Digital microfluidics for biological analysis and applications
Xing
Xu†
a,
Linfeng
Cai†
a,
Shanshan
Liang
a,
Qiannan
Zhang
a,
Shiyan
Lin
a,
Mingying
Li
a,
Qizheng
Yang
a,
Chong
Li
a,
Ziyan
Han
a and
Chaoyong
Yang
 *ab
*ab
aThe MOE Key Laboratory of Spectrochemical Analysis & Instrumentation, The Key Laboratory of Chemical Biology of Fujian Province, State Key Laboratory of Physical Chemistry of Solid Surfaces, Collaborative Innovation Center of Chemistry for Energy Materials, Department of Chemical Biology, Department of Chemical Engineering, College of Chemistry and Chemical Engineering, Xiamen University, Xiamen 361005, China. E-mail: cyyang@xmu.edu.cn
bInstitute of Molecular Medicine, Renji Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, 200127, China
First published on 16th January 2023
Abstract
Digital microfluidics (DMF) is an emerging liquid-handling technology based on arrays of microelectrodes for the precise manipulation of discrete droplets. DMF offers the benefits of automation, addressability, integration and dynamic configuration ability, and provides enclosed picoliter-to-microliter reaction space, making it suitable for lab-on-a-chip biological analysis and applications that require high integration and intricate processes. A review of DMF bioassays with a special emphasis on those actuated by electrowetting on dielectric (EWOD) force is presented here. Firstly, a brief introduction is presented on both the theory of EWOD actuation and the types of droplet motion. Subsequently, a comprehensive overview of DMF-based biological analysis and applications, including nucleic acid, protein, immunoreaction and cell assays, is provided. Finally, a discussion on the strengths, challenges, and potential applications and perspectives in this field is presented.
1. Introduction
Digital microfluidics (DMF) as an emerging liquid-handling technology has been established as a research field for over a decade. With the ability to precisely manipulate discrete droplets in an integrated microfluidic device, DMF allows the addressable picoliter-to-microliter-sized droplets to be dispensed, moved, merged and split according to a preset path programmatically.1,2 The actuation mechanism of DMF includes electrowetting on dielectric (EWOD),3–5 magnetic,6,7 and surface acoustic waves.8–11 Among them, although EWOD-based force requires relatively more complicated fabrication of multiple layers of structures and has less tolerance to different types of liquid,6 it is currently the most popular technology due to its fast response, simple system and high precision, which is the focus in this review. Specifically, compared to other DMF techniques, the EWOD-based DMF assay requires relatively complicated fabrication of multiple layers of structures including an electrode layer, dielectric layer and hydrophobic coating. Moreover, EWOD-based DMF is more sensitive to the surface tension, conductivity and permittivity of the fluid, and hence has less tolerance to different types of liquid. Alternatively, as one type of microfluidic technology, DMF offers comparable benefits with conventional microfluidics in terms of reduced use of reagents, rapid heat transformation, and high parallelism.1 Furthermore, it does not require the use of a micropump, microvalve and complicated three-dimension flow channel, which simplifies the device, enables a dynamic configuration and provides integrated capacity with external light,12,13 electron14 and magnet modules.15 Thus, due to these advantages, DMF is suitable for lab-on-chip manipulation and analysis, requiring high integration and complex operations.Over the past ten years, the tremendous development of DMF has facilitated biomolecule analysis and applications, including nucleic acids,16,17 proteins,18–20 and hormones.21 The tiny reaction volume in DMF can increase the biomolecule concentration to maximize the reaction efficiency, while the confined reaction space can exclude exogenous contaminants. Similarly, dramatic advances have been made in DMF-based cell assays. Due to the precise manipulation capability and picoliter-to-microliter reaction space of DMF, it is advantageous for cell culture,22,23 cell sorting24 and even single-cell analysis.25,26 The universal DMF bioassay has also given birth to a commercialized platform for biological applications.27 The Food and Drug Administration (FDA)-authorized ePlex system (GenMark diagnostics, CA) is a representative DMF platform for pathogenic bacterial detection.28 It integrates sample extraction, polymerase chain reaction (PCR) amplification and optical signal readout into a single cartridge to enable the qualitative and quantitative measurement of pathogenic DNA/RNA and has been applied for the detection of SARS-CoV-2, respiratory pathogens and bloodborne bacteria. The FDA has also cleared SEEKER (Baebies, NC) for clinical applications to screen lysosomal storage disorders (LSDs) in newborns.29 SEEKER implements enzymatic activity analysis in a single DMF device to complete screening within 3 h. Another commercialized system named Aqdrop (Oxford, UK) enables the manipulation of hundreds of droplets in one chip, managing to explore a wide range of biochemical applications in the study of cells, proteomes and genomes.30 Digifluidic Biotech (Zhuhai, CN) analysed the DNA melting curve on DMF for nucleotide detection, and is now working on a coronavirus detection platform.31 Commercial advances in the construction of a DNA library based on DMF have also been realized. For example, VolTRAX (Oxford Nanopore Technologies, UK),32 NeoPrep System (Illumina, CA)33 and DNBelab D series (MGI and CN)34 have integrated fully automated DNA or RNA library preparation for downstream sequencing. Consequently, DMF technology has fostered biological analysis and applications in both scientific research and industry.
Herein, we provide a comprehensive review of the state-of-the-art EWOD-actuated DMF bioassay (Fig. 1). Initially, we introduce the EWOD principle for droplet manipulation, together with the droplet motion types. Then, we overview the DMF-based biological analysis of nucleic acids, proteins, cells and immunoreaction, highlighting its strengths, challenges and potential applications. Finally, we summarize the benefits of DMF bioassay and provide perspectives in this field.
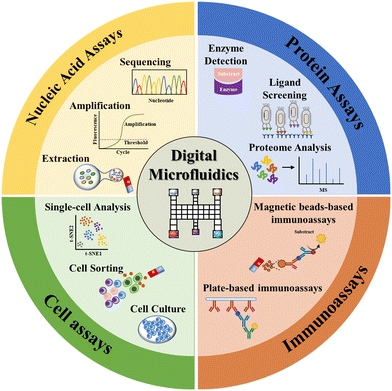 | ||
| Fig. 1 DMF-based bioassays including nucleic acid assays, protein assays, immunoassays and cell assays. | ||
2. The basal constitution of DMF devices
An EWOD-actuated DMF chip consists of several reservoirs for reagent loading and sufficient electrodes to implement biological and chemical operations. According to the architecture type, the DMF device can be classified as a passive matrix electrode array (PM-EWOD) and active matrix electrode array (AM-EWOD) (Fig. 2). In PM-EWOD, each electrode is directly connected to the driving signal generated by the peripheral control circuit, and thus the N electrodes need N signal lines. Differently, AM-EWOD integrates thin film transistors (TFTs) into each pixel electrode. Each TFT consists of a gate (G), source (S) and drain (D), which is used as an electronic switch. The gate manipulates the row signal, the source manipulates the column signal and the drain is connected to the pixel electrode. By controlling the TFT switch, each pixel can be addressed by row-column scanning, where the N (row) × M (column) array only needs N + M signal lines.35 AM-EWOD has advantages of the reduced use of signal lines, but its fabrication is difficult, thus it is less applied than PM-EWOD.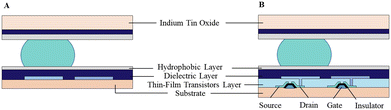 | ||
| Fig. 2 Different architectures of DMF device including A) passive matrix electrode array and B) active matrix electrode array. | ||
According to the plate number, the DMF device can be classified into the format of a single plate36 and two parallel plates37 (Fig. 3). In both formats, the bottom plate is typically comprised of four parts including a substrate, electrode, dielectric and hydrophobic layers. In this case, materials such as glass,38,39 printed circuit boards (PCB),40,41 and paper35 are widely utilized as the substrate. Among them, glass has the advantage of stable chemical properties, good optical properties, high chip fabrication precision, temperature resistance and electrical insulation; nevertheless it suffers from complicated fabrication processes and high cost. On the contrary, PCB and paper substrates benefit from low cost and batch fabrication, yet they both have a low fabrication precision.
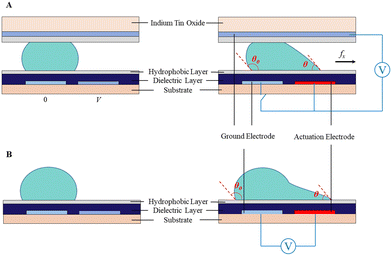 | ||
| Fig. 3 Schematic diagram of droplet actuation in A) two parallel plates and B) single plate due to the asymmetric contact angle across the droplet. | ||
The electrode layer is often metallic, such as chromium, aluminum, gold, copper and indium tin oxide (ITO), which can be fabricated on the substrate through photolithography. Then, the dielectric layer is deposited on the electrode layer using an SU-8 photoresist42,43 or parylene.44,45 Su-8 has excellent mechanical, insulation, optical and chemical properties, and thus has been widely used as the dielectric layer by the spin-coating strategy in recent years. Parylene is beneficial due to its good electrical properties and heat resistance, which is broadly used as coating layer by vapor deposition technology. Finally Teflon AF or fluoropolyer is coated on the surface as a hydrophilic layer to reduce the surface tension for the actuation of droplets.1
In the two-plate format, the top plate is made from ITO conductive glass coated with hydrophobic Teflon-AF46–48 or CYTOP38,49,50 to serve as the ground electrode. Teflon-AF has excellent chemical stability, light transmittance and electrical properties, which is usually dissolved by FC-40 for utilization. The contact angle in the Teflon-AF coating plate is larger than that in the CYTOP coating plate, and thus has higher efficiency in droplet actuation.51 The two-plate device is often operated in the air or filled with other filler media, such as silicone oil, to reduce the voltage necessary for droplet movement and prevent droplet volatilization. In the single-plate device, droplets are placed on the bottom plate with both driving and ground electrodes.
The DMF device is typically coated with Teflon-AF, which sometimes is susceptible to unwanted biomolecule adsorption (e.g., proteins), hence causing sample loss and cross contamination. Alternatively, careful modulation of the applied voltage polarity can reduce biomolecule adsorption by regulating the electrostatic attraction and repulsion.52 Specifically when the solution is negatively charged, the use of a positive voltage polarity can reduce biomolecule adsorption, whereas negative voltage polarity is beneficial in a positively charged solution. However, this strategy is less effective for complex solutions with different molecules presenting diverse positive or negative charges at physiological pH.53 Alternatively, the use of Pluronics as an additive to droplets is helpful, which consist of triblock copolymers of poly(ethylene oxide) (PEO) and poly(propylene oxide) (PPO) to reduce protein and cell adsorption to surfaces.
3. Basic droplet manipulations on DMF
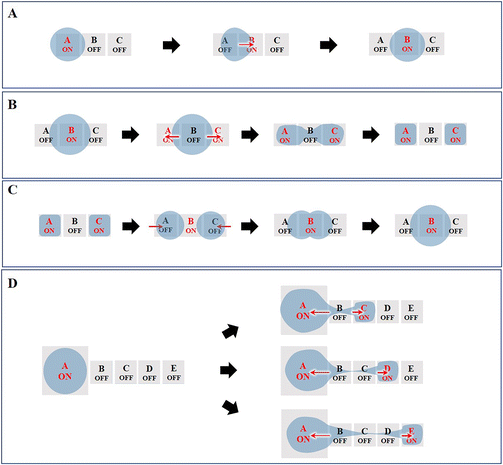
Currently, the most fundamental droplet manipulation types achievable by EWOD approaches include transporting, splitting, merging, and dispensing. The droplets on a two-plate DMF device can be dispensed, moved, split and merged, whereas those on a single-plate DMF device cannot be split and dispensed. However, one-plate devices facilitate the analyte observation and recovery and can manipulate droplets of submicrolitres to tens of microliters, while two-plate devices are suitable for droplet operation in the picolitre to microliter range.1,6,54 In this section, we describe the underlying principles of the four basic manipulations of droplets (Fig. 4).3.1 Droplet transporting
Droplet transport is accomplished by controlling the wettability of the droplets. When the droplet is located on the boundary between the actuation electrode (A) and the non-actuation electrode (B), the voltage applied on B actuates the droplet to move toward B due to the asymmetric contact angle in the droplet above A and B (Fig. 4A). Further, by switching the voltage “on” and “off” between a series of adjacent electrodes, the droplet can move along the trace of the actuated electrodes.3.2 Droplet splitting
As shown in Fig. 4B, to conduct droplet splitting, the droplet is firstly located on the actuation electrode unit (B), in contact with the electrode units (A and C), respectively. Based on EWOD, when a certain amplitude of voltage is applied to the electrode units (A and C), while switching off the intermediate electrode unit (B), the contact angle on two sides of the droplet will decrease. The driving force causes the droplet to form a “rugby ball” shape on both sides, sag in the middle to form a “bottle neck”, and eventually separate into two daughter droplets.3.3 Droplet merging
Droplet merging can be achieved by transporting two droplets to the same electrode. As shown in Fig. 4C, two droplets are transported to the electrode units (A and C), respectively, both in contact with electrode unit B. When applying a voltage to electrode unit B, the two droplets can merge to form a single larger droplet.In the lab-on-chip device, the experimental processes such as pretreatment, dilution, chemical reaction, and supply of cell culture medium all require mixing operations. However, achieving adequate mixing of the two droplets after merging requires further consideration. The liquid motion in traditional microfluidics is featured by laminar flow, which makes liquid mixing difficult. In contrast, DMF can realize the rapid and efficient mixing of droplets, which is addressed by three strategies, including droplet reciprocation, merge–split motion and loop motion (Fig. 5).55 In contrast, loop motion can provide the highest efficiency of droplet merging by inducing a stretching–folding pattern.55
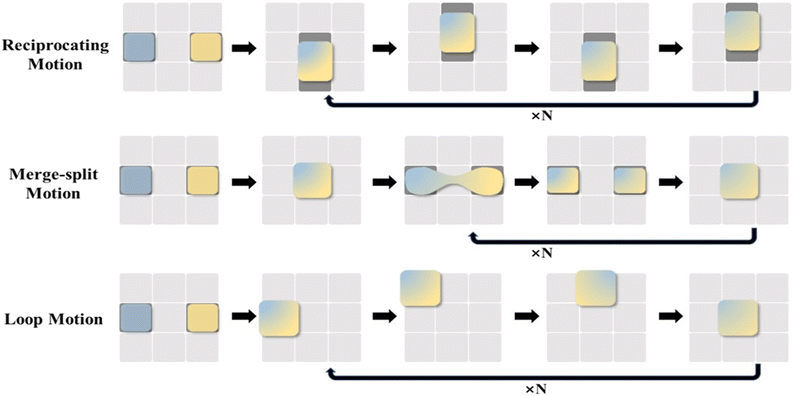 | ||
| Fig. 5 Schematic diagram of droplet merging by reciprocating motion, merge–split motion, and loop motion. | ||
3.4 Droplet dispensing
Typically, droplets are formed by dragging the liquid from a larger droplet, and then cutting it to generate a daughter droplet. As shown in Fig. 4D, the electrode units are sequentially switched on. Then, electrode unit (A) and electrode units (C)–(E) are energized, while the other electrode units are switched off. Consequently, the “liquid column” will be pinched to generate a daughter droplet. To increase the stability of the droplet generation process and improve the volume precision and accuracy of the dispensed droplets, an optimized electrode geometry in terms of shape or size is required. For example, Wang et al. proposed the introduction of circular arcs to eliminate the liquid tail after droplet pinch-off and to fix the position of the pinch-off point.56 Shih et al. optimized the size of the reservoir, middle electrode and destination electrode to improve the dispensing successful rate.574. DMF-based nucleic acid assays
Nucleic acid analysis has been utilized in a variety of fields including drug research, medical diagnosis, and forensic identification with high sensitivity, high accuracy, and rapid detection.58 In these applications, the nucleic acids extracted from tissues or cells are often in extremely minute quantities. Conventional bench-top manipulation may cause loss of nucleic acids due to nucleic acid adsorption during frequent transfer. Moreover, it will produce a large number of DNA amplicons during the amplification process, which are highly susceptible to aerosol contamination once exposed to air, resulting in a high background or false positive signals. DMF is especially well-suited for the preparation and analysis of trace or easily-contaminated nucleic acid samples due to its efficient droplet manipulation, outstanding low adsorption and excellent closure properties. In this section, we discuss DMF-based nucleic acid applications in extraction, amplification and sequencing (Fig. 6).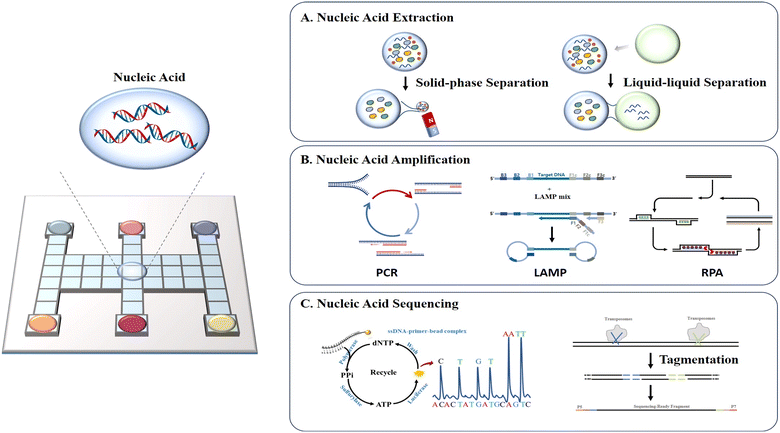 | ||
| Fig. 6 Schematic diagram of DMF-based nucleic acid assays including A) nucleic acid extraction, B) nucleic acid amplification, and C) nucleic acid sequencing. | ||
a) Nucleic acid extraction
Nucleic acid extraction and purification on a DMF device are primarily based on solid-phase and liquid–liquid separation methods, respectively. The solid-phase separation method utilizes magnetic beads for nucleic acid manipulation, with basic steps of nucleic acid binding, washing, buffer exchange, and elution. Sista et al.59 and Hung et al.60 extracted the whole genomic DNA from human whole blood using a DMF platform with magnetic beads. The whole blood droplets were mixed with lysis buffer to release DNAs, which were then captured by the magnetic beads. When fixing the magnetic beads by a magnet, the unbound supernatant droplets could be dragged away. Subsequently, the magnetic beads were washed several times to remove the residual cellular debris, followed by the release of the purified DNAs. Jebrail et al. developed a “world-to-DMF” integrated system to effectively collect target analytes from a macroscale sample and transport them to a DMF device for microscale volume manipulation (Fig. 7A).61 This system consisted of three distinct components including an extraction module for macroscale RNA extraction, a purification module for microscale RNA purification and concentration, and a module interface for mediating the interaction between modules. Pressure-driven samples of large volume were delivered from the extraction module through the 10 μL droplet region on the DMF purification module, and magnet-mediated recovery of bead-bound analytes was achieved. By using this system, RNAs were extracted from human whole blood (110–380 μL) and purified in microscale volumes of 5–15 μL. Compared to the traditional bench-top method, it saved 2-fold processing time and 12-fold reagent consumption. However, this solid-phase extraction method requires specialized magnetic beads and an external magnetic field, complicating the device to some extent.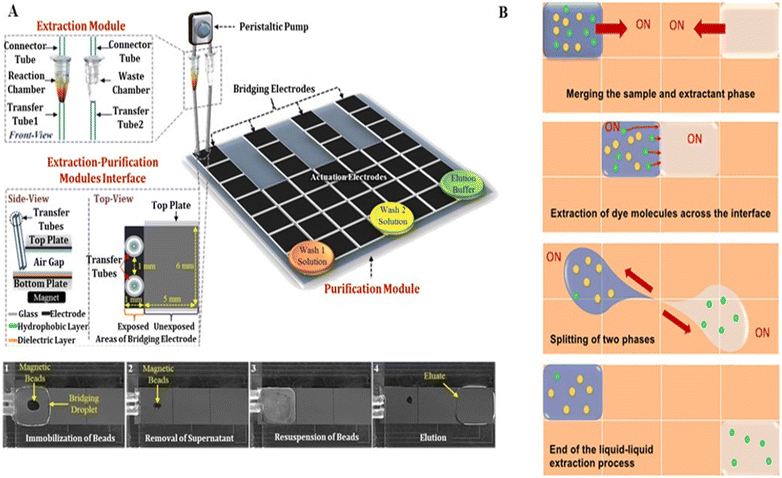 | ||
| Fig. 7 DMF-based nucleic acid extraction. A) “World-to-DMF” integrated system effectively collecting target analytes based on solid-phase separation. Copyright 2014, the American Chemistry Society.61 B) Liquid–liquid nucleic acid extraction by handling sample droplets and immiscible extractant phases. Copyright 2021, the American Institute of Physics.62 | ||
Alternatively, liquid–liquid nucleic acid extraction handles the sample droplets and immiscible extractant phases without the use of magnetic beads and an external magnetic field. Abdelgawad et al. purified the recombinant DNA from cell lysates by liquid–liquid extraction on DMF.40 After the sample droplet containing DNAs and proteins was driven into a water-immiscible phenolic solution, phase splitting was done to isolate the DNAs from proteins for further mass spectrometry analysis. To improve the extraction efficiency, Paul et al. optimized the mixing of the sample droplet and extractant phase (Fig. 7B).62 By keeping one phase stationary and moving the other phase continuously around, followed by circular motion of both phases, a higher extraction rate and better distribution of the extracted analytes could be simultaneously achieved. Using this strategy, plasmid DNA (pDNA) could be extracted from an aqueous sample into an ionic liquid in the presence of proteins with high selectivity. Nonetheless, liquid–liquid separation method is challenging to separate targets from the same biomolecules, such as specific DNA fragments. Therefore, the choice of nucleic acid extraction and purification method should consider both the sample and contents existing in the dispersion phase.
b) Nucleic acid amplification
The amount of nucleic acids extracted and separated from biological materials is typically extremely minute and difficult to detect directly using instruments. Therefore, it is necessary to amplify the extracted nucleic acids for downstream analysis. PCR is frequently employed for amplifying certain nucleic acid sequences, which can produce millions of copies from a tiny amount of starting templates.The integration of PCR technology with the DMF platform offers the following benefits: (1) approximately 100-fold decrease in reagent and sample consumption, reducing the reaction cost; (2) the small reaction volume improves the efficiency of solute diffusion and heat transfer, hence reducing the amplification time and increasing the reaction uniformity; and (3) higher amplification efficiency is achieved in small-volume PCR due to the enhanced template concentration. Nevertheless, performing PCR on DMF necessitates heat cycling management, which is often addressed through two strategies.
The first involves maintaining the droplet in situ, while varying the temperature in a single region. Chang et al. implemented the PCR process on a DMF chip using a single-zone thermal cycling approach.63 Droplets containing cDNAs and PCR reaction solution were dispensed individually from the reservoirs, and then mixed and transported to the PCR chamber. Two micro heaters and a micro temperature sensor were integrated in the PCR chamber for precise thermal cycling for PCR amplification. Ultimately, the products were collected and verified through gel electrophoresis off-chip. To further improve the processing throughput and avoid off-chip product verification, Norian et al. developed complementary-metal-oxide semiconductor (CMOS) integrated circuit technology to perform quantitative polymerase chain reaction (qPCR) on a DMF chip (Fig. 8A).64 Three electric heating modules and temperature sensor modules were incorporated in a DMF chip to enable three parallel qPCR processes, while single-photon avalanche diodes were used for fluorescence monitoring. The device was capable of quantifying DNA from Staphylococcus aureus in 1.2 nL, with the sensitivity of a single copy per droplet and a dynamic range exceeding four-orders-of-magnitude.
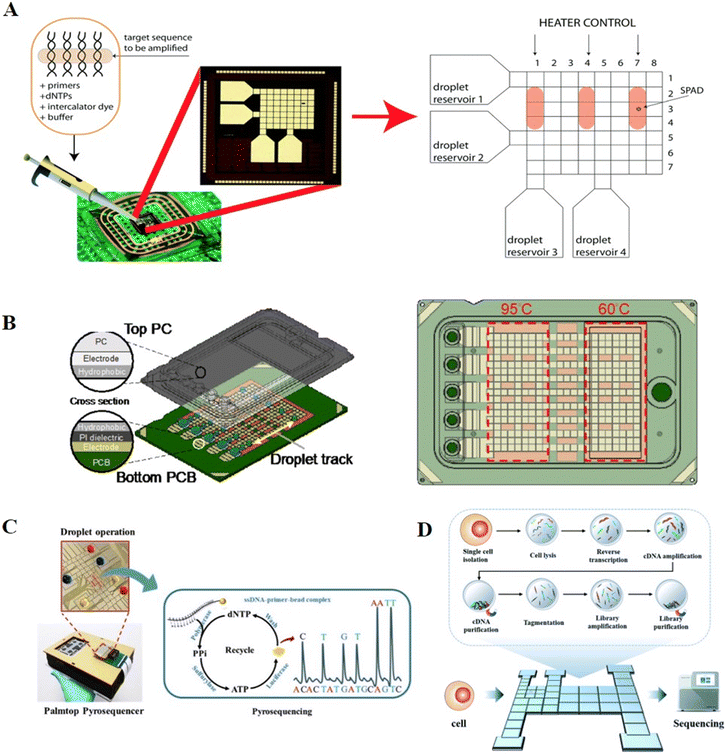 | ||
| Fig. 8 DMF-based nucleic acid amplification and sequencing methods. A) Three parallel DNA amplification reactions on DMF with varying temperatures in a single region. Copyright 2014, The Royal Society of Chemistry.64 B) Disposable point-of-care cartridge implementing qPCR on DMF with varying temperatures in different regions. Copyright 2022, MDPI (Basel, Switzerland).66 C) Palmtop sequencing platform based on pyrosequencing and DMF device. Copyright 2019, Elsevier B.V.13 D) “Cell-in-library-out” single-cell transcriptome sequencing library construction platform (Cilo-seq) integrating single-cell isolation, cell lysis, reverse transcription, cDNA amplification and final library preparation on a single DMF chip. Copyright 2022, The Royal Society of Chemistry.69 | ||
A second approach for PCR on chip involves varying the temperature in different zones, while actuating the reaction droplets in a closed-loop flow-through process. Sista et al. constructed two thermal zones with temperatures of 60 °C and 95 °C, respectively, and shuttled a droplet between the two zones. As a result of the fast heat transfer, the 40-cycle real-time PCR was completed in 12 min.59 Based on this technology, Hua et al. incorporated a small fluorimeter module for the fluorescence monitoring of qPCR and applied it for the amplification of pathogenic nucleic acids and the detection of infectious diseases.65 Ho et al. developed a disposable point-of-care cartridge to detect the SARS-CoV-2N gene by implementing qPCR on DMF (Fig. 8B).66 Consequently, 40 cycles were conducted in 17 min by actuating the droplets between two temperature zones of 60 °C and 95 °C, and the fluorescence after each cycle was automatically recorded by the optical module. This DMF cartridge featured multiple droplet tracks, enabling multiple tests and controls to be conducted simultaneously. Yehezkel et al. reported the de novo synthesis and cell-free cloning of custom DNA libraries using single-molecule PCR technology.67 Single molecules of the template were achieved by serial dilution, and subsequently subjected to PCR amplification through shuttling among three different temperature zones in the PCR lane including 62 °C, 72 °C and 95 °C. This system was designed with eight channels to support twenty-four parallel reactions, allowing a higher amplification throughput than the conventional tube-based method of amplification.
Although PCR technology has high amplification efficiency and specificity, the PCR process requires at least two temperatures to achieve amplification, increasing the complexity of the device and making it unsuitable for point-of-care testing (POCT). Isothermal nucleic acid amplification technology can address this problem by completing the entire amplification at a constant temperature, hence reducing the temperature requirements for devices, which is particularly applicable in resource-constrained areas. Hitherto, common isothermal nucleic acid amplification technologies such as loop-mediated isothermal amplification (LAMP), recombinase polymerase amplification (RPA) and multiple displacement amplification (MDA) have been effectively integrated with DMF.
Coelho et al. firstly verified the feasibility of LAMP amplification on DMF.68 Droplets of LAMP reaction reagent and target DNA were dispensed from reservoirs and mixed completely in the reaction zone for LAMP amplification at 65 °C. DMF-LAMP demonstrated increased sensitivity when amplifying target DNA at a concentration of 0.5 ng μL−1 in just 45 min with a reaction volume of 1.5 μL. The products validated by off-chip electrophoretic analysis had a higher normalized fluorescence intensity of the band than that from bench-top amplification reactions, indicating the higher amplification efficiency of DMF-LAMP. Wan et al. integrated LAMP amplification and real-time product detection on DMF using SYBR Green I, while implementing on-chip melting curve analysis to accurately identify the product specificity through the use of molecular beacon DNA probes with specific low melting temperature (Tm).70 Using purified Trypanosoma brucei DNA as the target, the entire amplification and detection processes were accomplished in 40 min with a detection limit of 10 copies per reaction.
Compared to LAMP, RPA permits nucleic acid amplification at a lower temperature of 37 °C and a shorter reaction time of 15 min. Kalsi et al. described a technique for the detection of the blaCTX-M-15 gene relying on RPA reaction on a DMF platform with fluorescence readout.71 Comparatively, this system enabled fewer than 10 copies of template to be detected with a hundred-fold improvement in the detection limit compared to a bench-top assay. Unlike LAMP and RPA, which could only amplify known DNA sequences, MDA could carry out whole genome amplification using a random primer and phi29 DNA polymerase. Liu et al. demonstrated the amplification of bacterial DNA in a DMF device for the rapid identification of bacterial species by MinION sequencing.72 Using this system, 10 fg C. glutamicum DNA could be detected within 3 h, resulting in a 100-fold reduction in the detection limit compared to the in-tube amplification approach. Overall, isothermal nucleic acid amplification combined with a DMF device is robust and sensitive, but its lower amplification specificity than PCR still needs to be addressed.
c) Nucleic acid sequencing
As an effective nucleic acid analysis method, sequencing technology has been widely adopted for applications in microbial community profiling, genetic abnormality identification and pathogen discovery. Pyrosequencing is a real-time sequencing-by-synthesis method with high sensitivity and speed. During DNA polymerization, the enzyme cascade reaction triggered by inorganic pyrophosphate (PPi) promotes the oxidation of luciferin for chemiluminescent readout. The sequence of the template is identified by the paired nucleotide, while the quantification of chemiluminescent detection of PPi is proportional to the number of bases incorporated in the synthesized strand of DNA. Traditionally, pyrosequencing involves multi-step incubation and washing steps, whereas DMF can automate these processes by programmatically processing the reaction droplets.Welch et al. constructed a DMF-based pyrosequencing system for the first time.73 Droplets with different concentrations of adenosine triphosphate (ATP) were mixed with luciferin to establish the working curve of the chemiluminescent intensity versus ATP concentration. This system achieved a low detection limit of 7 nM, which was much lower than that of ATP generated by single-base binding, verifying the feasibility of pyrosequencing on DMF. A further reduction in cost was achieved by Boles et al. by performing pyrosequencing on printed circuit boards.74 DNA templates were anchored to magnetic beads, allowing for thorough washing between nucleotide additions, and then the protocols and reagents were optimized to achieve a high signal-to-background ratio, which ultimately generated 100% accurate sequence determination of ∼60 bases. Zou et al. reported a palmtop sequencing platform based on pyrosequencing and DMF device for automatic, real-time and portable analysis of DNA mutations (Fig. 8C).13 During each round of nucleotide addition reaction, the washing buffer, enzymes, one type of dNTP, ammonium persulfate (APS) and the single-stranded target DNA immobilized on magnetic particles were placed in reservoirs, then dispensed, moved, and mixed for chemiluminescent detection by an integrated photomultiplier (PMT). After detection, the immobilized beads were washed to remove the supernatant and resuspended with reaction reagents for the next type of dNTP addition, and this step was repeated three times per round. This system enabled the sequencing of a DNA template of 53 bp with 100% accuracy in 2 h and was capable of detecting a mutation in the KRAS gene with a limit of detection (LOD) as low as 5% mutant level. Based on this palmtop sequencing platform, Ruan et al. expanded the application to DNA methylation detection.75 The unmethylated cytosine of the target DNA was transformed into uracil with the treatment of bisulfite. During PCR amplification, the methylated sample in the CpG sites remained as CG, while the unmethylated sample was converted to TG. By immobilizing the PCR products to magnetic beads and performing pyrosequencing on DMF, the methylated site could be detected with an LOD of 10 pg within 30 min.
A recent development in next-generation sequencing (NGS), such as Illumina sequencing, has facilitated massive parallel data collection, which allows genome or transcriptome sequencing to be detected for a few thousand dollars per run. However, the preparation of properly formatted sequencing libraries, including steps of DNA fragmentation, adaptor addition, library amplification and size selection, is lengthy and expensive, necessitating a streamlined method for preparing inexpensive sequencer-ready libraries. To facilitate the construction of a DNA library, Kim et al. demonstrated a droplet-based DMF platform for the preparation of an Illumina sequencing library.79 The DMF hub together with peripheral modules automated DNA fragmentation, PCR enrichment, magnetic bead-based clean-up and size-selection, yielding excellent sequencing coverage with >99% alignment when using 5 ng bacterial genomic DNA as the input. Recently, a commercial DMF platform named AQdrop™ with 41 thousand independently addressable electrodes was developed to prepare DNA samples for Illumina next-generation sequencing.80 In this platform, two variable temperature zones and 3/8 magnets were set for library reaction (e.g., amplification) and purification/elusion, which supported 3 or 8 parallel library constructions at once. Three different commercially available kits including Kapa Hyperplus (Roche), Collibri ES (Thermo Fisher) and NexteraFlex Enrichment kit (Illumina) were translated in this platform and produced a comparable library performance to the bench-top method. The 50-fold decrease in volume scale and increase in the number of samples processed per chip both contributed to reducing the cost per sample of library preparation.
Zhang et al. further integrated single-cell isolation, cell lysis, reverse transcription, cDNA amplification and final library preparation in DMF to achieve “cell-in-library-out” single-cell transcriptome analysis (Cilo-seq, Fig. 8D).69 After cDNA amplification, purification beads were utilized on chip to adsorb long-chain cDNAs, while leaving short-chain primers in the supernatant, facilitating the separation of cDNA products. Afterward, library preparation reagents were introduced in sequence for DNA fragmentation, adaptor addition, library amplification and magnet-aided product purification. The final library was recovered from the chip and processed by pair-end sequencing. The Cilo-seq platform presented a 1.4-fold enhancement in gene detection ability over the traditional pipetting-based method and was successfully applied for the analysis of heterogeneous genotypes in single circulating tumor cells. However, despite the remarkable advancements, the development of hybrid platforms integrating sequencer-ready library construction and in situ high-throughput sequencing remains a challenge.
5. DMF-based protein assays
Protein-related processing such as sample pretreatment and protein detection is typically time-consuming and inefficient, and thus an automated platform is urgently needed to improve the work efficiency. DMF is well suited for labor-intensive protein-associated operations and applications due to its high automation, which simplifies complex experimental operations. In recent years, DMF has been widely employed for enzyme detection,81 ligand screening75 and proteome purification and analysis82 (Fig. 9).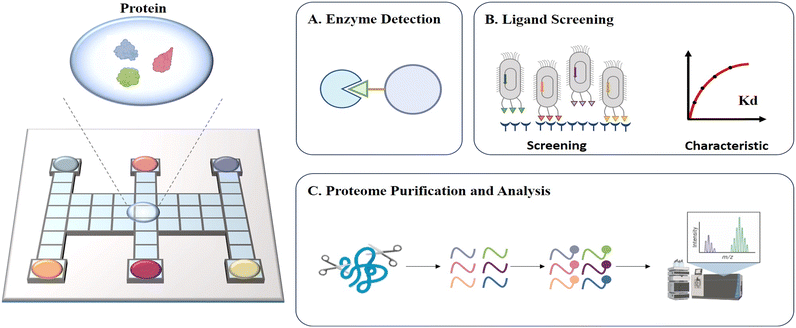 | ||
| Fig. 9 Schematic diagram of DMF-based protein assays including A) enzyme detection, B) ligand screening, C) proteome purification and analysis. | ||
5.1 Enzyme detection
Enzymes are special biochemical reaction catalysts that are essential to the normal metabolism of organisms. Diseases such as albinism and diabetes can be caused by abnormal enzyme expression or impaired enzyme activity, and thus enzyme detection is necessary. Enzyme assays usually rely on the catalyzed reactions of enzymes, which are challenged by stable conditions such as pH and temperature. Consequently, the reproducibility of assays is crucial for multiple measurements of enzymes. Accordingly, the uniform droplet generation and parallel analysis capacities of DMF offer a reliable platform for enzyme detection. Taniguchi et al. verified the feasibility of enzyme detection on DMF for the first time by moving and mixing droplets of luciferin and luciferase for fluorescence readout.81 By combining DMF and a photodiode, Srinivasan et al. reported the quantitative detection of glucose based on an enzyme reaction, whose results matched that produced by a spectrometer.83 Glucose was first converted into gluconic acid and hydrogen peroxide under the catalysis of glucose oxidase. Then, hydrogen peroxide was reacted with 4-amino antipyrine and N-ethyl-N-sulfopropyl-m-toluidine in the presence of peroxidase to form violet-colored quinoneimine with an absorbance peak at 545 nm for quantification. Sista et al. presented a DMF platform to perform the enzyme activity analysis of α-glucosidase acid (GAA) and α-galactosidase acid (GLA), screening for Pompe and Fabry disorders of lysosomal storage diseases (LSDs).84 The endpoint fluorescence was measured after the incubation of GAA/GLA with the substrate, and the enzymatic activities were identified as micromoles of substrate hydrolyzed per hour of incubation per liter of blood. By automating all the steps on a disposable cartridge, the lab-on-chip multiplex fluorometric enzymatic assays could reduce the incubation time from 6 h to 1 h but still produced the same detection results as the traditional bench-top 96-well microplate methods. The same group later expanded this system to a 5-plex fluorometric enzymatic assay for the rapid and high-throughput screening of 5 LSDs, including Pompe, Fabry, Hunter, Gaucher and Hurler diseases. Sample preparation to enzymatic activity could be completed within 3 h on each cartridge.85 Sathyanarayanan et al. reported a DMF-based methodology for the biopsy-scale analysis of human-derived hepatic cytochrome P450 (CYP) activity, which is responsible for the intrinsic clearance of most therapeutic drugs (Fig. 10A).76 Given that cytochrome P450 is mainly located in the smooth endoplasmic reticulum of hepatocytes and enriched in human liver microsomes (HLM), in this study, HLMs were first isolated and immobilized on magnetic beads. Then, the HLM-containing beads were actuated to incubate with CYP enzyme specific model compounds on DMF for fluorescence quantification when translating prefluorescent substrates or the co-substrate NADPH to fluorescein or resorufin to determine the CYP activity. This method consumed only about 15 μg microsomal protein per assay, and thus was technically adaptable to screen CYP enzyme function from biopsy-scale liver samples.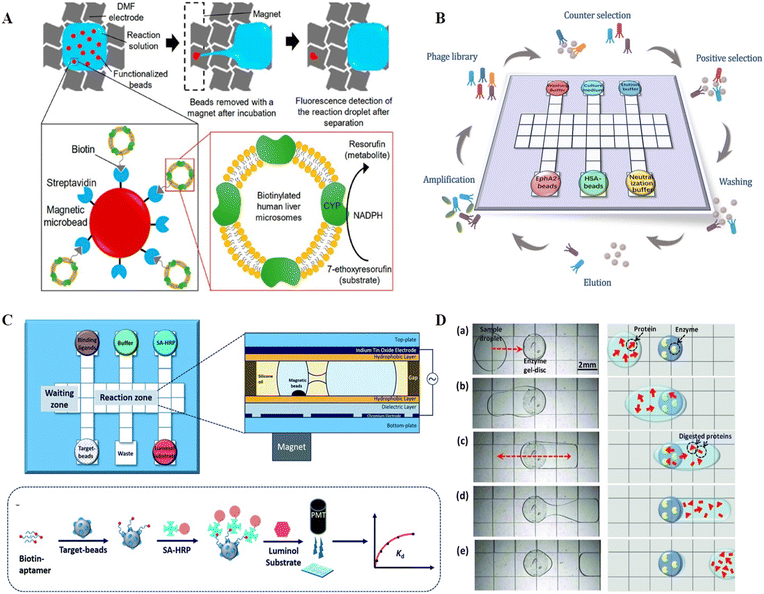 | ||
| Fig. 10 DMF-based protein analysis. A) A DMF-based methodology for biopsy-scale analysis of human-derived hepatic cytochrome P450 (CYP) activities. Copyright 2020, the American Chemistry Society.76 B) Automatic biopanning platform based on DMF for phage display (auto-panning). Copyright 2021, The Royal Society of Chemistry.77 C) Automated ligand binding affinity evaluation platform (Auto-Affitech). Copyright 2020, The Royal Society of Chemistry.12 D) Proteome pretreatment in a heterogeneous system by integrating cylindrical agarose discs coated with trypsin or pepsin on DMF. Copyright 2012, John Wiley and Sons.78 | ||
The Shih group utilized DMF for endoglucanase detection. Given that endoglucanase can catalyse the hydrolysis of the (1,4)-glycosidic bonds in 4-methylumbelliferyl β-D-cellobioside (MUC) to produce a methylbelliferone fluorescence product, droplets containing MUC and the endoglucanase enzyme were mixed, incubated, and then analysed via fluorescence to determine the enzyme activity.86 A similar strategy was also employed for the detection of fucosyltransferase.87 Overall, integrated DMF systems are capable of providing enzyme assays for a range of biochemical analyses, including glucose monitoring and enzyme activity evaluation, which offer substantial promise for medical and pharmaceutical research.
5.2 Ligand screening
Ligand screening in proteins plays a critical role in studying molecular recognition mechanisms and regulating protein functions. Established methods for screening ligands involve highly repetitive, labor-intensive, and time-consuming procedures. Biopanning based on phage display, for instance, requires rounds of positive selection, counter selection, washing, elution, and amplification to screen for a specific ligand, the complexity of which has hindered the efficient discovery of novel ligands. Thus, to address this problem, Wang et al. developed an automatic biopanning platform based on DMF for phage display (auto-panning, Fig. 10B).77 In this system, counter selection was firstly performed to remove non-specific binding phages, and then positive selection was carried out by mixing retained phages with target-coated beats. After multiple washings to remove low-affinity phages, high-affinity phages were collected. Following rounds of selection, enriched clones were picked and sequenced to achieve the affinity peptide sequence. Using this technology, a peptide ligand against the cancer marker ephrin type-A receptor 2 (EphA2) was effectively screened after three rounds of selection within 16 hours, demonstrating higher screening efficiency than the conventional bench-top method, which required nearly a week of manual operation. Besides ligand selection, DMF can also be applied for the evaluation of ligand binding ability. Guo et al. demonstrated a DMF-aided automated ligand binding affinity evaluation platform (Auto-affitech, Fig. 10C).12 Target-immobilized magnetic beads were employed for binding biotinylated ligands, followed by the signal translation using streptavidin-modified horseradish peroxidase enzyme (SA-HRP) and a luminol substrate. The chemiluminescence signal measured by a PMT was correlated with the number of ligands binding to the target-beads, allowing the characterization of the equilibrium dissociation constant (Kd) when introducing different concentrations of starting ligands. By using this platform, the Kd of an aptamer–protein system and an antigen–antibody system was evaluated with reduced manual labor, personal error and analysis time. With the high integration of DMF, it is anticipated that ligand screening integrated with binding ability evaluation on a single chip will be possible in the near future.5.3 Proteome purification and analysis
The analysis of proteome by mass spectrometry (MS) or other detectors usually involves tedious and complex pretreatment steps. For example, samples need to be dried and ionized before MS in matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS). Accordingly, to process samples and transfer targets directly to MS, attempts have been devoted to building protein sample preparation platforms on DMF, involving sample purification by precipitation,88 disulfide reduction,89,90 thiol alkylation,90,91 proteolytic digestion,91,92etc. Luk et al. demonstrated the compatibility of DMF to implement common protein sample preparation steps including reduction, alkylation, and enzymatic digestion.89 After off-chip desalting, the sample was subjected to MALDI-MS for quantification analysis. Chatterjee et al. performed lab-on-chip standard protein digestion, followed by crystallization with a MALDI matrix for in situ MALDI-MS analysis.93 However, the above-mentioned protein digestion was handled in homogenous solution conditions, where the enzyme concentration had to be kept low to avoid autolysis. These low enzyme conditions resulted in elevated temperatures and a lengthy incubation time, as well as an increase in nonspecific cleavage. Thus, to address this problem, Luk et al. further demonstrated a heterogeneous system by integrating cylindrical agarose discs coated with trypsin or pepsin on DMF (Fig. 10D).78 When the droplets containing proteome were exposed to the agarose disc, the proteins were enzymatically cleaved, followed by MALDI-MS for proteome analysis. After the optimization of enzyme density in the agarose discs to improve the digestion efficiency and speed, an integrated proteomic sample processing including reduction, alkylation and digestion was carried out by DMF. The MS analysis revealed that this system had higher coverage in model analytes than conventional homogenous processing.Despite the demonstrated suitability of DMF for proteome sample preparation prior to MS, the presence of surfactants, which is necessary for the facile movement of protein droplets, sometimes results in considerable ion suppression during MS analysis. Thus, to avoid the interference of surfactants, Aijian et al. replaced the surfactants with fluorinated solvents to facilitate droplet movement and limit protein adsorption on the chip surface.94 The fluorinated solvent could be evaporated so that it would not interfere with MALDI-MS analysis. Leipert et al. developed a DMF-based detergent-buffer system compatible with downstream LC-MS analysis by introducing the single-pot, solid-phase-enhanced sample preparation (SP3) approach.82 The SP3-bead mixture with 0.08% Pluronic F127 was firstly loaded on the DMF chip and mixed with the protein sample for the combination of protein. Then, the supernatant was separated from the beads after bead immobilization by a permanent magnet, which allowed the removal of salts and anti-fouling polymetric detergents. DMF-SP3 enabled proteomes from as little as 100 cells to be detected by LC-MS, demonstrating its great compatibility with low protein input. Overall, DMF has shown potential as a miniaturized proteome sample preparation platform and has been tailored for full device integration and low sample input, reducing the protein loss and improving detection sensitivity. Future work is anticipated to increase the throughput of parallel analysis and decrease the original input to lower levels, such as single-cell level.
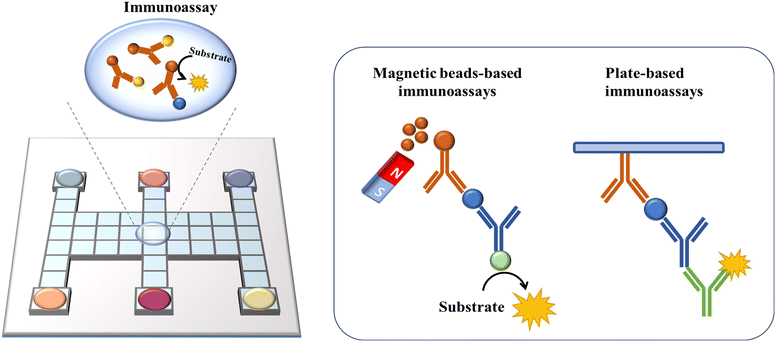
6. DMF-based Immunoassays
By utilizing the precise combination reaction between antigen and antibody, immunoreaction has been developed for the detection of trace analytes such as proteins,14 cytokines95 and hormones.59 Nevertheless, immunoassays are cumbersome and time-consuming to operate, and the high cost of detection makes them unsuitable for practical use. As a new discrete droplet manipulation technique, DMF can automate the steps and lower the reaction volume, which is frequently used for constructing immunoassays. For biomolecule detection, it is necessary to immobilize the biorecognition layer in DMF platforms for target molecule capture. Two different techniques are used, as follows: 1) employing microspheres as solid supports and 2) immobilizing the biorecognition agents on a surface (Fig. 11).Micron/nano-sized magnetic beads have been frequently utilized as microspheres due to their high surface-to-volume ratio, which can provide more binding sites and better adaptability for sample washing with a magnet. Sista et al. reported a DMF-based heterogeneous sandwich immunoassay based on magnetic beads for the detection of interleukin-6 and insulin.95 This approach allowed the magnetic beads to be retained up to 100% by a permanent magnet, while removing the supernatant containing unbound molecules. Then, the targets were resuspended from beads and reacted with APS-5 substrate for chemiluminescence signal readout. This type of immunoassay was also applied in whole-blood troponin I (cTnI) detection with high recovery.59 The Wheeler group performed DMF-based immunological assays in a remote setting to detect measles and rubella immunoglobulin G (IgG). In this study, paramagnetic particles coated with viral antigens were used to capture measles/rubella IgG from the sample. Then, anti-measles/anti-rubella IgG, anti-human IgG-horseradish peroxidase conjugate, luminol and H2O2 were introduced and incubated with magnetic particles on a DMF assay to generate a chemiluminescent product. Finally, the droplets containing the products were actuated to the detection zone for chemiluminescent detection to quantify the target IgG. This point-of-care system had sensitivities of 96% and specificities of 80% compared with the reference tests processed in a centralized laboratory, demonstrating the potential for global serological surveillance especially in areas with limited resources.96
Besides chemiluminescence, surface-enhanced Raman scattering (SERS)97 and electrochemistry98 are also leveraged as sensing techniques. Wang et al. demonstrated a highly sensitive immunoassay based on DMF and SERS sensing for quantifying the avian influenza virus H5N1 in human serum (Fig. 12A).97 A core(Au)–shell(Ag) nanostructure as an SERS tag was firstly constructed, and then embedded with 4-mercaptobenzoic acid as the Raman reporter. The target H5N1 virus was captured by magnetic beads, and then labelled by the synthesized SERS tags conjugated with detection antibodies. In this instance, by measuring the intensity of Raman scattering with a portable Raman spectrometer, the concentration of H5N1 was quantified. This system could detect as low as 74 pg mL−1 H5N1 in ∼30 μL human serum with excellent sensitivity. Shamsi et al. introduced the first DMF immunoassay based on electrochemical detection.98 In this system, gold sensing electrodes and silver counter/pseudoreference electrodes were modified on the top ITO plate. After conjugating the primary antibody with magnetic microparticles, antigen molecules were captured by microparticles, and then labelled with a secondary antibody modified with HRP. Given that HRP was capable of catalyzing the oxidation of 3,3′,5,5′-tetramethylbenzidine (TMB), the released electron could be detected amperometrically. This electrochemical immunoassay enabled the detection of thyroid stimulating hormone with an LOD of 2.4 IU mL−1, making it suitable for clinical applications.
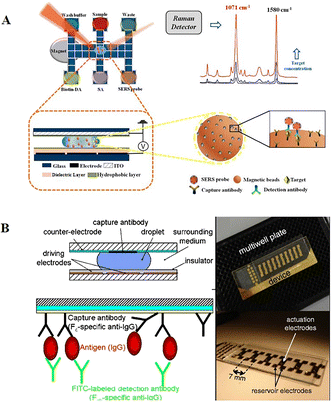 | ||
| Fig. 12 DMF-based immunoreaction analysis. A) Highly sensitive immunoassay based on DMF and SERS sensing for quantifying avian influenza virus H5N1 in human serum. Copyright 2018, the American Chemical Society.99 B) Heterogeneous immunoassays by immobilizing capture antibodies on the hydrophobic surface of the top ITO plate. Copyright 2011, Springer.14 | ||
Another type of immunoassay relies on immobilizing recognition molecules on the device surface. The transparent top plate can also facilitate the direct visualization by fluorescent readers or microscopy systems. Miller et al. developed heterogeneous immunoassays by immobilizing capture antibodies on the hydrophobic surface of the top ITO plate (Fig. 12B).14 In this device, the top ITO glass was coated with rugged Cytop instead of Teflon-AF, after which capture antibodies were pipetted onto the slide to generate antibody spots. These spots were allowed to air-dry, then rinsed with 0.1% Pluronic F-127 and left to air-dry again before use. After droplets containing human IgG were actuated across antibody-immobilized spots, the IgG could be captured by the antibodies immobilized on the top plate. Then, the target antigens were further labelled with fluorescein isothiocyanate-tagged secondary detection antibodies for fluorescence intensity measurement. This DMF-based immunoassay presented the rapid detection of IgG within 2.5 h and had a comparable dynamic range with well-plate assays. In summary, directly functionalizing recognition molecules on the device surface can eliminate the need for an external magnet to immobilize the magnetic beads, reducing manual intervention and simplifying DMF devices. However, the increased complexity of functionalizing antibody on the plate still needs to be addressed. Moreover, the antibody-coated hydrophilic surface on the top plate will impede droplet movement to some extent.
7. DMF-based cell assays
The manipulation of cells, including cell culture, sorting, and analysis, is an integral part of biological research and clinical practice (Fig. 13). Ideally, a cell manipulation platform should match the cell size, ensure accurate cell handling, and provide a contamination-free environment.100,101 Traditional cell manipulation relies on liquid-handling robotics, which is costly and consumes a great deal of reagents. As an advanced discrete droplet-handling technique, DMF can provide a tiny volume, support noncontact space, diminish complicated manipulation and integrate downstream analysis, and thus is well suited for diverse and multi-step cell manipulation.102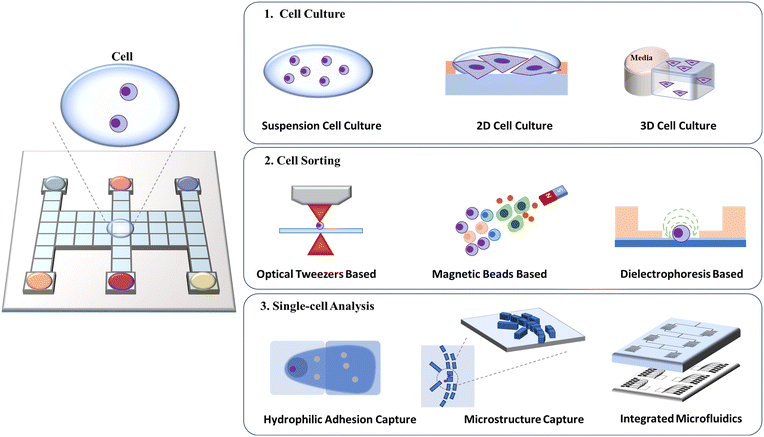 | ||
| Fig. 13 Schematic diagram of DMF-based cell assays including cell culture, cell sorting, and single-cell analysis. | ||
7.1 Cell culture
Unlike conventional cell culture, which utilizes cell culture dishes and well plates, DMF permits the introduction of various stimulation, the supplement of culture media, and real-time analysis and observation. With high biocompatibility and adaptability, DMF has been exploited in suspension cell culture, two-dimensional (2D) and three-dimensional (3D) adherent cell culture.By introducing droplets of culture medium, DMF can provide a static medium condition for suspension cells without undesirable shear stress.71 Barbulovic-Nad et al. developed a cell-based DMF assay for the first time and verified that the actuation by DMF had no adverse effects on the viability, proliferation and biochemistry of Jurkat T cells.103 The cytotoxicity test in the DMF platform presented ∼20-times higher sensitivity than that in a traditional well-plate assay, suggesting that DMF is a simple but versatile analytical tool for cytotoxicity tests. In addition to mammalian cells, DMF has also been used to cultivate bacteria104 and yeast cells.105 Gach et al. introduced a DMF platform for the culture, transformation, and recombinant proteins expression of bacteria, yeast and fungi. With a channel to continuously replenish oil for oxygen supply, the chip allowed approximately 5 days of cell culture and protein expression observation after plasmid transformation.104 In another study, Wang et al. created a DMF-based algae bioreactor to foster algae (Fig. 14A).105 The DMF chip patterned with a 24-electrode array was connected to the operation unit for position feedback and actuation and mounted on a microscope. To compensate for the system complexity and added cost, an electrode-saving strategy was described, where electrodes for algae culture were connected in a “finger-fringe” geometry instead of one or more interval electrodes. This system allowed for all processes of nutrient gradient generation, algae culturing and lipid accumulation analysis to be performed on chip with less effort and reduced time.
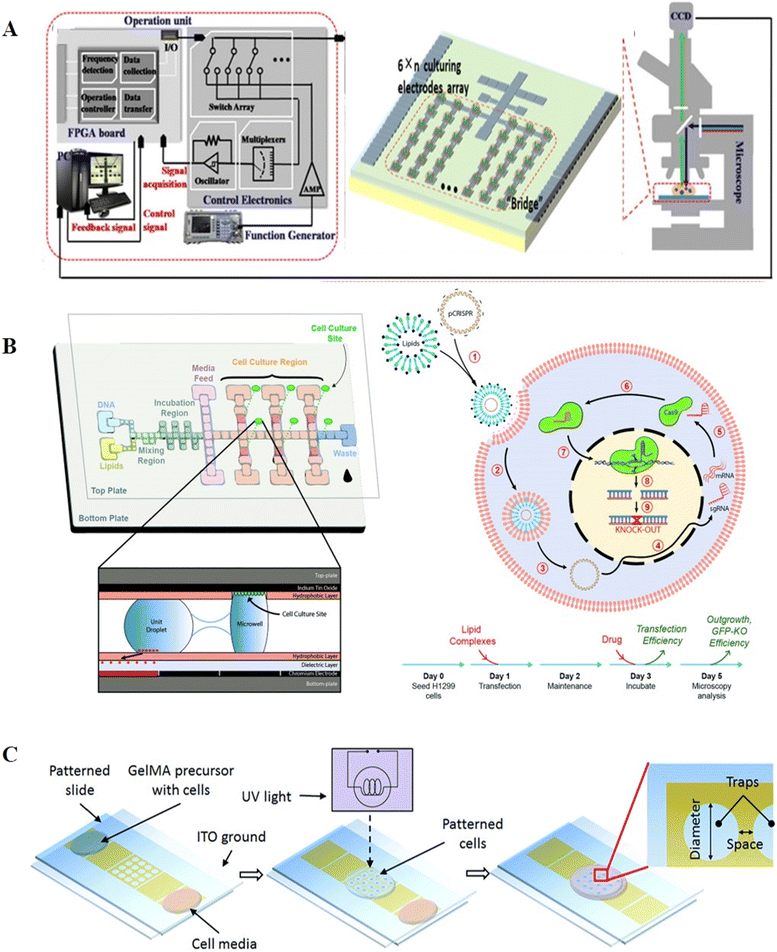 | ||
| Fig. 14 DMF-based cell culture. A) DMF-based algae bioreactor to cultivate algae. Copyright 2021, WILEY-BLACKWELL.105 B) Hydrophilic sites patterned on the top plate enabling 2D cell culture for gene editing. Copyright 2018, The Royal Society of Chemistry.110 C) 3D gelatin methacrylate hydrogel and planar trap-integrated DMF system developed for 3D cell culture. Copyright 2016, The Royal Society of Chemistry.118 | ||
Compared to suspension cells, the culture of adherent cells requires an appropriate hydrophilic-charged surface for cell proliferation and adhesion. The hydrophobic surface of DMF can be altered by adhesion factor adsorption,106 dry etching,107 and wet etching108 to produce suitable coating surfaces for 2D culture of adherent cells. Barbulovic-Nad et al. devised adhesion pads for adherent cell cultivation.106 The adhesion pads were prepared by dispensing the extracellular matrix (ECM) on the chip and allowing it to dry to the surface for producing a hydrophilic zone. Using these adhesion pads, Hela cells could be seeded and fed on the DMF surface. Witters et al. introduced a dry lift-off method by exploiting an easily-removable parylene-C mask to generate spatially controlled micropatches of biomolecules.107 With the micropatches, the cells could be arrayed as clusters on the DMF chip surface. Eydelnant et al. devised a Teflon-AF lift-off protocol for hydrophilic site patterning without the inclusion of additional biomolecules, resulting in the formation of virtual microwells, which enabled highly reproducible passive reagent dispensing and cell adhesion.108 The cells cultured in the virtual microwells were verified to have a comparable morphology and growth rate with that cultured in well plates. These virtual microwells could support over-week cell culture, and thus were further applied in cell apoptosis,109 immunohistochemistry26 and gene editing (Fig. 14B).110–112
Although 2D culture remains the most used technique in laboratories, 3D culture is gaining popularity given that it can recapitulate in vivo conditions and distribute cells in a three-dimensional skeleton to maintain higher viability.113,114 Hydrogels with high biocompatibility and flexibility are frequently utilized as culture skeletons, such as agarose alginate hydrogels,115 sol-phase alginate116,117 and gelatin methacrylate (GelMA) hydrogel.118 Fiddes et al. firstly provided the proof-of-concept of 3D culture on a DMF chip by introducing cylindrical agarose hydrogel discs. These discs allowed the systematically addressable droplets of reagent to pass through for effective medium exchange, and thus NIH-3T3 cells could be cultured for 7 days in the agarose hydrogel discs.115 George et al. demonstrated alginate gelation on the DMF platform and exploited alginate hydrogels for 3D cell culture.117 This design allowed the gels to be retained in situ during liquid delivery without the use of physical barriers or hydrophilic patterning. The effects of different concentrations of dimethyl sulfoxide on 3D cells in alginate hydrogels were investigated, suggesting the feasibility of chemical screening in this platform. Nestor et al. patterned planar traps in the electrodes and trapped cells in a 3D GelMA hydrogel (Fig. 14C).118 The cell-containing GelMA hydrogel droplet was immersed in mineral oil and transported to the trapping electrode. Upon reaching the trapping electrode, dielectrophoresis (DEP) was initiated for cell cluster patterning, followed by the use of ultra violet (UV) light to crosslink the hydrogel and the removal of mineral oil. With the specially designed trap, the cell survival of 78% after 4 days of culture was achieved.
Cell invasion can also be studied through 3D cell culture on DMF. Li et al. reported cell invasion in a digital microfluidic microgel system (CIMMS) with the ability to isolate subpopulations of invading cells for RNA-seq analysis (Fig. 15).23 In this system, microgels were first formed with collagen I core surrounded by a shell of basement membrane extract (BME). When droplets of cell suspension were actuated touching the edge of microgels, the device was rotated 90° to settle the cells on the microgel. As soon as the cells were attached, the device was returned to its original orientation, generating an invasion model for further analysis by confocal immunofluorescence microscopy and transcriptome sequencing. For example, breast cancer invasion models demonstrated differential expression of 244 genes between invading and non-invading cells. CIMMS was also used for assessing the effects of aryl hydrocarbon receptor (AHR) expression on invasion, where MDA-MB-231 cells with stable knockout of AHR presented enhanced invasive ability and reduced proliferation.119
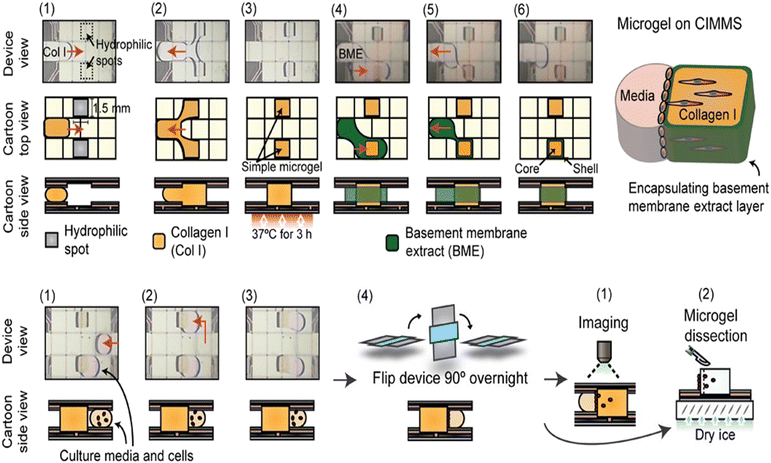 | ||
| Fig. 15 DMF-based cell invasion analysis by introducing microgels, which are formed with a collagen I core surrounded by a shell of basement membrane extract. Copyright 2020, Wiley-VCH Verlag GmbH & Co. KGaA.23 | ||
Therefore, DMF provides a variety of cell culture environments by automating media exchange, improving surface modification, and introducing 3D skeletons. The ease of observation and integration with other devices enables downstream tests and analysis such as reagent effects, protein expression, and behaviour states to be conducted on DMF chips during cell culture. Using DMF as a tool, new scientific questions related to cell growth, proliferation, and invasion are anticipated to be answered in the future.
7.2 Cell sorting
Sorting and purifying cells are essential components of cell analysis. Microfluidics has great value in cell sorting such as the enrichment of circulating tumor cells, separation of blood cells and isolation of stem-cell cells.120–122 Recently, the integration of optical,123,124 electrical,125 and magnetic126 approaches with DMF for cell sorting and control has been reported.Tewari Kumar et al. combined DMF and optical tweezers (OT) for the selective trapping and relocation of single bacterial cells.124 In this system, a single bacterium was captured in the microwell array of the DMF chip using magnetic bead-conjugated antibody. Then, OT was utilized to selectively capture the bacterium-bead complex of interest and relocate it to a new microwell array, allowing for the spatial organization of single bacteria. Fan et al. created dielectric-coated electrodes by integrating DEP and EWOD on a single chip to manipulate cells on different scales.125 The proposed chip was capable of selectively producing EWOD and DEP by applying electric signals with different frequencies to identical electrodes. With high-frequency signals, positive DEP and negative DEP were used to concentrate mammalian cells towards one side of a DMF droplet. Then, with low-frequency signals, EWOD was used to split the droplet into two daughter droplets with distinct densities. Shah et al. utilized antibody-conjugated magnetic beads (MB-ab) to separate CD8+ T-lymphocyte cells with high selectivity on the DMF chip.126 Droplets containing the sample and MB-abs were merged for the binding of CD8+ T-lymphocyte cells, and then MB-bound cells were separated with a magnet. Consequently, the collected CD8+ T-lymphocyte cells could be lysed chemically for the downstream detection of mRNA or proteins. In general, it is possible to selectively manage cells in EWOD-driven droplets when implementing fluidic functions such as mixing, splitting, and moving. However, the assistance of external force is required, which complicates the device. In the future, it is envisaged that new-type cell sorting methods will be developed using the DMF platform alone.
7.3 Single-cell analysis
Single cell analysis is an essential tool for analyzing cell heterogeneity, given that it may reveal critical markers that are obscured by conventional population-average analysis. Single-cell analysis is challenging due to the picoliter scale of cells and the presence of traces of cellular content in each cell. DMF platforms that can manipulate picoliter-to-microliter droplets with minimal loss of biomolecules are highly suitable for single-cell analysis. Based on different single-cell isolation strategies, we categorized single-cell DMF assays into hydrophilic adhesion capture, microstructure capture, and integrated microfluidic combination.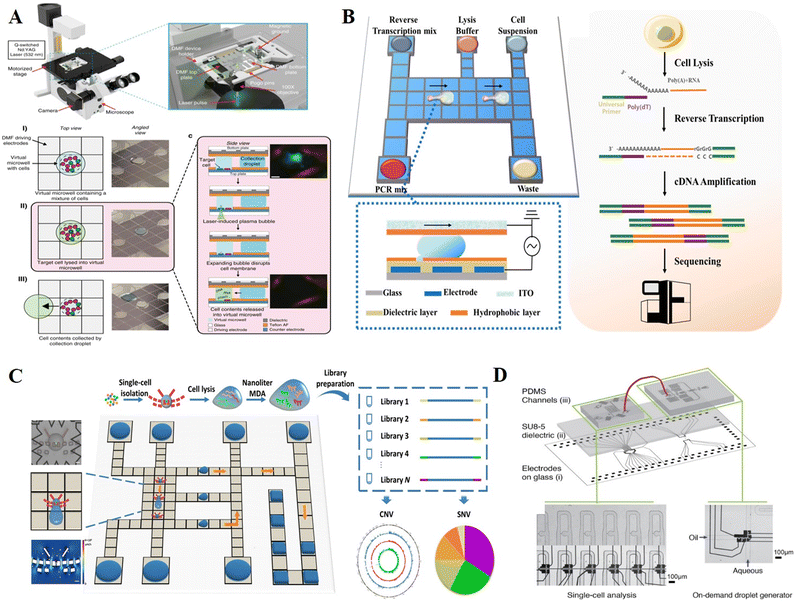 | ||
| Fig. 16 DMF-based single-cell analysis. A) Single-cell omics analysis platform (DISCO) featured by fabricating hydrophilic site for sub-droplet generation and utilizing high-energy laser pulse for targeted single-cell lysis. Copyright 2020, Nature Portfolio.127 B) Single-cell transcriptome sequencing platform with DMF based on Smart-seq2 strategy (Digital-RNA-seq) for highly sensitive single-cell gene expression analysis. Copyright 2020, the American Chemical Society.39 C) Single-cell genome sequencing platform with DMF based on MDA strategy (Digital-WGS) featured by a butterfly structure for single-cell capture. Copyright 2020, the American Association for the Advancement of Science.25 D) Hybrid microfluidic method to manipulate addressable droplets by a co-planar electrode system. Copyright 2020, Wiley-VCH Verlag GmbH & Co. KGaA.24 | ||
To further integrate single-cell sequencing library preparation into a single chip, Xu et al. demonstrated a DMF-dependent single-cell transcriptome sequencing platform (Digital-RNA-seq, Fig. 16B).39 On the bottom plate, a circular hydrophilic site was fabricated by lift-off technology for single-cell separation in the sub-droplet, while removing others. By optimizing the cell suspension concentration, hydrophilic site size and droplet movement pattern, this strategy enabled >95% single-cell isolation efficiency regardless of the cell type and size.128,129 Upon acquiring a single cell, reagents of Smart-seq2 were sequentially placed onto the DMF chip to conduct reverse transcription and cDNA amplification for single-cell transcriptome sequencing library construction. The increased template concentration in the confined nanoliter reaction space without exogenous contamination contributed to higher sensitivity and accuracy of transcriptome sequencing. A similar approach was also used for measuring single-cell DNA mutations.129 After isolating and lysing single cells, the whole genome was amplified using the MDA method, and multiplex mutation information was then obtained using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). This method allowed for the profiling of heterogeneous CTCs with Kirsten rat sarcoma viral oncogene mutations, which had tremendous promise for the direction of cancer therapy. Overall, hydrophilic site-based cell isolation is flexible, rapid and simple to integrate with downstream analysis. Nevertheless, isolating a single cell can be random and time intensive. The assistance of image processing based on artificial intelligence provides the opportunity to increase the efficiency of target cell isolation, saving manpower and achieving automation.
Furthermore, Ruan et al. combined hydrodynamics and surface wettability techniques to reach a high level of single-cell isolation efficiency (100%) in the Digital-WGS platform (Fig. 16C).25 In this system, the butterfly structure was designed in this system so that cells in the flow would be focused and funneled into the weir in the middle of the butterfly structure. When the weir was occupied by a single cell, the flow resistance was increased through the weir, redirecting subsequent cells to the slits on either side. Simultaneously, hydrophilic sites were fabricated beneath the butterfly microstructure, allowing the captured cell in the weir to be retained for further investigation. This platform was then coupled with single-cell whole genome sequencing and achieved a low coefficient of variation and high coverage due to the sufficient cell lysis and lossless amplicon recovery on the DMF chip. Nevertheless, despite the high efficiency of Digital-WGS, the sample throughput is limited to nine samples at one time. Therefore, it is intended that future development of microstructure capture methods is directed towards increasing single-cell manipulation and analysis throughput.
8. Conclusions and perspectives
In this review, we overviewed EWOD-actuated DMF bioassays by introducing the droplet movement mechanism and types, as well as a variety of biological applications in nucleic acid, protein, and cells. Compared with the traditional bench-top method, DMF offers a superior platform with higher sensitivity, accuracy, automation and integration. The overall advantages of DMF to provide biological research tools are summarized here, as follows: 1) a reduction in the consumption of reagents and samples. Costly reagents and limited samples pose challenges for biological analysis. Although conventional microfluidics can also lower the liquid requirement, the presence of dead volume leads to the waste of reagents and samples. DMF not only can produce picoliter-to-microliter level droplets but also manipulate them with minimized biomolecule adsorption due to its hydrophobic surface, which enhances the sample and reagent availability. 2) Shortened reaction and analysis time. A small-volume reaction is reported without limitation by the diffusion kinetics, resulting in a much shorter reaction time. Alternatively, DMF allows dynamic manipulation for faster and more effective mixing of droplets, which can enable in a significant reduction in detection and analysis time. 3) An improved analysis performance. Given that the tiny volume of droplets increases the contact between the liquid and the environment, including an increase in heat transfer, it will improve the reaction kinetics to achieve better analysis performance. 4) Isolation from exogenous contamination. DMF devices based on EWOD typically employ a closed two-plate structure for oil filling. This type of design enables excellent airtightness, thereby preventing direct contact with other contaminants. 5) Automation by use-programmed operation. With an electrical signal control system that automates the movement of droplets, the DMF device is suitable for reactions requiring repetitive and arduous steps, such as sample pretreatment, immunoreaction and nucleic acid extraction, making it an extremely versatile and practicable device. In addition, DMF eliminates the interference caused by random pipetting, thereby enhancing the analysis capability with excellent stability and reproducibility.Nevertheless, there are still issues that need to be resolved in future research (Fig. 17). Firstly, due to the limited electrode number, the parallel analysis throughput is still unsatisfactory. Thus, to further expand the scalability, the active matrix electrowetting-on-dielectric (AM-EWOD) strategy71 is appropriate to provide more addressable manipulation sites for the handling of thousands of droplets. Secondly, the possible presence of biofouling such as enzymes may induce sample contamination when reusing the chip, and thus optimized Pluronic additives or hydrophobic coating are required. Thirdly, the majority of contemporary DMF devices still require a large bulky external power source and a sophisticated circuit design, limiting further miniaturization and portability. Future devices are expected to be controlled by light, temperature or sound. Fourthly, flexible devices (e.g., wearable chip) provide great promise in POCT. Furthermore, current methods are primarily confined to optical technologies, which rely on expensive and cumbersome optical instrumentation and are unavailable in resource-limited areas. Consequently, portable signal readout devices integrated with DMF are necessary. For example, smartphones with high-quality camera lenses and imaging software make it possible to bring screening tests in resource-limited areas. Fifthly, advanced droplet routing algorithms are required to assist in automatic path selection and droplet scheduling to manage multiple droplet manipulation on a large scale from their source electrodes to their target electrodes without unexpected mixing.131 Overall, future work will witness the continuously updated engines for DMF bioassay, which will promote substantial advancements in the fields of biological, medical and clinical applications.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the National Natural Science Foundation of China (21927806, 22293031, and 22204132), the National Key R&D Program of China (2019YFA0905800), the Fundamental Research Funds for the Central Universities (20720210001, 20720220005) and the Innovative Research Team of High-Level Local Universities in Shanghai for their financial support.Notes and references
- K. Choi, A. H. Ng, R. Fobel and A. R. Wheeler, Annu. Rev. Anal. Chem., 2012, 5, 413–440 CrossRef CAS PubMed.
- H. Cheng, H. Liu, W. Li and M. Li, Electrophoresis, 2021, 42, 2329–2346 CrossRef CAS PubMed.
- R. B. Fair, Microfluid. Nanofluid., 2007, 3, 245–281 CrossRef CAS.
- E. M. Miller and A. R. Wheeler, Anal. Chem., 2008, 80, 1614–1619 CrossRef CAS PubMed.
- S. R. Barman, I. Khan, S. Chatterjee, S. Saha, D. Choi, S. M. Lee and Z. H. Lin, J. Food Drug Anal., 2020, 28, 595–621 CAS.
- Y. Zhang and N. T. Nguyen, Lab Chip, 2017, 17, 994–1008 RSC.
- H. Cheng, H. R. Liu, W. H. Li and M. Li, Electrophoresis, 2021, 42, 2329–2346 CrossRef CAS PubMed.
- L. Y. Yeo and J. R. Friend, Annu. Rev. Fluid Mech., 2014, 46, 379–406 CrossRef.
- Z. Guttenberg, H. Muller, H. Habermuller, A. Geisbauer, J. Pipper, J. Felbel, M. Kielpinski, J. Scriba and A. Wixforth, Lab Chip, 2005, 5, 308–317 RSC.
- J. Seo, S. K. Lee, J. Lee, J. S. Lee, H. Kwon, S. W. Cho, J. H. Ahn and T. Lee, Sci. Rep., 2015, 5, 12326 CrossRef PubMed.
- S. Y. Park, M. A. Teitell and E. P. Y. Chiou, Lab Chip, 2010, 10, 1655–1661 RSC.
- J. J. Guo, L. Lin, K. F. Zhao, Y. L. Song, M. J. Huang, Z. Zhu, L. J. Zhou and C. Y. Yang, Lab Chip, 2020, 20, 1577–1585 RSC.
- F. X. Zou, Q. Y. Ruan, X. Y. Lin, M. X. Zhang, Y. L. Song, L. J. Zhou, Z. Zhu, S. C. Lin, W. Wang and C. J. Yang, Biosens. Bioelectron., 2019, 126, 551–557 CrossRef CAS PubMed.
- E. M. Miller, A. H. C. Ng, U. Uddayasankar and A. R. Wheeler, Anal. Bioanal. Chem., 2011, 399, 337–345 CrossRef CAS PubMed.
- Q. Q. Zhang, X. Xu, L. Lin, J. Yang, X. Na, X. Chen, L. L. Wu, J. Song and C. Y. Yang, Lab Chip, 2022, 22, 1971–1979 RSC.
- M. Li, L. Wan, M. K. Law, L. Meng, Y. Jia, P. I. Mak and R. P. Martins, Lab Chip, 2022, 22, 537–549 RSC.
- S. Newman, A. P. Stephenson, M. Willsey, B. H. Nguyen, C. N. Takahashi, K. Strauss and L. Ceze, Nat. Commun., 2019, 10, 1706 CrossRef CAS PubMed.
- J. Leipert, M. K. Steinbach and A. Tholey, Anal. Chem., 2021, 93, 6278–6286 CrossRef CAS PubMed.
- D. Liu, Z. H. Yang, L. Y. Zhang, M. L. Wei and Y. Lu, RSC Adv., 2020, 10, 26972–26981 RSC.
- B. Seale, C. Lam, D. G. Rackus, M. D. Chamberlain, C. Liu and A. R. Wheeler, Anal. Chem., 2016, 88, 10223–10230 CrossRef CAS PubMed.
- S. Abdulwahab, A. H. C. Ng, M. D. Chamberlain, H. Ahmado, L. A. Behan, H. Gomaa, R. F. Casper and A. R. Wheeler, Lab Chip, 2017, 17, 1594–1602 RSC.
- S. C. Shih, P. C. Gach, J. Sustarich, B. A. Simmons, P. D. Adams, S. Singh and A. K. Singh, Lab Chip, 2015, 15, 225–236 RSC.
- B. B. Li, E. Y. Scott, M. D. Chamberlain, B. T. V. Duong, S. Zhang, S. J. Done and A. R. Wheeler, Sci. Adv., 2020, 6, eaba9589 CrossRef CAS PubMed.
- K. Samlali, F. Ahmadi, A. B. V. Quach, G. Soffer and S. C. C. Shih, Small, 2020, 16, e2002400 CrossRef PubMed.
- Q. Ruan, W. Ruan, X. Lin, Y. Wang, F. Zou, L. Zhou, Z. Zhu and C. Yang, Sci. Adv., 2020, 6, eabd6454 CrossRef CAS PubMed.
- A. H. C. Ng, M. D. Chamberlain, H. Situ, V. Lee and A. R. Wheeler, Nat. Commun., 2015, 6, 7513 CrossRef CAS PubMed.
- J. Li and C. J. Kim, Lab Chip, 2020, 20, 1705–1712 RSC.
- https://www.genmarkdx.com/solutions/systems/eplex-system/ .
- https://baebies.com/products/seeker/ .
- https://www.aqdrop.com/showcase/ .
- https://digifluidic.com/ .
- https://nanoporetech.com/prepare .
- https://omicsomics.blogspot.com/2017/02/illumina-drops-neoprep.html .
- https://en.mgi-tech.com/products/solution/4/ .
- B. Hadwen, G. R. Broder, D. Morganti, A. Jacobs, C. Brown, J. R. Hector, Y. Kubota and H. Morgan, Lab Chip, 2012, 12, 3305–3313 RSC.
- M. Washizu, IEEE Trans. Ind. Appl., 1998, 34, 732–737 CrossRef CAS.
- M. G. Pollack, A. D. Shenderov and R. B. Fair, Lab Chip, 2002, 2, 96–101 RSC.
- Y.-Y. Lin, R. D. Evans, E. Welch, B.-N. Hsu, A. C. Madison and R. B. Fair, Sens. Actuators, B, 2010, 150, 465–470 CrossRef CAS PubMed.
- X. Xu, Q. Zhang, J. Song, Q. Ruan, W. Ruan, Y. Chen, J. Yang, X. Zhang, Y. Song, Z. Zhu and C. Yang, Anal. Chem., 2020, 92, 8599–8606 CrossRef CAS PubMed.
- M. Abdelgawad, S. L. S. Freire, H. Yang and A. R. Wheeler, Lab Chip, 2008, 8, 672–677 RSC.
- J. Gong and C.-J. C. Kim, Lab Chip, 2008, 8, 898–906 RSC.
- Y. J. Chang, V. M. Bright, E. Schonbrun and K. Mohseni, A variable focus microlense using EWOD on a tapered SU-8 structure, 2006 Search PubMed.
- Y.-J. Chang, K. Mohseni and V. M. Bright, Sens. Actuators, A, 2007, 136, 546–553 CrossRef CAS.
- M. Bienia, F. Mugele, C. Quilliet and P. Ballet, Phys. A, 2004, 339, 72–79 CrossRef CAS.
- M. W. J. Prins, W. J. J. Welters and J. W. Weekamp, Science, 2001, 291, 277–280 CrossRef CAS PubMed.
- M. G. Pollack, A. D. Shenderov and R. B. Fair, Lab Chip, 2002, 2, 96–101 RSC.
- J.-h. Chang, D. Y. Choi, S. Han and J. J. Pak, Microfluid. Nanofluid., 2010, 8, 269–273 CrossRef CAS.
- D. P. Papageorgiou, A. Tserepi, A. G. Boudouvis and A. G. Papathanasiou, J. Colloid Interface Sci., 2012, 368, 592–598 CrossRef CAS PubMed.
- Y. Li, W. Parkes, L. I. Haworth, A. W. S. Ross, J. T. M. Stevenson and A. J. Walton, J. Microelectromech. Syst., 2008, 17, 1481–1488 CAS.
- L. Huang, B. Koo and C. Kim, J. Microelectromech. Syst., 2013, 22, 253–255 CAS.
- J. B. Chae, J. O. Kwon, J. S. Yang, D. Kim, K. Rhee and S. K. Chung, Sens. Actuators, A, 2014, 215, 8–16 CrossRef CAS.
- J. Y. Yoon and R. L. Garrell, Anal. Chem., 2003, 75, 5097–5102 CrossRef CAS PubMed.
- S. H. Au, P. Kumar and A. R. Wheeler, Langmuir, 2011, 27, 8586–8594 CrossRef CAS PubMed.
- J. H. Chang, D. S. Kim and J. J. Pak, J. Electr. Eng. Technol., 2011, 6, 402–407 CrossRef.
- J. Berthier, in Micro-Drops and Digital Microfluidics, ed. J. Berthier, William Andrew Publishing, Norwich, NY, 2008, pp. 225–294, DOI:10.1016/B978-081551544-9.50008-7.
- H. Wang and L. G. Chen, Nanotechnol. Rev., 2021, 10, 857–869 CrossRef CAS.
- S. C. C. Shih, N. S. Mufti, M. D. Chamberlain, J. Kim and A. R. Wheeler, Energy Environ. Sci., 2014, 7, 2366–2375 RSC.
- Y. Zhao, X. Zuo, Q. Li, F. Chen, Y. R. Chen, J. Deng, D. Han, C. Hao, F. Huang, Y. Huang, G. Ke, H. Kuang, F. Li, J. Li, M. Li, N. Li, Z. Lin, D. Liu, J. Liu, L. Liu, X. Liu, C. Lu, F. Luo, X. Mao, J. Sun, B. Tang, F. Wang, J. Wang, L. Wang, S. Wang, L. Wu, Z. S. Wu, F. Xia, C. Xu, Y. Yang, B. F. Yuan, Q. Yuan, C. Zhang, Z. Zhu, C. Yang, X. B. Zhang, H. Yang, W. Tan and C. Fan, Sci. China: Chem., 2021, 64, 171–203 CrossRef CAS PubMed.
- R. Sista, Z. Hua, P. Thwar, A. Sudarsan, V. Srinivasan, A. Eckhardt, M. Pollack and V. Pamula, Lab Chip, 2008, 8, 2091–2104 RSC.
- P. Y. Hung, P. S. Jiang, E. F. Lee, S. K. Fan and Y. W. Lu, Microsyst. Technol., 2017, 23, 313–320 CrossRef CAS.
- M. J. Jebrail, A. Sinha, S. Vellucci, R. F. Renzi, C. Ambriz, C. Gondhalekar, J. S. Schoeniger, K. D. Patel and S. S. Branda, Anal. Chem., 2014, 86, 3856–3862 CrossRef CAS PubMed.
- S. Paul and H. Moon, Biomicrofluidics, 2021, 15, 034110 CrossRef CAS PubMed.
- Y. H. Chang, G. B. Lee, F. C. Huang, Y. Y. Chen and J. L. Lin, Biomed. Microdevices, 2006, 8, 215–225 CrossRef CAS PubMed.
- H. Norian, R. M. Field, I. Kymissis and K. L. Shepard, Lab Chip, 2014, 14, 4076–4084 RSC.
- Z. Hua, J. L. Rouse, A. E. Eckhardt, V. Srinivasan, V. K. Pamula, W. A. Schell, J. L. Benton, T. G. Mitchell and M. G. Pollack, Anal. Chem., 2010, 82, 2310–2316 CrossRef CAS PubMed.
- K. L. Ho, H. Y. Liao, H. M. Liu, Y. W. Lu, P. K. Yeh, J. Y. Chang and S. K. Fan, Micromachines, 2022, 13, 196 CrossRef PubMed.
- T. Yehezkel, A. Rival, O. Raz, R. Cohen, Z. Marx, M. Camara, J. F. Dubern, B. Koch, S. Heeb, N. Krasnogor, C. Delattre and E. Shapiro, Nucleic Acids Res., 2016, 44, e35 CrossRef PubMed.
- B. J. Coelho, B. Veigas, H. Aguas, E. Fortunato, R. Martins, P. V. Baptista and R. Igreja, Sensors, 2017, 17, 2616 CrossRef PubMed.
- Q. Zhang, X. Xu, L. Lin, J. Yang, X. Na, X. Chen, L. Wu, J. Song and C. Yang, Lab Chip, 2022, 22, 1971–1979 RSC.
- L. Wan, T. Chen, J. Gao, C. Dong, A. H. Wong, Y. Jia, P. I. Mak, C. X. Deng and R. P. Martins, Sci. Rep., 2017, 7, 14586 CrossRef PubMed.
- S. Kalsi, M. Valiadi, M. N. Tsaloglou, L. Parry-Jones, A. Jacobs, R. Watson, C. Turner, R. Amos, B. Hadwen, J. Buse, C. Brown, M. Sutton and H. Morgan, Lab Chip, 2015, 15, 3065–3075 RSC.
- Y. Liu, P. Jeraldo, H. Mendes-Soares, T. Masters, A. E. Asangba, H. Nelson, R. Patel, N. Chia and M. Walther-Antonio, ACS Omega, 2021, 6, 25642–25651 CrossRef CAS PubMed.
- E. R. F. Welch, Y. Y. Lin, A. Madison and R. B. Fair, Biotechnol. J., 2011, 6, 165–176 CrossRef CAS PubMed.
- D. J. Boles, J. L. Benton, G. J. Siew, M. H. Levy, P. K. Thwar, M. A. Sandahl, J. L. Rouse, L. C. Perkins, A. P. Sudarsan, R. Jalili, V. K. Pamula, V. Srinivasarn, R. B. Fair, P. B. Griffin, A. E. Eckhardt and M. G. Pollack, Anal. Chem., 2011, 83, 8439–8447 CrossRef CAS PubMed.
- Q. Y. Ruan, F. X. Zou, Y. Wang, Y. K. Zhang, X. Xu, X. Y. Lin, T. Tian, H. M. Zhang, L. J. Zhou, Z. Zhu and C. Y. Yang, ACS Appl. Mater. Interfaces, 2021, 13, 8042–8048 CrossRef CAS PubMed.
- G. Sathyanarayanan, M. Haapala and T. Sikanen, Anal. Chem., 2020, 92, 14693–14701 CrossRef CAS PubMed.
- J. Wang, J. Guo, K. Zhao, W. Ruan, L. Li, J. Ling, R. Peng, H. Zhang, C. Yang and Z. Zhu, Lab Chip, 2021, 21, 2702–2710 RSC.
- V. N. Luk, L. K. Fiddes, V. M. Luk, E. Kumacheva and A. R. Wheeler, Proteomics, 2012, 12, 1310–1318 CrossRef CAS PubMed.
- H. Kim, M. J. Jebrail, A. Sinha, Z. W. Bent, O. D. Solberg, K. P. Williams, S. A. Langevin, R. F. Renzi, J. L. Van De Vreugde, R. J. Meagher, J. S. Schoeniger, T. W. Lane, S. S. Branda, M. S. Bartsch and K. D. Patel, PLoS One, 2013, 8, e68988 CrossRef CAS PubMed.
- S. Anderson, B. Hadwen and C. Brown, Lab Chip, 2021, 21, 962–975 RSC.
- T. Taniguchi, T. Torii and T. Higuchi, Lab Chip, 2002, 2, 19–23 RSC.
- J. Leipert and A. Tholey, Lab Chip, 2019, 19, 3490–3498 RSC.
- V. Srinivasan, V. K. Pamula and R. B. Fair, Anal. Chim. Acta, 2004, 507, 145–150 CrossRef CAS.
- R. S. Sista, A. E. Eckhardt, T. Wang, C. Graham, J. L. Rouse, S. M. Norton, V. Srinivasan, M. G. Pollack, A. A. Tolun, D. Bali, D. S. Millington and V. K. Pamula, Clin. Chem., 2011, 57, 1444–1451 CrossRef CAS PubMed.
- R. S. Sista, T. Wang, N. Wu, C. Graham, A. Eckhardt, T. Winger, V. Srinivasan, D. Bali, D. S. Millington and V. K. Pamula, Clin. Chim. Acta, 2013, 424, 12–18 CrossRef CAS PubMed.
- E. Moazami, J. M. Perry, G. Soffer, M. C. Husser and S. C. C. Shih, Anal. Chem., 2019, 91, 5159–5168 CrossRef CAS PubMed.
- L. M. Y. Leclerc, G. Soffer, D. H. Kwan and S. C. C. Shih, Biomicrofluidics, 2019, 13, 034106 CrossRef PubMed.
- M. J. Jebrail and A. R. Wheeler, Anal. Chem., 2009, 81, 330–335 CrossRef CAS PubMed.
- V. N. Luk and A. R. Wheeler, Anal. Chem., 2009, 81, 4524–4530 CrossRef CAS PubMed.
- D. Chatterjee, A. J. Ytterberg, S. U. Son, J. A. Loo and R. L. Garrell, Anal. Chem., 2010, 82, 2095–2101 CrossRef CAS PubMed.
- W. C. Nelson, I. Peng, G. A. Lee, J. A. Loo, R. L. Garrell and C. J. Kim, Anal. Chem., 2010, 82, 9932–9937 CrossRef CAS PubMed.
- V. N. Luk, G. Mo and A. R. Wheeler, Langmuir, 2008, 24, 6382–6389 CrossRef CAS PubMed.
- D. Chatterjee, A. J. Ytterberg, S. U. Son, J. A. Loo and R. L. Garrell, Anal. Chem., 2010, 82, 2095–2101 CrossRef CAS PubMed.
- A. P. Aijian, D. Chatterjee and R. L. Garrell, Lab Chip, 2012, 12, 2552–2559 RSC.
- R. S. Sista, A. E. Eckhardt, V. Srinivasan, M. G. Pollack, S. Palanki and V. K. Pamula, Lab Chip, 2008, 8, 2188–2196 RSC.
- A. H. C. Ng, R. Fobel, C. Fobel, J. Lamanna, D. G. Rackus, A. Summers, C. Dixon, M. D. M. Dryden, C. Lam, M. Ho, N. S. Mufti, V. Lee, M. A. M. Asri, E. A. Sykes, M. D. Chamberlain, R. Joseph, M. Ope, H. M. Scobie, A. Knipes, P. A. Rota, N. Marano, P. M. Chege, M. Njuguna, R. Nzunza, N. Kisangau, J. Kiogora, M. Karuingi, J. W. Burton, P. Borus, E. Lam and A. R. Wheeler, Sci. Transl. Med., 2018, 10, eaar6076 CrossRef PubMed.
- Y. Wang, Q. Ruan, Z. C. Lei, S. C. Lin, Z. Zhu, L. Zhou and C. Yang, Anal. Chem., 2018, 90, 5224–5231 CrossRef CAS PubMed.
- M. H. Shamsi, K. Choi, A. H. C. Ng and A. R. Wheeler, Lab Chip, 2014, 14, 547–554 RSC.
- X. Han, R. Wang, Y. Zhou, L. Fei, H. Sun, S. Lai, A. Saadatpour, Z. Zhou, H. Chen, F. Ye, D. Huang, Y. Xu, W. Huang, M. Jiang, X. Jiang, J. Mao, Y. Chen, C. Lu, J. Xie, Q. Fang, Y. Wang, R. Yue, T. Li, H. Huang, S. H. Orkin, G. C. Yuan, M. Chen and G. Guo, Cell, 2018, 172, 1091–1107 CrossRef CAS PubMed.
- X. Xu, M. Zhang, X. Zhang, Y. Liu, L. Cai, Q. Zhang, Q. Chen, L. Lin, S. Lin, Y. Song, Z. Zhu and C. Yang, Anal. Chem., 2022, 94, 8164–8173 CrossRef CAS PubMed.
- M. Zhang, Y. Zou, X. Xu, X. Zhang, M. Gao, J. Song, P. Huang, Q. Chen, Z. Zhu, W. Lin, R. N. Zare and C. Yang, Nat. Commun., 2020, 11, 2118 CrossRef CAS PubMed.
- J. L. He, A. T. Chen, J. H. Lee and S. K. Fan, Int. J. Mol. Sci., 2015, 16, 22319–22332 CrossRef CAS PubMed.
- I. Barbulovic-Nad, H. Yang, P. S. Park and A. R. Wheeler, Lab Chip, 2008, 8, 519–526 RSC.
- P. C. Gach, S. C. C. Shih, J. Sustarich, J. D. Keasling, N. J. Hillson, P. D. Adams and A. K. Singh, ACS Synth. Biol., 2016, 5, 426–433 CrossRef CAS PubMed.
- Y. H. Wang, H. Y. Zhao, X. M. Liu, W. Lin, Y. W. Jiang, J. F. Li, Q. Zhang and G. X. Zheng, Biotechnol. Bioeng., 2021, 118, 294–304 CrossRef CAS PubMed.
- I. Barbulovic-Nad, S. H. Au and A. R. Wheeler, Lab Chip, 2010, 10, 1536–1542 RSC.
- D. Witters, N. Vergauwe, S. Vermeir, F. Ceyssens, S. Liekens, R. Puers and J. Lammertyn, Lab Chip, 2011, 11, 2790–2794 RSC.
- I. A. Eydelnant, U. Uddayasankar, B. Li, M. W. Liao and A. R. Wheeler, Lab Chip, 2012, 12, 750–757 RSC.
- D. Bogojevic, M. D. Chamberlain, I. Barbulovic-Nad and A. R. Wheeler, Lab Chip, 2012, 12, 627–634 RSC.
- H. Sinha, A. B. V. Quach, P. Q. N. Vo and S. C. C. Shih, Lab Chip, 2018, 18, 2300–2312 RSC.
- A. B. V. Quach, S. R. Little and S. C. C. Shih, Anal. Chem., 2022, 94, 4039–4047 CrossRef CAS PubMed.
- M. C. Husser, P. Q. N. Vo, H. Sinha, F. Ahmadi and S. C. C. Shih, ACS Synth. Biol., 2018, 7, 933–944 CrossRef CAS PubMed.
- Y. J. Zhu, L. Wang, H. Yu, F. C. Yin, Y. Q. Wang, H. T. Liu, L. Jiang and J. H. Qin, Lab Chip, 2017, 17, 2941–2950 RSC.
- J. Kieninger, A. Weltin, H. Flamm and G. A. Urban, Lab Chip, 2018, 18, 1274–1291 RSC.
- L. K. Fiddes, V. N. Luk, S. H. Au, A. H. Ng, V. Luk, E. Kumacheva and A. R. Wheeler, Biomicrofluidics, 2012, 6, 14112–1411211 CrossRef PubMed.
- I. A. Eydelnant, B. Betty Li and A. R. Wheeler, Nat. Commun., 2014, 5, 3355 CrossRef PubMed.
- S. M. George and H. Moon, Biomicrofluidics, 2015, 9, 024116 CrossRef PubMed.
- B. A. Nestor, E. Samiei, R. Samanipour, A. Gupta, A. Van den Berg, M. D. D. Derby, Z. Wang, H. R. Nejad, K. Kim and M. Hoorfar, RSC Adv., 2016, 6, S7409–S7416 RSC.
- B. B. Li, E. Y. Scott, N. E. Olafsen, J. Matthews and A. R. Wheeler, Lab Chip, 2022, 22, 313–325 RSC.
- E. M. Darling and D. Di Carlo, Annu. Rev. Biomed. Eng., 2015, 17, 35–62 CrossRef CAS PubMed.
- X. Xu, J. Wang, L. Wu, J. Guo, Y. Song, T. Tian, W. Wang, Z. Zhu and C. Yang, Small, 2020, 16, e1903905 CrossRef PubMed.
- L. Wu, Y. Wang, X. Xu, Y. Liu, B. Lin, M. Zhang, J. Zhang, S. Wan, C. Yang and W. Tan, Chem. Rev., 2021, 121, 12035–12105 CrossRef CAS PubMed.
- D. Decrop, T. Brans, P. Gijsenbergh, J. Lu, D. Spasic, T. Kokalj, F. Beunis, P. Goos, R. Puers and J. Lammertyn, Anal. Chem., 2016, 88, 8596–8603 CrossRef CAS PubMed.
- P. Tewari Kumar, D. Decrop, S. Safdar, I. Passaris, T. Kokalj, R. Puers, A. Aertsen, D. Spasic and J. Lammertyn, Micromachines, 2020, 11, 308 CrossRef PubMed.
- S. K. Fan, P. W. Huang, T. T. Wang and Y. H. Peng, Lab Chip, 2008, 8, 1325–1331 RSC.
- G. J. Shah, J. L. Veale, Y. Korin, E. F. Reed, H. A. Gritsch and C. J. Kim, Biomicrofluidics, 2010, 4, 44106 CrossRef PubMed.
- J. Lamanna, E. Y. Scott, H. S. Edwards, M. D. Chamberlain, M. D. M. Dryden, J. Peng, B. Mair, A. Lee, C. Chan, A. A. Sklavounos, A. Heffernan, F. Abbas, C. Lam, M. E. Olson, J. Moffat and A. R. Wheeler, Nat. Commun., 2020, 11, 5632 CrossRef CAS PubMed.
- X. Xu, Q. Q. Zhang, J. Song, Q. Y. Ruan, W. D. Ruan, Y. J. Chen, J. Yang, X. B. Zhang, Y. L. Song, Z. Zhu and C. Y. Yang, Anal. Chem., 2020, 92, 8599–8606 CrossRef CAS PubMed.
- Q. Ruan, J. Yang, F. Zou, X. Chen, Q. Zhang, K. Zhao, X. Lin, X. Zeng, X. Yu, L. Wu, S. Lin, Z. Zhu and C. Yang, Anal. Chem., 2022, 94, 1108–1117 CrossRef CAS PubMed.
- J. Zhai, H. Li, A. H. Wong, C. Dong, S. Yi, Y. Jia, P. I. Mak, C. X. Deng and R. P. Martins, Microsyst. Nanoeng., 2020, 6, 6 CrossRef CAS PubMed.
- C. Jiang, R. Q. Yang and B. Yuan, Eng. Appl. Artif. Intell., 2022, 115, 105305 CrossRef.
Footnote |
| † These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2023 |