfluidic pressure system. DLD, first described by Huang et al.36 in 2004, leverages micropillar arrays to sort cells based on size and deformability with promising recovery rate and purity.44 The sorting criteria (critical diameter, DC) can be calculated by an empirical formula based on the geometrical parameters of the pillar arrays where particles larger than the DC are displaced laterally, away from their original stream.45 Particles smaller than the DC will remain in their original stream, causing a distinct separation between the larger and the smaller cells. DLD sorting using peripheral blood samples has produced high recovery and purity of nucleated cells.41–43 However, the cellular composition between peripheral blood and BMA differs. The latter comprises a higher amount of immature blood cells and nucleated cells with larger variations in cell biophysical properties, such as cell size, as studied by Xavier et al.46 Thus, the efficiency of direct MSC isolation from undiluted BMA samples remains unknown.
Three parameters, namely recovery rate, total experimental time, and MSC colony forming unit-fibroblast (CFU-F) count validated through FACS analysis, will be used to determine the effectiveness and efficiency of MSC isolation between DLD sorting and Ficoll DGC. To improve throughput scalability, we have also developed a novel multi-chip DLD system that can process 2.5 mL of raw BMA in 20 ± 5 minutes, achieving a 2-fold increase in MSC recovery compared to the Ficoll DGC protocol. Overall, the DLD platform shows excellent potential for fast and efficient isolation of MSCs from BMA in clinical settings.
II. Methods and materials
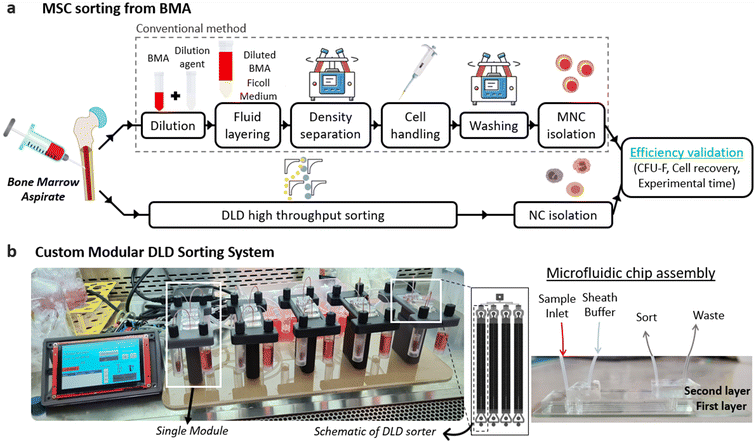
A. DLD device design
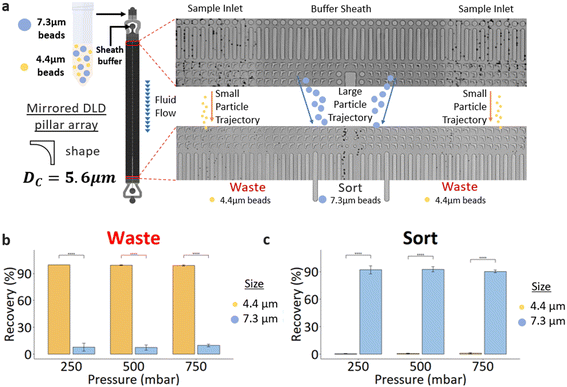
A high-throughput chip consisting of four DLD sorters (Fig. 1b), connected in parallel, was developed to increase the processing speed for BMA. This high-throughput chip involves layering two PDMS substrates where the second PDMS layer comprises channel connectors combining four channels into one, thereby reducing experimental complexity. Each DLD device design is based on a mirrored DLD pillar array and a centric sheath flow. Samples are injected from the sides of the device, and particles larger than the DC are displaced laterally into the center sheath stream and sorted into the collection outlet (Fig. 2a). Particles smaller than the DC follow their original stream, and they are not displaced, causing a separation between the smaller and the larger particles. Quantification of cells in BMA from flow cytometry in previous studies47–49 provides little information on biophysical properties such as size, shape, and deformability of MSCs in BMA. Thus, the effective DC of the device used in this study is approximately 5.6 μm with an unconventional inverted L-shaped pillar array instead of the original circle pillar array. This value is calculated empirically using an equation developed experimentally by Davis et al.50 and follows the form:DC = 1.4G![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) tan tan![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ0.48 θ0.48 | (1) |
B. DLD device fabrication and preparation
Fabrication of a silicon master mold follows a standard photolithographic process using a chromed quartz photomask (JD Photodata, UK). An SU-8 mold was made using SU-82020 spun at 4500 RPM, resulting in a final resist thickness of 18 μm. The fabrication of the master mold was made using standard photolithography techniques via exposure, post-exposure bake, and development. The master mold underwent a silanization step using trichloro(1H,1H,2H,2H-perfluorooctyl)silane (Sigma-Aldrich, Singapore) to ensure a hydrophobic coating and ease of downstream DLD chip manufacturing. The production of DLD chips follows a soft lithographic process using the master mold. PDMS (Dow Corning, USA) is mixed in a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio with its curing agent and poured over the master mold taped onto a Petri dish. The mixture will be degassed in a desiccator for 30 minutes before baking the degassed PDMS at 80 °C for 2 hours. After curing, a surgical knife is used to cut the PDMS devices from the mold.
1 ratio with its curing agent and poured over the master mold taped onto a Petri dish. The mixture will be degassed in a desiccator for 30 minutes before baking the degassed PDMS at 80 °C for 2 hours. After curing, a surgical knife is used to cut the PDMS devices from the mold.
Holes of different sizes are punched at the ingress and egress of the desired chip (1 mm for sample inlet, sheath flow, and outlet (sort), 1.5 mm for waste). The resulting PDMS product will be bonded onto a glass slide cleaned with 70% ethanol, using oxygen plasma treatment, and placed onto a hot plate at 140 °C for 10 minutes to ensure complete bonding. PDMS-to-PDMS bonding follows the same protocol as PDMS-to-glass bonding, with the only difference being placing the bonded product into an oven at 80 °C overnight instead of a hot plate.
Prior to experiments using biological samples, DLD chips are primed using 2% (w/v) Pluronic™ F-127 (Sigma, SG) with 20% isopropyl alcohol (IPA) for 30 minutes to prevent nonspecific adsorption and easy bubble removal due to its low surface tension properties from the ingress and 10 minutes from the egress using a pressure pump. Then, the chips will be washed with Dulbecco's phosphate-buffered saline (DPBS) (Thermo Fisher Scientific PTE Ltd, Singapore) or Gibco Dulbecco's modified Eagle's medium (GE Healthcare, USA) for 5 minutes from the ingress to cultivate a suitable cellular environment as Pluronic™ F-127 is toxic to cells.
C. Single-device characterization
To characterize sorting performance, the DLD chip is placed onto a microfluidic device platform with imaging capabilities powered by an Arduino microcontroller and a positive displacement pump (RS PRO, Singapore). Custom fittings are designed and 3D-printed using low-force stereolithography (Form 3, Formlabs), forming an airtight junction between the sample reservoir and the tubing (Cole-Parmer, USA) using O-rings. The tubing length is 30 cm. Microsphere counting techniques leverage computer vision technology, and cell counting is performed using a hemocytometer.D. Modular high-throughput sorting
For high-throughput sorting targeted for clinical applications, a new benchtop multi-chip microfluidic platform was developed, which allows multiple chips to operate concurrently. We have established a clear, easy-to-follow experimental workflow to complement our system. Tubing, fittings, and the prototype are sterilized in a biological safety cabinet (BSC) under UV light for 30 minutes before use. Experiments using BMA are also performed in the BSC to ensure sterility.A pressure sensor (AMS 5915, AMSYS, Germany) and pump setup was assembled to control the desired operating pressure. To offset the pressure drop at the tubing and connections, we integrated a simple pressure cutoff control in our PCB design that monitors the pressure continuously and activates the pump whenever the sensor detects a drop below the 10% threshold. Pneumatic solenoid valves (Parker Hannifin, USA) were connected to a manifold inside the pressure chamber to enable proper distribution of pressure to the correct ports. The pressure chamber with manifold, modular unit, and human–machine interface (HMI) stand is fabricated using 3D printing. The system components can be found in Fig. S3.†
E. Peripheral blood samples
Peripheral whole blood was obtained following informed consent under the approval of the ethics committee according to a protocol permitted by the SingHealth Centralised Institutional Review Board (CIRB Ref: 2022/2322). 100 μL of whole peripheral blood was processed from a 3 mL venous blood draw into a 3 mL EDTA tube. The blood sample was processed within 4 hours of the blood draw to prevent inherent aggregation and degradation of samples. The blood samples were stored at room temperature to ensure minimal biophysical changes to the nucleated cells.F. Bone marrow aspirate samples
Approximately 50 mL of bone marrow was harvested from the iliac crest of three healthy donors aged 26–52 under local anesthesia, following informed consent and strict adherence to ethical guidelines reviewed by the National Healthcare Group, Singapore (NHG DSRB 2019/00284). The aspiration was performed using a Jamshidi needle; the aspirate was collected into heparinized syringes and transferred to sterile containers. The bone marrow aspirate was transported to the National University of Singapore Tissue Engineering Programme (NUSTEP) for further processing. From the 50 mL aspirate, 5 mL was used for Ficoll DGC and DLD sorting at the Bioprocessing Technology Institute, A*STAR, Singapore. The remaining aspirate was not used in this study.G. BMA processing using Ficoll DGC
Heparinized bone marrow aspirate was first diluted with an equal amount of D10 culture medium. This culture medium consisted of Dulbecco's modified Eagle's medium (DMEM), 1000 mg ml−1 glucose supplemented with 10% fetal bovine serum and 2 mM L-glutamine. The diluted aspirate is carefully layered on top of 1.078 g ml−1 Ficoll-Paque PREMIUM (Cytiva, USA) at a ratio of 2.5 mL diluted aspirate to 2 mL Ficoll-Paque. The solution was centrifuged at 400g for 30 minutes at ambient temperature without brake mode. Next, the interface layer containing the mononuclear cells was collected and washed twice by centrifugation at 200g for 15 minutes at maximum brake mode using complete DMEM.H. Mononuclear cell culture
Following the user guide, cells obtained from the Ficoll DGC method and DLD sorting were counted using a NucleoCounter NC-250 analyzer (Chemometec, Denmark). Cells (3.5 × 106) were seeded into a 25 cm3 tissue culture flask (Thermo Fisher Scientific, Singapore) with 5 mL of D10 culture medium. DNase I (Roche, Germany) was added into the culture medium at 20 U mL−1 concentration. Culture medium change was done at three- to four-day intervals. The spent culture medium was centrifuged at 380 RCF for 5 minutes at each culture medium change. The supernatant was discarded, and the pellet was resuspended in 5 mL fresh culture medium. DNase I at 20 U mL−1 was added. The fresh medium with resuspended cells was seeded into the cell culture. Cells were cultured until 80% confluency was reached. These cells are termed passage 0 mesenchymal stem cells (P0 MSCs).I. Mesenchymal stem cell culture
Confluent P0 MSCs cultured in 25 cm3 tissue culture flasks were passaged using 0.25% trypsin-EDTA dissociation solution (Gibco, Singapore). The passaged cells were counted using NucleoCounter NC-250 and seeded into a 75 cm3 tissue culture flask (Thermo Fisher Scientific, Singapore) with 15 ml of D10 culture medium at a seeding density of 5000 cells per cm3. Cells were cultured until 80% confluency was reached. These cells are called passage 1 mesenchymal stem cells (P1 MSCs) at this stage. These P1 MSCs were then further passaged and seeded into (i) a 175 cm3 tissue culture flask (Thermo Fisher Scientific, Singapore) with 35 mL D10 culture medium at a seeding density of 5000 cells per cm3 and cultured until 80% confluent, and (ii) a 6-well tissue culture plate (Thermo Fisher Scientific, Singapore) with 2 mL D10 culture medium at a seeding density of 65 cells per well and cultured for 14 days with culture medium change on day 7 for CFU-F analysis. These cells were termed passage 2 mesenchymal stem cells (P2 MSCs).J. MSC quality analysis: colony-forming unit-fibroblast (CFU-F)
P2 MSCs cultured on a 6-well tissue culture plate at a seeding density of 65 cells per well for 14 days with culture medium change on day 7 were used for CFU-F analysis.51 The cells were washed with sterile phosphate-buffered saline (PBS) (Gibco, Singapore) and stained with 2 mL of CFU-F stain for 30 minutes at room temperature. The CFU-F stain was prepared by dissolving a 0.5% weight-to-volume ratio of crystal violet (Sigma-Aldrich, Singapore) in a 25% volume-to-volume ratio of methanol (Sigma-Aldrich, Singapore) in deionized water. The cells were then rinsed with PBS thrice to remove excess stain. The plate containing the stained cells was dried. Once the plate had dried, the number of colonies formed was counted. A colony is defined as a region with a minimum of 50 cells. Any areas with less than 50 cells were not considered a colony and thus were not counted.K. MSC quality analysis: cell surface marker (FACS)
P2 MSCs cultured in a 175 cm3 tissue culture flask at a seeding density of 5000 cells per cm3 until 80% confluency were used for cell surface marker (FACS) analysis. Firstly, cells were washed with sterile PBS, dissociated from the tissue culture flask using 0.25% trypsin-EDTA, and counted using NucleoCounter NC-250. 1 × 105 cells were used for each cell surface marker analysis. The positive selection markers used for the study were CD73, CD90, CD105, and CD146, and the negative selection markers were CD34 and CD45. Cells were washed in FACS buffer, which was made by dissolving a 1% weight-to-volume ratio of bovine serum albumin (Sigma-Aldrich, Singapore) in PBS. The washed cells were then added to a 96-well V-bottom plate (Greiner Bio-One, Germany) and individually incubated with each fluorophore-conjugated antibody of positive and negative selection markers (Biolegend, USA) on ice for 30 minutes. The cells were washed with FACS buffer to remove excess unbound antibodies and resuspended in FACS buffer. According to the user manual, cells were analyzed using Novocyte (ACEA Biosciences, USA).III. Results
A. DLD beads sorting characterization and optimization
To calibrate the DLD chip, polystyrene microspheres (Bangs Laboratory, USA) of sizes 4.4 μm and 7.3 μm were used at a stock 1.125% (w/v) concentration. This test illustrates the size-based sorting capabilities as the rigid beads are larger and smaller than the DC (5.6 μm). As the pressure and corresponding flow rate increased, the sorting of 7.3 μm beads showed a minor decrease in bead recovery (92.14 ± 3.85% to 90.25 ± 1.52%). Using the inverse L-shaped pillars could result in possible anisotropic effects as shown by Vernekar et al.52 By comparing the sorting efficiency of 4.4 μm beads, the DLD device with a DC of 5.6 μm performs as designed in the operating pressure range of 250 to 750 mbar. The 7.3 μm beads accounted for 7–10% of the total count at the waste outlet (Fig. 2b) and were not deflected via DLD (see ESI† Video S1 and S2). This is likely due to the functional design of the device, where the inlet sample flow interacts with the DLD array side walls, resulting in inefficient sorting near the walls.B. Peripheral blood sorting
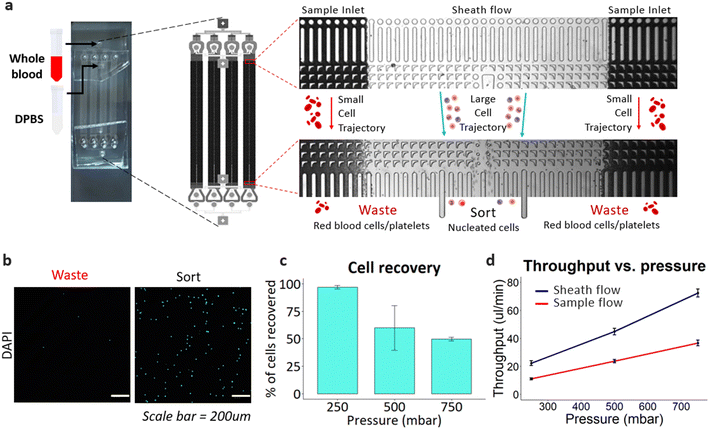
Peripheral blood was used to elucidate the cell deformability effects and sorting efficiency of the DLD device. Unlike rigid beads, it is known that cell deformability impacts the DLD sorting efficiencies.42,46,53,54 The same DLD device used in Fig. 2 was scaled linearly up to 4× by connecting the devices in parallel with a second layer microfluidic sample distribution channel at the input and output. Undiluted and unfiltered whole blood was processed using the device, as shown in Fig. 3a and ESI† Video S3 and S4.To facilitate the output cell count, cells are DNA stained with Hoechst 33342 fluorescent stain solution and counted with a hemocytometer using brightfield and DAPI microscopy. With fluorescence field imaging, the cell recovery rate is quantified through manual counting and carried out as follows:
C. MSC isolation from human bone marrow aspirate using DLD
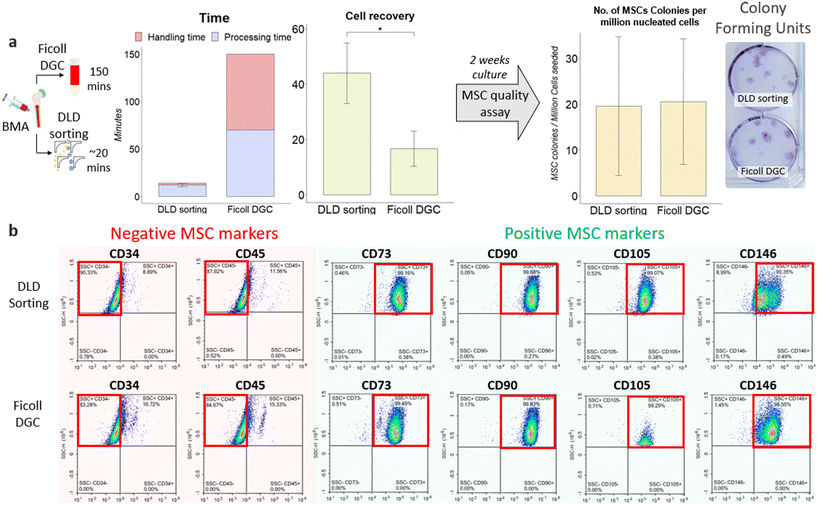
To validate our proof of concept that DLD sorting works better than Ficoll DGC, we performed two comparative sorting experiments: Ficoll centrifugation and DLD sorting. The Ficoll DGC procedure has been explained in section IIG. Microfluidic research on MSC cell sorting is commonly performed using cell lines, cultured cells which are spiked samples and not physiologically and clinically relevant.26,27,46,60,61 Therefore, it is critical to process BMA samples directly for clinical impact. The primary goal of this study is to develop a proof-of-concept sorting of BMA and evaluate the quality of sorted cells in comparison to Ficoll DGC. As part of efforts to increase throughput, five high-throughput chips were used, resulting in twenty parallel DLD devices operating concurrently. The workflow is described in Discussion S1.†Our undiluted BMA sample had an initial cell concentration of 8.59 ± 3 × 106 cells per mL and was subjected to a 20 μm filter before sorting to remove unwanted coagulation (see Fig. S5†). After sorting, we combined the outlet (sort) vials from each module into a 15 mL vial before data analysis. This was done likewise for the waste vials. The measured cell recoveries between peripheral blood and BMA at 750 mbar differ slightly (49.8 ± 1% and 43.9 ± 6.3%, respectively). Since the design of our DLD array is based on the static cell size, this illustrates the difference between nucleated cell sizes in BMA from that in the literature (Table 1). One of the reasons for the size disparity could be due to interactions with the DLD pillars causing cells to deform and change in size. Despite the lower cell recovery rate, the yield is better than that of Ficoll DGC by a factor of two (Fig. 4a).
| Myeloid cells | Monocytes | Blast cells | Lymphocytes | BM stem cells | |
|---|---|---|---|---|---|
| Size (μm) | 15 ± 5 | 16 ± 4 | 13 ± 5 | 6–15 | 9.1 ± 1.6 |
In contrast, Ficoll DGC has a fixed, established protocol, and the time taken is independent of sample volume. The Ficoll DGC method can take 2–3 hours (volume processed can range from 2 to 35 mL with dilution), and a series of centrifugation steps are needed to achieve better results. The time plot in Fig. 4a includes both processing and handling time. Processing time refers to the total sorting/isolation duration, and handling time refers to the time taken to handle the cells before/during the sorting, which includes washing and dilution. Studies have also shown that the Ficoll medium is toxic to cells,62 and multiple centrifugation might increase cell death due to prolonged exposure to the Ficoll medium. The results showed that DLD sorting reproduced a comparable number of colonies per million cells to that of Ficoll DGC at P0 (Fig. 4a). However, since the cell recovery is two times higher than that of Ficoll DGC, the total number of colonies and MSC recovery is significantly higher for DLD sorting. To verify that these colonies are MSCs, we performed a phenotypic assay after the second sub-cultivation (passage 2) to examine for MSC surface marker expression (CD73, CD90, CD105, CD146) and clear of the hematopoietic cell lineage (CD34−, CD45−) using FACS as iterated under section IIK. The findings (Fig. 4b) indicate that the MSC population is similar in the Ficoll DGC colonies. However, the total number of MSCs is still higher in DLD sorting due to higher cell yield. The quality and purity of the cells were also assessed using the same markers, showcasing that the DLD sorting method is able to effectively isolate the MSC population with minimal changes to the quality of MSCs (Fig. 4b and S6 and Table S2†). We also performed a CFU assay in passage 2 showing minimal differences in quality (Table S3†). These outcomes further substantiate the DLD method for successful isolation and enrichment of MSCs.
IV. Discussion
Stem cell therapy is the new disruptive innovation in regenerative medicine. Multiple biological tissues have been reported to contain MSCs, and identifying the most effective MSC isolation technique from a suitable source is critical for sorting functional MSCs. Despite having a low MSC yield, Ficoll DGC has long been the primary method for MSC isolation from BMA (Table 2).| Separation techniques | MSC harvesting methods/medium | Bone marrow aspirate processing | MNC recovery | Time taken | Label free | Ref. |
|---|---|---|---|---|---|---|
| Density gradient centrifugation | Ficoll-Paque™ | Centrifuged at 200g for 10 minutes | 13.5% | 70 minutes | Yes | 63 |
| Percoll | 15.7% | Yes | ||||
| Ficoll-Paque® PREMIUM | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 PBS dilution 2 PBS dilution |
30.9 ± 9.3% | 30 minutes | Yes | 64 | |
| BD Vacutainer® Cell Preparation Tube™ (CPT) | Undiluted | 22.9 ± 9.3% | 20 minutes | Yes | ||
| Cell lysis | Red blood lysis | Reported as 10 times better than Ficoll centrifugation | 10 minutes | Yes | 27 | |
| Immuno-depletion | MACS (removal of CD15+ PMNs) | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 PBS/3% FCS + 5 mM EDTA (RBC lysed) 10 PBS/3% FCS + 5 mM EDTA (RBC lysed) |
55% | 20 minutes | No | 26 |
| PluriBeads (removal of CD15+ PMNs) | 58% | 40 minutes | No | |||
| Filtration | Porous biological material | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 alpha-MEM containing 10% [v/v] fetal bovine serum (RBC lysed) 2 alpha-MEM containing 10% [v/v] fetal bovine serum (RBC lysed) |
41.3 ± 15.5% (carrier material dependent) | Not reported | Yes | 65 |
| Microfluidics | DLD sorting | Undiluted | 43.9 ± 6.3% | 13 minutes | Yes | NIL |
Comparing the recovery across all MSC harvesting methods, our current DLD design offers one of the best alternatives with no sample dilution or manual washing steps (handling time). “Time taken” in Table 2 refers to the reported processing time of these methods and does not include sample handling time such as pipetting, centrifuge braking or sample filtration which could plausibly increase the experimental timing significantly. However, what sets the DLD sorting technology apart from the rest of these methods is the absence of sample preparation and pre- and post-sort washing steps and potential scalability. The precision and continuous flow single-step sorting also eliminates variability associated with the cell handling steps. For the multistep Ficoll DGC process, the same personnel performing the technique will exhibit some form of variability due to human error, which is almost impossible to eliminate. DLD sorting also inherently integrates a washing step where sorted cells are migrated into the buffer stream, thus removing the need for additional washing steps.
The current sorting efficiency for MSCs is 43.9 ± 6.3%, and improvements on the nucleated cell recovery and processing time can be achieved by further optimizing microfluidic DLD specifications by reducing the critical size specification to sort out smaller ones or more deformable cells at the same or higher flow rate. It can be hypothesized that sorting smaller nucleated (mononuclear and polynuclear) cells will obtain a higher yield of MSCs than sorting only mononuclear cells via Ficoll DGC. Early evidence from this paper has shown that mononuclear cell sorting loses a large fraction of MSCs. This was also validated by Horn et al.,27 who employed a whole blood lysis protocol to extract all nucleated cells, showing that there exists a MSC population that is missed by mononuclear cell sorting methods. Given the similar cell recovery values between peripheral blood and BMA at 750 mbar, we are confident that our DLD device can achieve a higher cell recovery (∼80–90%) if deformability effects are negated (running at 250 mbar) at the expense of throughput. Also, expanding these cells to the desired cell quantity is often necessary to meet the requirements for MSC-based therapies. However, increased passaging is known to impair MSC plasticity and contribute to genomic instability, telomere shortening, and heterogeneity,66 which limits the therapeutic effects of MSCs, thereby reinforcing the need for an effective sorting method. While the CFU potential is comparable between the two approaches, DLD offers a higher cell recovery, translating to a higher MSC recovery. Given the rare occurrence of MSCs in BMA, a higher yield during sorting can lead to exponential cell numbers in downstream expansion.
While microfluidic chip stacking has been explored by various groups67,68 to enable scaling of sample processing, this paper investigates the possibility of scaling the sorting process using a modular sort unit that can be seamlessly integrated into a central pressure distribution setup. The microfluidic setup and processing workflow can also be used for leukapheresis with very high yield at lower flow rates. Future development of this prototype can focus on integrating portability, streamlined functionality, and user-friendly design to enable operation by non-trained users/clinicians. Without the need for multiple washing steps (Table 2), the major benefit of using the DLD sorting technique is the integration with bioreactors in a closed-loop processing of cell separation and manufacturing with the potential to reduce contamination and workforce cost and to improve consistency.
Isolating cells by label-free parameters such as size and deformability enables the enrichment of specific cell functions or phenotypes. The next questions to be answered following the sorting are: Do the sorted cells have differing growth potentials? What is the differentiation potential of MSCs to be of a particular phenotype for regenerative medicine? To quantify the functional qualities of these DLD-sorted MSCs, it is paramount that stemness and immunomodulation are validated through functional assays such as RT-PCR and differentiation assay, which serves as a motivation for future study. A previous study by Yin Lu et al. has ascertained that MSCs of sizes 17–21 μm have the most potential for chondrogenesis using inertial sorting.39 While downstream quantitative assays to test for the mesoderm cell lineage differentiation were not explored in this study, the utility of DLD sorting goes beyond a sample processing tool with the potential for specific phenotypical enrichment for more efficacious cell manufacturing.
V. Conclusion
In this study, we have developed a multi-chip DLD microfluidic platform for MSC sorting from BMA samples directly and demonstrated superior separation performance compared to Ficoll DGC (conventional standard). Improving the chip throughput and automation in our system makes this clinical workflow faster (∼20 min), more user-friendly and more robust for MSC isolation. With its label-free approach, this platform technology is low-cost, and we can envision it to be readily adapted to sorting other cell types by controlling DLD pillar shapes and geometries.Author contributions
N. T. K. Z., K. K. Z., H. W. H., S. O., S. M. C., and J. H. designed aspects of the project study. N. T. K. Z. and K. K. Z. designed and fabricated the chip. N. T. K. Z., K. K. Z., G. C. R., and Y. D. H. optimized and performed the experiments. N. T. K. Z., K. K. Z. and G. C. R. designed, fabricated, and calibrated the electronics and sorting setup. N. T. K. Z. and K. K. Z. analysed the data. N. T. K. Z. coded the algorithms for software and hardware. J. H. H. P. performed the bone marrow aspiration. T. K. L., M. L., J. L., and S. O. performed the patient recruitment and post-sort cell measurements and FACS profiling. N. T. K. Z., K. K. Z., S. M. C., H. W. H. and J. H. wrote the manuscript. All authors reviewed and approved the manuscript prior to submission.Conflicts of interest
We declare no conflict of interest.Acknowledgements
We thank Afizah Binte Mohd Hassan and Ren Xiafei for ensuring seamless coordination and operation during patient recruitment. We also like to express our gratitude to Dr. Francesca Lorraine Lim from the Singapore General Hospital for her assistance with the blood collection. This research is supported by the National Research Foundation, Prime Minister's Office, Singapore, under its Campus for Research Excellence and Technological Enterprise (CREATE) programme, through Singapore-MIT Alliance for Research and Technology (SMART): Critical Analytics for Manufacturing Personalized-Medicine (CAMP) Inter-Disciplinary Research Group.References
- A. S. Mao and D. J. Mooney, Regenerative medicine: Current therapies and future directions, Proc. Natl. Acad. Sci. U. S. A., 2015, 112(47), 14452–14459 CrossRef CAS PubMed.
- A. Hmadcha, et al., Therapeutic Potential of Mesenchymal Stem Cells for Cancer Therapy, Front. Bioeng. Biotechnol., 2020, 8, 43 CrossRef PubMed.
- M. F. Pittenger, et al., Multilineage potential of adult human mesenchymal stem cells, Science, 1999, 284(5411), 143–147 CrossRef CAS PubMed.
- C. T. van Velthoven, et al., Mesenchymal stem cell transplantation attenuates brain injury after neonatal stroke, Stroke, 2013, 44(5), 1426–1432 CrossRef CAS PubMed.
- B. Soria, et al., Human Mesenchymal Stem Cells Prevent Neurological Complications of Radiotherapy, Front. Cell. Neurosci., 2019, 13, 204 CrossRef CAS PubMed.
- C.-B. Liu, et al., Human Umbilical Cord-Derived Mesenchymal Stromal Cells Improve Left Ventricular Function, Perfusion, and Remodeling in a Porcine Model of Chronic Myocardial Ischemia, Stem Cells Transl. Med., 2016, 5(8), 1004–1013 CrossRef PubMed.
- Y. Si, et al., Infusion of Mesenchymal Stem Cells Ameliorates Hyperglycemia in Type 2 Diabetic Rats: Identification of a Novel Role in Improving Insulin Sensitivity, Diabetes, 2012, 61(6), 1616–1625 CrossRef CAS PubMed.
- V. Capilla-González, et al., PDGF Restores the Defective Phenotype of Adipose-Derived Mesenchymal Stromal Cells from Diabetic Patients, Mol. Ther., 2018, 26(11), 2696–2709 CrossRef PubMed.
- N. Ozeki, et al., Not single but periodic injections of synovial mesenchymal stem cells maintain viable cells in knees and inhibit osteoarthritis progression in rats, Osteoarthritis Cartilage, 2016, 24(6), 1061–1070 CrossRef CAS PubMed.
- N. Song, M. Scholtemeijer and K. Shah, Mesenchymal Stem Cell Immunomodulation: Mechanisms and Therapeutic Potential, Trends Pharmacol. Sci., 2020, 41(9), 653–664 CrossRef CAS PubMed.
- Y. C. Halvorsen, W. O. Wilkison and J. M. Gimble, Adipose-derived stromal cells—their utility and potential in bone formation, Int. J. Obes., 2000, 24(4), S41–S44 CrossRef CAS PubMed.
- O. K. Lee, et al., Isolation of multipotent mesenchymal stem cells from umbilical cord blood, Blood, 2004, 103(5), 1669–1675 CrossRef CAS PubMed.
- N. Rotter, et al., Isolation and characterization of adult stem cells from human salivary glands, Stem Cells Dev., 2008, 17(3), 509–518 CrossRef CAS PubMed.
- S. E. Haynesworth, et al., Characterization of cells with osteogenic potential from human marrow, Bone, 1992, 13(1), 81–88 CrossRef CAS PubMed.
- A. M. Mackay, et al., Chondrogenic differentiation of cultured human mesenchymal stem cells from marrow, Tissue Eng., 1998, 4(4), 415–428 CrossRef CAS PubMed.
- S. Muruganandan, A. A. Roman and C. J. Sinal, Adipocyte differentiation of bone marrow-derived mesenchymal stem cells: cross talk with the osteoblastogenic program, Cell. Mol. Life Sci., 2009, 66(2), 236–253 CrossRef CAS PubMed.
- J. Li, et al., Factors affecting mesenchymal stromal cells yield from bone marrow aspiration, Chin. J. Cancer Res., 2011, 23(1), 43–48 CrossRef PubMed.
- J. W. Tung, et al., Modern flow cytometry: a practical approach, Clin. Lab. Med., 2007, 27(3), 453–468 CrossRef PubMed.
- J. D. Adams, U. Kim and H. T. Soh, Multitarget magnetic activated cell sorter, Proc. Natl. Acad. Sci. U. S. A., 2008, 105(47), 18165–18170 CrossRef CAS PubMed.
- C. W. T. Shields, C. D. Reyes and G. P. López, Microfluidic cell sorting: a review of the advances in the separation of cells from debulking to rare cell isolation, Lab Chip, 2015, 15(5), 1230–1249 RSC.
- R. A. Denu, et al., Fibroblasts and Mesenchymal Stromal/Stem Cells Are Phenotypically Indistinguishable, Acta Haematol., 2016, 136(2), 85–97 CrossRef CAS PubMed.
- A. Andrzejewska, et al., Labeling of human mesenchymal stem cells with different classes of vital stains: robustness and toxicity, Stem Cell Res. Ther., 2019, 10(1), 187 CrossRef CAS PubMed.
- M. J. Gilmore, et al., A technique for rapid isolation of bone marrow mononuclear cells using Ficoll-Metrizoate and the IBM 2991 blood cell processor, Br. J. Haematol., 1982, 50(4), 619–626 CrossRef CAS PubMed.
- N. Y. Naung, et al., Comparative study of different centrifugation protocols for a density gradient separation media in isolation of osteoprogenitors from bone marrow aspirate, J. Oral Biol. Craniofac. Res., 2014, 4(3), 160–168 CrossRef PubMed.
- T. A. Juopperi, et al., Isolation of Bone Marrow–Derived Stem Cells using Density-Gradient Separation, Exp. Hematol., 2007, 35(2), 335–341 CrossRef CAS PubMed.
- C. Pösel, et al., Density gradient centrifugation compromises bone marrow mononuclear cell yield, PLoS One, 2012, 7(12), e50293 CrossRef PubMed.
- P. Horn, et al., Isolation of human mesenchymal stromal cells is more efficient by red blood cell lysis, Cytotherapy, 2008, 10(7), 676–685 CrossRef CAS PubMed.
- Z. Gudleviciene, et al., Quick and effective method of bone marrow mesenchymal stem cell extraction, Open Med., 2015, 10(1), 44–49 Search PubMed.
- N. Ahmadbeigi, et al., The aggregate nature of human mesenchymal stromal cells in native bone marrow, Cytotherapy, 2012, 14(8), 917–924 CrossRef CAS PubMed.
- J. El-Ali, P. K. Sorger and K. F. Jensen, Cells on chips, Nature, 2006, 442(7101), 403–411 CrossRef CAS PubMed.
- N. Lu, et al., Label-free microfluidic cell sorting and detection for rapid blood analysis, Lab Chip, 2023, 23(5), 1226–1257 RSC.
- F. Petersson, et al., Free Flow Acoustophoresis: Microfluidic-Based Mode of Particle and Cell Separation, Anal. Chem., 2007, 79(14), 5117–5123 CrossRef CAS PubMed.
- D. Li, Electrokinetics in Microfluidics, 2004 Search PubMed.
- P. Su, et al., Magnetophoresis in microfluidic lab: Recent advance, Sens. Actuators, A, 2021, 332, 113180 CrossRef CAS.
- D. Di Carlo, Inertial microfluidics, Lab Chip, 2009, 9(21), 3038–3046 RSC.
- R. Huang Lotien, et al., Continuous Particle Separation Through Deterministic Lateral Displacement, Science, 2004, 304(5673), 987–990 CrossRef PubMed.
- M. Yamada and M. Seki, Hydrodynamic filtration for on-chip particle concentration and classification utilizing microfluidics, Lab Chip, 2005, 5(11), 1233–1239 RSC.
- L. Yin, et al., Label-free separation of mesenchymal stem cell subpopulations with distinct differentiation potencies and paracrine effects, Biomaterials, 2020, 240, 119881 CrossRef CAS PubMed.
- L. Yin, et al., Microfluidic label-free selection of mesenchymal stem cell subpopulation during culture expansion extends the chondrogenic potential in vitro, Lab Chip, 2018, 18(6), 878–889 RSC.
- W. C. Lee, et al., Multivariate biophysical markers predictive of mesenchymal stromal cell multipotency, Proc. Natl. Acad. Sci. U. S. A., 2014, 111(42), E4409–E4418 CrossRef CAS PubMed.
- J. Choi, J. C. Hyun and S. Yang, On-chip Extraction of Intracellular Molecules in White Blood Cells from Whole Blood, Sci. Rep., 2015, 5, 15167 CrossRef CAS PubMed.
- D. Holmes, et al., Separation of blood cells with differing deformability using deterministic lateral displacement(†), Interface Focus, 2014, 4(6), 20140011 CrossRef PubMed.
- K. K. Zeming, S. Ranjan and Y. Zhang, Rotational separation of non-spherical bioparticles using I-shaped pillar arrays in a microfluidic device, Nat. Commun., 2013, 4(1), 1625 CrossRef PubMed.
- K. K. Zeming, et al., Microfluidic label-free bioprocessing of human reticulocytes from erythroid culture, Lab Chip, 2020, 20(18), 3445–3460 RSC.
- J. A. Davis, Microfluidic Separation of Blood Components through Deterministic Lateral Displacement in Department of Electrical Engineering, Princeton University, 2008 Search PubMed.
- M. Xavier, et al., Label-free enrichment of primary human skeletal progenitor cells using deterministic lateral displacement, Lab Chip, 2019, 19(3), 513–523 RSC.
- A. L. Kierszenbaum and L. L. Tres, Histology and cell biology : an introduction to pathology, 3rd edn, 2011 Search PubMed.
- W. K. Ovalle, P. C. Nahirney, F. H. Netter and H. FrankNetter's essential histology, Elsevier/Saunders, 2013 Search PubMed.
- J. S. Lowe, J. S. Anderson and A. Stevens, Stevens & Lowe's human histology, 4th edn, 2014 Search PubMed.
- J. A. Davis, Microfluidic Separation of Blood Components through Deterministic Lateral Displacement, 2008 Search PubMed.
- R. Pochampally, Colony forming unit assays for MSCs, Mesenchymal Stem Cells: Methods and Protocols, 2008, pp. 83–91 Search PubMed.
- R. Vernekar, et al., Anisotropic permeability in deterministic lateral displacement arrays, Lab Chip, 2017, 17(19), 3318–3330 RSC.
- J. P. Beech, et al., Sorting cells by size, shape and deformability, Lab Chip, 2012, 12(6), 1048–1051 RSC.
- K. K. Zeming, et al., Label-Free Biophysical Markers from Whole Blood Microfluidic Immune Profiling Reveal Severe Immune Response Signatures, Small, 2021, 17(12), 2006123 CrossRef CAS PubMed.
- P. R. Wheater and etc., Functional histology: Text and colour atlas, Churchill Livingstone, London, England, 1979 Search PubMed.
- J. P. Beech, Microfluidics Separation and Analysis of Biological Particles, 2011 Search PubMed.
- M. M. Frojmovic and R. Panjwani, Geometry of normal mammalian platelets by quantitative microscopic studies, Biophys. J., 1976, 16(9), 1071–1089 CrossRef CAS PubMed.
- S. Ranjan, et al., DLD pillar shape design for efficient separation of spherical and non-spherical bioparticles, Lab Chip, 2014, 14(21), 4250–4262 RSC.
- O. K. Baskurt, M. R. Hardeman and M. W. Rampling, Handbook of hemorheology and hemodynamics, IOS press, 2007, vol. 69 Search PubMed.
- S. Zia, et al., Effective Label-Free Sorting of Multipotent Mesenchymal Stem Cells from Clinical Bone Marrow Samples, Bioengineering, 2022, 9(2), 49 CrossRef CAS PubMed.
- I. Ullah, R. B. Subbarao and G. J. Rho, Human mesenchymal stem cells - current trends and future prospective, Biosci. Rep., 2015, 35(2), e00191 CrossRef PubMed.
- M. Alunni-Fabbroni and M. T. Sandri, Circulating tumour cells in clinical practice: Methods of detection and possible characterization, Methods, 2010, 50(4), 289–297 CrossRef CAS PubMed.
- K. Mareschi, et al., Multipotent Mesenchymal Stromal Stem Cell Expansion by Plating Whole Bone Marrow at a Low Cellular Density: A More Advantageous Method for Clinical Use, Stem Cells Int., 2012, 2012, 920581 Search PubMed.
- M. Pierini, et al., Efficient isolation and enrichment of mesenchymal stem cells from bone marrow, Cytotherapy, 2012, 14(6), 686–693 CrossRef CAS PubMed.
- W. Chu, et al., Comparison and characterization of enriched mesenchymal stem cells obtained by the repeated filtration of autologous bone marrow through porous biomaterials, J. Transl. Med., 2019, 17(1), 377 CrossRef CAS PubMed.
- T. Jiang, et al., In vitro expansion impaired the stemness of early passage mesenchymal stem cells for treatment of cartilage defects, Cell Death Dis., 2017, 8(6), e2851 CrossRef CAS PubMed.
- F. Fachin, et al., Monolithic Chip for High-throughput Blood Cell Depletion to Sort Rare Circulating Tumor Cells, Sci. Rep., 2017, 7(1), 10936 CrossRef PubMed.
- R. Campos-González, et al., Deterministic Lateral Displacement: The Next-Generation CAR T-Cell Processing?, SLAS Technol., 2018, 23(4), 338–351 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3lc00379e |
| ‡ Co-first authors. |
| This journal is © The Royal Society of Chemistry 2023 |