Transcriptomic and metabolomic analyses provide insights into the attenuation of neuroinflammation by nervonic acid in MPTP-stimulated PD model mice†
Xueqi
Wang
 a,
Xinliang
Zhu
abc,
Xu
Li
a,
Zhengdou
Li
a,
Ying
Mao
a,
Shunbin
Zhang
a,
Xiaoxiao
Liu
d,
Xingguo
Liu
d,
Yapeng
Liu
d,
Fuliang
Cao
*e and
Ji
Zhang
*abc
a,
Xinliang
Zhu
abc,
Xu
Li
a,
Zhengdou
Li
a,
Ying
Mao
a,
Shunbin
Zhang
a,
Xiaoxiao
Liu
d,
Xingguo
Liu
d,
Yapeng
Liu
d,
Fuliang
Cao
*e and
Ji
Zhang
*abc
aCollege of Life Science, Northwest Normal University, Lanzhou 730070, Gansu Province, China. E-mail: 2020119145@nwnu.edu.cn; xzhuaf@nwnu.edu.cn; wyylixu@163.com; 2020212640@nwnu.edu.cn; 201674040119@nwnu.edu.cn; nwnuzsb1995@163.com
bBioactive Products Engineering Research Center for Gansu Distinctive Plants, Lanzhou 730070, China
cInstitute of Rural Development and Research, Northwest Normal University, Lanzhou 730070, Gansu Province, China. E-mail: zhangj@nwnu.edu.cn
dLanzhou Institute of Food and Drug Control, Lanzhou 740050, China. E-mail: lxx0808@163.com; 49487298@qq.com; liuyplzu@163.com
eNanjing Forestry University, Nanjing 210037, Jiangsu Province, China. E-mail: fuliangcaonjfu@163.com
First published on 9th December 2022
Abstract
Nervonic acid is one of the most promising bioactive fatty acids, which is believed to be beneficial for the recovery of human cognitive disorders. However, the detailed neuroprotective effects and mode of action of nervonic acid have not yet been fully elucidated. In this study, we used an MPTP-stimulated mouse Parkinson's disease (PD) model as a target to investigate the neuroprotective effects by behavioral tests and integrative analysis of trancriptomes and metabolomes of PD mouse brain with nervonic acid injections. The KEGG pathway enrichment analysis of transcriptomes showed that the genes involved in neuroinflammation were significantly increased after MPTP induction and have been greatly inhibited by nervonic acid injection, while nervonic acid also greatly improved nerve growth and synaptic plasticity pathways which were significantly downregulated by MPTP. At the same time, the upregulation of oleic acid and arachidonic acid metabolism pathways and the downregulation of amino acid metabolism pathways in metabolomes were particularly highlighted in the nervonic acid protection groups compared with the PD model. Meanwhile, it was found that arachidonic acid, oleic acid and taurine play an important regulatory role in the neuroprotective mechanism of nervonic acid through fatty acid metabolism by integrative analysis. Therefore, our study laid a solid foundation for further studies on the specific role of nervonic acid in the inhibition of PD at the level of metabolic regulation.
1. Introduction
Parkinson's disease (PD) is one of the most common neurodegenerative diseases with a prevalence rate of 1% in the population above 60 years old.1 The estimated global population affected by PD is more than 6 million people worldwide, which is a result of an increased number of elderly people and age-standardized prevalence rates.2,3 Therefore, PD has caused a huge burden on human health, social economy and other aspects. Thus far, despite the increasing number of studies on PD, there are no neuroprotective therapies able to prevent or delay disease progression.4–6 PD is characterized by progressive degeneration of dopaminergic neurons in the substantia nigra pars compacta and loss of dopamine in the striatum.7 The typical symptoms of PD are static tremors, slow movements, and rigidity.8 In addition to the specific motor symptoms of PD, there are also various non-motor symptoms of neurobehavioral disorders (depression, anxiety), cognitive impairment (dementia), and autonomic nerve dysfunction (hyperhidrosis).9,10Nervonic acid is a 24-carbon cis-15-enoic acid, which is a long-chain monounsaturated fatty acid. It has attracted considerable attention because of its close relationship with brain development, improving memory, and delaying brain aging.11,12 Specifically, nervonic acid is the main fatty acid found in various sphingolipid species in the central nervous system and plays an important role in forming the plasma membrane's lipid bilayer and in maintaining normal myelin function.13,14 In addition, studies have found that the risk of NDs decreases with increasing serum nervonic acid levels, and oral nervonic acid supplementation can enhance learning memory in animal (normal mice and mice with experimental memory impairment) models as proven by water-maze swim and platform step-down technologies.15,16 Nervonic acid may also play the role of an antioxidant mediator in human brain cells to delay brain aging.17
Arachidonic acid (ARA) metabolism plays a crucial role during inflammatory responses. ARA, a key metabolite in the ARA metabolic pathway, is critical in neuron function, synaptic plasticity, and long-term potentiation in the hippocampus.18 After its release, free ARA is involved in synaptic function at several levels and it modulates the exocytosis of presynaptic neurotransmitters by stimulating soluble N-ethylmaleimide-sensitive factor receptor production.19 In addition, oleic acid has the ability to reduce the inflammatory effects of long-chain saturated fatty acids in human cells, and has been reported to alter Ca2+ homeostasis in several cell lines.20 In this sense, cells subjected to unsaturated fatty acid treatments result in an inhibition of different signal transduction pathways.21
In recent years, the development of omics technology has provided a powerful tool for studying pathological effects. Moreover, the integration of transcriptomics and metabolomics has been widely used to identify important genes and metabolites from the large interaction network of multisystem diseases such as NDs.22–24 In our article, we will reveal the neuroprotective effect of nervonic acid on MPTP-induced PD model mice. First, we evaluated the changes of motor skills and memory ability of PD model mice before and after treatment with nervonic acid by pole climbing, the Morris water maze and Y-maze tests. Second, metabolomics and transcriptomics were integrated to explore the correlations between metabolism and gene expression and to determine the key mechanism by which nervonic acid inhibits PD at the level of metabolic regulation.
2. Materials and methods
2.1 Chemicals
MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine) was purchased from Sigma-Aldrich (St Louis, MO, USA). Nervonic acid (24-carbon cis-15-enoic acid) was purchased from Sigma-Aldrich. The FastKing gDNA Dispelling RT SuperMix kit and the SYBR Green QuantiTect RT-PCR Kit were purchased from Tiangen (Beijing, China).2.2 Animals and drug treatment
A total of 60 C57BL/6 mice (8–12 weeks, 22–25 g) were obtained from the Lanzhou Institute of Animal Husbandry and Veterinary Medicine, Chinese Academy of Agricultural Sciences (China). All the experimental procedures were in accordance with the National Institutes of Health guidelines for the Care and Use of Laboratory Animals and approved by the Animal Ethics Committee of Northwest Normal University.Animals were randomly divided into the following six groups (n = 10 for each): (1) the control vehicle (normal saline) administered group; (2) the MPTP-treated (30 mg per kg per day) group; (3) the MPTP (30 mg per kg of body weight) + nervonic acid-treated (40 mg per kg of body weight) group (M + NA40); (4) the MPTP (30 mg per kg of body weight) + nervonic acid-treated (80 mg per kg of body weight) group (M + NA80); (5) the nervonic acid (40 mg per kg of bodyweight) alone group (NA40); and (6) the nervonic acid (80 mg per kg of bodyweight) alone group (NA80). The mice in the control group were injected with normal saline for 15 days; the mice in the MPTP group were injected with MPTP for 5 days, followed by normal saline for 10 days; the mice in the M + NA40 group and M + NA80 group were injected with MPTP for 5 days, followed by nervonic acid for 10 days; and the mice in the NA40 group and NA80 group were injected with normal saline for 5 days, followed by nervonic acid for 10 days. All injection methods used in this experiment were intraperitoneal injections. Nervonic acid was dissolved in DMSO, and the final injectable volume was prepared using normal saline. MPTP was dissolved in the same volume of normal saline used for nervonic acid.
2.3 Behavioral analysis
2.4 Transcriptome sequencing
Total RNA was extracted from each sample using the TRIzol reagent (Life technologies, USA) according to the protocol indicated by the manufacturer. A total of 18 cDNA libraries were prepared from the brain tissues of the group treated with normal saline (control), the group treated with MPTP (30 mg per kg per day), the group treated with MPTP (30 mg per kg body weight) + nervonic acid-treated (40 mg per kg body weight) group (MPTP + NA40), the MPTP (30 mg per kg body weight) + nervonic acid-treated (80 mg per kg body weight) group (MPTP + NA80), the nervonic acid (40 mg per kg body weight) alone group (NA40) and the nervonic acid (80 mg per kg body weight) alone group (NA80). After the samples passed the assay, the eukaryotic mRNA was enriched with magnetic beads containing oligo (DT). Then, the mRNA was randomly interrupted by adding a fragmentation buffer. Using the mRNA as a template, the first strand of cDNA was synthesized with random six-base primers (random hexamers), and then the second strand of cDNA was synthesized by adding buffer, dNTPs, and DNA polymerase I, and then the double-stranded cDNA was purified with AMPure XP beads. The purified double-stranded cDNA was subjected to final repair; a tail was added and the sequencing connector was connected, and then the fragment size was selected using AMPure XP beads, and finally the final cDNA library was obtained by PCR enrichment.2.5 Analysis of the transcriptomic data
Fragment per kilobase of transcripts per million mapped reads (FPKM) was used to calculate the gene expression values. For samples with biological duplication, we used the DEseq2 algorithm for differential gene expression analysis. The criterion was set for upregulation with log 2(fold change) > 1 or downregulation with log 2(fold change) < −1. The p-value of the altered genes was adjusted for a false discovery rate < 5%. Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses of differentially expressed genes (DEGs) were implemented using R,26 by using the clusterProfiler package. The terms with an adjusted P-value of less than 0.05 were considered significantly enriched.2.6 Metabolomic profiling
Mouse brain tissue samples of all groups were collected according to the RNA-seq procedure. 50 mg of the tissues were homogenized in liquid nitrogen and the total metabolites were extracted using 300 μL of methanol/acetonitrile/water solution (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v
1, v![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v
v![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v), and the total metabolites were subjected to ultrasonic extraction in an ice bath for 30 min, and then held for 10 min at −20 °C and centrifuged (14
v), and the total metabolites were subjected to ultrasonic extraction in an ice bath for 30 min, and then held for 10 min at −20 °C and centrifuged (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 20 min, 4 °C) to extract the supernatant. The supernatant was freeze-dried and stored at −80 °C for LC-MS analysis.
000g, 20 min, 4 °C) to extract the supernatant. The supernatant was freeze-dried and stored at −80 °C for LC-MS analysis.
The sample was analyzed using a Thermo Scientific™ Vanquish™ UHPLC system (Thermo) as previously described. An AC18 column (Thermo Hypersil Gold C18, 100 mm × 2.1 mm, 1.8 μm) coupled to Q Exactive mass spectrometer (Thermo) was used for the separation of metabolites. The mobile phase consisted of 0.1% formic acid in water (A) for the positive mode, 5 mM ammonium acetate in water (A) for the negative mode, and acetonitrile (B), using the following gradient conditions: 0 min–1 min, 1% B; 1 min–8 min, 1%–99% B; 8–10 min, 99% B; 10 min–10.1 min, 99%–1% B; 10.1–12 min, 1% B. The flow rate was 0.3 mL min−1, the column temperature was set as 35 °C and the sample injection volume was 4 μL. Mass spectrometric analysis was performed using a Q-Exactive mass spectrometer (Thermo Fisher Scientific, Bremen, Germany), using an electrospray ionization source in positive and negative ionization modes. The operating parameters were as follows: positive polarity; spray voltage – 4.0 kV (positive) or −3.6 kV (negative), the funnel RF lens value at 50, and a capillary temperature of 400 °C, and the flow rates for sheath gas, aux gas, and sweep gas were set to 45, 15, and 0, respectively. Except otherwise noted, data-dependent acquisition (DDA) was performed using the Full MS-ddMS2 setup. The full MS resolution was set to 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and the mass range was set to 100–1500. For the MS2 spectra, the resolution was set to 17
000 and the mass range was set to 100–1500. For the MS2 spectra, the resolution was set to 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500. The normalized collision energy was set to 20%, 40% and 60%. The dynamic exclusion values were set to 5 s.
500. The normalized collision energy was set to 20%, 40% and 60%. The dynamic exclusion values were set to 5 s.
2.7 Analysis of the metabolomic data
Firstly, the metabolic analysis 5.0 online server (https://www.metaboanalyst.ca/) was used to obtain the matched peak data from the original LC-MS data, and the peak area data were standardized using Microsoft Excel. Then, orthogonal partial least squares discriminant analysis (OPLS-DA) and volcanic maps were used to further analyze the standardized data; meanwhile, the differential metabolites (DEMs) were obtained based on the variable importance in projection (VIP) (VIP > 1) and t-test (P < 0.05). Lastly, the metabolic pathways of the potential biomarkers were analyzed by a bubble chart using MetaboAnalyst 5.0. Data analysis includes statistical analysis (one factor), enrichment analysis, differential metabolite screening, differential metabolite correlation analysis and KEGG pathway analysis.2.8 Combined transcriptomic and metabolomic analysis
DEGs were obtained from the transcriptomic data and DEMs from the metabolomic data. All DEGs and DEMs were identified from comparisons between the two different groups with three biological replicates per group. KEGG enrichment analysis was performed based on the corresponding omics data. For association analysis, a joint KEGG pathway analysis was performed for genes and metabolites.2.9 Real-time PCR assay
The total RNA of mouse brain tissues was obtained by using the TRIzol reagent according to the manufacturer's instructions. After the measurement of the RNA concentration using the absorbance ratio of the spectrophotometer at 260 nm and 280 nm, the RNA were reverse-transcribed (RT) into cDNA using the FastKing gDNA Dispelling RT SuperMix kit according to the manufacturer's instructions. The primer sequences are shown in Table S1.† Real-time PCR was then performed using a SYBR Green QuantiTect RT-PCR kit, and each sample was independently analyzed five times. PCR amplification was performed with 40 cycles: denaturation at 95 °C for 10 s, annealing at 55 °C for 20 s, and extension at 72 °C for 30 s. The relative values of the gene expression of each mRNA were calculated by normalization to β-actin mRNA expression according to the 2−ΔΔCT method.2.10 Statistical analysis
Statistical significance of the data was evaluated using single factor analysis of variance (one-tailed ANOVA) and Tukey's range test using Origin 8.0 software (San Diego, CA, USA). The results were considered significant when the p value was <0.05.3. Results
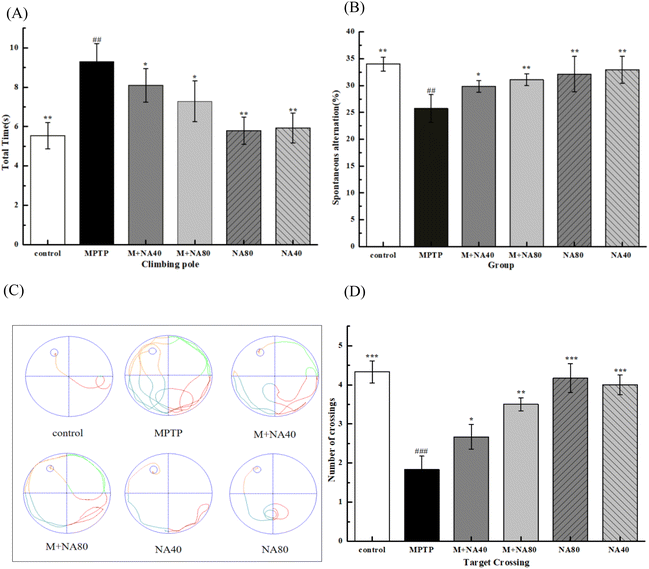
3.1 Nervonic acid ameliorates MPTP-induced memory, learning, and motor behavior impairment in C57BL/6 mice
Firstly, we used the pole-climbing experiment to systematically evaluate the motor function of mice in each group. The results of the pole climbing experiment showed that there was no difference in the total time of pole climbing between the control group and the simple nervonic acid group. Compared with the control group, the total climbing time of the model group (MPTP group) was significantly increased, indicating that the motor function of the mice was damaged. The total climbing time of mice in the nervonic acid group was significantly lower than that of the mice in the model group (P < 0.05) (Fig. 1A). In addition, the turn time of the model group mice was also significantly higher than that of the other groups, while the turn time of the nervonic acid group mice was significantly lower than that of the model group (P < 0.05) (Fig. S1†).Secondly, we performed the Y-maze test to assess spatial learning by the percentage of spontaneous switching behavior. The Y-maze data showed that the MPTP-treated mice had a lower percentage of spontaneous alternation behavior compared to the vehicle-treated control group, indicating impaired working memory (Fig. 1B). In addition, the total number of arm entries was also reduced in the model group compared with the control group (Fig. S2†). On the other hand, nervonic acid significantly increased the percentage of spontaneous changes compared with the MPTP alone-treated group.
Finally, to investigate the neuroprotective effect of nervonic acid on MPTP-induced memory impairment in vivo, an MWM assay was then performed to evaluate the cognitive function of MPTP-treated mice. The movement track of mice is shown in Fig. 1C. The control group mainly swam in a linear fashion, indicating that the mice showed a goal-directed search. Compared with the control group, the MPTP-treated mice exhibited an aimless search, indicating significant cognitive impairment in the MWM test. However, pretreatment with nervonic acid in the MPTP-induced mice showed a significant increase in the trend trajectory of the mice, especially in the high-dose group. There was no significant difference in the escape latency/distance of the mice in each group on the first day of training. As the number of training days increased, the escape latency/distance of mice in each group decreased (Fig. S3†). On the fifth day, the escape latency of mice in the MPTP group was significantly longer than that in the control group (P < 0.001), suggesting that model causes memory impairment in mice. However, pretreatment with nervonic acid in MPTP-induced mice showed a shorter escape latency (P < 0.05) (Fig. S4†). In this experiment, the platform was removed on the 6th day and the mice were allowed to explore the room for 60 s. As shown in Fig. 1D, the MPTP-induced mice exhibited a higher number of platform crossings under the pretreatment with nervonic acid compared to the model group (p < 0.05). The result is similar to the time spent in the target quadrant which has the hidden platform (Fig. S5†).
3.2 Transcriptomic analysis
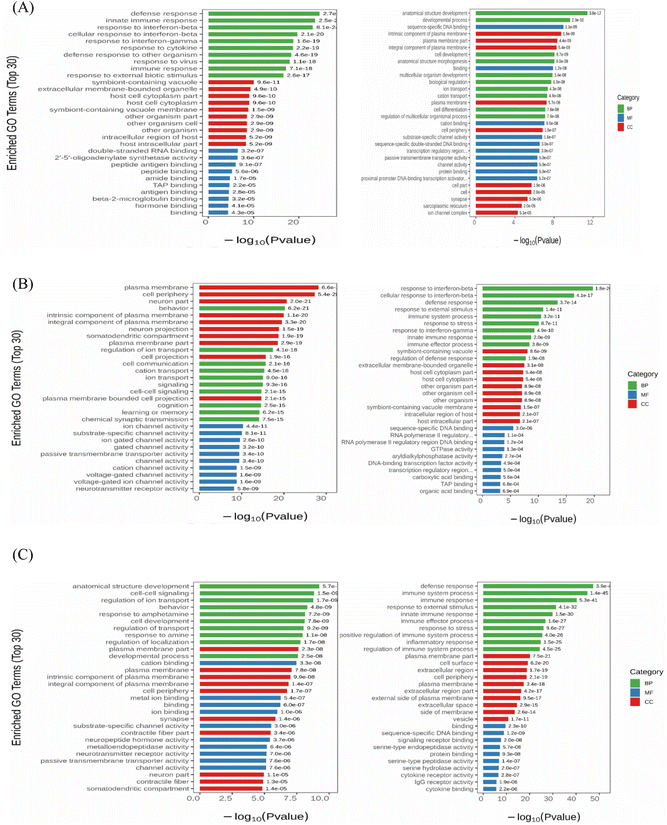
To investigate the ameliorative effect of nervonic acid on MPTP-induced brain injury in C57BL/6 mice, a gene expression pattern of nervonic acid was established as a potential neuroprotective mechanism by transcriptomic analysis. As shown in Fig. S6,† the correlation coefficient (R2) between the samples was greater than 0.85; and in the PCA diagram, the samples between groups were scattered and the samples within groups were clustered. There were 629 genes that were significantly upregulated between the model and control groups, whereas 424 genes were significantly downregulated, as shown in the column figure. Similarly, 522 genes were significantly upregulated and 439 genes were significantly downregulated between the model group and the MPTP + NA40 group. Between the model group and the MPTP + NA80 group, 619 genes were significantly upregulated and 616 genes were significantly downregulated.To better understand the gene expression patterns of the protective effect of nervonic acid in nerve injury, we enriched the GO function of differential genes. As shown in Fig. 2A, the functional enrichment of the significantly up-regulated DEGs in the model group mainly included a defense response, an innate immune response and a response to interferon-beta compared with the control group, while the down-regulated DEGs were mainly enriched in the development of the anatomical structure, developmental process and sequence-specific DNA. This result indicated that the immune process and neuroinflammatory response were significantly activated in the model group. When comparing the high-dose nervonic acid group with the model group, similar to the low-dose nervonic acid group, the defense response, immune system process, immune response, and inflammatory response were greatly downregulated; on the other hand, cell–cell signaling, regulation of ion transport, and development of the anatomical structure were also activated, and the trend toward the upregulation of the plasma membrane and neuronal part, the intrinsic component of the plasma membrane, and the behavioral response was particularly enriched (Fig. 2B and C).
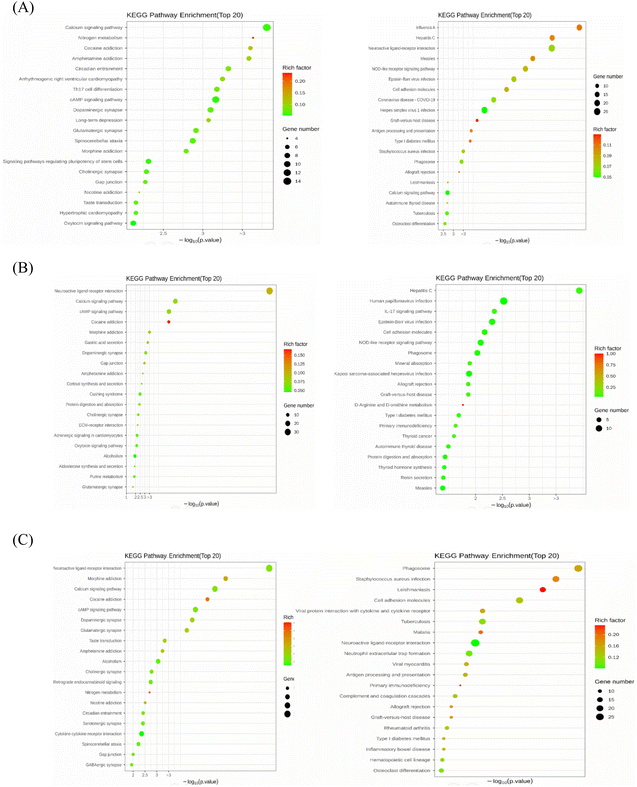
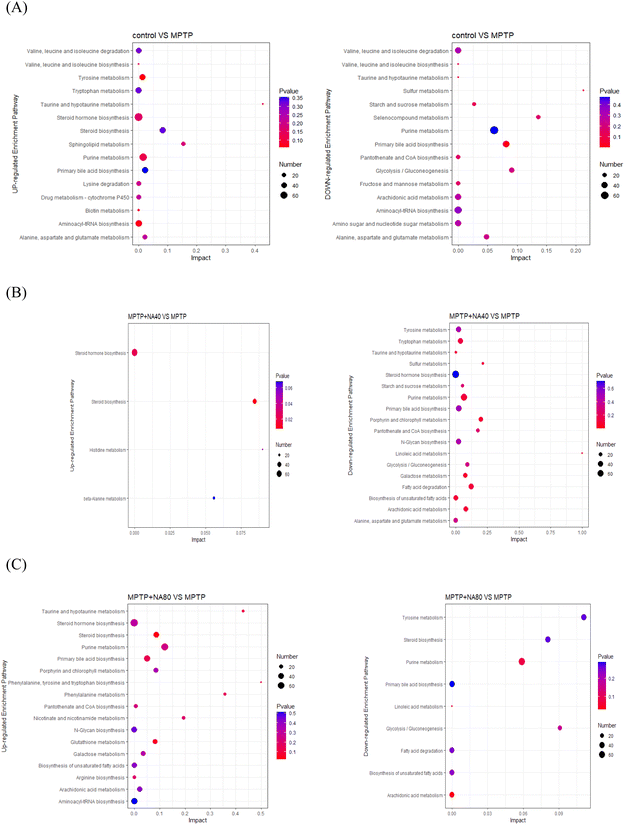
To further explore the mechanism of the signaling pathways of nervonic acid in the treatment of neuroinflammation, we compared the KEGG pathways (Fig. 3) between the MPTP vs. control, MPTP vs. MPTP + NA40, and MPTP vs. MPTP + NA80 groups. We found a total of 254, 266, and 264 signaling pathways that were significantly enriched, respectively. Compared with the control group, the up-regulated KEGG pathways in the model group were mainly the calcium signaling, cAMP signaling, dopaminergic synapse and glutamatergic synapse pathways, while the down-regulated KEGG pathways were mainly the neuroactive ligand–receptor interaction pathway, influenza A signaling pathway, cell adhesion molecule signaling pathway and NOD-like receptor signaling pathway (Fig. 3A). In the MPTP vs. MPTP + NA40 group, the up-regulated KEGG pathways were the neuroactive ligand–receptor interaction pathway, calcium signaling pathway and cAMP signaling pathway, while the down-regulated KEGG pathways were mainly human papillomavirus infection and NOD-like receptor signaling pathways (Fig. 3B). Similarly, in the MPTP vs. MPTP + NA80 group, the up-regulated DEGs were mainly enriched in the neuroactive ligand–receptor interaction pathway, calcium signaling pathway and cAMP signaling pathway, while the down-regulated DEGs were mainly enriched in the cell adhesion molecule pathway and cytokine–cytokine receptor interaction pathway (neutrophil extracellular trap formation) (Fig. 3C).
3.3 Metabolomic analysis
To identify the DEMs of the protective effect of nervonic acid on neuronal functional defects caused by MPTP, the mouse brain tissues were subjected to metabolomic profiling by an untargeted metabolomics approach. OPLS-DA showed a clear separation of the different sample groups. The metabolomic profiling data could be considered reliable to investigate the differentially accumulated metabolites (Fig. 4).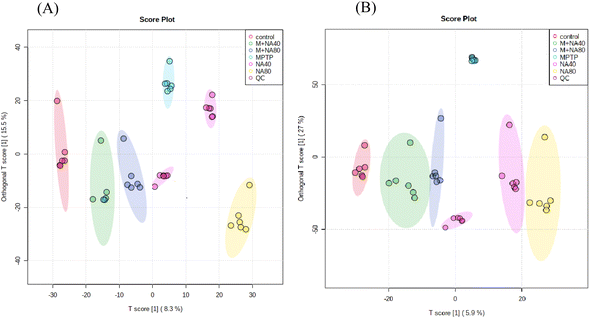 | ||
| Fig. 4 Score plots for the OPLS-DA analysis of data. (A) The positive OPLS-DA score map. (B) The negative OPLS-DA score map. | ||
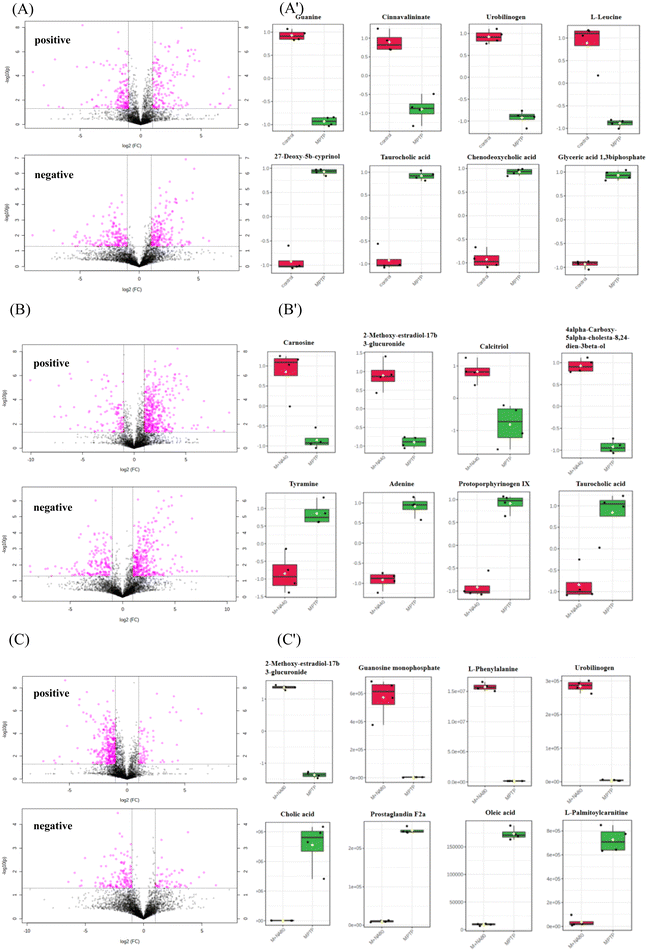
The DEMs obtained by the metabolomic analysis mainly contained organic compounds, such as amino acids, lipids, carbohydrates and nucleic acids. As shown in Fig. 5 and 6, MPTP (MPTP vs. control) induced significant downregulation of metabolites such as guanine, cinnavalininate, L-leucine, tetrahydrocorticosterone, taurine, urobilinogen, etc. Pathway analysis of DEMs indicated downregulation of primary bile acid biosynthesis, taurine and hypotaurine metabolism, starch and sucrose metabolism, purine metabolism, and other neurotransmitter-related pathways. In the high-dose nervonic acid group, adenosine monophosphate, L-phenylalanine, oxidized glutathione, guanosine monophosphate, and other metabolites closely related to taurine and hypotaurine metabolism, primary bile acid biosynthesis, purine metabolism, and steroid biosynthesis were significantly upregulated, with similar results to those in the low-dose nervonic acid group.
3.4 Integrated analysis of transcriptomics and metabolomics
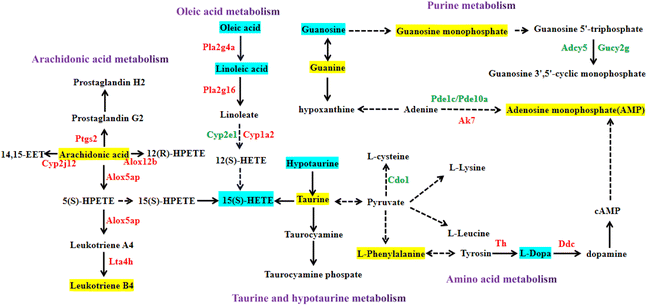
It can be seen from the previous data that the high-dose nervonic acid group has a significant improvement effect on MPTP-induced PD model mice. Therefore, we analyzed the key pathways, metabolites and regulatory genes of the M + NA80 vs. MPTP group through transcriptomics and metabolomics, and the correlation networks between them highlighted arachidonic acid metabolism, oleic acid metabolism, purine metabolism, taurine and hypotaurine metabolism and amino acid metabolism (Fig. 7).Firstly, the arachidonic acid metabolic pathway is the most significantly changed pathway obtained from the joint analysis of transcriptomics and metabolomics. In this pathway, genes were mainly up-regulated, most of which were catalytic enzymes, such as Ptgs2, Cyp2j12, Alox12b, Alox5ap and Lta4 h, and some were regulatory proteins, mainly including Casr, C5ar1 and Ryr1. In addition, ARA and leukotriene B4 were also up-regulated in the M + NA80 group compared with the MPTP group. Besides, oleic acid metabolism was also a key pathway regulated at the transcriptional and metabolic levels, of which the genes Pla2g4a, Pla2g16, Cyp1a2, Grp and Casr were up-regulated, while the genes Cyp2e1 and IL-22 were down-regulated. The down-regulated metabolites were mainly linoleic acid, oleic acid and 15(S)-HETE.
In addition to the two networks mentioned above, the correlation networks of metabolism of several amino acids were also constructed, mainly including tyrosine metabolism and lysine, leucine and phenylalanine biosynthesis. The related up-regulated catalytic enzymes were TH and DDC, and the metabolite L-phenylalanine was up-regulated and L-Dopa was down-regulated. At the same time, the up-regulated regulatory proteins were Slc6a3, Slc38a2 and Slc18a2, and the down-regulated ones were Gdnf, Penk, Drd2 and Drd3. Furthermore, in taurine and low taurine metabolism, Glr, IL8 and other regulatory protein genes were down-regulated, while Htr7 was up-regulated.
Lastly, purine metabolism was also a key pathway, in which the genes Adcy5, Gucy2g, Pde1c, Pde10a, Mpp3, Mpp4, Mpp5, Mpp7 and Cd300a were down-regulated and Ak7, Pmch, Pyr1, Pitx2, Cd38, Gbp2, Gbp5 and Gbp6 were up-regulated. The metabolites guanine, guanosine monophosphate and adenosine monophosphate were up-regulated, while guanosine was down-regulated.
3.5 Verification of the gene expression levels
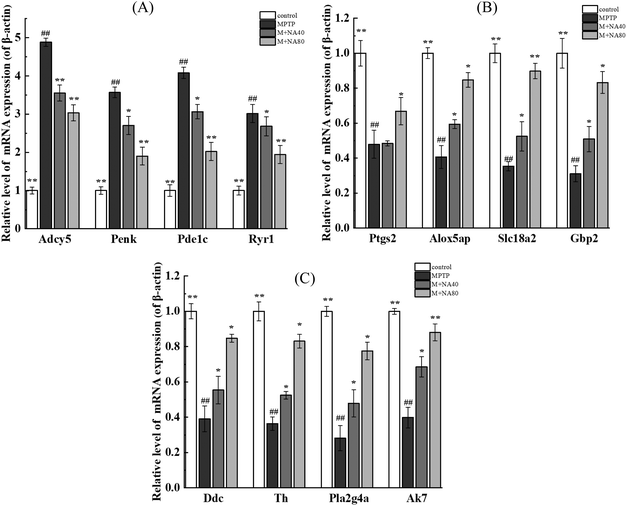
As shown in Fig. 8A, compared with the control group, the genes with increased mRNA expression in the MPTP group were regulatory proteins, such as Ryr1 and Penk, and a few catalytic enzymes, mainly including Adcy5 and Pde1c, but the expression of these genes decreased after nervonic acid treatment. Rather, the mRNA expressions of Ptgs2, Alox5ap, Slc18a2, Gbp2, Pla2g4a, Ddc, Th and Ak7 decreased in the MPTP group, while the expressions of these genes increased after nervonic acid treatment (Fig. 8B and C). In addition, the mRNA expressions of some key genes Mpp3, Mpp4, Mpp5, Mpp7, Zdhhc23, Cd300a, Rasd2, and Rasd1 were increased in the MPTP group, but the mRNA expression of these genes decreased after nervonic acid treatment (Fig. S7†). These results were the same as those observed in RNA-Seq analysis.4. Discussion
PD is an age-related neurodegenerative disease. Irrespective of etiology, all cases of PD show loss of dopaminergic neurons in the substantia nigra pars compacta, and most show pathology consisting of the Lewy bodies and Lewy neurites.27 In our study, mice were administered intraperitoneal injections of MPTP to mimic the lesions observed in PD patients. Previous studies have reported that environmental toxins can trigger PD, and MPTP is a suitable chemical to construct a PD animal model.28 Although the pathogenesis of PD is not yet clear, inflammation is thought to be one of the critical factors contributing to disease progression.29,30 Nervonic acid has potential anti-inflammatory effect in a number of human inflammatory diseases, but the mechanisms are still poorly understood. In this study, we first determined from animal behavior that nervonic acid has ameliorative effects on MPTP-induced C57BL/6 mice. Then, transcriptomics was used in combination with metabolomics to investigate the changes in the expressions of genes and metabolites after treatment with nervonic acid.Studies on metabolomics are crucial and can provide a more complete picture of changes in the body's metabolites under stress. Here, in our study, further enrichment analysis showed that the ARA metabolism pathway is significantly enriched in the control vs. MPTP, M + NA40 vs. MPTP and M + NA80 vs. MPTP groups. The pro- and anti-inflammatory effects were associated with the changes in ARA metabolites from different pathways.31 The three primary pathways by which ARA is metabolized are mediated by cyclooxygenase (COX) enzymes, lipoxygenase (LOX) enzymes, and cytochrome P450 (CYP) enzymes, and these three pathways produce eicosanoids, lipid signaling molecules that play roles in regulating lipid metabolism, biological processes and immune response.32 Leukotrienes (LTs) and hydroxyeicosatetraenoic acids (HETEs) are derived from the LOX metabolic pathway.33 Among them, HETEs have been shown to promote the proliferation, adhesion, and metastasis of inflammatory cells through their regulatory effects on different pathways.34 For example, 12-HETE can increase mitochondrial outer membrane permeability and release apoptosis-related proteins, thereby reducing apoptosis of cancer cells.35 Leukotriene B4 (LTB4) activates leukocyte chemotaxis36and inhibits the secretion of interleukin-10.37 ARA has also been shown to protect beta cells against the negative effects of palmitic acid exposure, and ARA could be beneficial not only for beta-cell function, but also for protecting or increasing beta-cell mass.38
Notably, in the taurine and hypotaurine metabolism pathways, taurine (2-aminoethanesulfonic acid) is the most abundant free amino acid in humans and plays a significant role in several essential biological processes such as bile acid conjugation, maintenance of calcium homeostasis, neuromodulation, osmoregulation and membrane stabilization.39 Moreover, attenuation of apoptosis and its antioxidant activity seem to be crucial for the cytoprotective effects of taurine.39 More noteworthy is taurine chloramine produced from taurine under inflammation exerts anti-inflammatory and cytoprotective effects.40 Taurine is regarded as a promising agent against numerous types of inflammatory injury (inflammatory bowel disease, pancreatitis, and gastric mucosal injury), due to its immunoregulatory importance.41 Accordingly, there is a growing body of preclinical data that shows the anti-inflammatory effects of taurine in both neural and systemic inflammation, including cardiovascular disease,42 traumatic brain injury,43 and lung injury.44 The anti-inflammatory effect of taurine has also been attributed to the reduced secretion of interleukins (ILs) (such as IL-8), as shown by experiments in Caco-2 cells, without any participation of polymorphonuclear leukocytes.45 We speculate that nervonic acid may be used in combination with taurine and its metabolites through the taurine and hypotaurine metabolism pathways, which can not only regulate the lasting response of various malignant inflammatory processes, but also circumvent the limitations pertaining to drugs, thus improving the therapeutic outcome.
The modulatory functions of nervonic acid towards amino acid-related metabolic pathways should not be ignored either. Among several amino acid metabolism pathways, tyrosine metabolism is the most representative pathway influenced by nervonic acid, in which a vital Slc18a2 gene is up-regulated to a great extent in the M + NA80 vs. MPTP group (Fig. 8). A previous study has suggested that neuroactive ligand–receptor interactions may play an important role in the process of PD; meanwhile, Slc18a2, GRIN1 and GRIN2D may be potential candidate genes for PD progression.46,47 Another metabolite, dopamine, an intermediate product of tyrosine metabolism should also be mentioned. The state of striatal dopamine release is of considerable importance in the diagnosis and research of PD.48 The anti-inflammatory effects of dopamine upon its binding to Drd1 and/or Drd2 receptors on microglia have been reported.49 Dopamine was shown to dampen inflammation in primary rat microglia by regulating the pro-inflammatory renin–angiotensin system through Drd1 and Drd2 signaling.50
In our study, through the combined analysis of transcriptomics and metabolomics, it was seen that ARA metabolism was significantly enriched, indicating that the ARA pathway is involved in the pharmacological mechanism of nervonic acid treatment in PD mice induced by MPTP. The genes Ptgs2, Cyp2j12, Alox12b, Lta4h, Casr, C5ar1 and Ryr1 involved in the ARA pathway are mainly related to the NF-κB signaling pathway, IL-17 signaling pathway, TNF signaling pathway, and serotonergic synapse pathway, and pathways of neurodegeneration – multiple diseases, inflammatory mediator regulation of TRP channels and the NOD-like receptor signaling pathway. Based on the existing literature,51–55 it has been confirmed that the above genes play a key role in neuroinflammation, synaptic plasticity and other pathways. ARA metabolism plays a crucial role during inflammatory responses, and notably, there have been strong functional links established between ARA metabolites and inflammation, both benign and malignant. Studies have shown that unsaturated fatty acids have vital roles in nerve cell function, neurotransmission and also in immune and inflammatory responses in neurocognitive diseases.56–58 Long-chain unsaturated fatty acids of the neuronal membrane structural components have an impact on cellular function and membrane characteristics, and serve as a source of lipid-derived messengers.59 Preclinical studies show that omega-3 polyunsaturated fatty acids (n-3 PUFAs) improve cognition, promote neuroprotection (and possibly neurorestoration), reduce neuroinflammation, and alter neuronal function.60 Based on the above, as a long-chain unsaturated fatty acid, nervonic acid may participate in the metabolism of arachidonic acid, oleic acid or some unsaturated fatty acids, and the decline of dopamine metabolites in this process indicates that inflammation is inhibited and neural plasticity is enhanced.
Our study integrated the metabolomic data and coupled massive transcriptomic data to explore the underlying neuroprotective mechanism of nervonic acid in MPTP-stimulated PD model mice. However, there are limitations in the present study. One of the limitations is that the relationship between metabolites and related genes is still in the prediction stage, and needs to be further verified by experiments. To comprehensively understand the mechanism of nervonic acid on neuroinflammation-mediated neurodegeneration, we will carry out corresponding experiments in the following work. In conclusion, the current results indicated that nervonic acid has a curative effect on PD, as shown by comprehensive metabolic and transcriptional alterations in PD mice, regulating the expression of genes (Mpp4, Cd300a, Adcy5, Slc18a2 and Ak7, etc.) to control the enzymes in purine metabolism, arachidonic acid metabolism, oleic acid metabolism, taurine and hypotaurine metabolism and amino acid metabolism, thereby reducing inflammation. These findings will facilitate further investigation on the metabolic pathway and its related genes in the treatment of PD with nervonic acid.
Author contributions
The contributions of X. W. and X. Z. are the same.Conflicts of interest
There is no conflict of interest.Acknowledgements
The financial assistance of this study was endorsed by the Science and Technology Planning Project of Gansu Province (22ZD1NA001), the Industrial Support and Guidance Project of Colleges and Universities in Gansu Province (No. 2021CYZC-37), and the Higher Education Industry Support Plan Project of Gansu Province (2020C-21). The Natural Science Foundation of Gansu Province in China (21JR7RA113).References
- E. Dorsey, T. Sherer and M. S. Okun, et al., The emerging evidence of the Parkinson pandemic, J. Parkinson's Dis., 2018, 8(1), 3–8 Search PubMed.
- D. Ararsland, L. Batzu and G. M. Halliday, et al., Parkinson disease-associated cognitive impairment, Nat. Rev. Dis. Primers, 2021, 7(1), 1–21 CrossRef PubMed.
- T. K. Karikari, A. Charway-Felli and K. Höglund, et al., Commentary: global, regional, and national burden of neurological disorders during 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015, Front. Neurol., 2018, 9(3), 201–211 CrossRef PubMed.
- I. Y. Abdi, S. S. Ghanem and O. M. El-Agnaf, Immune-related biomarkers for Parkinson's disease, Neurobiol. Dis., 2022, 170(12), 105771–105780 CrossRef CAS PubMed.
- Z. Mao, C. Liu and S. Ji, et al., The NLRP3 Inflammasome is Involved in the Pathogenesis of Parkinson's Disease in Rats, Neurochem. Res., 2017, 42(4), 1104–1115 CrossRef CAS PubMed.
- B. Brakedal, L. Toker and K. Haugarvoll, et al., A nationwide study of the incidence, prevalence and mortality of Parkinson's disease in the Norwegian population, npj Parkinson's Dis., 2022, 8(1), 1–8 CrossRef PubMed.
- J. Saravanan, Role of microgliosis, oxidative stress and associated neuroinflammation in the pathogenesis of Parkinson's disease: The therapeutic role of Nrf2 activators, Neurochem. Int., 2021, 145(10), 105014–105024 Search PubMed.
- D. Ararsland, L. Batzu and G. M. Halliday, et al., Parkinson disease-associated cognitive impairment, Nat. Rev. Dis. Primers, 2021, 7(1), 1–21 CrossRef PubMed.
- M. J. Farrer, S. Bardien and N. Hattori, et al., Celebrating the Diversity of Genetic Research to Dissect the Pathogenesis of Parkinson's Disease, Front. Neurol., 2021, 12(7), 648417–648426 CrossRef PubMed.
- X. R. Wang, S. X. Zhu and X. M. Wen, et al., Altered dopamine metabolism and its role in pathogenesis of Parkinson's disease, Shengli Xuebao, 2021, 73(1), 89–102 Search PubMed.
- F. Liu, P. Wang and X. Xiong, et al., A review of nervonic acid production in plants: Prospects for the genetic engineering of high nervonic acid cultivars plants, Front. Plant Sci., 2021, 12(6), 626625–626632 CrossRef PubMed.
- Q. Li, J. Chen and X. Yu, et al., A mini review of nervonic acid: Source, production, and biological functions, Food Chem., 2019, 301(4), 125286–125295 CrossRef CAS PubMed.
- M. R. Terluk, J. Tieu and S. A. Sahasrabudhe, et al., Nervonic Acid Attenuates Accumulation of Very Long-Chain Fatty Acids and is a Potential Therapy for Adrenoleukodystrophy, Neurotherapeutics, 2022, 19(5), 1007–1017 CrossRef CAS PubMed.
- M. A. Altinoz and A. Ozpinar, PPAR-δ and erucic acid in multiple sclerosis and Alzheimer's disease. Likely benefits in terms of immunity and metabolism, Int. Immunopharmacol., 2019, 69(13), 245–256 CrossRef CAS PubMed.
- S. Song, H. Wang and S. S. Jia, et al., Analysis of correlation between serum fatty acid profile and cognitive impairment in the elderly, Zhonghua Yufang Yixue Zazhi, 2018, 52(6), 636–641 CAS.
- C. Li, B. Yang and N. Tan, et al., Nervonic acid improves learning memory ability in normal mice and mice with experimental memory impairment, FASEB J., 2004, 18(4), 579–588 Search PubMed.
- H. Umemoto, S. Yasugi and S. Tsuda, et al., Protective Effect of Nervonic Acid Against 6-Hydroxydopamine-Induced Oxidative Stress in PC-12 Cells, J. Oleo Sci., 2021, 70(1), 95–102 CrossRef CAS PubMed.
- H. Tokuda, M. Kontani and H. Kawashima, et al., Differential effect of arachidonic acid and docosahexaenoic acid on age-related decreases in hippocampal neurogenesis, Neurosci. Res., 2014, 88(12), 58–66 CrossRef CAS PubMed.
- C. F. Latham, S. L. Osborne and M. J. Cryle, et al., Arachidonic acid potentiates exocytosis and allows neuronal SNARE complex to interact with Munc18a, J. Neurochem., 2007, 100(6), 1543–1554 CAS.
- Y. Zhou, Y. Song and H. Zhou, et al., Linoleic acid activates GPR40/FFA1 and phospholipase C to increase [Ca2+] i release and insulin secretion in islet beta-cells, Chin. Med. Sci. J., 2012, 27(1), 18–23 CrossRef CAS PubMed.
- C. Carrillo Pérez, M. M. Cavia Camarero and S. Alonso de la Torre, Role of oleic acid in immune system; mechanism of action; a review, Nutr. Hosp., 2012, 27(4), 978–990 Search PubMed.
- J. Wang, C. L. Li and B. J. Tu, et al., Integrated epigenetics, transcriptomics, and metabolomics to analyze the mechanisms of benzo [a] pyrene neurotoxicity in the hippocampus, Toxicol. Sci., 2018, 166(1), 65–81 CrossRef CAS PubMed.
- Z. Q. Li, L. L. Wang and J. Zhou, et al., Integration of transcriptomics and metabolomics profiling reveals the metabolic pathways affected in dictamnine-induced hepatotoxicity in mice, J. Proteomics, 2020, 213(8), 103603–103614 CrossRef CAS PubMed.
- Y. Zhu, X. Wu and Y. Liu, et al., Integration of transcriptomics and metabolomics reveals the responses of earthworms to the long-term exposure of TiO2 nanoparticles in soil, Sci. Total Environ., 2020, 719(5), 137492–137501 CrossRef CAS PubMed.
- A. Rajabian, H. R. Sadeghnia and S. Fanoudi, et al., Genus Boswellia as a new candidate for neurodegenerative disorders, Iran. J. Basic Med. Sci., 2020, 23(3), 277–289 Search PubMed.
- G. Yu, L. G. Wang and Y. Han, et al., ClusterProfiler: an R package for comparing biological themes among gene clusters, OMICS, 2012, 16(5), 284–287 CrossRef CAS PubMed.
- S. Huang, H. Liu and Y. Lin, et al., Berberine Protects Against NLRP3 Inflammasome via Ameliorating Autophagic Impairment in MPTP-Induced Parkinson's Disease Model, Front. Pharmacol., 2021, 11(6), 2509–2514 Search PubMed.
- M. K. Song, J. H. Lee and J. Kim, et al., Neuroprotective effect of NXP031 in the MPTP-induced Parkinson's disease model, Neurosci. Lett., 2021, 740(11), 135425–135434 CrossRef CAS PubMed.
- Y. Lee, S. Lee and S. C. Chang, et al., Significant roles of neuroinflammation in Parkinson's disease: therapeutic targets for PD prevention, Arch. Pharmacal Res., 2019, 42(5), 416–425 CrossRef CAS PubMed.
- M. Miranda, J. F. Morici and M. B. Zanoni, et al., Brain-derived neurotrophic factor: a key molecule for memory in the healthy and the pathological brain, Front. Cell. Neurosci., 2019, 13(7), 363–371 CrossRef CAS.
- Y. Lin, X. Lu and X. Qiu, et al., Arachidonic acid metabolism and inflammatory biomarkers associated with exposure to polycyclic aromatic hydrocarbons, Environ. Res., 2022, 212(13), 113498–113505 CrossRef CAS PubMed.
- K. J. Bosma, C. E. Kaiser and M. E. Kimple, et al., Effects of Arachidonic Acid and Its Metabolites on Functional Beta-Cell Mass, Metabolites, 2022, 12(4), 342–350 CrossRef CAS PubMed.
- M. Cordaro, D. Impellizzeri and R. Siracusa, et al., Effects of a co-micronized composite containing palmitoylethanolamide and polydatin in an experimental model of benign prostatic hyperplasia, Toxicol. Appl. Pharmacol., 2017, 329(4), 231–240 CrossRef CAS PubMed.
- X. Cao, Y. Shang and W. Kong, et al., Flavonoids derived from Anemarrhenae Rhizoma ameliorate inflammation of benign prostatic hyperplasia via modulating COX/LOX pathways, J. Ethnopharmacol., 2022, 284(1), 114740–114749 CrossRef CAS PubMed.
- A. A. Panagiotopoulos, K. Kalyvianaki and E. Castanas, et al., Eicosanoids in prostate cancer, Cancer Metastasis Rev., 2018, 37(2), 237–243 CrossRef CAS PubMed.
- B. C. Subramanian, R. Majumdar and C. A. Parent, The role of the LTB4-BLT1 axis in chemotactic gradient sensing and directed leukocyte migration, Semin. Immunol., 2017, 33(7), 16–29 CrossRef CAS PubMed.
- S. S. Landgraf, L. S. Silva and D. B. Peruchetti, et al., 5-Lypoxygenase products are involved in renal tubulointerstitial injury induced by albumin overload in proximal tubules in mice, PLoS One, 2014, 9(10), 107549–107552 CrossRef PubMed.
- D. C. Keane, H. K. Takahashi and S. Dhayal, et al., Arachidonic acid actions on functional integrity and attenuation of the negative effects of palmitic acid in a clonal pancreatic β-cell line, Clin. Sci., 2011, 120(5), 195–206 CrossRef CAS PubMed.
- J. Marcinkiewicz and E. Kontny, Taurine and inflammatory diseases, Amino Acids, 2014, 46(1), 7–20 CrossRef CAS PubMed.
- C. Kim and Y. N. Cha, Taurine chloramine produced from taurine under inflammation provides anti-inflammatory and cytoprotective effects, Amino Acids, 2014, 46(1), 89–100 CrossRef CAS PubMed.
- S. Baliou, A. M. Kyriakopoulos and D. A. Spandidos, et al., Role of taurine, its haloamines and its lncRNA TUG1 in both inflammation and cancer progression, On the road to therapeutics?, Int. J. Oncol., 2020, 57(3), 631–664 CrossRef CAS PubMed.
- Y. Yin, K. Wen and Y. Wu, et al., Inhibition of sodium current by taurine magnesium coordination compound prevents cesium chloride-induced arrhythmias, Biol. Trace Elem. Res., 2012, 146(2), 192–198 CrossRef CAS PubMed.
- Q. Wang, W. Fan and Y. Cai, et al., Protective effects of taurine in traumatic brain injury via mitochondria and cerebral blood flow, Amino Acids, 2016, 48(9), 2169–2177 CrossRef CAS PubMed.
- L. Guler, M. Tavlasoglu and O. Yucel, et al., Taurine attenuates lung ischemia–reperfusion injury after lung transplantation in rats, J. Anesth., 2014, 28(3), 347–353 CrossRef PubMed.
- Y. Shi, L. Zhong and H. Zhong, et al., Taurine supplements in high-carbohydrate diets increase growth performance of Monopterus albus by improving carbohydrate and lipid metabolism, reducing liver damage, and regulating intestinal microbiota, Aquaculture, 2022, 554(9), 738150–738156 CrossRef CAS.
- M. Yang, X. Q. Wu and C. B. Ding, et al., Weighted gene co-expression network analysis identifies specific modules and hub genes related to Parkinson's disease, NeuroReport, 2021, 32(13), 1073–1081 CrossRef CAS PubMed.
- E. Düz and T. Çakır, A co–expression network based molecular characterization of genes responsive for Braak stages in Parkinson's disease, Eur. J. Neurosci., 2022, 55(7), 1873–1886 CrossRef PubMed.
- A. F. Pike, I. Szabò and R. Veerhuis, et al., The potential convergence of NLRP3 inflammasome, potassium, and dopamine mechanisms in Parkinson's disease, npj Parkinson's Dis., 2022, 8(1), 1–9 CrossRef PubMed.
- Y. Yan, W. Jiang and L. Liu, et al., Dopamine controls systemic inflammation through inhibition of NLRP3 inflammasome, Cell, 2015, 160(1–2), 62–73 CrossRef CAS PubMed.
- A. Dominguez-Meijide, A. I. Rodriguez-Perez and C. Diaz-Ruiz, et al., Dopamine modulates astroglial and microglial activity via glial renin-angiotensin system in cultures, Brain, Behav., Immun., 2017, 62(4), 277–290 CrossRef CAS PubMed.
- Y. Jiang, M. Zhong and F. Long, et al., Deciphering the Active Ingredients and Molecular Mechanisms of Tripterygium hypoglaucum (Levl.) Hutch against Rheumatoid Arthritis Based on Network Pharmacology, Evidence-Based Complementary Altern. Med., 2020, 21(6), 356–383 Search PubMed.
- J. P. Graves, J. A. Bradbury and A. Gruzdev, et al., Expression of Cyp2c/Cyp2j subfamily members and oxylipin levels during LPS–induced inflammation and resolution in mice, FASEB J., 2019, 33(12), 14784–14797 CrossRef CAS PubMed.
- M. S. J. E. Neil, J. D. Lenn and M. B. Brown, et al., A new ex vivo skin model for mechanistic understanding of putative anti-inflammatory topical therapeutics, Int. J. Pharm., 2022, 617(5), 121610–121618 CrossRef PubMed.
- X. Wu, A. R. Sun and R. Crawford, et al., Inhibition of Leukotriene A4 Hydrolase Suppressed Cartilage Degradation and Synovial Inflammation in a Mouse Model of Experimental Osteoarthritis, Res. Square, 2022, 42(2), 145–173 Search PubMed.
- C. Yang, M. S. Rybchyn and W. G. M. De Silva, et al., UV–induced DNA Damage in Skin is Reduced by CaSR Inhibition, Photochem. Photobiol., 2022, 98(12), 1157–1166 CrossRef CAS PubMed.
- X. X. Li, J. D. Lee and N. L. Massey, et al., Pharmacological characterisation of small molecule C5aR1 inhibitors in human cells reveals biased activities for signalling and function, Biochem. Pharmacol., 2020, 180(6), 114156–114162 CrossRef CAS PubMed.
- S. Kotlyarov and A. Kotlyarova, Involvement of Fatty Acids and Their Metabolites in the Development of Inflammation in Atherosclerosis, Int. J. Mol. Sci., 2022, 23(3), 1308–1312 CrossRef CAS PubMed.
- H. Zhang, C. Zhao and Q. Liu, et al., Dysregulation of fatty acid metabolism associated with esophageal inflammation of ICR mice induced by nitrosamines exposure, Environ. Pollut., 2022, 297(8), 118680–118687 CrossRef CAS PubMed.
- N. Omgy, B. Anjana and V. Anusree, et al., Recent Advancements in Omega Fatty Acids to Treat Neurodegeneration[M]//Functional Foods and Therapeutic Strategies for Neurodegenerative Disorders, Springer, Singapore, 2022, vol. 14(6), pp. 121–134 Search PubMed.
- H. L. Fisk, C. E. Childs and E. A. Miles, et al., Modification of subcutaneous white adipose tissue inflammation by omega-3 fatty acids is limited in human obesity-a double blind, randomised clinical trial, EBioMedicine, 2022, 77(3), 103909–103915 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2fo02595g |
| This journal is © The Royal Society of Chemistry 2023 |