Organoids/organs-on-a-chip: new frontiers of intestinal pathophysiological models
Lei
Wu
,
Yongjian
Ai
,
Ruoxiao
Xie
,
Jialiang
Xiong
,
Yu
Wang
* and
Qionglin
Liang
 *
*
MOE Key Laboratory of Bioorganic Phosphorus Chemistry & Chemical Biology, Department of Chemistry, Laboratory of Flexible Electronics Technology, Center for Synthetic and Systems Biology, Tsinghua University, Beijing 100084, P.R. China. E-mail: liangql@tsinghua.edu.cn
First published on 16th January 2023
Abstract
Organoids/organs-on-a-chip open up new frontiers for basic and clinical research of intestinal diseases. Species-specific differences hinder research on animal models, while organoids are emerging as powerful tools due to self-organization from stem cells and the reproduction of the functional properties in vivo. Organs-on-a-chip is also accelerating the process of faithfully mimicking the intestinal microenvironment. And by combining organoids and organ-on-a-chip technologies, they further are expected to serve as innovative preclinical tools and could outperform traditional cell culture models or animal models in the future. Above all, organoids/organs-on-a-chip with other strategies like genome editing, 3D printing, and organoid biobanks contribute to modeling intestinal homeostasis and disease. Here, the current challenges and future trends in intestinal pathophysiological models will be summarized.
1. Introduction
1.1 Demand for new intestinal pathophysiological models in vitro
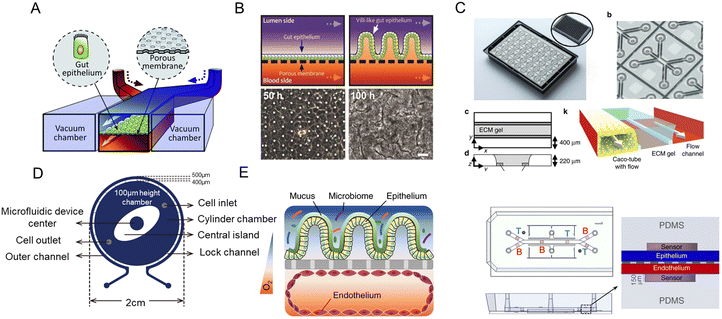
The intestine is vital for a variety of biological functions including food digestion, nutrient absorption, and the immune process. Besides, the intestine is home to a complex microbial community. Dysbacteriosis is associated with a series of intestinal diseases, such as enteritis, constipation, intestinal ulcers, inflammatory bowel disease (IBD), and irritable bowel syndrome (IBS).1–3 Moreover, the host–microbiota interactions are also critical to the flourishment of flora and human health.3,4 So the research on the intestinal microenvironment and its interaction with gut microbiota is significant to early diagnosis, treatment, and drug discovery of various diseases.Dramatic advances in new intestinal pathophysiological models have been seen over the years. However, current animal models are facing ethical issues and species-specific differences.5 As for cell lines, they have drawbacks concerning the transformed character and genomic instability.3 For this reason, organoids/organs-on-a-chip with better biomimetic complexity have drawn substantial attention. As a self-organized system generated from stem cells, intestinal organoids could simulate the genetic characteristics, in vivo architecture, and functions of the intestine. And advances in stem cell technology enable the creation of patient-specific preclinical models, distinguishing humans from other species.6–9 Intestine-on-a-chip is an emerging platform that integrates tissue engineering and microfluidic technology in vitro.10,11 By combining the best features of the above two approaches, a more powerful tool, organoids-on-a-chip, could be designed.12 And its preclinical potential is urgently required to be explored. For example, millions of children are suffering from environmental enteric dysfunction (EED), however, there is no human in vitro model of EED and a limited number of animal models at present. Thus, an EED patient-derived intestinal chip could facilitate the understanding of its mechanism and testing of candidate therapeutics.13 Besides, genome editing,14,15 3D printing,16 organoid biobanks,17 and more strategies are providing insights into the construction of new models. Currently, reviews are focusing on various human organ chips,18 human gut–microbe interactions,3 and mechanically active gut chips.19
Considering the urgent demand for new intestinal pathophysiological models in vitro, we summarize the indispensable role of organoids/organs-on-a-chip (Fig. 1) in this review. First, we will introduce the development of cutting-edge biotechnologies (organs-on-a-chip, organoids, and organoids-on-a-chip), and lay emphasis on how organs-on-a-chip/organoid models recapitulate tissue architectures and the key advantages of organoids-on-a-chip. At the same time, their technical features and magnificent prospects in intestinal disease research are reviewed. In the next part, we focus on the process from gut-x axes to human-on-a-chip. Subsequently, the construction of intestinal pathophysiological models and various therapeutic applications are also highlighted. As conclusions and prospects, we discuss current challenges and future trends in those models.
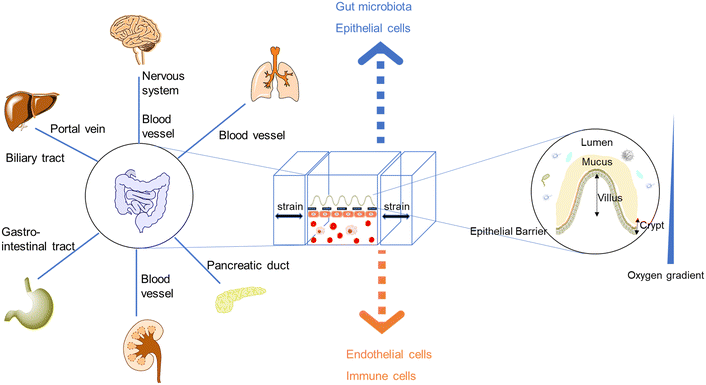 | ||
| Fig. 1 The demand for new intestinal pathophysiological models in vitro led to the development of organoids/organs-on-a-chip. Most intestine-on-a-chip models feature a design of apical and basal compartments: the apical layers could enable the co-culture of the intestinal epithelium and gut microbiota, while endothelial layers (capillary vasculatures or lymphatic vessels) play seminal roles in regulating molecular transport and immune cell recruitment.20–22 Moreover, intestinal organoids-on-a-chip models combine the strengths of chips and organoids, resulting in tubular mini-intestines with an extraordinary lifespan, cell-type diversity, architecture, and function.23,24 | ||
1.2 Cutting-edge technology – organoids/organs-on-a-chip – applied for intestinal pathophysiological models in vitro
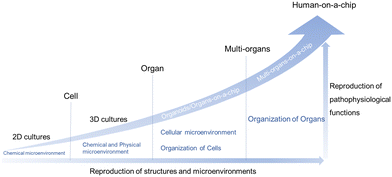
Cell-based in vitro platforms have always been key tools for the study of human biology. With the dramatic progress of the reproduction of structures and microenvironments, the reproduction of pathophysiological functions also reaches a higher level. Firstly, from 2D cultures to 3D cultures, the 3D matrix plays an important role. When stepping to the organ level, organoids/organs-on-a-chip are taking the stage and could provide insights into homeostasis and disease state modeling. On this level, the cellular microenvironment and organization of cells have drawn a lot of attention. Furthermore, multi-organ systems focus on some kind of interaction between organs. Last but not least, the ideal goal is the creation of “human-on-a-chip” with a more complex structure and more diverse function, mirroring the physiology of the entire human body (Fig. 2).25–28Traditional 2D cell culture systems couldn't produce natural cell morphology.39 Compared with traditional monolayer cell culture, a microfluidic system could offer dynamic microenvironments, such as mechanical stimulation, a 3D extracellular matrix (ECM) environment, cell–cell interactions and so on.40 Ingber's group fabricated a biomimetic microsystem and reconstituted organ-level lung functions on a chip firstly in 2010.41 Ingber's chip could reproduce complex integrated organ-level responses to bacteria and inflammatory cytokines. Since then, the term “organ-on-a-chip” was proposed and novel platforms were developed rapidly. A year later, Ingber and his collaborators proposed the concept of “human-on-a-chip”.25 Over the past years there has been continuing progress made in this interdisciplinary field, such as single-organ chips42,43 and tissue-chip systems.44 Up to now, organs-on-a-chip technology has been applied to solve many practical problems.45
Hans Clevers's group first demonstrated 3D, self-organized intestinal organoids built from stem cells.7,8 Based on recent studies, intestinal organoids are advancing our understanding of gastrointestinal inflammatory and infectious diseases. Intestinal organoids hold more clinical potential than traditional cell lines like Caco-2 cells. Moreover, organoids could replicate the complex interaction between microbes and the host epithelium.23,46–48
Organoids-on-a-chip systems have been applied for drug discovery, disease modeling, and regenerative medicine.12 Moreover, multi-organoid body-on-a-chip systems have been developed, which is appealing in improving the physiological relevance of every organoid part on the chip.49 Real-time tissue function sensors are also needed on the platform. The combination of organoids and real-time sensors promotes the performance of illustrating multi-tissue interactions, towards the goal of creating a “body-on-a-chip” platform.50,51
In recent years, attributed to the explosive development of microfluidics, microfabrication engineering, 3D printing, tissue function sensors, genome editing and organoid biobanks, organoids/organs-on-a-chip research has made great progress. We also summarized the typical advances focusing on the intestine (Fig. 3).
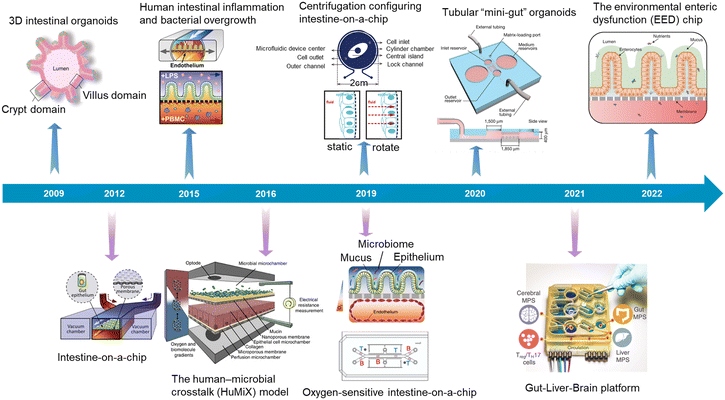 | ||
| Fig. 3 A timeline showing typical advances in intestinal organoids, intestine-on-a-chip, and intestinal organoids-on-a-chip. 3D intestinal organoids (Sato et al., 2009);8 intestine-on-a-chip (Kim et al., 2012);52 human intestinal inflammation and bacterial overgrowth (Kim et al., 2015);53 the human–microbial crosstalk (HuMiX) model (Shah et al., 2016);54 oxygen-sensitive intestine-on-a-chip (Jalili-Firoozinezhad et al., 2019);4 centrifugation configuring intestine-on-a-chip (Wang et al., 2019);55 tubular “mini-gut” organoids (Nikolaev et al., 2020);23 gut–liver–brain platform (Trapecar et al., 2021);56 the environmental enteric dysfunction (EED) chip (Bein et al., 2022).13 | ||
2. Basic technology in modeling intestinal pathophysiological states
2.1 Intestine-on-a-chip
Intestine-on-a-chip, like a miniaturized system, holds great potential in disease modeling, drug discovery, and personalized medicine.19 By providing dynamic physical stimuli like fluid flow and peristalsis-like motions, chemical stimuli, and cellular treatment, intestine-on-a-chip could obtain independent and exquisite control of various parameters in the intestinal microenvironment, to achieve a more physiological state.Villi are part of the intestinal mucosa. It is widely acknowledged that inter-tissue interfaces in the human body are indispensable for uncountable physiological functions. Their structural and functional integrity is very important to maintain normal physiological functions. Among them, the intestinal tract is the largest barrier tissue.61 The intestinal mucosa barrier plays an important role in the intestinal physiological and pathophysiological process.59 The damage to its integrity could induce serious intestinal dysfunction. Besides, the micro-villi create appropriate niches for different cell populations in the intestinal epithelium as well as different microbial communities.
Thus, the reproduction of intestinal surface topography is critical to constructing in vitro intestinal models.
Scaffolds could emulate the complex 3D architecture of the intestine and mimic growth factors, ECM, and more gradients along the crypt/villus axis. An intestine-on-a-chip with a ductal scaffold of collagen gel was fabricated by Nakajima and colleagues.62 The restricted 3D microspace induces buckling epithelium and the formation of apical polarity represented by vertical microvilli lining, columnar cell-like aspect ratio, and early tight junction formation. It also has advantages over conventional 2D models, especially in drug absorption (Fig. 4A). And a micropatterned collagen scaffold employing a suitable extracellular matrix and stiffness has been generated.63 Moreover, chemical gradients applied to the crypt–villus axis promoted the creation of a stem/progenitor-cell zone and supported cell migration to form a polarized crypt–villus architecture (Fig. 4B). And using 3D printing and polymeric scaffolds, Costello et al.64 integrated the scaffold into a bioreactor to produce 3D topography and dynamic fluid flow (Fig. 4C).
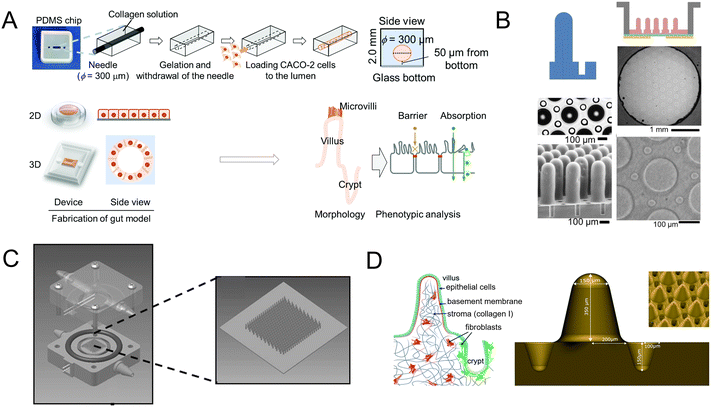 | ||
| Fig. 4 Mimicking intestinal surface topography on the chip. (A) Fabrication of the ductal scaffold of collagen gel.62 (B) Micromolding a collagen scaffold employing a suitable extracellular matrix and stiffness.63 (C) Scheme of the scaffold which is similar to the intestinal local villus surface topography.64 (D) Fabrication of the biomimetic scaffold of 3D collagen scaffold guided stromal cells into the organoids-on-a-chip platform.65 | ||
Stromal cells are important components, but they are absent in organoids. Verhulsel et al.65 proposed a novel intestinal organoids-on-a-chip design that mimicked the basement membrane and gut stroma by a 3D collagen scaffold. They observed that the organoids opened up and epithelialized the scaffold, generating a polarized epithelial monolayer; and the scaffold guided stromal cells into the platform for the first time, to our knowledge. The chip design allowed the co-culture of primary epithelial and stromal cells and their interactions to be studied (Fig. 4D).
Therefore, the chip device could more closely resemble intestinal surface topography. Moreover, a multiplex culture system that enables simultaneous recreation of multiple replications of the 3D microarchitecture of the human intestinal epithelium has been reported.66 In another work, a collagen scaffold bonded to a porous membrane in the chip could induce further improvement in gut functions.67 It not only facilitated the study of gut-related diseases but provided a step towards the integration of the gut and the liver as well.68
 | ||
| Fig. 5 The intestinal microenvironment on the chip. (A) Scheme of a biomimetic “human gut-on-a-chip” by Ingber.52 (B) Formation of villi structures in an intestine-on-a-chip device by trickling flow and peristalsis-like motions.69 (C) Schematic illustration of extracellular matrix-supported intestinal tubules.70 (D) Schematic illustration of flow stimulation perpendicular to the barrier.55 (E) Characterization of an oxygen-sensitive intestine-on-a-chip platform.4 | ||
The above compartmentalized design is important to mimic localized environments, especially the low oxygen gradients4,54 and microbial communities found on the apical side of the intestinal barriers as well as the transport of signaling molecules and secreted factors across the epithelium barrier.71,72
As for chip materials and cell types, polydimethylsiloxane (PDMS), which preserves advantages like transparency, flexibility, biocompatible and gas permeable, is the most widely applied material in microfluidic chips. However, the PDMS material usually adsorbs test molecules due to its hydrophobicity.73 As a robust human intestinal cancer cell line, Caco-2 cells are used most often for modeling healthy intestinal interactions.74 However, the absence of mucus, which provides chemical and physical defense mechanisms for the intestinal barrier, influences intestinal homeostasis and the study of absorption. So, several models have been presented including HT29-MTX cells, which are goblet-like cells for mucus production.75–78
Another typical example is extracellular matrix-supported intestinal tubules in the OrganoPlate platform (Fig. 5C).70 It consists of three channels: Caco-tube channel, ECM gel in the middle channel, and flow channel. And the sensitive, real-time investigation is expected to study compound effects on barrier integrity.
Now intestine-on-a-chip platforms feature a design of a combination of cyclic mechanical forces, controlled flow, connection with immune cells, microvascular endothelium, microbiome, and so on.19 Overall, to emulate the intestinal microenvironments and functions, intestine-on-a-chip should have the following properties:
Generation of flow. The generation of flow is a significant part of faithfully recapitulating gut physiology. Scientists usually use a pump system to generate the flow, like a pneumatic micropump,79 a syringe pump,52 or a peristaltic on-chip micropump;80 another type of flow is gravity-driven flow, like in an interval rocker platform.70 Gravity-driven flow is also applied for the simulation of 3D glomerulus.43 Similarly, there is centrifugal force-driven flow.55
It has been demonstrated that the low rate of fluid flow can produce low shear stress (0.02 dyne cm−2) and accelerate the formation of 3D villi-like structures.69 Another study by Wang et al.55 shows that flow stimulation perpendicular to the intestinal barrier could promote the biomimetic folded morphology of the intestinal epithelial cell layer and the cell cultivation lasts for several days (Fig. 5D).
Generation of the physiological oxygen gradient. When blood vessels are created, they start to supply oxygen. The physical oxygen level drops precipitously along the radial axis from the intestinal submucosa to the lumen, which is home to anaerobic microbes.81 A physiological oxygen gradient contributes to the flourishment of gut microbiota, and its construction in vitro remains a challenge.
The control of the oxygen tension in a microfluidic device could be achieved through engineering approaches or chemical approaches.82,83 In human–microbial crosstalk (HuMiX) model, Shah et al.54 developed a co-culture system with an oxygen gradient. The oxygen gradient was formed via PC enclosures, continuous perfusion of anoxic media, and consumption by Caco-2 cells and LGG. Besides, Ingber and his colleagues established a hypoxia gradient in the chip, allowing for the co-culture of anaerobic and aerobic human commensal gut microbiota in a highly complex community. Moreover, it increased intestinal barrier function and generated physiologically relevant anaerobic conditions (<0.5%) of microbial diversity (Fig. 5E).4
As for the monitoring of oxygen concentrations, oxygen-quenched fluorescent particles embedded in the channels have been used.4 And a novel method has been proposed to monitor spatially-distributed oxygen across a three-dimensional human intestinal epithelial layer in an intestine-on-a-chip, with a turn-on fluorescence probe.84 Embedded electrochemical sensors were also employed for monitoring hypoxia.85
Connection with vascular biology. Blood flow makes a crucial impact on proper vascular growth and remodeling. Endothelial cells could sense and respond to flow, while vascular networks expand via angiogenesis.86 Moreover, the perfusable channels can not only facilitate the transportation of cellular metabolisms but also are essential for realizing tissue/organ functions.87
Human induced pluripotent stem cells (HiPSCs) derived from healthy individuals and patients differentiate into endothelial cells, vascular smooth muscle cells, and pericytes. Combining HiPSCs with intestine-on-a-chip, in vitro models of blood vessels can be developed.88
Peripheral blood mononuclear cells (PBMCs) have been added to the vascular compartment to mimic immune functions. PBMCs isolated from the deidentified whole blood was an important part of capillary components in gut inflammation-on-a-chip.53,89 Morini et al.90 proposed a co-culture model in which Caco-2 cells could be grown in the presence/absence of PBMCs to study the interplay between tumoral cells and the immune system.
Taking the above mentioned into consideration, intestine-on-a-chip platforms could offer precise control of chemical, physical and cellular parameters, and then better create disease models.
2.2 Intestinal organoids
Intestinal organoids are self-organized from isolated crypts or intestinal stem cells (ISCs). Under the embedding of Matrigel, stem cells could proliferate, differentiate and self-assemble into organoid structures that present certain intestinal level functions.91 Organoids grown from the small intestine are known as enteroids, and colonoids are referred to as organoids grown from the large intestine.92In 2011, a robust and efficient method for directed differentiation of human pluripotent stem cells in vitro was created.9 Moreover, differentiation is a key step for disease models mimicked by human intestinal organoids.93–96 Adult intestinal stem cells represent properties like the role of critical drivers of epithelial homeostasis and regeneration.97 Moreover, engineering strategies also contribute to the development of intestinal organoids. By combining intestinal organoids and 3D bioprinting, scientists successfully constructed centimeter-scale tissues.16 And Gjorevski et al.98 generated more structurally complex and reproducible intestinal organoids through multiple bioengineering strategies, such as photochemistry and hydrogel microfabrication.
During the growing process, matrices support the expansion of intestinal organoids. However, due to the high variability of organoid cultures, their therapeutic potential and applications are limited. Ingredients of animal-derived matrices are complex and their quality is difficult to control. Efforts have been put to explore more appropriate matrices–designer matrices.99 They have engineered synthetic matrices and used the Arg-Gly-Asp (RGD) peptide to create the intestinal organoid microenvironment.
Although human intestinal organoids (HIOs) are widely used to study intestinal diseases such as inflammatory bowel disease, intestinal injury regeneration, and cancer,100 they cannot fully reproduce the multi-component structure of the intestine, especially the complex intestinal wall.65 The combination of organoids with microfluidic technology could help achieve the goal of in vitro model reconstruction, regeneration applications, and disease treatment exploration.
2.3 Intestinal organoids-on-a-chip
Microfluidic technology plays a crucial role in constructing organ models that integrate microfabrication and manipulation of fluids with micro-scale resolution.101 It is difficult to directly assay the 3D structure of human intestinal organoids owing to their complex and multi-component structures. Once incorporated into microfluidic platforms, intestinal organoids are more amenable to study.102 Intestinal organoids-on-a-chip could mimic the substrate transportation by blood vessels and overcome the limitations of the inability to metabolize in traditional platforms. It also measures enteroid swelling by trap region design, accelerating the research of secretory diarrheas.103Enteroid swelling measurement is an important part of the research on secretory diarrheas. And they could be achieved through the trap region design (Fig. 6A).103 When immobilized in a trap region, enteroids exclude fluorescent dye and their volume could be continuously measured. Microfluidic-based chips not only measure immobilized intestinal enteroids, but also emerge as platforms for intestinal organoids with an accessible lumen.
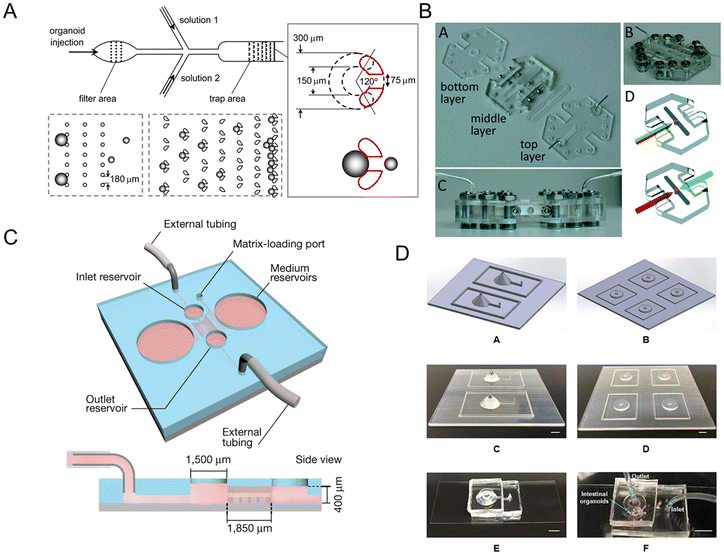 | ||
| Fig. 6 Intestinal organoids-on-a-chip platforms. (A) Microfluidic platform for the measurement of volume changes in immobilized intestinal enteroids.103 (B) Establishment of a gut organoid flow chip with laser-cut silicone gaskets.24 (C) Schematic of a 3D hydrogel-containing microdevice developed for the culture of tubular mini-guts and dimensions of a microchannel.23 (D) Scheme of long-term perfused intestinal organoids-on-a-chip with a PDMS pocket chamber.104 | ||
Long-term flow through the organoids is required in chip models, like a gut organoid flow chip, called GOFlowChip (Fig. 6B).24 The flow, which goes through the lumen of a gastrointestinal organoid for multiple days, successfully mimics long-term luminal flow, one of the most critical physiologic parameters of their in vivo counterparts. Lutolf et al.23 developed an intestinal organoids-on-a-chip platform guided by a 3D micro-structured biological scaffold (Fig. 6C). Due to the perfusion part, dead cells could be removed and the lifespan of the tissue could be extended to months. Furthermore, this study also demonstrates notable possibilities in modeling intestinal damage and regeneration.
However, air bubble accumulation remains a critical problem for PDMS-based long-term chips, which influences the microenvironment of adherent cells. A novel method to integrate a bubble pocket has been proposed to try to solve the problem (Fig. 6D).104 The air bubbles were trapped in an integrated PDMS pocket chamber and then diffused out.
Moreover, microfluidic devices could mimic the colon.105 The stomach and colon have bilayer mucus while the small intestine has one layer of it.106 Ingber and colleagues invented a colon-on-a-chip including colon organoids. Due to the dynamic fluid flow, the device has achieved high levels of goblet cell differentiation and the form of two-layered mucus, which is more physically relevant for mucus study.58 There are also colon tumor organoids on the microfluidic platform with periodic movement.107
2.4 Organoids/organs-on-a-chip for microbiome research
Human gut microbiota is composed of 1013 to 1014 microorganisms.108 The symbiosis of aerobic and anaerobic microorganisms in the intestine is closely associated with several functions. And they play a dual role in homeostasis and disease states.109 For one thing, healthy gut microbiota could promote immune homeostasis, immune responses, and protection against the pathogen.110 For another, they are involved in diverse diseases. Induced by gut microbiota, epithelial toll-like receptors also have relevance in healthy states while in colitis and tumorigenesis.111Gut and tumor microbiotas play emerging roles in response to cancer therapy.113 The chip could be used to analyze information on the microbiota and dissect disease mechanisms in a controlled manner.53 Schroeder et al.2 discussed the microbiota–host crosstalk and revealed the evidence for the pathogenic role of gut microbiota which is strongest in metabolic diseases.
Inspired by the balance of competition and positive interactions in natural microbial communities, a research team created an innovative chip (Fig. 7A).71 The three-layered chip was artificially constructed for the stability of gut microbiota: the independent upper layer is designed for bacterial growth. As the second layer, a semi-permeable membrane allowed the transportation of secreted substances. And the lower layer is for communication. New chip design has progressed through combined advances in synthetic biology. For the study of phenylketonuria, Nelson et al.75 fabricated a microfluidic platform with engineered live bacteria (Fig. 7B). And an engineering strain was co-cultured with human cancer cells on a chip,114 which proved that it can be used as a drug delivery platform.
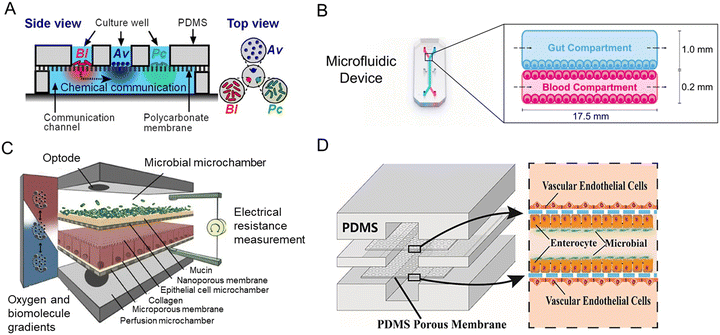 | ||
| Fig. 7 Intestine-on-a-chip platforms for the research of host–gut microbiota interactions. (A) A schematic of the three-layered chip device that artificially constructs a stable functional flora community.71 (B) Characterization of the intestine-on-a-chip for new treatment of phenylketonuria.75 (C) Design of the HuMiX model of three individual co-laminar microchambers.54 (D) A schematic drawing of the peristaltic human gut–vessel microsystem for studying host–microbial interaction.72 | ||
Human–microbial crosstalk (HuMiX) is an in vitro model focused on the human–microbe interface and the relationship between gut microbiota and human diseases (Fig. 7C).54 The device is composed of three individual co-laminar microchambers: medium perfusion, Caco-2 cells, and flora microchamber. While HuMiX permitted the study of LGG on the chip, a more recent platform containing more than 200 types of flora has been created.4
For the mechanism of the microbiome on intestinal diseases, a peristaltic human gut–vessel microsystem was invented. And it showed promising potential for investigating the host–microbial interaction, through intestinal damage and inflammatory responses caused by E. coli (Fig. 7D).72
However, intestine-on-a-chip platforms do have some limitations in terms of having micro-villi niches and spontaneously forming all the intestinal cell types. The absence of crypts in intestinal-on-chip also limits the investigation of gut homeostasis.65
Intestinal organoids don't have control and accessibility to the apical compartment of the intestinal barrier as intestine-on-a-chip. They can neither be cocultured with living microorganisms for more than ∼1 d nor sustain luminal oxygen levels below 0.5%, which is required for the coculture of certain obligate anaerobes.19,47
Furthermore, intestinal organoids-on-a-chip could provide a perusable environment, realizing long-term co-culture (30 days) between intestinal organoids and Cryptosporidium parvum as previously mentioned.23
3. From gut-x axes to human-on-a-chip
Tissue–tissue and multi-organ interactions are also essential for modeling. They form various gut-x axes, and experience homeostasis and disease state in the human body.19 So, the need for more comprehensive intestinal disease models has motivated researchers to explore improved platforms. In this section, we will discuss the in vitro modeling of the cross-talk between gut and other organs, and the pathophysiological information they reflect.3.1 Gut–brain axis
The brain controls the peristalsis of the intestine and the secretion of digestive juices through the autonomic nervous system (sympathetic and parasympathetic nerves). In addition, cutting-edge research has shown that the gut could feedback to affect human brain functions. This crosstalk between the intestine and the central nervous system is known as the gut–brain axis (GBA) (or brain–gut axis).1,120 Patients with several psychiatric and neurologic disorders (Parkinson's disease, autism spectrum disorders) have significant gastrointestinal comorbidities.121 And commensal flora metabolites can limit the pathogenic activity of microglia and astrocytes and suppress CNS inflammation.122Progress in organs-on-a-chip technology opens up new avenues for understanding the gut–brain axis, where two major barriers (gut barrier and blood–brain barrier) are exposed to flow. For the research on Parkinson's disease (PD), a pioneering team developed an integrated microfluidic system of the gut–liver–brain axis (Fig. 8A).56 The system connected the microenvironment of the gut and liver with brain organs derived from human-derived pluripotent stem cells, and during the process, CD4+ T cells were added to the culture medium to mimic the features of Parkinson's patients. The colon organoids HC176 of nondiseased tissue were from the Harvard Digestive Disease Center.
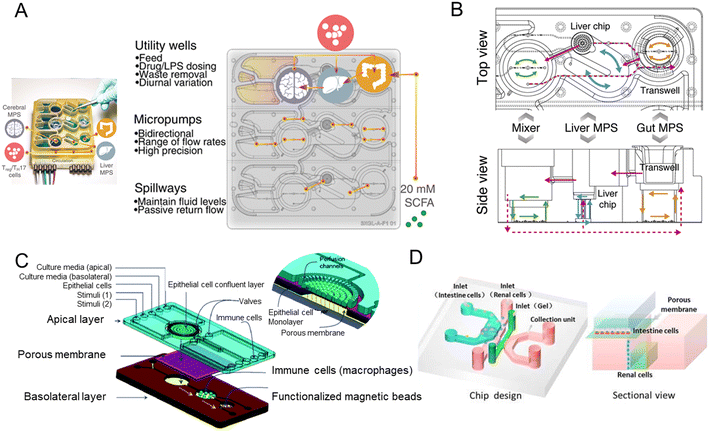 | ||
| Fig. 8 Microfluidic models of the gut-x axes. (A) Design, features, and parameters of the gut–liver–brain platform.56 (B) Top and side view of the model of the human gut–liver axis, including adaptive immune cells.117 (C) Schematic of the microfluidic chip forming the miniaturized gastrointestinal tract.118 (D) Schematic of the multi-layered intestine–kidney chip.119 | ||
Another gut-brain chip model has been constructed by Kim et al.123 Through the co-culture environment of brain endothelial and intestinal epithelial cells (Caco-2 cells), the transportation and communication between the two organs could be assessed. And extracellular vesicles play an important role in intercellular signaling and influence the pathophysiology of related parts of the body. Organs-on-a-chip technology also provides a platform of the gut–brain axis mediated by extracellular vesicles.124
Therefore, it is of great significance to deeply investigate the gut–brain axis and reveal its biological mechanism, for the research of abnormal intestinal function and neurological diseases.
3.2 Gut–liver axis
The liver and intestine highly connect through the biliary tract, portal vein, and systemic mediators. Liver products affect intestinal microbiota composition and its barrier integrity, while intestinal factors regulate bile acid synthesis, glucose, and lipid metabolism in the liver.125 Intestine–liver chip platforms have been used for hepatic steatosis modeling,126 PM2.5-induced metabolic diseases,127 gut–liver interaction in endotoxemia,128 IBD and its relation to liver disease,117 and metastasis of tumor cells.129In 2015, Maschmeyer et al.130 designed a multi-organ system for the modeling of liver–intestine and liver–skin. They used primary small intestine epithelial cells. And they achieved homeostatic long-term co-culture, which is 1/100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 the scale of their human counterparts in vivo. Later, Choe et al.68 invented a gut-liver chip that is capable of coculturing gut (Caco-2 cells) and liver cells, reproducing the dynamics of the first-pass metabolism. After that, it was used to demonstrate fatty acid absorption in the gut and lipid accumulation in the liver,126 which is called hepatic steatosis. This is the first demonstration of an in vitro model capable of lipid absorption in the gut and subsequent accumulation in the liver. As for the chip design, the expandable modular system has emerged as a plug-and-play approach for the co-culture of different tissues. Shuler's group designed a low-cost pumpless cell culture device131 and they re-designed it so that it can co-culture gastrointestinal tract (GI tract), epithelium (Caco-2 cells) and 3D primary liver tissues.132 In addition, Jie et al.133 created an intestine–liver–glioblastoma biomimetic chip platform to evaluate the drug combination therapy for glioblastoma. They used hollow fibers (HF) to culture Caco-2 cells in a tubular structure and transfer drugs.
000 the scale of their human counterparts in vivo. Later, Choe et al.68 invented a gut-liver chip that is capable of coculturing gut (Caco-2 cells) and liver cells, reproducing the dynamics of the first-pass metabolism. After that, it was used to demonstrate fatty acid absorption in the gut and lipid accumulation in the liver,126 which is called hepatic steatosis. This is the first demonstration of an in vitro model capable of lipid absorption in the gut and subsequent accumulation in the liver. As for the chip design, the expandable modular system has emerged as a plug-and-play approach for the co-culture of different tissues. Shuler's group designed a low-cost pumpless cell culture device131 and they re-designed it so that it can co-culture gastrointestinal tract (GI tract), epithelium (Caco-2 cells) and 3D primary liver tissues.132 In addition, Jie et al.133 created an intestine–liver–glioblastoma biomimetic chip platform to evaluate the drug combination therapy for glioblastoma. They used hollow fibers (HF) to culture Caco-2 cells in a tubular structure and transfer drugs.
For PM2.5-induced metabolic diseases, a novel membrane-free liver–gut-on-chip was used to study metabolic mechanisms exposed to PM2.5.127 Caco-2 cells were employed to represent the gut. And to model gut–liver interaction in endotoxemia, an integrated multiorgan microphysiological system (MPS) platform was created.128 The platform was under normal and inflammatory conditions. They achieved long-term (>2 weeks) maintenance of intestinal and hepatic functions in baseline interaction. In addition, IBD and its relation to liver disease can be studied on a microfluidic platform. Interestingly, Trapecar et al.117 created a multi-organ model of ulcerative colitis (UC). They showed that the interaction between gut MPS of UC (primary human UC epithelium, dendritic cells, and macrophages) and liver MPS increases metabolism and reduces inflammation. The system also contained adaptive immune cells and they circulated in the system connected in series (Fig. 8B).
Current animal and 2D cell cultures cannot fully mimic the migration and metastasis of tumor cells at the organ level. The need for improved methods has motivated Skardal and his colleagues129 to fabricate a “metastasis-on-a-chip” system that facilitated the tracking of colorectal carcinoma metastasizing from the colon to the liver. Furthermore, multiple organoids have been connected to study their communications (liver, pancreas, and gastrointestinal tract).134 Nevertheless, multiple-organ integration in stem cell culture remains a critical challenge.
3.3 Gastrointestinal tract
NutriChip, the integrated microfluidic platform like the miniaturized GIT, focused on postprandial inflammatory stress which potentially contributes to the development of chronic inflammatory diseases (Fig. 8C).118,135 The co-culture of Caco-2 cells and a monocytic cell line (U937 cells) differentiated into macrophages is the core biological component of the NutriChip. Another cell-free, miniaturized enzymatic digestive system (mouth, stomach, and small intestine) has been fabricated to mimic digestive functions.1363.4 Gut–kidney axis
There is a close relationship between the two organs, like the intestinal–renal syndrome proposed by Ritz,137 and intestinal flora is involved in the process of chronic kidney disease (CKD).138Meanwhile the absorption of drugs in the intestine is related to the subsequent drug-induced nephrotoxicity, which is a common side effect in clinical practice. Therefore, the development of highly accurate predictive models is badly needed in the early stages of drug discovery. Li et al.119 designed a microfluidic chip in which intestinal cells (Caco-2 cells) coculture with glomerular endothelial cells (Fig. 8D). The multi-layered chip not only mimics drug absorption, but completes the assessment of drug nephrotoxicity as well.
3.5 Gut–lung axis
Intestinal flora components were significantly changed in patients with COVID-19, and research suggested that it may associate with host immune responses.139Now in several IBD patients, their airways are also affected by the inflammatory process. Wang et al.140 studied the crosstalk between the gut and lungs through IBD. They hypothesized that each cell or molecule not only exerts its local role in its organ but also plays a “social” role, facilitating distant communication through the epithelium. On the other hand, changes in gut microbiome composition through diet, disease, or antibiotics are related to changes in immune responses and airway homeostasis.141
3.6 Gut–pancreas axis
Ahuja et al.142 reported that the secretion of pancreatic acinar cells plays a crucial role in intestinal innate immunity. On the other hand, gut dysbiosis may be involved in acute pancreatitis, a common disorder of the digestive system.1433.7 From multi-organs-on-a-chip to human-on-a-chip
Organ-on-a-chip could mimic organ-level pathophysiology, nevertheless, pharmacokinetic (PK) and pharmacodynamic (PD) analysis are in demand for multiple organ systems connected by vascular perfusion. For example, a PK study requires quantitative analysis of the entire process of drug absorption, distribution, metabolism, and excretion, involving interactions between multiple organs. The fabrication of multi-organs-on-a-chip enabled us to study organ interactions and systemic toxicity profiling.In 2015, Maschmeyer et al. established a multi-organs-on-a-chip for interconnected long-term co-culture of four human tissue types (intestine, liver, skin, and kidney equivalents) (Fig. 9A).80 They used primary human small intestinal epithelial cells to reconstruct the intestinal barrier. The system profiled ADME and repeated dose systemic toxicity testing of drug candidates over 28 days for the first time, as far as we know. After that, another system composed of four organs has been reported by Oleaga for toxicity analysis.147 The functional human model could assess multi-organ toxicity in a serum-free defined medium for 14 days using a pump-free platform under continuous flow conditions. There are also four autologous tissue models (intestine, liver, brain, and kidney) that last over 14 days.148 All differentiated tissues used for the model were derived from the same iPSC line.
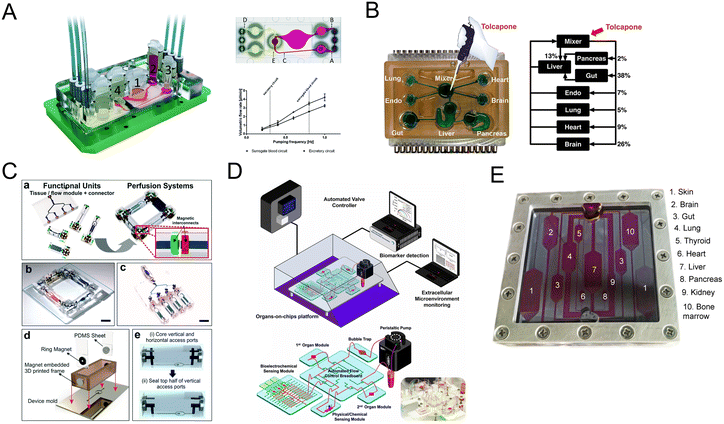 | ||
| Fig. 9 Schematic diagram of multi-organs-on-a-chip. (A) Schematic of the four-organ-chip model with a blood flow circuit (pink) and an excretory flow circuit (yellow). Numbers represent the four tissue culture compartments for intestine (1), liver (2), skin (3), and kidney (4) tissue.80 (B) The diagram of the 7 interacting MPS platform in a perfused capillary bed to mimic human physiological systems.144 (C) Scheme of the modular and functional perfusion systems with magnetic interconnections. They can form a serial recirculating perfusion system, and a one-pass parallel perfusion system.145 (D) Integrated automated multi-organs-on-a-chip and sensing platform.50 (E) Ten-organ-on-a-chip as part of a microphysiological system.146 | ||
When the above four-organ-chip come to a more complicated design, for example, drug efficacy and toxicity require multi-organ interactions. Through a perfused capillary bed, precise fluid control and differential flow distribution could be achieved (Fig. 9B).144 And an automated “Interrogator” instrument has been created,149 maintaining multiple human vascularized organ chips for weeks when fluidically linked.
Modular platforms are also in demand for the study of multi-organ interactions. A modular microfluidic platform was reported (Fig. 9C).145 Connected by a self-aligning magnetic device, it has broadened the potential applications for multi-organs-on-a-chip in fluidic connectivity and culture conditions. Furthermore, modular platforms can in situ monitor organoid behaviors automatically and continually (Fig. 9D).50
When modeling all 10 main body systems, challenges remain. Michael Shuler's group created a ten-organ-on-a-chip platform to accelerate drug testing (Fig. 9E).146 However, recapitulating the full immune system in vitro remains a big challenge.
The above platforms have the advantages of controllable flow, physiological shear stress, integration of biosensing, reproduction of tissue–tissue and multi-organ interactions, serving the purpose for systemic toxicity profiling and disease modeling. And they may contribute to inspiring a new frontier of research from multi-organs-on-a-chip to human-on-a-chip.
4. Construction of intestinal pathophysiological models and applications
4.1 Intestinal pathophysiological models
Intestinal pathophysiological models consist of infectious disease, autoimmune disease, cancer, and other disease models. The dynamic microfluidic platforms could be combined with intestinal organoids to investigate the mechanisms of the above various diseases.During the process of infection, a crucial step is the navigation of the pathogen through the commensal bacterial layer to attach to epithelial cells. Kim and colleagues invented a co-culture model and tested the effect of the commensal microenvironment on pathogen colonization.152 And sometimes the gut microbiota provides benefit to the host by limiting the expansion of enteric pathogens.153
As for viruses, enteroviruses are a major source of human disease, and the enterovirus genus includes poliovirus, coxsackieviruses, echoviruses, enterovirus 71, and enterovirus D68.154 Villenave et al.155 used coxsackievirus B1 (CVB1) as a proto-type enterovirus strain, and they demonstrated that human enterovirus infection, replication, and infectious virus production could be analyzed via a chip platform that supports the culture of highly differentiated human villus intestinal epithelium.
A biomimetic human intestine-on-a-chip was proposed by Qin and her colleagues to study SARS-CoV-2-induced intestinal responses in vitro (Fig. 10A).78 In this intestine-on-a-chip, Caco-2 cells and mucin-secreting HT-29 cells were co-cultured under continuous perfusion to build an intestinal epithelium. And they found that the disturbance of the mucus layer may be related to further invasion and infection by SARS-CoV-2. To the best of our knowledge, it provides the first proof-of-concept for building a human intestinal SARS-CoV-2 infection model via intestine-on-a-chip.
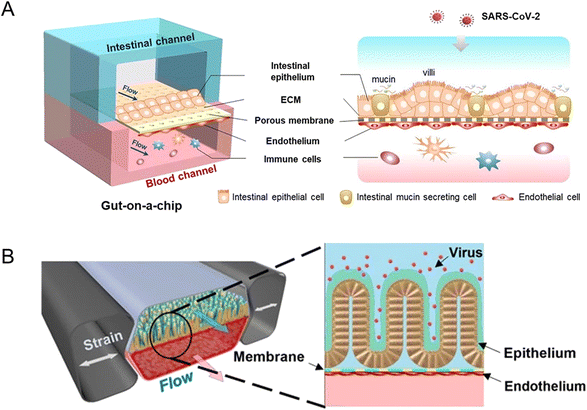 | ||
| Fig. 10 Organoids/organs-on-a-chip platforms to study intestinal infectious diseases. (A) The configuration of the multi-layered intestine-on-a-chip device infected with SARS-CoV-2.78 (B) Schematic of NL63 coronavirus inflection through the intestinal organoids-on-a-chip platform.151 | ||
Active SARS-Cov-2 infection and replication are possible in human and bat intestinal organoids.93 Human intestinal organoids derived from pluripotent stem cells (PSC-HIOs) serve as a faithful platform to study SARS-CoV-2 infection.94,156 As ACE2, the SARS-CoV-2 receptor,157 is highly expressed on differentiated enterocytes, SARS-CoV-2 can replicate and infect through gut organoids.94 In addition, PSC-HIOs are employed to dissect SARS-CoV-2 pathogenesis and its inhibition by remdesivir, one of the leading drugs discovered for the treatment of COVID-19.156 As another type of coronavirus, NL63 also uses ACE2 receptors. A microfluidic platform, together with patient organoid-derived epithelial cells, has been proposed for inflammatory response study (Fig. 10B).151 And for human norovirus, HSIOs have also allowed the first in vitro cultivation system of human norovirus and permitted host–pathogen studies.158
And for the investigation of pathogens, Grassart and his colleagues159 enable the molecular and mechanistic investigation of human-restricted enteric pathogens by creating a 3D colonic epithelium and modulating the mechanical forces of the microenvironment.
Modular chip platforms could study independent contributions of potential factors in the autoimmune disease process. Inflammatory bowel disease (IBD) results from a group of inflammatory conditions affecting the colon and small intestine.46 Kim et al.53 facilitated an intestine-on-a-chip and found that immune cells must be present with LPS or non-pathogenic bacteria to induce the production of a key set of pro-inflammatory cytokines, whose action led to the damage of villi and the disruption of the intestinal barrier (Fig. 11A). And through intestine-on-a-chip, DSS specifically induces epithelial barrier dysfunction (Fig. 11B).89 The chip successfully investigates intercellular host–microbiome cross-talk and dissects the mechanisms of gastrointestinal diseases. Moreover, interactions in epithelial organoids, immune components, and microbiota have a crucial role in the research of IBD.116
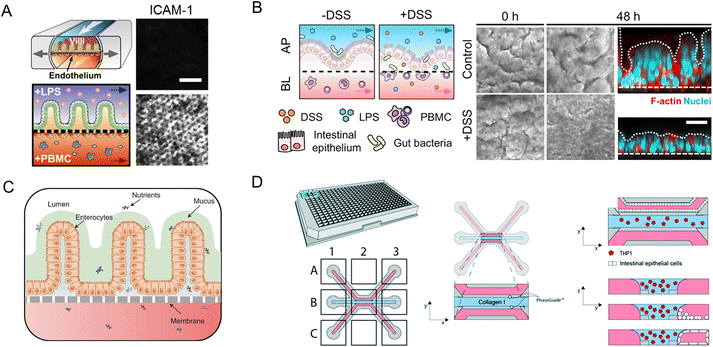 | ||
| Fig. 11 Organoids/organs-on-a-chip platforms to study intestinal autoimmune diseases. (A) Human intestinal inflammation and bacterial overgrowth.53 (B) The diagram of DSS-mediated epithelial barrier dysfunction and morphology of the villus epithelium.89 (C) The diagram of the EED chip model.13 (D) Schematic representation of the intestinal mucosal barrier with cellular innate immunity.160 | ||
The independent contributions couldn't be studied in animal models for their complexity. Besides versatile modularity for mimicking the intestinal pathophysiology, quantitation of intestinal barrier function is another unique advantage of chips in disease conditions. As a chronic inflammatory condition of the intestine, environmental enteric dysfunction (EED) affected millions of children. Ingber's group has created a chip that mimics key features of the transcriptome signature of EED patients (Fig. 11C).13
The neutrophilic infiltration and other critical immune responses have been increasingly presented in recent intestine-on-a-chip models. By adding immune elements into intestine-on-a-chip, Gijzen et al.161 successfully mimicked the decrease of mucous barrier function and increase in cytokines caused by intestinal inflammation. And Gjorevski et al.160 proposed a novel chip that first integrated a mucosal barrier, together with a 3D ECM microenvironment, and incorporated resident and infiltrating immune cells. Neutrophil invasion movement could be observed through the platform (Fig. 11D).
Organoids closely mimic several properties of the original tumor.17 Tumor organoids with a combination of immune cells and fibroblasts have been identified with essential tumor microenvironment features and present a new method for the development of immune therapies.163
Importantly, the co-culture of CRC patient-derived organoids (PDO) and cancer-associated fibroblasts (CAFs) contributes to the research of tumor progression and drug resistance. Luo et al.164 developed a co-culture strategy and further demonstrated that the co-culture models are fit for evaluating standard-of-care drugs (Fig. 12A). Besides CAFs, a vascularized micro-organ platform has also been developed by Sobrino and colleagues (Fig. 12B).165 The vascularized solid tumor platform has captured some of the complexity of tumors. Recently a more biomimetic vascularized platform that can be configured for tumor studies has been created (Fig. 12C).166 Its cellular heterogeneity, much higher than either monolayer or spheroid cultures, is a key property for drug discovery. And under peristalsis conditions, would the anti-tumor efficacy change? Fang and colleagues107 developed a colon tumor organoid chip platform to study the cellular response to nanomedicine treatment. The platform has advantages such as a mechanically responsive microenvironment and high-throughput culture of organoids (Fig. 12D).
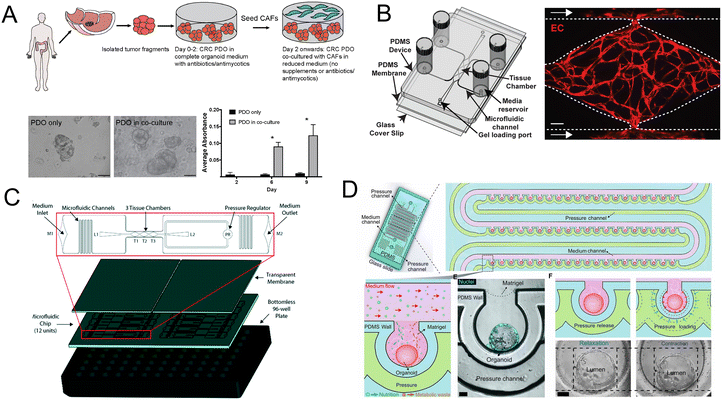 | ||
| Fig. 12 Organoids/organs-on-a-chip platforms for cancer research. (A) Schematic illustration of co-culture of CRC PDO and CAFs. Brightfield images and growth profile of PDO only and PDO in co-culture.164 (B) Schematic of the vascularized micro-organ platform.165 (C) The diagram of the vascularized micro-tumors microfluidic platform.166 (D) The diagram of colon tumor organoids on the microfluidic platform with periodic movement.107 | ||
To date, long-term organoid cultures have already been established from the primary colon.17,96,167 Remarkably, the first organoid biobank comprised the primary tumor and their matching normal organoids derived from colorectal carcinoma patients was generated by Van De Wetering and colleagues.17 And many living organoid biobanks have been seen over the years due to the genome stability of organoids, which is valuable for our understanding of cancer and other diseases. The above organoid technologies could open new avenues for the construction of advanced in vitro models, accelerating the process of cancer research.
4.2 Organoids/organs-on-a-chip platforms for drug discovery
Organoids/organs-on-a-chip platforms provide new strategies for disease mechanism research. In addition, they are also of special interest for drug permeability,170,171 high-throughput drug screening,172–174 drug absorption,175,176 drug concentration measurement177 and pharmaceutical analysis.178,179Drug permeability studies are of great importance. An integrated microfluidic platform has been fabricated,170 with Caco-2 cell lines and directly connected to mass spectrometry, a label-free detection tool. It shows promising potential for parallelization and increasing throughput (Fig. 13A). Another study focuses on the permeability of lipophilic prodrugs-SN38.171 In this study, intestine-on-a-chip acted as a test platform for oral SN38-based chemotherapy.
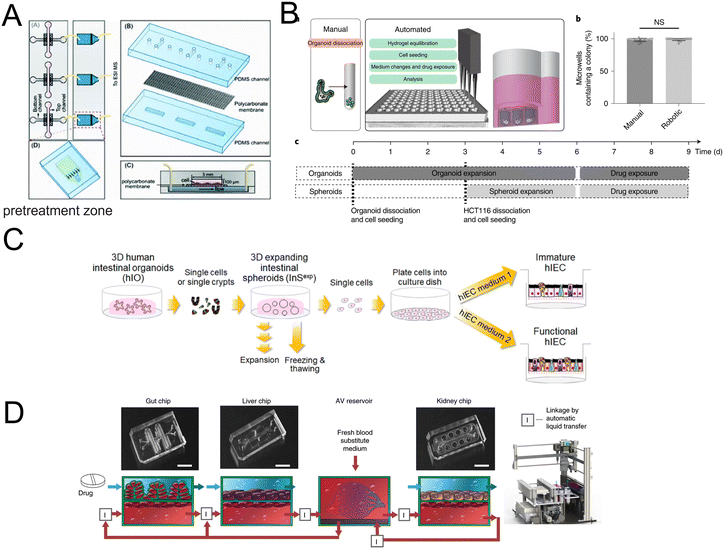 | ||
| Fig. 13 Organoids/organs-on-a-chip platforms for drug discovery. (A) Scheme of the microfluidic platform with membrane-based cell culture and pretreatment zone.170 (B) Automated fabrication of mouse intestinal organoid arrays and the timeline of the screening protocol.173 (C) Scheme of the 2D functional human intestinal epithelial cell (hIEC) monolayer from 3D intestinal organoids.175 (D) Development of a first-pass multi-organ-chip system: a linked gut–liver–kidney chip. Chips are fluidically linked to each other and the arteriovenous (AV) reservoir.177 | ||
High-throughput drug screening has aroused great interest. However, fully automated organoid culture for high-throughput and high-content organoid-based screening still is a challenge. Lukonin et al.172 designed an image-based screen to assay 2789 selected compounds. And the effects of these compounds on intestinal organoid growth and differentiation were observed. They identified an inhibitor of the retinoid X receptor that improves intestinal regeneration in vivo. An organoid array technology has been developed by Brandenberg and colleagues (Fig. 13B).173 Through the high-throughput automated organoid culture and patient-derived colorectal cancer organoids, they have screened for anticancer drug candidates. The method takes advantage of the versatility of customizing the physical and chemical properties of the hydrogel substrates, a reduction in the heterogeneity of the final tissues, as well as an increase in growth speed, single-organoid traceability from initial cell seeding to downstream analysis and accessibility. Moreover, it results in a considerable reduction in expensive reagent consumption.
For preclinical drug assessment, in vitro models with intestinal organoids have advantages in terms of a high level of CYP3A4 expression and induction over Caco-2 cell lines (Fig. 13C).175,176 Furthermore, a first-pass multi-organ system was applied to quantitatively predict the pharmacokinetic (PK) parameters of nicotine with a combination of gut, liver, and kidney (Fig. 13D).177 Importantly, the AV reservoir plays a significant role in the systemic circulation and the measurement of drug concentrations.
4.3 Organoids/organs-on-a-chip platforms for personalized medicine
Due to their ability to faithfully recapitulate the crucial features of intestinal diseases, organoids/organs-on-a-chip platforms are capable of advancing novel therapy development and amelioration of cancer treatments.HIPSCs or tissue-derived organoids could contribute to the development of personalized treatment strategies.100 Additionally, with the exciting breakthrough of genome editing techniques,14,15 the clinical potential of intestinal stem cells could be further realized. For example, in cystic fibrosis, patient-derived intestinal organoids are available to facilitate personalized medicine approaches.169 For the new therapy of ulcerative colitis, a research team developed an approach to transplant intestinal organoids into the colon of recipient mice.180 Moreover, as reported in Nature Medicine, children with intestinal failure (IF) are suffering from the lack of donor's intestines. Meran and colleagues181 successfully constructed jejunal mucosal grafts and expanded the target organoids.
When intestinal organoids meet microfluidic technology, a dynamic 3D mucosal interface-on-a-chip has been reported by Kim and his colleagues.182 They have built patient-specific models of major human chronic GI diseases. And through a small intestine-on-a-chip, primary epithelial cells are expanded as 3D organoids while continuous flow through the lumen can be collected and subsequently analyzed. The physical information from the organoids may contribute to the development of personalized medicine.183 Intestinal organoids derived from iPSCs can also be incorporated into the chip.102
Furthermore, the study of small intestine vasculature is of critical importance. And for personalized medicine, patient-derived intestinal subepithelial myofibroblasts (ISEMFs) and endothelial cells (ECs) have been incorporated into the chip platform, as endothelium and stroma are organ-specific markers to study the development and physiologic role of small intestine vasculature.184
All in all, the above intestinal pathophysiological models have a variety of unique advantages over conventional systems, providing potential for the advancement of personalized medicine.
Conclusions and prospects
Organoids/organs-on-a-chip, which are more similar to natural tissues, are like human “avatars”.18 Intestine-on-a-chip could realize the co-culture of cells and other elements through different compartments and layers. The precise chemical, physical and cellular microenvironments are more controllable via microfluidic technology, making them ideal for studying the pathophysiology of a variety of intestinal diseases. For example, besides establishing an oxygen gradient across the lumen through the intestine-on-a-chip,4 it is of great potential to establish an oxygen gradient from different regions of the intestine (e.g., duodenum, jejunum, ileum, colon) to recapitulate in vivo oxygen features. Moreover, the combination of intestinal organoids with microfluidic technology could form perfusable lumens, and bring a higher level of physiological relevance.23 Recent years have seen a number of remarkable intestinal models in vitro. On one hand, research on these diverse gut-x axes has opened up new approaches for the study of the host–microbiota, tissue–tissue, and multi-organ interactions, advancing the development of human-on-a-chip. On the other hand, further studies would focus on patient-derived intestinal organoids and the organ-specific microenvironment of diseased tissues, for the standardization of organoid-based therapy.However, currently, several challenges need to be addressed. For intestinal organoids, their lifespan requires to be increased. The lack of mechanics and reproducibility also limits their potential. For multiple systems, the limited flexibility under cultural conditions and fluidic connectivity remains a problem. The better utilization of engineering methods in organoid culture is essential for solving many of the above problems. And intestinal pathophysiological models may create possible breakthroughs in the proper vascularization, incorporation of immune elements, integrated real-time tissue function sensors, standardization and compatibility in multi-organs models, and so on.
Above all, future opportunities lie ahead, like advancing the utilization of organoids/organs-on-a-chip platforms for evaluation of drug efficacy, and systemic toxicity profiling. The therapeutic efficacy of intestinal organoids is beginning to be explored. New technologies may also extend their potential to generate a transplantable intestine.
In the near future, organoids/organs-on-a-chip technology may pave a promising avenue for intestinal disease modeling and therapeutic applications.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Key R&D Program of China (2022YFA1103403), National Natural Science Foundation of China (No. 82104128, 22074157, 81872835), Tsinghua-Foshan Innovation Special Fund (2022THFS6121), Postdoctoral Innovative Talent Support Program (BX20220160) and China Postdoctoral Science Foundation Funded Project (2022M711779).Notes and references
- J. F. Cryan, K. J. O'Riordan, C. S. M. Cowan, K. V. Sandhu, T. F. S. Bastiaanssen, M. Boehme, M. G. Codagnone, S. Cussotto, C. Fulling, A. V. Golubeva, K. E. Guzzetta, M. Jaggar, C. M. Long-Smith, J. M. Lyte, J. A. Martin, A. Molinero-Perez, G. Moloney, E. Morelli, E. Morillas, R. O'Connor, J. S. Cruz-Pereira, V. L. Peterson, K. Rea, N. L. Ritz, E. Sherwin, S. Spichak, E. M. Teichman, M. van de Wouw, A. P. Ventura-Silva, S. E. Wallace-Fitzsimons, N. Hyland, G. Clarke and T. G. Dinan, Physiol. Rev., 2019, 99, 1877–2013 CrossRef CAS PubMed.
- B. O. Schroeder and F. Backhed, Nat. Med., 2016, 22, 1079–1089 CrossRef CAS PubMed.
- J. Puschhof, C. Pleguezuelos-Manzano and H. Clevers, Cell Host Microbe, 2021, 29, 867–878 CrossRef CAS PubMed.
- S. Jalili-Firoozinezhad, F. S. Gazzaniga, E. L. Calamari, D. M. Camacho, C. W. Fadel, A. Bein, B. Swenor, B. Nestor, M. J. Cronce, A. Tovaglieri, O. Levy, K. E. Gregory, D. T. Breault, J. M. S. Cabral, D. L. Kasper, R. Novak and D. E. Ingber, Nat. Biomed. Eng., 2019, 3, 520–531 CrossRef CAS PubMed.
- C. Ma, Y. Peng, H. Li and W. Chen, Trends Pharmacol. Sci., 2021, 42, 119–133 CrossRef CAS PubMed.
- J. Kim, B. K. Koo and J. A. Knoblich, Nat. Rev. Mol. Cell Biol., 2020, 21, 571–584 CrossRef CAS PubMed.
- N. Barker, J. H. van Es, J. Kuipers, P. Kujala, M. van den Born, M. Cozijnsen, A. Haegebarth, J. Korving, H. Begthel, P. J. Peters and H. Clevers, Nature, 2007, 449, 1003–1007 CrossRef CAS PubMed.
- T. Sato, R. G. Vries, H. J. Snippert, M. van de Wetering, N. Barker, D. E. Stange, J. H. van Es, A. Abo, P. Kujala, P. J. Peters and H. Clevers, Nature, 2009, 459, 262–265 CrossRef CAS PubMed.
- J. R. Spence, C. N. Mayhew, S. A. Rankin, M. F. Kuhar, J. E. Vallance, K. Tolle, E. E. Hoskins, V. V. Kalinichenko, S. I. Wells, A. M. Zorn, N. F. Shroyer and J. M. Wells, Nature, 2011, 470, 105–109 CrossRef PubMed.
- D. Huh, Y. S. Torisawa, G. A. Hamilton, H. J. Kim and D. E. Ingber, Lab Chip, 2012, 12, 2156–2164 RSC.
- B. Jiang, W. Zheng, W. Zhang and X. Jiang, Sci. China: Chem., 2013, 57, 356–364 CrossRef.
- S. E. Park, A. Georgescu and D. Huh, Science, 2019, 364, 960–965 CrossRef CAS PubMed.
- A. Bein, C. W. Fadel, B. Swenor, W. Cao, R. K. Powers, D. M. Camacho, A. Naziripour, A. Parsons, N. LoGrande, S. Sharma, S. Kim, S. Jalili-Firoozinezhad, J. Grant, D. T. Breault, J. Iqbal, A. Ali, L. A. Denson, S. R. Moore, R. Prantil-Baun, G. Goyal and D. E. Ingber, Nat. Biomed. Eng., 2022, 6, 1236–1247 CrossRef PubMed.
- M. Matano, S. Date, M. Shimokawa, A. Takano, M. Fujii, Y. Ohta, T. Watanabe, T. Kanai and T. Sato, Nat. Med., 2015, 21, 256–262 CrossRef CAS PubMed.
- G. Schwank, B. K. Koo, V. Sasselli, J. F. Dekkers, I. Heo, T. Demircan, N. Sasaki, S. Boymans, E. Cuppen, C. K. van der Ent, E. E. Nieuwenhuis, J. M. Beekman and H. Clevers, Cell Stem Cell, 2013, 13, 653–658 CrossRef CAS PubMed.
- J. A. Brassard, M. Nikolaev, T. Hubscher, M. Hofer and M. P. Lutolf, Nat. Mater., 2021, 20, 22–29 CrossRef CAS PubMed.
- M. Van De Wetering, H. E. Francies, J. M. Francis, G. Bounova, F. Iorio, A. Pronk, W. Van Houdt, J. Van Gorp, A. Taylor-Weiner, L. Kester, A. McLaren-Douglas, J. Blokker, S. Jaksani, S. Bartfeld, R. Volckman, P. Van Sluis, V. S. W. Li, S. Seepo, C. Sekhar Pedamallu, K. Cibulskis, S. L. Carter, A. McKenna, M. S. Lawrence, L. Lichtenstein, C. Stewart, J. Koster, R. Versteeg, A. Van Oudenaarden, J. Saez-Rodriguez, R. G. J. Vries, G. Getz, L. Wessels, M. R. Stratton, U. McDermott, M. Meyerson, M. J. Garnett and H. Clevers, Cell, 2015, 161, 933–945 CrossRef CAS PubMed.
- D. E. Ingber, Nat. Rev. Genet., 2022, 23, 467–491 CrossRef CAS PubMed.
- A. Bein, W. Shin, S. Jalili-Firoozinezhad, M. H. Park, A. Sontheimer-Phelps, A. Tovaglieri, A. Chalkiadaki, H. J. Kim and D. E. Ingber, Cell. Mol. Gastroenterol. Hepatol., 2018, 5, 659–668 CrossRef PubMed.
- J. R. Pappenheimer and C. C. Michel, J. Physiol., 2003, 553, 561–574 CrossRef CAS PubMed.
- W. W. Agace, Trends Immunol., 2008, 29, 514–522 CrossRef CAS PubMed.
- W. Shin and H. J. Kim, Nat. Protoc., 2022, 17, 910–939 CrossRef CAS PubMed.
- M. Nikolaev, O. Mitrofanova, N. Broguiere, S. Geraldo, D. Dutta, Y. Tabata, B. Elci, N. Brandenberg, I. Kolotuev, N. Gjorevski, H. Clevers and M. P. Lutolf, Nature, 2020, 585, 574–578 CrossRef PubMed.
- B. Sidar, B. R. Jenkins, S. Huang, J. R. Spence, S. T. Walk and J. N. Wilking, Lab Chip, 2019, 19, 3552–3562 RSC.
- D. Huh, G. A. Hamilton and D. E. Ingber, Trends Cell Biol., 2011, 21, 745–754 CrossRef CAS PubMed.
- H. E. Abaci and M. L. Shuler, Integr. Biol., 2015, 7, 383–391 CrossRef PubMed.
- C. Zhang, Z. Zhao, N. A. Abdul Rahim, D. van Noort and H. Yu, Lab Chip, 2009, 9, 3185–3192 RSC.
- J. H. Sung, Y. I. Wang, N. Narasimhan Sriram, M. Jackson, C. Long, J. J. Hickman and M. L. Shuler, Anal. Chem., 2019, 91, 330–351 CrossRef CAS PubMed.
- G. M. Whitesides, Nature, 2006, 442, 368–373 CrossRef CAS PubMed.
- D. J. Harrison, A. Manz, Z. Fan, H. Luedi and H. M. Widmer, Anal. Chem., 1992, 64, 1926–1932 CrossRef CAS.
- R. Xie, Z. Liang, Y. Ai, W. Zheng, J. Xiong, P. Xu, Y. Liu, M. Ding, J. Gao, J. Wang and Q. Liang, Nat. Protoc., 2021, 16, 937–964 CrossRef CAS PubMed.
- Z. Su, W. Hu, L. Ye, D. Gao and J.-M. Lin, Chin. Chem. Lett., 2022 DOI:10.1016/j.cclet.2022.107790.
- R. Xie, P. Xu, Y. Liu, L. Li, G. Luo, M. Ding and Q. Liang, Adv. Mater., 2018, 30, 1705082 CrossRef PubMed.
- Y. Liu, P. Xu, Z. Liang, R. Xie, M. Ding, H. Liu and Q. Liang, RSC Adv., 2018, 8, 23475–23480 RSC.
- P. Xu, R. Xie, Y. Liu, G. Luo, M. Ding and Q. Liang, Adv. Mater., 2017, 29, 1701664 CrossRef PubMed.
- B. Zhang, M. Montgomery, M. D. Chamberlain, S. Ogawa, A. Korolj, A. Pahnke, L. A. Wells, S. Masse, J. Kim, L. Reis, A. Momen, S. S. Nunes, A. R. Wheeler, K. Nanthakumar, G. Keller, M. V. Sefton and M. Radisic, Nat. Mater., 2016, 15, 669–678 CrossRef CAS PubMed.
- L. Bai, Y. Fu, M. Yao and Y. Cheng, Chem. Eng. J., 2018, 332, 537–547 CrossRef CAS.
- Y. Ai, R. Xie, J. Xiong and Q. Liang, Small, 2020, 16, 1903940 CrossRef CAS PubMed.
- L. A. Low, C. Mummery, B. R. Berridge, C. P. Austin and D. A. Tagle, Nat. Rev. Drug Discovery, 2021, 20, 345–361 CrossRef CAS PubMed.
- K. Duval, H. Grover, L. H. Han, Y. Mou, A. F. Pegoraro, J. Fredberg and Z. Chen, Physiology, 2017, 32, 266–277 CrossRef CAS PubMed.
- D. Huh, B. D. Matthews, A. Mammoto, M. Montoya-Zavala, H. Y. Hsin and D. E. Ingber, Science, 2010, 328, 1662–1668 CrossRef CAS PubMed.
- L. D. Ma, Y. T. Wang, J. R. Wang, J. L. Wu, X. S. Meng, P. Hu, X. Mu, Q. L. Liang and G. A. Luo, Lab Chip, 2018, 18, 2547–2562 RSC.
- R. Xie, A. Korolj, C. Liu, X. Song, R. X. Z. Lu, B. Zhang, A. Ramachandran, Q. Liang and M. Radisic, ACS Cent. Sci., 2020, 6, 903–912 CrossRef CAS PubMed.
- K. Ronaldson-Bouchard, D. Teles, K. Yeager, D. N. Tavakol, Y. Zhao, A. Chramiec, S. Tagore, M. Summers, S. Stylianos, M. Tamargo, B. M. Lee, S. P. Halligan, E. H. Abaci, Z. Guo, J. Jackow, A. Pappalardo, J. Shih, R. K. Soni, S. Sonar, C. German, A. M. Christiano, A. Califano, K. K. Hirschi, C. S. Chen, A. Przekwas and G. Vunjak-Novakovic, Nat. Biomed. Eng., 2022, 6, 351–371 CrossRef CAS PubMed.
- M. L. Shuler, Lab Chip, 2020, 20, 3072–3073 RSC.
- D. Dutta, I. Heo and H. Clevers, Trends Mol. Med., 2017, 23, 393–410 CrossRef CAS PubMed.
- I. A. Williamson, J. W. Arnold, L. A. Samsa, L. Gaynor, M. DiSalvo, J. L. Cocchiaro, I. Carroll, M. A. Azcarate-Peril, J. F. Rawls, N. L. Allbritton and S. T. Magness, Cell. Mol. Gastroenterol. Hepatol., 2018, 6, 301–319 CrossRef PubMed.
- N. Sasaki, K. Miyamoto, K. M. Maslowski, H. Ohno, T. Kanai and T. Sato, Gastroenterology, 2020, 159, 388–390 e385 CrossRef CAS PubMed.
- A. Skardal, T. Shupe and A. Atala, Drug Discovery Today, 2016, 21, 1399–1411 CrossRef CAS PubMed.
- Y. S. Zhang, J. Aleman, S. R. Shin, T. Kilic, D. Kim, S. A. Mousavi Shaegh, S. Massa, R. Riahi, S. Chae, N. Hu, H. Avci, W. Zhang, A. Silvestri, A. Sanati Nezhad, A. Manbohi, F. De Ferrari, A. Polini, G. Calzone, N. Shaikh, P. Alerasool, E. Budina, J. Kang, N. Bhise, J. Ribas, A. Pourmand, A. Skardal, T. Shupe, C. E. Bishop, M. R. Dokmeci, A. Atala and A. Khademhosseini, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E2293–E2302 CAS.
- A. Skardal, S. V. Murphy, M. Devarasetty, I. Mead, H. W. Kang, Y. J. Seol, Y. Shrike Zhang, S. R. Shin, L. Zhao, J. Aleman, A. R. Hall, T. D. Shupe, A. Kleensang, M. R. Dokmeci, S. Jin Lee, J. D. Jackson, J. J. Yoo, T. Hartung, A. Khademhosseini, S. Soker, C. E. Bishop and A. Atala, Sci. Rep., 2017, 7, 8837 CrossRef PubMed.
- H. J. Kim, D. Huh, G. Hamilton and D. E. Ingber, Lab Chip, 2012, 12, 2165–2174 RSC.
- H. J. Kim, H. Li, J. J. Collins and D. E. Ingber, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E7–E15 CAS.
- P. Shah, J. V. Fritz, E. Glaab, M. S. Desai, K. Greenhalgh, A. Frachet, M. Niegowska, M. Estes, C. Jager, C. Seguin-Devaux, F. Zenhausern and P. Wilmes, Nat. Commun., 2016, 7, 11535 CrossRef CAS PubMed.
- Y. Wang, Z. Shao, W. Zheng, Y. Xie, G. Luo, M. Ding and Q. Liang, Biofabrication, 2019, 11, 045001 CrossRef PubMed.
- M. Trapecar, E. Wogram, D. Svoboda, C. Communal, A. Omer, T. Lungjangwa, P. Sphabmixay, J. Velazquez, K. Schneider, C. W. Wright, S. Mildrum, A. Hendricks, S. Levine, J. Muffat, M. J. Lee, D. A. Lauffenburger, D. Trumper, R. Jaenisch and L. G. Griffith, Sci. Adv., 2021, 7, eabd1707 CrossRef CAS PubMed.
- H. Clevers, Cell, 2013, 154, 274–284 CrossRef CAS PubMed.
- A. Sontheimer-Phelps, D. B. Chou, A. Tovaglieri, T. C. Ferrante, T. Duckworth, C. Fadel, V. Frismantas, A. D. Sutherland, S. Jalili-Firoozinezhad, M. Kasendra, E. Stas, J. C. Weaver, C. A. Richmond, O. Levy, R. Prantil-Baun, D. T. Breault and D. E. Ingber, Cell. Mol. Gastroenterol. Hepatol., 2020, 9, 507–526 CrossRef PubMed.
- P. Paone and P. D. Cani, Gut, 2020, 69, 2232–2243 CrossRef CAS PubMed.
- C. P. Gayer and M. D. Basson, Cell Signal., 2009, 21, 1237–1244 CrossRef CAS PubMed.
- C. L. Maynard, C. O. Elson, R. D. Hatton and C. T. Weaver, Nature, 2012, 489, 231–241 CrossRef CAS PubMed.
- T. Nakajima, K. Sasaki, A. Yamamori, K. Sakurai, K. Miyata, T. Watanabe and Y. T. Matsunaga, Biomater. Sci., 2020, 8, 5615–5627 RSC.
- Y. Wang, D. B. Gunasekara, M. I. Reed, M. DiSalvo, S. J. Bultman, C. E. Sims, S. T. Magness and N. L. Allbritton, Biomaterials, 2017, 128, 44–55 CrossRef CAS PubMed.
- C. M. Costello, M. B. Phillipsen, L. M. Hartmanis, M. A. Kwasnica, V. Chen, D. Hackam, M. W. Chang, W. E. Bentley and J. C. March, Sci. Rep., 2017, 7, 12515 CrossRef PubMed.
- M. Verhulsel, A. Simon, M. Bernheim-Dennery, V. R. Gannavarapu, L. Geremie, D. Ferraro, D. Krndija, L. Talini, J. L. Viovy, D. M. Vignjevic and S. Descroix, Lab Chip, 2021, 21, 365–377 RSC.
- H. B. Chong, J. Youn, W. Shin, H. J. Kim and D. S. Kim, Lab Chip, 2021, 21, 3316–3327 RSC.
- K. Y. Shim, D. Lee, J. Han, N. T. Nguyen, S. Park and J. H. Sung, Biomed. Microdevices, 2017, 19, 37 CrossRef PubMed.
- A. Choe, S. K. Ha, I. Choi, N. Choi and J. H. Sung, Biomed. Microdevices, 2017, 19, 4 CrossRef PubMed.
- H. J. Kim and D. E. Ingber, Integr. Biol., 2013, 5, 1130–1140 CrossRef CAS PubMed.
- S. J. Trietsch, E. Naumovska, D. Kurek, M. C. Setyawati, M. K. Vormann, K. J. Wilschut, H. L. Lanz, A. Nicolas, C. P. Ng, J. Joore, S. Kustermann, A. Roth, T. Hankemeier, A. Moisan and P. Vulto, Nat. Commun., 2017, 8, 262 CrossRef PubMed.
- H. J. Kim, J. Q. Boedicker, J. W. Choi and R. F. Ismagilov, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 18188–18193 CrossRef CAS PubMed.
- B. Jing, Z. A. Wang, C. Zhang, Q. Deng, J. Wei, Y. Luo, X. Zhang, J. Li and Y. Du, Front. Bioeng. Biotechnol., 2020, 8, 272 CrossRef PubMed.
- G. S. Park, M. H. Park, W. Shin, C. Zhao, S. Sheikh, S. J. Oh and H. J. Kim, Stem Cell Rev. Rep., 2017, 13, 321–334 CrossRef CAS PubMed.
- Y. Sambuy, I. De Angelis, G. Ranaldi, M. L. Scarino, A. Stammati and F. Zucco, Cell Biol. Toxicol., 2005, 21, 1–26 CrossRef CAS PubMed.
- M. T. Nelson, M. R. Charbonneau, H. G. Coia, M. J. Castillo, C. Holt, E. S. Greenwood, P. J. Robinson, E. A. Merrill, D. Lubkowicz and C. A. Mauzy, Nat. Commun., 2021, 12, 2805 CrossRef CAS PubMed.
- A. Béduneau, C. Tempesta, S. Fimbel, Y. Pellequer, V. Jannin, F. Demarne and A. Lamprecht, Eur. J. Pharm. Biopharm., 2014, 87, 290–298 CrossRef PubMed.
- F. Araújo and B. Sarmento, Int. J. Pharm., 2013, 458, 128–134 CrossRef PubMed.
- Y. Guo, R. Luo, Y. Wang, P. Deng, T. Song, M. Zhang, P. Wang, X. Zhang, K. Cui, T. Tao, Z. Li, W. Chen, Y. Zheng and J. Qin, Sci. Bull., 2021, 66, 783–793 CrossRef CAS PubMed.
- M. H. Wu, S. B. Huang, Z. Cui, Z. Cui and G. B. Lee, Biomed. Microdevices, 2008, 10, 309–319 CrossRef PubMed.
- I. Maschmeyer, A. K. Lorenz, K. Schimek, T. Hasenberg, A. P. Ramme, J. Hubner, M. Lindner, C. Drewell, S. Bauer, A. Thomas, N. S. Sambo, F. Sonntag, R. Lauster and U. Marx, Lab Chip, 2015, 15, 2688–2699 RSC.
- L. Zheng, C. J. Kelly and S. P. Colgan, Am. J. Physiol., 2015, 309, C350–C360 CrossRef CAS PubMed.
- V. Palacio-Castaneda, N. Velthuijs, S. Le Gac and W. P. R. Verdurmen, Lab Chip, 2022, 22, 1068–1092 RSC.
- L. Li, Y. Li, Z. Shao, G. Luo, M. Ding and Q. Liang, Anal. Chem., 2018, 90, 11899–11907 CrossRef CAS PubMed.
- H. Lee, W. Shin, H. J. Kim and J. Kim, Anal. Chem., 2021, 93, 16123–16132 CrossRef CAS PubMed.
- M. A. U. Khalid, K. H. Kim, A. R. Chethikkattuveli Salih, K. Hyun, S. H. Park, B. Kang, A. M. Soomro, M. Ali, Y. Jun, D. Huh, H. Cho and K. H. Choi, Lab Chip, 2022, 22, 1764–1778 RSC.
- C. J. Mandrycky, C. C. Howard, S. G. Rayner, Y. J. Shin and Y. Zheng, J. Mol. Cell. Cardiol., 2021, 159, 1–13 CrossRef CAS PubMed.
- R. Xie, W. Zheng, L. Guan, Y. Ai and Q. Liang, Small, 2020, 16, 1902838 CrossRef CAS PubMed.
- A. Cochrane, H. J. Albers, R. Passier, C. L. Mummery, A. van den Berg, V. V. Orlova and A. D. van der Meer, Adv. Drug Delivery Rev., 2019, 140, 68–77 CrossRef CAS PubMed.
- W. Shin and H. J. Kim, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E10539–E10547 CAS.
- J. Morini, G. Babini, S. Barbieri, G. Baiocco and A. Ottolenghi, Front. Immunol., 2017, 8, 223 Search PubMed.
- M. A. Lancaster and J. A. Knoblich, Science, 2014, 345, 1247125 CrossRef PubMed.
- T. E. Wallach and J. R. Bayrer, J. Pediatr. Gastroenterol. Nutr., 2017, 64, 180–185 CrossRef PubMed.
- J. Zhou, C. Li, X. Liu, M. C. Chiu, X. Zhao, D. Wang, Y. Wei, A. Lee, A. J. Zhang, H. Chu, J. P. Cai, C. C. Yip, I. H. Chan, K. K. Wong, O. T. Tsang, K. H. Chan, J. F. Chan, K. K. To, H. Chen and K. Y. Yuen, Nat. Med., 2020, 26, 1077–1083 CrossRef CAS PubMed.
- M. M. Lamers, J. Beumer, J. v. d. Vaart, K. Knoops, J. Puschhof, T. I. Breugem, R. B. G. Ravelli, J. P. v. Schayck, A. Z. Mykytyn, H. Q. Duimel, E. v. Donselaar, S. Riesebosch, H. J. H. Kuijpers, D. Schipper, W. J. v. d. Wetering, M. d. Graaf, M. Koopmans, E. Cuppen, P. J. Peters, B. L. Haagmans and H. Clevers, Science, 2020, 369, 50–54 CrossRef CAS PubMed.
- G. Noel, N. W. Baetz, J. F. Staab, M. Donowitz, O. Kovbasnjuk, M. F. Pasetti and N. C. Zachos, Sci. Rep., 2017, 7, 45270 CrossRef CAS PubMed.
- T. Sato, D. E. Stange, M. Ferrante, R. G. J. Vries, J. H. van Es, S. van den Brink, W. J. van Houdt, A. Pronk, J. van Gorp, P. D. Siersema and H. Clevers, Gastroenterology, 2011, 141, 1762–1772 CrossRef CAS PubMed.
- N. Barker, Nat. Rev. Mol. Cell Biol., 2014, 15, 19–33 CrossRef CAS PubMed.
- N. Gjorevski, M. Nikolaev, T. E. Brown, O. Mitrofanova, N. Brandenberg, F. W. DelRio, F. M. Yavitt, P. Liberali, K. S. Anseth and M. P. Lutolf, Science, 2022, 375, eaaw9021 CrossRef CAS PubMed.
- N. Gjorevski, N. Sachs, A. Manfrin, S. Giger, M. E. Bragina, P. Ordóñez-Morán, H. Clevers and M. P. Lutolf, Nature, 2016, 539, 560–564 CrossRef CAS PubMed.
- H. Clevers, Cell, 2016, 165, 1586–1597 CrossRef CAS PubMed.
- W. Zheng, R. Xie, X. Liang and Q. Liang, Small, 2022, 18, 2105867 CrossRef CAS PubMed.
- M. J. Workman, J. P. Gleeson, E. J. Troisi, H. Q. Estrada, S. J. Kerns, C. D. Hinojosa, G. A. Hamilton, S. R. Targan, C. N. Svendsen and R. J. Barrett, Cell. Mol. Gastroenterol. Hepatol., 2018, 5, 669–677 e662 CrossRef PubMed.
- B. J. Jin, S. Battula, N. Zachos, O. Kovbasnjuk, J. Fawlke-Abel, J. In, M. Donowitz and A. S. Verkman, Biomicrofluidics, 2014, 8, 024106 CrossRef PubMed.
- K. K. P. Lee, T. Matsu-Ura, A. E. Rosselot, T. R. Broda, J. M. Wells and C. I. Hong, Biomicrofluidics, 2021, 15, 014110 CrossRef CAS PubMed.
- A. Tovaglieri, A. Sontheimer-Phelps, A. Geirnaert, R. Prantil-Baun, D. M. Camacho, D. B. Chou, S. Jalili-Firoozinezhad, T. de Wouters, M. Kasendra, M. Super, M. J. Cartwright, C. A. Richmond, D. T. Breault, C. Lacroix and D. E. Ingber, Microbiome, 2019, 7, 43 CrossRef PubMed.
- G. C. Hansson, Curr. Opin. Microbiol., 2012, 15, 57–62 CrossRef CAS PubMed.
- G. Fang, H. Lu, R. Al-Nakashli, R. Chapman, Y. Zhang, L. A. Ju, G. Lin, M. H. Stenzel and D. Jin, Biofabrication, 2021, 14, 015006 CrossRef PubMed.
- S. R. Gill, M. Pop, R. T. DeBoy, P. B. Eckburg, P. J. Turnbaugh, B. S. Samuel, J. I. Gordon, D. A. Relman, C. M. Fraser-Liggett and K. E. Nelson, Science, 2006, 312, 1355–1359 CrossRef CAS PubMed.
- N. Kamada, S. U. Seo, G. Y. Chen and G. Nunez, Nat. Rev. Immunol., 2013, 13, 321–335 CrossRef CAS PubMed.
- J. M. Pickard, M. Y. Zeng, R. Caruso and G. Nunez, Immunol. Rev., 2017, 279, 70–89 CrossRef CAS PubMed.
- J. F. Burgueno and M. T. Abreu, Nat. Rev. Gastroenterol. Hepatol., 2020, 17, 263–278 CrossRef CAS PubMed.
- W. Hu, D. Gao, Z. Su, R. Qian, Y. Wang and Q. Liang, Chin. Chem. Lett., 2022, 33, 2096–2100 CrossRef CAS.
- B. A. Helmink, M. A. W. Khan, A. Hermann, V. Gopalakrishnan and J. A. Wargo, Nat. Med., 2019, 25, 377–388 CrossRef CAS PubMed.
- M. O. Din, T. Danino, A. Prindle, M. Skalak, J. Selimkhanov, K. Allen, E. Julio, E. Atolia, L. S. Tsimring, S. N. Bhatia and J. Hasty, Nature, 2016, 536, 81–85 CrossRef CAS PubMed.
- R. L. DiMarco, J. Su, K. S. Yan, R. Dewi, C. J. Kuo and S. C. Heilshorn, Integr. Biol., 2014, 6, 127–142 CrossRef CAS PubMed.
- Y. E. Bar-Ephraim, K. Kretzschmar and H. Clevers, Nat. Rev. Immunol., 2020, 20, 279–293 CrossRef CAS PubMed.
- M. Trapecar, C. Communal, J. Velazquez, C. A. Maass, Y. J. Huang, K. Schneider, C. W. Wright, V. Butty, G. Eng, O. Yilmaz, D. Trumper and L. G. Griffith, Cell Syst., 2020, 10, 223–239 e229 CrossRef CAS PubMed.
- Q. Ramadan, H. Jafarpoorchekab, C. Huang, P. Silacci, S. Carrara, G. Koklu, J. Ghaye, J. Ramsden, C. Ruffert, G. Vergeres and M. A. Gijs, Lab Chip, 2013, 13, 196–203 RSC.
- Z. Li, W. Su, Y. Zhu, T. Tao, D. Li, X. Peng and J. Qin, Biomicrofluidics, 2017, 11, 034114 CrossRef PubMed.
- J. Bienenstock, W. Kunze and P. Forsythe, Nutr. Rev., 2015, 73(Suppl 1), 28–31 CrossRef PubMed.
- C. R. Martin, V. Osadchiy, A. Kalani and E. A. Mayer, Cell. Mol. Gastroenterol. Hepatol., 2018, 6, 133–148 CrossRef PubMed.
- V. Rothhammer, D. M. Borucki, E. C. Tjon, M. C. Takenaka, C. C. Chao, A. Ardura-Fabregat, K. A. de Lima, C. Gutierrez-Vazquez, P. Hewson, O. Staszewski, M. Blain, L. Healy, T. Neziraj, M. Borio, M. Wheeler, L. L. Dragin, D. A. Laplaud, J. Antel, J. I. Alvarez, M. Prinz and F. J. Quintana, Nature, 2018, 557, 724–728 CrossRef CAS PubMed.
- M.-H. Kim, D. Kim and J. H. Sung, J. Ind. Eng. Chem., 2021, 101, 126–134 CrossRef CAS.
- M. H. Kim, D. van Noort, J. H. Sung and S. Park, Int. J. Mol. Sci., 2021, 22, 13513 CrossRef CAS PubMed.
- A. Tripathi, J. Debelius, D. A. Brenner, M. Karin, R. Loomba, B. Schnabl and R. Knight, Nat. Rev. Gastroenterol. Hepatol., 2018, 15, 397–411 CrossRef CAS PubMed.
- S. Y. Lee and J. H. Sung, Biotechnol. Bioeng., 2018, 115, 2817–2827 CrossRef CAS PubMed.
- X. Duan, L. Zheng, X. Zhang, B. Wang, M. Xiao, W. Zhao, S. Liu and G. Sui, ACS Sens., 2020, 5, 3483–3492 CrossRef CAS PubMed.
- W. L. K. Chen, C. Edington, E. Suter, J. Yu, J. J. Velazquez, J. G. Velazquez, M. Shockley, E. M. Large, R. Venkataramanan, D. J. Hughes, C. L. Stokes, D. L. Trumper, R. L. Carrier, M. Cirit, L. G. Griffith and D. A. Lauffenburger, Biotechnol. Bioeng., 2017, 114, 2648–2659 CrossRef CAS PubMed.
- A. Skardal, M. Devarasetty, S. Forsythe, A. Atala and S. Soker, Biotechnol. Bioeng., 2016, 113, 2020–2032 CrossRef CAS PubMed.
- I. Maschmeyer, T. Hasenberg, A. Jaenicke, M. Lindner, A. K. Lorenz, J. Zech, L. A. Garbe, F. Sonntag, P. Hayden, S. Ayehunie, R. Lauster, U. Marx and E. M. Materne, Eur. J. Pharm. Biopharm., 2015, 95, 77–87 CrossRef CAS PubMed.
- M. B. Esch, J. M. Prot, Y. I. Wang, P. Miller, J. R. Llamas-Vidales, B. A. Naughton, D. R. Applegate and M. L. Shuler, Lab Chip, 2015, 15, 2269–2277 RSC.
- M. B. Esch, H. Ueno, D. R. Applegate and M. L. Shuler, Lab Chip, 2016, 16, 2719–2729 RSC.
- M. Jie, S. Mao, H. Liu, Z. He, H. F. Li and J. M. Lin, Analyst, 2017, 142, 3629–3638 RSC.
- H. Koike, K. Iwasawa, R. Ouchi, M. Maezawa, K. Giesbrecht, N. Saiki, A. Ferguson, M. Kimura, W. L. Thompson, J. M. Wells, A. M. Zorn and T. Takebe, Nature, 2019, 574, 112–116 CrossRef CAS PubMed.
- G. Vergeres, B. Bogicevic, C. Buri, S. Carrara, M. Chollet, L. Corbino-Giunta, L. Egger, D. Gille, K. Kopf-Bolanz, K. Laederach, R. Portmann, Q. Ramadan, J. Ramsden, F. Schwander, P. Silacci, B. Walther and M. Gijs, Br. J. Nutr., 2012, 108, 762–768 CrossRef CAS PubMed.
- P. de Haan, M. A. Ianovska, K. Mathwig, G. A. A. van Lieshout, V. Triantis, H. Bouwmeester and E. Verpoorte, Lab Chip, 2019, 19, 1599–1609 RSC.
- E. Ritz, Blood Purif., 2011, 31, 70–76 CrossRef PubMed.
- T. Yang, E. M. Richards, C. J. Pepine and M. K. Raizada, Nat. Rev. Nephrol., 2018, 14, 442–456 CrossRef CAS PubMed.
- Y. K. Yeoh, T. Zuo, G. C. Lui, F. Zhang, Q. Liu, A. Y. Li, A. C. Chung, C. P. Cheung, E. Y. Tso, K. S. Fung, V. Chan, L. Ling, G. Joynt, D. S. Hui, K. M. Chow, S. S. S. Ng, T. C. Li, R. W. Ng, T. C. Yip, G. L. Wong, F. K. Chan, C. K. Wong, P. K. Chan and S. C. Ng, Gut, 2021, 70, 698–706 CrossRef CAS PubMed.
- H. Wang, J. S. Liu, S. H. Peng, X. Y. Deng, D. M. Zhu, S. Javidiparsijani, G. R. Wang, D. Q. Li, L. X. Li, Y. C. Wang and J. M. Luo, World J. Gastroenterol., 2013, 19, 6794–6804 CrossRef PubMed.
- A. T. Dang and B. J. Marsland, Mucosal Immunol., 2019, 12, 843–850 CrossRef CAS PubMed.
- M. Ahuja, D. M. Schwartz, M. Tandon, A. Son, M. Zeng, W. Swaim, M. Eckhaus, V. Hoffman, Y. Cui, B. Xiao, P. F. Worley and S. Muallem, Cell Metab., 2017, 25, 635–646 CrossRef CAS PubMed.
- B. K. Patel, K. H. Patel, M. Bhatia, S. G. Iyer, K. Madhavan and S. M. Moochhala, World J. Gastroenterol., 2021, 27, 5019–5036 CrossRef CAS PubMed.
- X. Wang, M. Cirit, J. S. Wishnok, L. G. Griffith and S. R. Tannenbaum, Anal. Chem., 2019, 91, 8667–8675 CrossRef CAS PubMed.
- L. J. Y. Ong, T. Ching, L. H. Chong, S. Arora, H. Li, M. Hashimoto, R. DasGupta, P. K. Yuen and Y. C. Toh, Lab Chip, 2019, 19, 2178–2191 RSC.
- A. Dance, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 6773–6775 CrossRef CAS PubMed.
- C. Oleaga, C. Bernabini, A. S. Smith, B. Srinivasan, M. Jackson, W. McLamb, V. Platt, R. Bridges, Y. Cai, N. Santhanam, B. Berry, S. Najjar, N. Akanda, X. Guo, C. Martin, G. Ekman, M. B. Esch, J. Langer, G. Ouedraogo, J. Cotovio, L. Breton, M. L. Shuler and J. J. Hickman, Sci. Rep., 2016, 6, 20030 CrossRef CAS PubMed.
- A. Ramme, L. Koenig, T. Hasenberg, C. Schwenk, C. Magauer, D. Faust, A. Lorenz, A.-C. Krebs, C. Drewell, K. Schirrmann, A. Vladetic, G. Lin, S. Pabinger, W. Neuhaus, F. Bois, R. Lauster, U. Marx and E. Dehne, Future Sci. OA, 2019, 5, FSO413 CrossRef CAS PubMed.
- R. Novak, M. Ingram, S. Marquez, D. Das, A. Delahanty, A. Herland, B. M. Maoz, S. S. F. Jeanty, M. R. Somayaji, M. Burt, E. Calamari, A. Chalkiadaki, A. Cho, Y. Choe, D. B. Chou, M. Cronce, S. Dauth, T. Divic, J. Fernandez-Alcon, T. Ferrante, J. Ferrier, E. A. FitzGerald, R. Fleming, S. Jalili-Firoozinezhad, T. Grevesse, J. A. Goss, T. Hamkins-Indik, O. Henry, C. Hinojosa, T. Huffstater, K. J. Jang, V. Kujala, L. Leng, R. Mannix, Y. Milton, J. Nawroth, B. A. Nestor, C. F. Ng, B. O'Connor, T. E. Park, H. Sanchez, J. Sliz, A. Sontheimer-Phelps, B. Swenor, G. Thompson, 2nd, G. J. Touloumes, Z. Tranchemontagne, N. Wen, M. Yadid, A. Bahinski, G. A. Hamilton, D. Levner, O. Levy, A. Przekwas, R. Prantil-Baun, K. K. Parker and D. E. Ingber, Nat. Biomed. Eng., 2020, 4, 407–420 CrossRef PubMed.
- Y. Wang, P. Wang and J. Qin, Acc. Chem. Res., 2021, 54, 3550–3562 CrossRef CAS PubMed.
- A. Bein, S. Kim, G. Goyal, W. Cao, C. Fadel, A. Naziripour, S. Sharma, B. Swenor, N. LoGrande, A. Nurani, V. N. Miao, A. W. Navia, C. G. K. Ziegler, J. O. Montanes, P. Prabhala, M. S. Kim, R. Prantil-Baun, M. Rodas, A. Jiang, L. O'Sullivan, G. Tillya, A. K. Shalek and D. E. Ingber, Front. Pharmacol., 2021, 12, 718484 CrossRef CAS PubMed.
- J. Kim, M. Hegde and A. Jayaraman, Lab Chip, 2010, 10, 43–50 RSC.
- F. Rivera-Chavez, C. A. Lopez and A. J. Baumler, Free Radical Biol. Med., 2017, 105, 93–101 CrossRef CAS PubMed.
- A. I. Wells and C. B. Coyne, Viruses, 2019, 11, 460 CrossRef CAS PubMed.
- R. Villenave, S. Q. Wales, T. Hamkins-Indik, E. Papafragkou, J. C. Weaver, T. C. Ferrante, A. Bahinski, C. A. Elkins, M. Kulka and D. E. Ingber, PLoS One, 2017, 12, e0169412 CrossRef PubMed.
- J. Kruger, R. Gross, C. Conzelmann, J. A. Muller, L. Koepke, K. M. J. Sparrer, T. Weil, D. Schutz, T. Seufferlein, T. F. E. Barth, S. Stenger, S. Heller, J. Munch and A. Kleger, Cell. Mol. Gastroenterol. Hepatol., 2021, 11, 935–948 CrossRef PubMed.
- Y. Imai, K. Kuba, S. Rao, Y. Huan, F. Guo, B. Guan, P. Yang, R. Sarao, T. Wada, H. Leong-Poi, M. A. Crackower, A. Fukamizu, C.-C. Hui, L. Hein, S. Uhlig, A. S. Slutsky, C. Jiang and J. M. Penninger, Nature, 2005, 436, 112–116 CrossRef CAS PubMed.
- K. Ettayebi, S. E. Crawford, K. Murakami, J. R. Broughman, U. Karandikar, V. R. Tenge, F. H. Neill, S. E. Blutt, X.-L. Zeng, L. Qu, B. Kou, A. R. Opekun, D. Burrin, D. Y. Graham, S. Ramani, R. L. Atmar and M. K. Estes, Science, 2016, 353, 1387–1393 CrossRef PubMed.
- A. Grassart, V. Malarde, S. Gobaa, A. Sartori-Rupp, J. Kerns, K. Karalis, B. Marteyn, P. Sansonetti and N. Sauvonnet, Cell Host Microbe, 2019, 26, 435–444 e434 CrossRef CAS PubMed.
- N. Gjorevski, B. Avignon, R. Gerard, L. Cabon, A. B. Roth, M. Bscheider and A. Moisan, Lab Chip, 2020, 20, 3365–3374 RSC.
- L. Gijzen, D. Marescotti, E. Raineri, A. Nicolas, H. L. Lanz, D. Guerrera, R. van Vught, J. Joore, P. Vulto, M. C. Peitsch, J. Hoeng, G. Lo Sasso and D. Kurek, SLAS Technol., 2020, 25, 585–597 CrossRef CAS PubMed.
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, Ca-Cancer J. Clin., 2021, 71, 209–249 CrossRef PubMed.
- D. Tuveson and H. Clevers, Science, 2019, 364, 952–955 CrossRef CAS PubMed.
- X. Luo, E. L. S. Fong, C. Zhu, Q. X. X. Lin, M. Xiong, A. Li, T. Li, T. Benoukraf, H. Yu and S. Liu, Acta Biomater., 2021, 132, 461–472 CrossRef CAS PubMed.
- A. Sobrino, D. T. Phan, R. Datta, X. Wang, S. J. Hachey, M. Romero-Lopez, E. Gratton, A. P. Lee, S. C. George and C. C. Hughes, Sci. Rep., 2016, 6, 31589 CrossRef CAS PubMed.
- S. J. Hachey, S. Movsesyan, Q. H. Nguyen, G. Burton-Sojo, A. Tankazyan, J. Wu, T. Hoang, D. Zhao, S. Wang, M. M. Hatch, E. Celaya, S. Gomez, G. T. Chen, R. T. Davis, K. Nee, N. Pervolarakis, D. A. Lawson, K. Kessenbrock, A. P. Lee, J. Lowengrub, M. L. Waterman and C. C. W. Hughes, Lab Chip, 2021, 21, 1333–1351 RSC.
- M. Fujii, M. Shimokawa, S. Date, A. Takano, M. Matano, K. Nanki, Y. Ohta, K. Toshimitsu, Y. Nakazato, K. Kawasaki, T. Uraoka, T. Watanabe, T. Kanai and T. Sato, Cell Stem Cell, 2016, 18, 827–838 CrossRef CAS PubMed.
- M. E. V. Johansson, H. Sjövall and G. C. Hansson, Nat. Rev. Gastroenterol. Hepatol., 2013, 10, 352–361 CrossRef CAS PubMed.
- J. F. Dekkers, C. L. Wiegerinck, H. R. de Jonge, I. Bronsveld, H. M. Janssens, K. M. de Winter-de Groot, A. M. Brandsma, N. W. de Jong, M. J. Bijvelds, B. J. Scholte, E. E. Nieuwenhuis, S. van den Brink, H. Clevers, C. K. van der Ent, S. Middendorp and J. M. Beekman, Nat. Med., 2013, 19, 939–945 CrossRef CAS PubMed.
- D. Gao, H. Liu, J. M. Lin, Y. Wang and Y. Jiang, Lab Chip, 2013, 13, 978–985 RSC.
- K. Pocock, L. Delon, V. Bala, S. Rao, C. Priest, C. Prestidge and B. Thierry, ACS Biomater. Sci. Eng., 2017, 3, 951–959 CrossRef CAS PubMed.
- I. Lukonin, D. Serra, L. Challet Meylan, K. Volkmann, J. Baaten, R. Zhao, S. Meeusen, K. Colman, F. Maurer, M. B. Stadler, J. Jenkins and P. Liberali, Nature, 2020, 586, 275–280 CrossRef CAS PubMed.
- N. Brandenberg, S. Hoehnel, F. Kuttler, K. Homicsko, C. Ceroni, T. Ringel, N. Gjorevski, G. Schwank, G. Coukos, G. Turcatti and M. P. Lutolf, Nat. Biomed. Eng., 2020, 4, 863–874 CrossRef CAS PubMed.
- L. Li, W. Wang, M. Ding, G. Luo and Q. Liang, Anal. Chem., 2016, 88, 6734–6742 CrossRef CAS PubMed.
- O. Kwon, K. B. Jung, K.-R. Lee, Y. S. Son, H. Lee, J.-J. Kim, K. Kim, S. Lee, Y.-K. Song, J. Jung, K. Park, D.-S. Kim, M. J. Son, M.-O. Lee, T.-S. Han, H.-S. Cho, S. J. Oh, H. Chung, S.-H. Kim, K.-S. Chung, J. Kim, C.-R. Jung and M.-Y. Son, Sci. Adv., 2021, 7, eabh1586 CrossRef CAS PubMed.
- M. Kasendra, R. Luc, J. Yin, D. V. Manatakis, G. Kulkarni, C. Lucchesi, J. Sliz, A. Apostolou, L. Sunuwar, J. Obrigewitch, K. J. Jang, G. A. Hamilton, M. Donowitz and K. Karalis, eLife, 2020, 9, e50135 CrossRef CAS PubMed.
- A. Herland, B. M. Maoz, D. Das, M. R. Somayaji, R. Prantil-Baun, R. Novak, M. Cronce, T. Huffstater, S. S. F. Jeanty, M. Ingram, A. Chalkiadaki, D. Benson Chou, S. Marquez, A. Delahanty, S. Jalili-Firoozinezhad, Y. Milton, A. Sontheimer-Phelps, B. Swenor, O. Levy, K. K. Parker, A. Przekwas and D. E. Ingber, Nat. Biomed. Eng., 2020, 4, 421–436 CrossRef CAS PubMed.
- X. Jia, X. Yang, G. Luo and Q. Liang, J. Pharm. Biomed. Anal., 2022, 209, 114534 CrossRef CAS PubMed.
- Y. Ai, F. Zhang, C. Wang, R. Xie and Q. Liang, TrAC, Trends Anal. Chem., 2019, 117, 215–230 CrossRef CAS.
- S. Watanabe, S. Kobayashi, N. Ogasawara, R. Okamoto, T. Nakamura, M. Watanabe, K. B. Jensen and S. Yui, Nat. Protoc., 2022, 17, 649–671 CrossRef CAS PubMed.
- L. Meran, I. Massie, S. Campinoti, A. E. Weston, R. Gaifulina, L. Tullie, P. Faull, M. Orford, A. Kucharska, A. Baulies, L. Novellasdemunt, N. Angelis, E. Hirst, J. Konig, A. M. Tedeschi, A. F. Pellegata, S. Eli, A. P. Snijders, L. Collinson, N. Thapar, G. M. H. Thomas, S. Eaton, P. Bonfanti, P. De Coppi and V. S. W. Li, Nat. Med., 2020, 26, 1593–1601 CrossRef CAS PubMed.
- Y. C. Shin, W. Shin, D. Koh, A. Wu, Y. M. Ambrosini, S. Min, S. G. Eckhardt, R. Y. D. Fleming, S. Kim, S. Park, H. Koh, T. K. Yoo and H. J. Kim, Micromachines, 2020, 11, 663 CrossRef PubMed.
- M. Kasendra, A. Tovaglieri, A. Sontheimer-Phelps, S. Jalili-Firoozinezhad, A. Bein, A. Chalkiadaki, W. Scholl, C. Zhang, H. Rickner, C. A. Richmond, H. Li, D. T. Breault and D. E. Ingber, Sci. Rep., 2018, 8, 2871 CrossRef PubMed.
- K. M. Seiler, A. Bajinting, D. M. Alvarado, M. A. Traore, M. M. Binkley, W. H. Goo, W. E. Lanik, J. Ou, U. Ismail, M. Iticovici, C. R. King, K. L. VanDussen, E. A. Swietlicki, V. Gazit, J. Guo, C. J. Luke, T. Stappenbeck, M. A. Ciorba, S. C. George, J. M. Meacham, D. C. Rubin, M. Good and B. W. Warner, Sci. Rep., 2020, 10, 3842 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |