Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Extracting structured seed-mediated gold nanorod growth procedures from scientific text with LLMs†
Nicholas
Walker
 *a,
Sanghoon
Lee
ad,
John
Dagdelen
ad,
Kevin
Cruse
bd,
Samuel
Gleason
ae,
Alexander
Dunn
*a,
Sanghoon
Lee
ad,
John
Dagdelen
ad,
Kevin
Cruse
bd,
Samuel
Gleason
ae,
Alexander
Dunn
 ad,
Gerbrand
Ceder
bd,
A. Paul
Alivisatos
bdef,
Kristin A.
Persson
ad,
Gerbrand
Ceder
bd,
A. Paul
Alivisatos
bdef,
Kristin A.
Persson
 cdf and
Anubhav
Jain
*a
cdf and
Anubhav
Jain
*a
aEnergy Technologies Area, Lawrence Berkeley National Laboratory, 1 Cyclotron Road, Berkeley, CA, USA. E-mail: walkernr@lbl.gov; ajain@lbl.gov
bMaterials Sciences Division, Lawrence Berkeley National Laboratory, 1 Cyclotron Road, Berkeley, CA, USA
cMolecular Foundry, Lawrence Berkeley National Laboratory, 1 Cyclotron Road, Berkeley, CA, USA
dDepartment of Materials Science and Engineering, University of California Berkeley, 210 Hearst Memorial Mining Building, Berkeley, CA, USA
eDepartment of Chemistry, University of California Berkeley, 419 Latimer Hall, Berkeley, CA, USA
fKavli Energy NanoScience Institute, University of California Berkeley, 101C Campbell Hall, Berkeley, CA, USA
First published on 20th September 2023
Abstract
Although gold nanorods have been the subject of much research, the pathways for controlling their shape and thereby their optical properties remain largely heuristically understood. Although it is apparent that the simultaneous presence of and interaction between various reagents during synthesis control these properties, computational and experimental approaches for exploring the synthesis space can be either intractable or too time-consuming in practice. This motivates an alternative approach leveraging the wealth of synthesis information already embedded in the body of scientific literature by developing tools to extract relevant structured data in an automated, high-throughput manner. To that end, we present an approach using the powerful GPT-3 language model to extract structured multi-step seed-mediated growth procedures and outcomes for gold nanorods from unstructured scientific text. GPT-3 prompt completions are fine-tuned to predict synthesis templates in the form of JSON documents from unstructured text input with an overall accuracy of 86% aggregated by entities and 76% aggregated by papers. The performance is notable, considering the model is performing simultaneous entity recognition and relation extraction. We present a dataset of 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 644 entities extracted from 1137 papers, resulting in 268 papers with at least one complete seed-mediated gold nanorod growth procedure and outcome for a total of 332 complete procedures.
644 entities extracted from 1137 papers, resulting in 268 papers with at least one complete seed-mediated gold nanorod growth procedure and outcome for a total of 332 complete procedures.
1 Introduction
Gold nanoparticles have been synthesized for centuries due to their interesting optical properties, dating back to the Lycurgus Cup from 4th century Rome,1 as well as imperial bowls and decorated dishes from the Qing dynasty.2 However, scientific interest did not develop until the work of Michael Faraday in the mid-19th century, when he accidentally synthesized colloidal gold while investigating the interaction between light and matter.3 In the last three decades, chemists have developed the ability to synthesize anisotropic metal nanoparticles in a controllable and reproducible fashion.4 Around the turn of the millennium, multi-step seed-mediated growth methods were developed to prepare gold nanorods with aspect ratios ranging from 8 to 20.4–6 This generated a great deal of interest in anisotropic gold nanoparticles due to a combination of the convenience of the wet-chemistry approach, as well as the ability to tune the shape of the synthesized nanorods. The anisotropic gold nanoparticles, in turn, provide access to shape-dependent optical phenomena not observed with spherical gold nanoparticles.7–10 Their applications are widespread across many domains, including semiconductor technology,11,12 biomedicine,13,14 and cosmetics.15 The suitability of a nanoparticle for a particular application depends on its morphology and size, which correspond to different plasmonic properties.16–18Despite the popularity of anisotropic gold nanoparticles, systematic investigation of the control of these properties has only recently been approached.19 Although some theories and models do exist for identifying and explaining the mechanisms of synthesis that determine nanoparticle morphology,4,20–22 most synthesis exploration is still guided by heuristics based on domain knowledge.
For gold nanorods, it is clear that the simultaneous presence of various reagents during the synthesis affects the characteristics of the resulting gold nanoparticles.4 To better understand these effects, computational simulation and analysis of the formation energetics of the nanoparticles or the nucleation and growth steps can be used. Density functional theory (DFT) can be used to investigate the energetic landscape of potential gold nanoparticle morphologies, including the effects of surface ligands that are vital for the solution-phase synthesis of noble metal nanoparticles.23–25 However, this approach does not account for the nuances of nucleation and growth competition in solution-based nanoparticle syntheses. These aspects can be addressed by modeling real-time growth and dispersity dynamics with continuum-level model, though this sacrifices access to small-scale energetics granted by DFT.26 Alternatively, direct experimentation can be used to explore the synthesis space by varying precursor amounts over many experiments, though this is impractical due to the both the number of experiments required to sample the synthesis space and the condition that a single experiment can take many hours to complete. Automated labs may address this problem in the future, though most are still in their infancy.
A third approach seeks to leverage the wealth of information contained in scientific literature. Many seed-mediated gold nanorod recipes have been published in the materials science and chemistry literature, but parsing them requires domain experts to manually read these articles to retrieve the relevant precursors, procedures, laboratory conditions, and target characterizations. This comes with its own complications, however, as over time, the body of materials science literature has grown to an unwieldy extent, preventing researchers from absorbing the full breadth of information contained in established literature or even reasonably following research progress as it emerges.27 Thus, it is unreasonable to expect domain experts in gold nanoparticle synthesis to manually read and parse the complete existing synthesis literature efficiently, motivating the development of high-throughput text-mining methods to extract this information.
The resulting databases built with these methods are the first steps toward developing data-driven approaches to understanding synthesis, which are being developed at an accelerating pace as a rapidly emerging third paradigm of scientific investigation. Generally speaking, these approaches involve the use of both conventional and machine learning methods to both build large databases and perform downstream analysis and inference over said databases. Natural language processing (NLP) has been successfully applied in the chemical, medical, and materials sciences to produce structured data from unstructured text using methods and models such as pattern recognition, recurrent neural networks, and language models.28,28–52
For applications specifically related to materials synthesis, data-driven approaches have been successful for tasks such as materials discovery, synthesis protocol querying, and simulation and interpretation of characterization results.53–57 However, these approaches are fundamentally limited by the quality of the data, such as the completeness and substance of the data source. To address this, careful data curation is necessary, as seen with the construction and maintenance of large databases of characteristic features of nanostructures.58
Recently, the wealth of unstructured information about gold nanoparticle synthesis and characterization in literature has been directly tapped through the combination of various NLP models and other text-mining techniques to produce a dataset of over five thousand codified gold nanoparticle synthesis protocols and outcomes.59 This general dataset contains a wealth of information, including detected materials, material quantities, morphologies, synthesis actions, and synthesis conditions, as well as tags for seed-mediated synthesis, synthesis paragraph classifications, and characterization paragraph classifications.
Despite the breadth of accurate information provided, the general dataset still suffers from a few pitfalls: (i) the inability to distinguish between seed and growth solution procedures in seed-mediated growth synthesis; (ii) the inability to detect references to materials that do not contain specific formulae or chemical names (e.g. “AuNP seed solution”); and (iii) the inability to detect target morphologies as opposed to incidentally mentioned morphologies. To address these issues, this work intends to use a large sequence-to-sequence language model to extract full synthesis procedures and outcomes in a single-step inference. Generally speaking, a sequence-to-sequence model in NLP maps an input sequence to an output sequence by learning to produce the most likely completion of the input by conditioning the output on the input.60
In this work, we leverage the capabilities of the latest language model in the Generative Pre-trained Transformer (GPT) family, GPT-3,61 to build a dataset of highly structured synthesis templates for seed-mediated gold nanorod growth. A similar approach using GPT-3 to build materials science datasets has been applied to extracting dopant-host material pairs, cataloging metal–organic frameworks, and extracting general chemistry/phase/morphology/application information for materials.62 We extracted these templates for seed-mediated gold nanorod growth from 2969 paragraphs across 1137 filtered papers, starting with using a question-answering framework aided by the zero-shot performance of GPT-3 to construct a small initial dataset. We then fine-tuned GPT-3 to produce complete synthesis templates for input paragraphs. Fine-tuning GPT-3 consists of using multiple examples of paragraph and synthesis template pairs to train GPT-3 to perform this specific task. Each synthesis template in the final dataset contains information on relevant synthesis precursors, precursor amounts, synthesis conditions, and characterization results, all structured in a JSON format. This dataset provides reproducible summaries of procedures and outcomes, explicitly establishing the relationships between the components of the recipe (e.g. accurately linking the correct volumes and concentrations with the correct precursors in the correct solution). However, this specificity comes at the cost of generality, as the dataset focuses on seed-mediated gold nanorod growth. The final dataset consists of 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 644 entities extracted from 1137 filtered papers, 268 of which contain least one complete seed-mediated gold nanorod growth procedure and outcome for a total of 332 complete procedures.
644 entities extracted from 1137 filtered papers, 268 of which contain least one complete seed-mediated gold nanorod growth procedure and outcome for a total of 332 complete procedures.
While our primary focus revolved around the application of a fine-tuned GPT-3 Davinci model, we further extended our research horizon by employing 13 billion parameter variant of Llama-2 (ref. 63) to undertake the same task for benchmark. Llama-2, an acronym for “Large Language Model Meta AI – 2′′, emerges from a lineage of language models that have been reported to exceed performance of much larger models (such as GPT-3 Davinci) on many NLP benchmarks.64 Compared to GPT-3, Llama utilizes different approaches to architecture including the use of SwiGLU activations instead of ReLU,65 rotary position embeddings instead of absolute position embeddings,66 and RMS layer-normalization67 instead of standard layer normalization.68 Additionally, Llama-2 boasts a 4192 token context window instead of the 2048 token context window provided by GPT-3.
2 Dataset
The relevant data for constructing the training, testing, and prediction data for this model was collected using the database of gold nanoparticle synthesis protocols and outcomes developed by Cruse et al.59 from the full-text database developed by Kononova et al.28 through text- and data-mining agreements with several major scientific journal publishers. The original full-text database contains more than 4.9 million materials science articles, and the pipeline for identifying and extracting gold nanoparticle synthesis articles consists of progressively finer-meshed filtering steps using text-mining tools adapted from Kononova et al.28 and Wang et al.69 These steps include regular expression matching to identify nanomaterial papers, document and vocabulary vectorization using term frequency-inverse document frequency (TF-IDF) to reveal papers related more to gold than other noble metals, BERT-based binary classifiers to identify paragraphs related to gold nanoparticle synthesis or characterization (particularly morphological information), a combination of BiLSTM-based named entity recognition (NER) and rules-based methods to extract synthesis procedure details from synthesis paragraphs, and MatBERT49 NER to extract morphology and size information from characterization paragraphs.Using the extracted information, 5145 papers were identified to contain gold nanoparticle synthesis protocols,70 of which 1137 filtered papers were found to contain seed-mediated recipes using the “seed_mediated” flag as well as rod-like morphologies (“rod or “NR” in “morphologies” under “morphological_information”) or aspect ratio measurements (“aspect” or “AR” in “measurements” under “morphological_information”). This was done to filter the total papers down to only those likely to contain seed-mediated synthesis recipes for gold nanorods.
3 Methods
At the core of the GPT-1 model was a focus on improving language understanding by generative pre-training involving the use of a large language model in conjunction with a very large and diverse pre-training corpus with long stretches of contiguous text, which greatly facilitated the model's ability to learn “world knowledge” alongside its ability to process long-range dependencies.71 For a sequence-to-sequence generative model, outputs are generated by maximizing the log probability of p(output|input).60 To further improve zero-shot performance for both learning and task transfer, GPT-2 modified the training objective to include task conditioning, p(output|input, task), thus establishing the model as an unsupervised multitask learner.72 With GPT-3, more extensions of the model size and the pre-training corpus have produced a model with considerable capacity for few-shot learning that is capable of producing text that is difficult to distinguish from human-written text or performing tasks it was not explicitly trained on, such as writing code or summing numbers.61 We employed the 175 billion parameter variant of GPT-3 (OpenAI Davinci) for this work.Of the 1137 filtered papers identified to contain information about seed-mediated gold nanorod synthesis, 240 (consisting of 661 relevant paragraphs) were randomly sampled and fully annotated with JSON-formatted recipes by a single annotator with machine assistance to serve as a training set. An additional 40 filtered papers (consisting of 117 relevant paragraphs) were annotated to serve as a testing set. Each relevant paragraph was separately annotated due to length constraints imposed by GPT-3, which limits the capability to process an entire article at once. A limit of 2048 tokens is shared between the input prompt and the output completion, corresponding to approximately 1500 words.61
3.1 Overall procedure
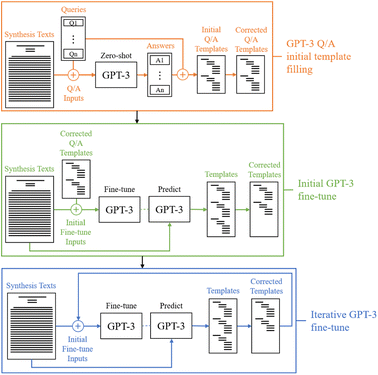
A diagram outlining the general process for producing the final fine-tuned model for template-filling is shown in Fig. 1. In the initial stage (orange), a simple question-answering framework is used to individually fill in templates for an initial set of paragraphs. These results are then corrected according to the described annotation procedure and used as an initial training set for fine-tuning GPT-3 to produce complete templates in the second stage (green). The final stage (blue) is an iterative training process in which new templates are predicted, corrected, and added to the training set to update the fine-tuned model, thus improving its performance with each iteration. Default settings through the OpenAI API (v0.13.0) are used for all fine-tunes of the GPT-3 Davinci model, and a temperature of zero is used for all model predictions with a double line break as the stop sequence. By using a temperature of zero, the results should be deterministic assuming that floating point errors in the GPU calculations are smaller than the differences between the log probabilities of the next token prediction candidates.To assess Llama-2-13B's efficacy in extracting two-step seed-mediated gold nanorod synthesis procedures, we adopted a fine-tuning approach using Low-Rank Adaptation (LoRA) as described in ref. 73, facilitated by the Parameter-Efficient Fine-Tuning library.74 The base model of Llama-2-13B75 with 8 bit quantization was fine-tuned with the identical training data on a single GPU (NVIDIA A100). Some of the fine-tuning parameters we used are as follows: 4 epochs, batch size of 1, learning rate of 0.0001, LoRA r of 8, LoRA alpha of 32 and LoRA dropout of 0.05.
3.2 Template structure and annotation scheme
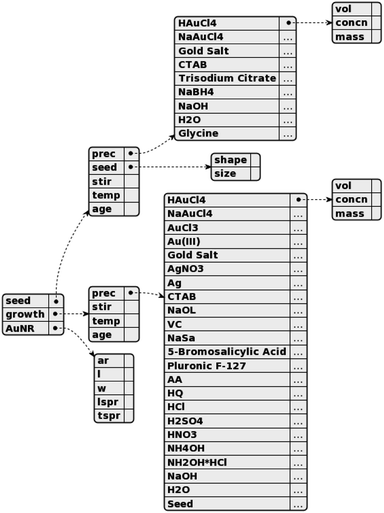
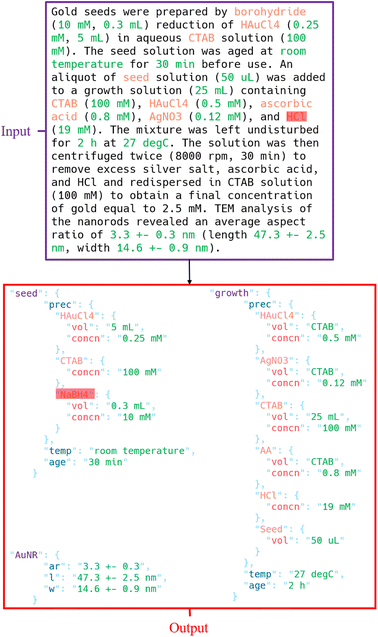
The structure and content of the synthesis templates are shown in Fig. 2. The synthesis templates are stored as JSON documents, which contain three components: the seed solution, the growth solution, and the resulting nanorods. For the seed and growth solutions, the precursors and their associated volumes (vol), concentrations (concn), and/or masses are recorded, as well as the ages of the respective mixed solutions at the time of use and the temperatures (temp) at which they are aged. Furthermore, the stirring rates when adding sodium borohydride (NaBH4) to the seed solution and when adding the seed solution to the growth solution are recorded. The shape and size of the gold seeds in the seed solution are also noted. For the gold nanorods (AuNR), the aspect ratios (ar), lengths (l), widths (w), and longitudinal/transverse surface plasmon resonances (SPRs) are recorded. The JSON documents have identical structures and thus contain an entry for every value that can be requested; any values not present in a given paragraph are filled with an empty string.When available, numerical quantities with units are extracted. For precursor volumes, the units are provided in variations of liters, though the concentrations may be measured in either molarity, molality, or weight percentage. In some cases, the total volume of a collection of precursors may be specified instead of the individual volumes of the precursors. In this case, the explicit volume is associated with the first precursor and the volumes for the remaining precursors refer to the name of the first precursor, implicitly communicating a shared volume. For temperatures, degrees Celsius are most commonly provided, though more qualitative descriptions such as “room temperature” will still be recorded if the explicit temperature is not provided in the text but a qualitative description is. Similarly, for solution ages, minutes or hours are most common, but sometimes only descriptions like “overnight” are provided and recorded. For stirring rates, the revolutions per minute (rpm) is preferred, but many papers will instead provide descriptions such as “gentle” or “vigorous” that are recorded. For the gold nanorod properties, aspect ratios are unitless while the other quantities (length, width, SPRs) are provided in units of length, with the exception of some cases where the LSPR is only provided as “NIR” (near-infrared). Throughout all stages of the annotation process, three additional researchers were consulted to reach a consensus on the appropriate annotations for various edge cases caused by unclear wording or other ambiguities.
3.3 Question answering completions
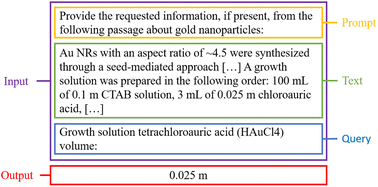
Unfortunately, the standard pre-trained GPT-3 Davinci model is not capable of providing consistent completed templates of high quality in one request. However, the model is capable of answering simple questions about synthesis paragraphs without any fine-tuning, which allows for the fields of the synthesis templates to be individually filled using answers from a simple question-answering framework using GPT-3. An example is shown in Fig. 3.76 This machine-assisted annotation approach avoids the laborious process of manually filling in each field of the templates by hand, as an annotator only needs to verify and correct the provided answers as-needed. However, this approach does not scale well to large numbers of papers, as each query is a separate model request, meaning that each paragraph in each paper would require a large number of requests in order to fill a single template. Therefore, this approach is used to construct an initial dataset consisting of synthesis templates for paragraphs from a small number of papers. Due to the small number of papers used, this initial dataset does not necessarily capture the variety of precursors or manners in which critical data can be communicated in text. As such, only information known to be commonly present in seed-mediated gold nanorod synthesis (e.g. the common precursor volumes/concentrations) were queried. Nevertheless, these initial templates, when corrected, provide a suitable starting point for fine-tuning GPT-3 to provide complete synthesis templates in single requests for each paragraph. Through an iterative process of fine-tuning GPT-3 on the available templates, predicting new templates, correcting them, and fine-tuning a new model using all of the corrected templates constructed thus far, a final fine-tuned model can be obtained.The initial synthesis template dataset was constructed using the zero-shot question-answering framework with 40 randomly sampled filtered papers. If a relevant precursor, condition, or characterization was identified with regular expression pattern matching in the paragraph, the framework would be to request the information using GPT-3. For example, if “ascorbic acid”, “AA”, “vitamin C”, or “C6H8O6” appeared in the paragraph, the framework would request the volume, concentration, and mass of ascorbic acid. This initial dataset only requested information about the eight most common precursors, including “HAuCl4”, “CTAB”, and “NaBH4” for the seed solution, and “HAuCl4”, “CTAB”, “AgNO3”, “AA”, and “seed solution” for the growth solution. To capture different ways of expressing each precursor, multiple aliases were checked to include variations on chemical names as well as the chemical formulae. Additionally, the framework requested information about the stir rate when adding NaBH4 to the seed solution, the age of the seed solution, the temperature of the seed solution during aging, the size and shape of the seeds, the stir rate when adding the seed solution to the growth solution, the age of the growth solution, and the temperature of the growth solution during aging. All request completions for each paragraph were aggregated into a single JSON entry according to the synthesis template scheme shown in Fig. 2.
The approach of using zero-shot GPT-3 question answering requests to fill the templates tended to produce poor results, but it offered an acceptable starting point for collecting structured recipes. Most of the templates only required correcting the incorrect entries, rather than filling them in manually from scratch, which greatly accelerated the creation of the initial dataset. However, some entries had to be added from scratch due to recipes including precursors outside the initial set of eight common precursors. Note that the static nature of the synthesis templates across all paragraphs means that when one paragraph requires the addition of a new precursor to the template, this is applied to all templates for all paragraphs. Additionally, annotation was done strictly, requiring that the synthesis method must be seed-mediated growth and the target gold nanoparticle morphology must be nanorods. This provides an important test for the model, as the difference between recipes that produce very similar morphologies can sometimes be subtle.
3.4 Fine-tuning procedure and dataset construction
These corrected templates derived from the question answering completions provided an initial training set for fine-tuning GPT-3 to produce the desired filled templates. From there, templates for paragraphs from 40 more randomly sampled filtered papers were iteratively predicted, corrected (adding new precursors as necessary), and added to the training set until templates for paragraphs from 240 filtered papers had been corrected in total. With each iteration, the correction process became much easier and faster. Initially, templates for information-dense paragraphs took approximately 4 minutes to validate and correct, whereas, by the end of the process, they took around a minute each. This is because GPT-3 largely predicted filled templates with high accuracy. The testing dataset was composed of paragraphs from an additional random sampling of 40 papers. Not all of the papers filtered from the original dataset were guaranteed to contain information that should be placed into synthesis templates. For example, seed-mediated growth or nanorod measurements and morphologies may only be incidentally mentioned in a given paragraph that is otherwise not relevant to a specific seed-mediated gold nanorod growth procedure. Of the 240 filtered papers in the training set and the 40 filtered papers in the testing set, 141 and 23 papers respectively contained at least one paragraph with information that could be placed into a synthesis template. The following command was used to perform the fine-tuning:4 Results
The described training dataset of synthesis templates was used to fine-tune a GPT-3 model to reproduce said synthesis templates from the unstructured text. Default parameters for the fine-tuning process were employed, incurring a cost of 85.30 USD (191![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 069 prompt tokens and 522
069 prompt tokens and 522![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 649 completion tokens). The predictions over the testing dataset (40 papers composed of 117 paragraphs) took around eighty minutes to complete and incurred a cost of 14.39 USD (27
649 completion tokens). The predictions over the testing dataset (40 papers composed of 117 paragraphs) took around eighty minutes to complete and incurred a cost of 14.39 USD (27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 327 prompt tokens and 92
327 prompt tokens and 92![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 126 completion tokens). The performance of the fine-tuned model was then evaluated using the testing dataset.
126 completion tokens). The performance of the fine-tuned model was then evaluated using the testing dataset.
4.1 Error evaluation examples and definitions
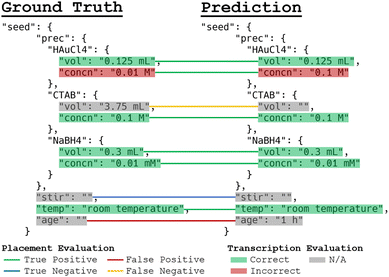
An example prediction is depicted in Fig. 4.77 Errors are highlighted in red. For this example, two errors were made. First, the quantities for “Borohydride” in the seed solution were instead placed under “NaBH4” in the seed solution. Arguably, this is not truly an error since sodium borohydride is often conventionally referred to as “borohydride”, possibly indicating “world knowledge” exhibited by GPT-3. However, there are technically other borohydrides, such as potassium borohydride, that can be used as a reducing agent for seed-mediated gold nanorod growth,78 so this was still marked as incorrect due to possible ambiguity. The second error was the failure to extract the HCl volume. Note the rather complex relationship in the growth solution precursor volumes, where CTAB, HAuCl4, ascorbic acid, AgNO3, and HCl all share the same 25 mL volume. To avoid confusion, the volume is explicitly associated with the first-mentioned precursor in the mixture, and the following precursors refer back to that first precursor. This ensures that downstream applications can unambiguously process the data to mean that the precursors are sharing a single volume. Other than these two errors, the model performs very well at extracting quantities in this example.For the 117 testing paragraphs, two types of errors are tracked: placement errors and transcription errors. This is done in order to evaluate the model's capability for separately identifying which fields of the synthesis templates should contain information, as well as how accurate the appropriately placed information is. To evaluate information placement, only the existence of information in the fields of the prediction and ground truth synthesis templates are considered. For example, if the same field contains information (as opposed to being empty) in both templates, that is considered a true positive prediction regardless of whether the information explicitly matches. If both fields are empty, then that is a true negative. If the prediction field contains information while the ground truth field is empty, then that is a false positive, while the reverse is a false negative. These categories of placement errors are used to calculate the precision, recall, and F1-score for information placement. Examples of these evaluations are shown in Fig. 5.
For evaluating transcription accuracy, only the agreement between the prediction and the annotation for true positive placements are considered, as the other types of errors are accounted for by the evaluations of information placement. For numerical values with units, the units must be exactly correct and the quantitative relative error was calculated according to the function s(p, q) = 2·min(p, q)/(p + q), which is derived from the absolute proportional difference r(p, q) = |p − q|/(p + q) and is bounded on [0,1] for non-negative numerical values p (predicted numerical value) and q (annotated numerical value). Some values may have modifiers attached, such as “>3 h”. If the prediction misses this information, e.g., gives “3 h”, the prediction is considered half-correct even if the quantity and unit are both correct. Some quantities will additionally be expressed as a range or list of values. In these cases, the range boundaries are split into a list as necessary, and the transcription accuracies are scored and aggregated across the values in the list with proper ordering enforced. For non-numerical predictions such as stir rates described as “vigorous” or gold seed morphologies, an exact string match is required for the prediction to be marked as correct. The combined accuracy (adjusted F1-score) is presented as the product of the F1-score for information placement and the transcription accuracy. This is the most meaningful metric to evaluate the overall performance of the model.
4.2 Model performance
The total performance of the model aggregated over each recipe component as well as all entries is shown in Table 1. The model appears to be proficient at generally identifying which information should be filled in the template based on the content of the text, with a rather high F1-score of 90% that favors neither precision nor recall. It additionally performs exceptionally at accurately transcribing the information with an accuracy of 95%. By taking the product of the placement F1-score and the transcription accuracy, this provides an impressive overall adjusted F1-score of 86%. This indicates a significant improvement over comparable efforts in solid-state synthesis text-mining, which report an overall accuracy of 51% for extracting all recipe items (chemistry, operations, and attributes of the operations).28 Direct comparison is, however, rather challenging, as some aspects of the two-step, seed-mediated growth synthesis are more complicated, such as the presence of two solutions with distinct precursor sets and a greater amount of precursor information needed due to the solution-based format. On the other hand, solid-state synthesis extraction carries its own challenges, considering the greater variation in procedural steps and conditions that must be considered.| Placement | Transcription | Combined | Support | ||||
|---|---|---|---|---|---|---|---|
| Precision | Recall | F1 | Accuracy | Adj. F1 | |||
| Seed solution | GPT-3 | 0.97 | 0.92 | 0.94 | 0.95 | 0.90 | 159 (142) |
| Llama-2 | 0.90 | 0.91 | 0.91 | 0.94 | 0.85 | 169 (140) | |
| Growth solution | GPT-3 | 0.90 | 0.94 | 0.92 | 0.96 | 0.88 | 244 (206) |
| Llama-2 | 0.88 | 0.92 | 0.90 | 0.94 | 0.84 | 247 (202) | |
| AuNR | GPT-3 | 0.79 | 0.74 | 0.76 | 0.95 | 0.72 | 96 (59) |
| Llama-2 | 0.75 | 0.70 | 0.72 | 0.97 | 0.70 | 99 (56) | |
| Overall | GPT-3 | 0.90 | 0.90 | 0.90 | 0.96 | 0.86 | 499 (407) |
| Llama-2 | 0.87 | 0.88 | 0.87 | 0.94 | 0.82 | 515 (398) | |
It is clear that the adjusted F1-scores for the recipe entities associated with the seed and growth solutions are very promising, indicating that the model is reliable for extracting the necessary information from the text for the component solutions to the synthesis procedure. However, the performance is worse overall for the gold nanorod properties, with an adjusted F1-score of approximately 72%. This is still an improvement over similar results, as the gold nanoparticle synthesis protocol and outcome database developed by Cruse et al.59 extracts morphology measurements, sizes, and units with F1-scores of 70%, 69%, and 91% via NER with MatBERT. However, these entities are not linked together, so while doing so would inevitably introduce additional sources of error and performance would be additionally constrained by the lowest performing extractions, a direct quantitative comparison is not applicable.
Table 2 shows the model performance for detecting precursors in the seed and growth solutions. Precursor detection is calculated implicitly based on which precursors the extracted volumes, concentrations, and masses are associated with. This is a clear improvement over the results in the gold nanoparticle synthesis protocol and outcome database developed by Cruse et al.59 The prior work detected precursors via a BiLSTM-based NER model with an F1-score of 90%. However, as mentioned earlier, this does not distinguish between seed and growth solution precursors and cannot detect precursors that do not contain specific formulae or chemical names, such as the seed solution that is added to the growth solution. This means that direct quantitative comparison is not applicable. The fine-tuned GPT-3 model missed cases where cationic surfactant, PP, BH4, and AuCl3 were used as well as a case where HCl was used in the seed solution. None of these cases occurred in the training set. Notably, the model correctly normalized “AsA” to “AA”, despite “AsA” never appearing in the training data.
| Seed solution | Growth solution | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Precision | Recall | F1 | Support | Precision | Recall | F1 | Support | ||
| Precursor | GPT-3 | 0.98 | 0.90 | 0.94 | 61 | 0.93 | 0.92 | 0.92 | 118 |
| Llama-2 | 0.95 | 0.90 | 0.92 | 63 | 0.91 | 0.91 | 0.91 | 120 | |
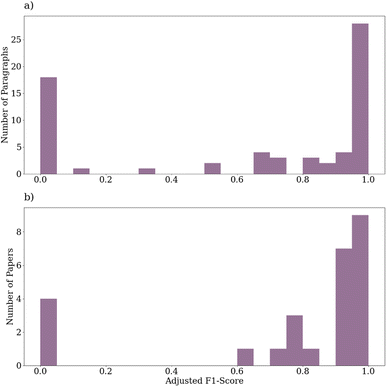
The adjusted F1-scores aggregated over extracted entities for the paragraph-wise and paper-wise predictions are shown in Fig. 6. Instances in which there were no entities present in either the ground truths or the predictions are omitted from the results, giving a total of 66 paragraphs and 26 papers. For the paragraphs, the average adjusted F1-score was approximately 64% with 22 (33%) perfect predictions and 32 (48%) predictions with >90% adjusted F1-score. For the papers, the average adjusted F1-score was approximately 76% with 4 (15%) perfect predictions and 16 (62%) predictions with >90% adjusted F1-score.
Comparative performance of Llama-2-13B against GPT-3 Davinci is also detailed in Tables 1 and 2. Although Llama-2 exhibits comparatively diminished performance, its viability is context-dependent. Its value arises from being a smaller model, amenable for non-commercial on-premise deployment without relying on an API. Moreover, its reduced size compared to GPT-3 Davinci makes it an economical choice from a computational standpoint.
4.3 Full filtered dataset
The fine-tuned GPT-3 model was applied to the full filtered dataset of 1137 filtered papers (2969 paragraphs) at a total cost of 384.31 USD (838![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 901 prompt tokens and 2
901 prompt tokens and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 332
332![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 796 completion tokens) over 33 hours. In total, 11
796 completion tokens) over 33 hours. In total, 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 644 entities were extracted from the paragraphs that contained information of interest. The dataset is presented as a JSON file containing a list with each element corresponding to a single article. Table 3 summarizes the structure of the JSON documents for each paper alongside a breakdown of how the total extracted entities across the entire dataset are distributed across the entity types. While the template extractions were performed paragraph-by-paragraph, the templates have been merged by article for convenience. However, this does mean that some conflicts and repetitions are present in the dataset. A conflict arises when a particular entity type in a paper (e.g. the volume of a particular precursor) is specified with different values across multiple paragraphs and a repetition arises when it is specified with the same value across multiple paragraphs. Of the 11
644 entities were extracted from the paragraphs that contained information of interest. The dataset is presented as a JSON file containing a list with each element corresponding to a single article. Table 3 summarizes the structure of the JSON documents for each paper alongside a breakdown of how the total extracted entities across the entire dataset are distributed across the entity types. While the template extractions were performed paragraph-by-paragraph, the templates have been merged by article for convenience. However, this does mean that some conflicts and repetitions are present in the dataset. A conflict arises when a particular entity type in a paper (e.g. the volume of a particular precursor) is specified with different values across multiple paragraphs and a repetition arises when it is specified with the same value across multiple paragraphs. Of the 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 644 extracted entities, 10
644 extracted entities, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 098 (∼87%) are uniquely identified, meaning there are no conflicts or repetitions (the associated value is extracted from exactly one paragraph). An additional 353 entries present at least one conflict without any repetitions, 251 with at least one repetition and no conflicts, and 57 with both conflicts and repetitions. Repetitions do not need to be manually resolved since this arises from the specification of identical information across multiple paragraphs (e.g. mentioning the gold nanorod aspect ratios in paragraphs about both the synthesis procedure as well as the nanorod characterization), but conflicts can be challenging to resolve in a consistent manner without manual inspection. For instance, if two separate volumes for a particular precursor are provided in two separate paragraphs, it can be ambiguous whether the volumes are part of the same synthesis procedure or distinct synthesis procedures in the same paper due to the lack of cross-paragraph context. With this in mind, of the 11
098 (∼87%) are uniquely identified, meaning there are no conflicts or repetitions (the associated value is extracted from exactly one paragraph). An additional 353 entries present at least one conflict without any repetitions, 251 with at least one repetition and no conflicts, and 57 with both conflicts and repetitions. Repetitions do not need to be manually resolved since this arises from the specification of identical information across multiple paragraphs (e.g. mentioning the gold nanorod aspect ratios in paragraphs about both the synthesis procedure as well as the nanorod characterization), but conflicts can be challenging to resolve in a consistent manner without manual inspection. For instance, if two separate volumes for a particular precursor are provided in two separate paragraphs, it can be ambiguous whether the volumes are part of the same synthesis procedure or distinct synthesis procedures in the same paper due to the lack of cross-paragraph context. With this in mind, of the 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 644 extracted entities, 10
644 extracted entities, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 349 (∼89%) can be safely extracted by automatically resolving repetitions and discarding entities with conflicts. Of the entities with conflicts, 341 have two distinct values, 47 have three, 12 have five, 9 have four, and 1 has five.
349 (∼89%) can be safely extracted by automatically resolving repetitions and discarding entities with conflicts. Of the entities with conflicts, 341 have two distinct values, 47 have three, 12 have five, 9 have four, and 1 has five.
| Root key | First subkey | Second subkey | Third subkey | Description | Total |
|---|---|---|---|---|---|
| a The “doi” key contains the article DOI and the “text” key contains index keys of the relevant paragraphs within that article which in turn contain the paragraph text. The “seed” and “growth” keys respectively contain the keys for the seed and growth solution information, including the “prec” key for precursors, the “stir” key for stir rates (when adding the reducing agent for the seed solution and when adding the seed solution for the growth solution), the “temp” key for the aging temperature, and the “age” key for the solution aging time. The “seed” key has an additional “seed” key that contains the “size” and “shape” keys for the size and shape of the seeds in the seed solution. The “prec” key for each solution contains multiple keys for each precursor in each solution, anonymized as “<precursor name>” in the table. For each precursor, there are three keys: “vol”, “concn”, and “mass” for the precursor volume, concentration, and mass, respectively. The “AuNR” key contains keys for measurements of gold nanorod dimensions: “ar”, “l”, “w”, “lspr”, and “tspr” for the aspect ratio, length, width, LSPR, and TSPR, respectively. Each extracted value is additionally stored as a key with a corresponding list of the paragraph indices that the value was extracted from in order to preserve information about entity sources. The final column displays the total number of entities extracted for each key (with no subkeys). | |||||
| doi | Article DOI | 1137 | |||
| text | <integer> | Paragraph text for <integer>th paragraph | 2969 | ||
| seed | prec | <precursor name> | volume | Seed solution precursor volume | 1347 |
| concentration | Seed solution precursor concentration | 1385 | |||
| mass | Seed solution precursor mass | 6 | |||
| seed | size | Seed solution seed size | 137 | ||
| shape | Seed solution seed shape | 24 | |||
| stir | Seed solution reducing agent stir rate | 266 | |||
| temp | Seed solution aging temperature | 284 | |||
| age | Seed solution aging time | 352 | |||
| growth | prec | <precursor name> | volume | Growth solution precursor volume | 2664 |
| concentration | Growth solution precursor concentration | 2178 | |||
| mass | Growth solution precursor mass | 65 | |||
| stir | Growth solution reducing agent stir rate | 134 | |||
| temp | Growth solution aging temperature | 322 | |||
| age | Growth solution aging time | 464 | |||
| AuNR | ar | Gold nanorod aspect ratio | 587 | ||
| l | Gold nanorod length | 443 | |||
| w | Gold nanorod width | 452 | |||
| lspr | Gold nanorod LSPR | 357 | |||
| tspr | Gold nanorod TSPR | 177 | |||
With post-processing applied (as was done for evaluation of the testing dataset), splitting lists of extracted values into distinct entities and resolving repetitions of identical information extracted across different paragraphs within the same papers results in a total of 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 770 unique entities. In the post-processed version of the dataset, each property contains a list of dictionaries with structures indicated in Table 4.
770 unique entities. In the post-processed version of the dataset, each property contains a list of dictionaries with structures indicated in Table 4.
| Key | Structure | Description |
|---|---|---|
| mod | <modifier> | A string indicating if a value is a range, approximate, bounding, or unprocessed |
| val | [<value>, …, <value>] | A list of the extracted values. Ranges will consist of two values for the range boundaries. Processed values will be numbers while unprocessed values will be strings |
| unit | <unit> | The units for the extracted values, if applicable, as a string |
| src | [[<index>, …, <index>], …, […]] | A list of lists of paragraph indices to indicate the source for the extracted information |
| index | [[<index>, …, <index>], …, […]] | A list of lists of positional indices to retain ordering for values that were split from a list during post-processing |
4.4 Full filtered dataset analysis
In order to evaluate the completeness of the components of the procedure and the outcome, for seed and growth solutions, only fully specified precursors were considered necessary for reproducibility. Auxiliary information, such as stirring rates, aging times, aging temperatures, and seed particle morphologies and sizes, while useful, was not considered necessary. The precursor information was considered to be full specified for a given paper if all of the precursor quantities were fully specified with either volume and concentration, mass, or a specific concentration within another solution for each precursor with extracted quantities. Exceptions were made for water and the seed solution that is added to the growth solution, which both only needed a reported volume or mass. Additionally, seed solution in the growth solution precursors was required for the growth solution precursors to be considered complete. For the gold nanorod dimensions to be considered complete, either the aspect ratio, length, or LSPR measurement had to be specified, with the latter two at least providing an avenue for estimation of the aspect ratio if reported alone.
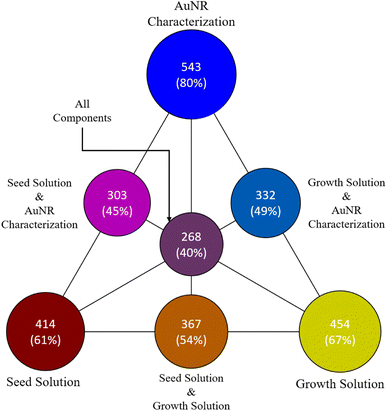
Fig. 7 shows how the papers in the full filtered prediction dataset are distributed across fully-specified synthesis procedure and outcome components according to these criteria. The vast majority of the papers reported gold nanorod dimensions, with 80% of the 678 papers with at least one fully specified synthesis component containing fully-specified gold nanorod dimensions. Additionally, the majority of the papers fully-specified the seed and growth solutions (respectively 61% and 67%). However, they are distributed such that 40% (268) of the papers fully specified all three components. This is a reasonable result considering that many papers will directly report the relevant gold nanorod dimensions without specifying a synthesis procedure, opting instead to reference the established recipe that the researchers used to produce the gold nanorods. Additionally, some researchers will opt to purchase gold seed solution instead of producing their own, which accounts for cases where some papers are missing information about seed solution preparation. Most of the papers with fully-specified synthesis procedures and outcomes (162) used the typical 8-precursor synthesis and an additional 49 use the same synthesis precursors with the addition of HCl in the growth solution. In the post-processed version of the dataset, it is determined that of the 268 papers that fully specified all three components, 233 contained exactly one procedure. An additional 16 contained two, 13 contained three, 3 contained four, 2 contained five, and 1 contained six for a total of 332 complete procedures. This final dataset should be suitable for downstream analysis and inference, given the overall model performance for extracting complete synthesis procedures and outcomes from the literature.
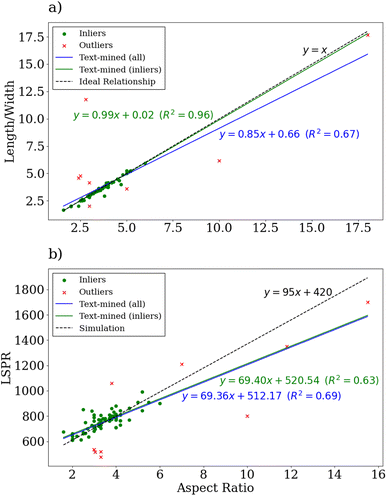 | ||
| Fig. 8 A diagram showing the relationships between the gold nanorod aspect ratios and other gold nanorod measurements extracted from the literature including the (a) ratio between length and width and (b) the LSPR peak. The inlier datapoints are shown in purple and the outlier datapoints in red. The linear regressions derived from the text-mined data using all of the available data and only the inlier data are respectively shown in red and purple on each sub-diagram. For the comparison to the ratio between length and width (a), the ideal relation is shown with a dashed black line and for the LSPR comparison (b), a simulated relationship is shown with a dashed black line.79 | ||
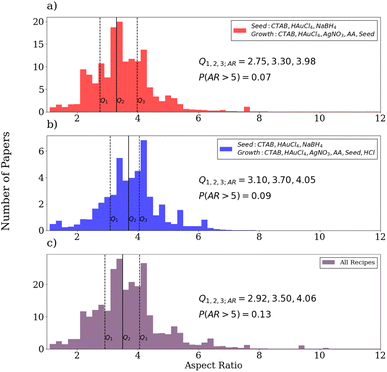
From the distribution of the standard recipe, it is readily apparent that the median nanorod aspect ratio is 3.3 with respective first and third quartiles of 2.75 and 3.98. Comparing with experiments reporting that varying the concentration of AgNO3 in the growth solution varies the resulting nanorod aspect ratios from 1.83 to 5.04,83 the distribution of gold nanorod aspect ratios text-mined from the literature is consistent with this range, though it is narrower. Notably, there is a non-negligible amount of samples with aspect ratios greater than 5 in the distribution for the standard procedure. This is not consistent with heuristic knowledge of the limitations of the standard procedure for producing large aspect ratio gold nanorods, usually due to shorter growth times compared to procedures that adjust the pH of the growth solution to retard the nanorod growth.84,85 This is primarily due to erroneous extractions of nanowire measurements from overgrowth experiments or missed precursors based on manual inspection of the data. However, the statistics are still dominated by the lower aspect ratios. Compared to the distribution for experiments using HCl in the growth solution, it is apparent that the addition produces a distribution shifted towards larger aspect ratios. This is consistent with experiments that have determined that the use of HCl in the growth solution grants broader tunability of the gold nanorod aspect ratios, allowing for more controlled growth of longer nanorods relative to the standard procedure.86,87 Notably, ∼7% of the procedures using the standard procedure and ∼9% of the procedures using HCl in the growth solution provide nanorods with aspect ratios of 5 or higher. However, when all recipes are considered, it is clear that even longer nanorods can be synthesized, though these recipes are not as popular in the literature.
5 Discussion
Overall, the model performs well at identifying and extracting relevant information specific to seed-mediated gold nanorod growth procedures in the literature. The model achieves an overall adjusted F1-score of 86% on the testing dataset, indicating that it performs rather well at the task of simultaneous entity recognition and relation extraction. However, due to the static nature of the relations provided by the synthesis template and the single inference step, the entity recognition and relation extraction tasks are not easily disentangled, which limits direct comparison with conventional two-step approaches. Instead, the model performance for information retrieval is evaluated according to its ability to place information into fields of the template where information should exist and then the accuracy of the information that is correctly placed. For information placement, the precision, recall, and F1-score are balanced at 90%, indicating notable performance with no preference for false positives or false negatives. Of the information that is correctly placed in the templates, the model predicts the specific values with 96% accuracy. Thus, the primary source of error is the accurate placement of information into the template rather than the accurate prediction of correctly placed information. However, the template model struggles with identifying new precursors that were not present in the training set.The dataset produced by the model provides a wealth of information about seed-mediated gold nanorod growth experiments and, to our knowledge, constitutes the largest structured database with this level of depth and completeness. The model's ability to distinguish between precursors in the seed and growth solutions provides an example of very useful information. The simultaneous identification of precursors alongside linking them to the appropriate solutions in the two-step seed-mediated procedure had proven difficult using established methods due to the propagation of errors introduced by the reliance on separate models for entity extraction and relation. However, with this model, if a researcher wants to quickly find papers that used a particular precursor in the seed solution for seed-mediated growth of gold nanorods, this task can be accomplished with high fidelity using the predicted templates. Access to this information can be expected to greatly improve tools for scientific literature searches, as conventional simple keyword searches do not offer this specific relational dependence for complicated multi-step procedures.
For a more ambitious goal, the full synthesis procedure data can be leveraged for multiple downstream tasks, which would require the creation of additional models for inference. One example would be a model that predicts gold nanorod dimensions conditioned on a specific synthesis procedure: p(properties|procedure). Such a model may be leveraged to predict the outcomes of proposed procedures without the need to perform them explicitly. Building on this, the inverse problem, p(procedure|properties), can also be modeled. This would be very useful for streamlining synthesis experiments, as the necessary procedures for synthesizing gold nanorods with the desired properties can be inferred to provide a starting point that reduces the number of experiments that must be conducted to synthesize the desired gold nanorods. However, in the most likely case, any model trained on literature data alone will be incomplete and require further data generation and fine tuning.
Furthermore, it is worth considering how these templates fit into a larger project for downstream synthesis outcome predictions and synthesis procedure recommendations. The data extracted from literature can be used to pre-train models used for these purposes, while explicit experimental data can be used to further train the models to produce better predictions. The new templates provided by the experimental results are expected to be of extremely high quality, which will mitigate the errors present in the pre-training data from literature over time as more experimental results are added to the template database.
While this dataset is restricted to seed-mediated gold nanorod growth, the flexibility and performance of the templating approach using GPT-3 motivates application to other tasks for structured information retrieval from unstructured scientific text as has been shown in recent literature.62 To this end, the dataset can be extended to accommodate seed-mediated growth of other gold nanoparticle morphologies, which may even improve overall model performance, as many errors were caused by the model erroneously extracting information from procedures that mentioned nanorod morphologies, but synthesized a different morphology. Additionally, more complex synthesis methods, such as three-step processes in which nanorods are first synthesized via seed-mediated growth to be used as seeds in a growth solution for overgrowth into nanowires, as well as other synthesis methods, such as citrate reduction, may require the creation of new templates and fine-tuning a separate model for each synthesis method to improve overall performance. Generally, it can be expected that more complex templates will require more examples for fine-tuning.
6 Conclusions
The presented model for static structured templating of seed-mediated gold nanorod growth procedures extracted from unstructured text using GPT-3 is demonstrated to be a promising approach for constructing high-quality structured databases of information from the scientific literature. This approach for extracting seed-mediated gold nanorod procedures and outcomes achieves an impressive adjusted F1-score of 86% for the simultaneous identification and linking of synthesis procedure components. We present a final dataset of 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 644 entities extracted from 1137 filtered papers, resulting in 268 papers with at least one complete seed-mediated gold nanorod growth procedure and outcome for a total of 332 complete procedures. This method can potentially be utilized for many downstream applications including procedure searches oriented around specific features, statistical analysis of synthesis outcomes, synthesis outcome predictions conditioned on procedures, and synthesis procedure recommendations conditioned on outcomes among others given the wealth of structured information present. Overall, we present this approach as a flexible candidate for general-purpose structured data extraction from unstructured scientific text and contribute a dataset that may serve as a useful tool for investigating synthesis pathways beyond heuristics.
644 entities extracted from 1137 filtered papers, resulting in 268 papers with at least one complete seed-mediated gold nanorod growth procedure and outcome for a total of 332 complete procedures. This method can potentially be utilized for many downstream applications including procedure searches oriented around specific features, statistical analysis of synthesis outcomes, synthesis outcome predictions conditioned on procedures, and synthesis procedure recommendations conditioned on outcomes among others given the wealth of structured information present. Overall, we present this approach as a flexible candidate for general-purpose structured data extraction from unstructured scientific text and contribute a dataset that may serve as a useful tool for investigating synthesis pathways beyond heuristics.
Data and code availability
The data composed of DOIs and associated structured JSON outputs can be found online at https://doi.org/10.6084/m9.figshare.19719310.v4.88 The texts for the paragraphs in each paper are excerpted due to copyright restrictions.Author contributions
A. J., G. C., and K. A. P. supervised the research. K. C. wrote the data collection infrastructure, performed the data collection, and wrote and applied the initial gold nanoparticle article classification and information extraction models. S. G. provided experimental domain knowledge necessary for the template design. J. D. introduced the GPT-3 sequence-to-sequence information extraction methodology and prepared the graphic representation of the extraction template. N. W. co-developed the GPT-3 sequence-to-sequence information extraction methodology, designed the extraction templates, wrote the code for interfacing with GPT-3, performed all annotations, performed all GPT-3 experiments, and prepared all results. S. L. performed fine-tuning on Llama-2 for the benchmark and provided additional result validation. All authors contributed to the discussion and writing of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded and intellectually led by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, Materials Sciences and Engineering Division under Contract No. DE-AC02-05CH11231 (D2S2 program KCD2S2). Additional funding used for data set generation via the OpenAI API was provided by Toyota Research Institute through the Accelerated Materials Design and Discovery program. This research used resources of the National Energy Research Scientific Computing Center (NERSC), a U.S. Department of Energy Office of Science User Facility operated under Contract No. DE-AC02-05CH11231. This work also used the Extreme Science and Engineering Discovery Environment (XSEDE) GPU resources, specifically the Bridges-2 supercomputer at the Pittsburgh Supercomputing Center, through allocation TG-DMR970008S.89Notes and references
- S. Mohan Bhagyaraj and O. S. Oluwafemi, Synthesis of Inorganic Nanomaterials, Woodhead Publishing, 2018, pp. 1–18 Search PubMed.
- P. Colomban, M. Gironda, G. Simsek Franci and P. d’Abrigeon, Materials, 2022, 15(16), 5747 CrossRef CAS PubMed.
- S. Szunerits and R. Boukherroub, Encyclopedia of Interfacial Chemistry, Elsevier, Oxford, 2018, pp. 500–510 Search PubMed.
- S. E. Lohse and C. J. Murphy, Chem. Mater., 2013, 25, 1250–1261 CrossRef CAS.
- N. D. Burrows, S. Harvey, F. A. Idesis and C. J. Murphy, Langmuir, 2017, 33, 1891–1907 CrossRef CAS PubMed.
- L. Gou and C. J. Murphy, Chem. Mater., 2005, 17, 3668–3672 CrossRef CAS.
- P. K. Jain, X. Huang, I. H. El-Sayed and M. A. El-Sayed, Acc. Chem. Res., 2008, 41, 1578–1586 CrossRef CAS PubMed.
- E. C. Dreaden, A. M. Alkilany, X. Huang, C. J. Murphy and M. A. El-Sayed, Chem. Soc. Rev., 2011, 41, 2740–2779 RSC.
- S. Eustis and M. A. El-Sayed, Chem. Soc. Rev., 2006, 35, 209–217 RSC.
- J. C. Hulteen and C. R. Martin, J. Mater. Chem., 1997, 7, 1075–1087 RSC.
- K. Sandeep, B. Manoj and K. G. Thomas, J. Chem. Phys., 2020, 152, 044710 CrossRef CAS PubMed.
- M. Lau, A. Ziefuss, T. Komossa and S. Barcikowski, Phys. Chem. Chem. Phys., 2015, 17, 29311–29318 RSC.
- L. A. Dykman and N. G. Khlebtsov, Acta Nat., 2011, 3, 34–55 CrossRef CAS.
- X. Huang and M. A. El-Sayed, J. Adv. Res., 2010, 1, 13–28 CrossRef.
- S. Kaul, N. Gulati, D. Verma, S. Mukherjee and U. Nagaich, J. Pharm., 2018, 2018, 3420204 Search PubMed.
- K. I. Requejo, A. V. Liopo, P. J. Derry and E. R. Zubarev, Langmuir, 2017, 33, 12681–12688 CrossRef CAS PubMed.
- Y. C. Dong, M. Hajfathalian, P. S. N. Maidment, J. C. Hsu, P. C. Naha, S. Si-Mohamed, M. Breuilly, J. Kim, P. Chhour, P. Douek, H. I. Litt and D. P. Cormode, Sci. Rep., 2019, 9, 14912 CrossRef PubMed.
- S. A. Ng, K. A. Razak, A. A. Aziz and K. Y. Cheong, J. Exp. Nanosci., 2014, 9, 64–77 CrossRef CAS.
- C. Daruich De Souza, B. Ribeiro Nogueira and M. E. C. Rostelato, J. Alloys Compd., 2019, 798, 714–740 CrossRef CAS.
- E. Agunloye, L. Panariello, A. Gavriilidis and L. Mazzei, Chem. Eng. Sci., 2018, 191, 318–331 CrossRef CAS.
- M. L. Personick and C. A. Mirkin, J. Am. Chem. Soc., 2013, 135, 18238–18247 CrossRef CAS PubMed.
- M. Grzelczak, J. Pérez-Juste, P. Mulvaney and L. M. Liz-Marzán, Colloidal Synth. Plasmonic Nanomet., 2020, 197–220 CAS.
- D. F. Mukhamedzyanova, N. K. Ratmanova, D. A. Pichugina and N. E. Kuz'menko, J. Phys. Chem. C, 2012, 116, 11507–11518 CrossRef CAS.
- M. Domingo, M. Shahrokhi, I. N. Remediakis and N. Lopez, Top. Catal., 2018, 61, 412–418 CrossRef CAS.
- I. Chakraborty and T. Pradeep, Chem. Rev., 2017, 117, 8208–8271 CrossRef CAS PubMed.
- D. V. Talapin, A. L. Rogach, M. Haase and H. Weller, J. Phys. Chem. B, 2001, 105, 12278–12285 CrossRef CAS.
- O. Kononova, T. He, H. Huo, A. Trewartha, E. A. Olivetti and G. Ceder, iScience, 2021, 24, 102155 CrossRef PubMed.
- O. Kononova, H. Huo, T. He, Z. Rong, T. Botari, W. Sun, V. Tshitoyan and G. Ceder, Sci. Data, 2019, 6, 203 CrossRef PubMed.
- S. Eltyeb and N. Salim, J. Cheminf., 2014, 6, 17 Search PubMed.
- P. Corbett and J. Boyle, J. Cheminf., 2018, 10, 59 Search PubMed.
- Z. Liang, J. Chen, Z. Xu, Y. Chen and T. Hao, Front. Artif. Intell., 2019, 2, 1 CrossRef PubMed.
- A. Sniegula, A. Poniszewska-Maranda and L. Chomatek, Procedia Comput. Sci., 2019, 160, 260–265 CrossRef.
- K. r. Kanakarajan, B. Kundumani and M. Sankarasubbu, Proceedings of the 20th Workshop on Biomedical Language Processing, 2021, pp. 143–154 Search PubMed.
- L. Weston, V. Tshitoyan, J. Dagdelen, O. Kononova, A. Trewartha, K. Persson, G. Ceder and A. Jain, J. Chem. Inf. Model., 2019, 59, 3692–3702 CrossRef CAS PubMed.
- T. He, W. Sun, H. Huo, O. Kononova, Z. Rong, V. Tshitoyan, T. Botari and G. Ceder, Chem. Mater., 2020, 32, 7861–7873 CrossRef CAS.
- K. Hatakeyama-Sato and K. Oyaizu, Commun. Mater., 2020, 1, 49 CrossRef.
- O. Kononova, T. He, H. Huo, A. Trewartha, E. A. Olivetti and G. Ceder, iScience, 2021, 24, 102155 CrossRef PubMed.
- E. A. Olivetti, J. M. Cole, E. Kim, O. Kononova, G. Ceder, T. Y.-J. Han and A. M. Hiszpanski, Applied Physics Reviews, 2020, 7, 041317 CrossRef CAS.
- T. Dieb, M. Yoshioka, S. Hara and M. Newton, Beilstein J. Nanotechnol., 2015, 6, 1872–1882 CrossRef CAS PubMed.
- M. Gaultois, T. Sparks, C. Borg, R. Seshadri, W. Bonificio and D. Clarke, Chem. Mater., 2013, 25, 2911–2920 CrossRef CAS.
- N. Pang, L. Qian, W. Lyu and J.-D. Yang, Transfer Learning for Scientific Data Chain Extraction in Small Chemical Corpus with BERT-CRF Model, 2019 Search PubMed.
- P. Corbett and A. Copestake, BMC Bioinf., 2008, 9, S4 CrossRef PubMed.
- M. Krallinger, O. Rabal, A. Lourenço, J. Oyarzabal and A. Valencia, Chem. Rev., 2017, 117, 7673–7761 CrossRef CAS PubMed.
- T. Rocktäschel, M. Weidlich and U. Leser, Bioinformatics, 2012, 28, 1633–1640 CrossRef PubMed.
- M. Krallinger, O. Rabal, F. Leitner, M. Vazquez and D. Salgado, et al. , J. Cheminformatics, 2015, 7, S2 CrossRef PubMed.
- R. Leaman, C.-H. Wei and Z. Lu, J. Cheminformatics, 2015, 7, S3 CrossRef PubMed.
- I. Korvigo, M. Holmatov, A. Zaikovskii and M. Skoblov, J. Cheminformatics, 2018, 10, 28 CrossRef PubMed.
- M. García-Remesal, A. García-Ruiz, D. Pérez-Rey, D. De La Iglesia and V. Maojo, Biomed Res. Int., 2013, 2013, 410294 CrossRef PubMed.
- A. Trewartha, N. Walker, H. Huo, S. Lee, K. Cruse, J. Dagdelen, A. Dunn, K. A. Persson, G. Ceder and A. Jain, Patterns, 2022, 3, 100488 CrossRef PubMed.
- A. M. Bran, S. Cox, A. D. White and P. Schwaller, ChemCrow: Augmenting large-language models with chemistry tools, 2023, https://arxiv.org/abs/2304.05376 Search PubMed.
- M. C. Ramos, S. S. Michtavy, M. D. Porosoff and A. D. White, Bayesian Optimization of Catalysts With In-context Learning, 2023, https://arxiv.org/abs/2304.05341 Search PubMed.
- A. D. White, G. M. Hocky, H. A. Gandhi, M. Ansari, S. Cox, G. P. Wellawatte, S. Sasmal, Z. Yang, K. Liu, Y. Singh and W. J. Peña Ccoa, Digital Discovery, 2023, 2, 368–376 RSC.
- F. Ren, L. Ward, T. Williams, K. J. Laws, C. Wolverton, J. Hattrick-Simpers and A. Mehta, Sci. Adv., 2018, 4, eaaq1566 CrossRef PubMed.
- C. C. Fischer, K. J. Tibbetts, D. Morgan and G. Ceder, Nat. Mater., 2006, 5, 641–646 CrossRef CAS PubMed.
- L. Weston, V. Tshitoyan, J. Dagdelen, O. Kononova, A. Trewartha, K. A. Persson, G. Ceder and A. Jain, J. Chem. Inf. Model., 2019, 59, 3692–3702 CrossRef CAS PubMed.
- X. Wang, J. Li, H. D. Ha, J. C. Dahl, J. C. Ondry, I. Moreno-Hernandez, T. Head-Gordon and A. P. Alivisatos, JACS Au, 2021, 1, 316–327 CrossRef CAS PubMed.
- N. J. Szymanski, C. J. Bartel, Y. Zeng, Q. Tu and G. Ceder, Chem. Mater., 2021, 33, 4204–4215 CrossRef CAS.
- X. Yan, A. Sedykh, W. Wang, B. Yan and H. Zhu, Nat. Commun., 2020, 11, 2519 CrossRef CAS PubMed.
- K. Cruse, A. Trewartha, S. Lee, Z. Wang, H. Huo, T. He, O. Kononova, A. Jain and G. Ceder, Sci. Data, 2022, 9, 234 CrossRef PubMed.
- I. Sutskever, O. Vinyals and Q. V. Le, Sequence to Sequence Learning with Neural Networks, 2014, https://arxiv.org/abs/1409.3215 Search PubMed.
- T. Brown, B. Mann, N. Ryder, M. Subbiah, J. D. Kaplan, P. Dhariwal, A. Neelakantan, P. Shyam, G. Sastry and A. Askell, et al. , Adv. Neural Inf. Process. Syst., 2020, 33, 1877–1901 Search PubMed.
- A. Dunn, J. Dagdelen, N. Walker, S. Lee, A. S. Rosen, G. Ceder, K. Persson and A. Jain, Structured information extraction from complex scientific text with fine-tuned large language models, 2022, https://arxiv.org/abs/2212.05238 Search PubMed.
- H. Touvron, L. Martin, K. Stone, P. Albert, A. Almahairi, Y. Babaei, N. Bashlykov, S. Batra, P. Bhargava, S. Bhosale, D. Bikel, L. Blecher, C. C. Ferrer, M. Chen, G. Cucurull, D. Esiobu, J. Fernandes, J. Fu, W. Fu, B. Fuller, C. Gao, V. Goswami, N. Goyal, A. Hartshorn, S. Hosseini, R. Hou, H. Inan, M. Kardas, V. Kerkez, M. Khabsa, I. Kloumann, A. Korenev, P. S. Koura, M.-A. Lachaux, T. Lavril, J. Lee, D. Liskovich, Y. Lu, Y. Mao, X. Martinet, T. Mihaylov, P. Mishra, I. Molybog, Y. Nie, A. Poulton, J. Reizenstein, R. Rungta, K. Saladi, A. Schelten, R. Silva, E. M. Smith, R. Subramanian, X. E. Tan, B. Tang, R. Taylor, A. Williams, J. X. Kuan, P. Xu, Z. Yan, I. Zarov, Y. Zhang, A. Fan, M. Kambadur, S. Narang, A. Rodriguez, R. Stojnic, S. Edunov and T. Scialom, Llama 2: Open Foundation and Fine-Tuned Chat Models, 2023 Search PubMed.
- H. Touvron, T. Lavril, G. Izacard, X. Martinet, M.-A. Lachaux, T. Lacroix, B. Rozière, N. Goyal, E. Hambro, F. Azhar, A. Rodriguez, A. Joulin, E. Grave and G. Lample, LLaMA: Open and Efficient Foundation Language Models, 2023 Search PubMed.
- J. Su, Y. Lu, S. Pan, A. Murtadha, B. Wen and Y. Liu, RoFormer: Enhanced Transformer with Rotary Position Embedding, 2022 Search PubMed.
- J. Su, Y. Lu, S. Pan, A. Murtadha, B. Wen and Y. Liu, RoFormer: Enhanced Transformer with Rotary Position Embedding, 2022 Search PubMed.
- B. Zhang and R. Sennrich, Root Mean Square Layer Normalization, 2019 Search PubMed.
- J. L. Ba, J. R. Kiros and G. E. Hinton, Layer Normalization, 2016 Search PubMed.
- Z. Wang, O. Kononova, K. Cruse, T. He, H. Huo, Y. Fei, Y. Zeng, Y. Sun, Z. Cai, W. Sun and G. Ceder, Dataset of Solution-based Inorganic Materials Synthesis Recipes Extracted from the Scientific Literature, 2021, DOI:10.48550/arXiv.2111.10874.
- K. Cruse, A. Trewartha, S. Lee, Z. Wang, H. Huo, T. He, O. Kononova, A. Jain and G. Ceder, Text-mined AuNP Synthesis Recipes Dataset, figshare, 2021, DOI:10.6084/m9.figshare.16614262.v3.
- A. Radford, K. Narasimhan, T. Salimans and I. Sutskever, OpenAI Assets Research Covers, 2018, https://s3-us-west-2.amazonaws.com/openai-assets/research-covers/language-unsupervised/language_understanding_paper.pdf Search PubMed.
- A. Radford, J. Wu, R. Child, D. Luan, D. Amodei, I. Sutskever, et al., OpenAI blog, 2019, vol. 1, p. 9 Search PubMed.
- E. J. Hu, Y. Shen, P. Wallis, Z. Allen-Zhu, Y. Li, S. Wang, L. Wang and W. Chen, LoRA: Low-Rank Adaptation of Large Language Models, 2021 Search PubMed.
- S. Mangrulkar, S. Gugger, L. Debut, Y. Belkada and S. Paul, PEFT: State-of-the-art Parameter-Efficient Fine-Tuning methods, https://github.com/huggingface/peft, 2022 Search PubMed.
- https://huggingface.co/meta-llama/Llama-2-13b-hf .
- M. Ma, H. Chen, Y. Chen, X. Wang, F. Chen, X. Cui and J. Shi, Biomaterials, 2012, 33, 989–998 CrossRef CAS PubMed.
- K. W. Smith, H. Zhao, H. Zhang, A. Sánchez-Iglesias, M. Grzelczak, Y. Wang, W.-S. Chang, P. Nordlander, L. M. Liz-Marzán and S. Link, ACS Nano, 2016, 10, 6180–6188 CrossRef CAS PubMed.
- M. Zareie, X. Xu and M. Cortie, Small, 2007, 3, 139–145 CrossRef CAS PubMed.
- X. Huang, S. Neretina and M. A. El-Sayed, Adv. Mater., 2009, 21, 4880–4910 CrossRef CAS PubMed.
- P. J. Rousseeuw, J. Am. Stat. Assoc., 1984, 79, 871–880 CrossRef.
- F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel, M. Blondel, P. Prettenhofer, R. Weiss, V. Dubourg, J. Vanderplas, A. Passos, D. Cournapeau, M. Brucher, M. Perrot and E. Duchesnay, J. Mach. Learn. Res., 2011, 12, 2825–2830 Search PubMed.
- L. Vigderman and E. R. Zubarev, Chem. Mater., 2013, 25, 1450–1457 CrossRef CAS.
- L. Feng, Z. Xuan, J. Ma, J. Chen, D. Cui, C. Su, J. Guo and Y. Zhang, J. Exp. Nanosci., 2015, 10, 258–267 CrossRef CAS.
- N. D. Burrows, S. Harvey, F. A. Idesis and C. J. Murphy, Langmuir, 2017, 33, 1891–1907 CrossRef CAS PubMed.
- Y. Wang, Y. Guo, Y. Shen, R. Chen, F. Wang, D. Zhou and D. Zhou, J. Nanosci. Nanotechnol., 2016, 16, 1194–1201 CrossRef CAS PubMed.
- Y. Wang, Y. Guo, Y. Shen, R. Chen, F. Wang, D. Zhou and S. Guo, J. Nanosci. Nanotechnol., 2016, 16, 1194–1201 CrossRef CAS PubMed.
- M.-Z. Wei, T.-S. Deng, Q. Zhang, Z. Cheng and S. Li, ACS Omega, 2021, 6, 9188–9195 CrossRef CAS PubMed.
- N. Walker, S. Leee, J. Dagdelen, K. Cruse, S. Gleason, A. Dunn, G. Ceder, A. P. Alivisatos, K. A. Persson and A. Jain, Seed-mediated AuNR Synthesis Extraction Dataset, figshare, 2023, DOI:10.6084/m9.figshare.19719310.v4.
- J. Towns, T. Cockerill, M. Dahan, I. Foster, K. Gaither, A. Grimshaw, V. Hazlewood, S. Lathrop, D. Lifka, G. D. Peterson, R. Roskies, J. R. Scott and N. Wilkins-Diehr, Comput. Sci. Eng., 2014, 16, 62–74 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3dd00019b |
| This journal is © The Royal Society of Chemistry 2023 |