Open Access Article
Open Access ArticleNano-structured hydrotrope-caged cytochrome c with boosted stability in harsh environments: a molecular insight†
Pranav
Bharadwaj‡
a,
Dheeraj Kumar
Sarkar‡
 bc,
Meena
Bisht§
d,
Sachin M.
Shet
a,
Nataraj Sanna
Kotrappanavar
bc,
Meena
Bisht§
d,
Sachin M.
Shet
a,
Nataraj Sanna
Kotrappanavar
 a,
Veeresh
Lokesh
d,
Gregory
Franklin
a,
Veeresh
Lokesh
d,
Gregory
Franklin
 *d,
Jan
Brezovsky
*d,
Jan
Brezovsky
 *bc and
Dibyendu
Mondal
*bc and
Dibyendu
Mondal
 *ad
*ad
aCentre for Nano and Material Sciences, Jain (Deemed-to-be University), Jain Global Campus, Kanakapura, Bangalore, Karnataka 562112, India. E-mail: dmtapu@gmail.com; dmon@igr.poznan.pl
bLaboratory of Biomolecular Interactions and Transport, Department of Gene Expression, Institute of Molecular Biology and Biotechnology, Faculty of Biology, Adam Mickiewicz University, Uniwersytetu Poznanskiego 6, 61-614 Poznan, Poland. E-mail: janbre@amu.edu.pl; jbrezovsky@iimcb.gov.pl
cInternational Institute of Molecular and Cell Biology in Warsaw, Ks Trojdena 4, 02-109 Warsaw, Poland
dInstitute of Plant Genetics (IPG) of the Polish Academy of Sciences, Strzeszyńska 34, 60-479 Poznań, Poland. E-mail: fgre@igr.poznan.pl
First published on 11th August 2023
Abstract
Green and nano-structured catalytic media are vital for biocatalysis to attenuate the denaturation tendency of biocatalysts under severe reaction conditions. Hydrotropes with multi-faceted physiochemical properties represent promising systems for sustainable protein packaging. Herein, the ability of adenosine-5′-triphosphate (ATP) and cholinium salicylate ([Cho][Sal]) ionic liquid (IL) to form nano-structures and to nano-confine Cytochrome c (Cyt c) enhanced the stability and activity under multiple stresses. Experimental and computational analyses were undertaken to explain the nano-structured phenomenon of ATP and IL, structural organizations of nano-confined Cyt c, and site-specific interactions that stabilize the protein structure. Both ATP and IL form nano-structures in aqueous media and could cage Cyt c via multiple nonspecific soft interactions. Remarkably, the engineered molecular nano-cages of ATP (5–10 mM), IL (300 mg mL−1), and ATP + IL surrounding Cyt c resulted in 9-to-72-fold higher peroxidase activity than native Cyt c with exceptionally high thermal tolerance (110 °C). The polar interactions with the cardiolipin binding site of Cyt c, mediated by hydrotropes, were well correlated with the increased peroxidase activity. Furthermore, higher activity trends were observed in the presence of urea, GuHCl, and trypsin without any protein degradation. Specific binding of hydrotropes in highly mobile regions of Cyt c (Ω 40–54 residues) and enhanced H-bonding with Lys and Arg offered excellent stability under extreme conditions. Additionally, ATP effectively counteracted reactive oxygen species (ROS)-induced denaturation of Cyt c, which was enhanced by the [Sal] counterpart of IL. Overall, this study explored the robustness of nano-structured hydrotropes to have a higher potential for protein packaging with improved stability and activity under extreme conditions. Thus, the present work highlights a novel strategy for real-time industrial biocatalysis to protect mitochondrial cells from ROS-instigated apoptosis.
Introduction
Adenosine-5′-triphosphate (ATP) has shown multi-faceted functions, including its participation in the electron transport chain, acting as the energy currency of cells, holding the key for caspase-9 activated apoptosis, and taking part in transcription by revamping chromatin complexes.1–3 The role of physiological concentrations of ATP in preventing thermally induced protein aggregation in mammalian cells was previously demonstrated by Nguyen and Benusaude,4 and the answer regarding the regulation of a high concentration of ATP (2–10 mM) in the cell was unveiled by Patel and co-workers, whereby ATP is claimed as a biological hydrotrope.5 The term ‘hydrotrope’ was coined by Neuberg for amphiphilic molecules having a characteristic range of minimum hydrotrope concentration, which augment the solubility of partially soluble organic or hydrophobic substances in water.6,7 This opened a new channel to explore ATP in the medical field to treat Parkinson's and Alzheimer-like diseases by preventing phase separation of biological fluids, which causes amyloid fibrillation.8,9 The solubilizing tendency of ATP was attributed to its nonspecific aggregation, charge reinforcement, and self-assembly through hydrophobic interactions (Fig. 1a).10,11 Thus, the hydrotropic mechanism of ATP follows both the Neuberg and Hofmeister effects.12 Self-aggregated ATP is found to provide thermal stability to lysozyme, malate dehydrogenase, and ubiquitin,13 and so it is important to understand how the self-aggregation property of ATP is useful in protein packaging under multiple stressors, which has not been previously studied.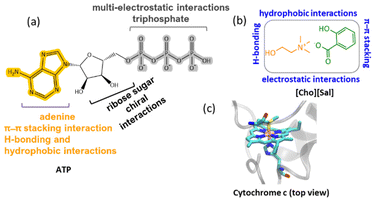 | ||
| Fig. 1 Chemical structures and structural features of (a) ATP and (b) [Cho][Sal] IL, with possible interaction sites. (c) The modeled active site structure of Cyt c, based on the crystallographic structure of bovine heart Cyt c (PDB code: 2B4Z) at 1.5 Å resolution. | ||
Unlike ATP, various amphiphilic nano-structured solvents have recently emerged with promising applications in energy-efficient bioprocesses.14 Due to their highly manipulative nature, ionic liquids (ILs) have gained interest for their ability to increase enzyme activity, solubility and thermal stability.15 Since the initial demonstration of ILs in biocatalysis, numerous research groups have focused on utilizing imidazolium, pyridium, tetraalkyl-ammonium, and tetraalkyl-phosphonium based ILs for catalysis involving various classes of enzymes.16,17 Further, with extended studies over ILs, bio-based ILs of the cholinium family have shown hydrotropic properties18 with greater hydrophobicity and ability to enhance the nano-structuring of amphiphilic solutes in aqueous media.19 Although earlier reports showed protein packaging with long-term stability and activity using cholinium ILs,15 but there is a lack of understanding of the underlying mechanisms by which hydrotropic ILs (potential amphiphilic molecules) can regulate protein stability and activity.20 In this regard, an IL with a salicylate counterpart caught our attention as it contains carboxylic and hydroxyl groups, along with a hydrophobic aromatic ring featuring the salicylate ion and showing multiple interaction sites in the aqueous medium (Fig. 1b).21,22 Additionally, salicylic acid is an important phytohormone that alters the mitochondrial processes by amplifying the concentration of reactive oxygen species (ROS) and induces cells to undergo programmed cell death.23,24 During apoptosis, ATP plays a crucial role in caspase-9 activation, a process stimulated by cytochrome c (Cyt c) release from mitochondria.25 Since both ATP and salicylate are coupled to Cyt c, the latter was chosen as a model protein for this study (Fig. 1c), and ATP and choline salicylate ([Cho][Sal]) IL as the nano-structured hydrotropes.
Cyt c is a mitochondrial-based 13 kDa monomeric protein comprising of approximately 104 residues and is widely distributed across eukaryotes, bacteria, and archaea.26 Despite its significant role in electron transfer, this protein is of great interest in many technological applications like biosensors, synthetic receptors, and molecular imprinting among others.26 Cyt c also shows peroxidase-like activity and serves as a bio-catalyst in several catalytic transformations.26 The protein is highly vulnerable to denaturation under prevailing stress conditions such as extreme temperatures, oxidative stress, and proteolytic and chemical denaturants.27 Recently, a surface modification strategy using quantum dots, metal–organic frame-works,28 and DNA was reported to enhance the stability of Cyt c.27,29 However, the stability of the protein when confined within a hydrotrope system has not yet been reported. Considering the hydrotropism and probability of nano-structuring,26 the present study aimed to cage Cyt c in ATP and [Cho][Sal] IL-based hydrotropes, and thereby improve the activity and stability of the protein in gentle and harsh environments (such as high temperature, chemical denaturants, protease digestion, and oxidative stress). Therefore, detailed experimental and in silico approaches were undertaken to study the structure–function relationship of nano-confined Cyt c in ATP, [Cho][Sal] and ATP + IL at a molecular level to develop a novel and sustainable solvent manipulation strategy for dynamic protein packaging and robust biocatalysis.
Results and discussion
ATP and [Cho][Sal] IL as nano-structured hydrotropes
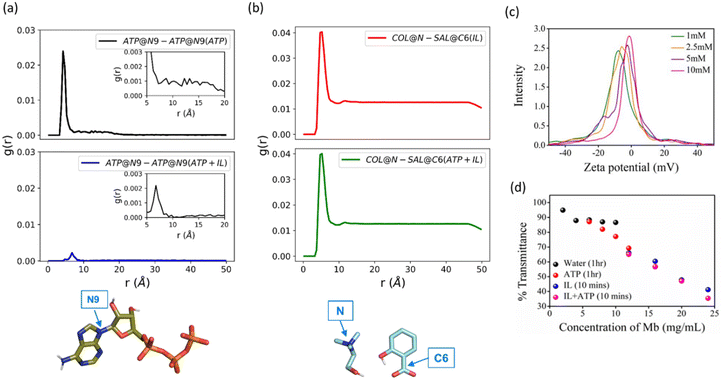
To understand the extent to which the IL affects the self-aggregation of ATP, which is vital for its role as a biological hydrotrope, we performed molecular dynamics (MD) simulations of 5 mM ATP, 300 mg mL−1 [Cho][Sal], and their mixture. The pair-wise interactions of ATP and [Cho][Sal] molecules in water and the mixture were investigated using radial distribution functions (RDF). As expected, ATP exhibited a self-aggregating tendency at 4-to-5 Å and up to 15 Å (Fig. 2a). The self-aggregating propensity of ATP molecules from a chemical perspective in the presence of an IL can occur in three possible ways, first the anion–π interactions between the aromatic ring and negatively charged oxygen moiety of phosphate groups, secondly through a non-bonding aromatic ring interaction (H-bonding, π–π stacking interactions, and cation–π interactions) and third, H-bond formation between the sugar and triphosphate group.11 It was evident from our analysis that two ATP molecules preferentially formed stack-like configurations in water (Fig. S1a†). In the presence of [Cho][Sal], the IL-mediated H-bonding (Fig. S1b†) markedly decreased the occurrence of the stack-like configurations of ATP (Fig. 2a). Notably, we found that the IL at a molecular concentration of 300 mg mL−1 forms dense H-bonding network mediated by cholinium and salicylate (Fig. S1c†). Unsurprisingly, we have not observed any perturbation of the [Cho][Sal] nano-structure upon the addition of ATP, given its low concentration (Fig. 2b). In addition to RDFs, the self-aggregating behavior of ATP and ATP + IL was further verified by a concentration-dependent ζ-potential study (Fig. 2c and Fig. S2†). At 1 mM concentration, which is below the critical hydrotropic concentration (2 mM), a single broad distribution peak was observed with maxima of −8.12 mV, indicating the existence of a single non-aggregated structure (Fig. 2c). At 2.5 mM, the distribution curve shifted to a lower negative value with peak maxima of −5.96 mV. This lowering in the ζ-potential indicates the beginning of aggregated structures similar to the self-aggregation of nano-particles as a result of the decrease in the electrophoretic mobility.30 This is further evidenced by the shift of maxima from −5.96 to −2.35 mV for 5 mM ATP. Finally, at 10 mM concentration, a broad single peak with maxima at −1.42 mV was obtained, which indicates the presence of a self-assembled, nano-structured ATP state. Conversely, 200 mg mL−1 of [Cho][Sal] shows a very broad Gaussian peak with a maximum at −9.12 mV (Fig. S3†), which shifts to a lower value of −4.02 mV in the presence of 5 mM ATP, justifying the hypothesis that the IL enhanced the self-assembly of amphiphiles.21 The hydrodynamic radius (Rh) of 5 mM ATP was found to be 0.83 nm (slightly higher than Rh of monomeric ATP, which is 0.57–0.65 nm),10 suggesting the existence of nano-structured/oligomeric forms and supporting our earlier evidence. In the presence of the IL, the Rh of ATP increased to 1.12 nm (Fig. S4†).To demonstrate the hydrotropic property of ATP and [Cho][Sal], solubility studies of myoglobin (Mb) were carried out in an aqueous medium at pH 7 in the presence and absence of 5 mM ATP, 100 mg mL−1 IL and a combination of both (Fig. 2d). After 1 h, the maximum solubility of Mb was found to be 6 mg mL−1. In contrast, ATP-induced solubility of Mb increased to 16 mg mL−1 during the same duration, which directly shows the hydrotropic nature of ATP. In the case of [Cho][Sal], the same solubility was achieved within 5 min. Thus, the concentration of Mb was further increased and examined until 24 mg mL−1. Interestingly, for ATP + IL, 24 mg mL−1 Mb showed higher solubility than the IL-only medium. These observations of enhanced solubility in the presence of two hydrotropes inspired us towards nano-confinement of Cyt c using ATP and [Cho][Sal]-based nano-structured hydrotropes to enable improved biological activity and stability of the protein.
Structural features of nano-structured hydrotrope-caged Cyt c
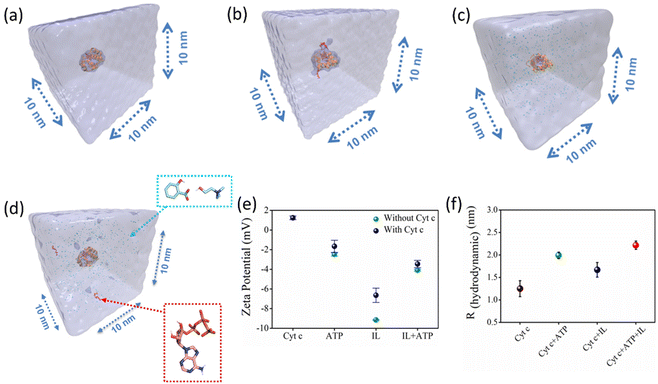
To understand the structural features of caged Cyt c, four molecular systems were prepared, namely keeping Cyt c in water, ATP, IL, and both ATP + IL at 26.85 °C (Fig. 3a–d). All molecular systems were solvated in a cubic box with a side length of 10 nm prepared using a PACKMOL package.31 The RDFs of hydrotropes in all systems with and without the presence of the protein were in agreement, indicating that the presence of Cyt c had a negligible effect on the adopted hydrotrope nano-structures (Fig. 2a, b & Fig. S5†). A lower dielectric constant of the protein or more negative zeta potential of ATP and IL can be related to the colloidal stability of caged Cyt C. Accordingly, ζ-potential analysis was employed to envisage the electrostatic interaction between Cyt c and nano-structured hydrotropes. We found the ζ-potential of native Cyt c solution (10 mM) to be +1.24 mV (Fig. 3e), and the ζ-potential of ATP (5 mM), IL (200 mg mL−1), and ATP + IL was −2.48 mV, −9.16 mV, and −4.09 mV, respectively. After Cyt c incubation with ATP, IL, and IL + ATP, the ζ-potential values decreased to −1.66 mV, −6.64 mV, and −3.46 mV, respectively (Fig. 3e and Fig. S6†). These changes in the ζ-potential demonstrate the possible electrostatic interaction between the protein and the nano-structured hydrotropes. Further Rh of native Cyt c was found to be 1.25 nm, which agrees with a previous report,32 and in the presence of ATP, the value increased to 1.99 nm showing self-assembly of ATP around the protein (Fig. 3f and Fig. S7†). In the presence of IL + ATP, Rh of Cyt c is further increased to 2.22 nm. Thus, the hydrotropic mechanism of ATP occurs initially by charge reinforcement, followed by the stacking phenomenon. Along with ζ-potential and DLS, structures of nano-confined Cyt c were further characterized by UV-vis spectra (Fig. S8†). In the presence of 5 mM ATP, a hyperchromic shift (a blue shift of 2 nm) in the Soret band (409 nm) was observed, suggesting that a non-polar environment dominates around the heme. In the case of IL (300 mg mL−1 [Cho][Sal]), absorbance in the Soret band enhanced considerably together with a blue shift, while the Q-band (520–550 nm) is broadened due to enhanced absorbance. This suggests changes in the tertiary conformations.33 Overall, it is evident that the self-aggregation propensities of the hydrotropes persisted even in the presence of Cyt c, and the structure of caged Cyt c was partially affected (without any unfolding, as discussed below) due to multiple interactions with nano-structured hydrotropes.Nano-structured hydrotrope-caged Cyt c with boosted peroxidase activity: molecular insights on the improved catalytic activity and enhanced stability
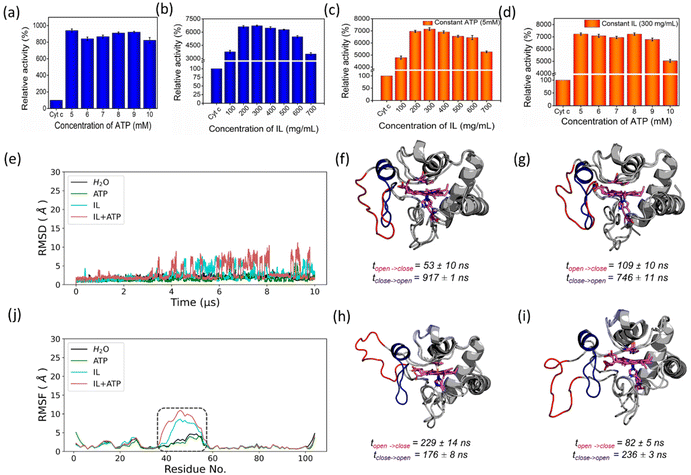
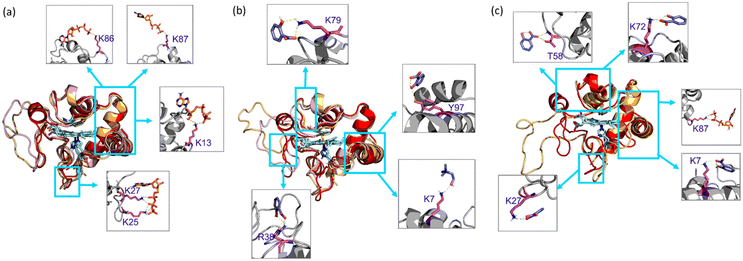
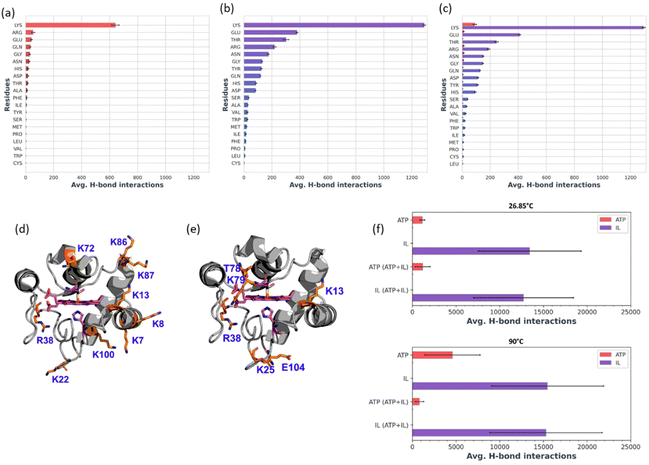
In the presence of H2O2, Cyt c undergoes covalent modifications, which facilitate peroxidase-like activity.34 The activity of Cyt c in the presence of 5–10 mM ATP showed a similar trend with an >8-fold higher activity than native Cyt c (Fig. 4a). Since a typical nano-structure is formed around Cyt c in the range of hydrotropic concentration (5–10 mM), similar relative activity is achieved rather than a maxima–minima trend. ATP acts as a hydrotrope initially by unwinding the protein and then stabilizing the extended chain through electrostatic interactions,11 which may be a reason for the high peroxidase activity. In the case of the [Cho][Sal] IL, nearly the same relative activities were seen from the 200-to-500 mg mL−1 range (Fig. 4b). A clear trend was observed with the highest relative activity of 67-fold at the concentration of the IL of 300 mg mL−1. The activity data agree with the UV-vis absorption spectra that inferred the transition of Cyt c towards penta-coordinated high spin complex. When the Met80 ligand is displaced, Cyt c naturally exhibits pronounced peroxidase-like activity.34 Interestingly, a replica of the IL optimization trend pattern was observed when using the ATP + IL mixture when [ATP] was kept constant (5 mM) and IL was varied (Fig. 4c). At 300 mg mL−1 IL and 5 mM ATP concentrations, the activity of Cyt c was enhanced 72-fold more than native Cyt c. This activity enhancement is indicating that the structure of Cyt c is still intact and combined hydrotropic systems boost the peroxidase activity further. Some ILs enhance the self-assembly of co-solutes in the media due to the virtue of their solvophobic effects.19 Accordingly, the concentration of ATP was varied with IL being constant (300 mg mL−1). A similar trend was observed with a >69-fold increase in activity (Fig. 4d). These trends demonstrate that both IL and ATP further enhance the conformational changes of Cyt c, followed by stabilization of the extended protein structure by the self-assembling of ATP to surround it. Similar inference can be observed from Fig. S5a (blue line),† in which ATP tends to self-aggregate, even in the presence of IL and Cyt c. To understand the behavior of nano-structured hydrotrope-caged Cyt c, adaptive sampling MD simulations with a total of 10 μs simulation time for each system were performed. In all systems, we observed reversible structural rearrangements of the protein backbone (Fig. 4e), which were more pronounced in mixtures containing IL and ATP + IL. Next, we traced these changes to the reversible opening of the loop in the Ω 40–54 residue region that was relatively more mobile than other parts of the protein for all four systems (Fig. 4j). This region corresponds to the mAb 1D3 binding site and is reported to be more dynamic,35 while having specificity towards Cytochrome c oxidase.36 To delineate the details of this process and its modulation by the action of nano-structured hydrotropes, we have constructed Bayesian Markov state models from the simulations. This inference revealed the presence of metastable states with an open conformation of the Ω 40–54 loop of Cyt c in all systems (Fig. 4f–i), which were characterized by their increased RMSD and elevated radius of gyration (Fig. S9†). The interactions of ATP and IL with Cyt c resulted in marked effects on the stability and dynamics of this region. In the presence of ATP, the closing process was approximately 2 times slower, whereas the opening became approximately 20% faster (Fig. 4g). Similarly, the presence of the IL resulted in a much faster opening process and the closing speed was slowed even more prominently than that observed for ATP (Fig. 4h). Finally, the dynamics of Cyt c in the mixture of ATP and IL exhibited ATP-like closing and IL-like opening times, enhancing the dynamics of this functionally important region (Fig. 4i). To understand if the hydrotropes molecules interact directly with the heme group of Cyt c, a structural analysis of was carried out by computational study. The simulations show infrequent interactions of hydrotropes with the heme group (Fig. S10†). Such interactions are unlikely to contribute to markedly improved activity observed with all three systems (ATP, IL, and ATP + IL), in particular, considering their absence with ATP. Since both ATP and IL were significantly affecting the dynamics of Cyt c, we identified the residues exhibiting the most frequent H-bonding with hydrotropes in the simulations (Fig. 5). In general, ATP interacted preferentially with Lys residues (Fig. S11a and S12a†), which is in accordance with an earlier report demonstrating that triphosphate group of ATP can deliver weak, nonspecific interactions with conserved Lys or Arg residues.10 Additionally, the IL had a wider range of residues as relevant binding partners, although Lys was the most populated in terms of H-bonding (Fig. S11b and S12b†). This trend was also maintained in the mixture of ATP and IL (Fig. S11c and S12c, d†). We observed marked overlap in particular interaction hot spots for IL and ATP on the surface of Cyt c (Fig. S11d and e†). ATP most often formed interactions with K13, K25, K27, K72, K86, and K87 (Fig. 5a and S11d†). Notably, K72 is involved in site A of Cyt c, which is crucial for interacting with cardiolipin,37 and was reported to have a significant role in the peroxidase activity of Cyt c.38 Also, IL demonstrated general preference to Lys, with somewhat higher affinity towards K7, K79, R38 (Fig. 5b and S11e†). Interestingly we also found considerable affinity towards Y97, which has been ascribed a significant role in keeping the terminal helix intact and hence providing structural stability to the protein.39Nano-structured hydrotrope-caged Cyt c with enhanced thermal stability for high-temperature bio-catalysis
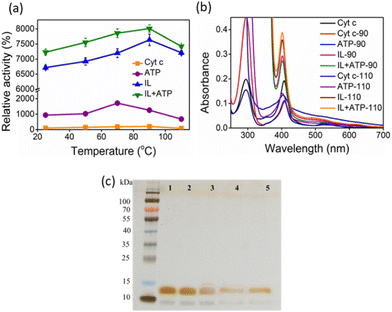
The activity of Cyt c increases with temperature up to 70 °C, above which the protein starts to denature gradually (Fig. 6a). Above 80 °C, the activity drops due to significant denaturation of the protein.27 A similar trend is observed in the presence of ATP, but the activity extends to approximately a 20-fold increase than for native Cyt c at 70 °C. In the case of IL and ATP + IL systems, the denaturation is prevented even up to extremes of 110 °C. At 90 °C, both Cyt c + IL and Cyt c + IL + ATP showed an accelerated activity value corresponding to 76–80-fold higher than native Cyt c. The stability aspects of Cyt c, when coupled to hydrotropes at elevated temperatures, were supported by UV-vis spectra (Fig. 6b). Enhancement in the Soret band intensity was observed for both IL and IL + ATP at 90 °C compared to the results obtained at 26.85 °C, which implies that the secondary conformation, formed due to weak interactions between the hydrotrope systems and Cyt c, was preserved. ATP + IL showed an intensified Soret peak compared to the other conditions at 110 °C, suggesting the advantage of engineering Cyt c with two different hydrotropes when aiming for high-temperature bio-catalysis. Additionally, as evident from non-reducing SDS-PAGE (Fig. 6c), improved activity trends at high temperatures were achieved without structural degradation of the protein.A systematic computational study was undertaken at 90 °C to gain structural insights into the higher stability of nano-structured-caged Cyt c at elevated temperatures. As expected, the Cyt c conformational ensemble was significantly altered at the higher temperature, with backbone RMSD being approximately 3–4 times higher than that at 26.85 °C for all four solvents (Fig. S13a†). No significant effects were observed between the mobility of the Ω 40–54 residue region in the four systems than that observed for 26.85 °C (Fig. 4e and S13b†). However, the thermal stability of the Ω 70–85 residues region was significantly enhanced with nano-structured hydrotropes compared to water (Fig. S13b†).
From Bayesian Markov state models constructed based on the high-temperature simulations (Fig. S13c–f†), we could observe that Cyt c in water exhibited severe conformational instability, underlined by a very high RMSD of the open state, as well as the presence of multiple secondary open-like states (Fig. S14†). In contrast, the presence of ATP or/and IL approximately halved the maximal range of displacements of the Cyt c backbone, providing heightened Cyt c resistance. Hence, it is evident that ATP and IL could thermally stabilize the functional component of Cyt c via multiple polar interactions.26 Affinity of hydrotrope molecules towards specific amino acids at the higher temperature (Fig. 7a–c and Fig. S15†) was found to be similar to that observed at 26.85 °C (Fig. S11 & S12†). Overall, Lys and Arg were found to have frequent H-bonding at 90 °C to ATP (Fig. 7a, c and d). Similarly to the lower temperature environment, the IL had a broader range of specificity, and more affinity with Lys, Glu and Thr in terms of H-bonding (Fig. 7b, c and e). The highest affinity was seen towards K7, K8, K13, K22, R38, K72, K86, K87 and K100 with ATP (Fig. 7d), and K13, K25, R38, T78, K79, and E104 with IL (Fig. 7e), all of which are crucial in maintaining thermal stability.40 It is important to highlight that the synergistic effects of ATP and IL were observed for the peroxidase activity of Cyt C (Fig. 4a–d) but not for stabilization, where IL and the mixture of IL with ATP grant similar enhancements to the Cyt C resistance towards higher temperature (Fig. 6a). Indeed, molecules of IL form numerous H-bonds with the protein at 26.85 °C as well as at 90 °C indicating their key role in the observed stabilisation (Fig. 7f). Moreover, the overall number of formed H-bonds is similar in IL and ATP + IL systems corresponding well with the observed enhancements in the Cyt C stability in the presence of IL as well as its mixture with ATP.
Improved stability of nano-structured hydrotrope-caged Cyt c when exposed to chemical denaturants
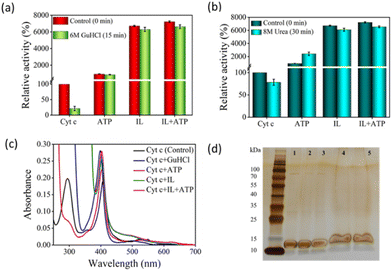
Above a particular concentration and on more prolonged exposure to some chaotropic chemicals like H2O2, guanidine hydrochloride (GuHCl), and urea cause amide bond disruption or overexpose the metallic core of the enzyme, causing a loss of structural integrity and function. To investigate the stability of Cyt c against chemical denaturants, the protein was incubated with the three nano-structured hydrotropic systems for 10 min, followed by the addition of a chemical denaturant. After the interaction with 6 M GuHCl for 15 min, there was a complete disruption in the heme pocket of native Cyt c and the activity severely declined, with the remaining activity approximately ∼20%. Conversely, hydrotrope-caged Cyt c maintained initial activity of 8.5-fold, 63-fold, and 66-fold in the presence of ATP, IL, and ATP + IL, respectively (Fig. 8a). While urea is known to be a hydrotrope, it is well recognized as a chemical denaturant at concentrations above 6 M. In the pre-denaturation concentrations of urea (1–6 M), a ∼30 fold increase in peroxidase-like activity was obtained for Cyt c.41 After 30 min incubation with 8 M urea, its activity was recorded at approximately 70% of the initial activity, while IL and ATP + IL hydrotrope-stabilized Cyt c showed similar activity (Fig. 8b).Native Cyt c showed a denaturing pattern in the UV-vis spectra with GuHCl (Fig. 8c), where the Q band structure was entirely lost and the Soret band was severely blue-shifted. In the case of ATP, changes in the 280 nm shoulder peak suggest partial dissociation of aromatic residues like Tyr and Trp. In contrast, the structures remained almost the same in the cases of IL and ATP + IL, except that the absorbance of the IL-based system trended lower. In the case of ATP, the activity of Cyt c almost doubled (∼24-fold higher than native Cyt c) in the presence of urea compared to the Cyt c + ATP system (Fig. 7b). This strange observation suggested that ATP cannot prevent further unfolding of Cyt c in the presence of urea, a behavior confirmed by the UV-vis spectra (Fig. S16†). Furthermore, SDS-PAGE analysis demonstrated that Cyt c did not degrade upon treatment with denaturing agents in the presence or absence of ATP and [Cho][Sal] (Fig. 8d), thus, confirming that the variation in the activity was due to the changes in the structural conformation of the protein.
Nano-structured hydrotrope-caged Cyt c is resistant to protease digestion
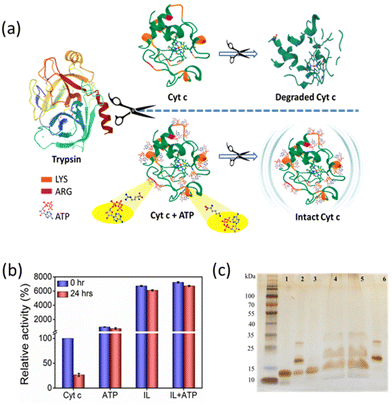
Trypsin, being a proteolytic enzyme, hydrolyses the peptide bond at basic amino acids such as Arg and Lys residues (Fig. 9a), thereby denaturing the target protein.42 Native and hydrotrope-stabilized Cyt c systems were incubated with 6 μM trypsin at 37 °C for 24 h. After the interaction, only 25% of activity was retained for native Cyt c, whereas the hydrotropic systems showed excellent performance and maintained Cyt c resistant to the action of trypsin (Fig. 9b). In the presence of ATP, Cyt c lost only ∼2-fold activity compared to activity at the 0 hour (∼700% retained), whereas IL and ATP + IL systems retained ∼61- and ∼67-fold relative activity, respectively. This excellent performance against protease digestion can be deduced by close examination of the interactions of ATP and IL with Cyt c. H-bonding analysis revealed that ATP has a higher tendency to interact with Lys, Thr, and Gln at both 26.85 °C and 90 °C temperatures, thus suggesting the efficacy of ATP to impart stability to Cyt c (Fig. 7a–c and Fig. S13†). Additionally, IL also greatly affected the dynamic nature by interacting with a wide range of residues. The affinity of ATP and IL towards Lys may be helpful to protect Cyt c by hindering the degradation mechanism of trypsin. The electrostatic and H-bond interactions that occur when Cyt c is nano-structured with ATP completely safeguard the protein from trypsin degradation (Fig. 9a). This was further shown by non-reducing SDS-PAGE analysis of the Cyt c before and after digestion with trypsin (Fig. 9c). Most of the native Cyt c in the presence of trypsin was found degraded (lane 2, Fig. 9c). However, the protein was intact when it was digested in the presence of ATP, IL and a combination of both. From UV-vis spectra (Fig. S17†), the Q band is lost in almost all the cases (except with ATP) after incubation with trypsin. However, only marginal changes were observed in the Soret band of Cyt c + ATP, and Cyt c + ATP + IL systems, indicating the stability induced by the nano-structured hydrotropes. In addition, the hydrotropic systems enhance the activity of trypsin (Fig. S18†) with 1.4 to 2 folds higher activity than control. These results give strong evidence that the higher activity of Cyt c in the presence of trypsin is due to the stabilisation provided by nanostructured hydrotropic systems and not by the inactivation of trypsin.Nano-structured hydrotrope-caged Cyt c reaction to oxidative stress
Unlike other stress conditions where the peptide back-bone is attacked, H2O2 initially targets the catalytic site, followed by a breakdown of peptide bonds. Thus, even though H2O2 is essential for the peroxidase activity of Cyt c, upon prolonged exposure it causes severe impairment to the protein structure (Fig. 10a). In the peroxidase activity studies mentioned above, 1 mM of H2O2 was added at the end, whereas for this study, Cyt c was incubated with H2O2 for 30 min, and the substrate was added afterwards. The activity of bare Cyt c following the induction of oxidative stress was found to be reduced to approximately 60% of its original activity (Fig. 10b). Conversely, in the presence of ATP, ∼219% of activity was retained. This observation can be correlated with the earlier findings in which ATP was found to prevent the oxidation of Cyt c from the Cyt c oxidase enzyme in the mitochondria by hampering the electron flow rate.43,44 Alternatively, there was a severe drop in the activity of Cyt c from 6700% to just 80% in the presence of IL. These results are not astonishing since the salicylate counterpart is known to enhance the formation of ROS.24 However, the presence of ATP was found to prevent the oxidation and Cyt c from IL-induced oxidative stress and demonstrated 192% activity. These findings demonstrate the stabilizing effect of nano-structured ATP surrounding Cyt c.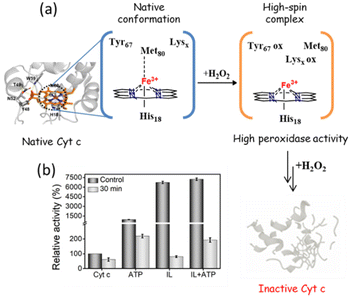 | ||
| Fig. 10 (a) Schematic mechanism showing H2O2 induced structural degradation of Cyt c. (b) Effect of H2O2 on the peroxidase activity of Cyt c in the presence of different hydrotropes. | ||
Conclusions
In summary, this study systematically demonstrated the robustness of ATP and IL-based mixed nano-structured hydrotropes and their utility in the improvement of protein packaging in extreme conditions. From the Mb solubility data, it was shown that the hydrotropic nature of ATP accelerated with the addition of a co-hydrotrope like [Cho][Sal] and vice versa. Through RDF, DLS, ζ-potential, and UV-vis data, we provided convincing data that ATP forms oligomeric nano-structures at concentrations above 2.5 mM around Cyt c, which can be augmented further with the addition of [Cho][Sal]. A 9-fold increase in peroxidase activity with ATP, 67-fold with IL, and 72-fold with IL + ATP suggested a ‘partial reversible unfolding-electrostatic stabilization’ mechanism. Reversible binding of ATP and IL with the Ω 40–54 residue loop region of Cyt c showed higher stability at 26.85 °C. Moreover, specific binding of nano-structured hydrotropes with Cyt c to Lys and Arg residues through H-bonding and polar interactions with Ω 70–85 region, presented an exceptionally high thermal tolerance with 80-fold activity even at 90 °C. Because of such binding specificity, the structure and activity of nano-confined Cyt c were found to be retained against protease digestion, which explicitly cleaves at Lys and Arg residues. Furthermore, ATP and IL-based nano-structured hydrotropes showed efficacy in retaining the functional integrity of Cyt c even in the presence of chemical denaturants like urea and GuHCl, which suggested the suitability of these solvent manipulation strategies for industrial bio-catalysis. Through oxidative stress studies, a stark observation was made where [Cho][Sal] was found to enhance ROS formation, which could be efficiently subdued by adding ATP. These results have the potential for counteracting oxidative stress inside the living cells. Thus, the novel strategy of protein confinement in nano-structured hydrotropes can find significant usage in protein packaging under biotic and abiotic stresses, high-temperature bio-catalysis, and cell protection against ROS.Experimental
Materials and characterization techniques, IL synthesis, Mb solubility study, SDS-PAGE analysis, peroxidase activity of Cyt c at room temperature and under harsh conditions, system setup and Molecular Dynamics simulation details, and Markov state models are provided in ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
D. M. acknowledges SERB, India (EEQ/2021/000059), for the research grant. D. K. S. was a recipient of a scholarship provided by POWER project POWR.03.02.00-00-I006/17. The computations were performed at the Poznan Supercomputing and Networking Center. D. M. and G. F. acknowledge the NANOPLANT project, which received funding from the European Union's Horizon 2020 research and innovation program under grant agreement no. 856961.References
- Y. Eguchi, A. Srinivasan, K. J. Tomaselli, S. Shimizu and Y. Tsujimoto, Cancer Res., 1999, 59, 2174–2181 CAS.
- D. V. Fyodorov and J. T. Kadonaga, Nature, 2002, 418, 896–900 CrossRef PubMed.
- M. Hochstrasser and J. Wang, Nat. Struct. Biol., 2001, 8, 294–296 CrossRef CAS PubMed.
- S.-J. Lee, Antioxid. Redox Signaling, 2003, 5, 337–348 CrossRef CAS PubMed.
- A. Patel, L. Malinovska, S. Saha, J. Wang, S. Alberti, Y. Krishnan and A. A. Hyman, Science, 2017, 356, 753–756 CrossRef CAS PubMed.
- C. Neuberg, Biochem. Z., 1916, 76, 107–108 CAS.
- K. Werner, H. Krister and Z. Thomas, Curr. Opin. Colloid Interface Sci., 2016, 22, 99–107 CrossRef.
- D. Mei, K. Jian, L. Liangzhong, L. Yifan, W. Lu and S. Jianxing, Biochem. Biophys. Res. Commun., 2020, 522, 247–253 CrossRef PubMed.
- V. G. Jack and G. Thomas, Exp. Eye Res., 2020, 190, 107862 CrossRef PubMed.
- M. Nishizawa, E. Walinda, D. Morimoto, B. Kohn, U. Scheler, M. Shirakawa and K. Sugase, J. Am. Chem. Soc., 2021, 143, 11982–11993 CrossRef CAS PubMed.
- S. Sarkar and J. Mondal, J. Phys. Chem. B, 2021, 125, 7717–7731 CrossRef CAS PubMed.
- M. Johannes, D. Tuan-Minh, T. Didier, H. Max, K. Ali, H. Dominik and K. Werner, Cell Rep. Phys. Sci., 2021, 2, 100343 CrossRef.
- X. Ou, Y. Lao, J. Xu, Y. Wutthinitikornkit, R. Shi, X. Chen and J. Li, JACS Au, 2021, 1, 1766–1777 CrossRef CAS PubMed.
- H. J. Jiang, R. Atkin and G. G. Warr, Curr. Opin. Green Sustainable Chem., 2018, 12, 27–32 CrossRef.
- A. Sindhu, S. Kumar, D. Mondal, I. Bahadur and P. Venkatesu, Phys. Chem. Chem. Phys., 2020, 22, 14811–14821 RSC.
- T. Itoh, Chem. Rev., 2017, 117, 10567–10607 CrossRef CAS PubMed.
- R. A. Sheldon, Green Chem., 2021, 23, 8406–8427 RSC.
- T. E. Sintra, D. O. Abranches, J. Benfica, B. P. Soares, S. P. M. Ventura and J. A. P. Coutinho, Eur. J. Pharm. Biopharm., 2021, 164, 86–92 CrossRef CAS PubMed.
- T. L. Greaves and C. J. Drummond, Chem. Soc. Rev., 2013, 42, 1096–1120 RSC.
- L. Bui-Le, C. J. Clarke, A. Bröhl, A. P. S. Brogan, J. A. J. Arpino, K. M. Polizzi and J. P. Hallett, Commun. Chem., 2020, 3, 55 CrossRef CAS PubMed.
- R. Hayes, G. G. Warr and R. Atkin, Chem. Rev., 2015, 115, 6357–6426 CrossRef CAS PubMed.
- A. Sanchez-Fernandez, A. E. Leung, E. G. Kelley and A. J. Jackson, J. Colloid Interface Sci., 2021, 581, 292–298 CrossRef CAS PubMed.
- Z. Xie and Z. Chen, Plant Physiol., 1999, 120, 217–226 CrossRef CAS PubMed.
- P. Poór, Biomolecules, 2020, 10, 341 CrossRef PubMed.
- (a) I. Martínez-Reyes and N. S. Chandel, Nat. Commun., 2020, 11, 102 CrossRef PubMed; (b) Z. Duan, Y. Ouyang, Y. Fu, F. Huang, F. Xia and I. Willner, Angew. Chem., Int. Ed., 2023, 62, e2023014 Search PubMed.
- (a) D. Alvarez-Paggi, L. Hannibal, M. A. Castro, S. Oviedo-Rouco, V. Demicheli, V. Tórtora, F. Tomasina, R. Radi and D. H. Murgida, Chem. Rev., 2017, 117, 13382–13460 CrossRef CAS PubMed; (b) N. Kapil, A. Singh and D. Das, Angew. Chem., Int. Ed., 2015, 54, 6492–6495 CrossRef CAS PubMed.
- S. M. Shet, P. Bharadwaj, M. Bisht, M. M. Pereira, S. K. Thayallath, V. Lokesh, G. Franklin, N. S. Kotrappanavar and D. Mondal, Int. J. Biol. Macromol., 2022, 215, 184–191 CrossRef CAS PubMed.
- Y. Chen, F. Jiménez-Ángeles, B. Qiao, M. D. Krzyaniak, F. Sha, S. Kato, X. Gong, C. T. Buru, Z. Chen, X. Zhang, N. C. Gianneschi, M. R. Wasielewski, M. Olvera de la Cruz and O. K. Farha, J. Am. Chem. Soc., 2020, 142, 18576–18582 CrossRef CAS PubMed.
- S. M. Shet, S. K. Thayallath, M. Bisht, M. M. Pereira, J. A. P. Coutinho, N. Sanna Kotrappanavar and D. Mondal, ACS Sustainable Chem. Eng., 2021, 9, 8327–8335 CrossRef CAS.
- M. Aurora, C. Ileana, T. Gheorghe, B. Liviu-Dorel, H. Ossi and T.-C. Maria, Colloids Surf., A, 2009, 338, 93–101 CrossRef.
- A.-R. Allouche, J. Comput. Chem., 2011, 32, 174–182 CrossRef CAS PubMed.
- I. Gomes, N. C. Santos, L. M. A. Oliveira, A. Quintas, P. Eaton, E. Pereira and R. Franco, J. Phys. Chem. C, 2008, 112, 16340–16347 CrossRef CAS.
- U. K. Singh, M. Kumari, S. H. Khan, H. B. Bohidar and R. Patel, ACS Sustainable Chem. Eng., 2018, 6, 803–815 CrossRef CAS.
- V. Yin, G. S. Shaw and L. Konermann, J. Am. Chem. Soc., 2017, 139, 15701–15709 CrossRef CAS PubMed.
- V. E. Kagan, V. A. Tyurin, J. Jiang, Y. Y. Tyurina, V. B. Ritov, A. A. Amoscato, A. N. Osipov, N. A. Belikova, A. A. Kapralov, V. Kini, I. I. Vlasova, Q. Zhao, M. Zou, P. Di, D. A. Svistunenko, I. V. Kurnikov and G. G. Borisenko, Nat. Chem. Biol., 2005, 1, 223–232 CrossRef CAS PubMed.
- W. B. Gordon, V. L. Gordon and D. B. Gary, J. Mol. Biol., 1990, 214, 585–595 CrossRef PubMed.
- J. D. Lambeth, J. Biol. Chem., 1981, 256, 4757–4762 CrossRef CAS PubMed.
- L. J. McClelland, T.-C. Mou, M. E. Jeakins-Cooley, S. R. Sprang and B. E. Bowler, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 6648–6653 CrossRef CAS PubMed.
- (a) B. San Francisco, E. C. Bretsnyder and R. G. Kranz, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, E788–E797 CrossRef CAS PubMed; (b) A. D. Savino, J. M. Foerster, T. L. Haye, A. Blok, M. Timmer, G. M. Ullmann and M. Ubbink, Angew. Chem., Int. Ed., 2020, 59, 23239–23243 CrossRef PubMed.
- W. Sato, S. Hitaoka, K. Inoue, M. Imai, T. Saio, T. Uchida, K. Shinzawa-Itoh, S. Yoshikawa, K. Yoshizawa and K. Ishimori, J. Biol. Chem., 2016, 291, 15320–15331 CrossRef CAS PubMed.
- T. Nataša, V. Rastislav, L. Veronika, M. Andrej and S. Erik, Biochim. Biophys. Acta, Proteins Proteomics, 2018, 1866, 1073–1083 CrossRef PubMed.
- M. Krieger, L. M. Kay and R. M. Stroud, J. Mol. Biol., 1974, 83, 209–230 CrossRef CAS PubMed.
- L. Smith, H. C. Davies and M. E. Nava, Biochemistry, 1980, 19, 1613–1617 CrossRef CAS PubMed.
- D. B. Craig and C. J. A. Wallace, Protein Sci., 1993, 2, 966–976 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3gc01704d |
| ‡ These authors have contributed equally to this work. |
| § Current address: Department of Chemistry, Sri Venkateswara college, University of Delhi, Dhaula Kuan, New Delhi 110021, India. |
| This journal is © The Royal Society of Chemistry 2023 |