Open Access Article
Open Access ArticleGeneralizing property prediction of ionic liquids from limited labeled data: a one-stop framework empowered by transfer learning†
Guzhong
Chen
 ab,
Zhen
Song
ab,
Zhen
Song
 *a,
Zhiwen
Qi
*a,
Zhiwen
Qi
 *a and
Kai
Sundmacher
*a and
Kai
Sundmacher
 bc
bc
aState Key Laboratory of Chemical Engineering, School of Chemical Engineering, East China University of Science and Technology, 130 Meilong Road, Shanghai 200237, China. E-mail: songz@ecust.edu.cn; zwqi@ecust.edu.cn
bProcess Systems Engineering, Max Planck Institute for Dynamics of Complex Technical Systems, Sandtorstr. 1, D-39106 Magdeburg, Germany
cProcess Systems Engineering, Otto-von-Guericke University Magdeburg, Universitätsplatz 2, D-39106 Magdeburg, Germany
First published on 12th May 2023
Abstract
Ionic liquids (ILs) could find use in almost every chemical process due to their wide spectrum of unique properties. The crux of the matter lies in whether a task-specific IL selection from enormous chemical space can be achieved by property prediction, for which limited labeled data represents a major obstacle. Here, we propose a one-stop ILTransR (IL transfer learning of representations) that employs large-scale unlabeled data for generalizing IL property prediction from limited labeled data. By first pre-training on ∼10 million IL-like molecules, IL representations are derived from the encoder state of a transformer model. Employing the pre-trained IL representations, convolutional neural network (CNN) models for IL property prediction are trained and tested on eleven datasets of different IL properties. The obtained ILTransR presents superior performance as opposed to state-of-the-art models in all benchmarks. The application of ILTransR is exemplified by extensive screening of CO2 absorbent from a huge database of 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 333
333![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 096 synthetically-feasible ILs.
096 synthetically-feasible ILs.
1 Introduction
Ionic liquids (ILs) are molten salts comprised fully of cations and anions, which can remain in liquid state around room temperature. In recent years, ILs have attracted remarkable attention in various applications, both in chemistry and engineering,1,2 due to their unique physicochemical properties such as negligible vapor pressure, high thermal and electrochemical stability, wide liquidus range, etc.2,3 More importantly, ILs also offer great potential to tune their physical and chemical properties by judicious selection of the cations and anions. For this reason, ILs could be designed to offer desirable properties to meet specific requirements for arbitrary given applications. The challenge, however, is to accurately evaluate various IL properties related to the target performance and identify optimal ILs from the nearly infinite combinations of possible cations and anions.4,5So far, IL selection toward a specific process mainly relies on laborious trial-and-error experiments. However, such approaches are not only very time-consuming but also limited to a small IL chemical space, leaving many potentially promising structures unexplored. Alternatively, computational methods can be used for estimating the properties of ILs and IL-involved mixtures.6 Traditional models such as equations of states (EoSs)7 and group contribution models (GCMs)8,9 have been widely employed for estimating the thermodynamic, transport, and EHS (environment, health, and safety) related properties of ILs. Nevertheless, both the two schemes are prone to the inherent weakness of limited predictive power and/or insufficient accuracy.8 Another computational method for IL property prediction is the quantitative structure–property relationship (QSPR) approach, wherein a property of interest is correlated quantitatively with certain descriptors of involved molecules9,10 (for which machine learning methods have recently gained popularity11–17). Notably, the availability of IL property databases like ILThermo18 has stimulated the use of ML methods for modeling IL properties, wherein diverse types of molecular descriptors were used as IL representation.19–26 However, despite the high accuracy achieved by these models, such models still suffer from the inherent weakness of molecular descriptors for IL representation as well as the relatively limited databases of IL properties available for model development. Moreover, manually engineered IL descriptors usually require expert knowledge of specific types of ILs and the properties to be modeled, which could work well for specific tasks but may not generalize well for others.27 In the past few years, there has been rapid progress in ML methods, particularly deep neural networks (DNNs). These DNN-based methods have garnered significant attention due to their ability to overcome the limitations of conventional models and achieve high accuracy in predicting complex tasks.28–31 The growth of deep learning (DL) has offered excellent flexibility and performance to learn molecular representations from data, without explicit guides from experts.32–34 Typically, a sufficiently large labeled training dataset is desirable for developing DL approaches.35 This is practical in areas like image classification as the number of labeled samples could easily reach several millions or even more. However, it is obviously not the case for IL properties prediction, for which the labeled datasets available are far smaller than such a scale and insufficient as opposed to the giant chemical space of potential ILs. DL models trained on such a limited dataset can easily get overfit while generalizing poorly on IL molecules dissimilar to the training set.
The issue of developing generalized models based on limited datasets is not unique to molecular property prediction, but is also encountered in natural language processing (NLP) where there exists a vast amount of unlabeled data but only a limited proportion of labeled data.36 To address this challenge in NLP, researchers have made extensive efforts, including the pre-training and fine-tuning approach.37 This approach works by deriving word representations from statistical analysis of large unlabeled text corpora during pre-training; the resultant pre-trained representations provide valuable distributional information about words that can improve the generalization of models trained on limited labeled data via fine-tuning. The structure of molecular sequences is inherently similar to that of natural language sentences when molecules are represented by the simplified molecular-input line-entry system (SMILES).38,39 Online databases like PubChem and ChEMBL contain millions of readily accessible molecules. By leveraging such large-scale unlabeled datasets, pre-training can enable the learning of molecular representations, which can be subsequently fine-tuned for downstream molecular property prediction tasks with a smaller set of labeled data. Winter et al.40 have developed a pre-trained sequence-to-sequence (seq2seq) model based on recurrent neural networks (RNNs) for predicting molecular properties. Gómez-Bombarelli et al.32 have utilized variational autoencoders (VAEs) to obtain continuous representations of molecules in a latent space, which are subsequently used to predict molecular properties by decoding SMILES from the learned representations. In addition to these approaches, the transformer model41 that features a more parallelizable encoder-decoder architecture (superior to the aforementioned seq2seq models) has also been employed for molecular property prediction42,43 and reaction prediction.44,45 This approach has demonstrated higher performance on small databases than other pre-training methods.27
As ILs are genetically distinct from conventional molecules, molecular representations derived from conventional molecules can hardly be expected to generalize well for ILs. However, until now, DL-based IL representations have not been considered. Despite numerous ML works reported on the property prediction of this limelighted class of molecules, these works have generally employed traditional molecular descriptors as input features. To bridge this gap, we propose ILTransR (IL transfer learning of representations), a pre-training and fine-tuning two-stage framework in this article (see Fig. 1). Importantly, ILTransR does not make use of any manually engineered molecular fingerprint. Instead, a self-attention mechanism is used to learn the high-dimensional structure of ILs from SMILES. First, a large (∼10 million) unlabeled SMILES dataset specifically composed of IL-like molecules is exploited for the unsupervised pre-training of the self-attention mechanism (i.e., IL transformer model), obtaining the encoder-decoder architecture that can well capture the structural information of an IL from its SMILES. Following that, the encoder of the IL transformer model is integrated with a convolutional neural network (CNN) architecture for the supervised training of predictive models of IL properties. By simply switching the labeled IL property dataset (and concatenating other necessary inputs such as temperature and pressure if needed), predictive models for various IL properties can be developed based on the proposed framework.
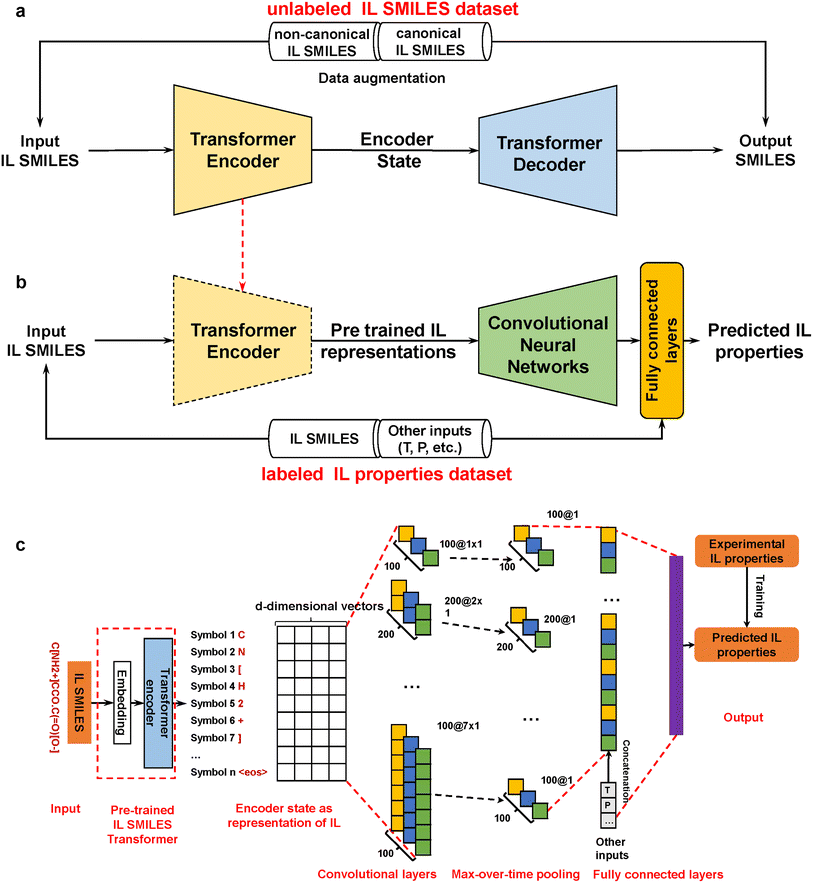 | ||
| Fig. 1 Overview of ILTransR. (a) Unsupervised pre-training of IL transformer. A large unlabeled SMILES database of IL-like molecules is taken for unsupervised pre-training to obtain the encoder-decoder architecture. (b) Supervised training of IL property prediction model. The encoder of the pre-trained IL transformer as learned IL representation is integrated with a CNN architecture (adding temperature/pressure in necessary cases). (c) Detailed ILTransR architecture for IL properties prediction. After IL SMILES is encoded by the pre-trained IL transformer, the CNN mainly uses a one-dimensional convolutional layer and a max-over-time pooling layer,46 giving rise to a fixed-length vector representation. This IL representation (concatenated with temperature/pressure if necessary) goes through fully connected layers and converts to the output layer for IL properties prediction. | ||
It is worth mentioning that very few molecular representation studies have been benchmarked with properties dependent on temperature and/or pressure.47 In this work, based on the modeling of eleven IL properties, we demonstrate that the ILTransR can well handle different types of inputs namely the IL structure and temperature and/or pressure. In comparison to literature-reported models trained by supervised learning, the ILTransR remarkably improves the performance in all these benchmark cases; on some of these IL properties, our model rivals or even exceeds the corresponding supervised learning baselines that have not taken rigorous dataset splitting strategy. Moreover, the one-stop ILTransR for predicting different IL properties enables high-throughput IL screening toward a specific task, as exemplified by the screening of the most promising CO2 absorbents from 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 333
333![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 096 synthetically-feasible ILs.26 Data and code involved in this work are publicly provided online at https://github.com/GuzhongChen/ILTransR.
096 synthetically-feasible ILs.26 Data and code involved in this work are publicly provided online at https://github.com/GuzhongChen/ILTransR.
2 Methods
2.1 Framework of ILTransR
The ILTransR proposed in this work is developed upon a pre-training and fine-tuning two-stage framework, wherein the pre-training stage is inherited from the transformer architecture originally constructed for neural machine translation (NMT) tasks41 and the fine-tuning stage is evolved from the text-CNN structure46 originally developed for sentence classification.Like NMT tasks, the IL transformer model is trained on a translation task from non-canonical SMILES to canonical SMILES. It is based on the encoder-decoder architecture, which is similar to the aforementioned seq2seq models used for molecular property prediction43,48, and reaction prediction.44 Its main architectural difference from the aforementioned seq2seq models is that the RNN component is removed and it is fully based on the attention mechanism combined with positional embedding for encoding sequential information. A more detailed description of the encoder-decoder architecture, attention mechanism, and positional encoding that comprise the building blocks of the IL transformer can be found in the ESI (Note 1†). In this work, the IL transformer is pre-trained on a large unlabeled SMILES database of more than 9 million IL-like molecules.
As different ILs have different length of SMILES, the input size of the downstream property prediction models (the output of the IL SMILES transformer encoder) can vary from case to case. Therefore, the convolutional neural network (CNN) structure originally developed for sentence classification46 is used for the downstream IL properties prediction model as such structure can conveniently deal with distinct input lengths. The pre-trained IL transformer is then fine-tuned for IL property prediction by CNN (see Fig. 1c). To be specific, the encoder of the pre-trained IL transformer is utilized to generate latent representations of input ILs, and afterward, the CNN mainly uses a one-dimensional convolutional layer and a max-over-time pooling layer.46 The input of the CNN model is a matrix of n × k, where n refers to the number of symbols in an IL SMILES and k denotes the dimension of the vector corresponding to each symbol. 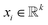 is used here to represent the k dimension embedding of the ith symbol in the IL SMILES string. On the input matrix n × k, a kernel
is used here to represent the k dimension embedding of the ith symbol in the IL SMILES string. On the input matrix n × k, a kernel 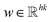 and a window xi:i+h−1 are used to perform convolution operations to generate a feature ci, that is, ci = f(w·xi:i+h−1 + b). Herein, xi:i+h−1 represents a window of h × k formed by row i to row i + h − 1 of the input matrix, which is formed by splicing xi, xi+1, …, xi+h−1; h denotes the number of symbols in the window; w is a h × k-dimensional weight matrix; b is the offset parameter and f is a non-linear function; w·xi:i+h−1 is the dot product operation. The filter is applied to the SMILES string, moving from top to bottom one step at a time (i = 1 … n − h + 1). Each convolution operation is equivalent to a feature vector extraction. By defining different windows, different feature vectors can be extracted to form the output of the convolutional layer. For the pooling layer, this work uses max-over-time pooling, and then the filtered largest features are spliced together to form a fixed-length vector representation.
and a window xi:i+h−1 are used to perform convolution operations to generate a feature ci, that is, ci = f(w·xi:i+h−1 + b). Herein, xi:i+h−1 represents a window of h × k formed by row i to row i + h − 1 of the input matrix, which is formed by splicing xi, xi+1, …, xi+h−1; h denotes the number of symbols in the window; w is a h × k-dimensional weight matrix; b is the offset parameter and f is a non-linear function; w·xi:i+h−1 is the dot product operation. The filter is applied to the SMILES string, moving from top to bottom one step at a time (i = 1 … n − h + 1). Each convolution operation is equivalent to a feature vector extraction. By defining different windows, different feature vectors can be extracted to form the output of the convolutional layer. For the pooling layer, this work uses max-over-time pooling, and then the filtered largest features are spliced together to form a fixed-length vector representation.
After a dropout layer to deal with overfitting, the pooling result is then concatenated with other inputs (i.e., temperature and/or pressure if necessary) for IL properties prediction. Finally, the data go through fully connected layers and convert to the one-neuron output layer for the prediction of IL properties. It is worth noting that as the prediction of the eleven IL properties involved in this work are all regression problems, only one neuron is needed in the output layer here; if there are IL related classification or multiple regression problems, one can also easily set the output layer neurons to the required number.
2.2 Datasets
For pre-training the IL transformer of ILTransR, the PubChem48 compound database (ftp://ftp.ncbi.nlm.nih.gov/pubchem/Compound/) is used. The original database contains a total of 108![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 923
923![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 995 molecules along with their canonical SMILES representations. Due to the limited memory size of the computer used, we cannot do the pre-training on the entire PubChem compound database. Therefore, considering the aim of learning IL representation, only the molecules containing ‘+’ and/or ‘−’ symbols in the SMILES are first screened to form a subset of IL-like molecules, retaining 10
995 molecules along with their canonical SMILES representations. Due to the limited memory size of the computer used, we cannot do the pre-training on the entire PubChem compound database. Therefore, considering the aim of learning IL representation, only the molecules containing ‘+’ and/or ‘−’ symbols in the SMILES are first screened to form a subset of IL-like molecules, retaining 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 243
243![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 410 structures. As illustrated in Fig. 2, 92.10% of IL-like molecules in the subset has SMILES strings with less than 100 characters. To avoid excessively long input length for the model, only molecules with SMILES strings of length less than or equal to 100 characters are used to form the pre-training dataset (retaining 9
410 structures. As illustrated in Fig. 2, 92.10% of IL-like molecules in the subset has SMILES strings with less than 100 characters. To avoid excessively long input length for the model, only molecules with SMILES strings of length less than or equal to 100 characters are used to form the pre-training dataset (retaining 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 434
434![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 070 structures). This dataset is then augmented 10-fold (as recommended by Tetko et al.49) using the SMILES enumerator to enhance the performance of DNN models that can be developed, resulting in a total of 94
070 structures). This dataset is then augmented 10-fold (as recommended by Tetko et al.49) using the SMILES enumerator to enhance the performance of DNN models that can be developed, resulting in a total of 94![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 340
340![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 non-canonical SMILES strings.50
700 non-canonical SMILES strings.50
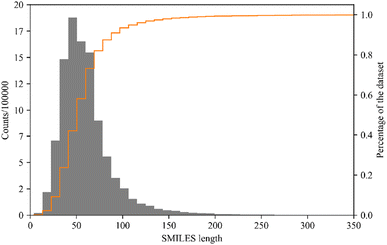 | ||
| Fig. 2 Distribution of the length of canonical SMILES representations of molecules in the IL-like dataset. | ||
For the fine-tuning of ILTransR, the datasets of eleven IL properties benchmarked in this work are derived from several recent refs. 20–22, 24–26 and 51 as listed in Table 1. From these datasets, only ILs with SMILES string length less than or equal to 100 characters (consistent with the pre-training dataset) are kept.
| Property | Number of data points | Number of ILs | Data source |
|---|---|---|---|
| Melting point Tm(K) | 2212 | 2212 | Low et al.20 |
| Glass transition temperature Tg(°C) | 609 | 609 | Venkatraman et al.25 |
| Thermal decomposition temperature Td(°C) | 1223 | 1223 | Venkatraman et al.25 |
| Heat capacity ln(Cp) | 9083 | 236 | Venkatraman et al.25 |
| Refractive index nD | 3009 | 464 | Venkatraman et al.25 |
| Density ρ | 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 167 167 |
2257 | Paduszyński21 |
| Viscosity ln(η) | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 368 368 |
1964 | Paduszyński24 |
| Surface tension γ | 2972 | 331 | Venkatraman et al.25 |
| CO2 solubility xCO2 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 116 116 |
124 | Song et al.22 |
| Cytotoxicity towards the leukemia rat cell line IPC-81 log10(EC50) | 326 | 326 | Wang et al.51 |
| Thermal conductivity λ | 454 | 73 | Venkatraman et al.26 |
2.3 Implementation details
This work applies RDKit (https://www.rdkit.org) for processing IL SMILES and for generating canonical SMILES used in the pre-training. For the implementation and training of the proposed ILTransR, the MXNet library52 with GPU acceleration (on a single RTX2080Ti and CUDA 10.1) and GluonNLP toolkit53 are employed.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 434
434![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 070 molecules in the pre-training dataset. The characters as well as their indexes in the vocabulary are detailed in the ESI (Note 3†).
070 molecules in the pre-training dataset. The characters as well as their indexes in the vocabulary are detailed in the ESI (Note 3†).
Considering the much smaller vocabulary required and the less complicacy of the SMILES canonicalization task than common NMT tasks, the numbers of transformer blocks, heads in multi-head attention, and units for the output are decreased from 6, 8 and 512 to 3, 4 and 128, respectively, with reference to the original paper.39 Identical to the original paper, a dropout rate of 0.1 is set for model regularization. The IL transformer is trained for 10 epochs by Adam optimizer57 with a base learning rate of 0.001. After four epochs of training, the learning rate is multiplied by a factor of 0.5 for each epoch.
The masked softmax cross entropy loss41 is used as the loss function for pre-training, which is implemented by the gluonnlp.loss.MaskedSoftmaxCELoss() function. From the pre-training dataset after augmentation (containing 94![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 340
340![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 SMILES), 100
700 SMILES), 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 100
000 and 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 SMILES are randomly split to form the validation set and test set, while the rest of the pre-training dataset is kept as the training set.
000 SMILES are randomly split to form the validation set and test set, while the rest of the pre-training dataset is kept as the training set.
3 Results and discussion
3.1 Performance of ILTransR
To evaluate the effectiveness of the IL transformer model, two standard evaluation metrics are utilized. The first metric is the bilingual evaluation understudy (BLEU) score,54 a standard measure used to assess the similarity between a given translation (i.e., the output canonical SMILES generated by the IL transformer model) and the reference translation (i.e., the original canonical SMILES). The second metric is the translation accuracy, which is calculated based on the number of perfect matches between the predicted and actual canonical SMILES. As illustrated in Fig. S2 (ESI Note 2†), the results of both metrics indicate that the pre-trained SMILES transformer model is highly effective in capturing key molecular features from IL SMILES.The performance of the proposed ILTransR for predicting IL properties is benchmarked on eleven different IL properties and compared with the state-of-the-art models in literature.20–22,24–26,51 The involved properties of ILs can be divided into two types. One type is the properties related only to IL molecular structure namely melting point (Tm), glass transition temperature (Tg), thermal decomposition temperature (Td), and cytotoxicity towards the leukemia rat cell line IPC-81 (log10EC50). The second type relates to not only IL molecular structure but also conditions such as temperature and/or pressure, including heat capacity (Cp), refractive index (nD), density (ρ), viscosity (η), surface tension (γ), CO2 solubility (xCO2), and thermal conductivity (λ). To make a fair comparison, this work trains ILTransR on the same IL properties datasets as used in the corresponding references. Moreover, it should be noted that the random splitting of the entire dataset as adopted in the references may cause overestimation of models by separating data points of the same ILs (with only difference in temperature and/or pressure) into both the training and test sets when dealing with the second type of IL properties. That is to say, data points of the same IL under different temperature and/or pressure conditions are likely to be distributed into both the training and test sets, leading to data leakage. Therefore, in such benchmark cases, two different dataset split strategies are also compared: one is the random split of all data points and the other is the more rigorous split of data points according to different ILs. By using the second strategy, data points of the same IL at different temperatures and pressures can only enter the same subset during the splitting of training and test sets, which can avoid data leakage and give an unbiased test score.
The comparative results for the eleven IL properties are summarized in Table 2. As can be seen, for the four properties related only to the molecular structure of ILs (namely Tm, Tg, Td, and log10EC50), the prediction error (MAE) of the proposed ILTransR is all notably lower than that of the reference models in the literature, decreasing by 62.56%, 43.58%, 23.24%, and 41.81%, respectively. These results demonstrate that the ILTransR is able to extract the molecular representations of ILs better than the various descriptors used in the literature, especially when the database of IL properties is limited. For the second type properties that are also related to temperature and/or pressure (namely Cp, nD, ρ, η, γ, xCO2, and λ), the ILTransR outperforms all the reference models when adopting the random dataset splitting by data points, with an improvement ratio ranging from 5.26% for Cp to 62.22% for λ. Notably, the ILTransR still has comparable and even lower MAE (for the properties of ρ, η, and λ) on the test set rigorously split by ILs than the reference models do on the test set split non-rigorously by data points. This comparison proves that, in addition to more informative IL representations, the ILtransR can well handle different types of input via the CNN structure, leading to higher prediction accuracy as opposed to the reference models. It should be mentioned that some of the above references have also tried to use neural network methods in their model development; however, the neural network methods constructed in these references cannot achieve better prediction accuracy compared with the models listed in Table 2. The reason is that the size of most of such IL properties datasets is not large enough to train a neural network model with a high enough prediction accuracy, leading to the final selection of other statistical ML methods as the best model in the references.
| Property | Number of data points | Number of ILs | Descriptor | Method | Test MAE (split by data points) | Test MAE (split by ILs) | Source |
|---|---|---|---|---|---|---|---|
| T m(K) | 2212 | 2212 | ECFP4 and CM | KRR | — | 29.78 | Low et al.20 |
| ILTransR | — | 11.15 | This work | ||||
| T g (°C) | 609 | 609 | Charge distributions and geometrical indices | Cubist | — | 12 | Venkatraman et al.25 |
| ILTransR | — | 6.77 | This work | ||||
| T d(°C) | 1223 | 1223 | Charge distributions and geometrical indices | RF | — | 25 | Venkatraman et al.25 |
| ILTransR | — | 19.19 | This work | ||||
| ln(η) | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 368 368 |
1964 | Group contributions | LSSVM | 0.42 | — | Paduszyński24 |
| ILTransR | 0.17 | 0.35 | This work | ||||
| ρ | 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 167 167 |
2257 | Group contributions | LSSVM | 29.76 | — | Paduszyński21 |
| ILTransR | 12.31 | 16.46 | This work | ||||
| ln(Cp) | 9083 | 236 | Charge distributions and geometrical indices | GBM | 0.19 | — | Venkatraman et al.25 |
| ILTransR | 0.18 | 0.28 | This work | ||||
| γ | 2972 | 331 | Charge distributions and geometrical indices | GBM | 0.0027 | — | Venkatraman et al.25 |
| ILTransR | 0.0014 | 0.0030 | This work | ||||
| n D | 3009 | 464 | Charge distributions and geometrical indices | GBM | 0.011 | — | Venkatraman et al.25 |
| ILTransR | 0.0047 | 0.015 | This work | ||||
| xCO2 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 116 116 |
124 | Group contributions | SVM | 0.024 | — | Song et al.22 |
| ILTransR | 0.022 | 0.057 | This work | ||||
| log10EC50 | 326 | 326 | Structural descriptors | SVM | — | 0.1935 | Wang et al.51 |
| ILTransR | — | 0.1126 | This work | ||||
| λ | 454 | 73 | Charge distributions and geometrical indices | GBM | 0.009 | — | Venkatraman et al.26 |
| ILTransR | 0.0034 | 0.0061 | This work |
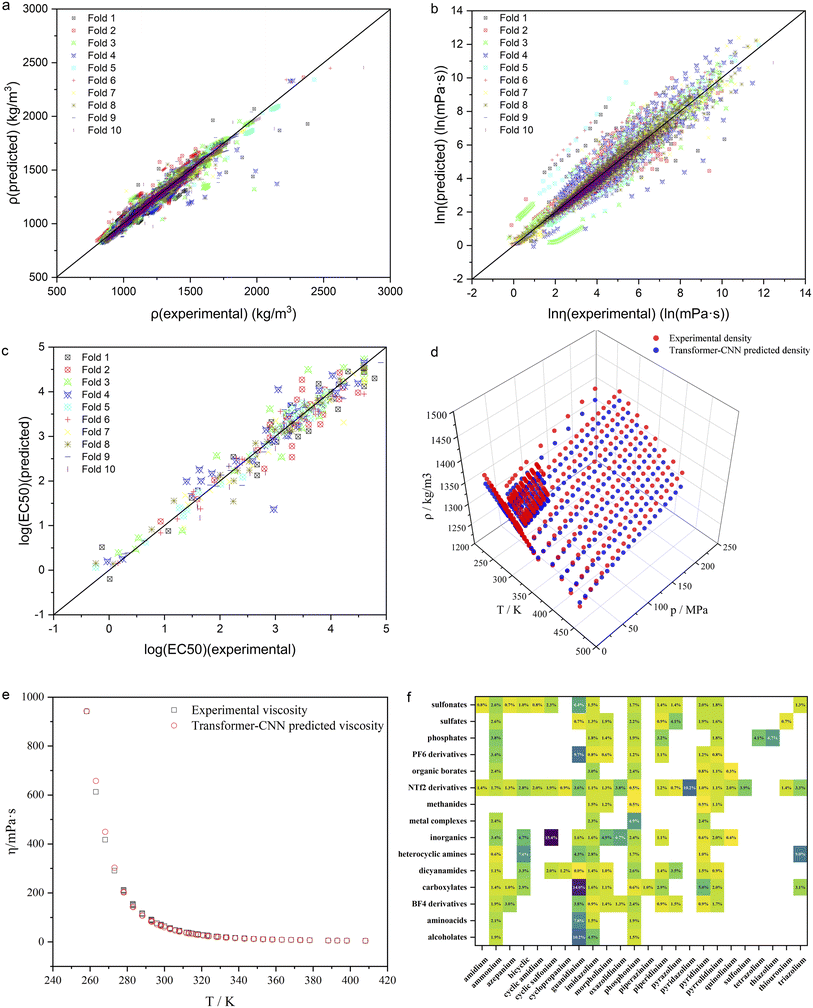
To show the predictive performance of the proposed ILTransR more vividly, the ρ, η, and log10EC50 of ILs are taken as examples to inspect the model test results in more detail. As seen in Fig. 3a–c, the test set points of each fold in the 10-fold cross-validation for the ρ, η, and log10EC50 are distributed almost evenly in a close region around the diagonal in the parity plot. These examples prove that the ILTransR can well predict different types of IL properties by fine-tuning on the corresponding IL properties dataset based on the IL representation learned by the pre-trained transformer encoder. To further illustrate that the ILTransR can well handle different inputs for IL property prediction, 1-hexyl-3-methylimidazolium bistriflamide ([C6C1Im][NTf2]) is selected as a representative to examine its predicted η-T and ρ-T–P relationship. As seen in Fig. 3d, the ILTransR model provides very satisfactory prediction for the density of [C6C1Im][NTf2] as compared to the experimental data over a wide temperature and pressure range (up to T = 450 K and P = 200 MPa, respectively). As for the viscosity of [C6C1Im][NTf2], the predictions by the ILTransR well resemble the experimental data over a wide range of temperature (Fig. 3e). It is worth mentioning that very few previously-reported ML models have scrutinized whether the temperature and/or pressure dependence of such IL properties could be correctly captured.
To gain more insight into the performance of the ILTransR, IL density is again selected as a representative to analyze the model predictions for each possible combination of cation and anion families. Corresponding AARE values are obtained by averaging the test set results in the 10-fold cross-validation. As shown in Fig. 3f, the AAREs for most of the involved anionic and cationic combinations are below 5%, which again proves that the ILTransR has a high prediction accuracy for IL density. Moreover, such prediction accuracy is found to be dependent on the moieties forming IL. For instance, the AAREs for the imidazolium-based ILs are all lower than 5%, with 13 of the 15 anionic families below 3%; low AAREs are also observed for carboxylates ILs, except that the paired cationic moiety is guanidinium. The highest AARE of 15.4% is obtained for the combination of cyclic sulfonium cations and common inorganics, as this combination only appears once in the entire dataset (the density prediction in this case in cross-validation is fully extrapolated). To wrap up, the detailed analyses of the density prediction well demonstrate that the ILTransR could reasonably predict IL properties for different IL families.
3.2 Application example of ILTransR: CO2 absorbent screening
From the ILTransR obtained above, the eleven IL properties can be reliably and quickly predicted, allowing for many model applications such as the high-throughput IL screening toward different processes. Herein, the screening of ILs as CO2 absorbent is presented as an illustrative case study.When screening ILs for CO2 capture, a set of thermodynamic and physical properties of ILs are of great importance. The capacity of IL to absorb CO2 can be evaluated by the gas solubility in ILs at the desired absorption temperature, while the desorption performance of IL can be estimated by the difference in the CO2 solubility at the desired absorption temperature and desorption temperature, respectively. The melting point, viscosity, thermal decomposition temperature, toxicity, and heat capacity of ILs should be considered as constraints because all these properties determine the feasibility and suitability of ILs as absorbents.55–57 To be specific, the melting point limits the lowest absorption temperature of ILs as liquid CO2 absorbents; the thermal decomposition temperature limits the highest temperature for CO2 desorption; the energy consumption of solvent regeneration can be assessed from the heat capacity of IL; the toxicity is a key factor related to the potential EHS impacts of ILs. All the above properties can be covered by the ILTransR developed in this work.
In this case study, a virtual library of 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 333
333![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 096 (219
096 (219![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 216 cations combined with 38 anions) synthetically feasible ILs as suggested by Venkatraman et al.26 is used as the initial candidate database. By using the ILTransR, the xCO2 of ILs at 298 K and 328 K (P = 1 bar) are calculated for evaluating the absorption and desorption performance of ILs; Cp, Tm, log10EC50, η, and Td under 1 bar and 298 K are also predicted. As the calculation speed of the ILTransR is very fast, a database of the seven properties for all the 8
216 cations combined with 38 anions) synthetically feasible ILs as suggested by Venkatraman et al.26 is used as the initial candidate database. By using the ILTransR, the xCO2 of ILs at 298 K and 328 K (P = 1 bar) are calculated for evaluating the absorption and desorption performance of ILs; Cp, Tm, log10EC50, η, and Td under 1 bar and 298 K are also predicted. As the calculation speed of the ILTransR is very fast, a database of the seven properties for all the 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 333
333![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 096 candidate ILs is obtained in only around 14 hours (2 hours per property for all the 8
096 candidate ILs is obtained in only around 14 hours (2 hours per property for all the 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 333
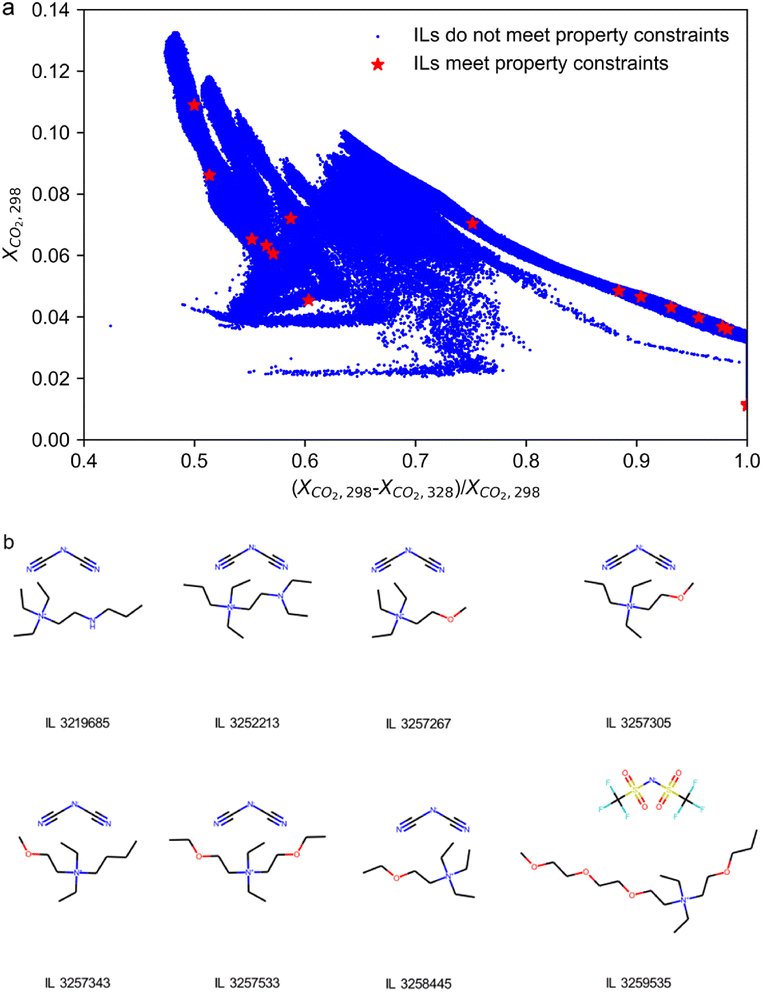
333![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 096 candidate ILs) on a laptop equipped with an RTX3070 GPU. Applying the constraints namely Tm < 298 K, Td > 150 °C, log10EC50 > 3, and η < 100 mPa s, a high-throughput screening over the entire IL database is performed, which retains 18 ILs meeting all the four constraints (as illustrated in Fig. 3a, see detailed information of these ILs in Table S1 in ESI Note 4†). Among them, eight ILs are basically located on the pseudo pareto front of all the candidate ILs in terms of the potential absorption and desorption performance. It should be noted that the four ILs in the lower right corner of Fig. 4a are excluded due to very low solubility of CO2 at the absorption temperature.
096 candidate ILs) on a laptop equipped with an RTX3070 GPU. Applying the constraints namely Tm < 298 K, Td > 150 °C, log10EC50 > 3, and η < 100 mPa s, a high-throughput screening over the entire IL database is performed, which retains 18 ILs meeting all the four constraints (as illustrated in Fig. 3a, see detailed information of these ILs in Table S1 in ESI Note 4†). Among them, eight ILs are basically located on the pseudo pareto front of all the candidate ILs in terms of the potential absorption and desorption performance. It should be noted that the four ILs in the lower right corner of Fig. 4a are excluded due to very low solubility of CO2 at the absorption temperature.
The molecular structures of the eight retained ILs are shown in Fig. 3b with their predicted properties of them tabulated in Table S1 in ESI Note 4.† These eight ILs are highly worth investigating in future studies as they are survivals from 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 333
333![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 096 candidates. It is worth mentioning that this case study is for the first time that such a huge database of ILs is considered for a high-throughput solvent screening toward a specific process, which benefits from both the high prediction accuracy and fast calculation speed of the ILTransR.
096 candidates. It is worth mentioning that this case study is for the first time that such a huge database of ILs is considered for a high-throughput solvent screening toward a specific process, which benefits from both the high prediction accuracy and fast calculation speed of the ILTransR.
4 Conclusion
In this work, we propose a pre-training and fine-tuning paradigm entitled ILTransR to generalize IL property prediction from limited labeled data. The ILTransR utilizes the power of unlabeled molecular data from a large-scale (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 434
434![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 070 IL-like molecules) pre-training through a translation task of non-canonical SMILES to canonical SMILES, and then can be easily fine-tuned on labeled IL properties datasets using CNN architecture. In experiments on eleven benchmark datasets of diverse IL properties, the proposed ILTransR surpasses all state-of-the-art ML models in literature, showing that not only better IL representations are learned but also different types of input features are well handled.
070 IL-like molecules) pre-training through a translation task of non-canonical SMILES to canonical SMILES, and then can be easily fine-tuned on labeled IL properties datasets using CNN architecture. In experiments on eleven benchmark datasets of diverse IL properties, the proposed ILTransR surpasses all state-of-the-art ML models in literature, showing that not only better IL representations are learned but also different types of input features are well handled.
The ILTransR provides a one-stop solution to accurately predict general properties of ILs, which could guide through the large IL chemical space even only limited labeled data are currently available. As an example, a high-throughput screening of CO2 absorbents from an enormous virtual library of 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 333
333![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 096 synthetically feasible ILs is performed, which identifies eight promising ILs based on calculating seven different properties by ILTransR. Moving beyond, it is highly expected that the proposed ILTransR could be a revolutionizing tool for the whole IL community for the quick discovery of the best candidate toward a specific task.
096 synthetically feasible ILs is performed, which identifies eight promising ILs based on calculating seven different properties by ILTransR. Moving beyond, it is highly expected that the proposed ILTransR could be a revolutionizing tool for the whole IL community for the quick discovery of the best candidate toward a specific task.
Code availability
The code accompanying this work is available in the GitHub repository at https://github.com/GuzhongChen/ILTransR.Data availability
The pre-training data can be downloaded from PubChem at ftp://ftp.ncbi.nlm.nih.gov/pubchem/Compound/ and can be processed using the code provided. The eleven IL property datasets used as benchmarks are available at https://github.com/GuzhongChen/ILTransR.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research is supported by the National Natural Science Foundation of China (NSFC) under the grants of 22278134, 22208098, and 21CAA01709. G. C. acknowledges the financial support of the China Scholarship Council (CSC) for his joint PhD program (No. 202106740022) with the Max Planck Institute for Dynamics of Complex Technical Systems, Magdeburg, Germany.References
- J. F. Brennecke and E. J. Maginn, AIChE J., 2001, 47, 2384–2389 CrossRef CAS.
- R. D. Rogers and K. R. Seddon, Science, 2003, 302, 792–793 CrossRef PubMed.
- F. Bezold, S. Roehrer and M. Minceva, Chem. Eng. Technol., 2019, 42, 474–482 CrossRef CAS.
- H. Niedermeyer, J. P. Hallett, I. J. Villar-Garcia, P. A. Hunt and T. Welton, Chem. Soc. Rev., 2012, 41, 7780–7802 RSC.
- N. V. Plechkova and K. R. Seddon, Chem. Soc. Rev., 2008, 37, 123–150 RSC.
- E. I. Izgorodina, Phys. Chem. Chem. Phys., 2011, 13, 4189 RSC.
- F. M. Maia, I. Tsivintzelis, O. Rodriguez, E. A. Macedo and G. M. Kontogeorgis, Fluid Phase Equilib., 2012, 332, 128–143 CrossRef CAS.
- S. M. Hosseini, A. Mulero and M. M. Alavianmehr, J. Chem. Thermodyn., 2019, 130, 47–94 CrossRef CAS.
- J. A. P. Coutinho, P. J. Carvalho and N. M. C. Oliveira, RSC Adv., 2012, 2, 7322 RSC.
- R. N. Das and K. Roy, Mol. Divers., 2013, 17, 151–196 CrossRef CAS PubMed.
- S. Stocker, G. Csányi, K. Reuter and J. T. Margraf, Nat. Commun., 2020, 11, 5505 CrossRef CAS PubMed.
- J. Townsend, C. P. Micucci, J. H. Hymel, V. Maroulas and K. D. Vogiatzis, Nat. Commun., 2020, 11, 3230 CrossRef CAS PubMed.
- J. Li, L. Li, Y. W. Tong and X. Wang, Green Chem. Eng., 2023, 4, 123–133 CrossRef.
- J. Li, L. Pan, M. Suvarna and X. Wang, Chem. Eng. J., 2021, 426, 131285 CrossRef CAS.
- B. Winter, C. Winter, J. Schilling and A. Bardow, Digit. Discov., 2022, 1, 859–869 RSC.
- E. I. S. Medina, S. Linke, M. Stoll and K. Sundmacher, Digit. Discov., 2022, 1, 216–225 RSC.
- S. Käser, L. I. Vazquez-Salazar, M. Meuwly and K. Töpfer, Digital Discov., 2023, 2, 28–58 RSC.
- Q. Dong, C. D. Muzny, A. Kazakov, V. Diky, J. W. Magee, J. A. Widegren, R. D. Chirico, K. N. Marsh and M. Frenkel, J. Chem. Eng. Data, 2007, 52, 1151–1159 CrossRef CAS.
- Y. Ding, M. Chen, C. Guo, P. Zhang and J. Wang, J. Mol. Liq., 2021, 326, 115212 CrossRef CAS.
- K. Low, R. Kobayashi and E. I. Izgorodina, J. Chem. Phys., 2020, 153, 104101 CrossRef CAS PubMed.
- K. Paduszyński, Ind. Eng. Chem. Res., 2019, 58, 5322–5338 CrossRef.
- Z. Song, H. Shi, X. Zhang and T. Zhou, Chem. Eng. Sci., 2020, 223, 115752 CrossRef CAS.
- D. Peng and F. Picchioni, J. Hazard. Mater., 2020, 398, 122964 CrossRef CAS PubMed.
- K. Paduszyński, Ind. Eng. Chem. Res., 2019, 58, 17049–17066 CrossRef.
- V. Venkatraman, S. Evjen, K. C. Lethesh, J. J. Raj, H. K. Knuutila and A. Fiksdahl, Sustain. Energy Fuels, 2019, 3, 2798–2808 RSC.
- V. Venkatraman, S. Evjen and K. Chellappan Lethesh, Data, 2019, 4, 88 CrossRef.
- S. Honda, S. Shi and H. R. Ueda, arXiv, 2019, preprint arXiv: 1911.04738.
- A. Sivaram, L. Das and V. Venkatasubramanian, Comput. Chem. Eng., 2020, 134, 106669 CrossRef CAS.
- L. Das, A. Sivaram and V. Venkatasubramanian, Comput. Chem. Eng., 2020, 139, 106895 CrossRef CAS.
- H. Wen, Y. Su, Z. Wang, S. Jin, J. Ren, W. Shen and M. Eden, AIChE J., 2022, 68, e17402 CrossRef CAS.
- Y. Xing, Y. Dong, C. Goergakis, Y. Zhuang, L. Zhang, J. Du and Q. Meng, AIChE J., 2022, 68, e17713 CrossRef CAS.
- R. Gómez-Bombarelli, J. N. Wei, D. Duvenaud, J. M. Hernández-Lobato, B. Sánchez-Lengeling, D. Sheberla, J. Aguilera-Iparraguirre, T. D. Hirzel, R. P. Adams and A. Aspuru-Guzik, ACS Cent. Sci., 2018, 4, 268–276 CrossRef PubMed.
- S. Kearnes, K. McCloskey, M. Berndl, V. Pande and P. Riley, J. Comput. Aided Mol. Des., 2016, 30, 595–608 CrossRef CAS PubMed.
- Z. Xu, S. Wang, F. Zhu and J. Huang, in Proceedings of the 8th ACM International Conference on Bioinformatics, Computational Biology,and Health Informatics, ACM, Boston Massachusetts USA, 2017, pp. 285–294 Search PubMed.
- Y. Wang, J. Wang, Z. Cao and A. Barati Farimani, Nat. Mach. Intell., 2022, 4, 279–287 CrossRef.
- T. Mikolov, E. Grave, P. Bojanowski, C. Puhrsch and A. Joulin, arXiv, 2017, preprint, arXiv:1712.09405, DOI:10.48550/arXiv.1712.09405.
- A. M. Dai and Q. V. Le, in Advances in Neural Information Processing Systems, Curran Associates, Inc., 2015, vol. 28 Search PubMed.
- D. Weininger, J. Chem. Inf. Model., 1988, 28, 31–36 CrossRef CAS.
- D. Weininger, A. Weininger and J. L. Weininger, J. Chem. Inf. Comput. Sci., 1989, 29, 97–101 CrossRef CAS.
- R. Winter, F. Montanari, F. Noé and D.-A. Clevert, Chem. Sci., 2019, 10, 1692–1701 RSC.
- A. Vaswani, N. Shazeer, N. Parmar, J. Uszkoreit, L. Jones, A. N. Gomez, Ł. Kaiser and I. Polosukhin, in Advances in Neural Information Processing Systems, Curran Associates, Inc., 2017, vol. 30 Search PubMed.
- P. Karpov, G. Godin and I. V. Tetko, J. Cheminf., 2020, 12, 17 Search PubMed.
- S. Wang, Y. Guo, Y. Wang, H. Sun and J. Huang, in Proceedings of the 10th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, ACM, Niagara Falls, NY, USA, 2019, pp. 429–436 Search PubMed.
- V. Mann and V. Venkatasubramanian, AIChE J., 2021, 67, e17190 CrossRef CAS.
- P. Schwaller, T. Laino, T. Gaudin, P. Bolgar, C. A. Hunter, C. Bekas and A. A. Lee, ACS Cent. Sci., 2019, 5, 1572–1583 CrossRef CAS PubMed.
- Y. Zhang and B. Wallace, arXiv, 2015, preprint, arXiv:1510.03820, DOI:10.48550/arXiv.1510.03820.
- F. Jirasek, R. A. S. Alves, J. Damay, R. A. Vandermeulen, R. Bamler, M. Bortz, S. Mandt, M. Kloft and H. Hasse, J. Phys. Chem. Lett., 2020, 11, 981–985 CrossRef CAS PubMed.
- S. Kim, P. A. Thiessen, E. E. Bolton, J. Chen, G. Fu, A. Gindulyte, L. Han, J. He, S. He, B. A. Shoemaker, J. Wang, B. Yu, J. Zhang and S. H. Bryant, Nucleic Acids Res., 2016, 44, D1202–D1213 CrossRef CAS PubMed.
- I. V. Tetko, P. Karpov, E. Bruno, T. B. Kimber and G. Godin, in Artificial Neural Networks and Machine Learning – ICANN 2019: Workshop and Special Sessions, eds. I. V. Tetko, V. Kůrková, P. Karpov and F. Theis, Springer International Publishing, Cham, 2019, pp. 831–835 Search PubMed.
- E. J. Bjerrum, arXiv, 2017, preprint arXiv:1703.07076.
- Z. Wang, Z. Song and T. Zhou, Processes, 2021, 9, 65 CrossRef.
- T. Chen, M. Li, Y. Li, M. Lin, N. Wang, M. Wang, T. Xiao, B. Xu, C. Zhang and Z. Zhang, arXiv, 2015, preprint arXiv: 1512.01274.
- J. Guo, H. He, T. He, L. Lausen, M. Li, H. Lin, X. Shi, C. Wang, J. Xie, S. Zha, A. Zhang, H. Zhang, Z. Zhang, Z. Zhang, S. Zheng and Y. Zhu, arXiv, 2020, preprint arXiv: 1907.04433.
- K. Papineni, S. Roukos, T. Ward and W.-J. Zhu, in Proceedings of the 40th Annual Meeting of the Association for Computational Linguistics, Association for Computational Linguistics, Philadelphia, Pennsylvania, USA, 2002, pp. 311–318 Search PubMed.
- Y. Zhao, R. Gani, R. M. Afzal, X. Zhang and S. Zhang, AIChE J., 2017, 63, 1353–1367 CrossRef CAS.
- S. Zheng, S. Zeng, Y. Li, L. Bai, Y. Bai, X. Zhang, X. Liang and S. Zhang, AIChE J., 2022, 68, e17500 CAS.
- M. Taheri, R. Zhu, G. Yu and Z. Lei, Chem. Eng. Sci., 2021, 230, 116199 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3dd00040k |
| This journal is © The Royal Society of Chemistry 2023 |