Open Access Article
Open Access ArticleNatural alternative heme-environments allow efficient peroxygenase activity by cytochrome P450 monooxygenases†
Ana C.
Ebrecht
 ,
Martha S.
Smit
,
Martha S.
Smit
 * and
Diederik J.
Opperman
* and
Diederik J.
Opperman
 *
*
Department of Microbiology and Biochemistry, University of the Free State, 205 Nelson Mandela Drive, Bloemfontein, 9300, South Africa. E-mail: opperdj@ufs.ac.za
First published on 29th September 2023
Abstract
Cytochrome P450 monooxygenases (CYPs) are biocatalysts able to catalyze a variety of regio- and stereoselective oxyfunctionalization reactions using an iron(IV)-oxo porphyrin pi-cation radical prosthetic group, commonly referred to as compound I (Cpd I). The formation of Cpd I is, however, dependent on molecular oxygen and the sequential transfer of electrons from expensive nicotinamide cofactors via additional redox partner proteins. Recently, CYPs have been engineered to introduce or enhance peroxygenase activity, whereby Cpd I is formed from H2O2. Here we explore the potential of natural CYPs containing an aspartate instead of the conventional threonine on the distal side of the heme, for peroxygenase activity. Peroxygenase activity was demonstrated with three new CYPs, with SscaCYP from Streptomyces scabiei demonstrating the highest activity towards the hydroxylation of trans-β-methyl styrene and the sulfoxidation of thioanisole. The X-ray crystal structure of SscaCYP revealed two potential acid–base catalysts (D241 and E284) in close proximity to the axial water molecule for the heterolytic O–O cleavage during H2O2 activation for Cpd I formation. Both side chain groups are also located at H2O channels to the bulk solvent. Mutagenesis of either side chain increased the peroxygenase activity, whereas removal of both abolished the peroxygenase activity of SscaCYP. Spectral analysis confirmed the tight binding of the axial water molecule when both carboxylate groups are present. With only one carboxylate group present lower concentrations of H2O2 are also required for catalysis, suggesting H2O and H2O2 exchange as a rate limiting factor in the wild-type SscaCYP. The improved peroxygenase activity and increased substrate scope of the SscaCYP variants, have identified two “hot-spots” for future protein engineering with one an alternative site for the introduction of an acid–base catalyst for engineering peroxygenase activity in other CYPs.
Introduction
Cytochrome P450 monooxygenases (CYPs/P450s) are a large group of heme-thiolate enzymes able to perform a variety of reactions, most notably the hydroxylation of inert C–H bonds and the epoxidation of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds.1–4 Moreover, these reactions are often highly regio- and stereoselective, making CYPs attractive catalysts for oxyfunctionalization reactions. Their impressive substrate scope and selectivity have also been expanded on and refined in recent years through directed evolution and rational design.5 One of their major drawbacks, however, is the requirement for redox partner proteins and the sequential transfer of two electrons to the heme centre from the expensive cofactor NAD(P)H to activate molecular oxygen for catalysis.6 Upon binding the substrate, the ferric (FeIII) iron in the heme cofactor is reduced to ferrous (FeII) iron in the first electron transfer. Molecular oxygen binds to the ferrous iron, and a second electron is delivered to yield the ferric-peroxo species. Protonation, mediated via an acid–alcohol pair in the active site, results in the ferric-hydroperoxo species compound 0 (Cpd 0). A second protonation step induces O–O bond scission and dehydration to form compound I (Cpd I), a ferryl (FeIV)-oxo porphyrin-pi-cation radical, regarded as the catalytically active species. For hydroxylation, Cpd I abstracts a hydrogen from the target C–H bond, to form a ferryl-hydroxy species (Cpd II) and a substrate radical, which, via radical rebound, hydroxylates the substrate (Fig. 1a).7 Mutation studies of the conventional acid–alcohol pair, typically an Asp and Thr residue, have demonstrated their importance in stabilization of Cpd 0 and formation of Cpd I.8
C bonds.1–4 Moreover, these reactions are often highly regio- and stereoselective, making CYPs attractive catalysts for oxyfunctionalization reactions. Their impressive substrate scope and selectivity have also been expanded on and refined in recent years through directed evolution and rational design.5 One of their major drawbacks, however, is the requirement for redox partner proteins and the sequential transfer of two electrons to the heme centre from the expensive cofactor NAD(P)H to activate molecular oxygen for catalysis.6 Upon binding the substrate, the ferric (FeIII) iron in the heme cofactor is reduced to ferrous (FeII) iron in the first electron transfer. Molecular oxygen binds to the ferrous iron, and a second electron is delivered to yield the ferric-peroxo species. Protonation, mediated via an acid–alcohol pair in the active site, results in the ferric-hydroperoxo species compound 0 (Cpd 0). A second protonation step induces O–O bond scission and dehydration to form compound I (Cpd I), a ferryl (FeIV)-oxo porphyrin-pi-cation radical, regarded as the catalytically active species. For hydroxylation, Cpd I abstracts a hydrogen from the target C–H bond, to form a ferryl-hydroxy species (Cpd II) and a substrate radical, which, via radical rebound, hydroxylates the substrate (Fig. 1a).7 Mutation studies of the conventional acid–alcohol pair, typically an Asp and Thr residue, have demonstrated their importance in stabilization of Cpd 0 and formation of Cpd I.8
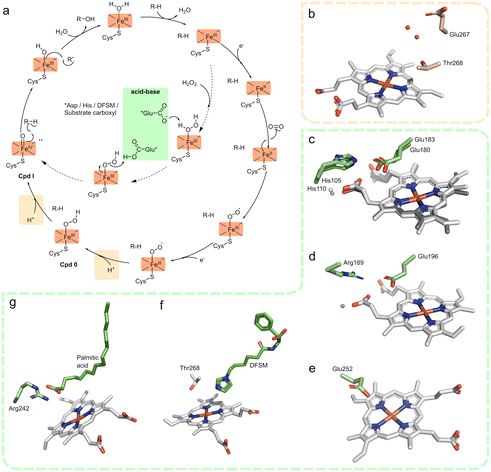 | ||
| Fig. 1 (a) Typical reaction cycle of cytochrome P450 monooxygenases for oxygenation using external electron donors and molecular oxygen (outer circle) or H2O2-shunt pathway with acid–base catalysis for heterolytic cleavage of O–O bond (inner circle). (b) Active site of a typical CYP with an acid–alcohol pair (Asp–Thr) to deliver protons to heme (CYP102A1, PDB 2HPD). (c) Active site overlay of short unspecific peroxygenases (UPOs) and chloroperoxidase with acid–base catalysts glutamate hydrogen bonded to His (CPO, PDB 1CPO; HspUPO, PDB 7O1R). (d) Active site of long UPOs with acid–base catalysts glutamate hydrogen bonded to Arg (AaeUPO, 2YP1). (e) Artificial CYP peroxygenase with catalytic Thr mutated to Glu for acid–base catalysis (CYP199A4_T252E, PDB 7TP5). (f) Active site of CYP102A1_F87A with DFSM (N-imidazolyl-hexanoyl-L-phenylalanine) bound (PDB 7EGN). (g) Substrate bound CYP152A1 peroxygenase with the substrate carboxylate group bonded to active site Arg (PDB 1IZO). | ||
Peroxidases, peroxygenases, and to a lesser extent some CYPs, can efficiently utilize H2O2via a peroxidase shunt mechanism to yield Cpd I.9 After H2O2 displaces the water molecule, Cpd 0 is formed by deprotonation of the iron bound oxygen and the reaction mechanism proceeding as usual (Fig. 1a). In peroxidases and peroxygenases, the deprotonation and protonation steps are mediated via general acid–base catalysis by an amino acid side chain positioned above the distal side of the heme. Unspecific peroxygenases (UPOs) utilizes a glutamate, hydrogen bonded to either a histidine or arginine in short and long UPOs, respectively10,11 (Fig. 1c and d). This is similar to chloroperoxidase (CPO) that utilizes a Glu-His as an acid–base pair for Cpd I formation12 (Fig. 1c) and horseradish peroxidase (HRP) that utilizes a His–Arg pair.13 More variation is observed with dye-decolorizing peroxidase (DyP) typically utilizing Asp or Glu, with Arg for charge stabilization.14 CYP152s is unique amongst CYPs, as it naturally exhibits peroxygenase activity due to its substrate, with the fatty acid substrate's carboxyl group, coordinated via an active-site Arg, acting as the general acid–base catalysts15,16 (Fig. 1g).
This acid–base concept has been explored using dual function small molecules (DFSMs)17–22 and site-directed mutagenesis of the catalytic Thr of CYPs to gain peroxygenase activity. DFSMs is an evolution of decoy molecules, whereby they not only occupy part of the substrate tunnel, but also has two end functional groups: an anchoring group and a basic imidazole group positioned close to the distal side of the heme which functions as a general acid–base catalyst in the heterolytic cleavage of the heme iron coordinated H2O2 for Cpd I formation19 (Fig. 1f). The introduction of a carboxylate (conserved Thr to Glu mutation, Fig. 1e) in CYP119A1 and CYP199A4 allowed for H2O2-driven styrene oxygenation23 and oxidative demethylation,24 respectively. Similarly, mutation of the corresponding Ala to Glu (the equivalent in structural space of the Thr in CYPs) in CYP152B1, allowed for peroxygenase activity during styrene oxygenation.23 Similar improvement or introduction of peroxygenase activity has been observed in CYP102A1 (P450BM3)25 and CYP101A1 (P450cam).23
Conservation of a conventional acid–alcohol pair is not absolute within CYPs, as a small number of CYPs do occur that lack the highly conserved Thr/Ser. These CYPs, however, typically catalyzes rearrangement reactions of the substrate hydroperoxide, exemplified by CYP74.26 In CYP177A1 (XplA) the acid–alcohol pair is replaced by Met-Ala, and catalyzes oxygen independent reductive denitration.27 Two notable exceptions are found in CYP107A1 (P450EryF)28,29 and (CYP176A1) P450cin30,31 that catalyzes the hydroxylation of deoxyerythronolide B and 1,8-cineole, respectively, despite an Ala and Asn occupying the place of the conserved Thr. In CYP107A1, a hydroxyl group of the substrate assumes the role of the active site alcohol, making it the only substrate accepted by CYP107A1,28 whereas the Asn in CYP176A1 is involved in substrate positioning, but does not participate in heterolytic O–O cleavage to form Cpd I. To date, only two families of CYPs naturally containing a Glu in the conserved Thr position of the acid–alcohol pair have been characterized. AgcA (CYP255A1) and GcoA (CYP255A2) catalyze the aryl O-demethylation of guaiacol and related O-methylated aromatic monomers, including 4-alkylguaiacols by AgcA.32,33 CYP1232A24 and CYP1232F1 catalyze the para O-demethylation of 3,4-dimethoxyphenylacetic acid, with CYP1232A24 also able to perform meta O-demethylation.34 Both these CYP families contain instead of the typical Asp–Thr acid–alcohol pair an Asn/Gln-Glu that could potentially indicate peroxygenase activity. However, both have physiological redox partner proteins that transfers electrons from NAD(P)H to the CYPs.32,34 Recently, two new members of the CYP255A family (GcoA and SyoA) were indeed shown to efficiently use H2O2 in the O-demethylation of guaiacol and syringol.35
However, to our knowledge, no CYP naturally containing an Asp has been found to hydroxylate C–H bonds through peroxygenase activity. This prompted us to explore other members of the CYP superfamily that contain an Asp in the conventional alcohol (Thr) position. In this study, we describe a novel CYP peroxygenase containing an Asp instead of the catalytic alcohol (Thr), able to catalyze C–H as well as heteroatom oxygenation using H2O2. The X-ray crystal structure and mutagenesis of active site residues revealed two potential carboxyl groups as candidates for heterolytic O–O cleavage during Cpd I formation, enabling efficient peroxygenase activity.
Results and discussion
Heterologous expression and characterization of peroxygenase activity
Three CYPs were identified from the 3DM database36 that contained an Asp as the structural equivalent of the conventional Thr of the typical acid–alcohol pair. All three CYPs showed more than 40% sequence identity to CYP177A1 (XplA) from Rhodococcus rhodochrous.37 CYP177A1, the prototypic CYP from the CYP177A family, is atypical as the acid–alcohol pair is replaced with Met-Ala and the CYP has a flavodoxin domain fused to the N-terminus.27 Together with its redox partner XplB, a flavodoxin reductase, it catalyzes reductive denitration, which does not require oxygen.Heterologous expression in E. coli was achieved for all three CYPs, with active soluble expression as evaluated through CO-difference spectra (Fig. S1a†). The CYPs were expressed with N-terminal hexahistidine tags and purified to near homogeneity through immobilized metal affinity chromatography (Fig. S1b†). Their peroxygenase activity was evaluated against a panel of aryl alkenes (1a–4a), alkyl benzenes (5a–6), linear (7a–12a) and cyclic alkenes (13a–19a), thioanisole (20a) and monoterpenes and monoterpenoids (21a–27a) using in vitro H2O2 production by the glucose oxidase (GOx) system. For most of the substrates screened (Table S2, Fig. S2†), only trace amounts of products were obtained. No conversion was observed for the oxyfunctionalization of the linear alkenes, monoterpenes and monoterpenoids. Despite the low activity, notable differences were observed between the CYPs with respect to substrate acceptance and regioselectivity (Scheme 1). Whereas SscaCYP showed no activity to either styrene (1a), ethylbenzene (5a) and propylbenzene (6a), MwolCYP and MconCYP showed low-level epoxidation of 1a to styrene epoxide (1b) and hydroxylation of 5a to 1-phenylethanol (5b). Only MwolCYP showed conversion of propylbenzene (6a) through benzylic hydroxylation (6b) which was further oxidized to the corresponding ketone (6c). Trace amounts of epoxide (4b) was, however, observed from allyl benzene (4a) with both SscaCYP and MwolCYP. Interestingly, regiodivergent oxyfunctionalization of trans-β-methyl styrene (3a) was observed, with SscaCYP yielding the allylic terminal alcohol (3b), which was further oxidized to cinnamaldehyde (3c), while Mwol and MconCYP yielded only the epoxide (3d). SscaCYP was also the only candidate to perform heteroatom oxygenation of thioanisole (20a) to form the sulfoxide (20b) (Fig. S3†).
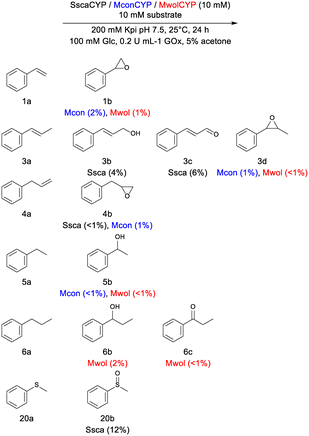 | ||
| Scheme 1 Representative transformations with new CYPs. Percentage conversion are shown in parenthesis. | ||
Reaction optimization
As SscaCYP showed the highest activity during our initial screening, it was selected for further optimization. SscaCYP was optimally active at pH 7.0 and 8.0, both for sulfoxidation and hydroxylation (Fig. S5†) and no difference between HEPES or potassium phosphate buffer was observed. At pH 5.0, the activity was drastically reduced with no activity at pH 4.0 for the hydroxylation of 3a. Sulfoxidation was still observed at these lower pH values but is attributed to background chemical oxidation of 20a (vide infra).As most of the substrates have low solubility in water, we tested different concentrations of acetone as a co-solvent to ultimately improve substrate loadings. SscaCYP activity was evaluated in reactions with different concentrations of acetone for 3a and 20a (Fig. S5†). In addition, the enzyme's stability was analyzed with different acetone concentrations (1–20% v/v). SscaCYP was incubated in the different conditions and its ability to still produce a CO-difference spectrum was evaluated over time (Fig. S6†). Similar results were observed with 2–10% (v/v) acetone, whereas at 20% (v/v) acetone, the ability to form CO-difference spectra rapidly decreased with only 30% remaining after 2 h, and no product formation observed for 3a, and substantially lowered activity for 20a. All further experiments were thus done with 5% (v/v) acetone.
Direct addition of H2O2 (2 times the molar equivalent to the substrate) was also compared to the in vitro GOx/glucose system for H2O2 production. Although the initial turnover frequencies (TOFs) increased with direct H2O2 (20 mM) addition, the total products formed after 24 h and turnover numbers (TONs) remained unchanged (Fig. S7†). Evaluation of the activities at different H2O2 concentrations showed the highest initial activity with 100 mM H2O2 for 3a (Fig. S9†). As CYPs can be deactivated through heme-bleaching at high H2O2 concentrations, we tested the operational stability of SscaCYP during catalysis. Again, the SscaCYP was evaluated through CO-difference spectra using 10 and 50 mM H2O2, both in the presence and absence of substrate. Both substrates (3a and 20a) appeared to have a protective capacity, as even the initial CO-difference spectra at the start of the reactions did not decrease as significantly in their presence, nor during the reactions (Fig. S10†). The rate of heme bleaching was similarly reduced in CYP199A4 and CYP255s by their respective substrates.35,38 In the absence of substrate and excess H2O2, a second H2O2 can react (proton abstraction) with Cpd I to form Cpd II, and via a “catalase malfunction” reaction to Cpd III, reacting again with H2O2, ultimately leading to radical formation and heme bleaching.39,40
Unfortunately, the background sulfoxidation of 20a with the direct H2O2 addition also increased when compared to the in vitro GOx system (Fig. S8†). As the direct sulfoxidation with H2O2 yields a racemic mixture, we determined the stereoselectivity of 20b after the SscaCYP reaction. The enzyme produced the (S)-sulfoxide enantiomer (ee > 60%), suggesting that during the reaction with SscaCYP, the majority of the sulfoxidation is due to the CYP and not directly to H2O2 (Fig. S4, Table S4†).
Crystal structure of SscaCYP
As natural and engineered CYP peroxygenases contain a Glu instead of an Asp as an acid–base catalyst in the conventional alcohol (Thr) position, or in some CYPs, such as CYP116B5 (ref. 41 and 42) as well as evolved CYPs as in the case of CYP102A1 21B3,43 display peroxygenase activity without a carboxylate at this position, we decided to determine the crystal structure of SscaCYP to investigate the potential determinants of the peroxygenase activity observed.The full-length SscaCYP was crystallized and solved at a resolution of 1.48 Å using CYP177A1 (XplA) for molecular replacement (Table S5†). Despite having only ca. 42% sequence identity, structural alignment between SscaCYP and XplA gave a RMSD of only 1.23 Å. SscaCYP shows the canonical CYP fold (Fig. 2a), with the heme cofactor's iron coordinated by Cys349. On the distal side, an axial water molecule is coordinated as the sixth ligand of the Fe. In addition to the expected Asp241 side chain above the distal side of the heme, a glutamate side chain (Glu284) from the bulge between the K-helix and subsequent β strand, extends inwards to the heme (Fig. 2b). In CYP177A1, this position is occupied by a glutamine (Gln438). Interestingly, a Glu is routinely found within many members of this CYP subfamily. These two carboxylate groups are in close proximity of the axial water, both of which can potentially take part in general acid–base catalysis during H2O2 activation, either directly or via another water molecule. The two carboxylate groups are also abnormally close to each other, but the distorted electron density and presence of ambiguous difference density around the side chains suggests the mobility of these side chains and alternative conformations. Apart from these two side chains, the active site of SscaCYP is lined with hydrophobic amino acids.
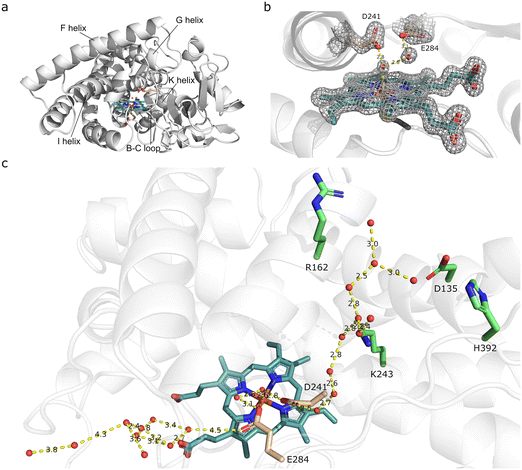 | ||
| Fig. 2 Crystal structure of SscaCYP (PDB: 8Q5J). (a) Overall fold of SscaCYP shown as a ribbon diagram with the heme cofactor displayed as a stick model. Potential catalytic residues D241 and E284 are displayed in pink with the bound waters. (b) Active site architecture with D241 and E284 shown as stick models and the coordinated water molecules as red spheres. Electron density is shown as mesh from the omit map 2Fo–Fc, contoured at σ = 1.5 Å. Hydrogen bonds are shown as dashed lines. (c) Water networks in access channels from D241 and E284 (pink) to bulk solvent/surface. Charged residues interacting with the water are displayed in green. | ||
CYPs display a level of structural plasticity which allows conformational changes between open and closed states, and even intermediate states as exemplified by CYP101A1 (P450cam),44,45 allowing different ligand access channels for substrate access and binding from the bulk solvent.46,47 This is often mediated through the movement of the F and G helices and the associated F/G-loop, as well as the B/C-loop. Despite the F and G helices and F/G-loop adopting an apparent closed conformation in SscaCYP, two access tunnels from the bulk solvent to the active site glutamate and aspartate were found (Fig. 2c). The smaller tunnel is located next to the I and F helices, perpendicular to the heme cofactor, with water molecules hydrogen bonded from D241 to the bulk surface solvent. The second wider access tunnel is located next to the B/C- and F/G-loops, approaching the heme closer to horizontal, containing multiple water molecules from the active site E284 to the surface. Whereas the smaller tunnel contains basic and polar residues hydrogen bonded with the water molecules (Fig. 2c), the second wider channel is mostly hydrophobic. Recently it was demonstrated that engineering H2O tunnels to H2O2 tunnels promote peroxygenase activity in CYPs.48 As SscaCYP was crystallized in the absence of substrate, we assume the second wider channel to be the substrate and H2O2 channel.
Aspartate D241 and glutamate E284 as potential acid–base catalysts
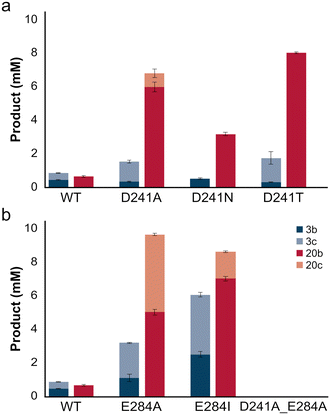
As the introduction of a carboxylate group to the distal side of the heme, through mutating the Thr of the typical acid–alcohol pair found in CYPs, have been found to either improve or introduce peroxygenase activity through acid–base catalysis, we rationalized that mutating this residue would impair the peroxygenase activity of SscaCYP. Aspartate 241 (D241), found in the equivalent position to the catalytic alcohol (Thr), was mutated to either an Ala, a Thr, as typically observed in CYPs, or an Asn, another common amino acid found in this position within the CYP177 family. Surprisingly, all the mutants displayed activity, which was in most cases even improved. Replacing D241 with a methyl group (Ala) or an alcohol (Thr) increased the allylic hydroxylation of 3a, with both also showing increased over-oxidation to the aldehyde (3c). The introduction of an Asn (D241N), however, did not improve the activity towards 3a, suggesting that removal of the Asp side chain potentially decreased steric hindrance. The sulfoxidation of 20a similarly improved the most with the Ala and Thr mutants. Also, whereas the wild-type showed no over-oxidation to the sulfone (20c) trace amounts were detected with D241A. Unlike with 3a, the D241N mutant also showed significantly improved activity, suggesting that not only steric influences, but also electrostatic interactions play a role in substrate binding.As the mutation of D241 did not negatively affect the peroxygenase activity of SscaCYP, we turned our attention to E284 as a potential acid–base catalyst. The glutamate was mutated to Ala as well as Ile, a bulkier hydrophobic amino acid that would not introduce electrostatic effects. In the case of the E284 mutants activity again increased significantly compared with the wild-type with both 3a and 20a (Fig. 3b), and was higher than what was observed with any of the D241 mutants. SscaCYP_E284I displayed the highest activity with 3a, forming near equal amounts of the allylic alcohol (3b) and the over-oxidized aldehyde product (3c). The sulfoxidation of 20a by both mutants similarly showed near-complete conversion and significant over-oxidation to the sulfone (20c). This would suggest that either E284 or D241 can act as the acid–base catalysts for heterolytic cleavage of the O–O bond in Cpd I formation. This was further substantiated by the creation of a double mutant with both D241 and E284 mutated to Ala, which showed no activity to either substrate. This suggested redundancy of D241 and E284 as acid–base catalysts does not eliminate the possibility that both carboxylate groups participate in H2O2 activation in a concerted manner. Potentially, D241 could abstract the proton from the proximal oxygen of the coordinated H2O2, and E284 located further away, donating a proton to Cpd 0 for heterolytic cleavage and H2O production, as both the carboxylate groups are hydrogen bonded to water molecules and accessible through their respective access channels to the bulk solvent.
Spectral analysis of SscaCYP and D241 and E284 mutants
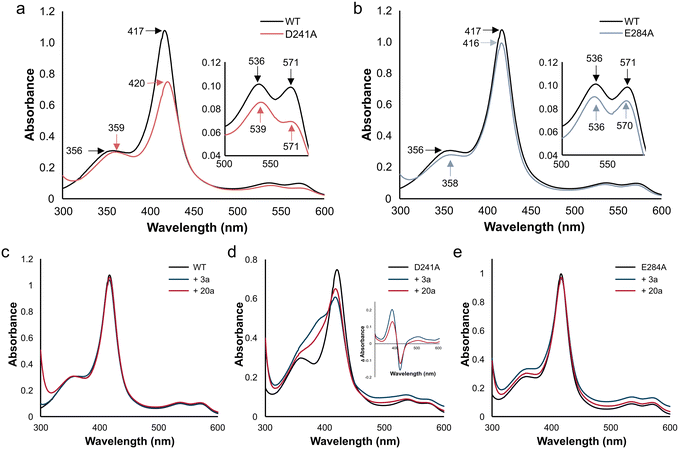
UV-vis spectroscopy was performed on the purified SscaCYP wild-type and the mutants to investigate changes to the heme environment. SscaCYP displayed a typical low-spin resting state corresponding to the water bound structure, with a Soret band at 417 nm and the minor α- and β-bands at 571 and 536 nm, respectively (Fig. 4). The removal of the distal carboxylate groups in the SscaCYP_D241A and E284A variants resulted in a red-shift to 420 nm and a blue shift to 416 nm of the Soret band, respectively (Fig. 4a and b). Not only was the red-shift also observed for the β-band in D241A, the α-band was significantly reduced relative to the β-band. Similar results were obtained for the other mutants (Fig. S11†).Type I binding was observed with both 3a and 20a (Fig. 4c–e). Whereas the WT and E284A showed minimal transition to the high-spin state, the D241A mutant yielded more high-spin state resulting from displacement of the axial coordinated water molecule. As this distal water molecule is directly hydrogen bonded to D241 in the wild-type, removal of the carboxylate group could facilitate type I binding. This general trend was also observed with the other mutants of D241 and E284 (Fig. S12†). A similar reduction in spin-state shifting was observed with CYP199A4 when the Thr from the acid–alcohol pair was mutated to a glutamate.24
Whereas the reduction of SscaCYP WT by excess sodium dithionite only showed a decrease in the Soret band at 417 nm, all the SscaCYP mutants displayed a significant blue-shift upon Na2S2O4 reduction (Fig. S13†), suggesting the ferric to ferrous heme reduction is accompanied by a shift to a high-spin state. This correlates with the reduced CO-spectra, with the SscaCYP WT showing the most prominent peak remaining at 420 not transitioning to 450 nm. These changes in ease of spin-state shifting and CO-complex formation suggests that the WT coordinates the axial water (sixth ligand) strongly, which is in agreement with the crystal structure that displayed the water molecule directly hydrogen bonded to D241, and hydrogen bonded to E284 via a water bridge.
Stability and heme-bleaching of SscaCYP and D241 and E284 mutants
The stability of SscaCYP and the respective D241 and E284 mutants were evaluated by incubating the purified proteins with 10 mM H2O2, with and without 3a, and determining their ability to yield productive CO-difference spectra. In the absence of substrate, all the mutants showed a decreased stability to the wild-type SscaCYP (Fig. S14a†). However, apart from the D241N mutant, protein stability towards H2O2 were increased during the reaction (in the presence of 3a, Fig. S14b†). This could potentially be attributed to the protective nature of a productive catalytic cycle, whereby Cpd I is not available for further reaction with H2O2. Additionally, productive reactions would decrease the effective H2O2 concentration over time. A similar trend was observed with heme bleaching experiments (Fig. S15†), with the SscaCYP wild-type being the most resistant to H2O2, whereas nearly no Soret absorbance peaks were detected after 120 min for all the mutants. This is consistent with results obtained with CYP199A4, where more rapid heme bleaching was observed with a Thr to Glu mutant than the wild-type enzyme.38 However, the equivalent mutation in CYP102A1 (T268E) was shown to result in slower heme bleaching in the presence of H2O2.25Next, the influence of H2O2 concentration on the initial reaction rates (TOFs at 1 h) of the mutants were evaluated (Table S5†). Whereas the wild-type gave the highest product concentrations at 100 mM H2O2, all the mutants only required 10–20 mM H2O2 for optimal activity (Fig. S16†). This requirement of lower H2O2 concentrations to drive the reactions is in agreement with our spectral, stability and heme-bleaching observations as tight binding of the axial water, although protecting against heme bleaching, requires higher H2O2 concentrations for efficient catalysis. By removing one of the carboxylate groups, the efficacy of Cpd I formation is increased. Although we cannot exclude the possibility of steric and electrostatic effects, the binding orientation of 20a was evaluated through the stereochemical outcome of its hydroxylation. The SscaCYP wild-type and all the mutants gave the same major (S)-enantiomer (Fig. S4†), suggesting similar binding poses. Furthermore, the mutants showed increased enantioselectivity, probably due to an increased reaction rate, leading to lower background oxidation.
Increased catalytic scope of SscaCYP variants
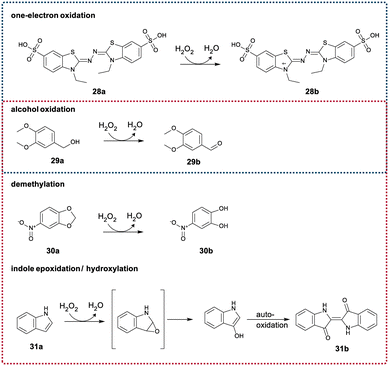
Peroxygenases such as the UPOs are routinely screened using convenient spectrophotometric assays for peroxidase and peroxygenase activity.49,50 These diverse reactions include the monitoring of the one-electron oxidation of 2,2′-azino-bis-(3-ethylbenzothiazoline-6-sulfonic acid) (28a, ABTS), the alcohol oxidation of veratryl alcohol (29a) to veratrylaldehyde (29b), the demethylation of 5-nitro-1,3-benzodioxole (30a, NBD) to nitrocatechol (30b) and the epoxidation/hydroxylation of indole (31a) to 3-hydroxyindole, which upon further autooxidation with molecular oxygen dimerizes to indigo (31b) (Scheme 2). Initial screening of SscaCYP yielded very little to no activity with these substrates. As the mutants showed significantly increased activity with 3a and 20a, we evaluated the mutants with these four substrates, as the wild-type SscaCYP only showed notable peroxidase activity with 28a. Increased activity towards all four substrates with all the mutants were observed, with D241A giving the highest initial activity towards 28a–30a, whereas D241N gave the highest activity towards hydroxylation of 31a. Similar to the activity with 3a and 20a, the activity with the D241 mutants in general gave the highest activity (Fig. S17†). Not only does this show that a single carboxylate group is better for peroxygenase and peroxidase activity, but that these two positions are also hot-spots for future protein engineering to improve or alter the substrate scope.Conclusions
CYPs have been evolved in recent years for peroxygenase activity by replacing the catalytic Thr found in the acid–alcohol pair with a glutamate. This eliminates the requirement of additional redox partner proteins as well as costly nicotinamide cofactors, with H2O2 serving as both the oxygen and electron donor. In this study, several members of the CYP177A sub-family, naturally containing an aspartate instead of the conventional threonine, were explored for peroxygenase activity. The CYP177As were found to catalyze the H2O2-driven epoxidation or hydroxylation of aryl alkenes, with SscaCYP also performing sulfoxidation of thioanisole. Unexpectedly, mutagenesis of the catalytic site aspartate (D241) in SscaCYP resulted in increased peroxygenase activity. Through structural analysis, a second carboxylate side chain, belonging to glutamate E284, was found on the distal side of the heme, hydrogen bonded to the axial water molecule via a water bridge. Similar to the mutagenesis of D241, replacing E284 with either an Ala or Ile, also increased the peroxygenase activity of SscaCYP. Peroxygenase activity was, however, completely abolished when both carboxylate groups were removed, suggesting that either could function as an acid–base catalyst for the heterolytic cleavage of O–O during H2O2 activation and Cpd I formation. The increased activity, by mutating either D241 or E284, can be explained by increased H2O2 and H2O exchange as the sixth ligand of the heme iron, with the mutants requiring substantially less H2O2 concentrations for optimal activity. These variants also showed higher TOFs than that typically observed with artificial Thr-to-Glu mutants of various CYPs (Table S5†). Not only was the activity increased with either D241 and E284, their substrate scope and catalytic versatility were also expanded on. Structural analysis also revealed two ligand and H2O/H2O2 access channels that potentially facilitate the peroxygenase activity, making SscaCYP an interesting scaffold for future protein engineering and directed evolution studies.In summary, data mining can be used for identification of natural CYP peroxygenases containing an acid–base catalyst on the distal side of the heme. Additionally, we have identified a second “hot-spot”, the equivalent position of E284 located on the active site bulge following the K-helix in CYPs, as a potential site for engineering CYPs for peroxygenase activity.
Materials and methods
Cloning, mutagenesis, heterologous expression and purification of CYPs
The genes encoding CYPs from Streptomyces scabiei (WP_059082811.1), Mycolicibacterium wolinskyi (WP_067848190), and Mycolicibacterium conceptionense (WP_019346461) were codon optimized for expression in Escherichia coli and synthesized by Twist Bioscience (USA). The open reading frames were cloned into the expression vector pET28a(+), via NdeI and BamHI. SscaCYP mutants were generated using the Megaprimer method.51 PCR reaction conditions and primers are summarized in Table S1.†The constructs were transformed into Escherichia coli BL21Gold-(DE3) (Agilent Technologies) and heterologous expression performed in auto-induction media (ZYP-5052)52 supplemented with 30 mg L−1 kanamycin, 0.5 mM δ-aminolevulinic acid hydrochloride and 50 μM FeCl3·6H2O (20 °C for 48 h, 200 rpm).
For purification, cells were harvested by centrifugation (5000 × g, 10 min, 4 °C) and resuspended (0.2 g wet weight per mL) in 25 mM Tris-HCl buffer (pH 8.0). Disruption of the cells was carried out by single passage through a One-Shot Cell disrupter (Constant Systems Ltd) at 30 kPsi, followed by centrifugation (30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, 45 min, 4 °C). The resulting soluble fraction was loaded onto a 1 mL His GraviTrap column (Cytiva), previously equilibrated with 25 mM Tris-HCl buffer (pH 8.0) containing 300 mM NaCl and 40 mM imidazole. The column was washed with 5 column volumes of the same buffer and the CYPs eluted in 25 mM Tris-HCl buffer (pH 8.0) containing 300 mM NaCl and 300 mM imidazole. CYP containing fractions were concentrated and desalted using a PD-10 column (GE Healthcare) equilibrated with 10 mM potassium phosphate buffer (pH 7.4) containing 150 mM NaCl.
000 × g, 45 min, 4 °C). The resulting soluble fraction was loaded onto a 1 mL His GraviTrap column (Cytiva), previously equilibrated with 25 mM Tris-HCl buffer (pH 8.0) containing 300 mM NaCl and 40 mM imidazole. The column was washed with 5 column volumes of the same buffer and the CYPs eluted in 25 mM Tris-HCl buffer (pH 8.0) containing 300 mM NaCl and 300 mM imidazole. CYP containing fractions were concentrated and desalted using a PD-10 column (GE Healthcare) equilibrated with 10 mM potassium phosphate buffer (pH 7.4) containing 150 mM NaCl.
Protein concentration was determined by Pierce BCA assay (ThermoFisher Scientific) using bovine serum albumin (BSA) as a standard. Active CYPs were confirmed using CO-difference spectra53 in microtiter plates (200 μL) as previously described.54
Biotransformation reactions
Reactions (1 mL) were performed, unless otherwise indicated, in 4 mL glass vials and consisted of 200 mM potassium phosphate buffer (pH 8.0), 10 μM CYP, 5% (v/v) acetone, 10 mM substrate, 20 mM H2O2 or 0.2 U mL−1 glucose oxidase (GOx) from Aspergillus niger (Sigma-Aldrich) with 100 mM glucose. Reactions were incubated at 25 °C with shaking (200 rpm), where after they were stopped and extracted by the addition of 150 μL HCl (5 M) followed by 1 mL ethyl acetate containing 2 mM internal standard (3-octanol or 1-undecanol). Samples were analysed by GC-FID (Shimadzu GC-2010) and GC-MS (Thermo Scientific Trace 1310 GC with ISQ 7000 Single Quadrupole MS) using a FactorFour VF-5 ms column (60 m × 0.32 mm × 0.25 μm, Varian) or a CHIRALDEX B-TA column (30 m × 0.25 mm × 0.12 μm) for chiral analysis. Product and remaining substrate concentrations were calculated using commercial standards. Temperature programs are described in Table S3.†Crystallization and structure determination
SscaCYP was purified as described above, followed by size exclusion chromatography on a Sephacryl HR-200 column (GE Healthcare) in 10 mM Tris-HCl (pH 8) containing 50 mM NaCl. Protein samples were concentrated to 6 and 8 mg mL−1 using an Amicon Ultra-15 Centrifugal Filter (30 kDa MWCO, Millipore). Crystals were grown using sitting drop vapor diffusion, by mixing 1 μL of protein and 1 μL of the reservoir solution (0.2 M magnesium formate dihydrate, 20% w/v polyethylene glycol 3350) at 16 °C. Crystals were soaked in reservoir solution containing 30% (v/v) glycerol before cryo-cooling. X-ray diffraction data were collected at Diamond Light Source (UK) on beamline I24 (0.9999 Å) at 93 K. Data were processed and scaled using XDS and XSCALE through the Xia2 pipeline55 for auto-index and integration, and automated molecular replacement was performed using Balbes.56 Iterative cycles of manual model building in COOT57 and refinement using Refmac58 were performed. Data collection and refinement statistics are summarised in Table S5.† The 3D structure of SscaCYP have been deposited at the PDB under accession ID 8Q5J.Spectral analysis
Spectral studies were performed at room temperature in 50 mM potassium phosphate (pH 8.0) in a final volume of 1 mL, containing 10 μM of CYP. UV-vis spectra (300–600 nm, Beckman Coulter DU 800 UV-vis) were recorded for the substrate-free enzymes, and after addition of 1 mM of substrates (trans-β-methyl styrene 3a or thioanisole 20a) stock solutions of substrates were prepared in acetone and added in volumes of 10 μL, to not exceed 1% of the total volume.Co-solvent, H2O2 stability and heme-bleaching
SscaCYP (10 μM) was incubated in 200 mM potassium phosphate buffer (pH 8.0) with different concentrations of acetone at 25 °C. Samples were taken at different time intervals and CO-difference spectra were measured. Residual CO-binding was calculated as the CO-difference spectrum measured/CO-different spectrum before adding acetone.For stability towards H2O2, 10 μM SscaCYP were incubated in 200 mM potassium phosphate buffer (pH 8.0), with 0, 10 or 50 mM H2O2, in the presence and absence of 10 mM substrate (trans-β-methyl styrene 3a or thioanisole 20a, in 5% (v/v) acetone) at 25 °C. Time resolved analysis of CO-difference spectra was performed with residual CO-binding calculated as the CO-difference spectrum measured/CO-different spectrum before adding H2O2. For mutants, stability assays were performed with 10 mM H2O2, in presence or absence of 10 mM of 3a. Additionally, heme bleaching assays were performed by monitoring the reduction of the Soret band of SscaCYP and mutants (10 μM CYP in 50 mM potassium phosphate buffer, pH 8.0) by incubating it at room temperature with 10 mM H2O2 with spectra (300–800 nm) recorded every 10 min. Time 0 is representative of the spectrum before the addition of H2O2.
The effect of H2O2 concentration on initial activity (1 h) was evaluated using reactions (1 mL) containing 10 mM trans-β-methyl styrene 3a with varying concentrations of H2O2.
Peroxidase and peroxygenase activity
Activity of SscaCYP and the mutants were evaluated towards typical peroxygenase and peroxidase substrates: 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (28a, ABTS, 0.1 mM), veratryl alcohol (29a, 5 mM), 5-nitro-1,3-benzodioxole (30a, NBD, 0.1 mM), and indole (31a, 10 mM). All reactions (200 μL) were carried out at room temperature in 50 mM potassium phosphate buffer (pH 8.0), except for reactions with ABTS where sodium acetate buffer (pH 5.0) was used. Reactions containing 20 μM CYP were started with the addition of 1 mM of H2O2 and followed spectrophotometrically using a Multiskan SkyHigh microtiter plate reader (Thermo Scientific) and concentrations determined using the following extinction coefficients at their respective wavelengths: ABTS (ε436 = 29.3 mM−1 cm−1), NBD (ε425 = 9.7 mM−1 cm−1), veratryl alcohol (ε310 = 9.3 mM−1 cm−1), indole (ε670 = 4.8 mM−1 cm−1).Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank the beamline scientists of Diamond Light Source beamline i24 for assisting with data collection under proposal mx28402, and Mr. Sarel Marais for GC analysis. This work was supported by the CSIR-IBH (Council for Scientific and Industrial Research – Industrial Biocatalysis Hub) initiative funded by the Department of Science and Innovation (DSI) and the Technology Innovation Agency (TIA). Protein X-ray crystallography was funded by the Global Challenges Research Fund (GCRF) through the Science & Technology Facilities Council (STFC), grant number ST/R002754/1: Synchrotron Techniques for African Research and Technology (START).References
- V. B. Urlacher and M. Girhard, Trends Biotechnol., 2012, 30, 26–36 CrossRef CAS PubMed.
- G. C. Schröder, M. S. Smit and D. J. Opperman, Curr. Opin. Green Sustainable Chem., 2023, 39, 100734 CrossRef.
- V. B. Urlacher and S. Eiben, Trends Biotechnol., 2006, 24, 324–330 CrossRef CAS.
- R. Bernhardt, J. Biotechnol., 2006, 124, 128–145 CrossRef CAS PubMed.
- Z. Li, Y. Jiang, F. P. Guengerich, L. Ma, S. Li and W. Zhang, J. Biol. Chem., 2020, 295, 833–849 CrossRef.
- K. J. McLean, D. Luciakova, J. Belcher, K. L. Tee and A. W. Munro, Adv. Exp. Med. Biol., 2015, 851, 299–317 CrossRef CAS PubMed.
- F. P. Guengerich, ACS Catal., 2018, 8, 10964–10976 CrossRef CAS.
- T. Coleman, J. E. Stok, M. N. Podgorski, J. B. Bruning, J. J. De Voss and S. G. Bell, J. Biol. Inorg. Chem., 2020, 25, 583–596 CrossRef CAS PubMed.
- O. Shoji and Y. Watanabe, JBIC, J. Biol. Inorg. Chem., 2014, 19, 529–539 CrossRef CAS PubMed.
- L. Rotilio, A. Swoboda, K. Ebner, C. Rinnofner, A. Glieder, W. Kroutil and A. Mattevi, ACS Catal., 2021, 11511–11525 CrossRef CAS.
- K. Piontek, E. Strittmatter, R. Ullrich, G. Gröbe, M. J. Pecyna, M. Kluge, K. Scheibner, M. Hofrichter and D. A. Plattner, J. Biol. Chem., 2013, 288, 34767–34776 CrossRef CAS.
- M. Sundaramoorthy, J. Terner and T. L. Poulos, Structure, 1995, 3, 1367–1378 CrossRef CAS PubMed.
- A. Henriksen, A. T. Smith and M. Gajhede, J. Biol. Chem., 1999, 274, 35005–35011 CrossRef CAS PubMed.
- T. Yoshida and Y. Sugano, Biochem. Biophys. Rep., 2023, 33, 101401 CAS.
- D. S. Lee, A. Yamada, H. Sugimoto, I. Matsunaga, H. Ogura, K. Ichihara, S. ichi Adachi, S. Y. Park and Y. Shiro, J. Biol. Chem., 2003, 278, 9761–9767 CrossRef CAS PubMed.
- O. Shoji, T. Fujishiro, S. Nagano, S. Tanaka, T. Hirose, Y. Shiro and Y. Watanabe, J. Biol. Inorg. Chem., 2010, 15, 1331–1339 CrossRef CAS PubMed.
- J. Chen, S. Dong, W. Fang, Y. Jiang, Z. Chen, X. Qin, C. Wang, H. Zhou, L. Jin, Y. Feng, B. Wang and Z. Cong, Angew. Chem., Int. Ed., 2023, 62, e202215088 CrossRef CAS PubMed.
- S. Di, S. Fan, F. Jiang and Z. Cong, Antioxidants, 2022, 11, 529 CrossRef CAS PubMed.
- N. Ma, Z. Chen, J. Chen, J. Chen, C. Wang, H. Zhou, L. Yao, O. Shoji, Y. Watanabe and Z. Cong, Angew. Chem., 2018, 130, 7754–7759 CrossRef.
- X. Qin, Y. Jiang, J. Chen, F. Yao, P. Zhao, L. Jin and Z. Cong, Int. J. Mol. Sci., 2022, 23, 7901 CrossRef CAS PubMed.
- S. J. P. Willot, F. Tieves, M. Girhard, V. B. Urlacher, F. Hollmann and G. de Gonzalo, Catalysts, 2019, 9, 1–9 CrossRef.
- J. Chen, F. Kong, N. Ma, P. Zhao, C. Liu, X. Wang and Z. Cong, ACS Catal., 2019, 9, 7350–7355 CrossRef CAS.
- O. Shoji, T. Fujishiro, K. Nishio, Y. Kano, H. Kimoto, S. C. Chien, H. Onoda, A. Muramatsu, S. Tanaka, A. Hori, H. Sugimoto, Y. Shiro and Y. Watanabe, Catal. Sci. Technol., 2016, 6, 5806–5811 RSC.
- M. N. Podgorski, J. S. Harbort, J. H. Z. Lee, G. T. H. Nguyen, J. B. Bruning, W. A. Donald, P. V. Bernhardt, J. R. Harmer and S. G. Bell, ACS Catal., 2022, 12, 1614–1625 CrossRef CAS.
- J. Akter, T. P. Stockdale, S. A. Child, J. H. Z. Lee, J. J. De Voss and S. G. Bell, J. Inorg. Biochem., 2023, 244, 112209 CrossRef CAS PubMed.
- A. R. Brash, Phytochemistry, 2009, 70, 1522–1531 CrossRef CAS PubMed.
- F. Sabbadin, R. Jackson, K. Haider, G. Tampi, J. P. Turkenburg, S. Hart, N. C. Bruce and G. Grogan, J. Biol. Chem., 2009, 284, 28467–28475 CrossRef CAS PubMed.
- H. Xiang, R. A. Tschirret-Guth and P. R. Ortiz de Montellano, J. Biol. Chem., 2000, 275, 35999–36006 CrossRef CAS.
- J. R. Cupp-Vickery and T. L. Poulos, Nat. Struct. Mol. Biol., 1995, 2, 144–153 CrossRef CAS PubMed.
- Y. Madrona, S. Tripathi, H. Li and T. L. Poulos, Biochemistry, 2012, 51, 6623–6631 CrossRef CAS.
- K. E. Slessor, A. J. Farlow, S. M. Cavaignac, J. E. Stok and J. J. De Voss, Arch. Biochem. Biophys., 2011, 507, 154–162 CrossRef CAS PubMed.
- S. J. B. Mallinson, M. M. Machovina, R. L. Silveira, M. Garcia-Borràs, N. Gallup, C. W. Johnson, M. D. Allen, M. S. Skaf, M. F. Crowley, E. L. Neidle, K. N. Houk, G. T. Beckham, J. L. Dubois and J. E. McGeehan, Nat. Commun., 2018, 9, 1–12 CrossRef CAS PubMed.
- M. M. Fetherolf, D. J. Levy-Booth, L. E. Navas, J. Liu, J. C. Grigg, A. Wilson, R. Katahira, G. T. Beckham, W. W. Mohn and L. D. Eltis, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 25771–25778 CrossRef CAS PubMed.
- J. M. Klenk, M. Fischer, P. Dubiel, M. Sharma, B. Rowlinson, G. Grogan and B. Hauer, J. Biochem., 2019, 166, 51–66 CrossRef CAS PubMed.
- A. C. Harlington, K. E. Shearwin, S. G. Bell and F. Whelan, Chem. Commun., 2022, 58, 13321–13324 RSC.
- H.-J. Joosten, R. Kuipers, T. van den Bergh, G. Vriend, P. Schaap and M. Smit, in 17th International conference on Cytochrome P450: biochemistry, biophysics and structure, 2011, pp. 25–36 Search PubMed.
- R. G. Jackson, E. L. Rylott, D. Fournier, J. Hawari and N. C. Bruce, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 16822–16827 CrossRef CAS.
- J. H. Z. Lee, M. N. Podgorski, M. Moir, A. R. Gee and S. G. Bell, Chem. – Eur. J., 2022, 28, e202201366 CrossRef CAS PubMed.
- A. Karich, K. Scheibner, R. Ullrich and M. Hofrichter, J. Mol. Catal. B: Enzym., 2016, 134, 238–246 CrossRef CAS.
- P. George, J. Biol. Chem., 1953, 201, 427–434 CrossRef CAS PubMed.
- A. Ciaramella, G. Catucci, G. Di Nardo, S. J. Sadeghi and G. Gilardi, New Biotechnol., 2020, 54, 71–79 CrossRef CAS.
- A. Ciaramella, G. Catucci, G. Gilardi and G. Di Nardo, Int. J. Biol. Macromol., 2019, 140, 577–587 CrossRef CAS PubMed.
- P. C. Cirino and F. H. Arnold, Angew. Chem., Int. Ed., 2003, 42, 3299–3301 CrossRef CAS PubMed.
- Y. T. Lee, E. C. Glazer, R. F. Wilson, C. D. Stout and D. B. Goodin, Biochemistry, 2011, 50, 693–703 CrossRef CAS.
- Y. T. Lee, R. F. Wilson, I. Rupniewski and D. B. Goodin, Biochemistry, 2010, 49, 3412–3419 CrossRef CAS PubMed.
- X. Yu, V. Cojocaru and R. C. Wade, Biotechnol. Appl. Biochem., 2013, 60, 134–145 CrossRef CAS.
- P. Urban, T. Lautier, D. Pompon and G. Truan, Int. J. Mol. Sci., 2018, 19, 1617 CrossRef PubMed.
- P. Zhao, F. Kong, Y. Jiang, X. Qin, X. Tian and Z. Cong, J. Am. Chem. Soc., 2023, 8–13 Search PubMed.
- A. Kinner, K. Rosenthal and S. Lütz, Front. Bioeng. Biotechnol., 2021, 9, 705630 CrossRef PubMed.
- R. Ullrich, M. Poraj-Kobielska, O. M. Herold-Majumdar, J. Vind and M. Hofrichter, Catalysts, 2021, 11, 1495 CrossRef CAS.
- J. Sanchis, L. Fernández, J. D. Carballeira, J. Drone, Y. Gumulya, H. Höbenreich, D. Kahakeaw, S. Kille, R. Lohmer, J. J.-P. Peyralans, J. Podtetenieff, S. Prasad, P. Soni, A. Taglieber, S. Wu, F. E. Zilly and M. T. Reetz, Appl. Microbiol. Biotechnol., 2008, 81, 387–397 CrossRef CAS PubMed.
- F. W. Studier, Protein Expression Purif., 2005, 41, 207–234 CrossRef CAS PubMed.
- F. P. Guengerich, M. V. Martin, C. D. Sohl and Q. Cheng, Nat. Protoc., 2009, 4, 1245–1251 CrossRef CAS PubMed.
- A. Pennec, C. L. Jacobs, D. J. Opperman and M. S. Smit, Adv. Synth. Catal., 2015, 357, 118–130 CrossRef CAS.
- G. Winter, J. Appl. Crystallogr., 2010, 43, 186–190 CrossRef CAS.
- F. Long, A. A. Vagin, P. Young and G. N. Murshudov, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2007, 64, 125–132 CrossRef PubMed.
- P. Emsley, B. Lohkamp, W. G. Scott and K. Cowtan, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 486–501 CrossRef CAS PubMed.
- G. N. Murshudov, P. Skubák, A. A. Lebedev, N. S. Pannu, R. A. Steiner, R. A. Nicholls, M. D. Winn, F. Long and A. A. Vagin, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2011, 67, 355–367 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental detail and results on the cloning, mutagenesis, expression and purification of the biocatalysts, GC and GC-MS data for all substrate conversions and products detected including chiral separation, spectral analysis of CYPs and variants, results on CYP stability and crystallographic data collection and refinement parameters. See DOI: https://doi.org/10.1039/d3cy01207g |
| This journal is © The Royal Society of Chemistry 2023 |