 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Genomic characterization supporting the development of new food and crop options from the Australian flora
Robert J.
Henry
 ab
ab
aQueensland Alliance for Agriculture and Food Innovation, University of Queensland, Brisbane, QLD 4072, Australia. E-mail: robert.henry@uq.edu.au
bARC Centre of Excellence for Plant Success in Nature and Agriculture, University of Queensland, Brisbane, QLD 4072, Australia
First published on 23rd February 2023
Abstract
Plant biodiversity is a key resource underpinning efforts to satisfy the food needs of growing human populations while coping with climate change. Crop wild relatives provide options for diversification and adaptation of existing crops while other species provide opportunities for domestication of completely new crops. Around 10% of the world’s flora is found in Australia with many unique taxa that evolved during a long period of isolation. Australian plant species have been used for human food for more than 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 years apparently without domestication and provide new options for domestication as crops adapted to new environments. However, the use of Australian species is very limited in modern agriculture. Australian crop wild relatives that remain poorly characterized include those for rice, Sorghum, soybean, mung bean, pigeon pea, citrus and cotton. Plants such as the Eucalyptus and Macadamia have been widely cultivated but remain only partially domesticated. Many species were used as food by indigenous Australians. The impact of humans on the evolution of these wild populations has not been established. Rapid advances in genomics are providing efficient tools to characterize plants, allowing their utility as food crops in agriculture to be assessed, supporting the utilization of novel genes in current crops and the acceleration of domestication of new species. This knowledge will also guide enhanced in situ and ex situ conservation of these critical genetic resources. Macadamia genomics provides a new platform for the use and conservation of this unique Australian group. Genomics of key Australian crop wild relatives such as those of rice and Sorghum is facilitating their use in global agriculture. Genome analysis reveals genes that can be used in crop improvement and identifying potential candidates for de novo domestication to provide completely new crop options with novel climate adaptation. Contributions of genomics to an improved understanding of traditional indigenous food production systems will also have wider environmental and social benefits contributing to sustainable environmental management and may improve appreciation of indigenous culture.
000 years apparently without domestication and provide new options for domestication as crops adapted to new environments. However, the use of Australian species is very limited in modern agriculture. Australian crop wild relatives that remain poorly characterized include those for rice, Sorghum, soybean, mung bean, pigeon pea, citrus and cotton. Plants such as the Eucalyptus and Macadamia have been widely cultivated but remain only partially domesticated. Many species were used as food by indigenous Australians. The impact of humans on the evolution of these wild populations has not been established. Rapid advances in genomics are providing efficient tools to characterize plants, allowing their utility as food crops in agriculture to be assessed, supporting the utilization of novel genes in current crops and the acceleration of domestication of new species. This knowledge will also guide enhanced in situ and ex situ conservation of these critical genetic resources. Macadamia genomics provides a new platform for the use and conservation of this unique Australian group. Genomics of key Australian crop wild relatives such as those of rice and Sorghum is facilitating their use in global agriculture. Genome analysis reveals genes that can be used in crop improvement and identifying potential candidates for de novo domestication to provide completely new crop options with novel climate adaptation. Contributions of genomics to an improved understanding of traditional indigenous food production systems will also have wider environmental and social benefits contributing to sustainable environmental management and may improve appreciation of indigenous culture.
Introduction
Human populations rely on a very small number of plant species for the bulk of food production.1 A key strategy for improving food security and adapting agriculture and food production to climate change is the diversification of food crops to include more variation within currently used species and to utilize new species.2 Advances in genomic technologies are facilitating the rapid assessment of the genetic relationships between plants and their potential value as sources of novel variation for food production.3,4 The diverse Australian flora has been largely overlooked in the domestication of modern food crops. Progress and prospects for applying genomics to the use of potential Australian food crops are reviewed here.The Australian flora
More than 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 species of vascular plants are native to Australia.5 The flora of Australia has a history that has included a long period of evolution in isolation and more recent extensive exchange of species with Asia in the north. The most unique elements of the Australian flora evolved in considerable isolation while the land mass was well separated from others for a long period of time. More recently Australia and Asia have been in closer contact allowing a more ready exchange of species6 resulting in a flora that contains elements that are from the Southeast Asian plate (Sahul) and others from the Australian continental plate (Sunder) that trace back to Gondwana. This suggests that the Australian flora represents a reservoir of novel genetics for crop improvement. The adaptation of many of these species to different environments indicates the potential value of their use in addressing the urgent need for climate adaptation.
000 species of vascular plants are native to Australia.5 The flora of Australia has a history that has included a long period of evolution in isolation and more recent extensive exchange of species with Asia in the north. The most unique elements of the Australian flora evolved in considerable isolation while the land mass was well separated from others for a long period of time. More recently Australia and Asia have been in closer contact allowing a more ready exchange of species6 resulting in a flora that contains elements that are from the Southeast Asian plate (Sahul) and others from the Australian continental plate (Sunder) that trace back to Gondwana. This suggests that the Australian flora represents a reservoir of novel genetics for crop improvement. The adaptation of many of these species to different environments indicates the potential value of their use in addressing the urgent need for climate adaptation.
Indigenous use
Humans have utilized indigenous plants for a very long time in Australia. The date humans reached Australia has continued to be re-assessed with current estimates placing it at 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 years ago.7 The long association of humans and plants in Australia had developed highly productive and sustainable methods for cultivation, harvesting, storage and processing of crops.8,9 However, modern agriculture in Australia is relatively recent and has been based largely upon imported plants. Europeans arriving in the late 18th century brought agricultural methods that rapidly displaced the traditional practises of the first Australians. The indigenous populations were widely considered to be hunter gatherers and not recognized as practising agriculture, something that puzzled those aware of the connections between continental Australian populations and those practising agriculture in the north (New Guinea).10 Recent research11 has uncovered archaeological and historical evidence for extensive food production systems that seem to satisfy at least some definitions of agriculture but the distinction between hunter gatherers and agriculturalists may be less important than understanding the food production systems used. Many of the species that were used as food are not part of any current food production system but may be found in the wild. Molecular evidence supports the involvement of humans in the prehistoric migration of food plants over large distances in Australia.12
000 years ago.7 The long association of humans and plants in Australia had developed highly productive and sustainable methods for cultivation, harvesting, storage and processing of crops.8,9 However, modern agriculture in Australia is relatively recent and has been based largely upon imported plants. Europeans arriving in the late 18th century brought agricultural methods that rapidly displaced the traditional practises of the first Australians. The indigenous populations were widely considered to be hunter gatherers and not recognized as practising agriculture, something that puzzled those aware of the connections between continental Australian populations and those practising agriculture in the north (New Guinea).10 Recent research11 has uncovered archaeological and historical evidence for extensive food production systems that seem to satisfy at least some definitions of agriculture but the distinction between hunter gatherers and agriculturalists may be less important than understanding the food production systems used. Many of the species that were used as food are not part of any current food production system but may be found in the wild. Molecular evidence supports the involvement of humans in the prehistoric migration of food plants over large distances in Australia.12
Current utilization
Australian and world agriculture do not utilize Australian plants to a significant extent. This is probably partially due to the relative isolation of Australia from the agriculture adopted elsewhere and the development of unique food production systems that apparently did not involve traditional domestication.10 The current dependence upon imported species for food has resulted in a lack of public awareness of the extent of plant genetic resources in Australia that have potential to contribute to agriculture. Recent research suggests that many local plants of great potential importance to global food security have been overlooked. The Australian flora contains many plants consumed by the first Australians and many relatives of crop plants (crop wild relatives (CWRs)) that were domesticated in other parts of the world but very few species have been domesticated for modern agriculture. Advances in genetic technology (sequencing, assembly, annotation and functional analysis of genomes),13 especially very recent developments,14 have made the use of these resources more feasible.Ownership and access
Utilization of the Australian flora in agriculture has become more complex as a result of greater recognition of intellectual property issues. The convention on biodiversity has enabled ownership of biodiversity to be claimed by Australian States and Territories. The Food and Agriculture Treaty on Plant Genetic Resources for Food and Agriculture has provided new arrangements for access to species related to major agricultural crops. The Nagoya Treaty has agreed with the need for prior informed consent and mutually agreed terms for access to biodiversity to be negotiated with indigenous people. This has created challenges in the utilization of historical collections15 and digital sequence information such as DNA sequences.16Advances in technology
Advances in DNA sequencing14 have driven rapid advances in the application of genomics to plant genetic resources3 and made the cost-effective analysis of large numbers of plant species possible.17 This has enabled many new opportunities for the utilization of wild plant genetic resources18 to be considered3 and effectively expanded the gene pool for many species.19 Improved long-read sequencing technology has reduced the cost of this key technology17 and, more recently, the development of techniques to re-sequence the same molecule many times has allowed long-read sequences with much greater accuracy to be generated, greatly improving the quality of genomes that can be assembled.20 Improvements in software for assembly have supported these advances.21 The application of RNA seq tools for transcriptome analysis allows gene annotations to be supported by transcript evidence. Together this enables the sequencing assembly and annotation of a genome for any plant to be achieved rapidly and at a relatively low cost. The genomes are no longer highly fragmented and are usually chromosome-level assemblies that are resolved into haplotypes. For example, recent assemblies of Australian wild citrus22 and Macadamia species23 have produced mostly whole chromosomes for both haplotypes of the diploid plants with a continuous sequence from telomere to telomere. These genomes have unprecedented accuracy allowing the identification of more than 99% of the conserved angiosperm genes.22 Genome analysis can reveal genes with potential use in food crops and define the evolutionary relationships between species that are useful guides to their potential for use in plant improvement.24 The ability to characterise the genomes of large numbers of wild plants in this way makes possible more extensive efforts to find sources of novel genetics and new potential domesticates. The availability of new high-quality genome sequences also makes efforts to rapidly domesticate a plant using gene editing a more viable option. For example, grasses could be domesticated as new cereals by targeting the genes that were selected in domestication of the small number of current cereal crop species.25 Selection of grasses from novel environments offers the potential to generate cereals adapted to novel or altered environments.This review is prompted by the very recent emergence of highly efficient and cost-effective techniques for whole genome characterization and the potential of these to greatly accelerate the evaluation and use of wild genetic resources for more sustainable and diverse food production. This provides a great opportunity to utilize the poorly exploited Australian flora in food production and may also be useful worldwide in expanding the use of genetic resources.
Examples of utility of Australian species
The Australian flora includes many key wild relatives of species (https://www.gbif.org/dataset/07044577-bd82-4089-9f3a-f4a9d2170b2e) that have been domesticated elsewhere (Fig. 1). A few Australian species have been domesticated or partially domesticated. Many species have the potential for domestication in the future. Genomics is beginning to be applied to the improved characterization of some of these species (Table 1).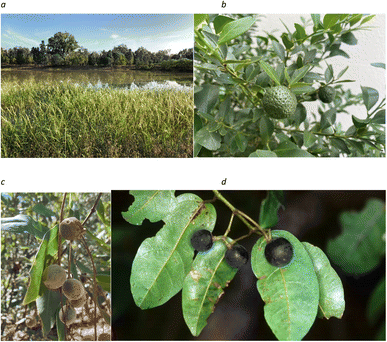 | ||
| Fig. 1 Crop wild relatives from Australia, (a) Oryza meridionalis, (b) Citrus australis, (c) Macadamia jansenii, and (d) Cissus antarctica. | ||
| Species | Applications of genomics | Reference |
|---|---|---|
| Cajanus | Sequencing for marker discovery | 100 |
| Citrus | Genome sequencing | 22 and 101 |
| Corymbia | Genome sequencing | 75 and 77 |
| Eucalyptus | Genome sequencing | 74 and 90 |
| Gossypium | Genome sequencing | 81 |
| Macadamia | Genome sequencing | 68 |
| Oryza | Genome sequencing | 36 and 41 |
| Sorghum | Genome sequencing | 51 |
Cereals and other grasses
Australia is home to a great diversity of grasses with around 1000 species representing the diversity of the Poaceae and about 10% of all grass species.5 They include relatives of many of the major crops and species that might prove new options for domestication (Table 2).| Species | Useful traits |
|---|---|
| Oryza australiensis | Rhizome32 |
| Oryza meridionalis | Close relative of domesticated rice, large seeds29 |
| Oryza rufipogon | Very close relative of domesticated rice43 |
| Oryza officinalis | Disease resistance, grain quality, yield, and abiotic stress tolerance |
| Potamophila parviflora | Separate sex flowers102 |
| Leersia hexandra | Distant relative of rice |
| Microlaena stipoides | Cold and drought tolerance44 |
| Sorghum macrospermum | Close relative of domesticated Sorghum49 |
| Sorghum laxiflorum | Close relative of domesticated Sorghum49 |
| Echinochloa turneriana | Close relative of domesticated millet103 |
| Astrebla spp | Dominant grasses |
| Themeda spp | Dominant grasses |
Rice
Rice is a major food crop with significant Australian wild relatives.26 Australian species of Oryza include close relatives of domesticated rice and more distant members of the genus.27 Rice was domesticated in Asia from O. rufipogon and in Africa from O. barthii.28 The Australian populations in the A genome clade (including domesticated rice) are O. meridionalis29 and Australian populations of O. rufipogon30 that are distinct in having a chloroplast genome closer to O. meridionalis.31 These Australian populations are a resource for rice globally representing a large gene pool that is directly accessible for rice breeders. The more distant O. australiensis32 has many attributes (disease resistance, photosynthesis and grain quality) that may be useful in rice breeding and adaptation to new climates.The genomes of the Oryza genus33 have been characterized by production of high-quality genome sequences.34 However, the quality of these genomes has continued to improve as sequencing and analysis techniques have improved in the more than 20 years since a rice genome was first reported.35 Sequencing of the Australian A genome species has increased understanding of their relationship with domesticated rice.36 The evolution of the Oryza species appears to have involved processes of divergence, dispersal (maybe by birds), hybridization and introgression.
The recent study of the evolution of the Oryza genus has been facilitated by the availability of many high-quality genome sequences in this genus.37 The chloroplast genomes of wild38 and domesticated rice have suggested the domestication of Asian rice from two maternal gene pools.39 The Australian populations are a useful resource in understanding the evolution of Oryza prior to domestication due to their isolation, in northern Australia, from the domesticated rice populations that have been grown widely for a long time in Asia.
Oryza australiensis is widely distributed across northern Australia. This species has a rhizome that allows regeneration after the dry season. The genome is larger than that of other Oryza (estimated at 965 MB) due to the presence of large amounts of repetitive sequences with unique retrotransposon sequences (Kangourou and Wallabi) accounting for much of the genome.40 A draft genome sequence was recently reported for this species.41
Oryza meridionalis is present in large populations in northern Australia with distinctive closed panicles and small anthers. This species is the most distant relative of domesticated rice with an AA genome in the primary gene pool.29 The genome is estimated to be 370 Mb with around 46% of the genome being gypsy-rich repetitive elements.36 Knowledge of this genome is being used to guide editing to produce a newly domesticated crop25 with the desirable disease resistance and quality characteristics of this species. This species has large grains suggesting the possibility that a long period of human use by wild harvest may have impacted the genetics of these populations.29,42
Evidence for the presence of hybrid populations in the wild in Australia43 suggests that reticulate evolution may have been a key process in the evolution of the progenitor species from which rice was domesticated.
More distant relatives of rice have been considered for domestication. Microlaena stipoides is a member of the Oryzeae tribe. Genomic approaches to characterization of domestication genes44 in this species have supported efforts to use mutagenesis to domesticate this as a new food crop.45Potamophila parviflora has separate sex flowers46 suggesting novel reproductive traits that might be useful in rice.
Application of more recent technology to the sequencing of wild rice relatives that is currently in progress will facilitate the more widespread use of these resources in rice improvement and may result in domestication of additional Oryza species.
Sorghum
Sorghum was domesticated in Africa, but the genus is largely found in Australia.47,48 Australian species such as S. macrospermum are close relatives of domesticated Sorghum49 that have traits that may be useful in Sorghum breeding. Domesticated Sorghum has a well-characterized genome50 but the sequencing of the remainder of the genus is only now in progress.48 Active sequencing of the Australian wild relatives is in progress and should provide a strong platform for Sorghum genetic improvement. The initial phase of this research has generated sequence data from the nuclear and chloroplast genomes and is defining the relationships within the genus, suggesting two distinct groups of species.51 Variation in genes controlling seed size, a key domestication trait, has recently been explored in these species.52 The group including domesticated Sorghum includes two Australian species S. macrospermum and S. laxiflorum as close relatives of domesticated Sorghum.49 The other Australian species are in a distinct clade including the species in the more poorly resolved Parasorghum and Stiposorghum subgenera within the Sorghum genus as currently defined. Analysis of genome sequences suggests that analysis of more accessions may be required to define the evolutionary relationships and taxonomy of this group.51 This knowledge will allow these species to be used to develop Sorghum genotypes with desirable abiotic and biotic stress tolerances from wild relatives.48Sugarcane
Sugarcane is harvested in greater quantities than any other crop globally. Modern sugarcane varieties are hybrids between Saccharum officinarum and S. spontaneum. S. officinarum was probably domesticated in New Guinea. S. spontaneum is found in Australia but has been considered an escape from populations used for sugarcane breeding or plantings to prevent riverbank erosion (Atlas of Living Australia). However, this does not explain herbarium records from the Northern Territory, suggesting that these populations may be native. Genome sequences are being produced for the complex polyploid genome of sugarcane.53 With this knowledge wild germplasm could be used to create new sugarcane hybrids widening the current narrow genetic base of sugarcane. Sorghum and sugarcane are closely related and their common genetic resources need to be considered together.54Pulses
Pulses are of growing importance in the sustainable supply of the protein required in human diets. The Australian flora contains many wild relatives of domesticated pulses and other species that may provide an opportunity to domesticate a completely new pulse.55,56 The Australian Fabaceae includes 1500 species from 136 genera (https://www.anbg.gov.au/cpbr/cd-keys/peakey/key/ThePeaKey/Media/Html/about_peas.html) with many that have the potential for food use (Table 3). The genera Canavalia, Crotalaria, Cullen, Desmodium, Glycine, Glycyrrhiza, Hardenbergia, Indigofera, Kennedia, Lotus, Rhynchosia, Swainsona, Trigonella and Vigna include 242 species that have been evaluated as potential new food crops.57 These species are adapted to a wide range of environments providing new opportunities to produce pulses in diverse or altered climates.| Genus | Number of species | Potential use | References |
|---|---|---|---|
| Cajanus | 13 | Insect resistance | 63 |
| Canavalia | 4 | New domesticates | 57 |
| Crotalaria | 19 | New domesticates | 57 |
| Cullen | 25 | Pharmaceuticals/forage | 57 |
| Desmodium | 21 | Forage | 57 |
| Glycine | 23 | Drought/disease resistance | 57 |
| Glycyrrhiza | 1 | New domesticate | |
| Hardenbergia | 3 | Limited use | 57 |
| Indigofera | 33 | Limited use | 57 |
| Kennedia | 15 | Limited use | 57 |
| Lotus | 2 | Forage | 57 |
| Rhynchosi | 6 | Forage | 57 |
| Swainson | 84 | Includes indigenous grain | 57 |
| Trigonella | 1 | New domesticate | 57 |
| Vigna | 5 | Tropical species/progenitor | 57 |
Mung bean (Vigna radiata) is an important pulse crop and Australian wild mung bean (the progenitor of cultivated mung bean)58 and other Australian Vigna species59,60 are an important genetic resource. Research to better characterize this resource is needed.
The pigeon pea (Cajanus cajan) genome has been characterized.61 The wild relatives of pigeon pea have a distribution including a strong concentration in northern Australia.62 The reference genome for cultivated pigeon pea has been used in re-sequencing (Vanambathina et al. 2021a) of Australian wild relatives63 that are being used as a source of insect resistance (Vanambathina et al. 2021b) in breeding of pigeon pea. The molecular basis of insect resistance has been explored in relation to polymorphisms in candidate genes encoding the production of compounds implicated in conferring insect resistance.
Soybean (Glycine max) is a major global pulse crop with relatively low genetic diversity64 and a well-characterized genome. Glycine species are abundant in the Australian flora but the Australian species remain poorly characterized.65 More species may be defined within the Australian glycine as little taxonomic work has been conducted on the herbarium samples already collected. They represent an important resource for soybean breeding representing sources of disease resistance and abiotic stress tolerance.
Rattlepods (Crotalaria sp.) may include options for domestication.
Genome sequencing and annotation of the poorly characterized but diverse wild Australian pulse relatives would greatly improve access to these diverse genetic resources. Prospects for these contributing to improved varieties of current pulses and to the domestication of completely new pulses are good.
Tree crops
Macadamia
Macadamia is a member of the predominantly southern hemisphere family, Proteaceae. Domestication of Macadamia was very recent and largely based upon genotypes developed in Hawaii from seeds obtained from Australia around 100 years ago. The current expanding production in Australia and internationally is largely based upon Hawaiian varieties. Active breeding in Australia is seeking to use the four wild species as germplasm to support improvement of the domesticated gene pool. All four are now rare and M. jansenii is endangered.Sequencing of the genomes of M. integrifolia66,67 and M. jansenii23 has been reported. The M. integrifolia genome is based upon a domesticated variety. This genome has been updated as genome sequencing technology has advanced. The original genome was based upon short read sequencing66 and was improved by the use of some long read sequences.67 A genome based upon additional and more accurate long read data provides a further major advance.20M. jansenii has been used as a model for the study of genome sequencing and assembly.68 Recently sequences for all four species were reported20 which should allow comparative genomics to be undertaken in these species. This should lead to a greater understanding of the evolutionary relationships between the species. All four species of Macadamia can be used as parents in breeding. The small stature of M. jansenii and M. ternifolia may support the breeding of smaller trees for more intense production of Macadamia. Sequence data have defined relationships among these species.69
Genomics of other members of the Proteaceae is progressing with the sequencing of the Waratah (Telopea speciosissima) supported by the Genomics of Australian Plants Program of Bioplatforms Australia (https://www.genomicsforaustralianplants.com/) and the Nightcap Oak (Eidothea hardenianna) supported by the Threatened Species Initiative and Rewarewa (Knightia excelsa) in New Zealand.70 These species are of value as ornamentals or for timber and the analysis of their genomes will advance knowledge of the evolution and biology of the Proteaceae. Other Proteaceae species with seeds that have been considered food sources include Hicksbeachia pinnatifolia and Athertonia diversifolia. The fruits of the many Persoonia species are another source of possible new foods.
Macadamia has not been greatly altered in the brief period of domestication indicating the potential of these techniques to support new domestications and accelerated genetic improvement.
Citrus
The distribution of the Citrus genus is centred in South-East Asia. Six species are recognized in Australia and are now being targeted for genome sequencing because of their value as genetic resources for citrus improvement. These species can be crossed with domesticated citrus and are important sources of disease resistance for citrus breeding. Citrus australasica (finger-lime) is in limited cultivation and selection has produced genotypes that are early domesticates. The Genomics for Australian Plants program is supporting analysis of the genomes of the Australian species Citrus inodora, Citrus garrawayi, Citrus australasica, Citrus australis, Citrus gracilis and Citrus glauca (Desert Lime). These species have a wide range of potentially useful traits including resistance to diseases.A high-quality haplotype-resolved genome sequence has recently been reported for C. australis.22 This is probably the highest-quality genome sequencereported for a citrus species to date. The sequence has revealed the sequences of citrus-specific genes including defence genes that may be the basis of the resistance of this species to major global citrus diseases and may facilitate accelerated use in breeding of citrus to combat major disease threats. Current efforts to sequence all the other species are in progress.
Coffee
The genomes of Robusta coffee (Coffea canephora)71 and Arabica coffee (C. arabica) have been characterized due to the high value of global coffee production. The single Australian member of the genus is C. brassii from northern Australia. Only chloroplast72 has been characterized in this species. This species is not closely related to domesticated coffee72 but comparative genomics may make it useful in understanding coffee quality or caffeine content.73Eucalyptus
The eucalyptus have spread worldwide in the last 20 years to become a major global tree crop with a wide range of uses including timber, pulp and essential oil production. The two largest genera, Eucalyptus74 and Corymbia75 have sequenced genomes demonstrating a divergence around 60 million years ago. Significant conservation of synteny was found despite this long period of divergence and large differences in genome size. Reticulate evolution is widespread in this group with chloroplast exchange between diverse species being found locally.76 This may be a key feature of this very large group of plants which dominate the Australian landscape. A better understanding of the very extensive reticulate evolution in these species may be derived from genomic analysis and should inform strategies for managing and sourcing genetics for improvement of these and other plant species. The evolution of terpene synthases has been studied in this group.77 Other plants in Myrtaceae also produce oils of value. Melaleuca alternifolia has been used as a source of tea-tree oil with antiseptic applications. The terpene synthases78 that determine the unique chemistry of this product have been characterized. The food use of these genera of Myrtaceae is limited partly by this chemistry and by the absence of significant fruits.Other crop wild relatives
The Australian flora included many other species that are related to plants of economic or social importance.Grapes
Australian wild relatives of grapes include species of Cissus that may be useful rootstocks and sources of disease-resistance genes for grape production. Genetic analysis of Australian Cissus suggests distinct groups including some that may be closely related to Vitis.79 Genome analysis may provide useful information on evolution in the family relative to the well-characterized grape genome.Cotton
The Australian flora includes a diversity of wild relatives of cotton species with 17 endemic Gossypium species found in Australia.80 The Australian diploid species, Gossypium australe, has been sequenced and has important disease resistance for cotton breeding.81Bananas
Bananas (Musa) originate from just north of Australia with some species reported from north Queensland. Williams et al. (2020)82 provided evidence for the cultivation of bananas on Mabuyag Island in Torres Straight starting more than 2000 years ago. Musa acuminata ssp. Banksia is native to northern Australia and may have been domesticated in New Guinea.83 Indigenous Australians are reported to have consumed fruits in some areas and stems in others.83 Genomics of this group84,85 may provide resources for breeding disease-resistant bananas.Tomato
There are many (approximately 140) indigenous Australian species in Solanaceae that represent a genetic resource for agriculture.5 Bush tomatoes (Solanum species) have been widely consumed by indigenous people86 and may have potential for domestication or use in breeding but the genomes remain uncharacterized. While several species are edible, some require removal of toxins before consumption and genetic selection may allow this requirement to be eliminated. The related Duboisia species grown for their alkaloids have been bred by inter-specific hybridization (D. myoporoides X D. leichhardtii) and selected for high alkaloid content.87 More widespread food uses of Solanum species will require selection for low alkaloids and this should be assisted by knowledge of the biochemical pathways and gene sequences for Duboisia (currently being sequenced).Ginger and turmeric
Zingiberaceae includes 6 Australian ginger (Alpinia) species. Curcuma australasica, known as the Cape York Lily has yellow rhizomes similar to those of the related Cucuma species that are consumed as turmeric. Native cardamom (Hornstedtia scottiana) is also part of this group. The genome of Zingiber officinale, (domesticated ginger), is being characterized and should provide resources for analysis of the Australian species.Yams
Yams (Dioscorea spp.) were either naturally distributed in Australia or introduced by humans before European arrival in the 18th century.83 Resolution of the origin of these poorly characterized populations may indicate the extent to which they represent a unique genetic resource. Populations of the greater yam (Dioscorea alata) have been found in Australia, indicating human introduction, as this is considered a domesticated plant with little genetic diversity. Other Dioscorea species are widespread in Australia but may be naturally dispersed. Yam genomes have been reported88 but the Australian populations have not been examined.Taro
Taro (Colocasia esculenta) has a distribution centred in South-East Asia and is another plant suspected to have a history of human dispersal. DNA analysis has been used to study the relationships between Australian wild populations but has not resolved the role that humans may have played in dispersal.89 The related Cunjevoi (Alocasia macrorrhiza) may also have value in the development of tuber crops.Lilly pilly
Myrtaceae has many species with edible fruits. The lillipillies are a group of Syzgium species with attractive fruits. Syzygium luehmannii, the riberry or small-leafed lilly pilly has been cultivated as a “bush food” and many other related species have similar uses. For example, Austromytus dulcis (Midyim Berry) is also cultivated for its fruits. Beyond the eucalyptus, genome analysis in Myrtaceae has been limited.90 Simple sequence repeats have been used to study genetic diversity in the tea tree (Melaleuca alternifolia).91Myrtaceae shows great potential as a source of novel fruit crops.Acacias
The Acacia genus includes a large number of species that are a source of edible seeds. These seeds are currently being used in some grain-based foods. The wide distribution of Acacia species across Australia suggests that careful species selection and genetic improvement might deliver a very useful food crop that could be produced in marginal environments.Others
The cheese fruit, Morinda citrifolia, of tropical areas is species with a long history of use. Tetragonia tetragonoides, commonly called New Zealand spinach, has been widely used in other countries.Unique Australian species
The Australian flora includes many species that are unique to Australia with potential as food crops.Davidsonia
Davidsonia species, found in the rainforest of eastern Australia, have fruits that have been produced commercially on a small scale. The genetics of this genus of three species has been explored to a very limited extent using microsatellite markers92 suggesting diverse reproductive biology.93 Genome-scale analysis of this genus could explain these differences. The most recently described species, Davidsonia johnsonii, is seedless making it a potentially attractive food crop.Other unique Australian species
Many other poorly characterized species with potential for use as crops include Kakadu plum (Terminala ferdinandiana), Bunya nut (Araucaria bidwillii), Mountain pepper (Tasmannia lanecolata), Lemon Myrtle (Backhousia citriodora), Quandong (Santalum acuminatum), Saltbush (Atriplex spp), Murrnong (Microseris lanceolata), Desert Banana (Leichhardtia australis), Cooper’s clover (Trigonella sauvissima), Nardo (Marsilea drummondii), Kurumi (Tetracornia arborea), Boab (Adansonia gregorii), Onion grass (Cyperus bubosus) and figs (Ficus species).Studies targeting improved understanding of plant adaptation
Analysis of genetic variation of plants across environmental gradients can reveal the genetic basis of plant adaptation.94 Variation in grass genomes in Australia has been used to infer that climate adaptation may be associated with adaptation to a greater range of pests and diseases at higher temperatures.95 Australian wild plant populations provide many opportunities to explore plant adaptation at the genome level. As genome sequencing becomes more accessible this approach is likely to yield many new insights into natural selection that will have implications for agricultural selection.96Conservation of these genetic resources
Conservation of these diverse populations as a resource for food security will require a combination of increased in situ and ex situ conservation. Norton97 reviewed the priorities for collection for ex situ conservation. Wild populations offer the added advantage of allowing ongoing evolution and adaptation to a changing environment. The hybrid rice found in wild populations in Australia43 indicates active reticulate evolution in progress. The critical need for in situ conservation of plants in Australia to support food security is not widely recognized and will require ongoing efforts to increase public awareness.Conclusions
The Genomics of Australia Plants initiative of Bioplatforms, Australia, has begun the systematic application of genomics to the flora. Genomic analysis of Australian crop wild relatives has already defined the importance of some wild populations as genetic resources for crops. Ongoing analysis of the genomics of Australian plant species will reveal many more ways in which this extensive biodiversity can be utilized in agriculture and food production. Recent advances in technology should greatly accelerate the application of genomics to the characterization of these diverse but underutilized genetic resources. New crop options include known relatives of current crops such as Oryza meridionalis, species that were used traditionally by early Australians such as wild millets98 (the identity of species used is not yet established but is under investigation) and completely new options that will be revealed by their genomes. Genomic analysis will facilitate the improved use of species, such as Macadamia, that have already been domesticated by supporting greater use of wild germplasm in breeding. Knowledge of the genomes of other species with potential for domestication (e.g. the rice, Sorghum, millet and pulse relatives) will greatly accelerate the process in the near future by allowing molecular analysis to guide domestication using rapid methods such as gene editing.25 Genomics will also enable novel gene discovery that can contribute genes to crops to improve performance, environmental adaptation or nutritional or functional traits.96,99 Many Australian species also have wide utility beyond food crops as ornamentals, pasture, fibre, and timber species that will be more accessible with genomic tools. The history of plant use and domestication will also be illuminated. For example, the role of humans in the distribution of bananas, taro and yam in northern Australia may be further resolved by genome analysis. This research will contribute greatly to an improved understanding of the food production systems and culture of early Australians and their impact upon the environment.9,10 Greater use of Australian plant genetic resources promises to make a significant contribution to global food security by both providing new genetics for current major crops and offering new crop options. Priorities for both in situ and ex situ97 conservation will also be informed by improved characterization of the resources. The efficient genome sequencing and annotation techniques that are emerging14 will rapidly provide the information necessary to fill the large gaps in knowledge of the genetics of wild plants and have the potential to be applied globally to enhance food diversity and sustainability.Data availability
Data sharing is not applicable as no new data were generated or analysed during this study.Conflicts of interest
The author declares no conflict of interest.Acknowledgements
This research did not receive any specific funding.References
- R. J. Henry, Plant Resources for Food, Fuel, and Conservation, Earthscan, London, Sterling, VA, 2010 Search PubMed.
- M. Abberton, J. Batley, A. Bentley, J. Bryant, H. Cai, J. Cockram, A. Costa de Oliveira, L. J. Cseke, H. Dempewolf, C. De Pace, D. Edwards, P. Gepts, A. Greenland, A. E. Hall, R. Henry, K. Hori, G. T. Howe, S. Hughes, M. Humphreys, D. Lightfoot, A. Marshall, S. Mayes, H. T. Nguyen, F. C. Ogbonnaya, R. Ortiz, A. H. Paterson, R. Tuberosa, B. Valliyodan, R. K. Varshney and M. Yano, Plant Biotechnol. J., 2015, 14, 1094–1098 Search PubMed.
- R. J. Henry, Plant Genet. Resour., 2014, 12, S9–S11 CrossRef.
- P. W. Wambugu, M.-N. Ndjiondjop and R. J. Henry, Briefings Funct. Genomics, 2018, 17, 198–206 CrossRef CAS PubMed.
- Australia, Bureau of Flora and Fauna and Australian Biological Resources Study, Flora of Australia, ed. A. S. George, Australian Govt. Pub. Service, Canberra, 1982 Search PubMed.
- J.-Y. S. Yap, M. Rossetto, C. Costion, D. Crayn, R. M. Kooyman, J. Richardson and R. Henry, J. Biogeogr., 2018, 45, 838–847 CrossRef.
- C. Clarkson, Z. Jacobs, B. Marwick, R. Fullagar, L. Wallis, M. Smith, R. G. Roberts, E. Hayes, K. Lowe, X. Carah, S. A. Florin, J. McNeil, D. Cox, L. J. Arnold, Q. Hua, J. Huntley, H. E. A. Brand, T. Manne, A. Fairbairn, J. Shulmeister, L. Lyle, M. Salinas, M. Page, K. Connell, G. Park, K. Norman, T. Murphy and C. Pardoe, Nature, 2017, 547, 306 CrossRef CAS PubMed.
- B. Gammage, The Biggest Estate on Earth : How Aborigines Made Australia, Allen & Unwin, Crows Nest, N.S.W., 2011 Search PubMed.
- B. Pascoe, Dark Emu : Black Seeds : Agriculture or Accident?, Magabala Books, Broome, Western Australia, 2014 Search PubMed.
- J. M. Diamond, Guns, Germs, and Steel, Spark Pub., New York, 2003 Search PubMed.
- M. C. Westaway, D. Williams, K. Lowe, N. J. Wright, R. Kerkhove, J. Silcock, J. Gorringe, J. Miszkiewicz, R. Wood, R. Adams, T. Manne, S. Adams, T. Miscamble, J. Stout, G. D. Wrobel, J. Kemp, B. Hendry, M. Gorringe, B. Gorringe, K. Lander, S. Gorringe, I. Andrews and M. Collard, Antiquity, 2021, 95, 1043–1060 CrossRef.
- M. Rossetto, E. J. Ens, T. Honings, P. D. Wilson, J. Y. S. Yap, O. Costello, E. R. Round and C. Bowern, PLoS One, 2017, 12, e0186663 CrossRef PubMed.
- R. J. Henry, Plant Genet. Resour., 2014, 12, S9–S11 CrossRef.
- R. J. Henry, Appl. Biosci., 2022, 1, 113–128 CrossRef.
- B. Sherman and R. J. Henry, Nat. Plants, 2020, 6, 430–432 CrossRef PubMed.
- B. Sherman and R. J. Henry, Mol. Plant, 2021, 14, 701–704 CrossRef CAS PubMed.
- V. Murigneux, S. K. Rai, A. Furtado, T. J. C. Bruxner, W. Tian, I. Harliwong, H. M. Wei, B. C. Yang, Q. Y. Ye, E. Anderson, Q. Mao, R. Drmanac, O. Wang, B. A. Peters, M. Y. Xu, P. Wu, B. Topp, L. J. M. Coin and R. J. Henry, Gigascience, 2020, 9, giaa146 CrossRef PubMed.
- R. Henry, Advances in Botanical Research Incorporating Advances in Plant Pathology, 2001, vol. 34, pp. 23–57 Search PubMed.
- M. Brozynska, A. Furtado and R. J. Henry, Plant Biotechnol. J., 2015, 15, 765–774 CrossRef PubMed.
- A. O. Sharma P, B. Alsubaie, I. Al-Mssallem, O. Nath, N. Mitter, A. M. G. Rodrigues, B. Topp, V. Murigneux, A. Kharabian Masouleh, A. Furtado and R. J. Henry, BioXriv, 2021,preprint, DOI:10.1101/2021.01.22.427724.
- P. Sharma, A. K. Masouleh, B. Topp, A. Furtado and R. J. Henry, Plant J., 2022, 109, 727–736 CrossRef CAS PubMed.
- U. Nakandala, A. K. Masouleh, M. W. Smith, A. Furtado, P. Mason, L. Constantin and R. J. Henry, bioRxiv, 2022, preprint, DOI:10.1101/2022.12.20.521315.
- P. Sharma, V. Murigneux, J. Haimovitz, C. J. Nock, W. Tian, A. K. Masouleh, B. Topp, M. Alam, A. Furtado and R. J. Henry, Plant Direct, 2021, 5, e364 CrossRef CAS PubMed.
- M. Brozynska, A. Furtado and R. J. Henry, Plant Biotechnol. J., 2016, 14, 1070–1085 CrossRef CAS PubMed.
- O. P. Abdulla M, A. Furtado and R. J. Henry, Front. genome ed., 2022, 875243, DOI:10.3389/fgeed.2022.875243.
- P. W. Wambugu, D. Nyamongo, M. N. Ndjiondjop and R. J. Henry, Compend Pl Genome, 2018, DOI:10.1007/978-3-319-71997-9_3, pp. 41–54.
- R. Henry, N. Rice, D. Waters, S. Kasem, R. Ishikawa, Y. Hao, S. Dillon, D. Crayn, R. Wing and D. Vaughan, Rice, 2009, 3, 235–241 CrossRef.
- P. W. Wambugu and R. J. Henry, Compend Pl Genome, 2018, DOI:10.1007/978-3-319-71997-9_6, pp. 67–74.
- A. M. Moner and R. J. Henry, Compend Pl Genome, 2018, DOI:10.1007/978-3-319-71997-9_16, pp. 177–182.
- D. L. E. Waters, C. J. Nock, R. Ishikawa, N. Rice and R. J. Henry, Ecol. Evol., 2012, 2, 211–217 CrossRef PubMed.
- M. Brozynska, E. S. Omar, A. Furtado, D. Crayn, B. Simon, R. Ishikawa and R. J. Henry, Trop. Plant Biol., 2014, 7, 111–120 CrossRef CAS PubMed.
- R. J. Henry, Compend Pl Genome, 2018, DOI:10.1007/978-3-319-71997-9_5, pp. 61–66.
- T. K. Mondal and R. J. Henry, Compend Pl Genome, 2018, DOI:10.1007/978-3-319-71997-9, pp. Xiii-Xv.
- J. C. Stein, Y. Yu, D. Copetti, D. J. Zwickl, L. Zhang, C. J. Zhang, K. Chougule, D. Y. Gao, A. Iwata, J. L. Goicoechea, S. R. Wei, J. Wang, Y. Liao, M. H. Wang, J. Jacquemin, C. Becker, D. Kudrna, J. W. Zhang, C. E. M. Londono, X. Song, S. Lee, P. Sanchez, A. Zuccolo, J. S. S. Ammiraju, J. Talag, A. Danowitz, L. F. Rivera, A. R. Gschwend, C. Noutsos, C. C. Wu, S. M. Kao, J. W. Zeng, F. J. Wei, Q. Zhao, Q. Feng, M. El Baidouri, M. C. Carpentier, E. Lasserre, R. Cooke, D. D. Farias, L. C. da Maia, R. S. dos Santos, K. G. Nyberg, K. L. McNally, R. Mauleon, N. Alexandrov, J. Schmutz, D. Flowers, C. Z. Fan, D. Weigel, K. K. Jena, T. Wicker, M. S. Chen, B. Han, R. Henry, Y. I. C. Hsing, N. Kurata, A. C. de Oliveira, O. Panaud, S. A. Jackson, C. A. Machado, M. J. Sanderson, M. Y. Long, D. Ware and R. A. Wing, Nat. Genet., 2018, 50, 285 CrossRef CAS PubMed.
- R. J. Henry, Mol. Plant, 2022, 15, 563–565 CrossRef CAS PubMed.
- M. Brozynska, D. Copetti, A. Furtado, R. A. Wing, D. Crayn, G. Fox, R. Ishikawa and R. J. Henry, Plant Biotechnol. J., 2017, 15, 765–774 CrossRef CAS PubMed.
- J. C. Stein, Y. Yu, D. Copetti, D. J. Zwickl, L. Zhang, C. Zhang, K. Chougule, D. Gao, A. Iwata, J. L. Goicoechea, S. Wei, J. Wang, Y. Liao, M. Wang, J. Jacquemin, C. Becker, D. Kudrna, J. Zhang, C. E. M. Londono, X. Song, S. Lee, P. Sanchez, A. Zuccolo, J. S. S. Ammiraju, J. Talag, A. Danowitz, L. F. Rivera, A. R. Gschwend, C. Noutsos, C.-c. Wu, S.-m. Kao, J.-w. Zeng, F.-j. Wei, Q. Zhao, Q. Feng, M. El Baidouri, M.-C. Carpentier, E. Lasserre, R. Cooke, D. d. R. Farias, L. C. da Maia, R. S. dos Santos, K. G. Nyberg, K. L. McNally, R. Mauleon, N. Alexandrov, J. Schmutz, D. Flowers, C. Fan, D. Weigel, K. K. Jena, T. Wicker, M. Chen, B. Han, R. Henry, Y.-i. C. Hsing, N. Kurata, A. C. de Oliveira, O. Panaud, S. A. Jackson, C. A. Machado, M. J. Sanderson, M. Long, D. Ware and R. A. Wing, Nat. Genet., 2018, 50, 1618 CrossRef CAS PubMed.
- A. M. Moner, A. Furtado and R. J. Henry, Mol. Phylogenet. Evol., 2018, 127, 475–487 CrossRef CAS PubMed.
- A. M. Moner, A. Furtado and R. J. Henry, BMC Plant Biol., 2020, 20, 472 CrossRef CAS PubMed.
- B. Piegu, R. Guyot, N. Picault, A. Roulin, A. Saniyal, H. Kim, K. Collura, D. S. Brar, S. Jackson, R. A. Wing and O. Panaud, Genome Res., 2006, 16, 1262–1269 CrossRef CAS PubMed.
- A. L. Phillips, S. Ferguson, N. S. Watson-Haigh, A. W. Jones, J. O. Borevitz, R. A. Burton and B. J. Atwell, Sci. Rep., 2022, 12, 10823 CrossRef CAS PubMed.
- R. J. Henry, Front. Plant Sci., 2019, 10, 10354 Search PubMed.
- A. M. Moner, A. Furtado, I. Chivers, G. Fox, D. Crayn and R. J. Henry, Ecol. Evol., 2018, 8, 4360–4366 CrossRef PubMed.
- S. Malory, F. M. Shapter, M. S. Elphinstone, I. H. Chivers and R. J. Henry, Plant Biotechnol. J., 2011, 9, 1131–1140 CrossRef CAS PubMed.
- F. M. Shapter, M. Cross, G. Ablett, S. Malory, I. H. Chivers, G. J. King and R. J. Henry, PLoS One, 2013, 8, e82641 CrossRef PubMed.
- M. Abedinia, R. Henry and S. Clark, Genet. Resour. Crop Evol., 1998, 45, 399–406 CrossRef.
- S. L. Dillon, P. K. Lawrence, R. J. Henry and H. J. Price, Plant Syst. Evol., 2007, 268, 29–43 CrossRef.
- G. K. S. Ananda, H. Myrans, S. L. Norton, R. Gleadow, A. Furtado and R. J. Henry, Front. Plant Sci., 2020, 11, 01108 CrossRef PubMed.
- S. Dillon, P. Lawrence and R. Henry, Plant Syst. Evol., 2004, 249, 233–246 CrossRef.
- Y. Chen, Y. B. Zhang, H. J. Wang, J. Sun, L. C. Ma, F. H. Miao, Z. X. Zhang, Y. Cheng, J. W. Huang, G. F. Yang and Z. Y. Wang, Front. Genet., 2022, 13, 844385 CrossRef CAS PubMed.
- G. Ananda, S. Norton, C. Blomstedt, A. Furtado, B. Moller, R. Gleadow and R. Henry, Plant Genome, 2021, e20123, DOI:10.1002/tpg2.20123.
- G. K. S. Ananda, S. L. Norton, E. Barnes, A. Furtado, B. L. Moller, R. Gleadow and R. J. Henry, Genet. Resour. Crop Evol., 2022, 558 Search PubMed.
- O. Garsmeur, G. Droc, R. Antonise, J. Grimwood, B. Potier, K. Aitken, J. Jenkins, G. Martin, C. Charron, C. Hervouet, L. Costet, N. Yahiaoui, A. Healey, D. Sims, Y. Cherukuri, A. Sreedasyam, A. Kilian, A. Chan, M.-A. Van Sluys, K. Swaminathan, C. Town, H. Berges, B. Simmons, J. C. Glaszmann, E. van der Vossen, R. Henry, J. Schmutz and A. D’Hont, Nat. Commun., 2018, 9, 2638 CrossRef PubMed.
- S. Dillon, F. Shapter and R. Henry, Ann. Bot., 2007, 100, 975–989 CrossRef PubMed.
- K. Robinson, L. W. Bell, R. G. Bennett, D. A. Henry, M. Tibbett and M. H. Ryan, Aust. J. Exp. Agric., 2007, 47, 170–176 CrossRef.
- L. W. Bell, M. H. Ryan, R. G. Bennett, M. T. Collins and H. J. Clarke, J. Sci. Food Agric., 2012, 92, 1354–1361 CrossRef CAS PubMed.
- L. W. Bell, R. G. Bennett, M. H. Ryan and H. Clarke, Renew. Agric. Food Syst., 2011, 26, 72–91 CrossRef.
- R. J. Lawn and G. J. Rebetzke, Aust. J. Agric. Res., 2006, 57, 119–132 CrossRef.
- R. J. Lawn and A. E. Holland, Aust. J. Bot., 2003, 51, 295–308 CrossRef.
- T. Grant, R. J. Lawn and L. M. Bielig, Aust. J. Agric. Res., 2003, 54, 243–250 CrossRef.
- R. K. Varshney, W. B. Chen, Y. P. Li, A. K. Bharti, R. K. Saxena, J. A. Schlueter, M. T. A. Donoghue, S. Azam, G. Y. Fan, A. M. Whaley, A. D. Farmer, J. Sheridan, A. Iwata, R. Tuteja, R. V. Penmetsa, W. Wu, H. D. Upadhyaya, S. P. Yang, T. Shah, K. B. Saxena, T. Michael, W. R. McCombie, B. C. Yang, G. Y. Zhang, H. M. Yang, J. Wang, C. Spillane, D. R. Cook, G. D. May, X. Xu and S. A. Jackson, Nat. Biotechnol., 2012, 30, 83–U128 CrossRef CAS PubMed.
- M. T. Kassa, R. V. Penmetsa, N. Carrasquilla-Garcia, B. K. Sarma, S. Datta, H. D. Upadhyaya, R. K. Varshney, E. J. B. von Wettberg and D. R. Cook, PLoS One, 2012, 7, e39563 CrossRef CAS PubMed.
- P. Vanambathina, R. C. N. Rachaputi, R. J. Henry and S. L. Norton, Genet. Resour. Crop Evol., 2019, 66, 1699–1712 CrossRef CAS.
- S. Chang, C. S. Thurber, P. J. Brown, G. L. Hartman, K. N. Lambert and L. L. Domier, PLoS One, 2014, 9, e99427 CrossRef PubMed.
- R. S. Harbert, A. H. D. Brown and J. J. Doyle, Am. J. Bot., 2014, 101, 710–721 CrossRef PubMed.
- C. J. Nock, A. Baten, B. J. Barkla, A. Furtado, R. J. Henry and G. J. King, BMC Genomics, 2016, 17, 937 CrossRef PubMed.
- C. J. Nock, A. Baten, R. Mauleon, K. S. Langdon, B. Topp, C. Hardner, A. Furtado, R. J. Henry and G. J. King, G3: Genes, Genomes, Genet., 2020, 10, 3497–3504 CrossRef CAS PubMed.
- V. Murigneux, S. K. Rai, A. Furtado, T. J. C. Bruxner, W. Tian, I. Harliwong, H. Wei, B. Yang, Q. Ye, E. Anderson, Q. Mao, R. Drmanac, O. Wang, B. A. Peters, M. Xu, P. Wu, B. Topp, L. J. M. Coin and R. J. Henry, Gigascience, 2020, 9, giaa146 CrossRef PubMed.
- T. Mai, M. Alam, C. Hardner, R. Henry and B. Topp, Plants, 2020, 9, 714 CrossRef CAS PubMed.
- A. M. McCartney, E. Hilario, S. S. Choi, J. Guhlin, J. M. Prebble, G. Houliston, T. R. Buckley and D. Chagne, Mol. Ecol. Resour., 2021, 21, 2125–2144 CrossRef PubMed.
- F. Denoeud, L. Carretero-Paulet, A. Dereeper, G. Droc, R. Guyot, M. Pietrella, C. Zheng, A. Alberti, F. Anthony, G. Aprea, J.-M. Aury, P. Bento, M. Bernard, S. Bocs, C. Campa, A. Cenci, M.-C. Combes, D. Crouzillat, C. Da Silva, L. Daddiego, F. De Bellis, S. Dussert, O. Garsmeur, T. Gayraud, V. Guignon, K. Jahn, V. Jamilloux, T. Joet, K. Labadie, T. Lan, J. Leclercq, M. Lepelley, T. Leroy, L.-T. Li, P. Librado, L. Lopez, A. Munoz, B. Noel, A. Pallavicini, G. Perrotta, V. Poncet, D. Pot, Priyono, M. Rigoreau, M. Rouard, J. Rozas, C. Tranchant-Dubreuil, R. VanBuren, Q. Zhang, A. C. Andrade, X. Argout, B. Bertrand, A. de Kochko, G. Graziosi, R. J. Henry, R. M. Jayarama, C. Nagai, S. Rounsley, D. Sankoff, G. Giuliano, V. A. Albert, P. Wincker and P. Lashermes, Science, 2014, 345, 1181–1184 CrossRef CAS PubMed.
- C. Guyeux, J. C. Charr, H. T. M. Tran, A. Furtado, R. J. Henry, D. Crouzillat, R. Guyot and P. Hamon, PLoS One, 2019, 14, e0216347 CrossRef CAS PubMed.
- B. Cheng, A. Furtado, H. E. Smyth and R. J. Henry, Trends Food Sci. Technol., 2016, 57, 20–30 CrossRef CAS.
- A. A. Myburg, D. Grattapaglia, G. A. Tuskan, U. Hellsten, R. D. Hayes, J. Grimwood, J. Jenkins, E. Lindquist, H. Tice, D. Bauer, D. M. Goodstein, I. Dubchak, A. Poliakov, E. Mizrachi, A. R. K. Kullan, S. G. Hussey, D. Pinard, K. Van der Merwe, P. Singh, I. Van Jaarsveld, O. B. Silva, R. C. Togawa, M. R. Pappas, D. A. Faria, C. P. Sansaloni, C. D. Petroli, X. H. Yang, P. Ranjan, T. J. Tschaplinski, C. Y. Ye, T. Li, L. Sterck, K. Vanneste, F. Murat, M. Soler, H. S. Clemente, N. Saidi, H. Cassan-Wang, C. Dunand, C. A. Hefer, E. Bornberg-Bauer, A. R. Kersting, K. Vining, V. Amarasinghe, M. Ranik, S. Naithani, J. Elser, A. E. Boyd, A. Liston, J. W. Spatafora, P. Dharmwardhana, R. Raja, C. Sullivan, E. Romanel, M. Alves-Ferreira, C. K. Lheim, W. Foley, V. Carocha, J. Paiva, D. Kudrna, S. H. Brommonschenkel, G. Pasquali, M. Byrne, P. Rigault, J. Tibbits, A. Spokevicius, R. C. Jones, D. A. Steane, R. E. Vaillancourt, B. M. Potts, F. Joubert, K. Barry, G. J. Pappas, S. H. Strauss, P. Jaiswal, J. Grima-Pettenati, J. Salse, Y. Van de Peer, D. S. Rokhsar and J. Schmutz, Nature, 2014, 510, 356 CrossRef CAS PubMed.
- A. L. Healey, M. Shepherd, G. J. King, J. B. Butler, J. S. Freeman, D. J. Lee, B. M. Potts, O. B. Silva-Junior, A. Baten, J. Jenkins, S. Shu, J. T. Lovell, A. Sreedasyam, J. Grimwood, A. Furtado, D. Grattapaglia, K. W. Barry, H. Hundley, B. A. Simmons, J. Schmutz, R. E. Vaillancourt and R. J. Henry, Commun. Biol., 2021, 4, 537 CrossRef CAS PubMed.
- A. Healey, D. J. Lee, A. Furtado and R. J. Henry, Aust. J. Bot., 2018, 66, 369–378 CrossRef.
- J. B. Butler, J. S. Freeman, B. M. Potts, R. E. Vaillancourt, D. Grattapaglia, O. B. Silva-Junior, B. A. Simmons, A. L. Healey, J. Schmutz, K. W. Barry, D. J. Lee, R. J. Henry, G. J. King, A. Baten and M. Shepherd, Heredity, 2018, 121, 87–104 CrossRef CAS PubMed.
- D. Shelton, D. Leach and P. Baverstock, Plant Sci., 2002, 162, 9–15 CrossRef CAS.
- M. Rossetto, B. Jackes and K. Scott, Syst. Bot., 2002, 27, 522–533 Search PubMed.
- P. A. Fryxell, L. A. Craven and J. M. Stewart, Syst. Bot., 1992, 17, 91–114 CrossRef.
- Y. F. Cai, X. Y. Cai, Q. L. Wang, P. Wang, Y. Zhang, C. W. Cai, Y. C. Xu, K. B. Wang, Z. L. Zhou, C. X. Wang, S. P. Geng, B. Li, Q. Dong, Y. Q. Hou, H. Wang, P. Ai, Z. Liu, F. F. Yi, M. S. Sun, G. Y. An, J. R. Cheng, Y. Y. Zhang, Q. Shi, Y. H. Xie, X. Y. Shi, Y. Chang, F. F. Huang, Y. Chen, S. M. Hong, L. Y. Mi, Q. Sun, L. Zhang, B. L. Zhou, R. H. Peng, X. Zhang and F. Liu, Plant Biotechnol. J., 2020, 18, 814–828 CrossRef CAS PubMed.
- R. N. Williams, D. Wright, A. Crowther and T. Denham, Nat. Ecol. Evol., 2020, 4, 1342 CrossRef PubMed.
- T. Denham, M. Donohue and S. Booth, Antiquity, 2009, 83, 634–648 CrossRef.
- A. D'Hont, F. Denoeud, J. M. Aury, F. C. Baurens, F. Carreel, O. Garsmeur, B. Noel, S. Bocs, G. Droc, M. Rouard, C. Da Silva, K. Jabbari, C. Cardi, J. Poulain, M. Souquet, K. Labadie, C. Jourda, J. Lengelle, M. Rodier-Goud, A. Alberti, M. Bernard, M. Correa, S. Ayyampalayam, M. R. McKain, J. Leebens-Mack, D. Burgess, M. Freeling, D. Mbeguie-A-Mbeguie, M. Chabannes, T. Wicker, O. Panaud, J. Barbosa, E. Hribova, P. Heslop-Harrison, R. Habas, R. Rivallan, P. Francois, C. Poiron, A. Kilian, D. Burthia, C. Jenny, F. Bakry, S. Brown, V. Guignon, G. Kema, M. Dita, C. Waalwijk, S. Joseph, A. Dievart, O. Jaillon, J. Leclercq, X. Argout, E. Lyons, A. Almeida, M. Jeridi, J. Dolezel, N. Roux, A. M. Risterucci, J. Weissenbach, M. Ruiz, J. C. Glaszmann, F. Quetier, N. Yahiaoui and P. Wincker, Nature, 2012, 488, 213 CrossRef PubMed.
- A. D'Hont, A. Paget-Goy, J. Escoute and F. Carreel, Theor. Appl. Genet., 2000, 100, 177–183 CrossRef.
- L. S. Lee, Rangel., 2012, 34, 359–373 CrossRef.
- S. F. Ullrich, N. J. H. Averesch, L. Castellanos, Y. H. Choi, A. Rothauer and O. Kayser, Phytochemistry, 2016, 131, 44–56 CrossRef CAS PubMed.
- Y. Sugihara, K. Darkwa, H. Yaegashi, S. Natsume, M. Shimizu, A. Abe, A. Hirabuchi, K. Ito, K. Oikawa, M. Tamiru-Oli, A. Ohta, R. Matsumoto, P. Agre, D. De Koeyer, B. Pachakkil, S. Yamanaka, S. Muranaka, H. Takagi, B. White, R. Asiedu, H. Innan, A. Asfaw, P. Adebola and R. Terauchi, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 31987–31992 CrossRef CAS PubMed.
- H. V. Hunt, H. M. Moots and P. J. Matthews, Genet. Resour. Crop Evol., 2013, 60, 1695–1707 CrossRef.
- D. Grattapaglia, R. E. Vaillancourt, M. Shepherd, B. R. Thumma, W. Foley, C. Kulheim, B. M. Potts and A. A. Myburg, Tree Genet. Genomes, 2012, 8, 463–508 CrossRef.
- M. Rossetto, R. Slade and P. Baverstock, Mol. Ecol., 1999, 8, 633–643 CrossRef CAS PubMed.
- F. G. Eliott, C. Connelly, M. Rossetto, M. Shepherd, N. Rice and R. J. Henry, Conserv. Genet. Resour., 2013, 5, 161–164 CrossRef.
- F. G. Eliott, M. Shepherd, M. Rossetto, P. Bundock, N. Rice and R. J. Henry, Aust. J. Bot., 2014, 62, 451–464 CrossRef.
- J. Cronin, P. Bundock and R. Henry, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2773–2778 CrossRef CAS PubMed.
- T. L. Fitzgerald, F. M. Shapter, S. McDonald, D. L. E. Waters, I. H. Chivers, A. Drenth, E. Nevo and R. J. Henry, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 21140–21145 CrossRef CAS PubMed.
- R. J. Henry and E. Nevo, Plant Biotechnol. J., 2014, 12, 655–662 CrossRef PubMed.
- S. L. Norton, C. K. Khoury, C. C. Sosa, N. P. Castaneda-Alvarez, H. A. Achicanoy and S. Sotelo, Aust. J. Bot., 2017, 65, 638–645 CrossRef.
- H. T. Stalker, M. Warburton and J. R. Harlan, Harlan's Crops and Man : People, Plants and Their Domestication, John Wiley & Sons American Society of Agronomy Crop Science Society of America Soil Science Society of America, Inc., Hoboken, NJ, Madison, 3rd edn, 2021 Search PubMed.
- R. J. Henry, Front. Plant Sci., 2014, 5, 00068 Search PubMed.
- P. Vanambathina, R. J. Henry, R. C. N. Rachaputi and A. Furtado, Ann. Appl. Biol., 2022, 180, 259–272 CrossRef CAS.
- G. A. Wu, J. Terol, V. Ibanez, A. Lopez-Garcia, E. Perez-Roman, C. Borreda, C. Domingo, F. R. Tadeo, J. Carbonell-Caballero, R. Alonso, F. Curk, D. L. Du, P. Ollitrault, M. L. Roose, J. Dopazo, F. G. Gmitter, D. S. Rokhsar and M. Talon, Nature, 2018, 554, 311 CrossRef CAS PubMed.
- M. WHEELER, L. LEE and R. HENRY, Genet. Resour. Crop Evol., 2001, 48, 483–497 CrossRef.
- D. G. Conover and D. R. Geiger, Aust. J. Plant Physiol., 1984, 11, 395–408 Search PubMed.
| This journal is © The Royal Society of Chemistry 2023 |
