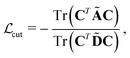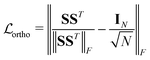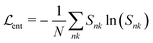Open Access Article
Open Access ArticleAutomatic identification of chemical moieties†
Jonas
Lederer
 *ab,
Michael
Gastegger
ab,
Kristof T.
Schütt
ab,
Michael
Kampffmeyer
*ab,
Michael
Gastegger
ab,
Kristof T.
Schütt
ab,
Michael
Kampffmeyer
 c,
Klaus-Robert
Müller
abdef and
Oliver T.
Unke
c,
Klaus-Robert
Müller
abdef and
Oliver T.
Unke
 *abd
*abd
aBerlin Institute of Technology (TU Berlin), 10587 Berlin, Germany. E-mail: jonas.lederer@tu-berlin.de; oliver.unke@googlemail.com
bBIFOLD - Berlin Institute for the Foundations of Learning and Data, Germany
cDepartment of Physics and Technology, UiT The Arctic University of Norway, 9019 Tromsø, Norway
dGoogle Deepmind, Germany
eDepartment of Artificial Intelligence, Korea University, Seoul 136-713, Korea
fMax Planck Institut für Informatik, 66123 Saarbrücken, Germany
First published on 30th August 2023
Abstract
In recent years, the prediction of quantum mechanical observables with machine learning methods has become increasingly popular. Message-passing neural networks (MPNNs) solve this task by constructing atomic representations, from which the properties of interest are predicted. Here, we introduce a method to automatically identify chemical moieties (molecular building blocks) from such representations, enabling a variety of applications beyond property prediction, which otherwise rely on expert knowledge. The required representation can either be provided by a pretrained MPNN, or be learned from scratch using only structural information. Beyond the data-driven design of molecular fingerprints, the versatility of our approach is demonstrated by enabling the selection of representative entries in chemical databases, the automatic construction of coarse-grained force fields, as well as the identification of reaction coordinates.
1 Introduction
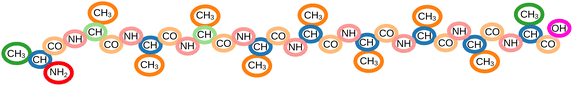
The computational study of structural and electronic properties of molecules is key to many discoveries in physics, chemistry, biology, and materials science. In this context, machine learning (ML) methods have become increasingly popular as a means to circumvent costly quantum mechanical calculations.1–37 One class of such ML methods are message passing neural networks (MPNNs),38 which provide molecular property predictions based on end-to-end learned representations of atomic environments.In contrast to such fine-grained representations, chemists typically characterize molecules by larger substructures (e.g. functional groups) to reason about their properties.39–41 This gives rise to the idea of using MPNNs for the automatic identification of “chemical moieties”, or characteristic parts of the molecule, to which its properties can be traced back. Since manually searching for moieties that explain (or are characteristic of) certain properties of molecules is a complex and tedious task, the capability of ML to find patterns and correlations in data could ease the identification of meaningful substructures drastically.
Previous work has introduced a variety of different approaches to identify substructures in molecules and materials, with objectives ranging from substructure mining42–47 over molecule generation48–52 and interpretability of machine learning architectures53–62 and coarse-graining63–65 to insights into atomistic simulations.66 While these methods offer substantial advantages for their specific tasks, it is important to note that most of them are tailored to address singular objectives, thereby limiting their overall applicability. In this work, the primary goal is to introduce a method that prioritizes versatility and generality, aiming to encompass a broader range of potential applications. Hence, to ensure the identification of meaningful moieties that can be utilized for a wide range of applications, a procedure is required (i) to be transferable w.r.t. molecule size, (ii) to provide a substructure decomposition of each molecule which preserves its respective global structure (required for, e.g., coarse-graining), and (iii) to allow for identifying several moieties of the same type in individual molecules (due to a common substructure often appearing multiple times). None of the methods mentioned above meets all of these criteria.
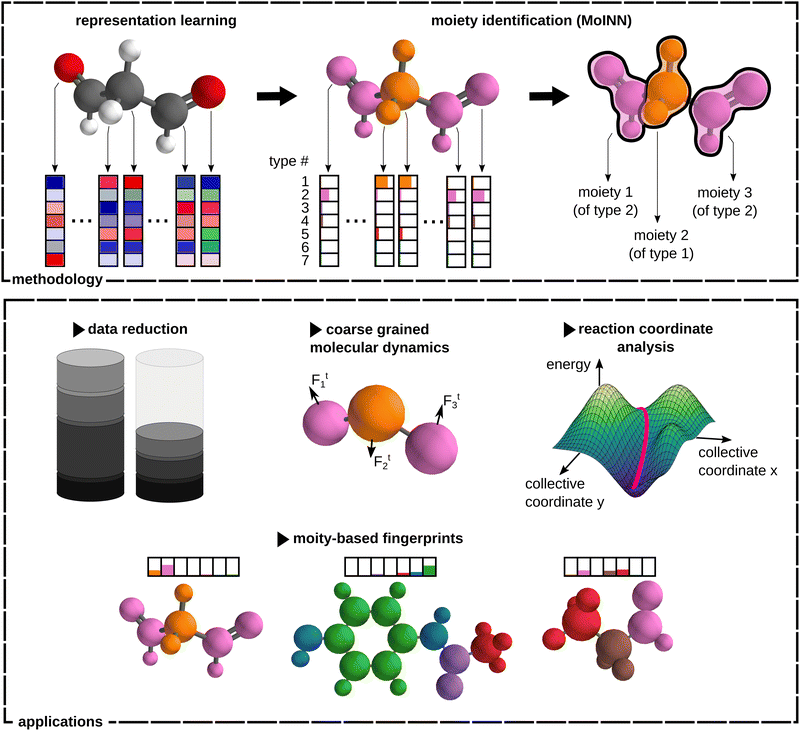
In this work, we propose MoINN (Moiety Identification Neural Network) – a method for the automatic identification of chemical moieties from the representations learned by MPNNs. This is achieved by constructing a soft assignment (or affinity) matrix from the atomic features, which maps individual atoms to different types of multi-atom substructures (Fig. 1, top). By employing representations from MPNNs pretrained on molecular properties, the identified moieties are automatically adapted to the chemical characteristics of interest. Alternatively, it is possible to find chemically meaningful substructures by training MPNNs coupled with MoINN in an end-to-end manner. Here, only structural information is required and ab initio calculations can be avoided. Crucially, MoINN is transferable between molecules of different sizes and automatically determines the appropriate number of moiety types. Multiple occurrences of the same structural motif within a molecule are recognized as the same type of moiety.
We demonstrate the versatility of MoINN by utilizing the identified chemical moieties to solve a range of tasks, which would otherwise require expert knowledge (Fig. 1, bottom). For example, the learned moiety types can serve as molecular fingerprints, which allow to extract the most representative entries from quantum chemical databases. Beyond that, moieties can be employed as coarse-grained representations of chemical structures, allowing to automatically determine beads for the construction of coarse-grained force fields. Finally, we use MoINN to identify reaction coordinates in molecular trajectories based on the transformation of detected moieties.
2 Method
The automated identification of moieties with MoINN corresponds to a clustering of the molecule into different types of chemical environments. Hence, atoms in comparable environments, i.e. with similar feature representations (see Section 2.1), are likely to be assigned to the same cluster. In the following, the term “environment types” or short “types” will be used, since each cluster is associated with a specific substructure that exhibits particular chemical characteristics. Note that atoms belonging to the same environment type are not necessarily spatially close, because similar substructures may appear multiple times at distant locations in a molecule. This is why, after atoms have been assigned to environment types (see Section 2.2), individual (spatially disconnected) chemical moieties can be found by introducing an additional distance criterion (see Section 2.3). Both steps are combined to arrive at an unsupervised learning objective for decomposing molecules into chemical moieties (see Section 2.4).2.1 Representation learning in message passing neural networks
Message passing neural networks (MPNNs)38 are able to learn atomic feature representations from data in an end-to-end manner (without relying on handcrafted features). They achieve state-of-the-art performance for molecular property prediction, solely taking atomic numbers and atom positions as inputs.7,11,12,14–17 The representation learning scheme of an MPNN can be described as follows. First, atomic features are initialized to embeddings based on their respective atomic numbers (all atoms of the same element start with the same representation). Subsequently, the features of each atom are iteratively updated by exchanging “messages” with neighboring atoms, which depend on their current feature representations and distances. After several iterations, the features encode the relevant information about the chemical environment of each atom. In this work, we use SchNet7,9,28 to construct atomic feature representations. In general, however, MoINN is applicable to any other representation learning scheme.2.2 Assigning atoms to environment types
Starting from F-dimensional atomic feature representations x1,…,xN of N atoms (e.g. obtained from an MPNN), a type assignment matrix S, which maps individual atoms to different environment types, is constructed. Following a similar scheme as Bianchi et al.,67 the type assignment matrix is given byS = softmax(SiLU(XW1)W2)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) , , | (1) |
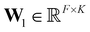 and
and 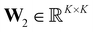 are trainable weight matrices, the n-th row of the feature matrix
are trainable weight matrices, the n-th row of the feature matrix 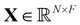 is the representation
is the representation 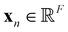 of atom n, and SiLU is the Sigmoid Linear Unit activation function.68 Here, K is a hyperparameter that denotes the maximum number of possible types. As will be shown later, a meaningful number of types is automatically determined from data and largely independent of the choice of K (see Section 2.4). The softmax function ensures that entries Snk of the N × K matrix S obey
of atom n, and SiLU is the Sigmoid Linear Unit activation function.68 Here, K is a hyperparameter that denotes the maximum number of possible types. As will be shown later, a meaningful number of types is automatically determined from data and largely independent of the choice of K (see Section 2.4). The softmax function ensures that entries Snk of the N × K matrix S obey 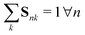 with Snk > 0. Thus, each row of S represents a probability distribution over the K environment types, with each entry Snk expressing how likely atom n should be assigned to type k. Even though assignments are “soft”, i.e. every atom is partially assigned to multiple environment types, the softmax function makes it unlikely that more than one entry in each row is dominant (closest to 1). The advantage of a soft type assignment matrix is that its computation is well suited for gradient-based optimization. In other contexts, however, it might be more natural to assign atoms unambiguously to only one environment type. For this reason, we also define a “hard” type assignment matrix
with Snk > 0. Thus, each row of S represents a probability distribution over the K environment types, with each entry Snk expressing how likely atom n should be assigned to type k. Even though assignments are “soft”, i.e. every atom is partially assigned to multiple environment types, the softmax function makes it unlikely that more than one entry in each row is dominant (closest to 1). The advantage of a soft type assignment matrix is that its computation is well suited for gradient-based optimization. In other contexts, however, it might be more natural to assign atoms unambiguously to only one environment type. For this reason, we also define a “hard” type assignment matrix 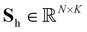 with entries
with entries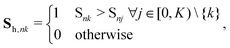 | (2) |
The atomic feature representations making up the matrix X can either be provided by a pretrained model, or learned in an end-to-end fashion. Depending on the use case, both approaches offer their respective advantages: since the type assignment matrix S is directly connected to Xviaeqn (1), pretrained features allow to find types adapted to a specific property of interest. End-to-end learned representations have the advantage that they do not rely on any reference data obtained from computationally demanding quantum mechanical calculations. Instead, they are found from structural information by optimizing an unsupervised learning problem (see Section 2.4).
2.3 Assigning atoms to individual moieties
Molecules may consist of multiple similar or even identical substructures. Consequently, distant atoms with comparable local environments can be assigned to the same type, even though they do not necessarily belong to the same moiety (see Fig. 1). To find the actual chemical moieties, i.e. groups of nearby atoms making up a structural motif, we introduce the N × N moiety similarity matrix given byC = SST○A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) , , | (3) |
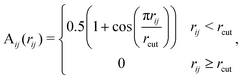 | (4) |
Analogous to the hard assignment matrix Sh (see eqn (2)), a hard moiety similarity matrix Ch, which unambiguously assigns atoms to a specific moiety, might be preferable over eqn (3) in some contexts. To this end, we define the matrix
C0h = ShSTh○Acov![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) . . | (5) |
2.4 Optimization of environment type assignments and moiety assignments
Chemical moieties are identified by minimizing the unsupervised loss function | (6) |
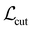 ,
,  , and
, and 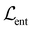 are cut loss, orthogonality loss, and entropy loss, and α is a trade-off hyperparameter. The cut loss
are cut loss, orthogonality loss, and entropy loss, and α is a trade-off hyperparameter. The cut loss 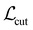 67 penalizes “cutting” the molecule, i.e. assigning spatially close atoms to different moieties. It is defined as
67 penalizes “cutting” the molecule, i.e. assigning spatially close atoms to different moieties. It is defined aswhere
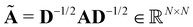 is a symmetrically normalized adjacency matrix (see eqn (4)). The degree matrix
is a symmetrically normalized adjacency matrix (see eqn (4)). The degree matrix 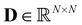 is diagonal with elements
is diagonal with elements 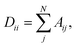 where Aij are the entries of A. Consequently,
where Aij are the entries of A. Consequently, ![[D with combining tilde]](https://www.rsc.org/images/entities/b_char_0044_0303.gif) is the degree matrix obtained from the entries of Ã.
is the degree matrix obtained from the entries of Ã.
To avoid converging to the trivial minimum of 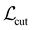 where all atoms are assigned to the same moiety and type, the orthogonality loss67
where all atoms are assigned to the same moiety and type, the orthogonality loss67
Finally, the entropy term70
In principle, the number of used types still depends on K and the entropy trade-off factor α. However, as is shown in Section S3 (ESI†), there is a regime of α where the number of used types is largely independent of K (as long as K is sufficiently large). Hence, we arbitrarily choose K = 100 in our experiments if not specified otherwise. In Section S4 (ESI†), we demonstrate the influence of different cutoff radii on the model output. The experiments show that for a cutoff radius rmin between one and two covalent bond lengths the number of environment types is largely independent of rmin.
3 Applications
This section describes several applications of MoINN. First, we use MoINN to identify common moieties in molecular data (Section 3.1). Leveraging these insights, we select representative examples from a database of structures to efficiently reduce the number of reference calculations required for property prediction tasks (Section 3.2). Next, an automated pipeline for coarse-grained molecular dynamics simulations built on top of MoINN is described (Section 3.3). Finally, we demonstrate how to utilize MoINN for automatically detecting reaction coordinates in molecular dynamics trajectories (Section 3.4).3.1 Identification of chemical moieties
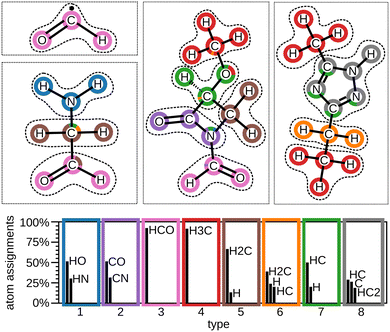
To demonstrate the automatic identification of chemical moieties, we apply MoINN to the QM9 dataset.71 Here, two different models are considered: one utilizes fixed feature representations provided by a SchNet model pretrained to predict energies, while the other model is trained in an end-to-end fashion on purely structural information. In the following, these will be referred to as the pretrained model and the end-to-end model, respectively. Details on the training of SchNet and MoINN, as well as, a comparison between pretrained model and end-to-end model can be found in Section S4 (ESI†). Also the impact of varying training data size on MoINN outputs is shown there.Fig. 2 depicts the results for the pretrained MoINN model evaluated on a test set of 1000 molecules that were excluded from the training procedure. The top shows four exemplary molecules with corresponding type assignments and moieties. As expected, we observe that moieties of the same type may occur across different molecules, as well as multiple times in a single molecule. The evaluation of environment types and corresponding moieties for all 1000 molecules (see bottom of Fig. 2) shows that each type is associated with a small set of similar moieties, i.e., the environment types form a “basis” of common substructures that can be combined to form all molecules contained in the dataset. In Section S5.1 (ESI†), we evaluate MoINN w. r. t. various ring systems and the largest identified moieties. We observe that while saturated rings are predominantly divided into several small moieties, MoINN tends to identify aromatic rings as individual entities.
3.2 Sampling of representative molecules
The quality of the reference dataset used to train ML models greatly impacts their generalization performance.72 Since the calculation of molecular properties at high levels of theory is computationally demanding, it is desirable to find ways to reduce the amount of reference data needed for training accurate machine learning models. One way to allow for more data efficient training is by sampling a representative subset of data points from chemical space (instead of choosing points randomly). Amongst other benefits, Huang et al. have shown that this can be achieved by utilizing small molecular building blocks (atom-in-molecule-based fragments) in the training data.35 Cersonsky et al. have evaluated supervised and unsupervised approaches based on low-rank approximation of the feature matrix and farthest point sampling (FPS).73 Browning et al. utilize a genetic algorithm to find an optimized training set,74 and Podryabinkin et al. proposed an approach based on the D-optimality criterion.75,76In contrast, we aim to find a representative set of molecules that can be understood as a “basis” of molecules that allows to reconstruct the features (fingerprints) of all the remaining molecules as closely as possible. To this end, we define type-based fingerprints from the assignments learned by the end-to-end MoINN model as
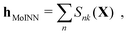 | (7) |
By stacking the fingerprints hMoINN of D molecules on top of each other we obtain the fingerprint matrix 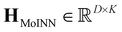 . To find a possible small subset of molecules (“basis”), which still represents the QM9 dataset sufficiently well, we minimize the loss function
. To find a possible small subset of molecules (“basis”), which still represents the QM9 dataset sufficiently well, we minimize the loss function
 | (8) |
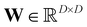 is a trainable weight matrix with entries {wij}. The first term in eqn (8) describes the reconstruction error. To avoid converging to the trivial solution, where the trainable matrix W is simply the identity matrix, we introduce a regularization term that enforces sparse rows in the weight matrix W. The trade-off between both terms can be tuned by the factor λ, i.e. larger values of λ will select a smaller subset of representative molecules. Intuitively, minimizing eqn (8) corresponds to selecting a small number of molecules as “basis vectors”, from which all other molecules can be (approximately) reconstructed by linear combination.
is a trainable weight matrix with entries {wij}. The first term in eqn (8) describes the reconstruction error. To avoid converging to the trivial solution, where the trainable matrix W is simply the identity matrix, we introduce a regularization term that enforces sparse rows in the weight matrix W. The trade-off between both terms can be tuned by the factor λ, i.e. larger values of λ will select a smaller subset of representative molecules. Intuitively, minimizing eqn (8) corresponds to selecting a small number of molecules as “basis vectors”, from which all other molecules can be (approximately) reconstructed by linear combination.
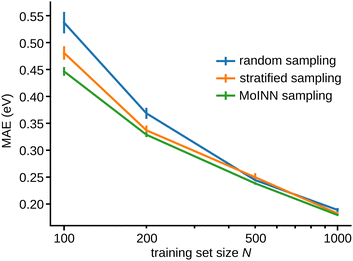
Based on this procedure, we select several QM9 subsets of different size as training sets and compare them to randomly sampled subsets, and subsets obtained by stratified sampling w. r. t. the number of atoms in each molecule. For each of these subsets, we train five SchNet models and evaluate their average performance (Fig. 3). Models trained on subsets chosen by MoINN perform significantly better than those trained on randomly sampled subsets and stratified sampled subsets. This effect is most pronounced for small training set sizes. Thus, selecting data with MoINN is most useful in a setting where only few data points can be afforded, e.g. when using a high level of theory to perform reference calculations. For more details on the experiment and a comprehensive discussion of the results please refer to Section S5.2 (ESI†).
3.3 Coarse-grained molecular dynamics
While ML force fields accelerate ab initio MD simulations by multiple orders of magnitude,13 the study of very large molecular structures is still computationally demanding. Coarse-graining (CG) reduces the dimensionality of the problem by representing groups of atoms as single interaction sites. Most approaches rely on systematically parametrized CG force fields,77,78 but also data driven approaches have been proposed.79–83 In both cases, however, the coarse-grained “beads” are usually determined manually by human experts.84Here, we propose an automated pipeline for coarse-grained molecular dynamics simulations (CG-MD), which comprises atomistic SchNet models for noise reduction, MoINN for reducing the molecule's degrees of freedom, and a SchNet model trained on the CG representation for simulating the CG dynamics. We apply this approach to the trajectory of alanine-dipeptide in water,85,86 which is a commonly used model system for comparing different CG methods.79–81
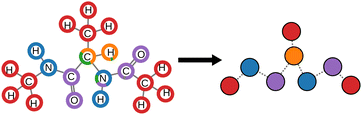
The CG representation, shown in Fig. 4, is inferred from the environment types and moieties provided by the pretrained MoINN model described in Section 3.1, which has been trained on the QM9 dataset. For a comparison to conventional CG representations such as, e.g., OPLS-UA,87,88 or an automated CG approach89 for the Martini force field,77 please refer to section S5.3 (ESI†). The original atomistic trajectory of alanine-dipeptide does not include reference energies. This is because the dynamics have been simulated in solvent, which introduces noise to the energy of the system if the solvent is not modeled explicitly. The data contains forces for all atoms in the alanine-dipeptide molecule, which implicitly include interactions with solvent molecules. However, sparsely sampled transition regions between conformers are challenging to learn with force targets only. Coarse-graining introduces additional noise on the energies and forces81 since some information about the atom positions is discarded.
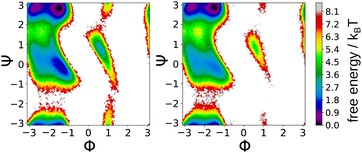
To reduce the noise, we train an ensemble of five SchNet models to provide a force field for alanine-dipeptide at atomic resolution. Subsequently, we use the corresponding forces ![[F with combining circumflex]](https://www.rsc.org/images/entities/b_char_0046_0302.gif) and energies Û as targets for training the CG SchNet model in a force-matching scheme adapted from ref. 79–81, 90, 91 (see Section S5.3 for details, ESI†). Based on the CG SchNet model, we run MD simulations in the NVT ensemble at room temperature (300 K). The trajectories are initialized according to the Boltzmann distribution at the six minima of the potential energy surface. For keeping the temperature constant, we use a Langevin thermostat. Fig. 5 shows that the free energy surfaces derived from the all-atom and CG trajectories are in good agreement.
and energies Û as targets for training the CG SchNet model in a force-matching scheme adapted from ref. 79–81, 90, 91 (see Section S5.3 for details, ESI†). Based on the CG SchNet model, we run MD simulations in the NVT ensemble at room temperature (300 K). The trajectories are initialized according to the Boltzmann distribution at the six minima of the potential energy surface. For keeping the temperature constant, we use a Langevin thermostat. Fig. 5 shows that the free energy surfaces derived from the all-atom and CG trajectories are in good agreement.
MoINN also allows for coarse-graining structures outside the scope of QM9. This is shown in Fig. 6 for decaalanine. The provided CG representation resembles the commonly used OPLS-UA representation.87,88 However it is striking that the type of terminal methyl groups differs from that of the backbone methyl groups, while in the OPLS-UA representation, by construction, those groups are considered to be interchangeable. For more details how the CG representation is derived from the provided environment types, see Section S5.3 (ESI†).
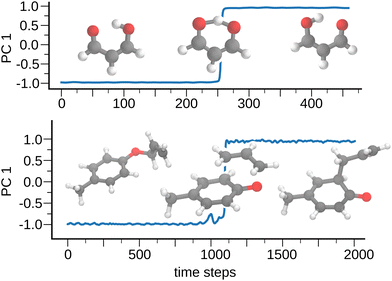
3.4 Dynamic clustering and reaction coordinate analysis
MoINN is also capable of learning environment types for molecular trajectories. In this case, the types describe “dynamic clusters”, which can be useful, e.g., for determining collective variables that describe chemical reactions. As a demonstration of this concept, we consider two chemical reactions, namely the proton transfer reaction in malondialdehyde and the Claisen rearrangement of allyl p-tolyl ether (see ref. 92 and 93 for details on how the trajectories were obtained). We train individual end-to-end MoINN models on each reaction trajectory.For each time step in the trajectory, we construct a high-dimensional coordinate vector
hdyn = (S11,S12,…,S1K,S21,…,S2K,…,SNK)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) , , |
4 Discussion
Owing to its computational efficiency, interpretability, and transferability, MoINN is applicable to a wide range of different tasks which otherwise rely on expert knowledge. MoINN involves a representation-based pooling approach which shares common ideas with the graph-pooling approaches DiffPool70 and MinCut.67 Latter describe the acquisition of a soft assignment matrix, which allows to pool graph nodes (atoms) to representative super-nodes (atom groups). In DiffPool the assignment matrix is learned utilizing GNNs, while MinCut employs a multilayer perceptron (MLP) architecture. Both methods introduce unsupervised loss functions to ensure a reasonable number of super-nodes while preferably grouping nearby nodes. Similar to MinCut, MoINN learns a mapping from atomic feature representations to type assignments by two dense layers (an MLP). The shallow network architecture results in computationally cheap training and inference. As the most prominent difference, MoINN distinguishes between individual moieties and environment types, while DiffPool and MinCut handle those entities interchangeably. As a result, MoINN stands out with regards to interpretability and transferability.The distinction between moieties and environment types allows for a more detailed analysis of the identified substructures. While the environment types explain the composition and conformation of the molecular substructures, the moieties represent individual molecular building blocks. Besides the benefits with regards to interpretability, the distinction between moieties and environment types allows to identify multiple identical moieties in the same molecule. This feature is particularly useful for molecular systems since those often possess atom groups (moieties) of the same type multiple times. In contrast, pooling distant nodes is penalized when utilizing MinCut or DiffPool. Hence, even though some distant nodes might exhibit similar feature representations, they are unlikely to be grouped together. This makes it difficult to find common moieties and might hamper transferability w.r.t. molecules of different size, since the mapping between atoms and atom groups becomes more sensitive to small changes in the feature representations. For more details on this problem, please refer to Section S6 (ESI†).
Another approach that allows for identifying reoccurring patterns in molecular data was proposed by Gasparotto et al. called PAMM.66 This method utilizes Gaussian mixture models to recognize reoccurring patterns in atomistic simulation data, and has shown to be of great use regarding the identification of hydrogen bonds. While MoINN and PAMM share the scheme of identifying motifs in molecular data, the underlying concepts are very different. One point that makes MoINN unique is its end-to-end framework. In contrast to PAMM, MoINN can be integrated into different neural network architectures and optimization tasks. This feature, inter alia, allows to obtain insight into the representation learning of neural networks. Furthermore, while the number of clusters needs to be specified for the Gaussian mixture model, MoINN does not require prior knowledge regarding the number of environment types but learns them. This makes MoINN well transferable (the transferability of MoINN regarding the molecule size is shown in Section S5.3, ESI†).
5 Conclusion and outlook
We have introduced MoINN, a versatile approach capable of automatically identifying chemical moieties in molecular data from the representations learned by MPNNs. Our method gives insight into the representation learning of MPNNs by identifying environment types based on the learned feature environments of the underlying MPNN. MoINN is differentiable, and thus, it can be exploited for a variety of applications beyond those showcased in this work. Depending on the task at hand, MoINN can be trained based on pretrained representations or in an end-to-end fashion. While pretrained representations may lead to moieties that are associated with a certain molecular property, training MoINN in an end-to-end fashion circumvents costly first principle calculations. By construction, MoINN allows to identify multiple moieties of the same type (e.g. corresponding to the same functional group) in individual molecules. This design choice also makes MoINN transferable w.r.t. molecule size and allows to automatically determine a reasonable number of moieties and environment types without relying on expert knowledge. The soft assignment of atoms to the respective environment types ensures a transparent identification of distinct moieties (compare Section S5.3, ESI†).Representing molecules as a composition of chemical moieties paves the way for various applications, some of which have been demonstrated in this work: human-readable and interpretable fingerprints can be directly extracted from the environment types identified by MoINN and they can be employed for selecting representative molecules from quantum mechanical databases to reduce the number of ab initio reference data necessary for training accurate models. Further, we have proposed a CG-MD simulation pipeline based on MoINN, which includes all necessary steps from the identification of CG representations, the machine learning of a CG force field, up to the MD simulation of the CG molecule. The pipeline is fully automatic and does not require expert knowledge. As another example, we have presented the dynamic clustering of chemical reactions, demonstrating that the environment types identified by MoINN capture conformational information that can be used to define reaction coordinates.
A promising avenue for future work is the application of MoINN in the domain of generative models.25,26 Jin et al. have shown that generating molecules in a hierarchical fashion can be advantageous.48,50 MoINN could help to identify promising motifs for molecule generation and hence facilitate the discovery of large molecules. Furthermore, MoINN could be utilized to analyze other interesting reactions. In summary, MoINN extends the applicability of MPNNs to a wide range of chemical problems otherwise relying on expert knowledge. In addition, we expect applications of MoINN to allow new insights into large-scale chemical phenomena, where MPNNs acting on individual atoms are prohibitively computationally expensive to evaluate.
Software and data availability
We provide the source code of MoINN on GitHub https://github.com/jnsLs/MoINN. For the experiments we solely utilized public datasets, which can be found at https://www.quantum-machine.org/.Author contributions
Jonas Lederer, Oliver T. Unke, Michael Gastegger, and Kristof T. Schütt conceived the work. Together with Michael Kampffmeyer they developed the method. Jonas Lederer, Oliver T. Unke, Michael Gastegger, Kristof T. Schütt and Klaus-Robert Müller collected promising application ideas. Jonas Lederer implemented the method and performed the experiments. All the authors contributed to the discussion, writing, editing, and revision of the manuscript.Conflicts of interest
The authors have no conflicts of interest to declare.Acknowledgements
MG has been at BASLEARN – TU Berlin/BASF Joint Lab for Machine Learning, co-financed by TU Berlin and BASF SE. JL, KTS, KRM and OTU acknowledge support by the Federal Ministry of Education and Research (BMBF) for the Berlin Institute for the Foundations of Learning and Data (BIFOLD) (01IS18037A,01IS14013A-E, 01GQ1115 and 01GQ0850). KRM acknowledges partial support by the Institute of Information & Communications Technology Planning & Evaluation (IITP) grants funded by the Korea government (MSIT) (No. 2019-0-00079, Artificial Intelligence Graduate School Program, Korea University and No. 2022-0-00984, Development of Artificial Intelligence Technology for Personalized Plug-and-Play Explanation and Verification of Explanation). OTU acknowledges funding from the Swiss National Science Foundation (Grant No. P2BSP2_188147).Notes and references
- J. Behler and M. Parrinello, Phys. Rev. Lett., 2007, 98, 146401 CrossRef PubMed.
- A. P. Bartók, M. C. Payne, R. Kondor and G. Csányi, Phys. Rev. Lett., 2010, 104, 136403 CrossRef PubMed.
- M. Rupp, A. Tkatchenko, K.-R. Müller and O. A. von Lilienfeld, Phys. Rev. Lett., 2012, 108, 058301 CrossRef PubMed.
- K. Schütt, P.-J. Kindermans, H. E. S. Felix, S. Chmiela, A. Tkatchenko and K.-R. Müller, Neural Information Processing Systems, 2017, pp. 991–1001 Search PubMed.
- S. Chmiela, A. Tkatchenko, H. E. Sauceda, I. Poltavsky, K. T. Schütt and K.-R. Müller, Sci. Adv., 2017, 3, e1603015 CrossRef PubMed.
- L. Zhang, J. Han, H. Wang, R. Car and W. E, Phys. Rev. Lett., 2018, 120, 143001 CrossRef CAS PubMed.
- K. T. Schütt, H. E. Sauceda, P.-J. Kindermans, A. Tkatchenko and K.-R. Müller, J. Chem. Phys., 2018, 148, 241722 CrossRef PubMed.
- L. Zhang, J. Han, H. Wang, R. Car and W. E, Phys. Rev. Lett., 2018, 120, 143001 CrossRef CAS PubMed.
- K. T. Schütt, P. Kessel, M. Gastegger, K. Nicoli, A. Tkatchenko and K. R. Müller, J. Chem. Theory Comput., 2018, 15, 448–455 CrossRef PubMed.
- H. E. Sauceda, S. Chmiela, I. Poltavsky, K.-R. Müller and A. Tkatchenko, Machine Learning Meets Quantum Physics, Springer, 2020, pp. 277–307 Search PubMed.
- O. T. Unke and M. Meuwly, J. Chem. Theory Comput., 2019, 15, 3678–3693 CrossRef CAS PubMed.
- K. Schütt, O. Unke and M. Gastegger, Proceedings of the 38th International Conference on Machine Learning, 2021, pp. 9377–9388 Search PubMed.
- O. T. Unke, S. Chmiela, H. E. Sauceda, M. Gastegger, I. Poltavsky, K. T. Schütt, A. Tkatchenko and K.-R. Müller, Chem. Rev., 2021, 121, 10142–10186 CrossRef CAS PubMed.
- O. T. Unke, S. Chmiela, M. Gastegger, K. T. Schütt, H. E. Sauceda and K.-R. Müller, Nat. Commun., 2021, 12, 7273 CrossRef CAS PubMed.
- J. Klicpera, J. Groß and S. Günnemann, International Conference on Learning Representations (ICLR), 2020.
- S. Batzner, A. Musaelian, L. Sun, M. Geiger, J. P. Mailoa, M. Kornbluth, N. Molinari, T. E. Smidt and B. Kozinsky, Nat. Commun., 2022, 13, 2453 CrossRef CAS PubMed.
- K. T. Schütt, F. Arbabzadah, S. Chmiela, K. R. Müller and A. Tkatchenko, Nat. Commun., 2017, 8, 13890 CrossRef PubMed.
- F. Noé, A. Tkatchenko, K.-R. Müller and C. Clementi, Annu. Rev. Phys. Chem., 2020, 71, 361–390 CrossRef PubMed.
- O. A. von Lilienfeld, K.-R. Müller and A. Tkatchenko, Nat. Rev. Chem., 2020, 4, 347–358 CrossRef PubMed.
- J. A. Keith, V. Vassilev-Galindo, B. Cheng, S. Chmiela, M. Gastegger, K.-R. Müller and A. Tkatchenko, Chem. Rev., 2021, 121, 9816–9872 CrossRef CAS PubMed.
- K. T. Butler, D. W. Davies, H. Cartwright, O. Isayev and A. Walsh, Nature, 2018, 559, 547–555 CrossRef CAS PubMed.
- S. Chmiela, H. E. Sauceda, K.-R. Müller and A. Tkatchenko, Nat. Commun., 2018, 9, 3887 CrossRef PubMed.
- J. S. Smith, O. Isayev and A. E. Roitberg, Chem. Sci., 2017, 8, 3192–3203 RSC.
- M. Popova, O. Isayev and A. Tropsha, Sci. Adv., 2018, 4, eaap7885 CrossRef CAS PubMed.
- N. W. Gebauer, M. Gastegger and K. T. Schütt, Proceedings of the 33rd International Conference on Neural Information Processing Systems, 2019, pp. 7566-7578.
- N. W. Gebauer, M. Gastegger, S. S. Hessmann, K.-R. Müller and K. T. Schütt, Nat. Commun., 2022, 13, 973 CrossRef CAS PubMed.
- S. Chmiela, H. E. Sauceda, I. Poltavsky, K.-R. Müller and A. Tkatchenko, Comput. Phys. Commun., 2019, 240, 38–45 CrossRef CAS.
- K. T. Schütt, S. S. P. Hessmann, N. W. A. Gebauer, J. Lederer and M. Gastegger, J. Chem. Phys., 2023, 158, 144801 CrossRef PubMed.
- S. Chmiela, V. Vassilev-Galindo, O. T. Unke, A. Kabylda, H. E. Sauceda, A. Tkatchenko and K.-R. Müller, Sci. Adv., 2023, 9, eadf0873 CrossRef PubMed.
- J. Lederer, W. Kaiser, A. Mattoni and A. Gagliardi, Adv. Theory Simul., 2019, 2, 1800136 CrossRef.
- A. Musaelian, S. Batzner, A. Johansson, L. Sun, C. J. Owen, M. Kornbluth and B. Kozinsky, Nat. Commun., 2023, 14, 579 CrossRef CAS PubMed.
- J. Gasteiger, J. Groß and S. Günnemann, International Conference on Learning Representations (ICLR), 2020.
- J. Gasteiger, S. Giri, J. T. Margraf and S. Günnemann, Machine Learning for Molecules Workshop, 2020 Search PubMed.
- S. Doerr, M. Majewski, A. Pérez, A. Kramer, C. Clementi, F. Noe, T. Giorgino and G. De Fabritiis, J. Chem. Theory Comput., 2021, 17, 2355–2363 CrossRef CAS PubMed.
- B. Huang and O. A. von Lilienfeld, Nat. Chem., 2020, 12, 945–951 CrossRef CAS PubMed.
- B. Huang and O. A. Von Lilienfeld, Chem. Rev., 2021, 121, 10001–10036 CrossRef CAS PubMed.
- K. Hansen, F. Biegler, R. Ramakrishnan, W. Pronobis, O. A. Von Lilienfeld, K.-R. Muller and A. Tkatchenko, J. Phys. Chem. Lett., 2015, 6, 2326–2331 CrossRef CAS PubMed.
- J. Gilmer, S. S. Schoenholz, P. F. Riley, O. Vinyals and G. E. Dahl, Proceedings of the 34th International Conference on Machine Learning-Volume 70, 2017, pp. 1263-1272.
- B. E. Evans, K. E. Rittle, M. G. Bock, R. M. DiPardo, R. M. Freidinger, W. L. Whitter, G. F. Lundell, D. F. Veber, P. S. Anderson, R. S. L. Chang, V. J. Lotti, D. J. Cerino, T. B. Chen, P. J. Kling, K. A. Kunkel, J. P. Springer and J. Hirshfield, J. Med. Chem., 1988, 31, 2235–2246 CrossRef CAS PubMed.
- C. D. Duarte, E. J. Barreiro and C. A. Fraga, Mini Rev. Med. Chem., 2007, 7, 1108–1119 CrossRef CAS PubMed.
- T. L. Lemke, Review of organic functional groups: introduction to medicinal organic chemistry, Lippincott Williams & Wilkins, 2003 Search PubMed.
- P. Ertl, J. Cheminf., 2017, 9, 1–7 Search PubMed.
- J. Klekota and F. P. Roth, Bioinformatics, 2008, 24, 2518–2525 CrossRef CAS PubMed.
- Y. Yamanishi, E. Pauwels, H. Saigo and V. Stoven, J. Chem. Inf. Model., 2011, 51, 1183–1194 CrossRef CAS PubMed.
- C. Borgelt and M. R. Berthold, 2002 IEEE International Conference on Data Mining, 2002. Proceedings., 2002, pp. 51-58.
- M. Coatney and S. Parthasarathy, Third IEEE Symposium on Bioinformatics and Bioengineering, 2003. Proceedings., 2003, pp. 336-340.
- A. T. Brint and P. Willett, J. Chem. Inf. Comput. Sci., 1987, 27, 152–158 CrossRef CAS.
- W. Jin, R. Barzilay and T. Jaakkola, International Conference on Machine Learning, 2020, pp. 4839-4848.
- T. S. Hy and R. Kondor, Multiresolution Graph Variational Autoencoder, 2021 Search PubMed.
- W. Jin, R. Barzilay and T. Jaakkola, ICML, 2018 Search PubMed.
- W. Jin, R. Barzilay and T. Jaakkola, International Conference on Machine Learning, 2020, pp. 4849-4859.
- M. Guarino, A. Shah and P. Rivas, 2017.
- G. Montavon, W. Samek and K.-R. Müller, Digital Signal Processing, 2018, 73, 1–15 CrossRef.
- W. Samek, G. Montavon, S. Lapuschkin, C. J. Anders and K.-R. Müller, Proc. IEEE, 2021, 109, 247–278 Search PubMed.
- T. Schnake, O. Eberle, J. Lederer, S. Nakajima, K. T. Schütt, K.-R. Müller and G. Montavon, IEEE Trans. Pattern Analysis Machine Intelligence, 2022, 44, 7581–7596 Search PubMed.
- E. Noutahi, D. Beani, J. Horwood and P. Tossou, arXiv:1905.11577 [cs, q-bio, stat], 2020.
- K. McCloskey, A. Taly, F. Monti, M. P. Brenner and L. J. Colwell, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 11624–11629 CrossRef CAS PubMed.
- B. Chen, T. Wang, C. Li, H. Dai and L. Song, International Conference on Learning Representations, 2020.
- A. Mukherjee, A. Su and K. Rajan, J. Chem. Inf. Model., 2021, 61, 2187–2197 CrossRef CAS PubMed.
- H. E. Webel, T. B. Kimber, S. Radetzki, M. Neuenschwander, M. Nazaré and A. Volkamer, J. Comput.-Aided Mol. Des., 2020, 34, 731–746 CrossRef CAS PubMed.
- A. H. Khasahmadi, K. Hassani, P. Moradi, L. Lee and Q. Morris, International Conference on Learning Representations, 2019.
- S. Letzgus, P. Wagner, J. Lederer, W. Samek, K.-R. Müller and G. Montavon, IEEE Signal Processing Magazine, 2022, 39, 40–58 Search PubMed.
- W. Wang and R. Gómez-Bombarelli, npj Comput. Mater., 2019, 5, 1–9 CrossRef.
- M. A. Webb, J.-Y. Delannoy and J. J. Pablo, J. Chem. Theory Comput., 2019, 15, 1199–1208 CrossRef CAS PubMed.
- M. Chakraborty, C. Xu and A. D. White, J. Chem. Phys., 2018, 149, 134106 CrossRef PubMed.
- P. Gasparotto and M. Ceriotti, J. Chem. Phys., 2014, 141, 174110 CrossRef PubMed.
- F. M. Bianchi, D. Grattarola and C. Alippi, International conference on machine learning, 2020, pp. 874-883.
- D. Hendrycks and K. Gimpel, arXiv, 2016, preprint, arXiv:1606.08415.
- S. S. Skiena, The Algorithm Design Manual, Springer Publishing Company, Incorporated, 2nd edn, 2008, pp. 162–166 Search PubMed.
- Z. Ying, J. You, C. Morris, X. Ren, W. Hamilton and J. Leskovec, Neural Information Processing Systems, 2018, pp. 4800–4810 Search PubMed.
- R. Ramakrishnan, P. O. Dral, M. Rupp and O. A. Von Lilienfeld, Sci. Data, 2014, 1, 1–7 Search PubMed.
- L. I. Vazquez-Salazar, E. Boittier, O. T. Unke and M. Meuwly, J. Chem. Theory Comput., 2021, 17, 4769–4785 CrossRef CAS PubMed.
- R. K. Cersonsky, B. A. Helfrecht, E. A. Engel, S. Kliavinek and M. Ceriotti, Machine Learning: Sci. Technol., 2021, 2, 035038 Search PubMed.
- N. J. Browning, R. Ramakrishnan, O. A. Von Lilienfeld and U. Roethlisberger, J. Phys. Chem. Lett., 2017, 8, 1351–1359 CrossRef CAS PubMed.
- E. V. Podryabinkin and A. V. Shapeev, Comput. Mater. Sci., 2017, 140, 171–180 CrossRef CAS.
- B. Settles, 2009.
- S. J. Marrink, H. J. Risselada, S. Yefimov, D. P. Tieleman and A. H. De Vries, J. Phys. Chem. B, 2007, 111, 7812–7824 CrossRef CAS PubMed.
- E. Brini, E. A. Algaer, P. Ganguly, C. Li, F. Rodríguez-Ropero and N. F. A. van der Vegt, Soft Matter, 2013, 9, 2108–2119 RSC.
- B. E. Husic, N. E. Charron, D. Lemm, J. Wang, A. Pérez, M. Majewski, A. Krämer, Y. Chen, S. Olsson and G. de Fabritiis, et al. , J. Chem. Phys., 2020, 153, 194101 CrossRef CAS PubMed.
- J. Wang, S. Chmiela, K.-R. Müller, F. Noé and C. Clementi, J. Chem. Phys., 2020, 152, 194106 CrossRef CAS PubMed.
- J. Wang, S. Olsson, C. Wehmeyer, A. Pérez, N. E. Charron, G. De Fabritiis, F. Noé and C. Clementi, ACS Cent. Sci., 2019, 5, 755–767 CrossRef CAS PubMed.
- L. Zhang, J. Han, H. Wang, R. Car and W. E, J. Chem. Phys., 2018, 149, 034101 CrossRef PubMed.
- Y. Chen, A. Krämer, N. E. Charron, B. E. Husic, C. Clementi and F. Noé, J. Chem. Phys., 2021, 155, 084101 CrossRef CAS PubMed.
- S. Riniker, J. R. Allison and W. F. van Gunsteren, Phys. Chem. Chem. Phys., 2012, 14, 12423–12430 RSC.
- F. Nüske, H. Wu, J.-H. Prinz, C. Wehmeyer, C. Clementi and F. Noé, J. Chem. Phys., 2017, 146, 094104 CrossRef.
- C. Wehmeyer and F. Noé, J. Chem. Phys., 2018, 148, 241703 CrossRef PubMed.
- W. L. Jorgensen and J. Tirado-Rives, J. Am. Chem. Soc., 1988, 110, 1657–1666 CrossRef CAS PubMed.
- W. L. Jorgensen, D. S. Maxwell and J. Tirado-Rives, J. Am. Chem. Soc., 1996, 118, 11225–11236 CrossRef CAS.
- T. D. Potter, E. L. Barrett and M. A. Miller, J. Chem. Theory Comput., 2021, 17, 5777–5791 CrossRef CAS PubMed.
- W. G. Noid, P. Liu, Y. Wang, J.-W. Chu, G. S. Ayton, S. Izvekov, H. C. Andersen and G. A. Voth, J. Chem. Phys., 2008, 128, 244115 CrossRef CAS PubMed.
- W. G. Noid, J.-W. Chu, G. S. Ayton, V. Krishna, S. Izvekov, G. A. Voth, A. Das and H. C. Andersen, J. Chem. Phys., 2008, 128, 244114 CrossRef CAS PubMed.
- K. Schütt, M. Gastegger, A. Tkatchenko, K.-R. Müller and R. J. Maurer, Nat. Commun., 2019, 10, 5024 CrossRef PubMed.
- M. Gastegger, K. T. Schütt and K.-R. Müller, Chem. Sci., 2021, 12, 11473–11483 RSC.
- H. Hotelling, J. Educ. Psy., 1933, 24, 498–520 Search PubMed.
- K. Pearson, London, Edinburgh Dublin Philos. Mag. J. Sci., 1901, 2, 559–572 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available: Additional details regarding the concept of MoiNN, training procedure, applications, and comparison to other graph-pooling approaches. See DOI: https://doi.org/10.1039/d3cp03845a |
| This journal is © the Owner Societies 2023 |