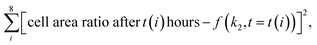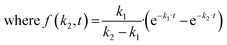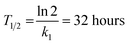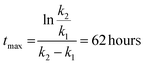Open Access Article
Open Access ArticleDigital biology approach for macroscale studies of biofilm growth and biocide effects with electron microscopy†
Konstantin S.
Kozlov‡
ac,
Daniil A.
Boiko‡
a,
Elena V.
Detusheva
b,
Konstantin V.
Detushev
 b,
Evgeniy O.
Pentsak
a,
Anatoly N.
Vereshchagin
b,
Evgeniy O.
Pentsak
a,
Anatoly N.
Vereshchagin
 a and
Valentine P.
Ananikov
a and
Valentine P.
Ananikov
 *ac
*ac
aZelinsky Institute of Organic Chemistry, Russian Academy of Sciences, Leninsky Prospekt 47, Moscow, 119991, Russia. E-mail: val@ioc.ac.ru
bState Research Center for Applied Microbiology and Biotechnology, Obolensk, Moscow Region, Russia
cLomonosov Moscow State University, Chemistry Department, Leninskie Gory 1/3, Moscow, 119991, Russia
First published on 13th September 2023
Abstract
Microbial interactions are one of the major topics of current research due to their great societal relevance. It is now established that biofilms—associations of microorganisms, exchanging various chemical compounds, including proteins and nucleic acids—are capable of promoting horizontal transfer of resistance genes. However, our understanding of the processes occurring in biofilms is rather limited. A possible method to partly overcome this problem is the implementation of highly efficient imaging and mapping of these structures. This work proposes a combination of automated scanning electron microscopy (SEM) and a comprehensive software system that uses deep neural networks to perform an in-depth analysis of biofilms. Time-dependent, high-throughput mapping of biofilm electron microscopy images was achieved using deep learning and allowed microscale data analysis of visible to the eye biofilm-covered area (i.e., at the macroscale). For this study, to the best of our knowledge, the first matrix and cell-annotated biofilm segmentation dataset was prepared. We show that the presented approach can be used to process statistical data investigation of biofilm samples in a volume, where automation is essential (>70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 separate bacterial cells studied; >1000 times faster than regular manual analysis). To evaluate the approach, multiple time steps of biofilm development were analyzed by first-to-date kinetic modeling of biofilms with SEM, revealing the complex dynamics of biofilm formation. Moreover, it was shown that the described procedure is capable of capturing differences between antibiotics and antimicrobial compounds applied to studied biofilms.
000 separate bacterial cells studied; >1000 times faster than regular manual analysis). To evaluate the approach, multiple time steps of biofilm development were analyzed by first-to-date kinetic modeling of biofilms with SEM, revealing the complex dynamics of biofilm formation. Moreover, it was shown that the described procedure is capable of capturing differences between antibiotics and antimicrobial compounds applied to studied biofilms.
Introduction
There is a growing crisis of antibiotic resistance that is causing several health issues worldwide.1 The reason for this may be both the irrational use of antimicrobials and their application to prevent a combined bacterial–viral infection. One of the ways for bacteria to endure antibiotic treatment is to form biofilms.2,3 Understanding the mechanisms of biofilm formation and the contribution of these processes to increasing levels of antibiotic resistance are critical public health priorities in developing new antibiofilm agents and methods to combat antimicrobial resistance.The implementation of high-tech innovations in biological research is currently making a significant contribution to the development of life science.4 Digital biology solutions provide insights into various scientific problems, such as climate change,5 genome annotation,6 biological image analysis,7,8 protein folding,9 drug discovery,10 cancer detection,11,12 biology laboratory virtualization,13 and the problem of antibiotic discovery.14,15 In fact, the combination of models, that predict antimicrobial activity of molecules or generate novel compounds with the high-throughput mapping of experimental microscopic data may become a powerful strategy for the accelerated discovery of antibiofilm agents in the near future.16–18
There are several definitions of biofilms.19–21 Generally, biofilms are highly structured associations of microorganisms attached to the surface (which can be both biotic or abiotic) or forming floating mats on liquid surfaces.22 These associations are contained within a self-producing matrix of extracellular polymeric substances (EPS) consisting of proteins, lipids, nucleic acids, and polysaccharides. These compounds play an important role in enhancing adhesion to the surface, aggregating microorganisms, and ensuring the structural integrity of the biofilm.23–25
Biofilms form on industrial production lines, heat exchangers, and work surfaces, leading to corrosion and damage to mechanisms and contamination of raw materials and products.26 For the food industry, contamination with biofilms can lead to much more severe effects,27 contributing to outbreaks of foodborne infections.28,29 Biofilms are among the drivers of the growing antibiotic resistance crisis and account for two-thirds of all infections.30 The National Institute of Health estimates that biofilms cause 65–80% of all microbial infections and 80–90% of all chronic infections, making biofilms a major public health problem.31 Diseases associated with biofilms include upper and lower respiratory tract diseases, endocarditis, chronic otitis media, eye infections, chronic wounds, diabetic foot ulcers, urinary tract infections, and periodontitis.32,33 Bacterial colonization of medical devices such as intravascular and urinary catheters, pacemakers, heart valves, contact lenses, breast implants, endotracheal tubes, and orthopedic implants can lead to device-associated infections.34,35
Various methods carry out the detection of biofilms: staining with further detection using photometric methods; various methods of microscopy, including staining, using fluorescent labels; molecular genetic techniques for detecting the expression of biofilm-forming genes, etc.36 There are several established techniques for the automated computational analysis of biofilm images using confocal microscopy,37–39 some of which can even handle statistical area analysis.40 Automated detection of stalked bacteria is also available.41 However, there is currently no single method or test system for the comprehensive AI-based automatic detection of biofilms and their conditions with Scanning Electron Microscopy (SEM), which can perform statistical analysis on a vast amount of imaging data, where manual analysis cannot be performed (including single cell counting and matrix quantification). In fact, there are prior studies that have measured cellular properties inside biofilms42 and the matrix distribution43 based on fluorescence images. However, the mechanisms and processes of biofilm formation are not well understood, and studies that detail the morphological changes in the bacterial population in the process of bacterial colonization are still limited.
With the development of automation techniques in electron microscopy, it has become an appealing imaging method. Currently, SEM imaging is widely used as a quality control method in semiconductor manufacturing and steel production.44 Furthermore, the combination of energy-dispersive X-ray spectroscopy with automated SEM imaging can be used for the elemental analysis of materials45 or pharmaceutical products.46 Contaminant identification, surface topography investigation, and defect detection are also offered with electron microscopy and find application in different areas of science and technology, e.g., solar panels.47
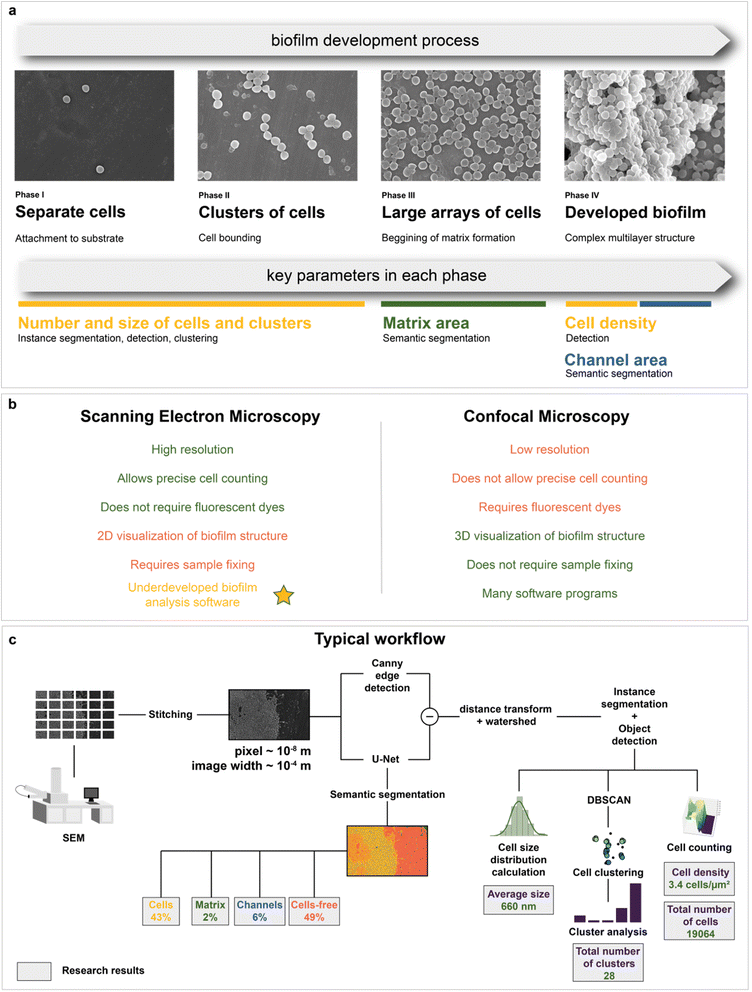
Microbiology is the area where electron microscopy can also be employed.48–50 SEM has been shown to be a suitable tool for tracking bacterial adhesion and biofilm formation on biotic51 and abiotic surfaces.52,53 SEM has the levels of magnification and resolution required to visualize individual cells of microorganisms and their communities or biofilms, as well as their spatial organization.54 In addition, SEM is a valuable tool for studying the effect of antimicrobial agents on the cell morphology of bacterial populations in various biofilms.55 The in-depth method to study biofilms would allow in silico high-throughput mapping and detection of the key features of the studied systems: separate cells, channels, and matrix (Fig. 1a). To the best of our knowledge, there are still no computer-aided solutions for SEM image analysis of biofilms that can provide the recognition of matrix and single cell detection without preliminary data annotation with similar or above quality.
The data acquisition rate is currently very high and would not allow a manual analysis of images without increasing research budgets and potential loss of analysis quality due to human errors. Recently, a solution to this problem—the application of deep learning algorithms—has become increasingly widely adopted. Examples of usages include biomedical applications,56 analysis of pharmaceutical powders,57 protein nanowires,58 catalysts,59 and analysis in a liquid phase.60 Besides segmentation and detection tasks, deep learning-based image inpainting has also performed.61 Despite the active application of deep learning algorithms in cell imaging,62–70 there is a significant lack of knowledge in AI-based biofilm SEM image analysis. State-of-the-art techniques only perform segmentation of whole biofilms without recognition of the matrix71 with the Trainable Weka Segmentation plugin,72 segmentation of cellular compartments,73–75 and cell segmentation on SBF-SEM images.76 The main reason for this small coverage of possible objects is most likely the unavailability of well-annotated large amounts of data for researchers compared to other types of microscopy.77 With the development of frameworks for semiautomatic microbiological data annotation78–80 or advanced segmentation deep learning architectures,81,82 it is possible to partially reduce the bottleneck with image annotation. However, higher quality data still significantly improve the results and allow other researchers to train their models more effectively.
An alternative method to study biofilm structures is using fluorescence confocal microscopy.83 Unlike SEM, it allows 3D visualization of biofilm architecture and can distinguish between living and dead microbial cells. Although spatial organization of bacterial cells and EPS in the biofilms can be determined by confocal laser scanning microscopy (CLSM), only SEM can show the architecture of biofilms at a single bacterial cell resolution level.84 In addition, CLSM lacks the necessary magnification and resolution for detailed observation of individual cells in a biofilm and their morphology.85 It also requires fluorescent dyes to evaluate biocide effects, which can affect biofilm physiology (e.g. reduction levels of resazurin that sometimes used in CLSM86 can be decreased in the presence of antibiotics,87 improve the breakthrough of cell membranes,88 affect the viability,89 lead to decrease of elongation rates90) or even damage living cells.87 Precise cell counting is a limiting factor for confocal microscopy, as most protocols, which use fluorescent pigments, allow only semiquantitative analysis.71,87 Since only 2D imaging with SEM is available, it is possible to calculate only the area density of cells. Nevertheless, it is likely to correlate with the bulk cell density, allowing evaluation of growth and biocidal effect (see “Kinetic modeling of biofilm growth” and “Automated mapping of large volumes of SEM images for the investigation of antimicrobial compound impact” in the Results and Discussion section for more information). A comparison of SEM and confocal microscopy is shown in Fig. 1b. Both methods have their own pros and cons91 and are often used together.92 However, there is much more software for quantitative analysis using confocal microscopy compared to electron microscopy.
Despite the fact that deep learning-based approaches are more efficient than the popular image thresholding, neural network solutions are not common at the moment. It slows down the digital transformation in biofilm studies. Implementation of novel computer vision techniques allows fast calculation of cells and matrix on images making possible to evaluate biofilm growth and biocide effects with a greater statistical accuracy.
Therefore, the main purpose of this work is to propose a digital approach to studying biofilm structures with SEM. It combines automated scanning electron microscopy with deep neural network analysis (Fig. 1c). As a result, it became possible to analyze biofilms with the use of machine intelligence at the macroscale and derive conclusions about their composition (cell, matrix, and channel area calculation) and morphology (cell size distribution, number, and size of cell clusters in the biofilm formation area, total number and density of bacterial cells in the area visible to the human eye). These data were used to study biofilm growth dynamics from SEM images. The composition of biofilms after antibiotic and antimicrobial compound application was also investigated with neural network image segmentation. This work not only proposes a method to perform automated image analysis but also shows how it can be implemented on case studies, where the analysis of large volumes of microscopic data is required.
Results and Discussion
Object of study
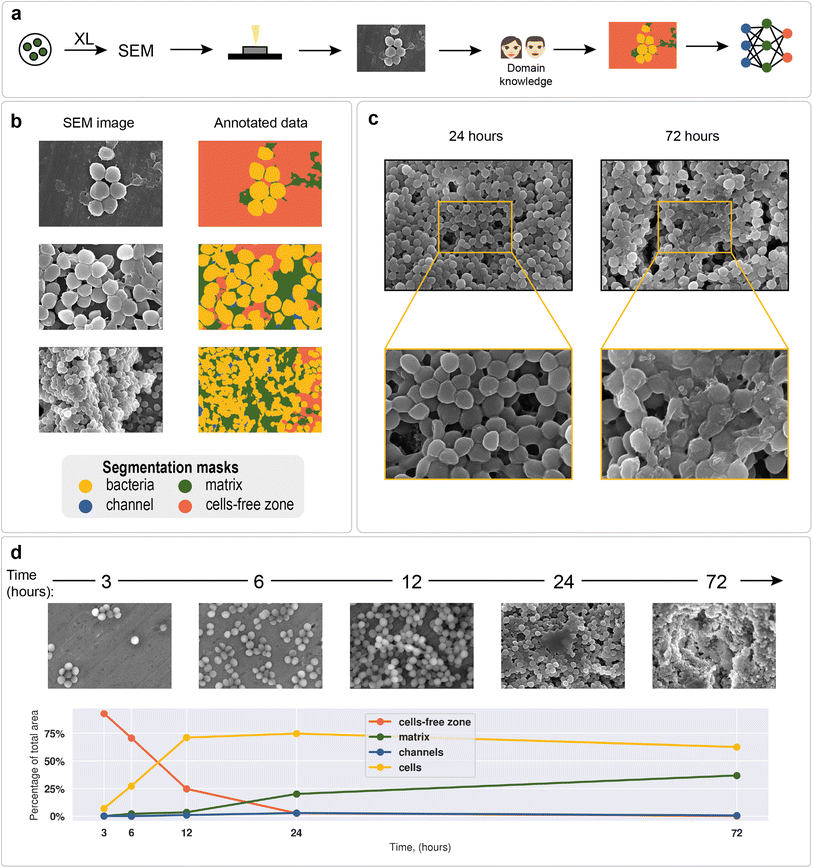
Staphylococcus aureus was chosen as a model microorganism, as it is one of the main etiological agents of nosocomial infections and is well known for its ability to form biofilms on host tissues and implants.93 Biofilm formation of S. aureus often leads to chronic infections in patients suffering from osteomyelitis, endocarditis, cystic fibrosis, or in patients undergoing medical procedures such as catheterization.94,95 All of the above makes S. aureus one of the main human pathogens and the major microorganism for biofilm research. Moreover, Staphylococcus is a convenient object of study due to its cell shape, which facilitates its image processing (Fig. 2a and b).To demonstrate differences between nascent and mature biofilms, Fig. 2c shows biofilms formed by the S. aureus ATCC 6538 strain at different cultivation times. After 24 hours of cultivation, spherical cell conglomerates with a small amount of matrix and clearly visible channels were observed. After 72 hours of biofilm cultivation, the cells were observed to be almost completely covered with matrix, and the outlines of individual cells were blurred.
The total biofilm formation dynamics are shown in Fig. 2d. After 3 hours of cultivation, single cells or groups of cells (cell clusters) were observed. After 6 hours, cell clusters connected by intercellular bridges appeared. After 12 hours of cultivation, connected cell layers formed multilayer structures with a clearly distinguishable matrix. A complex multilevel structure with a large amount of matrix was observed the following day. After 72 hours, the formed biofilm became a multidimensional structure, where numerous channels were observed. Cells immersed in the extracellular matrix were clearly distinguishable. Visible differences in biofilm morphology at different cultivation times allow us to recognize biofilm morphology without additional methods.
As an example of a typical biofilm development process, area analysis of example biofilm images was performed (see section “Model predictions” in the ESI† for the segmentation results). The results showed a sharp increase in bacterial area and a decrease in cell-free area. Matrix enlargement did not occur until 12 hours after the start of cultivation and reached a plateau after three days. Channel areas also showed an increase after 24 hours; however, their area share was insignificant compared with 2 dominant classes: cells and matrix.
Deep learning methodology
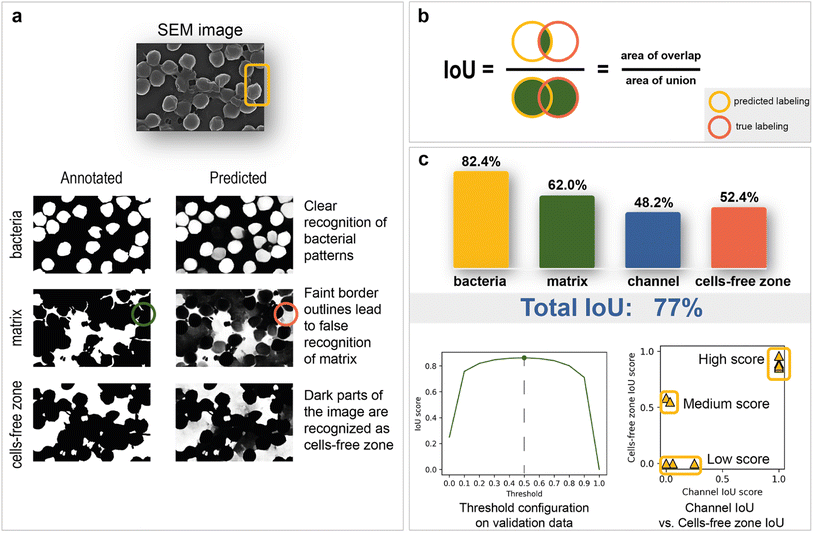
As mentioned above, deep learning is actively used in pattern recognition tasks, such as semantic segmentation and object detection. Convolutional neural networks have made remarkable progress in computer vision.96 They are based on convolution operations, which convert the original image into a set of feature maps. This set stores correlation information between closely located pixels and provides detection of distinctive local motifs, making it easier to train the model. Moreover, the nature of the convolution operation makes it easy to use graphical processing units to speed up the training process. Convolutional neural networks dominate computer vision tasks over classical machine learning and image processing algorithms.A typical neural network-aided computer vision pipeline includes the following steps: labeling the data, preprocessing the images, optimizing the network hyperparameters, and training the final model. Data labeling is a step of identifying specific objects in research data. SEM images of biofilms were manually labeled into masks using a special platform by research scientists with good domain knowledge (Fig. 2a). Segmentation masks included bacteria, matrix, channels, and support areas without cells (cell-free zone) (Fig. 2b). Channel zone differs from cell-free zone by relatively small size and specific location inside large clusters of cells (see section “Channel zone recognition” in the ESI† for detailed explanation). The main task of the segmentation network is to predict the areas of each class on images (Fig. 3a). The training dataset size was equal to 72. Although, it is possible to train a segmentation model using only 4 dissimilar high-resolution training images and proper augmentations with less than 10% loss in quality. High resolution and rich content of training images leads to convergence of the IoU score (Fig. 3b) vs. training dataset size curve despite the seeming lack of training images (see “Neural network implementation and training” in the Methods section for more information). It is worth mentioning that bacterial image labeling is not a clearly defined task. Different specialists can obtain different results, impacting the performance of the model. Image preprocessing includes modification techniques to facilitate model training (normalization, resizing, augmentations, etc.). Hyperparameter optimization is a step of choosing the best combination of hyperparameters (model architecture, encoder, learning rate, etc.) for the neural network. The final step is to train a neural network with optimal hyperparameters by minimizing the chosen loss function using a backpropagation algorithm (see “Neural network implementation and training” in the Methods for detailed information).
Model inference results
The total IoU score on the test data is 77%. The disparity between scores on validation (85%) and test is caused by the variations between image sets. The IoU scores for each segmentation class are shown in Fig. 3c. The best result of 82.4% is observed for bacterial cells, while channel areas are predicted to be the worst, with a result of 48.2%. The matrix obtained the second-best metric of 62%. It is worth noting that the IoU score is sensitive to the imbalance of segmentation classes, which can underestimate the true quality of the network. As the channels on images are not clearly defined and can easily be confused with areas without cells, the correlation between channel and cell-free zone segmentation quality was investigated with an IoU score scatter plot (Fig. 3c). Test data are divided into three groups (low score, medium score, high score):1. The low-score group is distinguished from the others by having no cell-free zones, which leads to many false positives.
2. The medium-score group—lack of channel zones along with a small area of support.
3. The high-score group tends to have a small channel area and thus reduces false negatives associated with the predisposition of the model to label segments as cell-free zones.
The relationship between the IoU score and binarization threshold was investigated on validation images. As seen in Fig. 3c, the optimal threshold has a value of 0.5.
The segmentation network also shows significantly better results on test images compared to classical computer vision algorithms (Table 1). In summary, this work's final model is devoid of significant systematic errors, making it a viable tool for morphology analysis.
| Algorithm | IOU score |
|---|---|
| Global thresholding (v = 127) | 36.3% |
| Otsu's thresholding | 45.6% |
| Adaptive Gaussian thresholding | 49.5% |
| Adaptive mean thresholding | 49.5% |
| Adaptive mean thresholding + closing | 51.2% |
| Edge-based segmentation | 27.9% |
| CNN-based segmentation (our approach) | 82.4% |
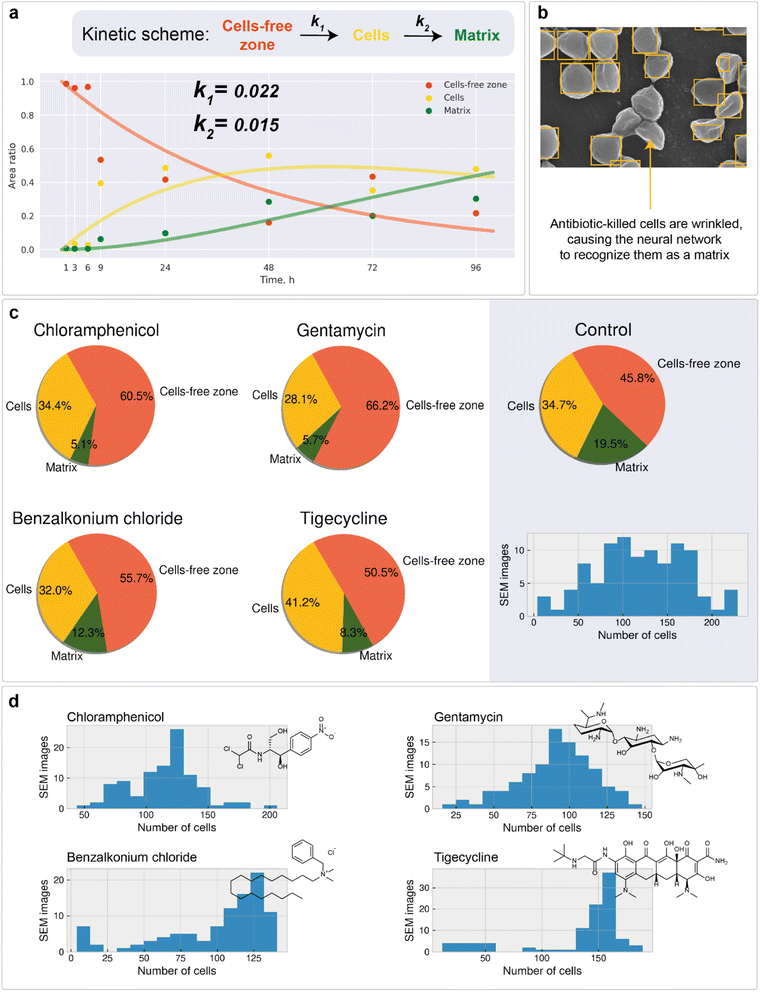
Microscale analysis of macroscale area
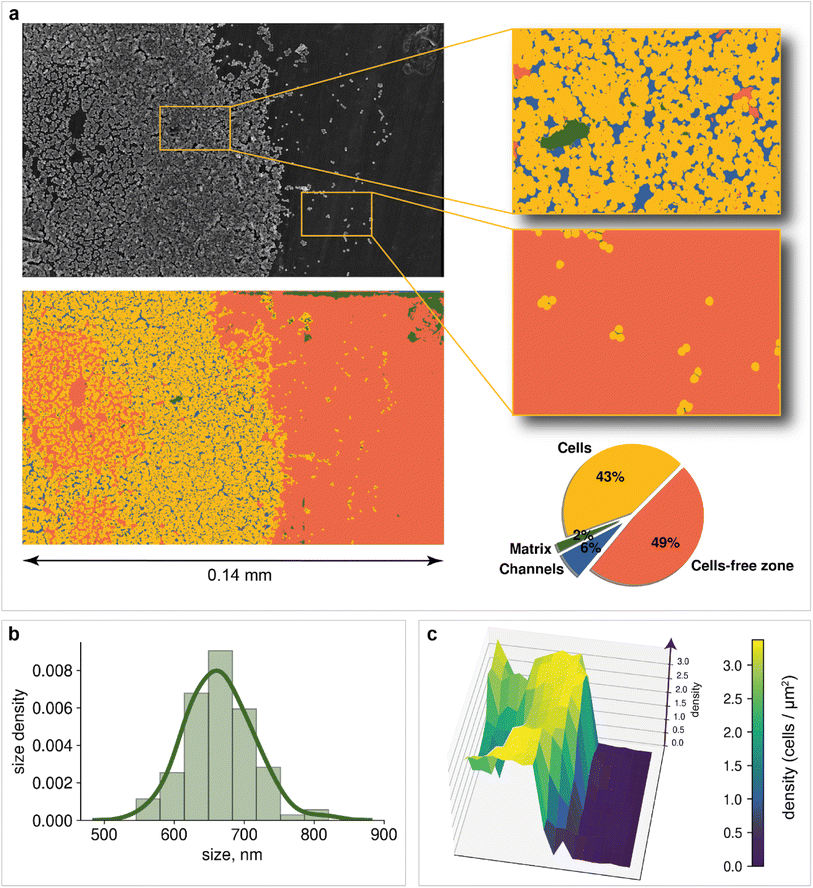
Microscopy is a local method, that is, the data obtained depend on the choice of the site for image recording. The site chosen may not be relevant to the characterization of the entire sample nor statistically representative. In turn, subjective factors, such as operator fatigue, aesthetic preferences, and many others, can influence the choice of an area for recording an image. The automatic digital operation mode has become available only recently, which makes it possible to eliminate the human factor and subjectivity in choosing the region for image registration. Automated scanning allows the sequential recording of multiple images according to a statistically relevant scheme. After each recording, the imaging area is shifted in a software-controlled manner (without human operator participation) to the neighboring region, followed by automatic adjustment of focus, brightness and contrast. The operator sets only the recording scheme: the required number of images along the x-axis and y-axis, as well as the overlapping of the recording areas, which can be negative. In contrast to manual human operation, software-controlled microscopy characterization can be performed nonstop for sufficient time (24–96 h, for instance). This is a great step forward, which opens new opportunities in studying biofilms. Here, in the present study, we used automatic image recording in connection with ML automation.Thus, automated scanning with further SEM image stitching provides the ability to scan macroscale areas (which are more than 0.05 mm in width and distinguishable by the naked eye) at the scale of 1 pixel being equal to 400 nm2. The developed neural network was used to characterize the morphology of the biofilm. Image analysis of the 0.14 × 0.08 mm2 area included the following:
1. Matrix, cells, channels, and cell-free zone mapping (Fig. 4a)
2. Cell detection (Fig. 5a and b)
3. Statistical analysis of bacterial cell size (Fig. 4b) and population (Fig. 4c)
4. Cell clustering in the region of biofilm formation (Fig. 5c)
The aim of the biofilm microscopy preparation in the work was to maintain the biofilm's structure to the greatest extent possible (see “Sample preparation” in the Methods section). But this technique leads to reduction of cell size when water is removed in the EM vacuum and to an underestimation of the amount of matrix in the biofilm.97 However, this distortion is invariant to biofilm samples. Considering this, kinetic modeling and biocide effect evaluation should remain valid, since the decrease of the matrix as a result of dehydration will be observed in all samples approximately equally. However, to achieve higher accuracy with the developed approach, correction factor should be included.
Mapping provides information on how matrix and bacterial cells are distributed on the support. Two dominant segmentation classes are depicted: cells (43% of the total area) and cell-free zones (49% of the total area). The slight presence of the matrix (2% of the total area) can be explained by the fact that before the registration of images, the cells have not yet had time to release a significant amount of matrix to form a cohesive biofilm. An additional possible reason is the dehydration of the obtained biofilm during sample preparation. Channels are observed over a large area of the image, confirming the process of incipient biofouling.
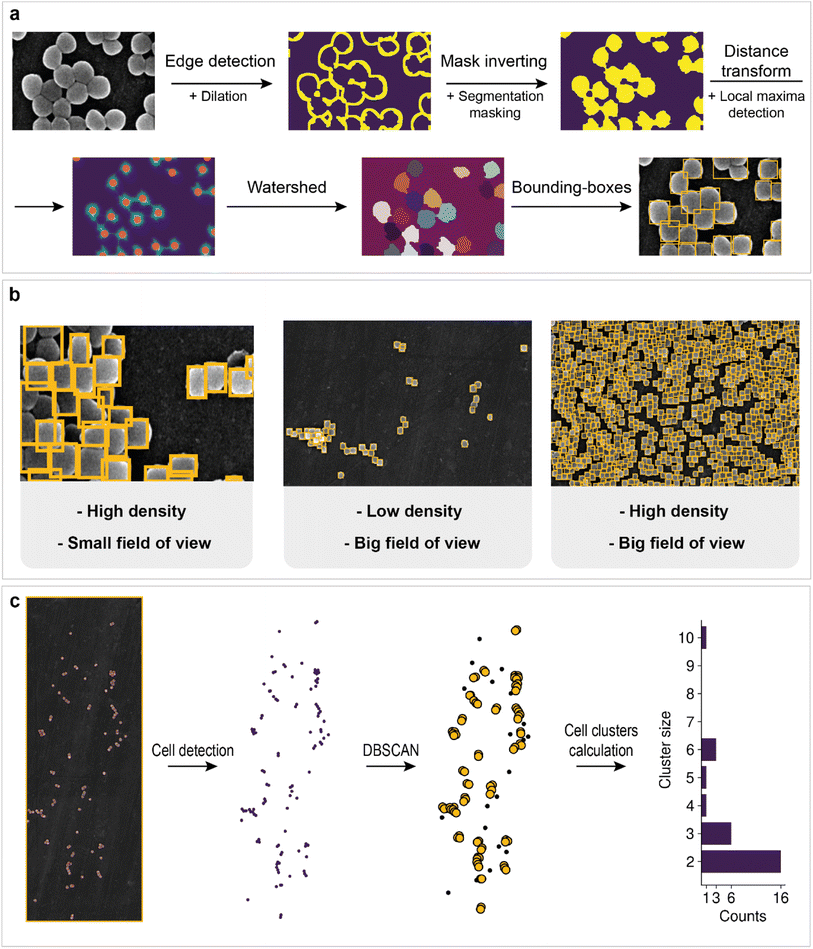
However, information about the total area of bacteria on the image does not allow us to estimate how many cells are located on support. Cell detection is a key problem for the statistical characterization of biofilms. Unfortunately, single-cell image annotation takes considerable time, as the density in biofilms may reach 3.4 cells per μm2. This disadvantage can be eliminated by applying mask postprocessing, which solves the problem of single-cell overlapping and obviates the need for annotation of individual bacteria. The approach is based on unsupervised edge detection with subsequent watershed algorithm implementation, which is actively used for separating different objects in an image, e.g., single cells98 (Fig. 5a).
First, Canny edge detection with additional dilation was employed to obtain an edge mask. Then, we inverted the mask and applied a cell semantic segmentation mask obtained with a neural network to ignore noncellular zones (matrix and channel areas are also ignored).
The distance transform and subsequent local maxima detection on the transformed image allows us to find the geometric centers of cells. Finally, watershed segmentation was used to estimate individual cell areas. The results for cell detection of regions with different fields of view are shown in Fig. 5b. Some cells are merged into one bounding box. This outcome is probably due to the low difference in intensities at the edges, which breaks the correct edge detection, or because of incomplete cell division. The overall procedure makes it possible to count the total number of cells in the biofilm and collect statistics.
For example, bacterial cell size analysis in the biofilm can be performed. It is important to avoid regions with severe cell overlap to eliminate the bias caused by the superimposition of one cell on another. The resulting histogram is shown in Fig. 4b. The bacterial size was normally distributed with an average of 660 nm.
Population density heat maps make it possible to estimate the number of cells per unit area in different parts of the biofilm. Automated creation of visualization maps may be useful for future biofilm formation studies. In the example, the 3D plot surface was constructed (Fig. 4c). The positive correlation between area cell density and channel zones helps to establish when and where the biofilm developed. The area cell density on the image varies from 0 cells per μm2 (bare support areas) to 3.4 cells per μm2 (central zones with an abundance of channels). The total number of cells visible in the image reaches 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 064, according to the model results.
064, according to the model results.
Although computer-aided counting of cells gives a lower bound, since it does not consider the multilayer structure of the biofilm, the approach could be scaled to 3D images, so the error will be corrected. Even though, the area and bulk cell densities should correlate. Thus, the biofouling trend should remain unchanged. The differences in ratios of cells, matrix and cells-free zone should have the same tendency either (see section “Method limitations” in the ESI†).
The creation of techniques for mining statistical data in microbiology research is important for the investigation of biofilm evolution over time. Cell clusters (groups of cells located close to each other) are initiators of biofilm creation, making their detection a particularly important task. The application of clustering algorithms on top of preprocessed cell detection allows the automatic calculation of the total number of cell clusters and their size (number of cells in a cluster). The approach in this work is based on DBSCAN (density-based spatial clustering of applications with noise). Its main advantage is that one does not need to specify a specific number of clusters to implement the algorithm. In addition, the possibility of filter noise makes it possible to separate single cells from cells in clusters. The workflow is shown in Fig. 5c. The number of clusters decreases as the cluster size increases, which may indicate the aggregation process in biofilms.
Kinetic modeling of biofilm growth
High-throughput analysis of biofouling requires quantitative characterization of the biofilm growth rate. The biofilm growth kinetics of Staphylococcus aureus were investigated by implementing a sequential first-order reaction kinetics scheme (Fig. 6a). 100 biofilm SEM images (square 10 × 10 with negative overlap) were registered and processed with the segmentation DL model for each period of time (1, 3, 6, 9, 24, 48, 72, and 96 hours, totaling 800 SEM images with a total analyzed area of 0.0832 mm2 and image pixel size near 100 nm2). Then, the median ratio of cells, matrix, and cell-free zone for each time period was calculated using segmentation data (median was chosen as a measure of central tendency due to stability towards statistical outliers).The calculated data show the expected tendency of cell-free zone ratio reduction and cell area enlargement. As the cells emerge, they begin to secrete matrix. The obtained results are then used to estimate the rate constants of cell and matrix formation (Fig. 6a). The rate constants of cell formation (k1 = 0.022) and cell coating with matrix (k2 = 0.015) were established. k1 is numerically equal to the rate (h−1) of cell area formation at the beginning of the biofilm development process. k2 is numerically equal to the rate (h−1) of cell coating with the matrix when the support substrate is fully covered with bacterial cells.
These constants can be applied to quantify how biofilms develop over time. For example, the half-life of the cell-free segmentation class (i.e., the time when 50% of the studied area is covered with cells and matrix) amounts to approximately 32 hours. In addition, the time at which the cell area ratio becomes close to the maximum possible is equal to 62 hours (all kinetic calculations can be found in “Kinetic calculations” in the Methods section).
Automated mapping of large volumes of SEM images for the investigation of antimicrobial compound impact
To ensure that the developed algorithms can be applied to study the antibiotic susceptibility of biofilms or the antibiotic activity of novel compounds, high-throughput mapping of SEM images with antibiotic-treated bacteria was performed. The work included the study of chloramphenicol (inhibitor of peptidyl transferase), gentamycin (disturbs tRNA and mRNA interactions), benzalkonium chloride (inhibitor of hyaluronidase), and tigecycline (protein synthesis inhibitor) impacts. For each compound, 100 SEM images of the developed biofilms treated with the antibiofilm agent were recorded. After that, segmentation with the following cell detection was performed on all images. The full data set under study was of the size of 500 images (including control sample) with an approximate total analyzed area of 0.052 mm2 having an image pixel size approximately equal to 100 nm2.Unlike normal cells, cells that have died as a result of antibiotic exposure have a curved shape with wrinkles. Despite this, the vast majority of such cells are correctly recognized by the system. However, there are examples where the shape deformation led to the recognition of cells as a matrix (Fig. 6b).
Area calculations (Fig. 6c) on SEM images of antibiotic-treated biofilms show insignificant (less than 1%) areas of channels and dominant cell-free zone areas, indicating an early stage of development. The distribution of cell counts on images (Fig. 6d) for chloramphenicol and gentamycin are symmetrical, with averages of 120 and 90 cells per image, respectively. The samples have almost similar matrix ratios. The distribution of cell counts on images for benzalkonium chloride is skewed to the right, indicating the presence of regions with uneven distribution of cell density. The tigecycline sample is also characterized by a rightly skewed cell per image distribution. In all samples, the number of images with a large number of cells (150+ units) is less than in the control sample. These results prove the applicability of the investigated antibacterial drugs in combating the formation of biofilms.
All samples showed a lower biofilm (cell + matrix) area than the control sample (developed biofilm, 72 hours after cultivation), indicating the presence of biocide effects. The value of the effect decreased consistently in the following series: gentamycin (−20.4%), chloramphenicol (−14.7%), benzalkonium chloride (−9.9%), and tigecycline (−4.7%). The protocol allows the differentiation of such samples, confirming the applicability of the described approach.
Considering the biofilm suppression effect, the following relative order can be suggested based on the reduced matrix area: gentamycin, chloramphenicol (∼5%), > tigecycline (∼8%) > benzalkonium chloride (∼12%). For the combined biofilm and individual cell suppression effect, gentamycin showed the best performance (Fig. 6c) with a more uniform suppression profile (Fig. 6d). It is important to note that these observations should be considered preliminary, since many more samples of different biofilms should be studied to make general conclusions. In addition, killing curve measurements should also be performed. However, the computer vision approach, which is the main topic of the article, will remain unchanged.
The present study developed a methodology for high-speed automatic analysis, including obtaining data for cell growth/inhibition kinetics over time (Fig. 6a), distinguishing individual cells and the matrix (Fig. 6c), and establishing the shape/uniformity of profiles (Fig. 6d). It is a versatile approach for complex analysis and comprehensive treatment of visualization abilities provided by electron microscopy.
Conclusions
In the present study, an automated biofilm SEM image analysis protocol was reported for the first time with direct biofilm characterization at the macroscale. The challenge of macroscale statistical characterization is not only the amount of data but also the depth-insight level needed to reveal morphology changes. All computational techniques used in the study can be scaled in the future to provide analysis of 3D data recorded with volume electron microscopy. To highlight the difference, we compared the automated approach developed here with a regular manual analysis.In summary, the pipeline allowed processing, mapping and detection of the following:
1. 1330 SEM images (800 images for kinetic modeling of biofilm growth, 500 images for investigation of biocide effects, and 30 images for mapping of the macroscale biofilm region)
2. 1.15 cm2 of total analysed area, registered with negative overlaps and 0.1472 mm2 of total SEM image area.
3. 74![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 736 separate bacteria cells with the establishment of the size for the fraction of the recognized cells.
736 separate bacteria cells with the establishment of the size for the fraction of the recognized cells.
The development of a baseline segmentation neural network takes approximately 24 hours for an experienced data scientist. The training procedure with known hyperparameters can be estimated at 5 hours.
Manual data annotation of one SEM image (without labeling of separate cells) takes an average of 25 minutes. Therefore, the annotation of all images that were used would have taken approximately 558 hours for one person, whereas one neural network can provide the mapping of one image with an approximate time of 1 second (1500 times faster than a specialist; see section “Neural network processing time” in the ESI†).
As a result, a combination of fully automated scanning electron microscopy measurements with the use of machine intelligence was developed. Automated SEM enables imaging and mapping of large sample areas, acquiring reliable statistics about sample composition with significantly less time compared to manual labeling. The developed protocol, which combines an image stitching algorithm, a deep neural network for image segmentation, and robust computer vision techniques for object detection and clustering, provides automated analysis of SEM images, yielding areas of cells, cell-free zones, channels, and a matrix, number of separate cells, their sizes, and the number of cell clusters.
The described approach was tested against more than 1000 images of Staphylococcus aureus, allowing us not only to study macroscale areas of the sample with nanoscale image resolution but also to study the complex dynamics of biofilm formation and antibiotic tolerance. The digital solution is supposed to be used in SEM biofilm studies, helping to validate novel antibiofilm compounds with high-throughput computing and statistical significance. Of course, it should be mentioned that the SEM used as an experimental technique and the computational software developed have their own limitations in terms of accuracy, reliability and algorithms. We expect that further studies will elucidate these limitations in more detail and lead to the development of improved techniques.
Methods
Sample preparation
Electron microscopy measurements
The microstructure of the samples was studied by field emission scanning electron microscopy (FE-SEM) on Hitachi SU8000 and Hitachi Regulus 8230 electron microscopes. The automatic acquisition mode of data was implemented using the “Zigzag function”. A 10 × 10 image square pattern (100 images for each sample; the average recording time for one sample was approximately 1.5 h) with negative overlap was used to study each sample. The images were recorded at a magnification of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 times in the secondary electron mode at an accelerating voltage of 10–30 kV and a working distance of 8–10 mm. SEM image stitching was performed using ImageJ software with a grid/collection stitching plugin based on the Fourier shift theorem.101
000 times in the secondary electron mode at an accelerating voltage of 10–30 kV and a working distance of 8–10 mm. SEM image stitching was performed using ImageJ software with a grid/collection stitching plugin based on the Fourier shift theorem.101
Neural network implementation and training
Data labeling was performed using Labelbox infrastructure.102 All networks were implemented with PyTorch103 and trained using the PyTorch Lightning python package.104 The training was performed on a single NVIDIA 1080 Ti graphics card. Each model was trained on 1000 epochs and validated on 8 SEM images.Segmentation model PyTorch implementations were used.105 Augmentation was performed using the Albumentations python package.106 Basic augmentations include flips, rotations, shifts, scaling, sharpening, blurring, grid distortion, and cropping.
Hyperparameters include model architecture, encoder, optimizer, batch size, learning rate, crop height in augmentations, and special parameters in the loss function (α). In our model, the loss function is in the form:
| L = α·BCE loss + (1 − α)·Dice loss |
The hyperparameter optimization consisted of sequential model training with different possible values of the selected hyperparameter and fixed values of other hyperparameters. The optimization scheme and optimal hyperparameters are shown in Fig. 7.
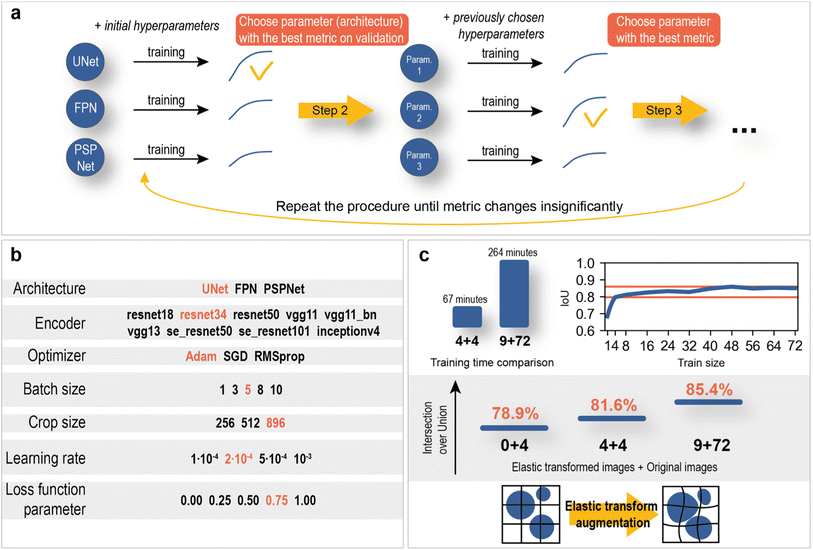 | ||
| Fig. 7 Neural network optimization. (a) hyperparameter optimization algorithm. (b) Possible hyperparameters (red – optimal parameters). (c) Train size and elastic transform augmentations effect. | ||
The training dataset size/validation IoU score relationship (Fig. 7c) was constructed to study the dependence of model quality on the number of training SEM images. As a result, an intersection over union score of 78.9% on validation was reached using only 4 training images. With further increases in the training size, the metric slowly reaches a value of approximately 85%. Such a good performance of the neural networks is most likely achieved due to the variety and large image size (2560 × 1760 px).
In addition, the use of elastic transformations (Fig. 7c) of the training images resulted in a 2.7% increase in the IoU score (0 + 4 vs. 4 + 4 elastic transformed images + original images). The computational cost of elastic transformation critically affects the learning time, which is why a set of pregenerated augmented images was used during training instead of real-time elastic transformations.
The final model consisted of 72 training images with 9 elastically transformed images. The training time averaged 4 hours and 24 minutes, which is almost 4 times slower than the training time on 4 original images with 4 elastic transformed images. The difference in IoU score was 3.8% (Fig. 7c). The results are statistically proven with the Mann–Whitney U test on two samples of models with a size of 4 (see section “Statistical significance of using larger amount of training data” in the ESI†). It can be concluded that segmentation models in this work do not demand significant amount of data, which greatly facilitates the data labeling process and reduces training time.
Computer vision methods
Image processing for detection was carried out with the OpenCV-Python107 package and included the following:1. Image reading.
2. Gaussian blurring for the following edge detection with a 3 × 3 kernel.
3. Canny edge detection with thresholds of 100 and 200.
4. One iteration of dilation with kernel 5 × 5.
5. Mask inversion.
6. Distance transform (distance type = L2)
7. Connected component computation (connectivity = 4, ltype = cv2. CV_32S)
8. Contour finding (cv2.RETR_LIST, cv2.CHAIN_APPROX_SIMPLE)
9. Ellipse fitting for bacterial size estimation.
Bacterial size data filtering was carried out with quantiles of 0.01 and 0.99.
Local maxima search (minimum distance between local maxima = 30, maximum_filter and minimum_filter with neighborhood_size = 25 and threshold difference of 5) and DBSCAN (eps = 0.1 after standard scaling procedure) were carried out with the SciPy Python package.108 Watershed segmentation was performed using the scikit-image Python package.109
Kinetic calculations
The neural network used for the computational study of biofilm development kinetics was trained with the same hyperparameters as for the main model but with additional images of an empty support and contrast and brightness augmentations to decrease model data uncertainty.The k1 rate constant was estimated by linear regression between the logarithm of cell-free zone ratios and time with sample weights (1/8, 1/8, 1/8, 1/8, 2/8, 1/8, 1/16, 1/16) as measures of prior knowledge certainties for each period of time.
The k2 rate constant was calculated using minimization of the function:
The half-life of the cell-free zone area was calculated with the following formula:
Time when the maximal cell area is reached:
Data availability
The code and a link for the files used for the analyses presented in this paper are available at https://github.com/Ananikov-Lab/cv4biofilms.Author contributions
Konstantin S. Kozlov, Daniil A. Boiko – algorithm development, programming, data processing, data analysis; Elena V. Detusheva, Konstantin V. Detushev, Anatoly N. Vereshchagin – microbiological study; Evgeniy O. Pentsak – electron microscopy study; Valentine P. Ananikov – conceptualization, algorithm development, data analysis, supervision. All authors – manuscript preparation and revisions.Conflicts of interest
The authors declare no competing financial or non-financial interests.Acknowledgements
Antimicrobial screening was performed in cooperation with the Molecular Microbiology Department of the Federal Budget Institution of Science State Research Center for Applied Biotechnology and Microbiology (FBSI SRC PMB, Obolensk, Russia) within the framework of the Sectoral Program of Rospotrebnadzor.References
- D. G. J. Larsson and C. F. Flach, Antibiotic resistance in the environment, Nat. Rev. Microbiol., 2022, 20(5), 257–269 CrossRef CAS PubMed.
- J. W. Costerton, P. S. Stewart and E. P. Greenberg, Bacterial Biofilms: A Common Cause of Persistent Infections, Science, 1999, 284(5418), 1318–1322 CrossRef CAS PubMed.
- D. Sharma, L. Misba and A. U. Khan, Antibiotics versus biofilm: an emerging battleground in microbial communities, Antimicrob. Resist. Infect. Control., 2019, 8(1), 76 CrossRef PubMed.
- M. Wainberg, D. Merico, A. Delong and B. J. Frey, Deep learning in biomedicine, Nat. Biotechnol., 2018, 36(9), 829–838 CrossRef CAS PubMed.
- E. C. McClure, M. Sievers, C. J. Brown, C. A. Buelow, E. M. Ditria and M. A. Hayes, et al., Artificial Intelligence Meets Citizen Science to Supercharge Ecological Monitoring, Patterns, 2020, 1(7), 100109 CrossRef PubMed.
- N. Beknazarov, S. Jin and M. Poptsova, Deep learning approach for predicting functional Z-DNA regions using omics data, Sci. Rep., 2020, 10(1), 19134 CrossRef CAS PubMed.
- H. Jeckel and K. Drescher, Advances and opportunities in image analysis of bacterial cells and communities, FEMS Microbiol. Rev., 2021, 45(4), fuaa062 CrossRef CAS PubMed.
- F. Piccinini, T. Balassa, A. Szkalisity, C. Molnar, L. Paavolainen and K. Kujala, et al., Advanced Cell Classifier: User-Friendly Machine-Learning-Based Software for Discovering Phenotypes in High-Content Imaging Data, Cell Syst., 2017, 4(6), 651–655 CrossRef CAS PubMed.
- J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov and O. Ronneberger, et al., Highly accurate protein structure prediction with AlphaFold, Nature, 2021, 596(7873), 583–589 CrossRef CAS PubMed.
- A. Zhavoronkov, Y. A. Ivanenkov, A. Aliper, M. S. Veselov, V. A. Aladinskiy and A. V. Aladinskaya, et al., Deep learning enables rapid identification of potent DDR1 kinase inhibitors, Nat. Biotechnol., 2019, 37(9), 1038–1040 CrossRef CAS PubMed . Available from: https://www.nature.com/articles/s41587-019-0224-x.
- B. H. Kann, A. Hosny and H. J. W. L. Aerts, Artificial intelligence for clinical oncology, Cancer Cell, 2021, 39(7), 916–927 CrossRef CAS PubMed . Available from: https://linkinghub.elsevier.com/retrieve/pii/S1535610821002105.
- X. Shen, X. Wu, R. Liu, H. Li, J. Yin and L. Wang, et al., Accurate segmentation of breast tumor in ultrasound images through joint training and refined segmentation, Phys. Med. Biol., 2022, 67(17), 175013 CrossRef PubMed.
- E. Check Hayden, The automated lab, Nature, 2014, 516(7529), 131–132 CrossRef CAS PubMed.
- J. M. Stokes, K. Yang, K. Swanson, W. Jin, A. Cubillos-Ruiz and N. M. Donghia, et al., A Deep Learning Approach to Antibiotic Discovery, Cell, 2020, 180(4), 688–702 CrossRef CAS PubMed.
- J. H. Yang, S. N. Wright, M. Hamblin, D. McCloskey, M. A. Alcantar and L. Schrübbers, et al., A White-Box Machine Learning Approach for Revealing Antibiotic Mechanisms of Action, Cell, 2019, 177(6), 1649–1661 CrossRef CAS PubMed.
- C. de la Fuente-Nunez, Antibiotic discovery with machine learning, Nat. Biotechnol., 2022, 40(6), 833–834 CrossRef CAS PubMed.
- J. Redshaw, D. S. J. Ting, A. Brown, J. D. Hirst and T. Gärtner, Krein support vector machine classification of antimicrobial peptides, Digital Discovery, 2023, 2, 502–511 RSC.
- S. Renaud and R. A. Mansbach, Latent spaces for antimicrobial peptide design, Digital Discovery, 2023, 2, 441–458 RSC.
- J. W. Costerton, G. G. Geesey and K. J. Cheng, How Bacteria Stick, Sci. Am., 1978, 238(1), 86–95 CrossRef CAS PubMed.
- R. M. Donlan, Biofilms: Microbial Life on Surfaces, Emerging Infect. Dis., 2002, 8(9), 881–890 CrossRef PubMed.
- M. Vert, Y. Doi, K. H. Hellwich, M. Hess, P. Hodge and P. Kubisa, et al., Terminology for biorelated polymers and applications (IUPAC Recommendations 2012), Pure Appl. Chem., 2012, 84(2), 377–410 CrossRef CAS.
- C. D. Nadell, K. Drescher and K. R. Foster, Spatial structure, cooperation and competition in biofilms, Nat. Rev. Microbiol., 2016, 14(9), 589–600 CrossRef CAS PubMed.
- O. Y. A. Costa, J. M. Raaijmakers and E. E. Kuramae, Microbial Extracellular Polymeric Substances: Ecological Function and Impact on Soil Aggregation, Front. Microbiol., 2018, 9, 1636 CrossRef PubMed.
- P. Di Martino, Extracellular polymeric substances, a key element in understanding biofilm phenotype, AIMS Microbiol., 2018, 4(2), 274–288 CAS.
- D. Campoccia, L. Montanaro and C. R. Arciola, Extracellular DNA (eDNA). A Major Ubiquitous Element of the Bacterial Biofilm Architecture, Int. J. Mol. Sci., 2021, 22(16), 9100 CrossRef CAS PubMed.
- Y. Baldera-Moreno, V. Pino, A. Farres, A. Banerjee, F. Gordillo and R. Andler, Biotechnological Aspects and Mathematical Modeling of the Biodegradation of Plastics under Controlled Conditions, Polymers, 2022, 14(3), 375 CrossRef CAS PubMed.
- X. Bai, C. H. Nakatsu and A. K. Bhunia, Bacterial Biofilms and Their Implications in Pathogenesis and Food Safety, Foods, 2021, 10(9), 2117 CrossRef CAS PubMed.
- C. Carrascosa, D. Raheem, F. Ramos, A. Saraiva and A. Raposo, Microbial Biofilms in the Food Industry—A Comprehensive Review, Int. J. Environ. Res. Public Health, 2021, 18(4), 2014 CrossRef CAS PubMed.
- E. M. Voglauer, B. Zwirzitz, S. Thalguter, E. Selberherr, M. Wagner and K. Rychli, Biofilms in Water Hoses of a Meat Processing Environment Harbor Complex Microbial Communities, Front. Microbiol., 2022, 13 Search PubMed.
- A. Y. An, K. Y. G. Choi, A. S. Baghela and R. E. W. Hancock, An Overview of Biological and Computational Methods for Designing Mechanism-Informed Anti-biofilm Agents, Front. Microbiol., 2021, 12 Search PubMed.
- M. Jamal, W. Ahmad, S. Andleeb, F. Jalil, M. Imran and M. A. Nawaz, et al., Bacterial biofilm and associated infections, J. Chin. Med. Assoc., 2018, 81(1), 7–11 CrossRef PubMed.
- S. Filardo, M. Di Pietro, G. Tranquilli and R. Sessa, Biofilm in Genital Ecosystem: A Potential Risk Factor for Chlamydia trachomatis Infection, Can. J. Infect. Dis. Med. Microbiol., 2019, 1–6 CAS.
- D. L. Hamilos, Biofilm Formations in Pediatric Respiratory Tract Infection, Curr. Infect. Dis. Rep., 2019, 21(2), 6 CrossRef PubMed.
- V. Folliero, G. Franci, F. Dell'Annunziata, R. Giugliano, F. Foglia and R. Sperlongano, et al., Evaluation of Antibiotic Resistance and Biofilm Production among Clinical Strain Isolated from Medical Devices, Int. J. Microbiol., 2021, 1–11 Search PubMed.
- N. C. T. Dadi, B. Radochová, J. Vargová and H. Bujdáková, Impact of Healthcare-Associated Infections Connected to Medical Devices—An Update, Microorganisms, 2021, 9(11), 2332 CrossRef PubMed.
- J. Goodwine, J. Gil, A. Doiron, J. Valdes, M. Solis and A. Higa, et al., Pyruvate-depleting conditions induce biofilm dispersion and enhance the efficacy of antibiotics in killing biofilms in vitro and in vivo, Sci. Rep., 2019, 9(1), 3763 CrossRef PubMed.
- A. Heydorn, A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov and B. K. Ersbøll, et al., Quantification of biofilm structures by the novel computer program comstat, Microbiology, 2000, 146(10), 2395–2407 CrossRef CAS PubMed.
- R. Hartmann, H. Jeckel, E. Jelli, P. K. Singh, S. Vaidya and M. Bayer, et al., Quantitative image analysis of microbial communities with BiofilmQ, Nat. Microbiol., 2021, 6(2), 151–156 CrossRef CAS PubMed.
- S. E. Mountcastle, N. Vyas, V. M. Villapun, S. C. Cox, S. Jabbari and R. L. Sammons, et al., Biofilm viability checker: An open-source tool for automated biofilm viability analysis from confocal microscopy images, npj Biofilms Microbiomes, 2021, 7(1), 44 CrossRef CAS PubMed.
- T. W. Hartman, E. Radichev, H. M. Ali, M. O. Alaba, M. Hoffman and G. Kassa, et al., BASIN: A Semi-automatic Workflow, with Machine Learning Segmentation, for Objective Statistical Analysis of Biomedical and Biofilm Image Datasets, J. Mol. Biol., 2023, 435(2), 167895 CrossRef CAS PubMed.
- R. Hartmann, M. C. F. Teeseling, M. Thanbichler and D. K. BacStalk, A comprehensive and interactive image analysis software tool for bacterial cell biology, Mol. Microbiol., 2020, 114(1), 140–150 CrossRef CAS PubMed.
- M. Zhang, J. Zhang, Y. Wang, J. Wang, A. M. Achimovich and S. T. Acton, et al., Non-invasive single-cell morphometry in living bacterial biofilms, Nat. Commun., 2020, 11(1), 6151 CrossRef CAS PubMed.
- S. Schlafer and R. L. Meyer, Confocal microscopy imaging of the biofilm matrix, J. Microbiol. Methods, 2017, 138, 50–59 CrossRef PubMed.
- C. L. Lin, F. S. Chen, L. J. Twu and M. J. J. Wang, Improving SEM Inspection Performance in Semiconductor Manufacturing Industry, Hum. Factors Ergon. Manuf., 2014, 24(1), 124–129 CrossRef.
- S. Rades, V. D. Hodoroaba, T. Salge, T. Wirth, M. P. Lobera and R. H. Labrador, et al., High-resolution imaging with SEM/T-SEM, EDX and SAM as a combined methodical approach for morphological and elemental analyses of single engineered nanoparticles, RSC Adv., 2014, 4(91), 49577–49587 RSC.
- S. Gupta, T. Omar and F. J. Muzzio, SEM/EDX and Raman chemical imaging of pharmaceutical tablets: A comparison of tablet surface preparation and analysis methods, Int. J. Pharm., 2022, 611, 121331 CrossRef CAS PubMed.
- P. Škarvada, R. Macků, D. S. Dallaeva, P. Sedlák, L. Grmela and P. Tománek, in SEM and AFM imaging of solar cells defects, ed. Tománek P., Senderáková D. and Páta P., 2015. p. 94501M Search PubMed.
- C. G. Golding, L. L. Lamboo, D. R. Beniac and T. F. Booth, The scanning electron microscope in microbiology and diagnosis of infectious disease, Sci. Rep., 2016, 6(1), 26516 CrossRef CAS PubMed.
- D. R. Beniac, S. L. Hiebert, C. G. Siemens, C. R. Corbett and T. F. Booth, A mobile biosafety microanalysis system for infectious agents, Sci. Rep., 2015, 5(1), 9505 CrossRef CAS PubMed.
- C. S. Goldsmith and S. E. Miller, Modern Uses of Electron Microscopy for Detection of Viruses, Clin. Microbiol. Rev., 2009, 22(4), 552–563 CrossRef PubMed.
- G. G. Anderson, J. J. Palermo, J. D. Schilling, R. Roth, J. Heuser and S. J. Hultgren, Intracellular Bacterial Biofilm-Like Pods in Urinary Tract Infections, Science, 2003, 301(5629), 105–107 CrossRef CAS PubMed.
- L. Kirchhoff, D. Arweiler-Harbeck, J. Arnolds, T. Hussain, S. Hansen and R. Bertram, et al., Imaging studies of bacterial biofilms on cochlear implants—Bioactive glass (BAG) inhibits mature biofilm, PLoS One, 2020, 15(2), e0229198 CrossRef CAS PubMed.
- T. Misra, M. Tare and P. N. Jha, Insights Into the Dynamics and Composition of Biofilm Formed by Environmental Isolate of Enterobacter cloacae, Front. Microbiol., 2022, 13 Search PubMed.
- C. Hannig, M. Follo, E. Hellwig and A. Al-Ahmad, Visualization of adherent micro-organisms using different techniques, J. Med. Microbiol., 2010, 59(1), 1–7 CrossRef CAS PubMed.
- L. C. Gomes and F. J. Mergulhão, SEM Analysis of Surface Impact on Biofilm Antibiotic Treatment, Scanning, 2017, 2017, 1–7 CrossRef PubMed.
- O. Ronneberger, P. Fischer and T. Brox. U-Net: Convolutional Networks for Biomedical Image Segmentation, 2015 Search PubMed.
- M. R. Wilkinson, L. Pereira Diaz, A. D. Vassileiou, J. A. Armstrong, C. J. Brown and B. Castro-Dominguez, et al., Predicting pharmaceutical powder flow from microscopy images using deep learning, Digital Discovery, 2023, 2, 459–470 RSC.
- S. Lu, B. Montz, T. Emrick and A. Jayaraman, Semi-supervised machine learning workflow for analysis of nanowire morphologies from transmission electron microscopy images, Digital Discovery, 2022, 1(6), 816–833 RSC.
- D. A. Boiko, E. O. Pentsak, V. A. Cherepanova, E. G. Gordeev and V. P. Ananikov, Deep neural network analysis of nanoparticle ordering to identify defects in layered carbon materials, Chem. Sci., 2021, 12(21), 7428–7441 RSC.
- A. S. Kashin, D. A. Boiko and V. P. Ananikov, Neural Network Analysis of Electron Microscopy Video Data Reveals the Temperature-Driven Microphase Dynamics in the Ions/Water System, Small, 2021, 17(24), 2007726 CrossRef CAS PubMed.
- I. Squires, A. Dahari, S. J. Cooper and S. Kench, Artefact removal from micrographs with deep learning based inpainting, Digital Discovery, 2023, 2, 316–326 RSC.
- E. Moen, D. Bannon, T. Kudo, W. Graf, M. Covert and D. Van Valen, Deep learning for cellular image analysis, Nat. Methods, 2019, 16(12), 1233–1246 CrossRef CAS PubMed.
- J. W. Johnson. Adapting Mask-RCNN for Automatic Nucleus Segmentation, 2018 Search PubMed.
- O. Z. Kraus, J. L. Ba and B. J. Frey, Classifying and segmenting microscopy images with deep multiple instance learning, Bioinformatics, 2016, 32(12), i52–i59 CrossRef CAS PubMed.
- J. J. Almagro Armenteros, C. K. Sønderby, S. K. Sønderby, H. Nielsen and O. Winther, DeepLoc: prediction of protein subcellular localization using deep learning, Bioinformatics, 2017, 33(21), 3387–3395 CrossRef PubMed.
- O. Z. Kraus, B. T. Grys, J. Ba, Y. Chong, B. J. Frey and C. Boone, et al., Automated analysis of high-content microscopy data with deep learning, Mol. Syst. Biol., 2017, 13(4), 924 CrossRef PubMed.
- D. P. Sullivan and E. Lundberg, Seeing More: A Future of Augmented Microscopy, Cell, 2018, 173(3), 546–548 CrossRef CAS PubMed.
- E. Gómez-de-Mariscal, C. García-López-de-Haro, W. Ouyang, L. Donati, E. Lundberg and M. Unser, et al., DeepImageJ: A user-friendly environment to run deep learning models in ImageJ, Nat. Methods, 2021, 18(10), 1192–1195 CrossRef PubMed.
- Z. Liu, H. Zhang, L. Jin, J. Chen, A. Nedzved and S. Ablameyko, et al., U-Net-based deep learning for tracking and quantitative analysis of intracellular vesicles in time-lapse microscopy images, J. Innov. Opt. Health Sci., 2022,(05), 15 CAS.
- H. Yang, J. Yu, L. Jin, Y. Zhao, Q. Gao and C. Shi, et al., A deep learning based method for automatic analysis of high-throughput droplet digital PCR images, Analyst, 2023, 148(2), 239–247 RSC.
- N. Vyas, R. L. Sammons, O. Addison, H. Dehghani and A. D. Walmsley, A quantitative method to measure biofilm removal efficiency from complex biomaterial surfaces using SEM and image analysis, Sci. Rep., 2016, 6(1), 32694 CrossRef CAS PubMed . Available from: https://www.nature.com/articles/srep32694.
- I. Arganda-Carreras, V. Kaynig, C. Rueden, K. W. Eliceiri, J. Schindelin and A. Cardona, et al., Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification, Bioinformatics, 2017, 33(15), 2424–2426 CrossRef CAS PubMed.
- A. J. Perez, M. Seyedhosseini, T. J. Deerinck, E. A. Bushong, S. Panda and T. Tasdizen, et al., A workflow for the automatic segmentation of organelles in electron microscopy image stacks, Front. Neuroanat., 2014, 8, 126 CrossRef PubMed.
- M. Žerovnik Mekuč, C. Bohak, S. Hudoklin, B. H. Kim, R. Romih and M. Y. Kim, et al., Automatic segmentation of mitochondria and endolysosomes in volumetric electron microscopy data, Comput. Biol. Med., 2020, 119, 103693 CrossRef PubMed.
- M. Žerovnik Mekuč, C. Bohak, E. Boneš, S. Hudoklin, R. Romih and M. Marolt, Automatic segmentation and reconstruction of intracellular compartments in volumetric electron microscopy data, Comput. Methods Programs Biomed., 2022, 223, 106959 CrossRef PubMed.
- H.-F. Yang and Y. Choe, Cell tracking and segmentation in electron microscopy images using graph cuts, in 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, IEEE, 2009. p. 306–309 Search PubMed.
- Z. Li, C. Li, Y. Yao, J. Zhang, M. M. Rahaman and H. Xu, et al., EMDS-5: Environmental Microorganism image dataset Fifth Version for multiple image analysis tasks, PLoS One, 2021, 16(5), e0250631 CrossRef CAS PubMed.
- A. D. Chakravarthy, P. Chundi, M. Subramaniam, S. Ragi and V. R. Gadhamshetty. A Thrifty Annotation Generation Approach for Semantic Segmentation of Biofilms, in 2020 IEEE 20th International Conference on Bioinformatics and Bioengineering (BIBE), IEEE, 2020, p. 602–607 Search PubMed.
- R. Hollandi, A. Szkalisity, T. Toth, E. Tasnadi, C. Molnar and B. Mathe, et al., nucleAIzer: A Parameter-free Deep Learning Framework for Nucleus Segmentation Using Image Style Transfer, Cell Syst., 2020, 10(5), 453–458 CrossRef CAS PubMed.
- R. Hollandi, Á. Diósdi, G. Hollandi, N. Moshkov and P. Horváth, AnnotatorJ: an ImageJ plugin to ease hand annotation of cellular compartments, Mol. Biol. Cell., 2020, 31(20), 2179–2186 CrossRef CAS PubMed.
- J. Zhang, C. Li, S. Kosov, M. Grzegorzek, K. Shirahama and T. Jiang, et al., LCU-Net: A novel low-cost U-Net for environmental microorganism image segmentation, Pattern Recognit., 2021, 115, 107885 CrossRef.
- A. Khadangi, T. Boudier and V. Rajagopal, EM-net: Deep learning for electron microscopy image segmentation, in 2020 25th International Conference on Pattern Recognition (ICPR), IEEE, 2021, p. 31–38 Search PubMed.
- R. J. Palmer and C. Sternberg, Modern microscopy in biofilm research: confocal microscopy and other approaches, Curr. Opin. Biotechnol., 1999, 10(3), 263–268 CrossRef CAS PubMed.
- R. P. Dassanayake, S. M. Falkenberg, J. A. Stasko, A. L. Shircliff, J. D. Lippolis and R. E. Briggs, Identification of a reliable fixative solution to preserve the complex architecture of bacterial biofilms for scanning electron microscopy evaluation, PLoS One, 2020, 15(5), e0233973 CrossRef CAS PubMed.
- D. O. Serra, A. M. Richter, G. Klauck, F. Mika and R. Hengge, Microanatomy at Cellular Resolution and Spatial Order of Physiological Differentiation in a Bacterial Biofilm, mBio, 2013, 4(2), e100103 CrossRef PubMed.
- A. P. Tiwari, D. P. Bhattarai, B. Maharjan, S. W. Ko, H. Y. Kim and C. H. Park, et al., Polydopamine-based Implantable Multifunctional Nanocarpet for Highly Efficient Photothermal-chemo Therapy, Sci. Rep., 2019, 9(1), 2943 CrossRef PubMed.
- F. Pantanella, P. Valenti, T. Natalizi, D. Passeri and F. Berlutti, Analytical techniques to study microbial biofilm on abiotic surfaces: pros and cons of the main techniques currently in use, Ann. Ig., 2013, 25(1), 31–42 CAS.
- Y. Deng, L. Wang, Y. Chen and Y. Long, Optimization of staining with SYTO 9/propidium iodide: interplay, kinetics and impact on Brevibacillus brevis, Biotechniques, 2020, 69(2), 88–98 CrossRef CAS PubMed.
- M. E. Fuller, S. H. Streger, R. K. Rothmel, B. J. Mailloux, J. A. Hall and T. C. Onstott, et al., Development of a Vital Fluorescent Staining Method for Monitoring Bacterial Transport in Subsurface Environments, Appl. Environ. Microbiol., 2000, 66(10), 4486–4496 CrossRef CAS PubMed.
- X. Han and C. K. Payne, Effect of Thioflavin T on the Elongation Rate of Bacteria, Bioelectricity, 2022, 4(1), 12–17 CrossRef.
- M. Relucenti, G. Familiari, O. Donfrancesco, M. Taurino, X. Li and R. Chen, et al., Microscopy Methods for Biofilm Imaging: Focus on SEM and VP-SEM Pros and Cons, Biology, 2021, 10(1), 51 CrossRef CAS PubMed.
- A. P. Jardine, F. Montagner, R. M. Quintana, I. M. Zaccara and P. M. P. Kopper, Antimicrobial effect of bioceramic cements on multispecies microcosm biofilm: a confocal laser microscopy study, Clin. Oral Investig., 2019, 23(3), 1367–1372 CrossRef PubMed.
- M. Idrees, S. Sawant, N. Karodia and A. Rahman, Staphylococcus aureus Biofilm: Morphology, Genetics, Pathogenesis and Treatment Strategies, Int. J. Environ. Res. Public Health, 2021, 18(14), 7602 CrossRef CAS PubMed.
- C. J. Lerche, F. Schwartz, M. Theut, E. L. Fosbøl, K. Iversen and H. Bundgaard, et al., Anti-biofilm Approach in Infective Endocarditis Exposes New Treatment Strategies for Improved Outcome, Front. Cell Dev. Biol., 2021, 9, 643335 CrossRef PubMed.
- N. Pant and D. P. Eisen, Non-Antimicrobial Adjuvant Strategies to Tackle Biofilm-Related Staphylococcus aureus Prosthetic Joint Infections, Antibiotics, 2021, 10(9), 1060 CrossRef CAS PubMed.
- Y. LeCun, Y. Bengio and G. Hinton, Deep learning, Nature, 2015, 521(7553), 436–444 CrossRef CAS PubMed.
- N. A. Sutton, N. Hughes and P. S. Handley, A comparison of conventional SEM techniques, low temperature SEM and the electro scan wet scanning electron microscope to study the structure of a biofilm of Streptococcus crista CR3, J. Appl. Bacteriol., 1994, 76(5), 448–454 CrossRef CAS PubMed.
- S. Ragi, M. H. Rahman, J. Duckworth, K. Jawaharraj, P. Chundi and V. Gadhamshetty, Artificial Intelligence-driven Image Analysis of Bacterial Cells and Biofilms, 2021 Search PubMed.
- F. Gomes, B. Leite, P. Teixeira, J. Azeredo and R. Oliveira, Farnesol in combination with N-acetylcysteine against Staphylococcus epidermidis planktonic and biofilm cells, Braz. J. Microbiol., 2012, 43(1), 235–242 CrossRef CAS PubMed.
- R. P. Dassanayake, S. M. Falkenberg, J. A. Stasko, A. L. Shircliff, J. D. Lippolis and R. E. Briggs, Identification of a reliable fixative solution to preserve the complex architecture of bacterial biofilms for scanning electron microscopy evaluation, PLoS One, 2020, 15(5), e0233973 CrossRef CAS PubMed.
- S. Preibisch, S. Saalfeld and P. Tomancak, Globally optimal stitching of tiled 3D microscopic image acquisitions, Bioinformatics, 2009, 25(11), 1463–1465 CrossRef CAS PubMed.
- Labelbox, Labelbox, 2023, available: https://labelbox.com Search PubMed.
- A. Paszke, S. Gross, F. Massa, A. Lerer, J. Bradbury and G. Chanan, et al., PyTorch: An Imperative Style, High-Performance Deep Learning Library, Advances in Neural Information Processing Systems 32, ed. Wallach H., Larochelle H., Beygelzimer A., d'textquotesingle Alché-Buc F., Fox E. and Garnett R., Curran Associates, Inc., 2019, p. 8024–8035 Search PubMed.
- W. A. Falcon, et al., PyTorch Lightning, GitHub, https://github.com/PyTorchLightning/pytorch-lightning. 2019.
- P. Yakubovskiy, Segmentation Models Pytorch, GitHub repository, GitHub, 2020 Search PubMed.
- A. Buslaev, V. I. Iglovikov, E. Khvedchenya, A. Parinov, M. Druzhinin and A. A. Kalinin, Albumentations: Fast and Flexible Image Augmentations, Information, 2020, 11(2), 125 CrossRef.
- G. Bradski, The OpenCV Library, Dr Dobb’s Journal of Software Tools, 2000 Search PubMed.
- P. Virtanen, R. Gommers, T. E. Oliphant, M. Haberland, T. Reddy, D. Cournapeau, E. Burovski, P. Peterson, W. Weckesser, J. Bright, S. J. van der Walt, M. Brett, J. Wilson, K. J. Millman, N. Mayorov, A. R. J. Nelson, E. Jones, R. Kern, E. Larson, C. J. Carey, İ. Polat, Y. Feng, E. W. Moore, J. VanderPlas, D. Laxalde, J. Perktold, R. Cimrman, I. Henriksen, E. A. Quintero, C. R. Harris, A. M. Archibald, A. H. Ribeiro, F. Pedregosa, P. van Mulbregt and SciPy 1.0 Contributors, SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python, Nat. Methods, 2020, 17(3), 261–272 CrossRef CAS PubMed.
- S. van der Walt, J. L. Schönberger, J. Nunez-Iglesias, F. Boulogne, J. D. Warner and N. Yager, et al., scikit-image: image processing in Python, PeerJ, 2014, 2, e453 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3dd00048f |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2023 |