Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The materials experiment knowledge graph†
Michael J.
Statt
*a,
Brian A.
Rohr
*a,
Dan
Guevarra
bc,
Ja'Nya
Breeden‡
c,
Santosh K.
Suram
 d and
John M.
Gregoire
d and
John M.
Gregoire
 *bc
*bc
aModelyst LLC, Palo Alto, CA 94306, USA. E-mail: brian.rohr@modelyst.io; michael.statt@modelyst.io
bDivision of Engineering and Applied Science, California Institute of Technology, Pasadena, CA 91125, USA. E-mail: gregoire@caltech.edu
cLiquid Sunlight Alliance, California Institute of Technology, Pasadena, CA 91125, USA
dToyota Research Institute, Los Altos, CA 94022, USA
First published on 28th June 2023
Abstract
Materials knowledge is inherently hierarchical. While high-level descriptors such as composition and structure are valuable for contextualizing materials data, the data must ultimately be considered in the context of its low-level acquisition details. Graph databases offer an opportunity to represent hierarchical relationships among data, organizing semantic relationships into a knowledge graph. Herein, we establish a knowledge graph of materials experiments whose construction encodes the complete provenance of each material sample and its associated experimental data and metadata. Additional relationships among materials and experiments further encode knowledge and facilitate data exploration. We illustrate the Materials Experiment Knowledge Graph (MekG) using several use cases, demonstrating the value of modern graph databases for the enterprise of data-driven materials science.
The materials community has envisioned a new paradigm in materials discovery wherein experiment automation and the integration of human and machine intelligence accelerate materials research to enable new technologies that address a range of societal needs.1–5 This vision is being realized in specific areas of materials research via advancements in high throughput computation, experiment automation, and artificial intelligence.6–9 Continued evolution of accelerated discovery efforts will require methods to aggregate data and knowledge from a diverse set of sources. Recent advancements for specific sources and domains of materials data include integration of computational databases via the JARVIS project10 and aggregation of perovskite solar cell data.11 Data management projects with a broader scope include the Materials Data Facility,12,13 which enables materials researchers to submit and annotate datasets.
Scientific knowledge and the discoveries that it provide are the result of cyclic learning. Scientific discovery can thus be accelerated by improving the quality and/or the frequency of learning cycles. Bolstered by the availability of machine learning methods to learn from an ever-expanding dataset, the autonomous or closed-loop approach to experiment automation focuses on increasing the frequency of learning cycles. Initial examples of autonomous operation of such learning cycles have been naturally limited to optimization of performance in a low-dimensional parameters space. Bolstered by these successes, the community is poised to broaden the purview of autonomous learning cycles, which places new constraints on both the breadth of knowledge that must be encoded and the speed of data exploration provided by the in-loop data store. The inherent challenges of managing a diverse set of data streams and establishing a performant data store for autonomous research are compounded by the historical dearth of research in establishing materials data infrastructure.2,14,15 Herein, we describe the use of graph databases to improve the management of data from materials experiments, provide scalability with respect to data diversity and quantity, and enable data exploration at a speed commensurate with autonomous execution of learning cycles.
Computational materials databases can track the origin of data entries via annotations of the code repository used to generate the data along with specific metadata describing the computational methods. The analogue of this metadata for experimental materials science is far more complex due to the broad range of instruments and their settings, reagents and their purities, etc. Perhaps most foundationally, the data resulting from materials experiments is often sensitive to the order of the experimental steps. Consequently, data management schema must encode the experiment provenance to uniquely represent a piece of experimental data. Recording experiment provenance is inherent to automated experiment workflows that track samples and record timestamps of experiments.16–19 Other strategies for provenance management have been introduced for spectroscopy experiments20 and augmented with facile metadata management.21 Our approach to this challenge is to recognize the experimental events as the data source, resulting in the Event Sourced Architecture for Materials Provenance Management (ESAMP).22
To facilitate ingestion of a variety of data sources and automate some aspects of data validation, we implemented ESAMP with a Structured Query Language (SQL) database. The sequence of experimental steps is most naturally modelled as a directed graph, and in the present work we demonstrate a graph database that encodes experiment provenance along with a variety of other relationships. The graph approach to modelling experiment sequences has been primarily applied in the field of chemical synthesis.23–26 The MekG extends this concept to span synthesis, processing, and characterization experiments, while additionally encoding other relationships that facilitate knowledge representation in general, and data exploration in particular. Every node, edge, and node tuple in the database follows the structure of subject, relationship, object, where the relationship is generally presented as a verb unless such representation would make it overly verbose or unclear.
We recently published the Materials Provenance Store (MPS),27 a database built with the ESAMP SQL schema based on the file-system organization of experimental provenance data from MEAD.18 In the present work, we ingested MPS into a neo4j database (see Code availability), in which there is a node for each material “Sample”, for each experiment “Process”, and for each “Sample-Process”, which is the application of a Process to a Sample. The experiment provenance for a given sample is encoded through directed edges of type “Next” that connect Sample-Process nodes. Additional nodes for collections of samples, details of each process, data files produced by processes, and analysis results are linked with edges derived from foreign keys in the SQL-based MPS database. We then add additional relationships, such as edges between Element nodes and Sample nodes as well as between pH nodes and electrochemical Process nodes. The encoded knowledge can be further expanded via additional relationships to facilitate data exploration, and relationships can extend to organizational knowledge such as project funding, intended research goal, and relevance to a publication.
The MekG contains a total of 52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 263
263![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 968 nodes and 111
968 nodes and 111![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 430
430![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 058 edges, a scale of data enabled by high throughput experimental synthesis of 11
058 edges, a scale of data enabled by high throughput experimental synthesis of 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 243
243![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 172 unique Samples, execution of 30
172 unique Samples, execution of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 656
656![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 368 Sample-Processes, and ensuing data analysis, as summarized for MPS.27 MekG contains 10 types of nodes (entity types) and 10 types of edge (relationship types), which are summarized along with the respective number of occurrences in the ESI.† The Samples were primarily synthesized by either combinatorial sputter deposition or inkjet printing, In addition to these synthesis Processes, a suite of optical, electrochemical, and standard materials characterization techniques were performed, with the most populous Process for performance characterization being the electrochemical evaluation of catalytic activity for the oxygen evolution reaction (OER). To illustrate the performance and utility of MekG, we present 4 use cases, commencing with the most general applications, (i) graphical exploration of data and (ii) data retrieval via queries. We then describe specific implementation of database queries to (iii) automate design of experiments and (iv) evaluate a hypothesis from crowd-sourced data.
368 Sample-Processes, and ensuing data analysis, as summarized for MPS.27 MekG contains 10 types of nodes (entity types) and 10 types of edge (relationship types), which are summarized along with the respective number of occurrences in the ESI.† The Samples were primarily synthesized by either combinatorial sputter deposition or inkjet printing, In addition to these synthesis Processes, a suite of optical, electrochemical, and standard materials characterization techniques were performed, with the most populous Process for performance characterization being the electrochemical evaluation of catalytic activity for the oxygen evolution reaction (OER). To illustrate the performance and utility of MekG, we present 4 use cases, commencing with the most general applications, (i) graphical exploration of data and (ii) data retrieval via queries. We then describe specific implementation of database queries to (iii) automate design of experiments and (iv) evaluate a hypothesis from crowd-sourced data.
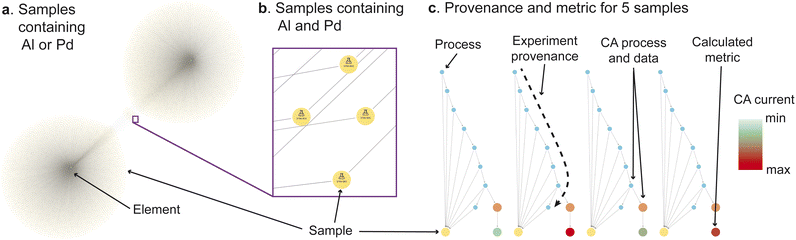
Human researchers possess domain expertise combined with intuition from their aggregated prior knowledge, both of which are unrivaled by machine learning to-date. Machine learning thrives in its scalability to large datasets that exceed the memory capabilities of a typical human. The MekG can assist the human in exploration of such large datasets through intuitive visualizations. Fig. 1 shows images of the MekG at select moments during a graphical data exploration exercise, for which the full video is available in the MekG-migrations repository (see Code availability). This interactive visualisation demo commences with viewing all samples that contain Pd or Al (Fig. 1a), focusing on samples that contain both (Fig. 1b), and then viewing their experiment provenances (Fig. 1c). In this last step, the sub-graph for each sample is expanded to show the analyzed electrochemical current density, for which a color legend is assigned to demonstrate simultaneous visualization of performance and experiment provenance.
Another mode of exploration, applicable to equally to human and machine users, is data exploration via queries. We developed the following set of queries to include a synthesis-based search, a synthesis and measurement-based search, a provenance-based search, and a provenance-based search conditioned on analysis results: (1) find samples annealed at 350 °C; (2) find all electrochemistry measurements performed on a sample that contains both Bi and V; (3) find all provenances wherein a sample was synthesized by inkjet printing and whose first 2 electrochemistry measurements were chronopotentiometry measurements at 0.03 and 0.1 mA, respectively, each with a duration between 7 and 15 s; and (4) find all provenances that contain a sequence of 5 electrochemistry experiments in NaOH-based electrolyte wherein the first 4 experiments were each chronoamperometry measurements that produced a measured current above 10−7, 10−8, 10−9, and 10−10 A, respectively, and the final electrochemistry experiment was a cyclic voltammogram that produced a maximum measured current above 10−6 A. The query execution times are summarized in Table 1, demonstrating the excellent performance of the graph-based query across a breadth of query types. For query 1, where the requisite data is indexed in a single SQL table, the SQL-based query is naturally the fastest. For provenance-based queries, the graph-based queries are several times faster than the SQL-based queries. More drastically, the complexity of query 4 revealed a marked difference in query preparation time. While the graph-based query was written in a matter of minutes, initial attempts at writing the SQL query resulted in query timeout after 104 s. Multiple days of human effort were required to obtain a query time within a factor of 5 of the graph-based query, which is reflected in the relative complexity of the queries (see ESI†). Our conclusion from this exercise is not that graph databases universally outperform the other data management methods with respect to query execution, but rather that the graph-based queries are sufficiently fast for real-time data exploration and can be achieved with intuitive query expressions that avoid complex query engineering. Furthermore, even though the underlying schema in the graph database and SQL database are nearly identical, we found the graph schema more intuitive than the SQL schema, both with respect to visual and computational exploration of the data and with respect to the insertion of additional entities and relationships to further encode knowledge.
| Query description: (type, criteria) | Execution time (s) | Num. results | ||
|---|---|---|---|---|
| Graph | SQL | File-Sys | ||
| a Query times were in excess of 104 s prior to extension query optimization. | ||||
| Sample, annealed at 350 °C | 54 | 12 | 306 | 5 × 105 |
| Process, echem on Bi–V samples | 15 | 36 | 365 | 9 × 104 |
| Provenance, process criteria | 12 | 83 | 480 | 2 × 104 |
| Provenance, many criteria | 108 | 523a | N/A | 2 × 102 |
As a moderately complex provenance-based query, query 3 was chosen to characterize how query time scales with data size. To achieve representative databases of smaller size, 3 sub-databases were created using the earliest 1/8, 1/4, and 1/2 of the Sample-Processes in the MPS, followed by removal of all orphaned samples, processes, analyses, etc. (see ESI†). Running query 3 on these databases informs us of how long the query would have taken if it had been performed at these various points in the lab's sequence of experiments. The results for graph and SQL-based version of query 3 are shown in Fig. 2, which illustrates the excellent relative performance of the graph-based query across all data sizes as well as a favorable power-law scaling relationship for the graph-based query. Extrapolating to a database with a billion Sample-Processes, the scaling law provides a projected query execution time of 65 s, illustrating the promise of graph database for aggregating large swaths of materials chemistry data while maintaining operability for both humans and machines.
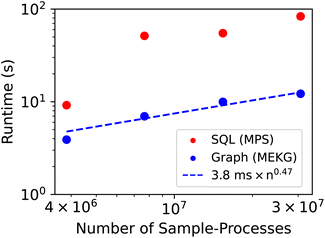 | ||
| Fig. 2 Using query 3 from Table 1, the query times for the graph-based query (MekG) and SQL-based query (MPS) are shown using each full database as well as 3 sub-databases with 1/8, 1/4, and 1/2 of the Sample-Processes. The dashed line shows the scaling law from the graph-based query determined via linear regression of the log-scaled data points, where n is the number of Sample-Processes. | ||
Our third use case involves the automated design of experiments, in particular the selection of OER catalysts that merit further investigation based on prior data. Sequential learning in closed-loop experimentation typically involves the design of a single acquisition from a collection of available experiments, a small-scope experiment design intended to iterate many times per day. Traditional human-executed learning cycles have a broad scope, typically occurring over the course of many days. Here, we consider the automated planning of experiments for a single batch of high throughput experiments that can be executed in a few hours. Electrocatalytic activity for the OER varies substantial with not only the catalyst composition and structure, but also the electrolyte, especially the electrolyte pH. While high throughput experimentation has amassed catalyst screening data, these cover a small fraction of all possible combinations of catalysts and electrolytes. We thus consider a automated design of experiments for choosing which catalysts available in the lab should be tested in a given electrolyte. While machine learning models could be invoked for this prediction, we simplify the design process to keep focus on the role of the MekG. We previously demonstrated a correlation of OER activity in pH 3 and pH 7 electrolytes among metal oxide catalysts,28 which helps define a simple design-of-experiments strategy. We conduct 2 queries, one to establish the catalysts screened in pH 7 but not pH 3 electrolyte, and a second to establish which catalysts have already been synthesized but not yet electrochemically tested. Evaluating the query results provides a set of composition libraries that are candidate for pH 3 OER screening, ranked by the expected activity based on prior pH 7 experiments. Running on the lab's notebook server (see ESI†), the initial query used criteria spanning experiment provenance, process details, and analysis details, identifying the 69 K activity measurements of interest from the set of 2.5 M electrochemistry measurements (Sample-Processes) with a query execution time of 70 s. In total, the design of experiment notebook runs in under 3 min, enabling human-guided, data-driven design of high throughput experiments.
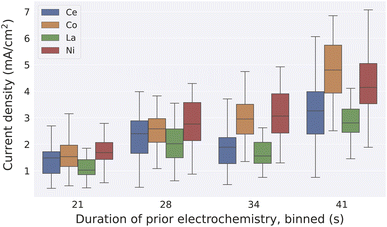
Our final use case involves the evaluation of a human-derived hypothesis based on existing data. Trotochaud and coworkers demonstrated that the activity of electrocatalysts for the OER may be enhanced due to incorporation of trace Fe impurities in standard electrolytes.29 Meanwhile, high throughput experiments revealed the broad range of compositions that are active OER catalysts in alkaline electrolytes.28 From these reports, a scientist may hypothesize that catalyst conditioning, perhaps through Fe incorporation, improves the activity of OER catalysts regardless of initial catalyst composition. This would imply that even poor catalysts will become competent catalysts upon aging, which has not been evaluated in the literature. Querying the MekG for experiments of the type reported in ref. 28 produces a dataset of catalyst activity, where we group measurements by the primary element of the catalyst (concentration at least 70%) and consider the total duration of prior electrochemistry. Fig. 3 summarizes the results, revealing that all catalysts experience conditioning over 10's of seconds of electrochemical operation, and while transition-metal-rich catalysts exhibit the highest activity, the conditioning results in high activity for rare-earth-rich catalysts that otherwise may not exhibit such activity. A similar analysis in Fig. S1† shows that the same conditioning trend is observed in an alternate measurement of catalytic activity (catalyst overpotential at 3 mA cm−2) in pH 13 electrolyte, while an opposite trend is observed in pH 7 electrolyte, indicating that catalyst instabilities outweigh any catalyst conditioning at near-neutral pH and demonstrating that evaluation of the aforementioned hypothesis pH-dependent. While the underlying high throughput experiments were not designed based on a catalyst conditioning hypothesis, the management of catalyst activity data in the context of experiment provenance enables rapid evaluation of such hypotheses using the MekG.
The MekG extends the rich use of graph and network models in materials science. Networks have been used to model all known inorganic materials30 and their interrelationships established with structural and electronic features.31 Materials knowledge graphs have been established for materials properties and their symbolic or data-driven relationships,32 for representing interrelationships among various sources of materials data,33 for integrating multiple data streams,34 and for encoding relationships among factual knowledge, analytical models, and domain experts.35 Knowledge graphs for specific domains of materials science have been established for common industrial metals,36 nanocomposites,37 metal organic frameworks,38 and battery materials.39,40 The value proposition for expanding the purview of such knowledge graphs has been made,41 and the present work builds towards a global materials knowledge graph by establishing best practices for representing experiments and their associated (meta)data in a scalable manner. With the proliferation of graph neural networks, causal modeling, and attention based networks such as transformer models in machine learning writ large, and the expectation that increased deployment for materials discovery is imminent, we believe the elevation of experimental data management to graph databases will pave the way for a new era of artificial intelligence for materials science.
Author contributions
M. J. S., B. A. R., D. G., S. K. S., and J. M. G. designed the MekG and the use cases. M. J. S. and B. A. R. implemented MekG with assistance from D. G. and J. M. G. J. B. and D. G. implemented the design of experiments use case.Data availability
The MPS SQL database from which MekG is built and the three sub-databases are available at https://data.caltech.edu/records/aeffy-dcr62 (doi: https://doi.org/10.22002/aeffy-dcr62). The MekG neo4j database is available at https://data.caltech.edu/records/h88fq-dk449 (doi: https://doi.org/10.22002/h88fq-dk449).Code availability
The code for the query time use cases and MekG migration from MPS is available at https://github.com/modelyst/MekG-migrations. The code for the design of experiments and hypothesis evaluation use cases is available at https://data.caltech.edu/records/m4mpa-4mt17 (doi: https://doi.org/10.22002/m4mpa-4mt17).Conflicts of interest
Modelyst LLC implements custom data management systems in a professional context.Acknowledgements
This material is primarily based on work performed by the Liquid Sunlight Alliance, which is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, Fuels from Sunlight Hub under Award DE-SC0021266. Development of the graph database schema was supported by Toyota Research Institute. Much of the underlying data was generated by research in the Joint Center for Artificial Photosynthesis, a DOE Energy Innovation Hub, supported through the Office of Science of the U.S. Department of Energy (Award No. DE-SC0004993). Storage is provided by the Open Storage Network via XSEDE allocation INI210004.Notes and references
- M. M. Flores-Leonar, L. M. Mejía-Mendoza, A. Aguilar-Granda, B. Sanchez-Lengeling, H. Tribukait, C. Amador-Bedolla and A. Aspuru-Guzik, Curr. Opin. Green Sustain. Chem., 2020, 25, 100370 CrossRef.
- J. Yano, K. J. Gaffney, J. Gregoire, L. Hung, A. Ourmazd, J. Schrier, J. A. Sethian and F. M. Toma, Nat. Rev. Chem., 2022, 6, 357–370 CrossRef PubMed.
- C. P. Gomes, B. Selman and J. M. Gregoire, MRS Bull., 2019, 44, 538–544 CrossRef.
- E. Stach, B. DeCost, A. G. Kusne, J. Hattrick-Simpers, K. A. Brown, K. G. Reyes, J. Schrier, S. Billinge, T. Buonassisi, I. Foster, C. P. Gomes, J. M. Gregoire, A. Mehta, J. Montoya, E. Olivetti, C. Park, E. Rotenberg, S. K. Saikin, S. Smullin, V. Stanev and B. Maruyama, Matter, 2022, 4, 2702–2726 CrossRef.
- J. H. Montoya, M. Aykol, A. Anapolsky, C. B. Gopal, P. K. Herring, J. S. Hummelshøj, L. Hung, H.-K. Kwon, D. Schweigert, S. Sun, S. K. Suram, S. B. Torrisi, A. Trewartha and B. D. Storey, Appl. Phys. Rev., 2022, 9, 011405 CAS.
- A. Jain, S. P. Ong, G. Hautier, W. Chen, W. D. Richards, S. Dacek, S. Cholia, D. Gunter, D. Skinner, G. Ceder and K. A. Persson, APL Mater., 2013, 1, 011002 CrossRef.
- K. T. Butler, D. W. Davies, H. Cartwright, O. Isayev and A. Walsh, Nature, 2018, 559, 547–555 CrossRef CAS PubMed.
- M. L. Green, C. L. Choi, J. R. Hattrick-Simpers, A. M. Joshi, I. Takeuchi, S. C. Barron, E. Campo, T. Chiang, S. Empedocles, J. M. Gregoire, A. G. Kusne, J. Martin, A. Mehta, K. Persson, Z. Trautt, J. V. Duren and A. Zakutayev, Appl. Phys. Rev., 2017, 4, 011105 Search PubMed.
- K. Alberi, M. B. Nardelli, A. Zakutayev, L. Mitas, S. Curtarolo, A. Jain, M. Fornari, N. Marzari, I. Takeuchi, M. L. Green, M. Kanatzidis, M. F. Toney, S. Butenko, B. Meredig, S. Lany, U. Kattner, A. Davydov, E. S. Toberer, V. Stevanovic, A. Walsh, N.-G. Park, A. Aspuru-Guzik, D. P. Tabor, J. Nelson, J. Murphy, A. Setlur, J. Gregoire, H. Li, R. Xiao, A. Ludwig, L. W. Martin, A. M. Rappe, S.-H. Wei and J. Perkins, J. Phys. D: Appl. Phys., 2018, 52, 013001 CrossRef PubMed.
- K. Choudhary, K. F. Garrity, A. C. E. Reid, B. DeCost, A. J. Biacchi, A. R. Hight Walker, Z. Trautt, J. Hattrick-Simpers, A. G. Kusne, A. Centrone, A. Davydov, J. Jiang, R. Pachter, G. Cheon, E. Reed, A. Agrawal, X. Qian, V. Sharma, H. Zhuang, S. V. Kalinin, B. G. Sumpter, G. Pilania, P. Acar, S. Mandal, K. Haule, D. Vanderbilt, K. Rabe and F. Tavazza, npj Comput. Mater., 2020, 6, 1–13 CrossRef.
- T. J. Jacobsson, A. Hultqvist, A. García-Fernández, A. Anand, A. Al-Ashouri, A. Hagfeldt, A. Crovetto, A. Abate, A. G. Ricciardulli, A. Vijayan, A. Kulkarni, A. Y. Anderson, B. P. Darwich, B. Yang, B. L. Coles, C. A. R. Perini, C. Rehermann, D. Ramirez, D. Fairen-Jimenez, D. Di Girolamo, D. Jia, E. Avila, E. J. Juarez-Perez, F. Baumann, F. Mathies, G. S. A. González, G. Boschloo, G. Nasti, G. Paramasivam, G. Martínez-Denegri, H. Näsström, H. Michaels, H. Köbler, H. Wu, I. Benesperi, M. I. Dar, I. Bayrak Pehlivan, I. E. Gould, J. N. Vagott, J. Dagar, J. Kettle, J. Yang, J. Li, J. A. Smith, J. Pascual, J. J. Jerónimo-Rendón, J. F. Montoya, J.-P. Correa-Baena, J. Qiu, J. Wang, K. Sveinbjörnsson, K. Hirselandt, K. Dey, K. Frohna, L. Mathies, L. A. Castriotta, M. H. Aldamasy, M. Vasquez-Montoya, M. A. Ruiz-Preciado, M. A. Flatken, M. V. Khenkin, M. Grischek, M. Kedia, M. Saliba, M. Anaya, M. Veldhoen, N. Arora, O. Shargaieva, O. Maus, O. S. Game, O. Yudilevich, P. Fassl, Q. Zhou, R. Betancur, R. Munir, R. Patidar, S. D. Stranks, S. Alam, S. Kar, T. Unold, T. Abzieher, T. Edvinsson, T. W. David, U. W. Paetzold, W. Zia, W. Fu, W. Zuo, V. R. F. Schröder, W. Tress, X. Zhang, Y.-H. Chiang, Z. Iqbal, Z. Xie and E. Unger, Nat. Energy, 2022, 7, 107–115 CrossRef CAS.
- B. Blaiszik, K. Chard, J. Pruyne, R. Ananthakrishnan, S. Tuecke and I. Foster, J. Mater., 2016, 68, 2045–2052 Search PubMed.
- B. Blaiszik, L. Ward, M. Schwarting, J. Gaff, R. Chard, D. Pike, K. Chard and I. Foster, MRS Commun., 2019, 9, 1125–1133 CrossRef CAS.
- M. K. Horton and R. Woods-Robinson, Patterns, 2021, 2, 100411 CrossRef PubMed.
- J. Amici, P. Asinari, E. Ayerbe, P. Barboux, P. Bayle-Guillemaud, R. J. Behm, M. Berecibar, E. Berg, A. Bhowmik, S. Bodoardo, I. E. Castelli, I. Cekic-Laskovic, R. Christensen, S. Clark, R. Diehm, R. Dominko, M. Fichtner, A. A. Franco, A. Grimaud, N. Guillet, M. Hahlin, S. Hartmann, V. Heiries, K. Hermansson, A. Heuer, S. Jana, L. Jabbour, J. Kallo, A. Latz, H. Lorrmann, O. M. Løvvik, S. Lyonnard, M. Meeus, E. Paillard, S. Perraud, T. Placke, C. Punckt, O. Raccurt, J. Ruhland, E. Sheridan, H. Stein, J.-M. Tarascon, V. Trapp, T. Vegge, M. Weil, W. Wenzel, M. Winter, A. Wolf and K. Edström, Adv. Energy Mater., 2022, 12, 2102785 CrossRef CAS.
- A. Zakutayev, N. Wunder, M. Schwarting, J. D. Perkins, R. White, K. Munch, W. Tumas and C. Phillips, Sci. Data, 2018, 5, 180053 CrossRef PubMed.
- K. R. Talley, R. White, N. Wunder, M. Eash, M. Schwarting, D. Evenson, J. D. Perkins, W. Tumas, K. Munch, C. Phillips and A. Zakutayev, Patterns, 2021, 2, 100373 CrossRef PubMed.
- E. Soedarmadji, H. S. Stein, S. K. Suram, D. Guevarra and J. M. Gregoire, npj Comput. Mater., 2019, 5, 1–9 CrossRef.
- I. M. Pendleton, G. Cattabriga, Z. Li, M. A. Najeeb, S. A. Friedler, A. J. Norquist, E. M. Chan and J. Schrier, MRS Commun., 2019, 9, 846–859 CrossRef CAS.
- J. Popp and T. Biskup, Chem.: Methods, 2022, 2, e202100097 CAS.
- B. Paulus and T. Biskup, Digit. Discov., 2023, 2, 234–244 RSC.
- M. Statt, B. A. Rohr, K. S. Brown, D. Guevarra, J. S. Hummelshøj, L. Hung, A. Anapolsky, J. Gregoire and S. Suram, Digit. Discov., 2023 10.1039/D3DD00054K.
- F. Friedler, K. Tarján, Y. W. Huang and L. T. Fan, Chem. Eng. Sci., 1992, 47, 1973–1988 CrossRef CAS.
- S. Mysore, E. Kim, E. Strubell, A. Liu, H.-S. Chang, S. Kompella, K. Huang, A. McCallum and E. Olivetti, Automatically Extracting Action Graphs from Materials Science Synthesis Procedures, 2017, http://arxiv.org/abs/1711.06872 Search PubMed.
- D. Barter, E. W. C. Spotte-Smith, N. S. Redkar, A. Khanwale, S. Dwaraknath, K. A. Persson and S. M. Blau, Digit. Discov., 2023, 2, 123–137 RSC.
- A. C. Vaucher, F. Zipoli, J. Geluykens, V. H. Nair, P. Schwaller and T. Laino, Nat. Commun., 2020, 11, 3601 CrossRef PubMed.
- M. J. Statt, B. A. Rohr, D. Guevarra, S. K. Suram, T. E. Morrell and J. M. Gregoire, Sci. Data, 2023, 10, 184 CrossRef PubMed.
- H. S. Stein, D. Guevarra, A. Shinde, R. J. R. Jones, J. M. Gregoire and J. A. Haber, Mater. Horiz., 2019, 6, 1251–1258 RSC.
- L. Trotochaud, S. L. Young, J. K. Ranney and S. W. Boettcher, J. Am. Chem. Soc., 2014, 136, 6744–6753 CrossRef CAS PubMed.
- V. I. Hegde, M. Aykol, S. Kirklin and C. Wolverton, Sci. Adv., 2020, 6, eaay5606 CrossRef CAS PubMed.
- O. Isayev, D. Fourches, E. N. Muratov, C. Oses, K. Rasch, A. Tropsha and S. Curtarolo, Chem. Mater., 2015, 27, 735–743 CrossRef CAS.
- D. Mrdjenovich, M. K. Horton, J. H. Montoya, C. M. Legaspi, S. Dwaraknath, V. Tshitoyan, A. Jain and K. A. Persson, Matter, 2020, 2, 464–480 CrossRef.
- R. Choudhury, M. Aykol, S. Gratzl, J. Montoya and J. Hummelshøj, J. Open Source Softw., 2020, 5, 2105 CrossRef.
- K. Hatakeyama-Sato and K. Oyaizu, Commun. Mater., 2020, 1, 1–10 CrossRef.
- K. S. Aggour, A. Detor, A. Gabaldon, V. Mulwad, A. Moitra, P. Cuddihy and V. S. Kumar, Integr. Mater. Manuf. Innov., 2022, 11, 467–478 CrossRef.
- X. Zhang, X. Liu, X. Li and D. Pan, Comput. Phys. Commun., 2017, 211, 98–112 CrossRef CAS.
- J. P. McCusker, N. Keshan, S. Rashid, M. Deagen, C. Brinson and D. L. McGuinness, The Semantic Web – ISWC 2020, 2020, pp. 144–159 Search PubMed.
- Y. An, J. Greenberg, X. Zhao, X. Hu, S. McCLellan, A. Kalinowski, F. J. Uribe-Romo, K. Langlois, J. Furst, D. A. Gómez-Gualdrón, F. Fajardo-Rojas and K. Ardila, Building Open Knowledge Graph for Metal-Organic Frameworks (MOF-KG): Challenges and Case Studies, 2022, http://arxiv.org/abs/2207.04502 Search PubMed.
- Z. Nie, Y. Liu, L. Yang, S. Li and F. Pan, Adv. Energy Mater., 2021, 11, 2003580 CrossRef CAS.
- Z. Nie, S. Zheng, Y. Liu, Z. Chen, S. Li, K. Lei and F. Pan, Adv. Funct. Mater., 2022, 32, 2201437 CrossRef CAS.
- X. Zhao, J. Greenberg, S. McClellan, Y.-J. Hu, S. Lopez, S. K. Saikin, X. Hu and Y. An, 2021 IEEE International Conference on Big Data (Big Data), 2021, pp. 4628–4632 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3dd00067b |
| ‡ Present address: Materials Science Division, Lawrence Livermore National Laboratory, Livermore, CA 94550, USA. |
| This journal is © The Royal Society of Chemistry 2023 |